Abstract
Capturing the diverse microbiota from healthy and/or stress resilient plants for further preservation and transfer to unproductive and pathogen overloaded soils, might be a tool to restore disturbed plant–microbe interactions. Here, we introduce Aswan Pink Clay as a low-cost technology for capturing and storing the living root microbiota. Clay chips were incorporated into the growth milieu of barley plants and developed under gnotobiotic conditions, to capture and host the rhizospheric microbiota. Afterward, it was tested by both a culture-independent (16S rRNA gene metabarcoding) and -dependent approach. Both methods revealed no significant differences between roots and adjacent clay chips in regard total abundance and structure of the present microbiota. Clay shaped as beads adequately supported the long-term preservation of viable pure isolates of typical rhizospheric microbes, i.e. Bacillus circulans, Klebsiella oxytoca, Sinorhizobium meliloti, and Saccharomyces sp., up to 11 months stored at −20°C, 4°C, and ambient temperature. The used clay chips and beads have the capacity to capture the root microbiota and to long-term preserve pure isolates. Hence, the developed approach is qualified to build on it a comprehensive strategy to transfer and store complex and living environmental microbiota of rhizosphere toward biotechnological application in sustainable plant production and environmental rehabilitation.
Keywords: clay beads, clay chips, long-term microbial preservation, microbiota capturing, microbiota transfer, plant microbiota
This study evaluates the ability of clay chips and beads as a practical and cheap methodology to capture the microbiota from barley roots and preserve pure isolates up to 11 months.
Introduction
Microbiota play an essential role in many ecosystem functions, e.g. enhancing the stability, resilience, and well-being of their hosts; plants, animals, and humans (Trivedi et al. 2020, Joseph and Curtis 2021). Management of these microbiota can offer an opportunity to improve plant resilience against biotic and abiotic stresses, including ecological restoration via approaches such as synthetic microbial community (SynCom) and microbiome engineering (Castrillo et al. 2017, Valliere et al. 2020, Ke et al. 2021). For instance, the transplantation of tomato rhizosphere microbiota from a resistant variety to a pathogen susceptible variety successfully suppressed the soil-borne pathogen Ralstonia solanacearum (Kwak et al. 2018). However, transfer and recruitment of the desirable microbiome and SynCom to restore microbiome functions requires suitable delivery systems for living microbe assemblages (Compant et al. 2019). Such systems should be able not only to capture the diverse microbiota, but also to maintain them during their transfer into the target ecosystems (Sessitsch et al. 2019). In spite of their importance, there is only a limited number of studies available focusing on developing effective microbiota delivery strategies (Qiu et al. 2019). Methods such as magnetic beads and nano-filters are used to capture and trap microbial isolates; however, these methods are limited to their use in clinical diagnosis so far (Sande et al. 2020).
Clay beads have shown to be more efficient in capturing the most significant parts of bacterial diversity in water, i.e. groundwater, surface water, and wastewater, when compared with gravel particles and glass beads (Voisin et al. 2016). Also, clay beads succeeded to act as trapping matrices for capturing the representative bacterial communities developing in stormwater infiltration systems (SIS) to assess the microbial quality of aquifers (Mermillod-Blondin et al. 2019). Such approaches inspired us to use clay as a natural matrix to capture and host the plant root-associated microbiota. A clay, known as ‘Pink Kaolinite Clay’, is tentatively connected to kaolinitic sediments (Rodziewicz 1992).
Most microbial product delivery methods have been developed as liquid formulations for soil applications, foliar sprays, seed and pellet coatings, or granules and powders that meet challenges such as limited shelf life, and depend on farmers' storage capacity and handling skills (Sessitsch et al. 2019). In fact, the effective microbial delivery strategy should support the long-term survival of microbes and persistence in the environment to prevent the rapid decline of introduced inoculants (Qiu et al. 2019, Batista and Singh 2021). Moreover, different studies refer to the importance to preserve the microbial community's diverse and beneficial microbiota from undergoing changes (Bello et al. 2018, West et al. 2019).
We aimed to develop a practical and low-cost methodology for comprehensively capturing and hosting the root microbiota. The Egyptian local type of clay used for pottery making, as an inexpensive and available natural material, is presented as an efficient trapping material, in the form of clay chips. To prove if clay chips, together mixed with quartz sand used as plant growth substrate, are able to capture the root-inhabiting microbiota, the tested barley plants were grown in a gnotobiotic plant growth system. Then, two distinct approaches have been employed: (i) the total bacterial microbiota originating from the plant roots and adjacent clay chips were analyzed based on 16S rRNA genes and (ii) the culturable bacterial microbiota of plant roots and adjacent clay chips, which grew on agar plates of plant (barley)-based culture media were compared to the standard R2A culture medium. In addition, MALDI-TOF Mass Spectrometry was used for the phyloproteomics analysis of the spectral profiles of the bacterial isolates represented the plant growing system. The other part of the study tested the ability of clay beads to preserve and long-term maintain microbial pure cultures stored at three different storage temperatures (freezing at −20°C, cooling at 4°C, and ambient room temperature).
