Abstract
When bound to thyroid hormone, the nuclear receptor TRα1 activates the transcription of a number of genes in many cell types. It mainly acts by binding DNA as a heterodimer with retinoid X receptors at specific response elements related to the DR4 consensus sequence. However, the number of DR4-like elements in the genome exceed by far the number of occupied sites, indicating that minor variations in nucleotides composition deeply influence the DNA-binding capacity and transactivation activity of TRα1. An improved protocol of synthetic self-transcribing active regulatory region sequencing was used to quantitatively assess the transcriptional activity of thousands of synthetic sites in parallel. This functional screen highlights a strong correlation between the affinity of the heterodimers for DNA and their capacity to mediate the thyroid hormone response.
Keywords: thyroid hormone, nuclear receptors, hormone response elements
Nuclear receptors mediate the action of a variety of natural and artificial ligands by regulating gene transcription (1). They bind DNA response elements by their N-terminal domain as homodimers or heterodimers (2). The binding of ligand in their C-terminal domain induces a conformational change, and, depending on the agonist or antagonist nature of the ligand, recruit transcription coactivators or corepressors (3). Response elements are composed of 2 half sites, which DNA sequence and arrangement dictate the type of dimers that can bind (4). Recent genome wide analyses revealed that, whereas there are more than 50 000 putative response elements in the genome for each nuclear receptor, only a few thousands are actually occupied in chromatin (5-11). The number of genes that transcription is responsive to the cognate ligand is further limited to a few hundreds. One hypothesis to explain this restricted response is that the DNA/nuclear receptor/ligand complex has allosteric properties (12, 13). Under this hypothesis, the exact nucleotide sequence of the DNA response element indirectly would not only determine its affinity for receptor binding but also influence the capacity of the distant ligand-binding domain to recruit transcription corepressors or coactivators. It is therefore important to provide a functional characterization of the response elements, in addition to assess their capacity to bind the nuclear receptors in vitro.
As an example, we selected the well-studied TRα1/retinoid X receptor (RXR)α heterodimer, which activates transcription when TRα1 bind the thyroid hormone (3,3′,5-triiodo-L-thyronine or T3) (2). In vitro studies (14) and motif analysis of chromatin immunoprecipitation sequencing (ChIP-seq) datasets (5, 7, 9, 11) previously established that this heterodimer preferentially binds the so-called DR4 element, in which 2 half-sites are separated by a 4-nucleotides spacer (5′AGGTCANNNNAGGTCA3′). By contrast, although they have the capacity to support TRα1 transactivation in transfected cells (2), other arrangement, notably everted repeat separated by 6 nucleotides, or inverted repeats without spacer, are not enriched in chromatin sites occupied by TRα1 (5, 9). Structural analysis indicates that the heterodimer orients so that RXR binds to the upstream half-site, and TRα1 to the downstream site. It also shows that the contact between DNA and TRα1 amino acids extend to the last 2 nucleotides of the spacer (15). To assess the functional consequences of variations around this consensus, we performed a functional screen of variant DR4 elements, using an adaptation of the recently developed self-transcribing active regulatory region sequencing (STARR-seq) method (16, 17).
Material and Methods
Library Construction
A 105 nucleotides oligomer containing a degenerated DR4-element 5′ATACTAGTCGCACTACGATCCTGCCGGGTGGNGGTCANNNNRGGNNAATCCCCTCCCACACCTAATGCAGAGCTAGCCA3′ (DR4 half-sites are underlined) was amplified with the following primers: 5′GGCTAACCGGTGCTAGCATACTAGTCGCACTACGATC3′ and 5′TGAAAGTCGACGCTAGCTCTGCATTAGGTGT3′. The nucleotides flanking the degenerated consensus in the template oligonucleotide were copied from the flanking sequence of a DR4 element located 1520 nucleotides upstream to the transcription start site of Hairless, which is a well-characterized TRα1 target gene (chr14:70552167-70552187 on the mouse genome; GRCm38/mm10 assembly). The resulting amplicon was purified using the QIAquick purification kit (Qiagen), cut with the AgeI and SalI restriction enzymes, and purified again. The cloning vector was prepared by cutting the hSTARR-seq-ORI plasmid (18) with the AgeI and SalI restriction enzymes. Extremities were dephosphorylated with rAPid Alkaline Phosphatase (Sigma-Aldrich) and the 2.5-kb fragment was purified after agarose gel electrophoresis using the QIAquick Gel Extraction Kit (Qiagen). Sixty nanograms of this vector were ligated (T4 DNA ligase Biolabs) to 60 ng of the amplicon insert in 30 μL of buffer. The ligation product was used to transform XL10-Gold ultra-competent cells (Agilent). Library DNA was purified from the entire bacteria library grown on agarose plates. DNA was extracted from the scrapped colonies using the QIAprep Spin Miniprep Kit (Qiagen) and further purified after a phenol/CHCl3 extraction by ethanol precipitation.
