Abstract
Nucleotide sequences of the Cryptosporidium oocyst wall protein (COWP) gene were obtained from various Cryptosporidium spp. (C. wrairi, C. felis, C. meleagridis, C. baileyi, C. andersoni, C. muris, and C. serpentis) and C. parvum genotypes (human, bovine, monkey, marsupial, ferret, mouse, pig, and dog). Significant diversity was observed among species and genotypes in the primer and target regions of a popular diagnostic PCR. These results provide useful information for COWP-based molecular differentiation of Cryptosporidium spp. and genotypes.
The gene coding for the Cryptosporidium oocyst wall protein (COWP) is one of the commonly used targets of molecular tools for genotyping Cryptosporidium parasites. Characterization of the COWP gene revealed genetic differences among human and bovine C. parvum isolates and C. wrairi. Based on the sequence diversity, a simple PCR-restriction fragment length polymorphism (PCR-RFLP) technique was developed to differentiate three genotypes of Cryptosporidium parasites (10, 12). Since then, this technique has been widely used in the genotyping of Cryptosporidium parasites in clinical samples (4, 8, 9, 11, 16).
There is a lack of COWP sequence information from other Cryptosporidium spp. During the evaluation of C. parvum genotyping tools, we found that the COWP-based PCR-RFLP tools also amplified DNA from purified oocysts of C. muris and C. serpentis, indicating that PCR primers used in COWP-based diagnostic tools are probably not C. parvum specific (13). A novel COWP genotype of Cryptosporidium has been found in one human patient recently (4). To expand the database on the Cryptosporidium COWP gene for diagnostic studies, we characterized the COWP genes of a variety of Cryptosporidium spp. and C. parvum genotypes.
Fecal samples used in this study were collected from animals or humans infected with C. baileyi (from a quail), C. felis (from an AIDS patient), C. meleagridis (from a turkey), C. muris (from a rock hyrax), C. andersoni (from a calf), C. serpentis (from a snake), C. wrairi (from a guinea pig), an unknown Cryptosporidium species (from a desert monitor), and the bovine, human, monkey, mouse, ferret, dog, pig, and marsupial (from a red kangaroo) genotypes of C. parvum. Almost all samples were used in our previous studies of Cryptosporidium parasites, and the sources of these samples were described in detail elsewhere (14, 17, 18). Cryptosporidium oocysts and DNA were isolated as described before (17, 18). The identity of Cryptosporidium species and genotypes was established based on morphologic examinations and sequence analysis of the small-subnuit (SSU) rRNA and 70-kDa heat shock protein (HSP70) genes (14, 17, 18).
Two sets of primers were used to amplify fragments of the COWP gene. All isolates used in this study were initially analyzed using primers 5′-CCCAACATTCCTGGTGTAGCTTCC-3′ and 5′-GAACGCACCTGTTCCCACTCAATG-3′. These primer sequences were based on the published COWP sequence (GenBank accession no. Z22537) obtained from a bovine C. parvum isolate and amplify a 1,033-bp fragment from the region flanking the sequence targeted by the method of Spano et al. (12). The isolates that failed to yield positive amplification by this primer set were further analyzed with primers (5′-GTAGATAATGGAAGAGATTGTG-3′ and 5′-GGACTGAAATACAGGCATTATCTTG-3') designed by Spano et al. (11), which amplify a 553-bp region located inside the 1033-bp fragment. The PCR conditions used for both primer sets were identical to those used in the technique developed by Spano et al. (12). The PCR product was analyzed by agarose gel electrophoresis and visualized after ethidium bromide staining. RFLP analysis of PCR products generated from the Spano primers was conducted with the restriction enzyme RsaI as previously described (12).
PCR products of both the small and large fragments were sequenced on an ABI 377 automated sequencer (Perkin Elmer, Foster City, Calif.). Sequence accuracy was confirmed by two-directional sequencing and by sequencing of a second PCR product. Multiple alignments of the DNA sequences were done using the Wisconsin package, version 9.0 (Genetics Computer Group, Madison, Wis.). Phylogenetic analysis was carried out on the aligned sequences to assess genetic relationships between various Cryptosporidium species and genotypes as previously described (14, 17, 18).
