Abstract
The zinc-finger transcription factor, GATA-3, plays a crucial role during early T-cell development and also dictates later T-cell differentiation outcomes. However, its role and collaboration with the Notch signaling pathway in the induction of T-lineage specification and commitment have not been fully elucidated. We show that GATA-3 deficiency in mouse hematopoietic progenitors results in an early block in T-cell development despite the presence of Notch signals, with a failure to up-regulate Bcl11b expression, leading to a diversion along a myeloid, but not a B-cell, lineage fate. GATA-3 deficiency in the presence of Notch signaling results in the apoptosis of early T-lineage cells, as seen with inhibition of CDK4/6 function, and dysregulated Cdkn2b expression. We also show that GATA-3 induces Bcl11b, and together with Bcl11b represses Cdkn2b expression, however loss of Cdkn2b failed to rescue the developmental block of GATA-3 deficient T cell progenitor. Our findings provide a signaling and transcriptional network by which the T-lineage program in response to Notch signals is realized.
Introduction
The T cell lineage has distinct developmental requirements that distinguish it from other hematopoietic lineages, and T progenitors must migrate to the thymus to fulfill their T cell potential (1, 2). In the thymus, T cell development proceeds through well-characterized stages that are defined by the expression of cell surface markers. The early thymic progenitors (ETPs) are found among the CD4− CD8− (double negative, DN) CD44+ CD25− cells (DN1), which derive predominantly from a thymic seeding cell (TSP) that is negative for lineage markers (Lin−), Sca-1+ and CD117 (Kit)+ (LSK) (3–6). ETPs, which retain non-T cell potential, are heterogeneous and can be further subdivided by the expression of Flt3 (7), CCR9 (5, 8–10), CD24 (11) or CD27 (10). T-lineage commitment occurs during the later CD44+ CD25+ (DN2) and CD44− CD25+ (DN3) stages, coincident with the rearrangement of the T cell receptor (TCR) β, γ and δ chains (12).
T cell commitment is dependent on Notch signals, as Notch1 deficiency leads to intra-thymic B lymphopoiesis and constitutive Notch activation enforces ectopic T cell development (13, 14). Other transcription factors that are indispensable for T cell development include TCF-1 and Bcl11b, which are both under the direct control of Notch signals (15–19). The zinc-finger transcription factor GATA-3 appears to be required from the very earliest stages of T cell development (20–24) and both Notch and GATA-3 are also required for later stages of T lymphopoiesis (25–27). Of note, the best characterized role for GATA-3 is in peripheral responses, where GATA-3 is a known Notch target and master regulator of the TH2 program (28–30).
In contrast to the well-defined role of GATA-3 in regulating peripheral T-helper responses (31), the role of GATA-3 during early T cell development is less well characterized. There is a profound requirement for GATA-3 during early T cell development, as Gata3−/− cells do not contribute to the DN1 thymocyte compartment in conventional knockouts, blastocyst chimeras or fetal thymic organ cultures (20–22, 32, 33). The existing data suggest that either prior Notch signals in the fetal liver are required for GATA-3 function during fetal T-lymphopoiesis, or that GATA-3 function critically depends on Notch expression (32). More recent findings established a role for GATA-3 prior to the DN3 stage of development (23, 24, 33). However, none of the available data clearly distinguish whether GATA-3 is required at earlier stages, such as in specification and/or commitment to the T cell fate or for survival following T-lineage commitment, nor do they elaborate the mechanism by which GATA-3 collaborates with Notch to drive T cell development.
Although evidence from the periphery indicates that GATA-3 can be a direct target of Notch, the interaction is more complex during thymocyte development. Specifically, while ectopic Notch activation enforces the T cell fate (34), ectopic GATA-3 overexpression in thymocytes blocks T cell development (35–39). The early embryonic lethality that results in the absence of GATA-3 precludes either in vivo studies or ex vivo assays from conventional GATA-3 knockouts and further complicates attempts to address the role of GATA-3 in T cell development (20, 21).
Here, we have taken advantage of Gata3−/− embryonic stem cells (ESCs) and conditional GATA-3 deletion in bone marrow derived-progenitors to elaborate on the interaction between Notch and GATA-3 at the inception of T cell development, and to identify the mechanism by which GATA-3 acts in early thymocytes. Specifically, we demonstrate that Gata3 deficient progenitors respond to Notch signals and exclude B-lineage differentiation, but fail to express the T lineage-associated gene Bcl11b and thus favor myeloid lineage outcomes. We also find that GATA-3 is absolutely required for the survival of developing thymocytes, and gene expression analysis of Gata3−/− in comparison to Gata3+/− progenitors revealed specific genes targets that are involved in cell cycle regulation and apoptosis. Finally, we show that GATA-3 directly induces and collaborates with Bcl11b to repress the expression of the pro-apoptotic tumor-suppressor cyclin dependent kinase inhibitor 2b (Cdkn2b), an inhibitor of cyclin dependent kinases-4 and −6 (CDK4/6). This genetic interplay may allow for the survival, proliferation and T-lineage commitment of ETPs in response to Notch signals. Thus, our results imply that Notch signals do not require GATA-3 to exclude the B cell fate, but collaborates with GATA-3 to induce T-lineage commitment and concomitant thymocyte survival and proliferation.
Material and Methods
Mice
The generation of Gata3f/f mice is described elsewhere (26). Gata3f/f mice were back-crossed to either Mx-Cre (Jackson Laboratory, Bar Harbor, ME) (40) or Vav-Cre transgenic mice [(41); a kind gift from Dr. Nancy Speck, University of Pennsylvania]. To allow for an inducible deletion of the Gata3 floxed alleles, Gata3f/f-Mx-Cre+ mice were injected with 400 μg of poly (I:C) i.p. 3 times (Sigma-Aldrich) over 6 days. Cdkn2bf/f mice (42) were obtained from the NCI mouse repository (Frederick, MD), and backcrossed with Gata3f/f Vav-Cre+ transgenic mice to obtain double-conditional knockout dcKO (Cdkn2bf/f;Gata3f/f;Vav-Cre+) mice. Bone marrow was harvested 14 to 30 days later. All animal procedures were approved by the Sunnybrook Health Science Centre Animal Care Committee.
Culture of OP9, OP9-C and OP9-DL1 or OP9-DL4 cells
OP9 cells were originally obtained from Dr. T. Nakano (Osaka University, Japan) and the Riken cell bank (Tsukuba, Japan). The OP9-C (control), OP9-DL1, OP9-DL4 cell lines were generated by infecting the BM stromal cell line OP9 with either the empty MigR1 retroviral vector or with the MigR1 retroviral vector engineered to express the Delta-like 1 or Delta-like 4 gene 5’ of the internal–ribosomal entry site, allowing the bicistronic expression of Delta-like 1 or Delta-like 4 and GFP as previously described (43). OP9-C cells, OP9-DL1 and OP9-DL4 cells were cultured as a monolayer in OP9 media (αMEM supplemented with 20% FCS (HyClone), 10 U/ml penicillin, 100 μg/ml streptomycin, and 2.2 g/liter sodium bicarbonate) (43).
Flow cytometry
Flow cytometry was performed using a FACScalibur or LSRII instrument (BD Biosciences, San Diego, CA). All antibodies were purchased from BD Biosciences: CD44, CD25, CD4, CD8, CD11b, CD11c, NK1.1, Ter119, Gr1, Sca-1, CD117, CD45R, CD19, CD90.2, AnnexinV. Anti-GATA3 (Thermo-Fisher), anti-TCF1 (Cell Signaling Technologies) were used for intracellular staining. For analysis of hematopoietic cells, live cells were gated based on forward- and side-scatter and lack of propidium iodide or DAPI uptake. Cells were sorted using a FACSDiVa or FACSAria instrument (BD Biosciences, San Diego, CA). Sorted cells were >99% pure, as determined by post-sort analysis. Magnetic assisted cell sorting (MACS) was performed according to the manufacturer’s instructions (Miltenyi Biotech, Bergisch Gladbach, Germany).
Confocal analysis of GATA-3 intracellular expression.
Freshly sorted DN cells were dropped on polylysine coated slides in a humid chamber at room temperature for 30 min followed by fixation for 10 min in PBS 2% formaldehyde, permeabilization for 10 min in PBS/0.2% TritonX and blocking for 20 min in PBS containing 10% goat serum. Fixed and permeabilized cells were stained for 1h at room temperature with a mouse anti-human GATA3 antibody (HG3–31, IgG, Santa Cruz Biotechnology, or mouse IgG control (Santa Cruz Biotechnology) followed by labeling with Alexa Fluor 555-conjugated goat anti-mouse IgG antibodies for 45 minutes at room temperature in the dark. Slides were mounted with Mowiol 4–88 medium (Calbiochem-Merck Chemicals, Darmstadt, Germany) containing DAPI. Cells were imaged by conventional (Zeiss AxioImager) or confocal (Zeiss LSM700) fluorescence microscopy. Fluorescence signal was quantified in individual cell images using ImageJ (http://rsb.info.nih.gov/ij/index.html). Frequencies of whole cell intensity were normalized to the IgG control to a mode of 100. Results were aggregated from 2 independent experiments (n=20 to 30 cells for each value). For nuclear localization analysis, nuclear and cytoplasmic fluorescence intensities were measured, and results represented as the mean ratio of nuclear to cytoplasmic fluorescent intensities.
Cell culture and differentiation of ESCs and bone marrow-derived progenitors
The ESC line R1 was obtained from G. Caruana (Mt. Sinai Hospital, Toronto). ESCs were maintained by culture in ES media (DMEM, supplemented with 15% FCS, 10 U/ml penicillin, 100 μg/ml streptomycin, 100 μg/ml gentamicin, 2 mM glutamine, 110 μg/ml sodium pyruvate, 50 μM 2-mercaptoethanol, and 10 mM HEPES) containing 1 ng/ml leukemia inhibitory factor (R&D Systems, Minneapolis, MN) on irradiated mouse embryonic fibroblasts. Embryonic fibroblasts were generated from day 15–18 embryos as previously described, and cultured in ES media (44). ESC/OP9 differentiation cocultures, using either OP9-DL1 or OP9-C cells, and ESR1, Rbpj+/−, Rbpj−/−, Gata3+/− and Gata3−/− ESCs were performed as previously described (45, 46). Briefly, 104 ES cells were seeded onto OP9 cell monolayers in 6-well plates, or 5 × 104 ES cells were seeded onto OP9 monolayers in 10 cm dishes. After 5 or 6 days of coculture, cells were harvested and made into single-cell suspensions by 0.25% Trypsin treatment and vigorous pipetting. Cells were then washed and directly reseeded onto new OP9 cell monolayers, with the addition of Flt3L at a final concentration of 5 ng/ml (R&D Systems, Minneapolis, MN). Alternatively, cells were either sorted for the expression of Flk1, or enriched for Flk1-expressing cells by magnetic assisted cell sorting (MACS) (Miltenyi Biotec), prior to reseeding onto OP9 cell monolayers. For continued ESC/OP9 coculture, non-adherent cells were passed to fresh OP9 monolayers again at day 8 of coculture (since the start of the coculture), and thereafter ESC/OP9 cocultures were maintained in the presence of 5 ng/ml of Flt3L and 1 ng/ml of IL-7 (R&D Systems Minneapolis, MN) by changing media or passaging (without trypsin) to fresh OP9 monolayers every 2 days. Gata3+/− and Gata3−/− ESCs were similarly cultured on OP9-C for the first 8 days to control the time of exposure to T-inducing Notch signals. For microarray experiments, hemangioblast formation was enhanced by culturing progenitors in 5 −10 μg/ml BMP4 (R&D Systems Minneapolis, MN) between day 1 and day 5 of coculture.