Materials and methods
General structure and overview of the experiments
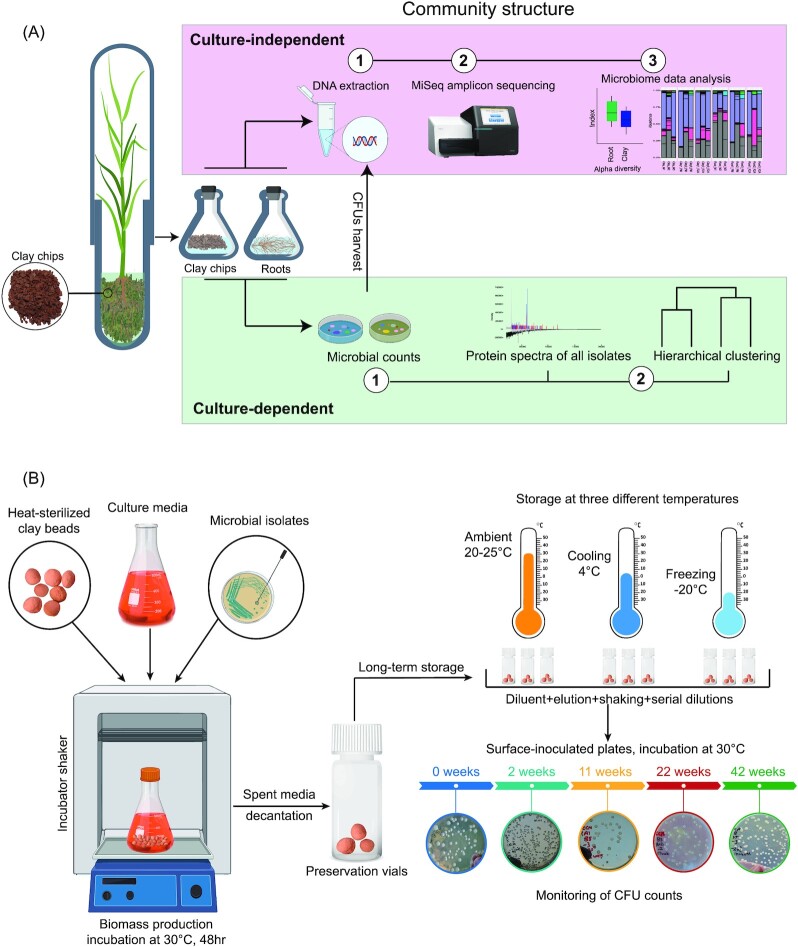
The workflow in Fig. 1 illustrates the main experimental setup of the two experiments. In the first part of the study, plants were grown in glass tubes to test the capability of clay chips, as a growth milieu, to capture the microbiota associated with developed barley roots (Fig. 1A). Culture-independent (MiSeq 16S rRNA sequencing) and culture-dependent (CFU counts and MALDI-TOF) methods were used to compare the microbiota load and structure of barley roots with the adjacent clay chips. The second part of the study (Fig. 1B) aimed to test the efficiency of clay beads to preserve and long-term maintain microbial pure cultures. Such efficiency was assessed at three different storage temperatures (freezing at −20°C, cooling at 4°C, and ambient room temperature) by measuring cell viability (CFU counts) over time up to 11 months.
Figure 1.
Workflow of the experiments: (A) capturing bacterial community on clay chips and (B) long-term preservation of different microbial pure strains (Bacillus circulans, Klebsiella oxytoca, Sinorhizobium meliloti, and Saccharomyces sp.) on clay beads.
Clay origin and physicochemical composition
Samples of Aswan Pink/red Kaolinitic Clays pastes were obtained from a local pottery handcraft at Old Cairo, Egypt. This type of clay, known as ‘Pink Kaolinite Clay’ was historically, and since the Pharos and Greco-Roman time, used in the manufactures of different kinds of ceramic artifacts of table wares and vessels forms. It derives from the Modern Aswan region, which is synonymous with the ancient cities of Syene and Elephantine (Rembart and Betina 2021). Such ‘Pink Clay’ (Rodziewicz 1992) is the term used in literature for this type of Egyptian clay tentatively connected to kaolinitic sediments. The Aswan Pink Clay sediments/quarries, are supposed to derive from the decomposition of pinkish feldspar minerals found in the local rose granite, available abundantly in the entire broader area (Soliman 1985). The physico-chemical profiles of the tested ‘Aswan Pink Clay’ reveal its suitability as good quality and environment-friendly raw material. This specifically includes: clay size fractions, low-order kaolinite, and illite, absence of I/S minerals, low fluxing agents such as alkali oxides (Na2O and K2O) and alkaline earth oxides (CaO and MgO), low S and Cl contents, and low contents of toxic elements (As, Cd, Hg, and Pb) (Baioumy and Ismael 2014). This resulted in the extensive use of such clays along the Egyptian history in manufacturing table wares and vessels for safe human consumption, a strong argument and justification for its use for housing microorganisms as well.
The chemical analysis of major and trace elements of the tested clays was carried out using X-ray fluorescence and performed by the Central laboratories of the Egyptian Mineral Resources Authority (EMRA), Giza, Egypt. Further, physico-chemical properties, e.g. saturation point (SP), porosity, water holding capacity (WHC), CaCO3, pH, EC, and soluble cations and anions, were also carried out by the Soils, Water, and Environment Research Institute, Agricultural Research Center, ARC, Giza, Egypt, using standardized methods (Jackson 1958, Richards 1965; Table 1).
Table 1.
Chemical analysis and nutritional profile of representative samples of Aswan Pink Clay and of prepared clay beads and chips.1,2
| Chemical properties2 | Mineral salts (wt %)1 | ||
|---|---|---|---|
| CaCO3 wt % | 4.07 | SiO2 | 49.87 |
| *pH (suspension 1:25) | 9.39 | TiO2 | 1.26 |
| EC (dSm−1) | 11.29 | Al2O3 | 18.87 |
| SP (saturated paste extract) % | 57.25 | Fe2O3 | 14.48 |
| Soluble cations and anions (mmol L−1) | MnO | 0.09 | |
| MgO | 0.63 | ||
| Ca++ | 28.90 | CaO | 4.01 |
| Mg++ | 5.40 | Na2O | 0.19 |
| Na+ | 77.50 | K2O | 0.71 |
| K+ | 0.53 | P2O5 | 0.03 |
| HCO3− | 1.32 | Cl | < 0.01 |
| Cl− | 77.90 | SO3 | < 0.01 |
| SO4−− | 33.15 | LOI | 9.52 |
| Physical properties of prepared 2 : porosity (%) | Water holding capacity (WHC %) | ||
| Clay beads | 64–71 | 21.82 | |
| Clay chips | 62–66 | 36.68 | |
The pH was measured in H2O.
Analyzed at the Central laboratories of the Egyptian Mineral Resources Authority (EMRA), Giza, Egypt; and
Analyzed at the Soils, Water, and Environment Research Institute, Agricultural Research Center (ARC), Giza, Egypt.
Manual production of clay chips and clay beads
The freshly prepared clay paste was used for the production of both clay chips and clay beads. Manually, the clay paste was shaped into beads of > 5–8 mm diameter. On the other hand, a pastry wooden roller was used to prepare the fresh flattened sheets of clay paste (> 1–2 mm thick); then with the help of a sharp cutter, chips of ca. 4–6 mm × 4–6 mm were prepared. Both clay beads and chips were subjected to hard-firing at 800–900°C in an electrical furnace to increase their hardness and decrease shrink–swell capacity in aqueous solutions. Before use, the prepared clay chips and clay beads were packed into metal boxes and were dry heat sterilized in a volcano thermal heater at 300°C for 3 h.