The PCR primers designed by us amplified the COWP genes from isolates of the C. parvum human, monkey, bovine, mouse, ferret, pig, and marsupial genotypes, C. wrairi, and C. meleagridis. However, they failed to amplify DNA from the C. parvum dog genotype, C. felis, C. baileyi, C. serpentis, C. andersoni, C. muris, and the Cryptosporidium parasite from the desert monitor. For the amplification of the COWP genes of parasites that could not be amplified by these primers, we used primers described by Spano et al. (12) which amplify a smaller fragment within the region covered by our primers. The efficiency of the latter in amplification of these divergent Cryptosporidium parasites was still low. However, light bands of PCR products were generated using these primers and purified oocysts (DNA from the equivalent of more than 100 oocysts per PCR for C. parvum dog genotype, C. serpentis, C. andersoni, and C. muris). This strategy allowed us to examine the sequence diversity in the region of the diagnostic primers of Spano et al. among different Cryptosporidium spp. and C. parvum genotypes.
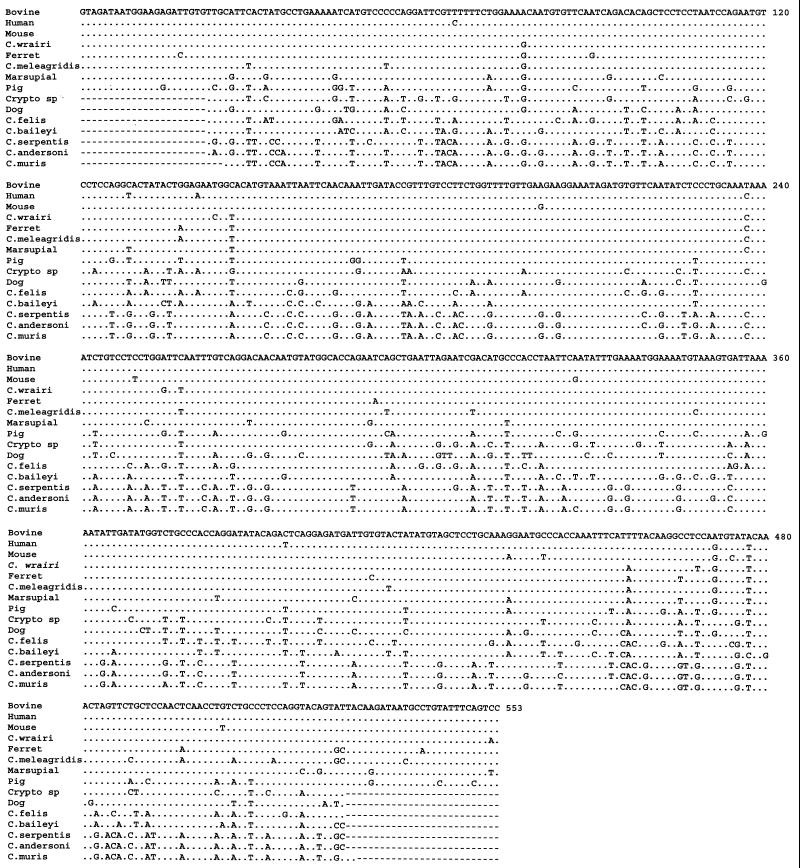
We sequenced the 1,033-bp PCR products from C. wrairi, C. meleagridis, and the human, monkey, bovine, mouse, ferret, pig, and marsupial genotypes of C. parvum and the 553-bp PCR products from the C. parvum dog genotype, C. felis, C. baileyi, C. serpentis, C. andersoni, C. muris, and the unknown Cryptosporidium sp. Nucleotide sequences obtained from the C. parvum human and bovine genotypes and C. wrairi were identical to those previously published (12), but different sequences were obtained for all the Cryptosporidium spp. and C. parvum genotypes studied (Fig. 1).
FIG. 1.
