Abstract
Enzymatic activities that could be involved in methanethiol generation in five cheese-ripening bacteria were assayed, and the major sulfur compounds produced were identified. l-Methionine and α-keto-γ-methyl-thio-butyric acid demethiolating activities were detected in whole cells and cell extracts (CFEs) of all the bacteria tested. No l-methionine deaminase activity could be detected in any of the ripening bacteria and l-methionine aminotransferase was detected in CFEs of Brevibacterium linens, Micrococcus luteus, and Corynebacterium glutamicum. The results suggest that several pathways for l-methionine catabolism probably coexist in these ripening bacteria.
l-Methionine degradation has been discussed recently (15) in regard to quantitatively minor volatile sulfur compounds (VSC) that are of major importance when they play a role in flavoring. A starting point of these studies is the low olfactory threshold of VSC (15). They are present in numerous cheeses, as is evident from the analysis of cheddar, Limburger, Camembert, and blue cheeses (1, 6, 12, 13), and make a significant contribution to the distinctive aroma of these cheeses. Most of them arise from the degradation of the sulfur-carbon bound of l-methionine to form methanethiol (MTL), which gives rise to a variety of compounds, including dimethyldisulfide (DMDS) and dimethyltrisulfide (DMTS) (2, 5), 2,4-dithiapentane (9), and S-methylthioesters (6). This is why the biosynthesis of MTL has been investigated in lactic acid bacteria such as lactococci and lactobacilli, as well as in the ripening bacterium Brevibacterium linens (7, 10).
The production of MTL is generally believed to involve the one-step degradation of l-methionine by the versatile enzyme l-methionine γ-lyase (EC 4.4.1.11) (14). This is a pyridoxal 5′-phosphate-dependent enzyme catalyzing the α,γ elimination of l-methionine to form α-ketobutyrate, MTL, and ammonia. It has been found in bacteria, including Pseudomonas putida, an Aeromonas sp., and Clostridium sporogenes (14). To date, B. linens is the only ripening bacterium for which a demethiolating activity has been shown (10). An l-methionine γ-lyase in B. linens has recently been purified and characterized (8). MTL can also be generated from l-methionine in a two-step degradation pathway initiated by an aminotransferase, also called transaminase. This enzyme requires the presence of an amino acceptor (e.g., α-ketoglutarate), yielding α-keto-γ-methyl-thio-butyric acid (KMBA) that is then transformed to MTL by an as-yet-unknown mechanism. This two-step sequence was recently demonstrated in lactococci (11) by 13C nuclear magnetic resonance (NMR) using [13C]methionine. An aminotransferase was recently identified in the lactic acid bacterium Lactococcus lactis (16). It is a pyridoxal 5′-phosphate-dependent enzyme that can catalyze the transamination of l-methionine to KMBA. To date, the transamination of l-methionine has never been described for cheese-ripening bacteria. Another two-step mechanism for the conversion of l-methionine to MTL is the oxidative deamination of l-methionine to KMBA and ammonia. KMBA in turn is converted to MTL. The oxidative deamination of sulfur amino acids, including l-methionine, by an l-amino acid oxidase from Proteus rettgeri has been demonstrated (4).
The primary objective of this work was to elucidate the enzymatic pathways of l-methionine degradation to MTL in five bacteria of technological importance in the ripening process (2). The capacities of these microorganisms to produce sulfur compounds were determined, and the metabolic pathways used are discussed.