Clay chips as substrate to capture barley root microbiota under gnotobiotic conditions
As growth substrate, clay chips and coarse sand particles/grains (sieved to 2–4 mm diameter and carefully washed several times with distilled water) were used. Aliquots of 30 g of a mixture (1:1, v/v) of clay chips and sand grains were filled into glass tubes (diameter 3 cm, height of 21 cm). Each tube was supplemented with 22 ml of semi-solid solution previously prepared from autoclaved barley-straw/distilled water infusion (0.1 g straw + 1.7 g agar L−1 distilled water), and then covered with tubes bigger in size (4 cm diameter and 11 cm length). The prepared growth tubes were autoclaved, then thoroughly mixed by vortex mixer prior transfer of barley seedlings.
The system was prepared and adjusted according to Youssef et al. (2004). Barley seeds (Hordeum vulgare L., Giza 127) were obtained from the Field Crops Research Institute, Agricultural Research Center (ARC), Giza. Healthy and intact seeds were germinated at room temperature (> 20–25°C) on water agar plates for 3 days. A total of three healthy seedlings were planted into the clay/sand mixture of each culture tube. A total of three sets, each of nine tubes, were prepared representing three biological replicates. The growth tubes were kept in an environmentally controlled growth chamber having a day/night cycle of 8 h and 16 h, respectively; with a temperature of 20–22°C for 45 days.
A total of three tubes were randomly selected from each of the three sets of tested plant growth tubes. Contents of each of the tubes were aseptically sorted to obtain two composite samples representing roots and clay chips. To prepare the original suspension, the entire intact roots (ca. 0.1 g), and clay chips (ca. 40 g) were separately suspended into 20 ml saline solution, half strength basal salts of CCM, and shaken for 1 h at 120 rpm. Thereafter, the prepared original suspensions were further used for culture-dependent and culture-independent analyses.
Culture-dependent bacterial community characterization of barley root and clay chips
Original root and clay chips suspensions were further serial diluted (1:10) in the saline solution (half strength basal salts of CCM). Aliquots of 200 µl from suitable dilutions (10−4:10−7) were spread onto agar plates of tested culture media; R2A and plant (barley)-based culture medium (PM) in three replicates. CFUs developed on agar plates (> 30–300 CFUs plate–1), were counted after 14 days of incubation at 25°C. We used bound water, which is the semi-solid water agar content, i.e. found between fresh and dry weights of root and clay chips. We used it as unit to compare the settlement of microorganisms of surface area. The simplified workflow of the applied procedures is graphically illustrated in Fig. 1(A).
MALDI-TOF MS analysis and protein profiling of bacterial isolates
The MALDI-TOF MS via MALDI Biotyper platform on intact cells was used for the construction of the phyloproteomics of the secured bacterial isolates as described previously by Nemr et al. (2020). A total of 417 bacterial colonies, representing all emerged morphotypes of CFUs of each milieu was picked (Table S1, Supporting Information). They were inoculated into the respective culture medium, and then incubated at 25°C for 1 week. The 1-week-old cultures were centrifuged at 4000 rpm for 10 min. Thereafter, cell pellets were subjected to protein extraction by mixing with 30 µl distilled water. A portion of 90 µl absolute ethanol (EtOH) was added, and then centrifuged at 4000 rpm for 10 min. The supernatants were discarded, and the remaining cell pellets were dried at room temperature for 10 min to increase the extraction efficiency. A volume of 10 µl of 70% formic acid was added to the dried pellets and mixed thoroughly for 5 min. An equal volume of acetonitrile was added, mixed for 5 min, and then centrifuged at 2000 rpm for 2 min. A volume of 1 µl of supernatant was deposited directly onto a polished steel MALDI target plate (Bruker Daltonik, GmbH, Bremen, Germany) and dried, then overlaid with 1 μl of matrix (Bruker Daltonik), which was a saturated solution of α-cyano-4-hydroxycinnamic acid (HCCA) in 50% acetonitrile 2.5% trifluoroacetic acid. Intact protein masses were acquired using ultrafleXtreme MALDI–TOF/TOF mass spectrometer (Bruker Daltonik GmbH, Germany) equipped with the smart beam-II laser-positive mode. Acquisition of the mass spectra was done in the range of 2.000–20.000 Da and at a sample rate of 1.2 GS s−1. The mass spectrometer was controlled by FlexControl version 3.4 software (Bruker Daltonik). For calibration, Bruker Bacterial Test Standard (BTS) was used.
For the analysis, we initially imported the protein spectra of all samples into a MALDI Protein and Small Molecule Bioinformatics Platform (IDBac; Clark et al. 2018), and aligned the spectra, and clustered them based on presence/absence. Peaks with a signal-to-noise ratio above 3 and occurring in greater than 70% of replicate spectra were considered present. While, peaks occurring below 3000 m/z or above 15000m/z were removed from the analyses for consistency throughout all samples. Then, the dendrogram was constructed based on Ward. D hierarchical clustering. Based on the previous clustering we dereplicated the spectra into proteotypes at a similarity level of > 75%. Using these proteotypes, we constructed a weighted matrix per proteotypes and culture media, and further conducted hierarchical clustering.
Culture media
Plant-only-based culture media (PM): The plant (clover, maize, and barley)-teabags culture media was prepared according to Sarhan et al. (2016). Standard chemically synthetic culture media: N-deficient combined carbon-sources medium; CCM (Hegazi et al. 1998), modified Reasoner's 2A agar; R2A ( Reasoner and Geldreich 1985), and yeast extract mannitol agar medium; YMA (Somasegaran and Hoben 1994).
DNA extraction and Illumina MiSeq sequencing
In culture-dependent treatments, all CFUs developed on respective agar plates, originating from root and clay chips and grown on two different culture media, PM and R2A (each in three replicates, 12 samples in total), were harvested and carefully washed using 7 ml of buffer solution (0.05 M NaCl). Then, cells were collected by centrifugation at 10,000 rpm for 15 min. DNA was extracted from the resulting pellets using QIAGEN DNeasy Plant Mini Kit (Qiagen Inc., Hilden, Germany) according to the manufacturer's instructions.