Bone marrow-derived LSKs were isolated from 6–8 week old Gata3f/f Vav-Cre+ and Gata3f/f Cdkn2bf/f Vav-Cre+ mice or following poly I:C induction from Gata3f/f Mx-Cre+ mice by staining for Lineage markers (Ter119, CD11b, CD11c, Gr-1, CD8, CD45R, CD19, NK1.1), CD117 and Sca-1 and sorting based on a Lin−CD117+Sca-1hi phenotype. Equal numbers of CD117+Sca-1hi hematopoietic progenitors were seeded onto a ~80% confluent monolayer of either OP9-DL1 or OP9-C cells and cultured in the presence of 1 ng/ml IL-7 and 5 ng/ml Flt3L.
For experiments using PD0332991, bone marrow LSKs were grown on OP9-DL4 for 5 days, followed by sorting of CD44+CD25− DN1 or CD44+CD25+ DN2 cells, which were re-seeded onto fresh OP9-DL4 and treated with 2 μg/ml of PD0332991 for the duration of 3 days.
Retroviral transduction
Gata3+/− or Gata3−/− total bone marrow cells were retrovirally transduced on pMigR-, or pMig-Bcl11b-expressing GP+e.86 packaging cells for 24 hours in the presence of 1 ng/ml IL-7, 5 ng/ml Flt3L, 1 ng/ml SCF, 8 mg/ml Polybrene, and GFP+ LSKs were purified by cell sorting as described above. Wild type LSK-OP9-DL4 co-cultured cells were transduced with either pMIY (provided by Dario Vignali, (47)) or pMIY-Cdkn2b GP+e.86-derived supernatants on day 5 for 24 hours. YFP+ DN1 cells were subsequently sorted and re-plated on the fresh OP9-DL4 monolayer and cultured for the additional 3 days before assessing for cell death and T cell development. Gata3+/− or Gata3−/− bone marrow-derived progenitors cultured on OP9-DL4 for 4 days were retrovirally transduced on pMigR-, or pMig-GATA-3-expressing GP+e.86 cells for 24 hours. GFP+ CD45+ cells were sorted, re-seeded on OP9-DL4 monolayer, and cultured for additional 3 days before GFP+ CD45+ DN cells were once again purified by cell sorting.
Genomic and Qualitative Real Time PCR
Genomic DNA was purified using the EasyDNA kit (Invitrogen) from Lin+ bone marrow. Assessment of deletion of the GATA-3 floxed allele was as described previously (26). Qualitative Real Time PCR (qPCR) was performed after TRIzol RNA extraction according to the manufacturer’s protocol (Life Technologies) and cDNA prepared using either a QuantiTect Reverse Transcription Kit (Qiagen) or SuperScript® VILO cDNA Synthesis Kit (Life Technologies). Transcripts were amplified using SYBRGreenER™ qPCR Supermix (Life Technologies) and data normalized to β-actin.
Microarray analysis
For microarray analysis of ESC-derived hematopoietic progenitors, co-cultures were performed as described above. On Day 9, 23 hours after transfer from OP9-C to OP9-DL1 stroma, CD45+ cells were collected by FACS. RNA was isolated using the Qiagen RNAeasy columns, as per the manufacturer’s instructions. Double-stranded cDNA was prepared from ~1 μg of total RNA using the Affymetrix cDNA synthesis kit (Affymetrix, Santa Clara, Ca), and a single round of in vitro transcription was performed using an IVT labeling kit (Affymetrix). cRNA product was purified using a GeneChip Sample Cleanup Module (Affymetrix) and 20 μg of biotin-labeled cRNA was fragmented and hybridized to Affymetrix Mouse MOE430_2.0 microarrays for 16 hr in the Affy 640 hybridization oven at a speed of 60 rpm. Microarrays were washed and stained using and Affymetrix FS400 Fluidics Station. GeneChip arrays were scanned using a GeneChip Scanner 3000 (Affymetrix). Initial probe set intensities were quantified using the GeneChip Operating Software (GCOS). All hybridized chips exceeded standard quality control criteria as recommended by the manufacturer, and as previously described (48, 49). Microarray data are available from the Gene Expression Omnibus under accession number GSE199279 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE199279).
Apoptosis assay
OP9-DL1 or OP9-DL4-cultured cells were stained for AnnexinV according to manufacturer’s recommendations (BD Bioscience). Briefly, cells were surface-stained with anti-CD45 antibody in FACS buffer, followed by two washes in cold PBS and resuspended in 1X Binding Buffer. Five microliters of AnnexinV APC, along with 2.5 μg/ml of PI, were added to a 100 μl of cell aliquot and incubated at room temperature in the dark for 15 minutes. The reaction was stopped with an addition of 1X Binding Buffer and Annexin V staining analyzed by flow cytometry.
ChIP assay
Predicted GATA-3 RESs were identified using rVista, genomatix MatlInspector and promoter scan in VectorNTI. ChIP was performed on 1.5×106 DN1, DN2 and DN3 thymocytes purified by flow cytometry by crosslinking in 1% of formaldehyde (Fluka) for 10 minutes with gentle rocking at room temperature. Crosslinking reaction was stopped by adding 125 mM of glycine and incubating cells for the additional 5 minutes at room temperature with rocking. Cells were pelleted and washed three times in ice-cold PBS containing protease inhibitors 1 mM PMSF (Sigma), 1 μg/ml Aprotinin (Sigma), and 1 μg/ml Leupeptin (Sigma). Final cell pellet was resuspended in 300 μl of Cell Lysis Buffer (5 mM PIPES (KOH), pH 8.0; 85 mM KCl; 0.5% NP-40) containing the above-mentioned protease inhibitors and incubated on ice for 10 minutes. Nuclei were pelleted by centrifugation at 5000 rpm for 5 minutes and nuclei resuspended in 500 μl of Nuclear Lysis Buffer (50 mM Tris, pH 8.1; 10 mM EDTA; 1% SDS; protease inhibitors). After a 10 min. incubation on ice, chromatin was sheared by sonication using a Branson Sonifier 450 in six 8-second bursts with incubation on ice in-between each cycle. The debris was cleared by centrifugation at 14,000 rpm for 10 minutes at 4°C and supernatant diluted 5-fold in ChIP Dilution Buffer (0.01% SDS; 1.1% Triton X-100; 1.2 mM EDTA; 16.7 mM Tris, pH 8.1; 167 mM NaCl; protease inhibitors). To reduce non-specific background, sample was precleared with 80 μl of salmon sperm DNA/protein G agarose slurry (Millipore) for 30 minutes at 4°C with nutation. Beads were pelleted by a brief (30 sec) centrifugation. One percent of the supernatant was saved as a total input control. The rest was divided into two fractions: one incubated with 5 μg of anti-GATA-3 antibody (HG3–31) (SantaCruz) or 5 μg of IgG (SantaCruz). Both were incubated overnight at 4°C with nutation. Immune complexes were collected with 60 μl of salmon sperm DNA/protein G agarose beads for 1 h at 4°C with nutation. Beads were washed for 5 minutes in 1 ml of each of the following: Low Salt Buffer (0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris, pH 8.1; 150 mM NaCl), High Salt Buffer (0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris, pH 8.1; 500 mM NaCl), LiCl Wash Buffer (0.25 M LiCl; 1% NP-40; 1% deoxycholate; 1 mM EDTA; 10 mM Tris, pH 8.0), twice in 1x TE buffer. Complexes were eluted by adding 250 μl of Elution Buffer (1% SDS; 0.1 M NaHCO3) to pelleted beads two times for the total amount of 500 μl of eluate. Crosslinking of protein to DNA was reversed with 20 μM of RNase I and 0.3 M NaCl at 65°C for 4–5 hours and DNA precipitated with 2.5 volumes of 100% ethanol overnight at −20°C. DNA was pelleted by centrifugation and resuspended in Proteinase K Buffer (10 μM EDTA, 40 μM Tris, pH 6.5 and 200 μM Proteinase K) and incubated for 2 hours at 45°C. DNA was purified using a QiaQuick spin columns (Qiagen) and eluted in 50 μl/column as per manufacturer’s recommendation. Two μl of DNA were used per qPCR reaction. Bcl11b and H3K273me ChIP assays followed the same protocol as above using anti-CTIP2 antibody (abcam) and anti-histone H3 (tri methyl K27) antibody (abcam), respectively.
ChIP Sequencing
Primary thymocytes were isolated from 3–5 week old mice and cultured for 4 hours prior to formaldehyde crosslinking. Crosslinked chromatin was prepared for analysis as previously described (50) and immunoprecipitated with agarose-coupled antibody to Bcl11b, which was generated in goat from KLH-coupled synthetic peptide (EATILEEDEGLEIEEPAAL) corresponding to residues 26–44 of the mouse Bcl11b sequence. ChIP and input samples were sequenced on an Illumina Genome Analyzer and the resulting sequence reads were aligned to the mouse mm10 reference genome assembly. Aligned read data were then processed to identify genomic regions enriched in Bcl11b associations. ChIP-seq data and detailed analytical methods are available from the Gene Expression Omnibus under accession number GSE59826 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE59826).
Results
Intracellular GATA-3 expression varies at different stages of T cell development.
GATA-3 expression varies across hematopoietic progenitors and thymocyte subsets and is indispensable for early T cell development (24, 49, 51–53). We assessed subsets of wild-type DN cells for the presence and intracellular distribution of GATA-3 and whether it coincided with the acquisition of T-lineage potential to more directly examine the levels and localization of GATA-3 expression in early thymocytes. We fractionated the CD44+ CD25− DN1 subset by CD117 (Kit) and CD24 expression to differentiate between the CD117hi CD24−/lo ETPs (DN1a/b) and the cells with the capacity for alternative fates (DN1c-e) (54). In DN1a/b cells, GATA-3 protein was expressed at moderate levels and localized predominantly inside the nucleus (Figure 1A–C). GATA-3 expression was decreased in CD117lo CD24hi DN1c cells. GATA-3 was highly expressed in CD117− CD24+ DN1d cells, but the protein was limited to the cytoplasm. In contrast, GATA-3 was observed in both the cytoplasm and the nucleus in CD117− CD24− DN1e cells. GATA-3 protein expression peaked in DN2 cells, concomitant with T-lineage commitment, and was present predominantly in the nucleus at this stage. GATA-3 levels fell drastically in pre-β-selected CD44− CD25+ CD27lo DN3a cells, but rose again and remained confined to the nucleus of CD27hi DN3b cells following β-selection. By the DN4 stage, GATA-3 was expressed diffusely throughout the nucleus and cytoplasm. Similar overall expression pattern was observed by intracellular staining for GATA-3 in DN thymocyte subsets, which mirrored TCF1 expression (Supplemental Figures 1A and 1B). Thus, GATA-3 protein levels fluctuate in early thymocytes, and GATA-3 nuclear localization during the DN stages of T cell development coincides with molecular events restricting progenitors towards the T-lineage.
Figure 1. Analysis of GATA-3 expression in double negative thymocytes.

(A) Immunofluorescence analysis of GATA-3 expression in DN thymocyte subsets. Cells were sorted, fixed, permeabilized and stained for GATA-3, or with IgG isotype control, and DNA counterstained with DAPI. Representative images are shown. (B) Whole cell and nuclear fluorescence pixel intensities were summed for individual cells in microscopy images. The nuclear perimeter was traced in the DAPI-stained images and duplicated in the GATA3-stained images, as detailed in (53). The measurements are plotted as the mean ratio of nuclear to cytoplasmic fluorescence signal (n=6 or 7 cells for each value) with SEMs and *p ≥0.05 or **p≥0.005 (relative to DN1d), indicated. (C) Whole cell fluorescence pixel intensities were summed for individual cells in microscopy images. Each bar represents a number of cells at a given fluorescence level. Statistical analyses are relative to DN1d population, *p < 0.05 and **p < 0.005 (one-tailed Student t-test).