Plasmids Construct
Reporter construct with luciferase reporter were based on the pGL4.12 [Luc2CP] (GB Acc# AY738224) (Promega), in which luciferase expression is driven by a minimum promoter derived from the HSV thymidine kinase gene. A single DR4 element was cloned upstream to this minimal promoter between the XhoI and BglII restriction enzymes.
Cell Transfection
All transfection experiments were performed in HEK293 cells seeded in 24-well plates (105 cells/well). Cells were transfected the following day with plasmid or library DNA using the TransIT-Lt1 transfection reagent (Mirus Corporation, Madison WI, USA). Because the cells express endogenous receptors at low level pSG5TRα1 (150 ng/well) and pSG5RXRα (150 ng/well) derived from pSG5 (Agilent Technology) were included in all experiments. T3 was added 6 hours later and cells were harvested the following day for either luciferase activity measurement or RNA analysis. Luciferase activity was measured with the luciferase assay reagent (Promega) and quantified with the Centro luminometer (Berthold Technologies). RNA was extracted using the NucleoSpin RNA kit (Macherey Nagel). Contaminant DNA was eliminated by 2 steps of DNase I treatment performed on the extraction column according manufacturer instructions. Before reverse transcription, an additional DNase I treatment was performed on 1 µg of RNA using the RNase-Free DNase set (Qiagen). After 15 minutes of incubation at room temperature, 25 mM EDTA was added and enzyme inactivated by 10 minutes of incubation at 65°C.
Deep Sequencing
RNA was reverse transcribed with Moloney Murine Leukemia Virus reverse transcriptase (Promega) and the DNA sequences corresponding to the 3′ end of the vector-driven transcript were amplified by polymerase chain reaction with 3 pairs of primers (pair 1: 5′TCGTCGGCAGCG TCAGATGTGTATAAGAGACAGTCACTGGAGTTGTCCCAATTCTTG3′ + 5′GTCTCGTGGGCTCGGAGATG TGTATAAGAGACAGCGTCGACGCTAGCTCTGCAT3′; pair 2: 5′TCGTCGGCAGCGTCAGATGTGTATAAGA GACAGNTCACTGGAG TTGTCCCAATTCTTG3′ + 5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGA CAGNCGTCGACGCTAGCTCTGCAT3′; pair3: 5′TCGTCGGCAGCGTCAGATGTGTATAAGAGA CAGNNTCACTGG AGTTGTCCCAATTCTTG3′, + 5′GTCTCGTGGGC TCGGAGATGTGTATAAGAGACAGNNCGTCGA CGCTAGCTCTGCAT3′) suitable to directly prepare an Illumina sequencing library (Illumina Nextera V2 index kit). A control was performed in which reverse transcriptase was omitted to ascertain the absence of contaminating DNA. Amplicons were sequenced both by Sanger sequencing or by deep sequencing (Illumina Microkit V2 300 cycles MiSeq). Paired-end reads contained in the.fastq files were merged (fastq-join; Galaxy Version 1.1.2-806.1) and converted to tabular format using FASTQ Groomer (Galaxy Version 1.0.4) and FASTQ to Tabular converter (Galaxy Version 1.1.5). Tab-delimited table files were curated from sequences that differ from the expected sequences at the conserved positions around the DR4 elements (20/20 identity). An R-script was used to produce counts tables from the sequence tables.
Statistics
The χ 2 test was used to detect significant changes in the observed distribution of nucleotides at specific positions, with Bonferroni correction. Specific response elements with differential response to T3 were identified using count tables and Deseq2 Galaxy Version 2.11.40.7 (19). Overlap with ChIP-seq datasets were analyzed by searching for Intervals intersection (Galaxy Version 2.30.0).