Variation in the COWP nucleotide sequences among nine Cryptosporidium spp. and eight C. parvum genotypes in the region targeted by the PCR-RFLP diagnostic tool (11). Dots denote sequence identity to the bovine genotype of C. parvum. Dashes indicate sequence ubiquity. The monkey and human genotypes of C. parvum had identical sequence in this region.
Certain Cryptosporidium parasites were more related to each other than others, as reflected in the number of base pair difference among them and the genetic distances calculated. The C. parvum human and monkey genotypes had only 2 bp of differences in the 1,033-bp fragment and had identical sequences in the region covered by the Spano primers. These and the C. parvum mouse and ferret genotypes, C. wrairi, and C. meleagridis, had only 7- to 25-bp differences from the bovine genotype of C. parvum in the 553-bp region (Fig. 1). This is also reflected in the genetic distance calculated, with <6.5% of nucleotide changes among them (data not shown). Other parasites, such as the pig and dog genotypes of C. parvum, C. felis, the unnamed Cryptosporidium sp., and C. baileyi, were much more distant from the first group of parasites and from each other, exhibiting ≥57-bp differences from the C. parvum bovine genotypes in the 553-bp region and genetic distances of 10 to 20% between each other. The third group, i.e., C. muris, C. andersoni, and C. serpentis, had small genetic differences between each other (2.23 to 3.29% of nucleotide changes) but large differences from other Cryptosporidium parasites (24.33 to 30.38% of nucleotide changes).
Neighbor-joining analysis of the COWP nucleotide sequences supported the above-described observations. All Cryptosporidium parasites analyzed formed two groups, with C. muris, C. andersoni, and C. serpentis separating from the rest (100% of bootstrapping). Within the other group the C. parvum human, monkey, bovine, mouse, and ferret genotypes, C. wrairi, and C. meleagridis clustered together with full statistical reliability (100% of bootstrapping), whereas C. baileyi, C. felis, the C. parvum dog and pig genotypes, and the Cryptosporidium parasite from desert monitors were placed at the bottom of the clade. Furthermore, the C. parvum human, monkey, bovine, and mouse genotypes formed a secondary monophyletic cluster (data not shown).
In the COWP-based genotyping tool, RFLP analysis of the PCR product with RsaI was used to differentiate C. parvum bovine and human genotypes and C. wrairi (11). Thus, the COWP sequences covered by the Spano primers obtained from various Cryptosporidium parasites were searched for the RsaI restriction site. This analysis revealed multiple band patterns for the Cryptosporidium parasites used in the analysis (Table 1). Unique RFLP patterns were predicted for the C. parvum pig, marsupial, and dog genotypes, C. meleagridis, C. felis, C. baileyi, and the Cryptosporidium parasite from desert monitors. The following Cryptosporidium parasites, however, would have RFLP patterns identical to each other: (i) C. muris, C. andersoni, and C. serpentis; (ii) C. parvum ferret genotype and C. wrairi; (iii) C. parvum bovine and mouse genotypes; and (iv) C. parvum human and monkey genotypes. Digestion of PCR products with RsaI produced RFLP patterns in agreement with predicted patterns for C. parvum human, monkey, bovine, mouse, ferret, marsupial, pig, and dog genotypes, C. wrairi, C. meleagridis, C. felis, C. baileyi, and C. muris (data not shown). Despite multiple attempts, the amount of PCR products generated from other Cryptosporidium parasites was not enough for RFLP analysis.
TABLE 1.
Predicted RsaI RFLP patterns of COWP PCR products of the diagnostic tool of Sapno et al. (11) for various Cryptosporidium parasites
| Species (genotype) | Sizes (kb) of predicated RFLP bandsa |
|---|---|
| C. parvum (bovine) | 413, 106, 34 |
| C. parvum (mouse) | 413, 106, 34 |
| C. parvum (human) | 284, 129, 106, 34 |
| C. parvum (monkey) | 284, 129, 106, 34 |
| C. wrairi | 266, 147, 106, 34 |
| C. parvum (ferret) | 266, 147, 106, 34 |
| C. meleagridis | 372, 147, 34 |
| C. parvum (marsupial) | 266, 140, 129, 18 |
| C. parvum (pig) | 266, 129, 106, 34, 18 |
| C. parvum (dog)b | 195, 106, 86, 71, 43, 34, 18 |
| Cryptosporidium sp.b | 413, 140 |
| C. felisb | 406, 86, 61 |
| C. baileyib | 486, 67 |
| C. murisb | 327, 140, 86 |
| C. andersonib | 327, 140, 86 |
| C. serpentisb | 327, 140, 86 |
Numbers in boldface are the sizes of bands visible on electrophoresis gel.