For culture-independent measurements, total DNA was extracted from the original suspensions prepared for roots and adjacent clay chips (in three replicates each, six samples in total). DNA quality was assessed using NanoPhotometer (NanoPhotometer NP80 Touch, Implen GmbH, Munich, Germany). A total of 18 DNA samples was subjected to paired-end read Illumina MiSeq platform targeting the V4 region of the 16S rRNA gene using the 515f/806r primer set by ATLAS Biolabs GmbH, Berlin, Germany.
Amplicon sequence data analysis
Raw sequences from the bacterial 16S rRNA gene were processed using a script by a combination of tools including USEARCHv11.0.667 (Edgar 2010), and VSEARCH v2.15.2 (Rognes et al. 2016). The paired-end 16S reads of each sample were merged into a single sequence ( 2,166,570 merged sequences) using USEARCH (Edgar 2010). Subsequently, quality of merged sequences was filtered based on maximum expected error (maxee = 1) and sequences with maxee values higher than 1 discarded by VSEARCH (Rognes et al. 2016). In this step, 61,118 sequences were discarded and 2,105,452 quality-filtered sequences kept. Then, sequences were dereplicated using VSEARCH (Rognes et al. 2016) and assigned as amplicon sequence variants (ASVs) via the denoising method as well as removed chimeric sequences using USEARCH (Edgar 2010). This yielded 130 ASVs. All sequences were clustered at 99% similarity when mapping the merged sequences to ASVs for generating the count table ( 1,952,148 of 2,105,452 (92.72%)). This clustering step was performed with VSEARCH (Rognes et al. 2016). The ASVs were taxonomically classified using USEARCH with RDP classifier using RDP database (Edgar 2010, Cole et al. 2014). To determine a phylogenetic affiliation, all ASVs were aligned using MAFFT (Katoh et al. 2002). ASVs detected in clay chips and not in the root were filtered; additionally, ASVs found on the three replicates of clay chips and only in one replicate of the root were excluded. As a result of this filtering step, 17 ASVs were discarded and 113 ASVs were further used in the downstream statistical analysis.
Long-term preservation of pure isolates of microorganisms on clay beads
Tested pure isolates of microorganisms: pure isolates representing various genera of rhizobacteria were included, i.e. Bacillus sp. (Bacillus circulans 8E), Klebsiella sp. (Klebsiella oxytoca En11/2), and Sinorhizobium sp. (Sinorhizobium meliloti KP765325.1), in addition to a single isolate of yeast (Saccharomyces sp. B1). All isolates were obtained from the culture collection of the Environmental Studies and Research Unite (ESRU), Department of Microbiology, Faculty of Agriculture, Cairo University. Stock cultures of B. circulans and K. oxytoca were kept on N-deficient combined carbon sources culture media (CCM), while S. meliloti and Saccharomyces sp. were grown on yeast extract mannitol agar media (YMA).
Microbial application onto clay beads: to impregnate clay beads with growing cells of tested isolates, 1.0 ml culture of ca. 106 cells of B. circulans, K. oxytoca, or Saccharomyces sp. were inoculated into flasks that were filled with 100 ml of standard CCM or plant (clover)-teabags culture media. In the case of S. meliloti, standard YEM and plant (maize)-teabags culture media were used. A number of ca. 250 previously heat-sterilized clay beads were aseptically immersed into each of the prepared flasks of liquid culture media, followed by gentle shaking of 100 rpm at 30°C for 48 h. Thereafter, clay beads were aseptically and gently transferred into empty sterile Petri dishes, and kept for 48 h at 30°C incubator for biofilm stabilization and drying. Portions of such air-dried and stabilized beads were stored in sterile vials and kept at three temperature regimes: −20°C, 4°C, and ambient temperature for up to 42 weeks.
Microbial survival and density of cells on/in clay beads were periodically monitored by counting CFUs on the corresponding agar culture media. For this purpose, three of the stored beads were transferred and immersed into 20 ml saline solution (half strength basal salts of CCM), and soaked overnight at room temperature, then shaken for 2–4 h for elution and retrieval of microbial cells. Out of the resulting suspensions, successive serial dilutions were prepared, and aliquots of 200 µl of suitable dilutions (10−2:10−6) were spread onto agar plates of the corresponding culture medium. The developed colony-forming units (CFUs) were counted after 72 h of incubation at 30°C. A simplified workflow of the applied procedure is graphically illustrated in Fig. 1(B).
Statistical analysis
All statistical analyses were performed with R v.4.1.1 (https://www.r-project.org/) and R-studio (https://www.rstudio.com/). The downstream analyses of the four generated files, i.e. ASVs count table, taxonomy table, phylogenetic tree, and metadata sheet, were carried out using phyloseq package (McMurdie and Holmes 2013). The rarefaction curve was generated for all samples to demonstrate the sequencing depth of each sample (Figure S1, Supporting Information). Alpha diversity estimates were computed for observed ASVs and Shannon diversity. Adonis statistical test was applied to detect significant differences among the microbiota based on UniFrac distances.
Results
Growth of barley seedlings reported in the assembled gnotobiotic plant culture tubes
The physico-chemical properties of the used natural Aswan Pink Clay (Table 1) revealed its suitability as good quality and environment-friendly natural raw material. As a result, barley seedlings exhibited normal growth and differentiation of healthy roots and shoots (Figure S2, Supporting Information). As well, it appeared that the natural nutrients provided through the diluted infusion of barley straw further supported such normal growth.
Microbiota composition-comparison between barley roots and adjacent clay chips
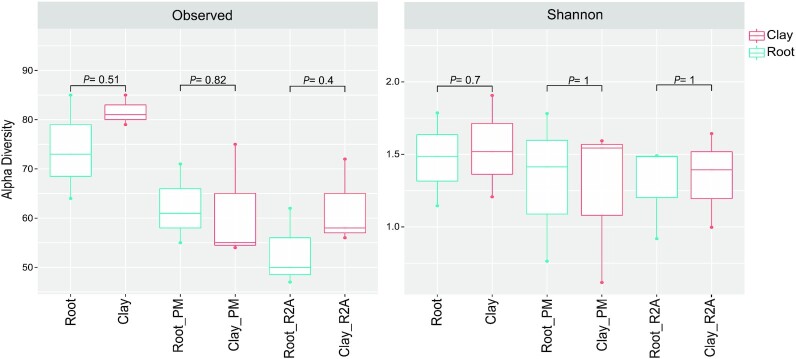
The Adonis test based on Unifrac distance was used to investigate variance among the whole existing microbiota of barley roots and adjacent clay chips based on detected ASVs (Table 2). Across all samples, no significant differences were reported for overall microbial compositions between clay chips and barley roots (Adonis test, P-values > 0.05). In both analyses of total microbiota and the cultivable part of the community, no significant differences were reported between clay chips and roots, in both Observed and Shannon diversity index (Wilcoxon test, P-values > 0.05; Fig. 2). Microbiota composition on genus level is shown in (Figure S3, Supporting Information). The highest abundance of Pseudomonas and Sphingomonas was reported in all tested samples, both of cultured communities and total DNA analysis of barley roots and clay chips.