GATA-3 is required at the earliest stages of T cell development.
We have previously demonstrated that ESC-derived hematopoietic progenitors undergo T-lymphopoiesis when cultured on OP9-DL1 cells (55, 56). Here, we employed the OP9-DL1 co-culture system and Gata3−/− ESC-derived hematopoietic progenitors to address the role of GATA-3 during early T cell development. Like wild-type ESC-derived CD45+ progenitors, which were generated after an 8 d coculture on OP9-C cells (55), the control Gata3+/− ESC-derived CD45+ progenitors progressed through the DN subsets, as defined by CD44 and CD25 (Figure 2A), to the CD4+ CD8+ double positive (DP) stage (Figure 2B). In contrast, Gata3−/− CD45+ progenitor cells showed an early block at the DN1 stage, produced very few DN2 cells, and did not give rise to DP cells even at later time points (Figures 2A and 2B). This demonstrates that Gata3−/− ESC-derived hematopoietic progenitors recapitulate the early developmental T cell defect that has been observed in vivo by others (21, 22).
Figure 2. GATA-3 deficiency abrogates early T-lymphopoiesis in ESC-derived progenitors cultured in the presence of Notch signals and results in gene expression changes.

(A) Flow cytometry analysis for cell surface expression of CD44 and CD25 and (B) CD8 and CD4 of Gata3+/− and Gata3−/− ESC-derived hematopoietic progenitors cultured, for the indicated time points, on OP9-DL1, after an initial 8 d co-culture on OP9-C cells to induce hematopoiesis. Live cells were gated as CD45+. Results are representative of at least three independent experiments. (C) Gata3+/− and Gata3−/− ESC-derived progenitors were cultured on OP9-C cells until inception of hematopoiesis and then transferred to and maintained on OP9-DL1 cells for the remainder of the co-culture. Flow cytometry analysis was performed at indicated time points for presence of CD45+ Annexin V+ cells. Data shows fold difference in Annexin V staining in CD45+ cells from each culture (mean + SEM; n = 3). (D) Co-cultures of Gata3+/− and Gata3−/− ESC and OP9-DL1 cells were sorted 36 hours after the initial transfer from OP9-C cells and CD45+ cells analyzed for global gene expression changes by microarray. Present/absent/moderate calls were established by GCOS and probe sets where Affy mas5 detection were present in 2 out of 3 or 3 out of 3 independent experiments were selected. Genes with absent/moderate calls were excluded from further analysis. Probe sets mapping to designated EntrezGene IDs were identified and multiple probe sets mapping to a single gene were collapsed. Results are shown as z-score. Student t-test, p-value < 0.05 (Gata3+/− vs Gata3−/−). (E) Heat map showing cell cycle regulators and pro-apoptotic genes that were differentially expressed between Gata3+/− and Gata3−/− ESC-derived progenitors cultured on OP9-DL1 cells.
The analysis of Gata3−/− ESC-derived progenitors cultured in the presence of OP9-DL1 cells revealed similarities to Rbpj−/− progenitors (data not shown), which are incapable of signaling through the canonical Notch pathway and do not commit to the T cell fate (56). The similarity between Gata3−/− and Rbpj−/− co-cultures in failing to progress to the earliest T cell-specified DN compartment confirms that GATA-3 is necessary downstream of Notch signals to initiate T-lineage differentiation.
GATA-3-deficient progenitors undergo apoptosis despite the presence of Notch signals
Despite a Notch-inductive environment, Gata3−/− ESC-derived hematopoietic progenitors were not only arrested at a DN1 stage of development but their survival was also compromised (Figure 2C). An analysis of cultured progenitors revealed an increase in AnnexinV+ Gata3−/− cells on OP9-DL1 cells as compared to Gata3+/− ESC-derived progenitors. The loss of Gata3−/− cells cultured on OP9-DL1 cells suggested that these cells might not be able to survive Notch-induced specification to the T cell fate.
The lack of GATA-3-deficient DN2 cells in OP9-DL1 cocultures led us to examine the targets of GATA-3 transcriptional regulation in early T cell development. Three sets of Gata3+/− and Gata3−/− RNA samples were generated independently from ESCs co-cultured with OP9-control cells to induce the hematopoietic lineage followed by 36 hours on OP9-DL1 cells, and two-colour gene expression microarray analyses were performed on each set. Affymetrix GeneChip Operating Software (GCOS) was used to determine absent/present/moderate calls. This program generated a list of 4,244 genes that were differentially regulated in Gata3−/− versus Gata3+/− progenitors in all three experiments (Figure 2D). The data were further grouped based on enrichment by scanning manually curated gene sets: KEGG, REACTOME, BIOCARTA Pathways, and custom user created gene sets. The most striking changes in gene expression were in genes implicated in cell cycle regulation and apoptosis pathways (Figure 2E, Supplemental Figure 2A, Supplemental Table 1). Genes involved either in DC maturation, Lmp1 response, IL-23- and IL-27-mediated signaling, enterotoxin and inflammatory responses, or targeted by the Notch signaling pathway were also enriched (Supplemental Table 1).
Because we had observed a survival defect in Gata3−/− progenitors cultured on OP9-DL1 cells, we were particularly interested in the genes associated with cell cycle arrest and apoptosis that were significantly up-regulated in Gata3−/− progenitors, such as Bcl2l11 (Bim), Nr4a1, Nr4a2, Gadd45, Cdkn1a, Cdkn2a, and Cdkn2b (Figure 2E, Supplemental Figure 2A). Trp53 and some of the components of the TGFβ regulatory pathway (Ppp2ca, Ppp2r1a), the TNF pathway (Traf1, Tnfaip3 and Tnip1), and the NF-κB pathway (Nfkb1a, Nfkb1b), which have been linked to cell cycle arrest and apoptosis, were also up-regulated in Gata3−/− progenitors (Supplemental Table 1). Thus, the lack of GATA-3 in ESC-derived progenitors cultured in the presence of Notch signals leads to the up-regulation of various cell cycle inhibitors and pro-apoptotic factors and culminates in a loss of viability and an early T cell developmental arrest.
Early T cell development block in Gata3−/− bone marrow-derived progenitor cells
To extend our findings from the ESC/OP9-DL1 co-cultures, Gata3f/f mice were initially crossed with Mx1-Cre+ mice to induce GATA-3 deletion in all bone marrow cells upon polyinosinic:polycytidylic acid (poly I:C) administration (data not shown). However, to avoid the effects of the poly I:C treatment on early hematopoietic cells (57), the Gata3f/f mice were also bred to Vav-Cre+ animals to conditionally delete GATA-3 in all hematopoietic lineage cells (Figure 3). The bone marrow LSK cells were isolated from 6 to 8 week old mice and cultured on either OP9-DL1 or OP9-Control (C) cells.
Figure 3. GATA-3 is required for T-lymphopoiesis and inhibits myeloid fate in developing thymocytes.

Bone marrow-derived LSK progenitors from 6–8 week old GATA-3+/f-, GATA-3f/f-, or GATA-3f/f Cdkn2bf/f-Vav-Cre+ mice were isolated by flow cytometry. (A) Cells were cultured on OP9-DL4 cells for 8 days and analyzed by flow cytometry for the expression of the indicated markers, with the cellularity of DN2, DN3 and CD11b+ cells indicated in the side bar graphs. (B) Cells were cultured on OP9-DL4 cells for 14 days and further analyzed for the expression of the indicated markers, with the analysis of CD44 and CD25 expression on CD4−CD8− DN gated cells. Results are representative of at least three independent experiments, and cellularity for the indicated subsets are shown in the side bar graphs.
The LSK cells from Gata3f/f Vav-Cre+ (Gata3−/−) mice showed a block in T cell development at the DN2 stage, in comparison to cells from Gata3+/f Vav-Cre+ (Gata3+/−) littermate controls that progressed to later stages of differentiation (Figures 3A and 3B). The difference between these results and the Gata3−/− ESC-derived co-cultures, which lacked the DN2 subset (Figure 2A), most likely reflects the more robust lymphopoiesis from ex vivo bone marrow-derived precursors, compared to ESC-derived, hematopoietic progenitors. The presence of CD25+ DN2 cells from Gata3f/f Vav-Cre+ LSK cells did not reflect incomplete deletion of GATA-3, as demonstrated by intracellular staining for GATA3 expression in thymocytes from Gata3f/f Vav-Cre+ mice, which also showed the presence of a small number of DN2/DN3 cells, as previously reported (33), but displayed lower levels of TCF1 (Supplemental Figures 1A and 1C). Gata3f/f Vav-Cre+ LSK cells also cells failed to progress to the later stages (DN3, DN4, and DP) and failed to increase in cellularity (Figures 3A and 3B). Like their ESC-derived Gata3−/− counterparts, OP9-DL1 co-cultured Gata3f/f Mx1-Cre+ (Gata3−/−) LSK cells exhibited a higher frequency of AnnexinV+ apoptotic cells (Figure 4A). This increased tendency to die was specific to the presence of Notch signals because there was no difference in apoptosis between Gata3+/− and Gata3−/− LSK cells co-cultured in the absence of Notch-inducing signals (OP9-C cells). These results suggest that GATA-3 deficient progenitors are induced to undergo apoptosis in response to Notch signals.
Figure 4. GATA-3 is required to inhibit apoptosis but is not required to inhibit B-lymphopoiesis in the presence of Notch signals.

(A) Gata3+/− and Gata3−/− bone marrow LSK progenitors were cultured on either OP9-DL1 or OP9-C cells and analyzed for presence of CD45+AnnexinV+ cells by flow cytometry at designated time points. All data is representative of at least three independent experiments. The AnnexinV+ PI+ cell frequencies of the day 8 cultures are indicated in the side bar graph (n=3). (B-C) ESCs of indicated genotypes were cultured on OP9-C cells until day 8 of co-culture, and thereafter placed on OP9-DL1 cells (B) or on OP9-C cells (C). Gata3+/− and Gata3−/− cultures were analyzed by flow cytometry on day 19 and Rbpj+/− and Rbpj−/− cultures, on day 17. Results are representative of at least three independent experiments.
We next assessed whether the absence of GATA-3 in hematopoietic progenitors cultured in the presence of Notch signals influenced T-lineage commitment by examining their ability to up-regulate CD90 (Thy1) cell surface expression during the early stages of T cell development. In contrast to the abundant CD90hi cells in Gata3+/− cultures, only a fraction of the cells expressed low levels of CD90 in Gata3−/− LSK/OP9-DL4 co-cultures (Figure 3B). Nevertheless, CD90+ cells were present in the Gata3−/− LSK/OP9-DL4 co-cultures, providing evidence for the initiation of a Notch-induced T-lineage differentiation program (58). However, in the absence of GATA-3 a large fraction of cells did not reach the CD90hi (DN2) stage. Instead, GATA-3 deficiency led to an increase in the frequency of CD11b+ cells in these co-cultures (Figures 3A and 3B), suggesting an important role for GATA-3 in inhibiting progenitors from adopting the myeloid fate. Thus, the absence of GATA-3 in hematopoietic progenitors cultured in the presence of Notch signals diverts the progenitors away from the T and renders them unable to survive Notch-induced lineage specification events, while allowing the selective survival of myeloid lineage outcomes.