Can be amplified efficiently only with DNA from purified oocysts.
The COWP-based genotyping tool is widely used in the diagnosis of Cryptosporidium parasites because of the use of a target unique to Cryptosporidium parasites and the presumed specificity. This is supported by the results of a recent evaluation study (13). In addition, unlike most other genotyping tools that are based on sequences of antigen genes, the COWP technique was shown to have the ability to amplify and detect Cryptosporidium parasites other than the human (genotype 1) and bovine (genotype 2) genotypes (4, 13), thus, it has been suggested that this tecnique may have potential in the differentiation of a broader range of C. parvum genotypes and Cryptosporidium spp. (13).
In the present study distinct COWP nucleotide sequences were obtained from nine Cryptosporidium species and eight different C. parvum genotypes. Restriction analysis revealed multiple electrophoresis band patterns for the Cryptosporidium parasites used in the analysis, although some parasites, such as the bovine and mouse genotypes of C. parvum or C. wrairi and the ferret genotype of C. parvum, had identical patterns. Difficulties were experienced, however, with the PCR amplification of DNA from Cryptosporidium parasites that are genetically more distant from the C. parvum bovine genotype. Thus, no amplification were achieved with the C. parvum dog genotype, C. felis, C. baileyi, C. muris, C. andersoni, C. serpentis, and the Cryptosporidium parasites from desert monitors using the primers designed by us, and only weak PCR amplifications were obtained from highly purified DNAs of these parasites with the primers of Spano et al. (12). This is expected judging by the extent of COWP sequence divergence of these parasites from the C. parvum bovine genotype, which the primer sequences were based on. Although sequence information for the primer regions was not available for the C. parvum dog genotype, C. felis, C. baileyi, C. muris, C. andersoni, C. serpentis, and the unnamed Cryptosporidium parasite, other C. parvum or C. parvum-related parasites exhibited sequence polymorphism, especially in the reverse primer region. This was likely the cause of poor PCR amplification of DNA from these divergent Cryptosporidium parasites.
The findings of this study have important implications for the use of the COWP-genotyping tool in the diagnosis of Cryptosporidium parasites. Based on the characterization of the rRNA gene, five or six Cryptosporidium parasites (the human, bovine, and dog genotypes of C. parvum, C. meleagridis, C. felis, and possibly C. muris) have thus far been found in humans (2, 19). Previous analyses of human samples with the COWP-based PCR-RFLP technique mostly revealed the presence of the human (genotype 1) and bovine (genotype 2) genotypes of C. parvum (4, 8, 9, 11, 16). A recent study, however, showed the presence of a third genotype (genotype 3) in one patient in the United Kingdom (4). The PCR-RFLP pattern or nucleotide sequence was not available for the third genotype; thus, its identity could not be established. However, because it is extremely difficult to amplify the COWP gene in DNA isolated from the C. parvum dog genotype, C. felis, and C. muris in fecal samples, it is conceivable that the third COWP genotype in humans could be C. meleagridis. Presently neither of the primer pairs investigated has the ability to detect efficiently all human-pathogenic Cryptosporidium parasites in clinical samples. Modifications will be needed for the diagnostic COWP primers to effectively detect these divergent Cryptosporidium parasites in clinical samples. Unfortunately, several other COWP primer pairs that we designed based on the sequence of the C. parvum bovine genotype failed to achieve positive amplification for these Cryptosporidium parasites (data not shown). This was probably because of the random distribution of mutations across the entire COWP gene in these distantly related Cryptosporidium parasites (Fig. 1). Presently the utility of the COWP-based PCR-RFLP technique in the analysis of environmental samples is probably limited because of the narrow spectrum of Cryptosporidium parasites detected.