Table 2.
Adonis test between ASVs observed of clay chips and barley roots microbiota, analyzed culture-independently from root (Root_M) and clay chips (Clay_M) or cultured on plant based medium (Root_PM, Clay_PM) or cultured on R2A medium (Root_R2A, Clay_R2A) based on UniFrac distances (Lozupone et al. 2011).
| Column_name | Comparison_between | Sample_counts | P-values |
|---|---|---|---|
| Root vs. Clay | Root | Clay | 9 | 9 | 0.753 |
| Milieu | Root_M | Clay_M | PM | R2A | 6 | 6 | 6 | 0.229 |
| All samples | Root_M | Clay_M | Root_PM | Clay_PM | Clay_R2A | Root_R2A | 3 | 3 | 3 | 3 | 3 | 3 | 0.924 |
Figure 2.
Alpha diversity indices to compare microbiota structure of barley roots and adjacent clay chips, using culture-independent analysis (Clay and Root), and cultured microbiota on R2A and plant-only-based (PM) culture media (Clay_PM, Root_PM, and Clay_R2A, Root_R2A). Alpha diversity boxplots were determined using observed ASVs and Shannon diversity index, using Amplicon 16S rRNA MiSeq sequencing. P-values between the different groups were shown using the Wilcoxon test.
Culture-dependent quantification of microbiota associated with barley roots and adjacent clay chips
Numbers of CFUs were quantified on two different culture media, the plant (barley)-teabags culture media and the synthetic R2A culture media. This is to evaluate if the same bacterial counts reported for barley roots could also be recovered from adjacent clay chips. Additionally, we wanted to test the efficiency of in vitro rhizobacteria cultivation when using a culture medium based on plant materials providing plant-adapted nutrient spectra (Sarhan et al. 2016) compared to a synthetic culture medium (R2A). No significant differences in CFU counts were observed between the root and adjacent clay chips when using the plant-only-based culture medium. This was not the case when using the synthetic R2A culture medium, where the differences were significant (Figure S4, Supporting Information).
Protein profiling of bacterial isolates associated with barley roots and clay chips on both culture media
The protein profile data from bacterial isolates were used to assess the differences between the cultured microbiota of barley roots and adjacent clay chips. The presented dendrogram (Figure S5, Supporting Information) has been constructed from the weighted matrix of all isolates proteotypes recovered from CFUs grown on plant-only-based (PM) and R2A culture media. These CFUs originated from the root and adjacent clay chips. Interestingly, the hierarchical clustering revealed that clay chips remarkably clustered together with plant roots. And, the two distinctive clusters of protein profiles indicated that the two culture media support growth of different bacterial communities (Figure S5, Supporting Information).
Assessment of clay beads for long-term microbial preservation of tested pure isolates
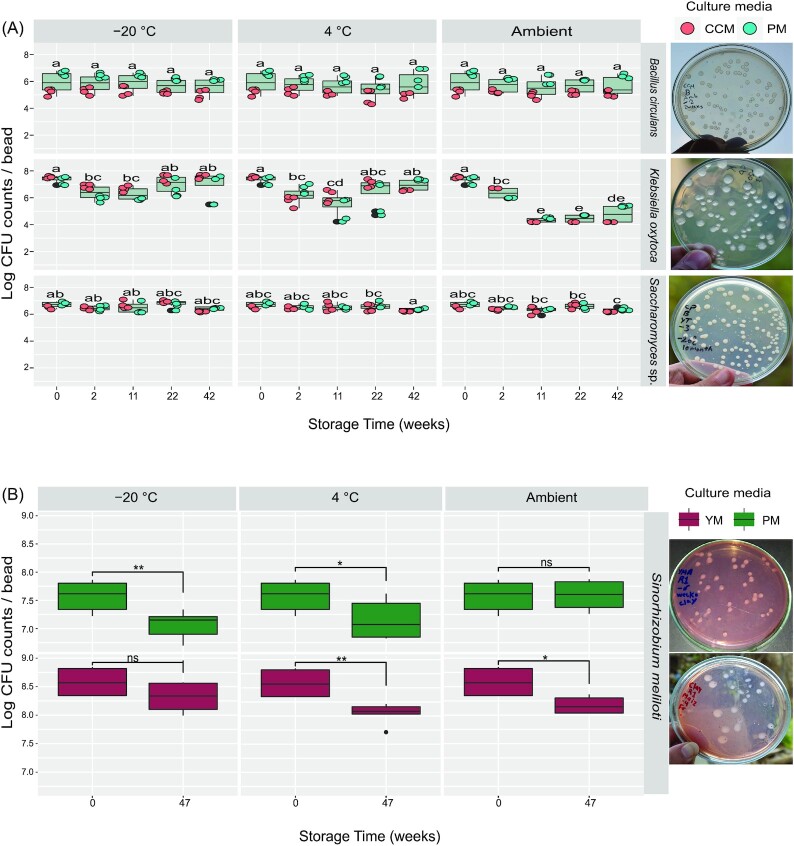
For achieving the potential aim to capture, preserve, and transfer microbial communities using clay material, in the first trial we checked three pure isolates of different bacterial families (B. circulans, K. oxytoca, and S. meliloti), and one yeast isolate (Saccharomyces sp.) for their long-term survival on clay beads stored on three different temperatures (−20°C, 4°C, and ambient). In general, all the tested microbes successfully survived at the three different storage temperatures in clay beads for more than 11 months.