GATA-3 is dispensable for excluding the B cell fate but required to inhibit myelopoiesis in developing thymocytes
Developing thymocytes undergo lineage restriction events as they migrate through the thymus. The majority of the B cell potential is lost by the DN1 stage, and myeloid and NK potentials are lost by the DN2b stage of development. As Gata3−/− progenitors could not effectively commit to the T-lineage despite the presence of Notch signals, we examined whether GATA-3 restricts alternative lineage outcomes in developing thymocytes. An initial analysis of Gata3−/− ESC-derived progenitors revealed that these cells did not give rise to a B220+CD19+ cell population when co-cultured with OP9-DL1 cells, unlike the Rbpj−/− cells, and similar to Gata3+/− and Rbpj+/− progenitors (Figure 4B). This was not due to any defect in the ability of Gata3−/− progenitors to undergo early B cell differentiation because both ESC- and bone marrow-derived Gata3−/− progenitors produced B220+CD19+ B cells when cultured on OP9-C cells (Figures 4C).
To extend the above findings, we generated early DN subsets from Gata3+/− or Gata3−/− LSKs cultured on OP9-DL1 cells. CD44+CD25−CD117+ DN1 and CD44+CD25+CD117+ DN2 cells were sort purified and plated on either OP9-DL1 or OP9-C cells to examine the non T-lineage potential of Gata3−/− progenitors. Lympho/myelopoiesis were assessed by flow cytometry 7 days later. The GATA-3 deficient DN1 cells cultured on OP9-DL1 cells were impeded in their ability to become T cells and gave rise to CD11b+ myeloid cells, in contrast to their GATA-3 sufficient counterparts (Figure 5A, upper panel). Unlike Gata3+/− DN2 cells, GATA-3−/− DN2 cells failed to either differentiate or survive (Figure 5B, upper panel). In the absence of Notch signals, Gata3+/− DN1 and DN2 cells gave rise to a few CD25+ cells. These CD25+ cells were absent from Gata3−/− OP9-C co-cultures (Figures 5A and 5B, lower panel) and most likely represent GATA-3-dependent CD90+ innate lymphoid cells type 2 (ILC2) (59, 60). The absence of GATA-3 also allowed a greater frequency of CD11b+ cells to develop from DN1 and DN2 progenitors in the absence of Notch signals (Figures 5A and 5B, lower panel). In contrast to the tendency of Gata3−/− progenitors to adopt a myeloid fate, GATA-3-deficient DN1 and DN2 cells lacked any B-lineage potential, as illustrated by the absence of CD19+ cells in both OP9-DL1 and OP9-C co-cultures (Figures 5A and 5B, lower panel) versus Gata3−/− LSKs (Figure 5C). Thus, although GATA-3 may be sufficient to block B-cell lineage outcomes (23, 24), it is not required to inhibit B-lymphopoiesis in differentiating thymocytes, whereas it is indispensable for restraining myeloid potential.
Figure 5. GATA-3 inhibits myeloid but not B-lineage fate in response to Notch signals.

(A, B) Co-cultures of Gata3+/− and Gata3−/− LSKs with OP9-DL1 cells were sorted for CD44+CD25−CD117+ DN1 or CD44−CD25+CD117+ DN2 cells and re-seeded on either OP9-DL1 or OP9-C cells for 7 days. Lympho- and myelopoiesis were assessed by flow cytometry. (C) Bone marrow-derived LSKs from Gata3+/− and Gata3−/− mice were cultured on OP9-C cells and presence of CD11b+ and CD19+ cells analyzed by flow cytometry on day 11 of co-culture. All plots in (A-C) were gated on CD45+ cells. Numbers within each plot indicate the percentage of cells in the indicated gates or quadrants. Representative plots from at least 3 independent experiments are shown. (D) qPCR for transcripts expressed in DN2 cells derived from Gata3+/− (white bars) and Gata3−/− (black bars) bone marrow LSK-OP9-DL1 co-cultures sorted on day 8. All samples were normalized to β-actin. Mean and SEM were calculated from 3 independent qPCR analyses.
The myeloid potential of GATA-3−/− progenitors was reflected by the up-regulation of Cebpa, Spib and Id2 in Gata3−/− co-culture-derived DN2 cells, while the expression levels of other genes such as Bcl11a and Spi1 remained unchanged (Figure 5D). Perhaps most importantly, the expression of the T-lineage factor Bcl11b was completely abrogated in the absence of GATA-3 (Figure 5D). Notch signaling in Gata3−/− DN2 cells arising in OP9-DL1 co-cultures did not appear to be hindered, as illustrated by the expression of the classical Notch target gene, Ptcra, while both known GATA-3 targets, Rag1 and Rag2 (61), were down-regulated in the absence of GATA-3 in these cells. Taken together, GATA-3 appears to induce Bcl11b expression at the DN2a to DN2b transition in collaboration with Notch, leading to the repression of myeloid lineage outcomes, while Notch signaling alone is sufficient to inhibit B lymphopoiesis in developing thymocytes.
Cdkn2b is up-regulated in Gata3−/− progenitors cultured in the presence of Notch signals and mediates their apoptosis
Our observation that Gata3−/− cells failed to progress along the T-lineage developmental pathway due to increased apoptosis led us to examine whether the failure in T-lymphopoiesis was mediated by the de novo expression of cell cyclin-dependent kinase inhibitors (Cdkn). Similar to Gata3−/− ESC-derived progenitors, Gata3f/f Vav-Cre+ LSK-OP9-DL1-derived DN2 cells showed an increased expression of the Cdkn2a transcript variant p19Arf, Cdkn2b, Cdkn1b and Nr4a2 (Figure 6A), all of which can induce cell cycle arrest and apoptosis. In contrast to the ESC-derived progenitors, Cdkn1a, Cdkn1c, Gadd45b and Trp53 were not up-regulated in these cells (Figure 6A). However, this could reflect the different developmental stage of analysis between the ESC- and bone marrow LSK-derived cultured progenitors: ESC progenitors were subjected to gene expression analysis after only 36 hours of co-culture, while bone marrow-derived progenitors were interrogated at the DN2 stage.
Figure 6. GATA-3-deficient progenitors express cell cycle inhibitory and pro-apoptotic genes, which block early T cell development.

(A) qPCR for transcripts expressed by DN2 cells sorted after 8 days of co-culture of Gata3+/− (white bars) and Gata3−/− (black bars) bone marrow LSKs with OP9-DL1. All samples were normalized to β-actin. Mean and SEM were calculated from 3 independent qPCR analyses. (B) Wild type bone marrow sorted LSKs co-cultured with OP9-DL4 were retrovirally transduced with either MIY or MIY-Cdkn2b vector on day 5. Cells were sorted 24 hours post-transduction as CD45+ YFP+ CD44+CD25− DN1, cultured on OP9-DL4 cells for 3 days. Cultures were analyzed by flow cytometry for expression of CD44 and CD25 as well as (C) Annexin V and PI. (D) Percentage of Annexin V+ CD45+ cells present in the wells of wild type YFP+ DN1 progenitors expressing MIY (white bar) or MIY-Cdkn2b (black bar) co-cultured with OP9-DL4 for 3 days. (E) Wild type bone marrow LSKs were cultured with OP9-DL4 cells for 5 days and DN1 (CD44+CD25−) or DN2 (CD44+CD25+) cells purified by flow cytometry. Sorted DN1 or DN2 cells were cultured on OP9-DL4 cells in the presence or absence of PD0332991 (2 μg/ml) and co-cultures analyzed after 3 days for presence of CD44 and CD25 as well as (F) Annexin V+ and PI+ cells. (G) Percentage of Annexin V+ CD45+ untreated (white bar) or PD0332991-treated (black bar) DN1 and DN2 cells 3 days after the DN1/DN2 sort. All data is represented as mean ± SEM; n = 2, *p < 0.05 (Student t-test).
Both p19Arf and Cdkn2b are known inducers of cell cycle arrest and apoptosis and potent tumor suppressors (62–64). While p19Arf activates p53 by inhibiting Mdm2, Cdkn2b causes G1 phase arrest by inhibiting CDK4/6 (65). We ectopically expressed either p19Arf or Cdkn2b in wild-type LSK-OP9-DL4 co-culture-derived DN1 cells to confirm that the Cdkn2a and Cdkn2b gene products, which are expressed in the absence of GATA-3, impose cell cycle arrest. We switched to OP9-DL4 co-cultures in lieu of OP9-DL1 to more closely mimic the thymic environment.
The enforced expression of p19Arf had no appreciable effect on the survival and differentiation of DN1 cells (data not shown). However, enforced Cdkn2b expression prohibited cells from developing past the DN1 stage (Figure 6B) and resulted in apoptosis (Figure 6C) with nearly 100% of cells staining positive for AnnexinV (Figure 6D). Thus, although GATA-3 deficiency caused developing thymocytes to up-regulate their expression of both p19Arf and Cdkn2b mRNA, Cdkn2b appeared to be the main driving force in the ensuing cell cycle arrest and apoptosis. In agreement with this hypothesis, the treatment of wild-type bone marrow-OP9-DL4 co-culture-derived DN1 or DN2 cells with PD0332991, which inhibits cyclin-dependent kinases 4 and 6 (CDK4/6) similarly to Cdkn2b, (66), arrested T cell development in OP9-DL4 co-cultures at their respective stages in contrast to control treated cells (Figure 6E). PD0332991 treatment also induced apoptosis in DN1 and DN2 cells (Figure 6F), resulting in 66% and 75% AnnexinV+ cells in these co-cultures, respectively (Figure 6G). Thus, treatment of early DNs with a CDK4/6 inhibitor, which mimics Cdkn2b activity, phenocopies the activity of ectopically-expressed Cdkn2b. In DNs, CDK4/6 inhibition, Cdkn2b expression and GATA-3-deficiency all induce apoptosis, making dysregulation of Cdkn2b expression an attractive candidate to induce DN2 cell death.
Notwithstanding the results showing that overexpression of Cdkn2b or CDK4/6 inhibition leads to apoptosis of developing T cells, we endeavored to test whether loss of Cdkn2b expression in the absence of GATA-3 would rescue their differentiation block and apoptosis (Figure 3). LSK from Gata3f/f Cdkn2bf/f Vav-Cre+ mice cultured on OP9-DL4 cells showed a similar block in T cell development at the DN2 stage as Gata3f/f Vav-Cre+ derived cells (Figures 3A and 3B). These results suggest that although dysregulated Cdkn2b expression can lead to an arrest in T cell development, it is not uniquely required for the observed block in differentiation and loss of survival of GATA-3 deficient DN2 cells, and likely due to the compensatory function of the other cell cycle inhibitors and apoptosis inducing genes regulated by GATA-3.
GATA-3 binds and represses the Cdkn2b locus in developing thymocytes
We hypothesized that successful T-lymphopoiesis would involve GATA-3 repressing the closely linked Cdkn2a and Cdkn2b loci, preventing the expression of p19Arf and Cdkn2b, and allowing for thymocyte proliferation and T-lineage differentiation. In the absence of GATA-3, Cdkn2a and Cdkn2b loci become de-repressed and the genes transcribed, resulting in apoptosis. We performed chromatin immunoprecipitation (ChIP) assays on wild-type thymus-derived DN1–3 cells to examine the association of GATA-3 with regulatory elements in DNA sequences (RES) in the Cdkn2b promoter regions to establish whether GATA-3 directly inhibits the expression of Cdkn2a and Cdkn2b. Thirty-six possible GATA-3 RES were identified within the 5,768 bp of the readable promoter sequence and intronic regions of the Cdkn2a/Cdkn2b loci using a combination of rVista, genomatix MatlInspector and promoter scan in VectorNTI (Figure 6A). Most of the identified sites corresponded to the previously reported WGATAA motif, which represents the most prevalent GATA-3 RES in developing thymocytes (67). These were present on either a direct or a complementary DNA strand (Supplemental Table 2). As previously shown, the genetic loci bound by GATA-3 in developing thymocytes are also closely occupied by other transcription factors, some of which help to orchestrate T-lineage programs (67). An initial ChIP scan was performed over the area marked by the GATA-3 RES in close proximity to the sites potentially occupied by Bcl11b, TCF-1, Runx, Ets, AP1, Runx and Areb6 to identify the most relevant GATA-3 binding sites involved in Cdkn2a/Cdkn2b regulation. Similar to a known GATA-3 RES within the TCRβ enhancer region (68), anti-GATA-3 ChIP showed a statistically significant enrichment over the IgG control at RES #14 located 566 bp upstream of the Cdkn2b transcription start site (TSS) (Figures 7B and 7C). The remaining RES sites, such as site #7, found 2,938 bp upstream of TSS, were not enriched for GATA-3 (Figures 7B and 7C). The Cdkn2b locus was previously reported to be marked by a repressive H3K273me chromatin signature in developing thymocytes (69), and significant H3K273me enrichment was found at RES #14 and at other GATA-3 RES, such as RES #7, but was absent at the TCRβ enhancer RES (Figure 7D). Thus, GATA-3 appears to occupy a site located very close to the Cdkn2b TSS, which is marked by a repressive signature during T cell development.