Four Actinomycetales bacteria, i.e., Corynebacterium glutamicum D13, Arthrobacter sp. strain 72, B. linens ATCC 9175, Micrococcus luteus 790, and Staphylococcus equorum strain 1265, were used. Strains were stored in 5% glycerol–nonfat dry milk at −80°C. The preculture medium (TSYE) was composed of tryptone peptone (Difco, Detroit, Mich.) (22.7 g/liter), papaic digest of soybean meal (Biokar Diagnostics, Beauvais, France) (4 g/liter), yeast extract (Labosi, Oulchy-le-Château, France) (6 g/liter), glucose (3.33 g/liter), K2HPO4 (3.33 g/liter), and NaCl (6.67 g/liter) (pH 7.5). Five-hundred-milliliter conical flasks containing 100 ml of medium were inoculated with 1 ml of thawed cells. With the exception of B. linens, for which the growth temperature was 25°C, all bacteria were grown at 30°C with mixing (200 rpm, 5-cm diameter stroke) for 3 days. Cultures were carried out in TSYE medium supplemented with 1 g of l-methionine/liter inoculated (1%) with the preculture cells and grown for various time periods. Two milliliters of cells was harvested by centrifugation (20,000 × g, 5 min, 4°C) and washed twice with 1 ml of 50 mM Tris-HCl (pH 8) plus 1 mM EDTA (an inhibitor for cation-dependent proteases). Cells were suspended in 1 ml of this buffer, and the enzyme assays were carried out on the suspension. For cell extracts (CFE) preparation, 90 ml of culture medium was harvested by centrifugation (12,000 × g, 10 min, 4°C). The cell pellet was washed twice with Tris-HCl-EDTA buffer. Two hundred to 400 mg of cells was suspended in 1 ml of the Tris-HCl-EDTA buffer and lysed by mixing with 0.6 g of glass beads (diameter, 100 μm, PolyLabo, Strasbourg, France) using a FP120 FastPrep cell disruptor (Savant Instruments Inc., Holbrook, N.Y.). Three 10-s mixing sequences (speed, 6.5 m/s) were successively applied. Samples were cooled in ice for 5 min between each mixing sequence. After centrifugation (20,000 × g, 5 min, 4°C), the supernatant (CFE) was collected and enzymatic activities were determined. Protein content was determined by the method of Bradford (3) using bovine serum albumin as a standard.
The demethiolating activity of each microorganism was determined on whole cells or CFE as previously described (10). l-Methionine or KMBA was used as the substrate. Controls without cells or CFE were included. Specific demethiolating activity was expressed as nanomoles of MTL · gram of protein−1 · second−1. Total demethiolating activity was expressed as nanomoles of MTL · liter of culture−1 · second−1. l-Methionine aminotransferase activity was determined by measuring the formation of glutamate as previously described (16), using l-methionine as the substrate. Activity was expressed as nanomoles of glutamate formed from α-ketoglutarate · gram of protein−1 · second−1. l-Methionine deaminase was determined by measuring the release of ammonia from l-methionine at 30°C. Ammonia was assayed by reaction with the Nessler reagent (Prolabo, Paris, France). Absorbance was read at 430 nm at room temperature, using ammonium chloride as a standard. Activity was expressed in nanomoles of ammonia formed · gram of protein−1 · second−1. When activities were measured in CFE, it was added to the reaction mixture to give a final protein concentration of 1 to 1.2 mg/ml.
VSC were analyzed with a headspace analyzer (HP 7695A purge and trap concentrator, Hewlett-Packard, Avondale, Pa.) (1). The identity and concentration of sulfur compounds were determined using calibrated standards of the pure products.
In all experiments, the measurements were carried out in triplicate and repeated at least once. The reported values are means ± standard deviations.
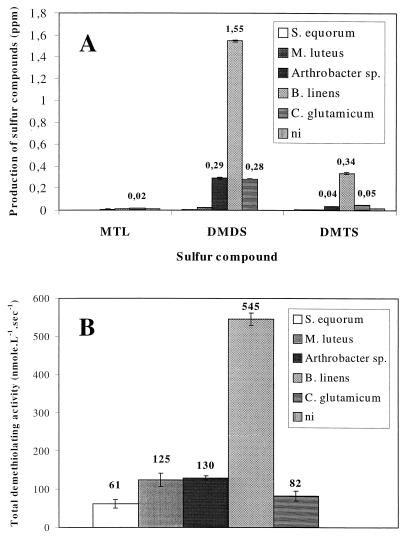
All of the strains produced MTL from l-methionine, while heat-treated (95°C, 20 min) cells could not, demonstrating that l-methionine was enzymatically converted to MTL by the bacteria but not chemically degraded. The production of VSC by l-methionine-supplemented cultures was investigated together with total demethiolating activity (Fig. 1). MTL production by C. glutamicum was highest, but levels remained low (20 ppb), probably because MTL is rapidly auto-oxidized to DMDS and DMTS (Fig. 1A). B. linens produced the highest levels of DMDS (1.55 ppm) and DMTS (0.34 ppm), while producing 10 ppb of MTL (Fig. 1A), correlating with the fact that B. linens also exhibited maximum total demethiolating activity (545 nmol of MTL · liter−1 · s−1) (Fig. 1B). In comparison, the Arthrobacter sp. and C. glutamicum produced 0.28 to 0.29 ppm of DMDS and 40 to 50 ppb of DMTS. Production of these VSC by these two microorganisms was accompanied by demethiolating activity. In contrast to the Arthrobacter sp. and C. glutamicum, although S. equorum and M. luteus possessed demethiolating activity, these microorganisms produced only trace amounts of VSC.