According to ANOVA analysis of CFU counts of B. circulans, there were no significant differences attributed either to the independent effect of storage temperature (P = 0.812) or time (P = 0.451), nor to the interaction between them (P = 0.980). The initial mean viable counts of the tested B. circulans were log 5.92 ± 0.76 CFU bead−1. After 42 weeks of storage, still nearly identical numbers were recovered, log 5.56 ± 0.63, 5.78 ± 0.89, and 5.66 ± 0.71 at −20°C, 4°C, and ambient temperature, respectively (Fig. 3A).
Figure 3.
Viable cell counts (CFUs) of the tested pure isolates of microorganisms, which were preserved on clay beads and kept at different storage temperatures (−20°C, 4°C, and ambient temperatures): (A)B. circulans, K. oxytoca, and Saccharomyces sp. as developed on standard CCM and plant-only-based (PM) culture media. Different letters indicate significant differences based on Tukey's Honestly Significant Differences (HSD), P < 0.05, n = 8. (B)Sinorhizobium meliloti developed on agar plates of YEM and plant-only-based culture media. Student's t-test was used in comparison and levels of significance are ns: P > 0.05, *P ≤ 0 .05, **P ≤ 0.01, and ***P ≤ 0.001. The far-right panels show examples of CFUs morphologies developed.
As to K. oxytoca, and throughout 11 months of preservation, the viability was comparable at storage temperatures of −20°C and 4°C. According to ANOVA analysis and HSD test, there were no significant differences between initial viable counts (log 7.39 ± 0.26 bead−1) and those reported after 42 weeks of storage (log 7.04 ± 0.95 and 6.95 ± 0.42 bead−1, at −20°C and 4°C, respectively). For ambient room storage, the CFUs counts significantly decreased (P < 0.05) over time, down to log 4.79 ± 0.64 bead−1 after 42 weeks (Fig. 3A).
The tested isolate of Saccharomyces sp. successfully maintained survivability after 42 weeks of preservation (Fig. 3A). At the three tested storage conditions, no significant reduction in viable counts was reported at −20°C and 4°C, compared with the initial counts of 6.72 ± 0.21 bead−1. Whereas, at the ambient room temperature, the clay beads maintained Saccharomyces sp. counts comparable to the initials until 22 weeks of storage, with slight but significant decreases after 42 weeks (log 6.35 ± 0.12 bead−1).
In the second trial, clay beads were efficient as well in the long-term preservation of S. meliloti. Irrespective of storage temperature and tested culture media, the decrease in the number of viable cell counts did not exceed a single log after 47 weeks of storage (Fig. 3B).
Discussion
Exploiting and manipulating the plant microbiome is an opportunity to improve plant growth and act against current ecosystem degradation (Toju et al. 2018). Although microbiome transfer has revealed the potential to restore the functionality of degraded environments, it faces challenges in terms of storage, handling, and delivery systems (Tosi et al. 2020). We proved the efficacy of clay chips to capture and host the living microbiota of barley roots. We also demonstrated the usability of clay for long-term preservation of rhizospheric bacterial and fungal pure strains at three different storage temperatures and even under ambient conditions.
The interaction of clay chips with barley roots during growth in a gnotobiotic plant growth system was successful in capturing and trapping most of microbiota associated with barley roots (Fig. 2). Correspondingly, Voisin et al. (2016) and Mermillod-Blondin et al. (2019) demonstrated that clay beads supported biofilm development, and were able to capture bacterial communities in different aqueous environments. This capability of clay chips is likely due to the roughness and porosity of their surface that supports microbial adhesion to the porous structure, which concurred with a number of reports (Yakub and Soboyejo 2012, Al-Amshawee et al. 2021). Taoka et al. (2021) also revealed that pottery-shard (PS) of ceramic is capable of adhering lactic acid bacteria in its porous structure. Additionally, a number of studies showed the suitability of kaolinite for adhesion and biofilm formation of different types of microbes (Wu et al. 2014, Huang et al. 2015). An explanation for the ability of clay chips to capture barley root microbes is their naturality that allows intimate interaction with growing plant roots, thus served as an extended habitat for the dwelling rhizobacteria. Further, the heterogeneity of pores in the clay chips support the accumulation of nutrients and water, while at the same time maintain an airflow that well simulates the environmental conditions within the plant–soil system. In the present study and by employing culture-dependent and culture-independent techniques, we reported the indistinguishable composition of microbiota associated with barley roots and adjacent clay chips. This confirmed the ability of clay chips to successfully capture and host barley root microbiota. Furthermore, the protein profiling of representative pure isolates prevailing in the tested plant system supported this finding.
Clay in the form of beads was also tested for long-term preservation of diverse pure cultures of rhizospheric microorganisms under different storage temperatures, −20°C, 4°C, and ambient room temperature. Microbial cells were successfully accommodated and preserved in such clay beads for up to 11 months at the three tested storage temperatures (Fig. 3). The success of survival pattern of B. circulans is consistent with the findings of Krumnow et al. (2009), who recorded appropriate preservation of B. subtilis on the natural polymers of acacia gum for 615 days without significant reduction. Although the clay beads kept K. oxytoca up to 11 months, there was a significant reduction in the survival pattern when preserved at ambient temperature. This is likely due to progressive reduction in water content; thus, strengthening the attachment of bacterial glycocalyx, which consists of extracellular polysaccharides (EPS), and clay particles/cavities. A phenomenon that possibly requires a longer agitation time to loosen such attachment and retrieve the adhered bacterial cells. A similar behavior was reported for the EPS-producing Deinococcus geothermalis, which was subjected to air drying that resulted in a reversible loss of flexibility and more firmness in attachment with different surfaces, thus yielding in better immobilization (Kolari et al. 2002). In general, and regardless of the type of preserved microorganisms, microbes can adapt to starvation and energy-limited conditions through activating survival strategies that allow them to persist for years. This has been demonstrated by Shoemaker et al. (2021) who reported that 21 bacterial taxa were able to survive for 1000 days in a closed system with zero resources. This is probably how microbes persisted on clay beads for such a long period of time in our study.
If the nearly intact bacterial community, which we captured from the barley rhizosphere under gnotobiotic conditions, will also survive on these clay chips for a long time has to be proven in the next step. Furthermore, following experiments will answer the question if these captured microorganisms can be transferred to a new donor plant by incorporating these clay chips into the rhizosphere.