Figure 7. GATA-3 and Bcl11b bind to and mediate repression of the Cdkn2b locus in developing thymocytes.

(A) Location of primers spanning all predicted GATA-3 bindings sites upstream of Cdkn2b and Cdkn2a TSS as identified by rVista, genomatix MatlInspector and manual scan in VectorNTI. (B) Putative GATA-3 RES were identified by rVista, genomatix MatlInspector and manual scan in VectorNTI. ChIP assay was performed on wild type DN1, DN2 and DN3 thymocytes with anti-GATA-3 and control IgG antibodies. Enrichment was quantified by qPCR, normalized to β-actin and calculated as percentage of input. Enrichment was further compared to a known GATA-3 DNA target located in a Tcrb enhancer region. Data is presented as fold increase of GATA-3 enrichment over the control IgG. (C) Chromatin was immunoprecipitated with anti-GATA-3, anti- H3K273me or control IgG antibodies from a combination of wild type DN1, DN2 and DN3 thymocytes. Enrichment for GATA-3 (mean ± SEM, n = 3) and (D) repressive H3K273me epigenetic signature (mean ± SEM, n = 2) at promoter regions of Cdkn2a/b and percentage of input samples before ChIP (1% of total starting cell population) were quantified by qPCR and data normalized to β-globin gene expression (*p < 0.05, Student t-test). (E) Location of primers spanning Bcl11b RES upstream of Cdkn2b as identified by ChIP-seq (See Supplemental Figure 2B) and USC genome browser. (F) Chromatin immunoprecipitation from wild type DN1, DN2 and DN3 cells with anti-CTIP2 (Bcl11b) or control IgG antibodies. Enrichment for Bcl11b (mean ± SEM, n = 3) at promoter region of Cdkn2b, along with 1% of percentage input, were quantified by qPCR and data normalized to β-globin gene expression (*p < 0.05, Student t-test).
GATA-3 collaborates with Bcl11b at the Cdkn2b promoter
GATA-3-deficient bone marrow-derived progenitors failed to up-regulate Bcl11b and repress the Cdkn2b locus in a Notch-responsive environment, thus preventing them from realizing their T-lineage program. Bcl11b is a known repressor of Cdkn1a (70) and Cdkn1c (71), and has been implicated in regulating Cdkn2b (17). Moreover, GATA-3 has been previously shown to bind the Bcl11b enhancer region (72) and is involved in inducing Bcl11b activation in T cell differentiation (73) (Figure 5D). Because GATA-3 appears to require binding partners to occupy various DNA sites throughout the genome (67), and the transcriptional control of Foxp3 expression in Tregs relies on the presence of both GATA-3 and Bcl11b in close juxtaposition to each other (74, 75), we examined whether GATA-3 both induces and collaborates with Bcl11b to repress Cdkn2b during T-lymphopoiesis. A ChIP-seq analysis of all thymocytes revealed that Bcl11b binding was enriched at a number of sites over the Cdkn2b locus (Supplemental Figure 2B). We validated this enrichment in wild-type DN1, DN2 and DN3 thymocytes using site-specific primers to scan the regions of the positive peaks (Figure 7E). Anti-Bcl11b ChIP analysis showed statistically significant binding within the Cdkn2b locus at two of the sites, which had been identified in DP thymocytes by ChIP-seq (Figure 7F and Supplemental Figure 2B). In particular, RES #11 spanned a region located 287 to 1000 bp upstream of the Cdkn2b TSS, was located in close proximity to the GATA-3 RES #14, and showed significant enrichment for Bcl11b in DN cells (Figure 7F). These findings are consistent with GATA-3 and Bcl11b cooperating to ensure the elaboration of the T-lineage developmental program by jointly binding the TSS-proximal promoter sequences of Cdkn2b and repressing this locus in the presence of Notch signals.
The ectopic expression of GATA-3 in Gata3−/− progenitors represses Cdkn2b expression and rescues the developmental block
GATA-3 expression is tightly regulated during T cell development because high concentrations of GATA-3 are detrimental to the T-lineage program and impart alternative lineage fates (35, 39). With this in mind, we initially enforced Bcl11b expression in Gata3−/− LSKs expecting that this would bypass the requirement for GATA-3 and rescue the T cell developmental arrest. While Gata3−/− progenitors expressing MIG-Bcl11b remained arrested at the DN2 stage of development, they gave rise to a more robust CD25+ population than Gata3−/− progenitors expressing MIG alone (Figure 8A, left panel). More importantly, MIG-Bcl11b allowed a greater number of cells to acquire CD90 expression than MIG alone (Figure 8A, right panel). However, the ectopic expression of Bcl11b in GATA-3-deficient progenitors did not rescue the GATA-3-deficient phenotype, increase cellularity (Figure 8B), or down-regulate Cdkn2b expression (Figure 8C). Our data indicate that although Bcl11b occupies regulatory sites in the Cdk2b promoter, it appears to require the presence of GATA-3 to act at this site.
Figure 8. GATA-3, in collaboration with Bcl11b, induces cell survival and repression of the Cdkn2b locus in early T cell development in the presence of Notch signals.
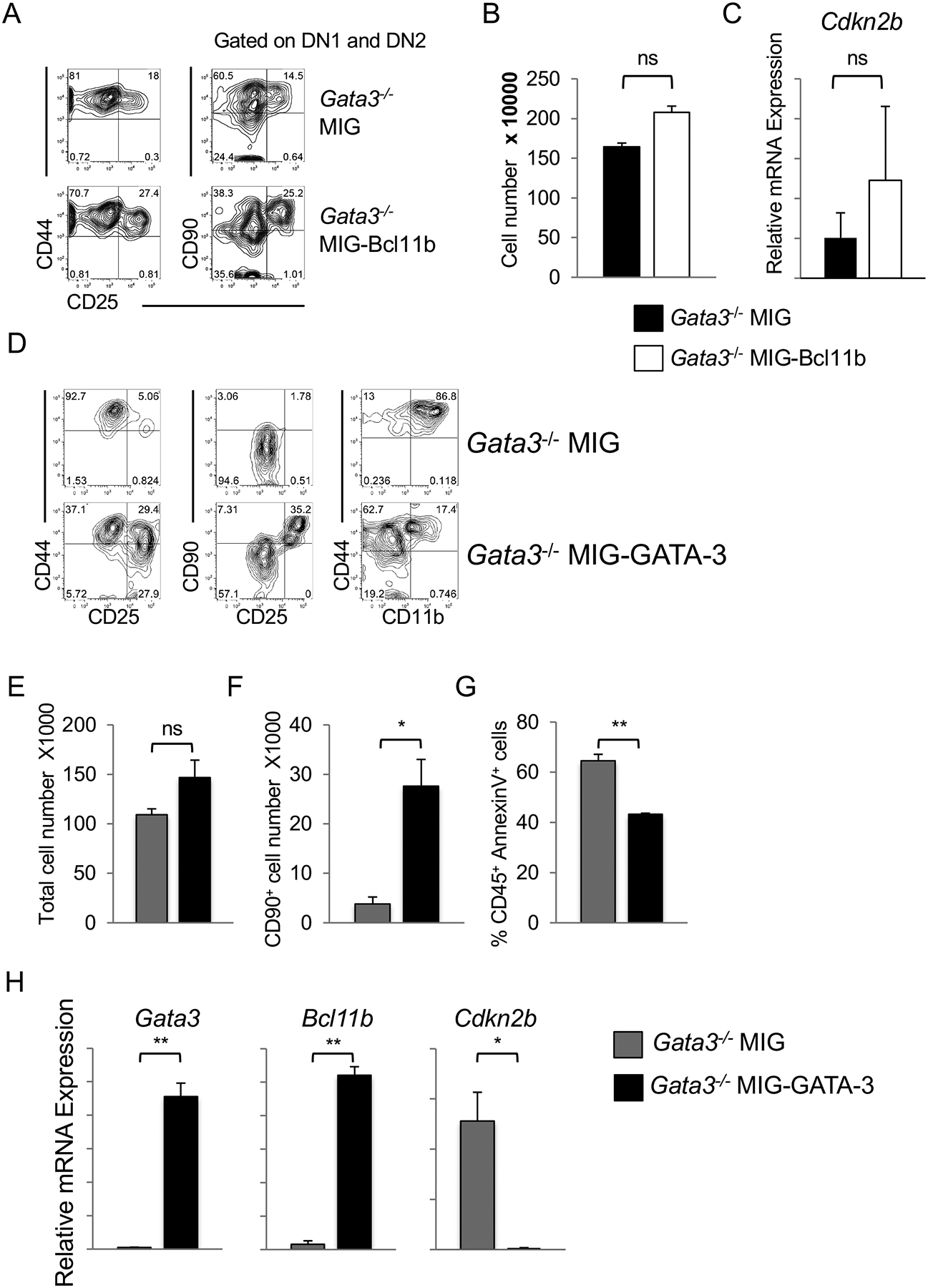
(A) Gata3−/− bone marrow-derived LSKs were retrovirally transduced with MIG or MIG-Bcl11b vector for 24 hours and sorted GFP+ cells cultured on OP9-DL4. GFP+ CD45+ cells were analyzed by flow cytometry on day 9 of co-culture for presence of CD44 and CD25 expression (left panels) with additional gating on CD44+CD25− DN1 and CD44−CD25+ DN2 cells (right panels) to assess CD90 expression. Plots are representative of at least three independent experiments. (B) Cellular fold expansion of MIG (black bar) or MIG-Bcl11b (white bar) Gata3−/−-OP9-DL4 co-cultures on day 9 (mean ± SEM; n = 2). (C) qPCR for Cdkn2b transcript expression by CD45+ GFP+ cells sorted from Gata3−/−-MIG or –MIG-Bcl11b cultured on OP9-DL4 cells for 9 days (mean ± SEM; n = 2). (D) Gata3−/− bone marrow-derived LSKs cultured on OP9-DL4 for 4 days were retrovirally transduced with either MIG or MIG-GATA-3 construct for 24 hours. CD45+ GFP+ cells were sorted and cultured on OP9-DL4 cells and GFP+ CD45+ cells analyzed by flow cytometry 3 days later with additional gating on CD44+CD25− DN1 and CD44−CD25+ DN2 cells (middle panels). Plots are representative of at least three separate experiments. (E) Cellular fold expansion of MIG (grey bar) or MIG-GATA-3 (black bar) Gata3−/−-OP9-DL4 co-cultures 3 days post-transduction (mean ± SEM; n = 3). (F) Percentage of GFP+ CD45+ CD90+ CD25+ and (G) Annexin+ cells arising from MIG- or MIG-GATA-3 transduced Gata3−/− cells cultured on OP9-DL4 for 3 and 6 days, respectively (mean ± SEM; n = 3). (H) qPCR for transcripts expressed by Gata3−/− MIG- or MIG-GATA-3-transduced cells (CD45+ GFP+) 3 days post-transduction and co-cultured with OP9-DL4 cells (mean ± SEM; n = 3), *p < 0.05, **p < 0.01; Student t-test.