Results
The STARR-seq method consists in inserting a putative enhancer in the 3′ noncoding sequence of an expression vector carrying a minimal transcription promoter. The abundance of the mRNA carrying the putative enhancer at its 3′ end then becomes an estimate of the enhancer activity. The method is sensitive, quantitative, and can be adapted to screen libraries of putative hormone responsive enhancers and response elements (20). We used here the most recent version of the method, which overcomes several technical pitfalls (18). A synthetic DR4 consensus element with degenerated nucleotides was used to generate a plasmid library. The degenerate positions were chosen based on previous definition of the optimal consensus derived from several previous ChIP-seq analyses. The library putatively contains 32 768 different DR4 sequences (Fig. 1A). We amplified the library and transfected HEK293 cells in duplicate with either 0.6μg, 1.2μg, or 2.4μg of DNA. T3 (10-8 M) was added to the culture medium of half of the transfected cells and RNA was extracted 36 hours later. We prepared cDNA and amplified the vector encoded RNA and perform a direct Sanger sequencing of the amplicons (Fig. 1B). Tide DNA (21) was used to quantify signal intensity on the Sanger electrophoretograms and provides a first indication that T3 has an influence on the representation of the different nucleotides at degenerate positions.
Figure 1.
Library description. (A) Logo of consensus DR4-like sequence discovered by de novo motif search (25) from a TRα1 ChIP-seq dataset obtained from mouse striatum neurons (11). According to structural data (15) the 5′ half-site (5′RGGTCA3′) is occupied by RXR. The 2 3′ nucleotides of the 4 nucleotides spacer and the 3′ half-site (5′NNRGGNCA3′) contact TRα1. (B) The expression vector is transcribed in eukaryotic cells from a cryptic minimal promoter present in the Ori sequence, spliced, and contains the cloned amplicon. Sequences flanking the DR4 element with degenerated nucleotides are flanked by sequences present in the Hairless gene promoter. The small arrows indicate the position of the primer used for cDNA amplification. (C) Sanger sequencing of the input library (left) indicates the presence of a mixture of different nucleotides at the expected positions. Sequencing of cDNA prepared from transfected cells (right) confirm the heterogeneity of the library and suggests that T3 has an influence on the respective abundance of different mRNA/DR4-like elements in the amplified cDNA.
We then performed deep sequencing for amplicons prepared from either the library DNA, or the 6 different cDNA. After curation for sequencing errors, the different DR4 sequences were counted. A total of 9129 different DR4 sequences were identified among 1 078 919 reads obtained from the plasmid library, with the expected balance between nucleotides at the variable positions (Table S1) (22). Based on these 9129 sequences, which represent only 28% of the 32 768 possible combinations, we calculated the frequency of each nucleotide at each variable position and addressed whether T3 can influence this distribution (Table S2) (22). A significant enrichment, indicating an influence on transactivation, was observed for specific nucleotides at each position, which was highly significant according to χ 2 tests. Because the enrichment was more pronounced when 1.2 μg of DNA was transfected, we concentrated the analysis on this experiment using the other datasets only to confirm the main results. The conclusion from this analysis is that there is a clear preference for specific nucleotides at several positions. In particular, A or G at position 1, T or C at position 9, A at position 11, and C at position 15 are clearly favorable to transactivation (Table 1).
Table 1.
Enrichment of specific nucleotides in DR4 elements occupied by retinoid X receptor α/TRα1 heterodimers according to self-transcribing active regulatory region sequencing analysis
| Position | 1 | 7 | 8 | 9 | 10 | 14 | 15 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Seq 5′ to 3′ | N | GGTCA | N | N | N | N | A/G | GG | N | N | A |
| A | 1.47 | 1.04 | 1.01 | 0.67 | 1.07 | 1.05 | 1.00 | 0.78 | |||
| C | 0.81 | 0.97 | 1.00 | 1.12 | 1.01 | 0.89 | 1.26 | ||||
| G | 1.25 | 0.88 | 0.97 | 0.79 | 1.12 | 0.90 | 0.85 | 0.89 | |||
| T | 0.68 | 1.14 | 1.01 | 1.30 | 0.83 | 1.24 | 0.91 |
Bold characters correspond to nucleotide which are clearly overrepresented.
