Abstract
A rapid and reliable mating type assay for Fusarium circinatum was created by applying primers specific for the MAT1-1 and MAT1-2 mating type alleles to genomic DNA in a single PCR. A similar approach may be applied to fungi not previously shown to reproduce sexually, thus enabling studies of population structure and inheritance.
In heterothallic ascomycetes, the mating type (MAT) locus determines sexual compatibility between haploid individuals. The locus exists as two alternate alleles, MAT1-1 and MAT1-2. The individuals participating in a cross must contain opposite alleles in order to reproduce sexually (17). The alleles at the MAT locus consist of unrelated sequences and thus have been termed idiomorphs to suggest that the two structurally unrelated forms of the gene evolved from different ancestral sequences (14). Identification of conserved regions within, or flanking, the idiomorphs has made it possible to clone these genes from many filamentous ascomycetes (1, 17).
The traditional approach for determining the mating type of any heterothallic individual is to attempt to cross it with each of two tester isolates which are already known to differ at the mating type locus. The mating type of the isolate being tested is the opposite of that with which it crosses successfully (i.e., produces ascospores). This is a time-consuming assay because sexual crosses in many heterothallic fungi take 4 to 8 weeks to complete (3, 7, 9, 11, 16). It also relies on established tester isolates, which are unavailable for species that have not yet been successfully crossed. For such species, finding compatible pairs of opposite mating types can be challenging. All potential partners must be intercrossed, and the number of crosses that must be attempted increases as the square of the number of isolates being tested (12). Furthermore, the likelihood of identifying sexually compatible pairs in many heterothallic species is reduced by the high proportion of wild field isolates that are either female sterile (e.g., unable to make perithecia) or completely sterile (i.e., unable to mate with either tester isolate) (2, 4, 13, 18, 19). In addition to thwarting genetic analysis, the commonness of sterility in the laboratory setting makes it difficult to accurately assess mating type frequencies in wild populations. To date, published molecular tests of mating type have relied upon PCR amplification of the MAT1-2 idiomorph (1, 6, 10). Because it does not assay the MAT1-1 idiomorph directly, this approach assigns mating type unambiguously only when applied to established mating type testers. A more reliable, rapid method for mating type determination would be beneficial in laboratories undertaking genetic analysis, especially of new species, or in laboratories with an interest in traits affecting population structure.
The heterothallic ascomycete Fusarium circinatum Nirenberg and O'Donnell (formerly Fusarium subglutinans f. sp. pini) causes pitch canker disease on many species of pine (8, 15, 20). Our objective in this study was to develop a rapid and reliable assay for determining which mating type idiomorph is present in any given F. circinatum isolate. Idiomorph-specific primers which amplify approximately 190 bp from MAT1-2 were designed previously (6). Use of only these primers to assign mating type is not ideal because a MAT1-1 isolate (indicated by the absence of a PCR product) cannot be distinguished from a failed PCR. In addition, contamination of a MAT1-1 sample with MAT1-2 DNA gives a false-positive result. Therefore, to obtain an unambiguous result in a PCR-based mating type assay it was essential that specific primers for the MAT1-1 idiomorph be designed. Meeting this objective required cloning and sequencing a portion of the MAT1-1 idiomorph and designing MAT1-1-specific primers. Combining these MAT1-1 primers with MAT1-2 primers in a PCR in which F. circinatum DNA serves as the template quickly gives unambiguous results. This assay will facilitate studies of F. circinatum inheritance and population structure.
Growth conditions and DNA extraction.
Mycelia from the isolates listed in Table 1 were grown as described by Covert et al. (6). The resulting hyphae were harvested on Miracloth (Calbiochem), and the DNA was extracted with the DNeasy Plant Mini Kit (Qiagen, Inc., Valencia, Calif.).
TABLE 1.