The COWP gene also provides an alternative target for molecular taxonomy and phylogenetic analysis of Cryptosporidium parasites. Controversy exists in the taxonomy of Cryptosporidium parasites (1, 6, 15, 19). To date, 23 species of Cryptosporidium have been named, but fewer than 10 are considered valid by some researchers (1, 3, 6, 15, 19). Results of recent studies of the SSU rRNA and HSP70 loci indicate that what we know now as C. parvum is probably a multispecies complex, because various host-adapted strains are polyphyletic in phylogenetic analysis and have genetic differences greater than those between C. parvum and some other Cryptosporidium spp., such as C. wrairi and C. meleagridis (5, 14, 18, 19). Results of phylogenetic analysis of the COWP sequences are in agreement with these observations. In addition, the genetic relationship among Cryptosporidium parasites revealed by the COWP phylogenetic tree is largely congruent to the one produced by the analysis of the rRNA gene and HSP70 gene. The only exception is the placement of C. andersoni, a new species recently named (3) from a Cryptosporidium parasite formerly known as the C. muris bovine genotype (7, 17). In the COWP phylogenetic tree, it clustered with C. serpentis, in comparison with a closer relationship to C. muris in the SSU rRNA- and HSP70-based phylogenetic analyses (5, 14, 17). The genetic distances among these three parasites, however, are very small at all three genes.
In conclusion, various Cryptosporidium spp. and host-adapted C. parvum strains have extensive sequence polymorphism in the COWP gene, which seems to reflect the genetic relatedness of different Cryptosporidium parasites. Thus, if genus-specific primers are found, the COWP gene can be a good target for species differentiation and genotyping of Cryptosporidium parasites. The sequences generated from this study have revealed potential problems in the current COWP-based geneotyping tool. It is likely that the efficiency of the primers used in amplifying DNA from some human pathogenic Cryptosporidium parasites may be compromised because of the heterogeneity in the primer regions.
Nucleotide sequence accession numbers.
The nucleotide sequences of the COWP genes of C. baileyi, C. felis, C. meleagridis, C. muris, C. andersoni, C. serpentis, C. wrairi, the unknown Cryptosporidium sp., and eight genotypes of C. parvum (human, bovine, dog, ferret, marsupial, monkey, mouse, and pig) were deposited in the GenBank database under accession no. AF266262 to AF266277.
Acknowledgments
This work was supported in part by an interagency agreement (DW75937730-01-0) between the U.S. Environmental Protection Agency and Centers for Disease Control and Prevention and funding from the Opportunistic Infectious Diseases program of the Centers for Disease Control and Prevention.
We thank Ron Fayer for providing the C. meleagridis sample used in the study.