We then asked if nucleotides at each position operate independently or rather if distinct combinations of nucleotides are more favorable to promote T3 response. At the 3′ end of the motif, the nonrandom distribution of dinucleotides enrichment was obvious (Fig. 2A). We used the χ 2 test to ask whether the frequency of some dinucleotides deviates from the expected frequency, assuming that nucleotide changes act independently (Table S3) (22). The deviations did not originate from the oligonucleotide synthesis or library preparation as the same χ 2 test did not reveal any significant bias in the input sequencing library. Although some dinucleotides combinations were clearly favored the most active dinucleotides were the ones predicted from the previous single nucleotide analysis (5′AC, 5′TC).
Figure 2.
Features of the DR4 response element favoring T3 response. (A) Frequency of specific dinucleotides in the downstream half-site contacted by TRα1 (positions 14 and 15). The table indicates the enrichment in cDNA library prepared from T3 treated cells compared with the input library. (B) Frequency of specific tetranucleotides in the spacer (positions 7 to 10). The table indicates the enrichment in cDNA library prepared from T3-treated cells compared with the input library for the 5 most favorable spacer tetranucleotides. (C) Global analysis of all tested DR4 (Deseq2). Each gray dot represents a specific sequence. Black triangles are for the 455 sequences significantly enriched in the cDNA library prepared from T3-treated cells compared with the input library (adjusted P < 0.05). The 5 most efficient DR4 sequences are indicated and listed in the table.
A similar analysis was applied to the spacer regions (Fig. 2B) in which 5 combinations stand out as most favorable (fold-change > 2), the 5′GATA sequence being the most enriched after T3 treatment. This result could not be predicted from the analysis of each position, as the expected frequency of this sequence, based on the individual nucleotides frequency, ranks at the 124 positions of 256 (Table S4) (22).
We then used a more global analysis to address the influence of combining favorable nucleotides at variable positions to optimize the transactivation capacity of the DR4 element (Table S5) (22). Differential analysis using Deseq2 analysis (Fig. 2C) showed that 455 of the tested sequences had a positive effect on mRNA level on T3 treatment. Among the sequences that were represented more than 100 times in each library, 5 sequences stand out as being clearly more efficient for transactivation (Fig. 2C).
To reinforce this conclusion and ascertain that the result can be generalized to other nucleotide contexts, we selected few of the tested DR4, cloned them in a luciferase reporter vector, and addressed their capacity to transactivate the reporter gene expression. Nine representative DR4 elements were tested confirming the main conclusions of the STARR-seq analysis. Interestingly, the response elements differed not only by their capacity to drive luciferase expression upon T3 stimulation, but also by their ability to repress reporter expression. Therefore, the DR4 elements ranking differed, depending on the chosen criterion: induction rate or maximal expression in presence of T3 (Fig. 3).
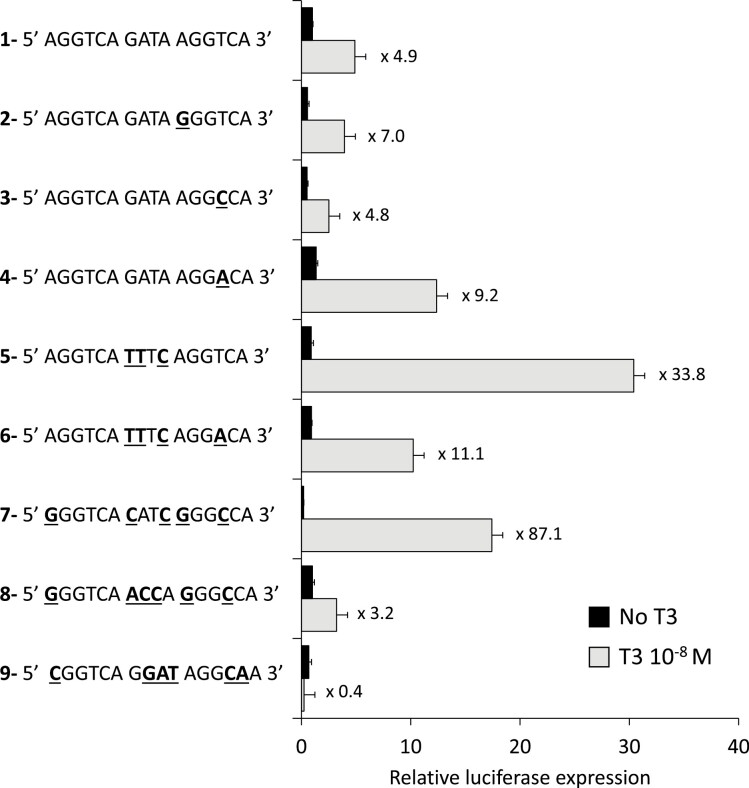
Figure 3.