F. circinatum isolates used in this study
| Isolatea | MAT idiomorph | Origin |
|---|---|---|
| NRRL 25331 | MAT1-1 | California |
| NRRL 25333 | MAT1-2 | South Africa |
| NRRL 25621 | MAT1-2b | South Africa |
| NRRL 25707 | MAT1-1b | North Carolina |
| NRRL 25708 | MAT1-1b | North Carolina |
| A 362 | MAT1-1 | California |
| Fsp 52 | MAT1-2b | California |
| Fsp 118 | MAT1-1b | California |
| GB 232 | MAT1-1 | Florida |
| GB 286 | MAT1-1 | Florida |
| GB 298 | MAT1-1 | Florida |
| GB 311 | MAT1-1 | Florida |
| GB 346 | MAT1-1 | Florida |
NRRL isolates are from the Agricultural Research Service Culture Collection, NCAUR, Peoria, Ill. A and Fsp isolates are from Tom Gordon, Department of Plant Pathology, University of California, Davis. GB isolates are from George Blakeslee, School of Forest Resources and Conservation, University of Florida, Gainsville.
Mating type was determined by PCR only. (Mating types of all other isolates were determined by PCR and by successful sexual crossing.)
Cloning a portion of MAT1-1.
Primer FMATp1 [(GT)ACCTAGTGCAACAA(GT)AAACAAAGCGAGTG] was designed by Sung-Hwan Yun and Gillian Turgeon (Cornell University; personal communication) from an alignment of the DNA flanking F. circinatum MAT1-2 (6), Fusarium oxysporum MAT1-1 (accession no. AB011379), and F. oxysporum MAT1-2 (AB011378). Primer MAT1p1 (CCTAGGAACTGCTGGCTTCT) was designed from an alignment of the F. oxysporum MAT1-1 and Gibberella fujikuroi mating population A MAT1-1 (AF100925) idiomorphs. The TaKaRa PCR kit (Panvera Corporation, Madison, Wis.) was used with a 0.2 mM concentration of each deoxynucleoside triphosphate, 30 pmol of each primer, and approximately 0.1 μg of F. circinatum isolate NRRL 25331 (Table 1) genomic DNA in a 50-μl final volume. After denaturing at 93°C for 5 min, the reaction was thermocycled 35 times (93°C, 45 s; 45°C, 1 min; 72°C, 1 min 30 s), extended for 10 min at 72°C, and then held at 4°C. The product was cloned into vector pNoTA/T7 using the Prime PCR Cloner cloning system (5Prime→3Prime, Inc., Boulder, Colo.).
Mating type assay.
Approximately 0.1 μg of genomic DNA from various F. circinatum isolates (Table 1) was used as the PCR template. Four primers (30 pmol of each) were included in each 50-μl reaction mixture: GcHMG1 (6), GcHMG2 (6), MAT1p2 (AGAAACTGACTGATACATCAAGGGG), and MAT1p3 (TCATAAGAAGTGTTGAAGGAATCACAG). HotStarTaq polymerase and the reagents accompanying it (Qiagen, Inc.) were used with a 2 mM concentration of MgCl2. Cycling conditions were as described by Covert et al. (6).
Nucleotide analysis.
Primer synthesis and DNA sequencing were done at the University of Georgia Molecular Genetics Instrumentation Facility. Sequences were analyzed with Lasergene software (DNAStar, Madison, Wis.). Amino acid alignments were constructed with Wisconsin Package version 10.0 (Genetics Computer Group, Madison, Wis.).
Partial MAT1-1 cloning and primer design.
A portion of the MAT1-1 idiomorph was amplified by PCR from F. circinatum isolate NRRL 25331. Primer FMAT1p1 annealed to the DNA flanking both MAT idiomorphs, while primer MAT1p1 annealed to MAT1-1-specific DNA. The 600-bp product was cloned and sequenced (Fig. 1). It contains one predicted intron, which possesses conserved 5′ and 3′ splice signals and a putative lariat sequence (5). When the intron is spliced out, the predicted amino acid sequence is 93.5% identical to a portion of the MAT1-1-3 protein from G. fujikuroi mating population A (data not shown). The end of the MAT1-1-specific DNA (Fig. 1) was determined by aligning this sequence with the F. circinatum MAT1-2 idiomorph (6). Two MAT1-1-specific primers (MAT1p2 and MAT1p3) were designed from the sequence in Fig. 1.