We retrovirally transduced Gata3+/− or Gata3−/− LSK-OP9-DL4-derived progenitors on day 4 of co-culture to express either MIG or MIG-GATA-3 and sorted GFP+ CD45+ DNs to culture further on OP9-DL4 cells. In contrast to the Gata3−/− cells expressing MIG alone that were arrested at an early DN2 stage of T lymphopoiesis, those transduced with MIG-GATA-3 gave rise to DN3 cells similarly to Gata3+/− progenitors (Figure 8D, left and middle panels). Of note, the ectopic expression of GATA-3 in deficient progenitors also hindered their ability to give rise to myeloid CD11b+ cells (Figure 8D, right panel). Although the total cell numbers were not significantly different between the MIG and MIG-GATA-3-transduced groups (Figure 8E), there was a significant increase in the percentage of CD25+ CD90+ cells in co-cultures seeded with Gata3−/−-MIG-GATA-3 progenitors (Figure 8F). The ectopic expression of GATA-3 in Gata3−/− CD45+ progenitors enhanced cell survival, as evidenced by a striking decrease in the percentage of CD45+ AnnexinV+ cells (Figure 8G), and, critically, resulted in a significant up-regulation of Bcl11b and down-regulation of Cdkn2b expression over Gata3−/− MIG-expressing cells (Figure 8H). Our findings indicate that GATA-3 acts downstream of instructive Notch signals both to induce the expression of Bcl11b to inhibit a myeloid developmental program, and to collaborate with Bcl11b to repress the Cdkn2b gene locus and allow for thymocyte survival and progression along the T-lineage differentiation program.
Discussion
The essential requirement for GATA-3 during early T-lymphopoiesis is widely acknowledged, but the mechanism by which GATA-3 allows for T-lineage specification and commitment has not been fully elucidated. In this study, we show that GATA-3 is differentially expressed in thymocyte subsets that are actively cycling and poised to fully commit to the T cell fate, and that GATA-3 deficiency abrogates T cell development at an early DN2 stage despite ongoing Notch responsiveness. Further, we provide evidence that GATA-3 induces the expression of Bcl11b, joins it at the Cdkn2b locus, and collaborates with it to mediate the repression of this tumor suppressor gene in early thymocytes. As a result, GATA-3 ensures cell survival, proliferation and the realization of the T-lineage commitment program in thymocytes responding to Notch signals.
Characterizing the role of GATA-3 during T cell development has been complicated by the early embryonic lethality that occurs in Gata3−/− mice (20). Gata3−/− ESCs and conditional knockout mice both provide the means to circumvent this lethality, and each approach has strengths and weaknesses. The former approach, using in vitro-differentiated ESCs, allows the use of completely GATA-3-deficient cells to be compared with Gata3+/− progenitors under identical culture conditions. However, it uses progenitors that are propagated in vitro, requires the in vitro induction of hematopoiesis from ESCs, and may not fully recapitulate the in vivo phenotype. The latter model allows ex vivo cells to be used but deletion using Mx1-Cre may be incomplete, and the inductive stimulus activates the type I interferon (IFN) pathway and may have deleterious effects on either hematopoietic cells or their environment (57). Because of these caveats, we focused on a third approach by using a Vav-Cre-mediated deletion of GATA-3. This genetic approach allowed for a complete deletion of Gata3-floxed alleles and, together with the GATA-3-deficient ESC system, enabled us to perform a focused examination of the role of GATA-3 in T cell development.
A profound requirement for GATA-3 from the earliest stages of T cell development has been previously shown; however, whether GATA-3 is required for commitment to the T cell fate or survival following T cell commitment, and how the Notch pathway and GATA-3 collaborate during T cell development are still unresolved. In agreement with work by Rothenberg and colleagues (24), we found that GATA-3 expression in thymic DNs rose in cells undergoing T-lineage commitment. Further, we found that the protein was differentially shuttled between the cytoplasm and nucleus during either differentiation or proliferative bursts, with nuclear location corresponding to active cell cycling, as previously noted in hematopoietic stem cells (53). GATA3 localization may be cell cycle-coupled which linked to the suppression in the expression of inhibitors of cell cycling. Nuclear-located, and thus presumably transcriptionally active (53), GATA-3 was found in the DNs most likely poised for or having just passed one of the sequential T-lineage commitment steps. Interestingly, the nuclear localization of GATA-3 was largely precluded in DN1c and DN1d cells, which have been previously shown to harbor multi-lineage potentials (54, 76, 77). Therefore, it appears that GATA-3 is a key player in T cell fate acquisition and regulation of cell cycle progression in thymic progenitors.
Notch/Dll interactions are sufficient to induce T cell specification even in the absence of GATA-3: Gata3−/− bone marrow-derived progenitors generate a population of CD25+ DN2 cells that express T-lineage genes, such as Ptcra. However, Notch alone could not fully impose T-lineage commitment without GATA-3. The gradual loss of GATA-3-deficient DN2 cells in OP9-DL co-cultures suggests that Gata3−/− progenitors respond to Notch signals by initiating T cell development, including the upregulation of TCF1 expression, but cannot survive this specification event. Both Rag-1 and Rag-2 were completely absent in Gata3−/− DN2 cells, consistent with the literature implicating GATA-3 in the regulation of the Rag genes in T lineage cells (61). However, the lack of RAG expression does not account for the early requirement for GATA-3 because Rag−/− thymocytes are not impaired until the β-selection checkpoint at the DN3 stage of development (78, 79). The early DN2 block from Gata3−/− progenitors resembles that observed in the absence of Bcl11b (16, 80), which is consistent with the loss of Bcl11b in GATA-3 deficient thymocytes (81). However, Bcl11b alone cannot rescue the Gata3−/− DN2 block and loss of cell viability, as ectopic expression of Bcl11b in the absence of GATA-3 did not re-establish T lymphopoiesis, despite a modest increase in the number of CD90+ cells. By contrast, early T cell development from Gata3−/− progenitors could be partly rescued by enforced GATA-3 expression, which prompted Bcl11b up-regulation. These results indicate that both GATA-3 and Bcl11b are required downstream of Notch signaling. Recently, a similar conclusion was reached when progenitors lacking the Notch target gene Tcf7 cultured on OP9-DL1 cells failed to upregulate both GATA-3 and Bcl11b expression, leading to a DN2 block in T cell development (82).
The role of GATA-3 in cementing T-lineage specification and/or commitment has been previously addressed, and GATA-3 was implicated in shutting off B-lineage potential in DN cells (23, 24). We did not observe a shift towards a B cell fate from GATA-3-deficient progenitors responding to Notch signals, although we do observe that Rbpj−/− progenitors undergo robust B cell development in the presence of Notch/Dll interactions. Furthermore, Gata3−/− DN1 or DN2 cells, which arise in the presence of Notch signals, could not be diverted to a B cell fate even in the absence of Notch signals, likely due to the induction of T lineage associated genes, such as TCF1. This supports the hypothesis that Notch signals are sufficient to block B lymphopoiesis even in the absence of GATA-3 (32). The discrepancy between our findings and those of others may stem from employing different animal models and means of generating GATA-3-deficient progenitors. Generating Gata3−/− fetal liver HSC chimeras in Rag−/− recipients might skew the ability of GATA-3-deficient cells to undergo B lymphopoiesis (23). Similarly, employing in vitro retrovirally-mediated Cre-deletions of the GATA-3 floxed alleles in either fetal liver hematopoietic progenitors or DN2 cells might change the lineage potential of the fetal cells (24). The use of fetal progenitor may be another difference from our approach.
Although Notch signaling was able to prevent B-lymphopoiesis in the absence of GATA-3, it could not inhibit a myeloid lineage outcome. Together with an up-regulation of myeloid lineage-promoting transcription factors, we found increased numbers of CD11b+ cells arising in Gata3−/−-OP9-DL1 cocultures, and a greater propensity of GATA-3-deficient DN1 and DN2 cells to adopt a myeloid phenotype. This phenomenon most likely stems from the inability of Gata3−/− progenitors to up-regulate Bcl11b, which normally represses myeloid fates (80, 82). Similarly, the ectopic expression of GATA-3 in Gata3−/− DNs cultured on OP9-DL4 induced Bcl11b expression and led to a decrease in CD11b+ cells in these co-cultures. This further supports a role for Bcl11b in blocking myeloid outcomes, albeit under the control of GATA-3 (83, 84).
It is clear that GATA-3 not only plays a role in the induction of T cell lineage genes but also regulates progenitor differentiation and survival in response to Notch signaling. Compared to Gata3+/− cells, Gata3−/− progenitors underwent apoptosis on OP9-Dll, but not OP9-C, cells. This coincided with the increased expression of a number of cell cycle inhibitors and pro-apoptotic factors in these cells, such as p19Arf and Cdkn2b. Not surprisingly, loss of either p19Arf or Cdkn2b in GATA-3 deficient progenitors did not lead to a rescue in their differentiation beyond the DN2 stage of development. This is likely due to compensation by other cell cycle inhibitors and apoptotic genes that become dysregulated in the absence of GATA-3.
ChIP analysis showed both GATA-3 and Bcl11b were enriched immediately upstream of the Cdkn2b TSS in a region marked by an H3K273me signature. This suggests that both transcription factors are required to repress the Cdkn2b, and possibly the Cdkn2a, locus in thymocytes. The up-regulation of p19Arf was previously shown to block early T cell development and induce apoptosis (85). Although we observed increased p19Arf expression in Gata3−/− DN2 cells, ectopically expressed p19Arf failed to induce any significant developmental arrest or apoptosis in DN thymocytes. In contrast, the effects of ectopic Cdkn2b positioned Cdkn2b as a potential executor of apoptosis in GATA-3-deficient thymocytes.
GATA-3 has been previously shown to mediate transcriptional repression of the Ifng locus and the Cdkn2c locus to promote Th2 lineage development (86, 87). Furthermore, GATA-3 also binds Cdkn2c in mammary luminal cells (88), and GATA-3-deficient ILC2s were shown to have higher levels of Cdkn2b than their wild type counterparts (60). Interestingly, GATA-3 repression of Cdkn2c in Th2 cells was shown to require the interacting partner Ruvbl2 (86), a finding that is reminiscent of a GATA-3-Bcl11b partnership for the regulation of Cdkn2b locus.
The lack of GATA-3 and consequent loss of Bcl11b leads to the apoptosis of differentiating thymocytes that may involve the de-repression and activation of Cdkn2b, an inhibitor of CDK4/6. This is consistent with the important role of CDK4/6 during early T cell development (89), and its regulation by Notch signals (90). The up-regulation of Cdkn2b expression in the absence of GATA-3-mediated repression could result via multiple pathways. The gene expression profile of Gata3−/− progenitors provides some clues and implicates the activation of the TGFβ signaling pathway. GATA-3 has been previously shown to inhibit the activation of TGFβ (91), and some TGFβ signaling components were up-regulated in the absence of GATA-3. Interestingly, TGFβ, either directly or via Miz-1, can activate Cdkn2b by inhibiting a Myc-mediated repression of this locus (92, 93). This is achieved via subsequent activation of SMAD proteins, including SMAD2 (94, 95), which was up-regulated in Gata3−/− progenitors. Other clues pointing to the involvement of the TGFβ pathway in the activation of Cdkn2b are the up-regulation of Protein Phosphatase 2A Catalytic subunits (Ppp2ca and Ppp2r1a), key components of the TGFβ signaling and cell cycle arrest pathways (96, 97) and the TGFβ target Bim (Bcl2l11) (98, 99).