Conclusions
Clay chips have the capacity to capture, host, and memorize microbiota. Such nature-inspired technology opens up new avenues for further progress in manipulating and delivery of environmental microbiomes toward soil rehabilitation. In addition, microbial preservation on clay beads is a low-cost, efficient methodology even suitable for less-equipped laboratories, and has the potential to easily transport and preserve microbes at various temperatures for several months, which in the present study was proven for microbial pure cultures. If such a long-term preservation will also be possible for complex microbial communities has to be attested in subsequent experiments.
Data availability statement
Raw sequence data were deposited into the NCBI Sequence Read Archive (SRA) under BioProject accession PRJNA816545.
Authors' contributions
Conceptualization: N.A.H., S.R., and M.R.A. Validation: M.R.A., M.A.H., H.H.Y., and M.A. M.E.S. and G.H.Y. supervised clay material analysis. M.E., M.H.T., and M.R.A. carried out the bioinformatics analysis. K.W. supervised the MALDI-TOF experiments. M.K., A.R.H., R.A.N., H.E., and G.V. provided the technical support. Writing original draft preparation: M.R.A., M.F., N.A.H., and S.R. Writing review and editing: M.R.A., S.R., N.A.H., M.F., S.K., and M.A. All authors have read and agreed to the published version of the manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Sascha Patz for his bioinformatics training during the summer school ‘IGZ-ESRU, 2019’. Thanks are also extended to Kerstin Fischer and Mandy Heinze (IGZ) for excellent technical support. With gratitude, we acknowledge the lab support of Mohamed S. Sarhan, Mohamed Y. Saleh, Elhussein F. Mourad, Mennatullah Abdou, Bishoy Sameh, Essam Adel, Saif Khodary, Abdul-Karim Noah, and Saad M. Abdelwakeel.
Contributor Information
Mohamed R Abdelfadil, Thaer-Institute, Faculty of Life Sciences, Humboldt University of Berlin, 10115 Berlin, Germany; Department of Microbiology, Faculty of Agriculture, Cairo University, 12613 Giza, Egypt; Department of Plant Microbe Systems, Leibniz Institute of Vegetable and Ornamental Crops, 14979 Großbeeren, Germany; RA Landscape Functioning, Leibniz Centre for Agricultural Landscape Research (ZALF), Eberswalder Str. 84, D-15374 Müncheberg, Germany.
Manar H Taha, Bioinformatics Group, Center of Informatics Sciences (CIS), Nile University, 12677 Giza, Egypt.
Mohamed El-Hadidi, Bioinformatics Group, Center of Informatics Sciences (CIS), Nile University, 12677 Giza, Egypt.
Mervat A Hamza, Department of Microbiology, Faculty of Agriculture, Cairo University, 12613 Giza, Egypt.
Hanan H Youssef, Department of Microbiology, Faculty of Agriculture, Cairo University, 12613 Giza, Egypt.
Mohab Khalil, Department of Microbiology, Faculty of Agriculture, Cairo University, 12613 Giza, Egypt.
Ahmed R Henawy, Department of Microbiology, Faculty of Agriculture, Cairo University, 12613 Giza, Egypt.
Rahma A Nemr, Department of Microbiology, Faculty of Agriculture, Cairo University, 12613 Giza, Egypt.
Hend Elsawey, Faculty of Organic Agriculture, Heliopolis University, 11785 Cairo, Egypt.
Gylaine Vanissa Tchuisseu Tchakounte, Department of Plant Microbe Systems, Leibniz Institute of Vegetable and Ornamental Crops, 14979 Großbeeren, Germany.
Mohamed Abbas, Department of Microbiology, Faculty of Agriculture and Natural Resources, Aswan University, 81528 Aswan, Egypt.
Gehan H Youssef, Department of Soil Chemistry and Physics, Soil, Water and Environment Research Institute, Agricultural Research Centre (ARC), 12112 Giza, Egypt.
Katja Witzel, Department of Plant Microbe Systems, Leibniz Institute of Vegetable and Ornamental Crops, 14979 Großbeeren, Germany.
Mohamed Essam Shawky, Department of Soil Science, Faculty of Agriculture, Cairo University, 12613 Giza, Egypt.
Mohamed Fayez, Department of Microbiology, Faculty of Agriculture, Cairo University, 12613 Giza, Egypt.
Steffen Kolb, Thaer-Institute, Faculty of Life Sciences, Humboldt University of Berlin, 10115 Berlin, Germany; RA Landscape Functioning, Leibniz Centre for Agricultural Landscape Research (ZALF), Eberswalder Str. 84, D-15374 Müncheberg, Germany.
Nabil A Hegazi, Department of Microbiology, Faculty of Agriculture, Cairo University, 12613 Giza, Egypt.
Silke Ruppel, Department of Plant Microbe Systems, Leibniz Institute of Vegetable and Ornamental Crops, 14979 Großbeeren, Germany.
Funding
The work was supported by Alexander von Humboldt Foundation (AvH) through equipment subsidy and financial support, and the German Academic Exchange Service (DAAD) for funding the Summer School ‘IGZ-ESRU, 2019’. We acknowledge the financial support of Yousef Jameel Academic Program at the Humboldt-University of Berlin, Germany.
Conflict of interest statement. None declared.