FIG. 1.
Partial DNA sequence of F. circinatum MAT1-1. A predicted intron is indicated by lowercase letters. The putative stop codon is overlined, and the end of the idiomorph is marked by an arrowhead. Primers MAT1p2 and MAT1p3 are overlined with arrows.
Mating type assay.
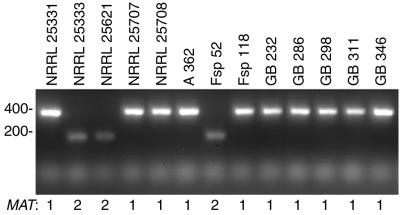
The PCR-based mating type assay for F. circinatum used four primers, two for MAT1-1 and two for MAT1-2, in a single reaction tube. The different amplification products were distinguished by their sizes; an approximately 380-bp band was amplified from MAT1-1 isolates, and an approximately 190-bp band was amplified from MAT1-2 isolates (Fig. 2). The mating types of several of the isolates tested and shown in Fig. 2 were previously determined by successful sexual crosses (Table 1) (6). In all cases the molecular mating type assay agreed with the previous biological assay. The presence of primers for both mating types in the PCR assay ensures that a positive result is recorded for each isolate. It also ensures that contaminated DNA is revealed by the amplification of both MAT-specific products. This assay will enable future studies on mating type frequencies in wild F. circinatum populations, as well as any genetic experiments in which a series of crosses is required.
FIG. 2.
PCR-based mating type assay. Four primers (GcHMG1, GcHMG2, MAT1p2, and MAT1p3) were used in combination with genomic DNA from F. circinatum isolates as labeled. The MAT idiomorph in each is indicated at the bottom. Sizes, in base pairs, are indicated on the left.
The recent cloning and sequencing of the mating type idiomorphs from several Fusarium species indicate that these genes are highly conserved across species boundaries. For example, the MAT1-1 idiomorph of F. circinatum is 88 and 86% identical at the nucleotide level to the MAT1-1 idiomorphs of G. fujikuroi mating population A and F. oxysporum, respectively. The MAT1-2 idiomorphs in these three fungi display even higher levels of identity (data not shown). It should be especially straightforward, therefore, to work from these known sequences to quickly develop unambiguous PCR-based mating type assays for additional Fusarium species.
Nucleotide sequence accession number.
The GenBank accession number for the F. circinatum MAT1-1 partial coding sequence is AF194868.
Acknowledgments
We thank G. Turgeon and S.-H. Yun for supplying unpublished primer sequences, D. Brown for assistance with figure preparation, and L. Tredway for helpful comments on the manuscript.
A graduate assistantship from the Warnell School of Forest Resources and a grant from the Warnell School of Forest Resources Alumni Fund supported this work.