Our data indicate that GATA-3 is required to repress Cdkn2b via the activation of Bcl11b expression, providing a new mechanistic understanding of how GATA-3 may promote the survival of thymocytes undergoing early T-lineage differentiation. These observations clarify the key role of GATA-3, alongside Notch, in T-lymphopoiesis. Taken together, our findings elucidate how GATA-3 both induces Bcl11b expression and collaborates with it in the repression of the Cdkn2b locus to permit the realization of the Notch-induced T cell developmental program.
Supplementary Material
Highlights.
GATA3 is required for a Notch induced program of T-lineage differentiation.
GATA3 is required for Bcl11b expression, and together repress the Cdkn2 locus
Dysregulated Cdkn2 expression leads to apoptosis of early T-lineage cells.
Acknowledgments
We thank Gisele Knowles, Courtney MacIntosh and Arian Khadani for their expert flow cytometry support, Roxanne Holmes for her technical support with the ESC/OP9 cell co-cultures, Drs. Hiroshi Nakase for RNA labeling and generating the microarray data, and Philippe Kastner for generating the Bcl11b ChIP-seq data set. We sincerely appreciate Drs. I. Cheng Ho and Sun Yun Pai for providing us the Gata3f/f mice, and Dr. I. Cheng Ho for the Gata3−/− ESCs, Drs. Thomas Graf and Nancy Speck for the Vav-Cre mice. We are grateful to Dr. Howard T. Petrie for his assistance in the analysis of the microarray data.
Footnotes
This work was supported by funds from the Canadian Institutes of Health Research (CIHR, MOP-119538, FDN-154332 to JCZ-P), Canadian Cancer Society Research Institute (JCZ-P), and a CIHR Banting and Best Doctoral Research Award (PKT). JCZ-P is a recipient of a Canada Research Chair in Developmental Immunology.
References
- 1.Petrie HT, and Zuniga-Pflucker JC. 2007. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol 25: 649–679. [DOI] [PubMed] [Google Scholar]
- 2.Bhandoola A, von Boehmer H, Petrie HT, and Zuniga-Pflucker JC. 2007. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity 26: 678–689. [DOI] [PubMed] [Google Scholar]
- 3.Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, and Bhandoola A. 2005. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol 6: 663–670. [DOI] [PubMed] [Google Scholar]
- 4.Heinzel K, Benz C, Martins VC, Haidl ID, and Bleul CC. 2007. Bone marrow-derived hemopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. J Immunol 178: 858–868. [DOI] [PubMed] [Google Scholar]
- 5.Benz C, Martins VC, Radtke F, and Bleul CC. 2008. The stream of precursors that colonizes the thymus proceeds selectively through the early T lineage precursor stage of T cell development. J Exp Med 205: 1187–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen ELY, Thompson PK, and Zuniga-Pflucker JC. 2019. RBPJ-dependent Notch signaling initiates the T cell program in a subset of thymus-seeding progenitors. Nat Immunol 20: 1456–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karsunky H, Inlay MA, Serwold T, Bhattacharya D, and Weissman IL. 2008. Flk2+ common lymphoid progenitors possess equivalent differentiation potential for the B and T lineages. Blood 111: 5562–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benz C, and Bleul CC. 2005. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J Exp Med 202: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinzel K, Benz C, Martins VC, Haidl ID, and Bleul CC. 2007. Bone marrow-derived hemopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. J Immunol 178: 858–868. [DOI] [PubMed] [Google Scholar]
- 10.Serwold T, Ehrlich LIR, and Weissman IL. 2009. Reductive isolation from bone marrow and blood implicates common lymphoid progenitors as the major source of thymopoiesis. Blood 113: 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, and Petrie HT. 2004. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity 20: 735–745. [DOI] [PubMed] [Google Scholar]
- 12.Hayday AC, and Pennington DJ. 2007. Key factors in the organized chaos of early T cell development. Nat Immunol 8: 137–144. [DOI] [PubMed] [Google Scholar]
- 13.Radtke F, Fasnacht N, and Macdonald HR. 2010. Notch signaling in the immune system. Immunity 32: 14–27. [DOI] [PubMed] [Google Scholar]
- 14.Yuan JS, Kousis PC, Suliman S, Visan I, and Guidos CJ. 2010. Functions of notch signaling in the immune system: consensus and controversies. Annu Rev Immunol 28: 343–365. [DOI] [PubMed] [Google Scholar]
- 15.Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, and Bhandoola A. 2011. A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476: 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Leid M, and Rothenberg EV. 2010. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 329: 89–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li P, Burke S, Wang J, Che X, Ortiz M, Lee S-C, Lu D, Campos D, Goulding D, Ng BL, Dougan G, Huntly B, Gottens B, Jenkins NA, Copeland NG, Colucci F, and Liu P. 2010. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 329: 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, Aizawa S, and Kominami R. 2003. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol 4: 533–539. [DOI] [PubMed] [Google Scholar]
- 19.Germar K, Dose M, Konstantinou T, Zhang J, Wang H, Lobry C, Arnett KL, Blacklow SC, Aifantis I, Aster JC, and Gounari F. 2011. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A 108: 20060–20065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandolfi PP, Roth ME, Karis A, Leonard MW, Dzierzak E, Grosveld FG, Engel JD, and Lindenbaum MH. 1995. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat Genet 11: 40–44. [DOI] [PubMed] [Google Scholar]
- 21.Ting CN, Olson MC, Barton KP, and Leiden JM. 1996. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature 384: 474–478. [DOI] [PubMed] [Google Scholar]
- 22.Hendriks RW, Nawijn MC, Engel JD, van Doorninck H, Grosveld F, and Karis A. 1999. Expression of the transcription factor GATA-3 is required for the development of the earliest T cell progenitors and correlates with stages of cellular proliferation in the thymus. Eur J Immunol 29: 1912–1918. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Ojeda ME, Klein Wolterink RG, Lemaitre F, Richard-Le Goff O, Hasan M, Hendriks RW, Cumano A, and Di Santo JP. 2013. GATA-3 promotes T-cell specification by repressing B-cell potential in pro-T cells in mice. Blood 121: 1749–1759. [DOI] [PubMed] [Google Scholar]
- 24.Scripture-Adams DD, Damle SS, Li L, Elihu KJ, Qin S, Arias AM, Butler RR 3rd, Champhekar A, Zhang JA, and Rothenberg EV. 2014. GATA-3 dose-dependent checkpoints in early T cell commitment. J Immunol 193: 3470–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, and Alberola-Ila J. 2003. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity 19: 83–94. [DOI] [PubMed] [Google Scholar]
- 26.Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, and Ho IC. 2003. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity 19: 863–875. [DOI] [PubMed] [Google Scholar]
- 27.Tanigaki K, and Honjo T. 2007. Regulation of lymphocyte development by Notch signaling. Nat Immunol 8: 451–456. [DOI] [PubMed] [Google Scholar]
- 28.Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, Busslinger M, McCright B, Gridley T, and Flavell RA. 2007. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity 27: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, and Pear WS. 2007. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity 27: 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, and Flavell RA. 2004. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell 117: 515–526. [DOI] [PubMed] [Google Scholar]
- 31.Tindemans I, Serafini N, Di Santo JP, and Hendriks RW. 2014. GATA-3 Function in Innate and Adaptive Immunity. Immunity 41: 191–206. [DOI] [PubMed] [Google Scholar]
- 32.Hozumi K, Negishi N, Tsuchiya I, Abe N, Hirano KI, Suzuki D, Yamamoto M, Engel JD, and Habu S. 2008. Notch signaling is necessary for GATA3 function in the initiation of T cell development. Eur J Immunol 38: 977–985. [DOI] [PubMed] [Google Scholar]
- 33.Hosoya T, Kuroha T, Moriguchi T, Cummings D, Maillard I, Lim KK, and Engel JD. 2009. GATA-3 is required for early T lineage progenitor development. J Exp Med 206: 2987–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pui JC, Allman D, Xu L, DeRocco S, Karnell FG, Bakkour S, Lee JY, Kadesch T, Hardy RR, Aster JC, and Pear WS. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11: 299–308. [DOI] [PubMed] [Google Scholar]
- 35.Anderson MK, Hernandez-Hoyos G, Dionne CJ, Arias AM, Chen D, and Rothenberg EV. 2002. Definition of regulatory network elements for T cell development by perturbation analysis with PU.1 and GATA-3. Dev Biol 246: 103–121. [DOI] [PubMed] [Google Scholar]
- 36.Ling KW, van Hamburg JP, de Bruijn MJ, Kurek D, Dingjan GM, and Hendriks RW. 2007. GATA3 controls the expression of CD5 and the T cell receptor during CD4 T cell lineage development. Eur J Immunol 37: 1043–1052. [DOI] [PubMed] [Google Scholar]
- 37.Nawijn MC, Ferreira R, Dingjan GM, Kahre O, Drabek D, Karis A, Grosveld F, and Hendriks RW. 2001. Enforced expression of GATA-3 during T cell development inhibits maturation of CD8 single-positive cells and induces thymic lymphoma in transgenic mice. J Immunol 167: 715–723. [DOI] [PubMed] [Google Scholar]
- 38.Taghon T, De Smedt M, Stolz F, Cnockaert M, Plum J, and Leclercq G. 2001. Enforced expression of GATA-3 severely reduces human thymic cellularity. J Immunol 167: 4468–4475. [DOI] [PubMed] [Google Scholar]
- 39.Taghon T, Yui MA, and Rothenberg EV. 2007. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol 8: 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhn R, Schwenk F, Aguet M, and Rajewsky K. 1995. Inducible gene targeting in mice. Science 269: 1427–1429. [DOI] [PubMed] [Google Scholar]
- 41.Stadtfeld M, and Graf T. 2005. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development 132: 203–213. [DOI] [PubMed] [Google Scholar]
- 42.Bies J, Sramko M, Fares J, Rosu-Myles M, Zhang S, Koller R, and Wolff L. 2010. Myeloid-specific inactivation of p15Ink4b results in monocytosis and predisposition to myeloid leukemia. Blood 116: 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitt TM, and Zúñiga-Pflücker JC. 2002. Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 in vitro. Immunity 17: 749–756. [DOI] [PubMed] [Google Scholar]
- 44.Robertson EJ 1997. Derivation and maintenance of embryonic stem cell cultures. Methods Mol Biol 75: 173–184. [DOI] [PubMed] [Google Scholar]
- 45.Cho SK, Webber TD, Carlyle JR, Nakano T, Lewis SM, and Zúñiga-Pflücker JC. 1999. Functional characterization of B lymphocytes generated in vitro from embryonic stem cells. Proc Natl Acad Sci U S A 96: 9797–9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakano T, Kodama H, and Honjo T. 1994. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science 265: 1098–1101. [DOI] [PubMed] [Google Scholar]
- 47.Holst J, Szymczak-Workman AL, Vignali KM, Burton AR, Workman CJ, and Vignali DA. 2006. Generation of T-cell receptor retrogenic mice. Nat Protoc 1: 406–417. [DOI] [PubMed] [Google Scholar]
- 48.Grifith AV, Fallahi M, Nakase H, Gosink M, Young B, and Petrie HT. 2009. Spatial mapping of thymic stromal microenvironments reveals unique features influencing T lymphoid differentiation. Immunity 31: 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tabrizifard S, Olaru A, Plotkin J, Fallahi-Sichani M, Livak F, and Petrie HT. 2004. Analysis of transcription factor expression during discrete stages of postnatal thymocyte differentiation. J Immunol 173: 1094–1102. [DOI] [PubMed] [Google Scholar]