FIG. 1.
Production of sulfur compounds (A) and total demethiolating activity (B) in bacterial cultures. Cells were harvested and volatile sulfur compounds were analyzed after 16 h (S. equorum 1265), 40 h (M. luteus 790, B. linens ATCC 9175), and 64 h (Arthrobacter sp. strain 72 and C. glutamicum D13) of growth in a medium supplemented with l-methionine. Demethiolating activities were determined using l-methionine as the substrate. ni, not inoculated.
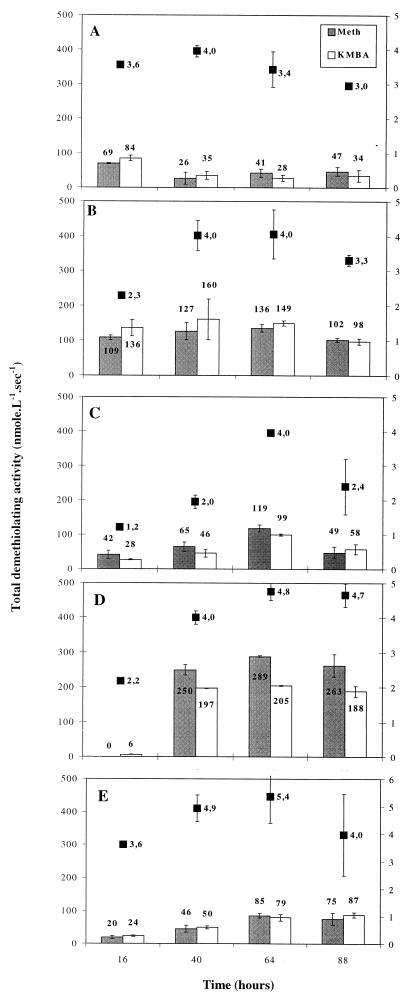
The time courses of demethiolating activities assayed with l-methionine or KMBA as substrates, and cell growth estimated by biomass dry weight, were monitored for every strain (Fig. 2). All bacteria formed MTL, regardless of the substrate. Among the five strains tested, B. linens had the highest demethiolating activities, reaching 289 nmol of MTL · liter−1 · s−1 with l-methionine and 205 nmol of MTL · liter−1 · s−1 with KMBA. With the exception of S. equorum, maximum dry weight (4 to 5.4 g · liter−1) coincided with maximum total demethiolating activity (Fig. 2B to 2E).
FIG. 2.
Time course of total demethiolating activity and dry weight (■) of whole bacterial cells grown in a medium supplemented with l-methionine. Demethiolating activities were measured using l-methionine ( ) or KMBA (□) as substrate. (A) S. equorum 1265; (B) M. luteus 790; (C) Arthrobacter sp. strain 72; (D) B. linens ATCC 9175; (E) C. glutamicum D13.
) or KMBA (□) as substrate. (A) S. equorum 1265; (B) M. luteus 790; (C) Arthrobacter sp. strain 72; (D) B. linens ATCC 9175; (E) C. glutamicum D13.
MTL production (i) from l-methionine or KMBA and (ii) by whole cells or CFEs of the five bacterial strains were monitored for 1 h. Whole cells could produce MTL from l-methionine or KMBA. B. linens was the most efficient for both precursors, with activities of 66 and 52 nmol of MTL · g of biomass−1 · s−1, respectively. With l-methionine, the demethiolating activities of M. luteus and the Arthrobacter sp. were 50% of that of B. linens cells, while the activities of S. equorum and C. glutamicum were 35 and 27%, respectively. With KMBA, the demethiolating activities of S. equorum, M. luteus, and C. glutamicum were comparable to those obtained when l-methionine was the substrate. B. linens and the Arthrobacter sp. exhibited substantially lower demethiolating capacities with KMBA than with l-methionine. When CFEs were used, degradation efficiencies for both substrates varied considerably with the strain. l-Methionine was efficiently converted to MTL by CFEs of B. linens (102 nmol of MTL · g of protein−1 · s−1), the Arthrobacter sp. (35 nmol of MTL · g of protein−1 · s−1), and C. glutamicum (27 nmol of MTL · g of protein−1 · s−1), while it was poorly converted (6 nmol of MTL · g of protein−1 · s−1) by S. equorum and M. luteus CFEs. C. glutamicum CFE could convert KMBA to MTL as efficiently as l-methionine (26 nmol of MTL · g of protein−1 · s−1). CFEs from the other microorganisms converted KMBA to MTL to a slight extent, since KMBA demethiolating activity was consistently lower than 7 nmol of MTL · g of protein−1 · s−1.