REFERENCES
- 1.Arie T, Christiansen S K, Yoder O C, Turgeon B G. Efficient cloning of ascomycete mating type genes by PCR amplification of the conserved MAT HMG box. Fungal Genet Biol. 1997;21:118–130. [PubMed] [Google Scholar]
- 2.Beremand M N, Desjardins A E, Hohn T M, Vanmiddlesworth F L. Survey of Fusarium sambucinum (Gibberella pulicaris) for mating type, trichothecene production, and other selected traits. Phytopathology. 1991;81:1452–1458. [Google Scholar]
- 3.Britz H, Coutinho T A, Wingfield M J, Marasas W F O, Gordon T R, Leslie J F. Fusarium subglutinans f. sp. pini represents a distinct mating population in the Gibberella fujikuroi species complex. Appl Environ Microbiol. 1999;65:1198–1201. doi: 10.1128/aem.65.3.1198-1201.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britz H, Wingfield M J, Coutinho T A, Marasas W F O, Leslie J F. Female fertility and mating type distribution in a South African population of Fusarium subglutinans f. sp. pini. Appl Environ Microbiol. 1998;64:2094–2095. doi: 10.1128/aem.64.6.2094-2095.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruchez J, Eberle J, Russo V. Regulatory sequences in the transcription of Neurospora crassa genes: CAAT box, TATA box, introns, poly(A) tail formation sequences. Fungal Genet Newsl. 1993;40:89–96. [Google Scholar]
- 6.Covert S F, Briley A, Wallace M M, McKinney V T. Partial MAT-2 gene structure and the influence of temperature on mating success in Gibberella circinata. Fungal Genet Biol. 1999;28:43–54. doi: 10.1006/fgbi.1999.1161. [DOI] [PubMed] [Google Scholar]
- 7.Desjardins A E, Beremand M. A genetic system for trichothecene toxin production in Gibberella pulicaris (Fusarium sambucinum) Phytopathology. 1987;77:678–683. [Google Scholar]
- 8.Dwinell L D, Barrows-Broaddus J B, Kuhlman E G. Pitch canker: a disease complex. Plant Dis. 1985;69:270–276. [Google Scholar]
- 9.Hsieh W H, Smith S N, Snyder W C. Mating groups in Fusarium moniliforme. Phytopathology. 1977;67:1041–1043. [Google Scholar]
- 10.Kerényi Z, Zeller K, Hornok L, Leslie J F. Molecular standardization of mating type terminology in the Gibberella fujikuroi species complex. Appl Environ Microbiol. 1999;65:4071–4076. doi: 10.1128/aem.65.9.4071-4076.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence E B, Nelson P E, Toussoun T A. Inheritance of compatibility and sex in Gibberella baccata. Phytopathology. 1985;75:322–324. [Google Scholar]
- 12.Leslie J F. Gibberella fujikuroi: available populations and variable traits. Can J Bot. 1995;73(Suppl. 1):S282–S291. [Google Scholar]
- 13.Leslie J F, Klein K K. Female fertility and mating type effects on effective population size and evolution in filamentous fungi. Genetics. 1996;144:557–567. doi: 10.1093/genetics/144.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metzenberg R L, Glass N L. Mating type and mating strategies in Neurospora. Bioessays. 1990;12:53–59. doi: 10.1002/bies.950120202. [DOI] [PubMed] [Google Scholar]
- 15.Storer A J, Gordon T R, Dallara P L, Wood D L. Pitch canker kills pines, spreads to new species and regions. Calif Agric. 1994;48:9–13. [Google Scholar]
- 16.Tegtmeier K J, Vanetten H D. Genetic studies on selected traits of Nectria haematococca. Phytopathology. 1982;72:604–607. [Google Scholar]
- 17.Turgeon B G. Application of mating type gene technology to problems in fungal biology. Annu Rev Phytopathol. 1998;36:115–137. doi: 10.1146/annurev.phyto.36.1.115. [DOI] [PubMed] [Google Scholar]
- 18.Valent B, Chumley F G. Molecular genetic analysis of the rice blast fungus, Magnaporthe grisea. Annu Rev Phytopathol. 1991;29:443–467. doi: 10.1146/annurev.py.29.090191.002303. [DOI] [PubMed] [Google Scholar]
- 19.VanEtten H D. Identification of additional habitats of Nectria haematococca mating population VI. Phytopathology. 1978;68:1552–1556. [Google Scholar]
- 20.Viljoen A, Wingfield M J. First report of Fusarium subglutinans f. sp. pini on pine seedlings in South Africa. Plant Dis. 1994;78:309–312. [Google Scholar]