Individual tests of 9 DR4 sequences in transient expression assays. DR4-1 is the most efficient of the tested DR4, according to Fig. 2C. Comparisons between DR4-1, -2, -3, and -4 highlight the most favorable nucleotide in the 3′ half-site occupied by TRα1. Comparisons with DR4, -5, -6, -7, and -8 demonstrate that spacer nucleotides have an important influence on transactivation and that a T at the third position of the spacer (position 9) is more favorable to transactivation. DR4-9 was predicted to be inactive by STARR-seq analysis, which is confirmed in this assay. Note that the negative influence exerted by unliganded TRα1, and that induction rate is maximal for DR-7. Two to 4 repeats of DR4-6 were previously used in reporter constructs to provide maximal transactivation capacity (18).
We then addressed the possibility that this functional screen reflects the capacity of specific sequences to bind TRs, as defined by genome-wide analysis of chromatin occupancy. We used for this published ChIP-seq datasets describing chromatin occupancy by TRα1 in the GABAergic neurons of mouse striatum (11) and in mouse cardiomyocytes (10). We also included a ChIP-seq dataset obtained for TRβ1 in liver (6). Using FIMO (Find Individual Motif Occurrences) search, from the MEME suite (23), we listed 51 231 matches in the mouse genome (GRCm38/mm10) for the DR4 motif (5′NGGTCANNNNRGGNNA3′) used for our STARR-seq screen. A small fraction of these were occupied by TRα1 (2099 in neurons, 1266 in cardiomyocytes) or TRβ1 (499 in liver). We then asked whether the nucleotide composition of the occupied and unoccupied DR4 motives differ. Table 2 reveals striking similarities between the outcome of this differential analysis and of the STARR-seq screen. There is therefore a strong correlation between the capacity of a specific DR4 element to recruit TRα1 in chromatin, and the capacity of this DR4 to mediate transactivation in transfected cells. We pursue the comparison between the outcome of ChIP-seq and STARR-seq analyses and tested the influence of combination of nucleotides for the largest dataset (GABAergic neurons). We also found that some specific combinations at position 14 and 15 are also more favorable to chromatin occupancy by TRα1 (Table 3). With only 2099 TRα1 binding sites in chromatin, this statistical comparison was not applicable to the spacer combinations, or to the whole sequence.
Table 2.
Enrichment of specific nucleotides in DR4 elements occupied by TRα1 and TRβ1 according to chromatin immunoprecipitation sequencing analyses
| TRα1 in GABAergic neurons | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Seq 5′ to 3′ | N | GGTCA | N | N | N | N | A/G | GG | N | N | A |
| A | 2.33 | 0.67 | 1.10 | 0.32 | 1.17 | 1.40 | 1.11 | 0.39 | |||
| C | 0.05 | 1.00 | 1.25 | 1.23 | 0.80 | 0.00 | 0.68 | 2.52 | |||
| G | 1.54 | 1.22 | 0.99 | 0.48 | 1.54 | 0.60 | 0.45 | 0.55 | |||
| T | 0.08 | 1.11 | 0.66 | 1.97 | 0.49 | 0.00 | 1.76 | 0.54 | |||
| TRα1 in cardiomyocytes | |||||||||||
| Seq 5′ to 3′ | N | GGTCA | N | N | N | N | A/G | GG | N | N | A |
| A | 2.18 | 0.58 | 0.92 | 0.49 | 1.04 | 1.35 | 1.31 | 0.43 | |||
| C | 0.04 | 1.01 | 1.32 | 1.29 | 0.96 | 0.00 | 0.80 | 2.52 | |||
| G | 1.58 | 1.49 | 0.90 | 0.52 | 1.43 | 0.66 | 0.48 | 0.54 | |||
| T | 0.20 | 0.92 | 0.86 | 1.70 | 0.57 | 0.00 | 1.40 | 0.51 | |||
| TRβ1 in liver | |||||||||||
| Seq 5′ to 3′ | N | GGTCA | N | N | N | N | A/G | GG | N | N | A |
| A | 2.31 | 0.51 | 0.95 | 0.53 | 1.09 | 1.30 | 1.16 | 0.44 | |||
| C | 0.08 | 0.87 | 1.30 | 1.28 | 1.05 | 0.00 | 0.82 | 2.23 | |||
| G | 1.34 | 1.71 | 0.91 | 0.54 | 1.10 | 0.70 | 0.54 | 0.67 | |||
| T | 0.27 | 0.91 | 0.84 | 1.65 | 0.76 | 0.00 | 1.48 | 0.67 |
Table 3.