In light of the above results, activities possibly involved in the initial degradation step of l-methionine, i.e., aminotransferase and deaminase, were sought. l-Methionine deaminase could not be detected in any of the microorganisms in CFEs. l-Methionine aminotransferase activity was detected in M. luteus (77 nmol · g of protein−1 · s−1), C. glutamicum (108 nmol · g of protein−1 · s−1), and B. linens (193 nmol · g of protein−1 · s−1). l-Methionine aminotransferase activity was undetectable in CFEs of the Arthrobacter sp. and S. equorum.
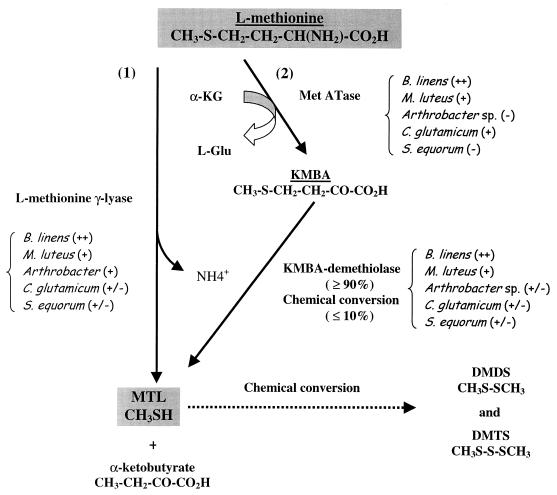
Our results show that not only demethiolating activities but also l-methionine aminotransferase are involved in l-methionine degradation in the cheese-ripening bacteria tested. It was shown that MTL can be produced from either l-methionine or KMBA by whole cells and that maximum demethiolating capacities coincided with maximum biomass. l-Methionine demethiolating activity has already been demonstrated in B. linens, and the enzyme l-methionine γ-lyase was subsequently purified and characterized (8). Although this enzyme can produce MTL from l-methionine, it cannot degrade KMBA to MTL (8). Our results show that whole B. linens ATCC 9175 cells can produce MTL not only from l-methionine, most probably via an l-methionine γ-lyase, but also via a KMBA demethiolating activity which was first demonstrated in B. linens. As previously demonstrated in L. lactis S3 (11), the degradation of l-methionine to MTL follows different patterns in B. linens ATCC 9175 and the Arthrobacter sp., depending on whether it is in CFEs or whole cells. An l-methionine aminotransferase activity was first reported in B. linens ATCC 9175, while it was not detected in B. linens BL2 (7). On the other hand, high l-methionine demethiolating activities induced by the addition of l-methionine were detected in CFEs of strain BL2. l-methionine aminotransferase activity was also first detected in M. luteus and C. glutamicum. l-Methionine deaminase activity was not detected in any of the bacteria. Our results show that demethiolating activities and l-methionine aminotransferase activity can coexist not only in B. linens ATCC 9175 but also in M. luteus and C. glutamicum. No more than 10% of KMBA could be spontaneously converted to MTL (data not shown). Although suggested for lactococci (11), the involvement of a KMBA demethiolating activity in the l-methionine→KMBA→MTL biodegradation sequence was first demonstrated in cheese-ripening bacteria. Two pathways for the degradation of l-methionine to MTL are thus proposed (Fig. 3): (i) a one-step pathway involving an l-methionine γ-lyase and (ii) a two-step pathway initiated by an l-methionine aminotransferase, with the transamination product (KMBA) being mainly (≥90%) converted enzymatically to MTL by a KMBA demethiolase. An attempt was made to estimate the extent of each bioconversion step in each strain (Fig. 3).
FIG. 3.
Proposed pathways of degradation of l-methionine to MTL in cheese-ripening bacteria. For each strain, an attempt to determine the relative importance of each bioconversion step was also carried out (++, very active; +, active; +/−, weakly active; −, inactive).