References
- Al-Amshawee S, Yunus MYB, Lynam JGet al. Roughness and wettability of biofilm carriers: a systematic review. Environ Technol Inno. 2021;21:101233. [Google Scholar]
- Baioumy HM, Ismael IS. Composition, origin and industrial suitability of the Aswan ball clays, Egypt. Appl Clay Sci. 2014;102:202–12. [Google Scholar]
- Batista BD, Singh BK. Realities and hopes in the application of microbial tools in agriculture. Microb Biotechnol. 2021;14:1258–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello MGD, Knight R, Gilbert JAet al. Preserving microbial diversity. Science. 2018;362:33–4. [DOI] [PubMed] [Google Scholar]
- Castrillo G, Teixeira PJPL, Paredes SHet al. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017;543:513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CM, Costa MS, Sanchez LMet al. Coupling MALDI-TOF mass spectrometry protein and specialized metabolite analyses to rapidly discriminate bacterial function. Proc Natl Acad Sci. 2018;115:4981–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JAet al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant S, Samad A, Faist Het al. A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J Adv Res. 2019;19:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. [DOI] [PubMed] [Google Scholar]
- Hegazi N, Hamza MA, Osman Aet al. Modified combined carbon N-deficient medium for isolation, enumeration and biomass production of diazotrophs. In: Nitrogen Fixation with Non-legumes. Berlin: Springer, 1998, 247–53. [Google Scholar]
- Huang Q, Wu H, Cai Pet al. Atomic force microscopy measurements of bacterial adhesion and biofilm formation onto clay-sized particles. Sci Rep. 2015;5:16857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M. Soil Chemical Analysis. Vol. 498. Englewood Cliffs: Prentice Hall Inc, 1958, 183–204. [Google Scholar]
- Joseph S, Curtis MA. Microbial transitions from health to disease. Periodontol 2000. 2021;86:201–9. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma Ket al. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J, Wang B, Yoshikuni Y. Microbiome engineering: synthetic biology of plant-associated microbiomes in sustainable agriculture. Trends Biotechnol. 2021;39:244–61. [DOI] [PubMed] [Google Scholar]
- Kolari M, Schmidt U, Kuismanen Eet al. Firm but slippery attachment of Deinococcus geothermalis. J Bacteriol. 2002;184:2473–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumnow AA, Sorokulova IB, Olsen Eet al. Preservation of bacteria in natural polymers. J Microbiol Methods. 2009;78:189–94. [DOI] [PubMed] [Google Scholar]
- Kwak MJ, Kong HG, Choi Ket al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol. 2018;36:1100–09.. DOI: 10.1038/nbt.4232. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights Det al. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. Phyloseq: an r package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermillod-Blondin F, Voisin J, Marjolet Let al. Clay beads as artificial trapping matrices for monitoring bacterial distribution among urban stormwater infiltration systems and their connected aquifers. Environ Monit Assess. 2019;191:58. [DOI] [PubMed] [Google Scholar]
- Nemr RA, Khalil M, Sarhan MSet al. “In situ similis” culturing of plant microbiota: a novel simulated environmental method based on plant leaf blades as nutritional pads. Front Microbiol. 2020;11:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Egidi E, Liu Het al. New frontiers in agriculture productivity: optimised microbial inoculants and in situ microbiome engineering. Biotechnol Adv. 2019;37:107371. [DOI] [PubMed] [Google Scholar]
- Reasoner DJ, Geldreich EE. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rembart L, Betina L. On the tracks of Aswan pink clay. New studies on the local clay deposits in the region of Aswan/Upper Egypt. Interdisc Archaeol. 2021;XII:37–44. [Google Scholar]
- Richards L. Physical condition of water in soil. In: Methods of Soil Analysis: Part 1 Physical and Mineralogical Properties, Including Statistics of Measurement and Sampling. Vol. 9. Madison: American Society of Agronomy, 1965, 128–52. [Google Scholar]
- Rodziewicz M. Field notes from Elephantine on the early Aswan pink clay pottery. In: Cahiers De La Céramique Égyptienne, Ateliers De Potiers Et Productions Céramiques En Égypte. Vol. 3. Cairo: Institut Français d'archéologie Orientale, 1992, 103–7. [Google Scholar]
- Rognes T, Flouri T, Nichols Bet al. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sande MG, Caykara T, Silva CJet al. New solutions to capture and enrich bacteria from complex samples. Med Microbiol Immunol. 2020;209:335–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarhan MS, Mourad EF, Hamza MAet al. Plant powder teabags: a novel and practical approach to resolve culturability and diversity of rhizobacteria. Physiol Plant. 2016;157:403–13. [DOI] [PubMed] [Google Scholar]
- Sessitsch A, Pfaffenbichler N, Mitter B. Microbiome applications from lab to field: facing complexity. Trends Plant Sci. 2019;24:194–8. [DOI] [PubMed] [Google Scholar]
- Shoemaker WR, Jones SE, Muscarella MEet al. Microbial population dynamics and evolutionary outcomes under extreme energy limitation. Proc Natl Acad Sci. 2021;118:e2101691118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman. Some textural patterns and their hearing on the origin of the granitic rocks of the Aswan region, south Egypt. J Univ Kuwait. 1985;12:299–308. [Google Scholar]
- Somasegaran P, Hoben HJ. Quantifying the growth of rhizobia. In: Handbook for Rhizobia. Berlin: Springer, 1994, 47–57. [Google Scholar]
- Taoka Y, Sakai K, Kinoshita Het al. Evaluation of rate of adhesion of Lactobacillus namurensis strain GYP-74 to porous fine ceramics. Processes. 2021;9:658. [Google Scholar]
- Toju H, Peay KG, Yamamichi Met al. Core microbiomes for sustainable agroecosystems. Nat Plants. 2018;4:247–57. [DOI] [PubMed] [Google Scholar]
- Tosi M, Mitter EK, Gaiero Jet al. It takes three to tango: the importance of microbes, host plant, and soil management to elucidate manipulation strategies for the plant microbiome. Can J Microbiol. 2020;66:413–33. [DOI] [PubMed] [Google Scholar]
- Trivedi P, Leach JE, Tringe SGet al. Plant-microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18:607–21. [DOI] [PubMed] [Google Scholar]
- Valliere JM, Wong WS, Nevill PGet al. Preparing for the worst: utilizing stress-tolerant soil microbial communities to aid ecological restoration in the anthropocene. Ecol Sol Evid. 2020;1:e12027. [Google Scholar]
- Voisin J, Cournoyer B, Mermillod-Blondin F. Assessment of artificial substrates for evaluating groundwater microbial quality. Ecol Indic. 2016;71:577–86. [Google Scholar]
- West AG, Waite DW, Deines Pet al. The microbiome in threatened species conservation. Biol Conserv. 2019;229:85–98. [Google Scholar]
- Wu HY, Chen WL, Rong XMet al. Adhesion of Pseudomonas putida onto kaolinite at different growth phases. Chem Geol. 2014;390:1–8. [Google Scholar]
- Yakub I, Soboyejo WO. Adhesion of E-coli to silver- or copper-coated porous clay ceramic surfaces. J Appl Phys. 2012;111:124324. [Google Scholar]
- Youssef HH, Fayez M, Monib Met al. Gluconacetobacter diazotrophicus: a natural endophytic diazotroph of nile delta sugarcane capable of establishing an endophytic association with wheat. Biol Fertil Soils. 2004;39:391–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data were deposited into the NCBI Sequence Read Archive (SRA) under BioProject accession PRJNA816545.