Preferential association of nucleotides at position 14 and 15
| Position 15 | A | C | G | T | |
|---|---|---|---|---|---|
| Position 14 | A | 0.23 | 2.18 | 0.36 | 0.94 |
| C | 0.45 | 1.59 | 0.98 | 0.29 | |
| G | 0.20 | 0.82 | 0.26 | 0.28 | |
| T | 1.18 | 3.66 | 1.57 | 0.99 |
Bold characters correspond to dinucleotide combinations which are clearly overrepresented.
Discussion
Our report represents an unprecedented wide analysis of the transactivation capacity of TRα1/RXRα heterodimers. We used an improved version of the synthetic STARR-seq protocol, which proves to reduce background transcription and facilitates the detection of hormonal response. Compared with the previous similar study, performed with the glucocorticoid receptor (20), we observed higher induction rates and enrichments. The protocol described previously should thus be directly applicable for other nuclear receptors response elements. In the case of TRα1/RXRα heterodimers, we first identified 455 efficient DR4 elements, and confirmed in independent transient expression assays that some of them outperform the one previously used in reporter constructs (24).
The main conclusion of our study is the striking concordance between functional tests and chromatin occupancy analyses. Previous low-throughput analyses that used naked DNA are also concordant, indicating for example that a 5′TGTA or 5′TGAG spacer is much more favorable than 5′TGAT or 5′TGAC and that the AGGTCA is also optimal for the 5′ half-site (25). Our main hypothesis is that the transactivation capacity of a given DR4 essentially reflects the affinity of the TRα1/RXRα heterodimer for its DNA sequence. As expected from the structural data, the 2 last nucleotides of the DR4 spacer participate in defining this affinity. An alternative possibility would be that nucleotide changes influence the competition between TRα1/RXRα heterodimers and other transcription factors able to bind on overlapping motives. This is an unlikely explanation, however, because, in the condition of STARR-seq, TRα1 and RXRα are overexpressed. Also, such a competition would influence expression in a T3 independent manner, whereas our statistical analysis in focused on T3 response.
The relationship between chromatin binding and T3-mediated transactivation remains difficult to establish. Notably, the repertoire of genes which transcription is activated by T3 is highly dependent on the cell type, which contrasts with the extended overlap between the cistrome of different cell types. Here for example, 60% (460/766) of the DR4 elements occupied by TRα1 in cardiomyocytes are also occupied in GABAergic neurons. This question cannot be easily addressed with a statistical approach because the number of T3 responsive genes because regulatory sequences occupied by TRα1 are small. Here for example, only 14 of the tested DR4 elements were found in the TRα1 binding site located within 30 kb of a transcription start site of a T3 responsive gene in GABAergic neurons. The explanation for the cell-specific response to T3, and the limited number of T3-responsive genes in a given cell type, is likely to be found in the cellular context, for example as a result of variations in the repertoire of transcription coactivators.
Acknowledgments
The authors thank Sandrine Hughes and Benjamin Gillet of the Plateau de Séquencage de l’IGFL facility for financial support, technical help, and discussions and Sophia Belkhir for gathering preliminary data.
Glossary
Abbreviations
- ChIP-seq
chromatin immunoprecipitation sequencing
- RXR
retinoid X receptor
- STARR-seq
self-transcribing active regulatory region sequencing
Contributor Information
Frédéric Flamant, Institut de Génomique Fonctionnelle de Lyon, Université Claude Bernard Lyon I, CNRS UMR 5242, INRAE USC 1370 Ecole Normale Supérieure de Lyon, 69364 Lyon, France.
Yanis Zekri, Institut de Génomique Fonctionnelle de Lyon, Université Claude Bernard Lyon I, CNRS UMR 5242, INRAE USC 1370 Ecole Normale Supérieure de Lyon, 69364 Lyon, France.
Romain Guyot, Institut de Génomique Fonctionnelle de Lyon, Université Claude Bernard Lyon I, CNRS UMR 5242, INRAE USC 1370 Ecole Normale Supérieure de Lyon, 69364 Lyon, France.