It has been shown that demethiolating activities are a prerequisite for the synthesis of VSC but are not systematically sufficient. This is especially obvious in S. equorum and M. luteus, which possess consistent demethiolating activities but produce minute quantities of sulfur compounds, even when the culture medium is supplemented with l-methionine. The degradation of l-methionine could therefore follow various pathways, the extent of each being strongly influenced by factors such as lysis of the microorganisms, transport, and accessibility of the substrates. These are crucial parameters when control of traditional fermented foods, such as in sausages and ripened cheeses, is considered.
Acknowledgments
We thank Mireille Yvon (INRA de Jouy, UBSP) and Carmen Lapadatescu (INRA de Grignon, LGMPA) for assaying methionine aminotransferase and methionine deaminase activities.
REFERENCES
- 1.Berger C, Khan J A, Molimard P, Martin N, Spinnler H E. The production of sulfur flavors by 10 strains of Geotrichum candidum. Appl Environ Microbiol. 1999;65:5510–5514. doi: 10.1128/aem.65.12.5510-5514.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloes-Breton S, Bergère J L. Production de composés soufrés volatils par des Micrococcaceae et des bactéries corynéformes d'origine fromagère. Lait. 1997;77:543–559. [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Chen S S, Hudspeth Walgate J, Duerre J A. Oxidative deamination of sulfur amino acids by bacterial and snake venom l-amino acid oxidase. Arch Biochem Biophys. 1971;146:54–63. doi: 10.1016/s0003-9861(71)80040-1. [DOI] [PubMed] [Google Scholar]
- 5.Chin H W, Lindsay R C. Ascorbate and transition-metal mediation of methanethiol oxidation to dimethyl disulfide and dimethyl trisulfide. Food Chem. 1994;49:387–392. [Google Scholar]
- 6.Cuer A, Dauphin G, Kergomard A, Roger S, Dumont J P, Adda J. Flavour properties of some sulfur compounds isolated from cheeses. Lebensm Wiss Technol. 1979;12:258–261. [Google Scholar]
- 7.Dias B, Weimer B. Conversion of methionine to thiols by lactococci, lactobacilli, and brevibacteria. Appl Environ Microbiol. 1998;64:3320–3326. doi: 10.1128/aem.64.9.3320-3326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dias B, Weimer B. Purification and characterization of l-methionine γ-lyase from Brevibacterium linens BL2. Appl Environ Microbiol. 1998;64:3327–3331. doi: 10.1128/aem.64.9.3327-3331.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumont J P, Roger S, Adda J. L'arôme du camembert: autres composés mineurs mis en évidence. Lait. 1976;56:595–599. [Google Scholar]
- 10.Ferchichi M, Hemme D, Nardi M, Pamboukdjian N. Production of methanethiol from methionine by Brevibacterium linens CNRZ 918. J Gen Microbiol. 1985;131:715–723. doi: 10.1099/00221287-131-4-715. [DOI] [PubMed] [Google Scholar]
- 11.Gao S, Mooberry E S, Steele J L. Use of 13C nuclear magnetic resonance and gas chromatography to examine methionine catabolism by lactococci. Appl Environ Microbiol. 1998;64:4670–4675. doi: 10.1128/aem.64.12.4670-4675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grill H, Patton S, Cone J F. Aroma significance of sulfur compounds in surface-ripened cheese. J Dairy Sci. 1966;49:409–412. [Google Scholar]
- 13.Molimard P, Spinnler H E. Compounds involved in the flavour of surface mold-ripened cheeses: origins and properties. J Dairy Sci. 1996;79:169–184. [Google Scholar]
- 14.Tanaka H, Esaki N, Soda K. A versatile bacterial enzyme: l-methionine γ-lyase. Enzyme Microb Technol. 1985;7:530–537. [Google Scholar]
- 15.Weimer B, Seefeldt K, Dias B. Sulfur metabolism in bacteria associated with cheese. Antonie Leeuwenhock. 1999;76:247–261. [PubMed] [Google Scholar]
- 16.Yvon M, Thirouin S, Rijnen L, Fromentier D, Gripon J C. An aminotransferase from Lactococcus lactis initiates conversion of amino acids to cheese flavor compounds. Appl Environ Microbiol. 1997;63:414–419. doi: 10.1128/aem.63.2.414-419.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]