- 50.Kastner P, Chan S, Vogel WK, Zhang LJ, Topark-Ngarm A, Golonzhka O, Jost B, Le Gras S, Gross MK, and Leid M. 2010. Bcl11b represses a mature T-cell gene expression program in immature CD4(+)CD8(+) thymocytes. Eur J Immunol 40: 2143–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tydell CC, David-Fung ES, Moore JE, Rowen L, Taghon T, and Rothenberg EV. 2007. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J Immunol 179: 421–438. [DOI] [PubMed] [Google Scholar]
- 52.David-Fung ES, Butler R, Buzi G, Yui MA, Diamond RA, Anderson MK, Rowen L, and Rothenberg EV. 2009. Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev Biol 325: 444–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frelin C, Herrington R, Janmohamed S, Barbara M, Tran G, Paige CJ, Benveniste P, Zuniga-Pflucker JC, Souabni A, Busslinger M, and Iscove NN. 2013. GATA-3 regulates the self-renewal of long-term hematopoietic stem cells. Nat Immunol 14: 1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zúñiga-Pflücker JC, and Petrie HT. 2004. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity 20: 735–745. [DOI] [PubMed] [Google Scholar]
- 55.Schmitt TM, de Pooter RF, Gronski MA, Cho SK, Ohashi PS, and Zuniga-Pflucker JC. 2004. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat Immunol 5: 410–417. [DOI] [PubMed] [Google Scholar]
- 56.de Pooter RF, Schmitt TM, de la Pompa JL, Fujiwara Y, Orkin SH, and Zuniga-Pflucker JC. 2006. Notch signaling requires GATA-2 to inhibit myelopoiesis from embryonic stem cells and primary hemopoietic progenitors. J Immunol 176: 5267–5275. [DOI] [PubMed] [Google Scholar]
- 57.Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, and Trumpp A. 2009. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 458: 904–908. [DOI] [PubMed] [Google Scholar]
- 58.Taghon TN, David ES, Zuniga-Pflucker JC, and Rothenberg EV. 2005. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev 19: 965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, Voehringer D, Busslinger M, and Diefenbach A. 2012. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity 37: 634–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, Hu G, Barron L, Sharma S, Nakayama T, Belkaid Y, Zhao K, and Zhu J. 2014. The transcription factor GATA3 is critical for the development of all IL-7Ralpha-expressing innate lymphoid cells. Immunity 40: 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kishi H, Wei XC, Jin ZX, Fujishiro Y, Nagata T, Matsuda T, and Muraguchi A. 2000. Lineage-specific regulation of the murine RAG-2 promoter: GATA-3 in T cells and Pax-5 in B cells. Blood 95: 3845–3852. [PubMed] [Google Scholar]
- 62.Lanigan F, Geraghty JG, and Bracken AP. 2011. Transcriptional regulation of cellular senescence. Oncogene 30: 2901–2911. [DOI] [PubMed] [Google Scholar]
- 63.Kim WY, and Sharpless NE. 2006. The regulation of INK4/ARF in cancer and aging. Cell 127: 265–275. [DOI] [PubMed] [Google Scholar]
- 64.LaPak KM, and Burd CE. 2014. The molecular balancing act of p16(INK4a) in cancer and aging. Mol Cancer Res 12: 167–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gil J, and Peters G. 2006. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol 7: 667–677. [DOI] [PubMed] [Google Scholar]
- 66.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, Albassam M, Zheng X, Leopold WR, Pryer NK, and Toogood PL. 2004. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther 3: 1427–1438. [PubMed] [Google Scholar]
- 67.Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, Zhu J, and Zhao K. 2011. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity 35: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang HC, Han L, Jabeen R, Carotta S, Nutt SL, and Kaplan MH. 2009. PU.1 regulates TCR expression by modulating GATA-3 activity. J Immunol 183: 4887–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang JA, Mortazavi A, Williams BA, Wold BJ, and Rothenberg EV. 2012. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149: 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cherrier T, Suzanne S, Redel L, Calao M, Marban C, Samah B, Mukerjee R, Schwartz C, Gras G, Sawaya BE, Zeichner SL, Aunis D, Van Lint C, and Rohr O. 2009. p21(WAF1) gene promoter is epigenetically silenced by CTIP2 and SUV39H1. Oncogene 28: 3380–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Topark-Ngarm A, Golonzhka O, Peterson VJ, Barrett B Jr., Martinez B, Crofoot K, Filtz TM, and Leid M. 2006. CTIP2 associates with the NuRD complex on the promoter of p57KIP2, a newly identified CTIP2 target gene. J Biol Chem 281: 32272–32283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L, Zhang JA, Dose M, Kueh HY, Mosadeghi R, Gounari F, and Rothenberg EV. 2013. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood 122: 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fang D, Cui K, Hu G, Gurram RK, Zhong C, Oler AJ, Yagi R, Zhao M, Sharma S, Liu P, Sun B, Zhao K, and Zhu J. 2018. Bcl11b, a novel GATA3-interacting protein, suppresses Th1 while limiting Th2 cell differentiation. J Exp Med 215: 1449–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vanvalkenburgh J, Albu DI, Bapanpally C, Casanova S, Califano D, Jones DM, Ignatowicz L, Kawamoto S, Fagarasan S, Jenkins NA, Copeland NG, Liu P, and Avram D. 2011. Critical role of Bcl11b in suppressor function of T regulatory cells and prevention of inflammatory bowel disease. J Exp Med 208: 2069–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y, Su MA, and Wan YY. 2011. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity 35: 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bell JJ, and Bhandoola A. 2008. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature 452: 764–767. [DOI] [PubMed] [Google Scholar]
- 77.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, and Kawamoto H. 2008. Adult T-cell progenitors retain myeloid potential. Nature 452: 768–772. [DOI] [PubMed] [Google Scholar]
- 78.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, and et al. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68: 855–867. [DOI] [PubMed] [Google Scholar]
- 79.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, and Papaioannou VE. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68: 869–877. [DOI] [PubMed] [Google Scholar]
- 80.Ikawa T, Hirose S, Masuda K, Kakugawa K, Satoh R, Shibano-Satoh A, Kominami R, Katsura Y, Kawamoto H. 2010. An essential developmental checkpoint for production of the T cell lineage. Science 329: 93–96. [DOI] [PubMed] [Google Scholar]
- 81.Kueh HY, Yui MA, Ng KK, Pease SS, Zhang JA, Damle SS, Freedman G, Siu S, Bernstein ID, Elowitz MB, and Rothenberg EV. 2016. Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment. Nat Immunol 17: 956–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garcia-Perez L, Famili F, Cordes M, Brugman M, van Eggermond M, Wu H, Chouaref J, Granado DSL, Tiemessen MM, Pike-Overzet K, Daxinger L, and Staal FJT. 2020. Functional definition of a transcription factor hierarchy regulating T cell lineage commitment. Sci Adv 6: eaaw7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hosokawa H, and Rothenberg EV. 2021. How transcription factors drive choice of the T cell fate. Nat Rev Immunol 21: 162–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Longabaugh WJR, Zeng W, Zhang JA, Hosokawa H, Jansen CS, Li L, Romero-Wolf M, Liu P, Kueh HY, Mortazavi A, and Rothenberg EV. 2017. Bcl11b and combinatorial resolution of cell fate in the T-cell gene regulatory network. Proc Natl Acad Sci U S A 114: 5800–5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miyazaki M, Miyazaki K, Itoi M, Katoh Y, Guo Y, Kanno R, Katoh-Fukui Y, Honda H, Amagai T, van Lohuizen M, Kawamoto H, and Kanno M. 2008. Thymocyte proliferation induced by pre-T cell receptor signaling is maintained through polycomb gene product Bmi-1-mediated Cdkn2a repression. Immunity 28: 231–245. [DOI] [PubMed] [Google Scholar]
- 86.Hosokawa H, Tanaka T, Kato M, Shinoda K, Tohyama H, Hanazawa A, Tamaki Y, Hirahara K, Yagi R, Sakikawa I, Morita A, Nagira M, Poyurovsky MV, Suzuki Y, Motohashi S, and Nakayama T. 2013. Gata3/Ruvbl2 complex regulates T helper 2 cell proliferation via repression of Cdkn2c expression. Proc Natl Acad Sci U S A 110: 18626–18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hosokawa H, Tanaka T, Suzuki Y, Iwamura C, Ohkubo S, Endoh K, Kato M, Endo Y, Onodera A, Tumes DJ, Kanai A, Sugano S, and Nakayama T. 2013. Functionally distinct Gata3/Chd4 complexes coordinately establish T helper 2 (Th2) cell identity. Proc Natl Acad Sci U S A 110: 4691–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pei XH, Bai F, Smith MD, Usary J, Fan C, Pai SY, Ho IC, Perou CM, and Xiong Y. 2009. CDK inhibitor p18(INK4c) is a downstream target of GATA3 and restrains mammary luminal progenitor cell proliferation and tumorigenesis. Cancer Cell 15: 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu MG, Deshpande A, Schlichting N, Hinds EA, Mao C, Dose M, Hu GF, Van Etten RA, Gounari F, and Hinds PW. 2011. CDK6 kinase activity is required for thymocyte development. Blood 117: 6120–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Joshi I, Minter LM, Telfer J, Demarest RM, Capobianco AJ, Aster JC, Sicinski P, Fauq A, Golde TE, and Osborne BA. 2009. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood 113: 1689–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sun J, He H, Pillai S, Xiong Y, Challa S, Xu L, Chellappan S, and Yang S. 2013. GATA3 transcription factor abrogates Smad4 transcription factor-mediated fascin overexpression, invadopodium formation, and breast cancer cell invasion. J Biol Chem 288: 36971–36982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, and Massague J. 2001. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol 3: 400–408. [DOI] [PubMed] [Google Scholar]
- 93.Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Moroy T, Bartek J, Massague J, Hanel F, and Eilers M. 2001. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol 3: 392–399. [DOI] [PubMed] [Google Scholar]
- 94.Feng XH, Liang YY, Liang M, Zhai W, and Lin X. 2002. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-beta-mediated induction of the CDK inhibitor p15(Ink4B). Mol Cell 9: 133–143. [DOI] [PubMed] [Google Scholar]
- 95.Feng XH, Lin X, and Derynck R. 2000. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-beta. EMBO J 19: 5178–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Janssens V, and Rebollo A. 2012. The role and therapeutic potential of Ser/Thr phosphatase PP2A in apoptotic signalling networks in human cancer cells. Curr Mol Med 12: 268–287. [DOI] [PubMed] [Google Scholar]
- 97.Seshacharyulu P, Pandey P, Datta K, and Batra SK. 2013. Phosphatase: PP2A structural importance, regulation and its aberrant expression in cancer. Cancer Lett 335: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wiener Z, Band AM, Kallio P, Hogstrom J, Hyvonen V, Kaijalainen S, Ritvos O, Haglund C, Kruuna O, Robine S, Louvard D, Ben-Neriah Y, and Alitalo K. 2014. Oncogenic mutations in intestinal adenomas regulate Bim-mediated apoptosis induced by TGF-beta. Proc Natl Acad Sci U S A 111: E2229–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ohgushi M, Kuroki S, Fukamachi H, O’Reilly LA, Kuida K, Strasser A, and Yonehara S. 2005. Transforming growth factor beta-dependent sequential activation of Smad, Bim, and caspase-9 mediates physiological apoptosis in gastric epithelial cells. Mol Cell Biol 25: 10017–10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