REFERENCES
- 1.Fayer R, Spear C A, Dubey J P. The general biology of Cryptosporidium. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press; 1997. pp. 1–41. [Google Scholar]
- 2.Katsumata T, Hosea D, Ranuh I G, Uga S, Yanagi T, Kohno S. Possible Cryptosporidium muris infection in humans. Am J Trop Med Hyg. 2000;62:70–72. doi: 10.4269/ajtmh.2000.62.70. [DOI] [PubMed] [Google Scholar]
- 3.Lindsay D S, Upton S J, Owens D S, Morgan U M, Mead J R, Blagburn B L. Cryptosporidium andersoni n. sp (Apicomplexa: Cryptosporiidae) from cattle, Bos taurus. J Eukaryot Microbiol. 2000;47:91–95. doi: 10.1111/j.1550-7408.2000.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 4.McLauchlin J, Pedraza-Diaz S, Amar-Hoetzeneder C, Nichols G L. Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J Clin Microbiol. 1999;37:3153–3158. doi: 10.1128/jcm.37.10.3153-3158.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan U M, Monis P, Fayer R, Deplazes P, Thompson R C A. Phylogenetic relationships among isolates of Cryptosporidium: evidence for several new species. J Parasitol. 1999;85:1126–1133. [PubMed] [Google Scholar]
- 6.Morgan U M, Xiao L, Fayer R, Lal A A, Thompson R C A. Variation in Cryptosporidium: towards a taxonomic revision of the genus. Int J Parasitol. 1999;29:1733–1751. doi: 10.1016/s0020-7519(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 7.Morgan U M, Xiao L, Monis P, Sulaiman I, Pavlasek I, Blagburn B, Olson M, Upton S J, Khramtsov N V, Lal A, Elliot A, Thompson R C A. Molecular and phylogenetic analysis of Cryptosporidium muris from various hosts. Parasitology. 2000;120:457–464. doi: 10.1017/s0031182099005703. [DOI] [PubMed] [Google Scholar]
- 8.Patel S, Pedraza-Diaz S, McLauchlin J. The identification of Cryptosporidium species and Cryptosporidium parvum directly from whole faeces by analysis of a multiplex PCR of the 18S rRNA gene and by PCR/RFLP of the Cryptosporidium outer wall protein (COWP) gene. Int J Parasitol. 1999;29:1241–1247. doi: 10.1016/s0020-7519(99)00079-x. [DOI] [PubMed] [Google Scholar]
- 9.Patel S, Pedraza-Diaz S, McLauchlin J, Casemore D P. Molecular characterisation of Cryptosporidium parvum from two large suspected waterborne outbreaks. Commun Dis Public Health. 1998;1:231–233. [PubMed] [Google Scholar]
- 10.Spano F, Puri C, Ranucci L, Putignani L, Crisanti A. Cloning of the entire COWP gene of Cryptosporidium parvum and ultrastructural localization of the protein during sexual parasite development. Parasitology. 1997;114:427–437. doi: 10.1017/s0031182096008761. [DOI] [PubMed] [Google Scholar]
- 11.Spano F, Putignani L, Crisanti A, Sallicandro P, Morgan U M, Leblancq S M, Tchack L, Tzipori S, Widmer G. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J Clin Microbiol. 1998;36:3255–3259. doi: 10.1128/jcm.36.11.3255-3259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spano F, Putignani L, McLauchlin J, Casemore D P, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 13.Sulaiman I M, Xiao L, Lal A A. An evaluation of Crypytosporidium parvum genotyping techniques. Appl Environ Microbiol. 1999;65:4431–4435. doi: 10.1128/aem.65.10.4431-4435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sulaiman I M, Morgan U M, Thompson R C A, Lal A A, Xiao L. Phylogenetic relationships of Crypytosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl Environ Microbiol. 2000;66:2385–2391. doi: 10.1128/aem.66.6.2385-2391.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tzipori S, Griffiths J K. Natural history and biology of Cryptosporidium parvum. Adv Parasitol. 1998;40:5–36. doi: 10.1016/s0065-308x(08)60116-5. [DOI] [PubMed] [Google Scholar]
- 16.Widmer G, Tchack L, Spano F, Tzipori S. A study of Cryptosporidium parvum genotypes and population structure. Mem Inst Oswaldo Cruz. 1998;93:685–686. doi: 10.1590/s0074-02761998000500021. [DOI] [PubMed] [Google Scholar]
- 17.Xiao L, Escalante L E, Yang C, Sulaiman I M, Escalante A A, Montali R, Fayer R, Lal A A. Phylogenetic analysis of Cryptosporidium parasites on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao L, Morgan U, Limor J, Escalante A, Arrowood M, Shulaw W, Thompson R C A, Fayer R, Lal A A. Genetic diversity within Cryptosporidium parvum and related species of Cryptosporidium. Appl Environ Microbiol. 1999;65:3386–3391. doi: 10.1128/aem.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao L, Morgan U M, Fayer R, Thompson R C A, Lal A A. Cryptosporidium systematics and implications for public health. Parasitol Today. 2000;15:287–292. doi: 10.1016/s0169-4758(00)01699-9. [DOI] [PubMed] [Google Scholar]