Abstract
COVID-19 is a highly transmissible disease caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), affects 226 countries and continents, and has resulted in >6.2 million deaths worldwide. Despite the efforts of all scientific institutions worldwide to identify potential therapeutics, no specific drug has been approved by the FDA to treat the COVID-19 patient. SARS-CoV-2 variants of concerns make the potential of publicly known therapeutics to respond to and detect disease onset highly improbable. The quest for universal therapeutics pointed to the ability of RNA-based molecules to shield and detect the adverse effects of the COVID-19 illness. One such candidate, miRNA (microRNA), works on regulating the differential expression of the target gene post-transcriptionally. The prime focus of this review is to report the critical miRNA molecule and their regular expression in patients with COVID-19 infection and associated comorbidities. Viral and host miRNAs control the etiology of COVID-19 infection throughout the life cycle and host inflammatory response, where host miRNAs are identified as a double-edged showing as a proviral and antiviral response. The review also covered the role of viral miRNAs in mediating host cell signaling expression during disease pathology. Studying molecular interactions between the host and the SARS-CoV-2 virus during COVID-19 pathogenesis offers the chance to use miRNA-based therapeutics to reduce the severity of the illness. By utilizing an appropriate delivery vehicle, these small non-coding RNA could be envisioned as a promising biomarker in designing a practical RNAi-based treatment approach of clinical significance.
Keywords: MiRNA, SARS coronavirus 2, Biomarker, Pathogen-host interaction, RNA silencing
Graphical abstract
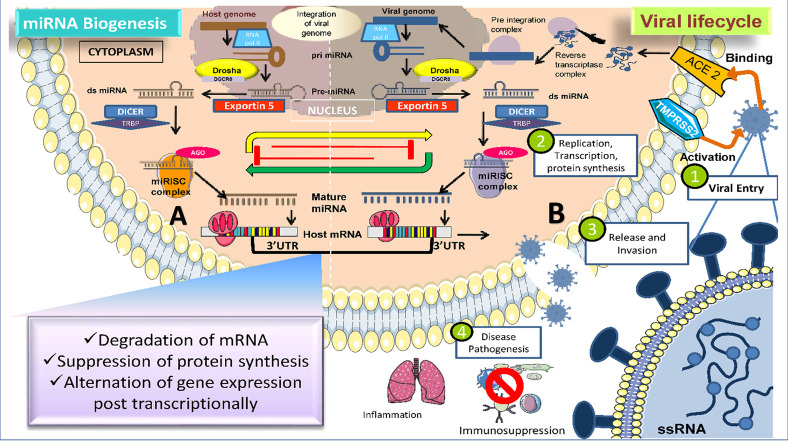
Schematic mode of the interplay between human miRNA and SARS CoV-2 viral miRNA in COVID-19 infected individuals. (A) Conventional mechanism of eukaryotic host miRNA biogenesis and regulation of host gene expression. (B) The life cycle of SARS-CoV-2 virus with possible means of SARS-CoV-2 genome integration in the host cell and contribution of SARS-CoV-2 virus miRNAs to the manifestation of viral infection 1) Virus attachment to host cell receptors (ACE-2) via its spike glycoprotein to penetrate the host cell. 2) Viral replication and host miRNA response alternation, as well as the maintenance of miRNA targeting sites in the viral genome 3) Synthesis and discharge of new viruses to invade unaffected host tissues. 4) Ultimately, triggering the immunosuppression followed by hyperinflammatory response in the host body.
1. Introduction
Recently the world went through a serious outburst of the deadly SARS Coronavirus 2 (severe acute respiratory syndrome coronavirus 2). It was started in the December of 2019 when there was a cluster of pneumonia cases in the city of Wuhan in China. Corona Viruses are the genera of single-stranded RNA viruses circulating within a population ranging from animals to humans for a very long time. These viruses are grouped under the subfamily Coronavirinae in the Coronaviridae family. This subfamily consists of four main genera of coronaviruses- Alpha, Beta, Gamma, and Delta (2020).
To date, we know seven coronaviruses viz 229E(Alpha), NL63(Alpha), OC43(Beta), HKU1(Beta), along with MERS-CoV(Beta coronavirus causing Middle East Respiratory Syndrome), SARS-CoV(Beta coronavirus causing severe acute respiratory syndrome) and the latest SARS-CoV-2(novel coronavirus causing COVID-19) which have a history of infecting humans [1]. Undoubtedly it could be quoted as the primary cause of human mortality. The symptoms of the novel Coronavirus could range from fever and cough to severe pneumonia with an incubation period of 2 weeks. Despite being equipped with advanced technology, it's a big challenge to keep pace with the quick spread of infectious diseases. The cause of this rapid reach could be quite diverse; majorly varied geographical zones and globalization have contributed to the emergence of new variants as potential sources of infection. In order to identify a potential therapy against this global situation, one approach could be finding the likeliness of SARS CoV-2 with other viral infections under the same family of viruses [2]. The currently hyped vaccine technology has proven successful in tackling the previous outbreaks. In 2020, vaccines for SARS CoV-2 had shattered previous records going from development to its approval in a matter of months, but still, it might not be able to cope with the newly emerging SARS-CoV-2 mutants and the durability of immunity post-vaccination [3]. That being the case, few novel diagnostic and therapeutic strategies targeting these RNA viruses is the need for an hour.