Financial Support
This work was supported by the European Union’s Horizon 2020 research and innovation program under grant agreement no. 825753 (ERGO).
Conflict of Interest
The authors have nothing to disclose.
Data Availability
Raw data and processed tables are available at the Gene Expression Omnibus (GSE196145). Supplementary data contain the complete datasets are available at figshare DOI: 10.6084/m9.figshare.19722988.
References
- 1. Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature of nuclear receptors. Pharmacol Rev. 2006;58:685-704. [DOI] [PubMed] [Google Scholar]
- 2. Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev. 2001;81:1097-1142. [DOI] [PubMed] [Google Scholar]
- 3. Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol. 2005;6: 542-554. [DOI] [PubMed] [Google Scholar]
- 4. Judelson C, Privalsky ML. DNA recognition by normal and oncogenic thyroid hormone receptors. Unexpected diversity in half-site specificity controlled by non-zinc-finger determinants. J Biol Chem. 1996;271:10800-10805. [DOI] [PubMed] [Google Scholar]
- 5. Chatonnet F, Guyot R, Benoit G, Flamant F. Genome-wide analysis of thyroid hormone receptors shared and specific functions in neural cells. Proc Natl Acad Sci USA. 2013;110:E766-E775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shabtai Y, Nagaraj NK, Batmanov K, et al. A coregulator shift, rather than the canonical switch, underlies thyroid hormone action in the liver. Genes Dev. 2021;35:367-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ramadoss P, Abraham BJ, Tsai L, et al. Novel mechanism of positive versus negative regulation by thyroid hormone receptor beta1 (TRbeta1) identified by genome-wide profiling of binding sites in mouse liver. J Biol Chem. 2014;289:1313-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ayers S, Switnicki MP, Angajala A, Lammel J, Arumanayagam AS, Webb P. Genome-wide binding patterns of thyroid hormone receptor Beta. PLoS One. 2014;9:e81186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grontved L, Waterfall JJ, Kim DW, et al. Transcriptional activation by the thyroid hormone receptor through ligand-dependent receptor recruitment and chromatin remodelling. Nat Commun. 2015;6:7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hirose K, Payumo AY, Cutie S, et al. Evidence for hormonal control of heart regenerative capacity during endothermy acquisition. Science. 2019;364:184-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Richard S, Guyot R, Rey-Millet M, et al. A pivotal genetic program controlled by thyroid hormone during the maturation of GABAergic neurons. iScience. 2020;23:100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coons LA, Hewitt SC, Burkholder AB, McDonnell DP, Korach KS. DNA sequence constraints define functionally active steroid nuclear receptor binding sites in chromatin. Endocrinology. 2017;158:3212-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Phan TQ, Jow MM, Privalsky ML. DNA recognition by thyroid hormone and retinoic acid receptors: 3,4,5 rule modified. Mol Cell Endocrinol. 2010;319:88-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203-211. [DOI] [PubMed] [Google Scholar]
- 16. Neumayr C, Pagani M, Stark A, Arnold CD. STARR-seq and UMI-STARR-seq: assessing enhancer activities for genome-wide-, high-, and low-complexity candidate libraries. Curr Protoc Mol Biol. 2019;128:e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074-1077. [DOI] [PubMed] [Google Scholar]
- 18. Muerdter F, Boryn LM, Woodfin AR, et al. Resolving systematic errors in widely used enhancer activity assays in human cells. Nat Methods. 2018;15:141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schone S, Bothe M, Einfeldt E, et al. Synthetic STARR-seq reveals how DNA shape and sequence modulate transcriptional output and noise. PLoS Genet. 2018;14:e1007793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flamant, F., Zekri, Y. and Guyot, R. Analysis of STARR-seq data.2022. doi: 10.6084/m9.figshare.19722988. [DOI]
- 23. Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Res. 2015;43:W39-W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chan IH, Privalsky ML. Thyroid hormone receptors mutated in liver cancer function as distorted antimorphs. Oncogene. 2006;25:3576-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Quack M, Frank C, Carlberg C. Differential nuclear receptor signalling from DR4-type response elements. J Cell Biochem. 2002;86:601-612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data and processed tables are available at the Gene Expression Omnibus (GSE196145). Supplementary data contain the complete datasets are available at figshare DOI: 10.6084/m9.figshare.19722988.