MicroRNA or miRNA is a 19–24 nucleotide long, evolutionary conserved endogenic RNA categorized under the family of small non-coding RNA. There is a prospective role of miRNA in the SARS-CoV2-Host dialogue [4]. It modulates its role by targeting complementary mRNA and silencing its function by either inhibiting mRNA translation or degradation of mRNA [5].
There are several ways by which SARS-CoV-2 infection could be hampered: (1) Inhibition of Viral Replication, (2) Blockage of cellular receptors, and (3) Hindering the function of viral proteins. The miRNA-based therapy is a novel approach and could be a possible diagnostic, prognostic, and therapeutic target that could be incorporated in addition to the conventional COVID-19 treatment regime. The miRNA can inhibit the viral replication by attaching itself to the 3′UTR region of the viral genome or the cellular targeting receptor or obstructing the structural and non-structural proteins of SARS-CoV-2 without perturbing the expression of the human genome [6]. This review encompasses the role of miRNA during different stages of the interaction of human and viral miRNAs during the course of SARS-CoV-2 infection. Considering the vital involvement of miRNA as the spoke in the wheel of pathogens, novel strategies inculcating the use of miRNA and the use of modern technological advancements, their delivery, associated shortcomings, and insight can be incorporated into future research for the management of COVID-19 pandemic.
2. Epidemiological update on COVID-19
As of 27th March 2022, the World Health Organization provides a comprehensive epidemiological state of COVID-19 prevailing around the globe. It suggests a sharp declining trend in the overall cases by 14 % compared to the prior week. In addition to that, the new death toll strikes the previous record by 43 %. It is worth mentioning that taking the six regions of WHO under consideration; all the regions have reached the landmark of reducing the number of new cases. Despite this fact, countries such as Chile, the USA, India, the Russian Federation, and the Republic of Korea show the maximum number of new deaths, which is alarming. The key observation from this report includes consistent fluctuation in COVID-19 tests leading to perform less number of tests and hence a reduction in the number of new cases. The represented data thus loses its accuracy and reliability, is less robust, and insufficient surveillance, thereby limiting the assessment of the spread of the COVID-19 virus and the Variants of Concern (VOC). This will clearly restrict the routine treatment and precautionary measures that may account for future COVID-19 waves. Hence, it is crucial to identify a universal vaccine tailored to appropriate social practices [7].
An epidemiological update on the Omicron-Delta recombinant variant (XD Pango lineage) has triumphed in recent times due to the high death rates. Further evidence is still needed from the WHO's end. A massive study had been performed on the trends of SARS CoV-2 on the basis of gender parity, suggesting a higher prevalence of morbidity and mortality in male than female. On a similar line of work, Giada Pontecorvi et al. mentioned the possible role of miRNAs in exhibiting the COVID-19 gender parity. Their objective was to extract the miRNAs which are mainly revolving around the prime interacting components; ACE-2 receptor, TMPRSS2, ADAM 17, and Furin expand the scope of post-transcriptional miRNA regulation in the COVID-19 pathogenesis [8].
3. COVID-19: the enemy
3.1. Genomic architecture of SARS-CoV-2 virus
SARS-CoV-2 genome is known to have a single-stranded enveloped positive-sense RNA (+ssRNA) virus. According to the NCBI genome database for the novel SARS CoV-2, it constitutes 13–15 open reading frames (ORF), of which 12 are functional with the GC content of about 38 %, the genomic arrangement of the ORFs is remarkably similar to that of SARS-CoV and MERS-CoV. In addition to that, the genetic makeup also involves 16 nonstructural proteins (nsp), four structural proteins viz. the Spike glycoprotein (S), an envelope protein (E), the membrane protein (M), and nucleocapsid phosphoprotein(N), and six accessory proteins (viz 3a, 6, 7a, 7b, 8, 10) [9]. The family of CoV is known to hold 16 non-structural proteins, which have a role mainly in their RNA synthesis. The CoVs have a unique and unprecedented feature where the CoV nsp14 codes for a special exoribonuclease (3′-5′) known as ExoN [10]. This assist in the persistence of infection, giving them an added advantage based on its stability. The Spike proteins are responsible for constructing spikes that aid in the attachment of the virus to the host receptor present on the surface of the viral coating. The electron microscopy images results suggest that the S proteins are made up of S1 and S2 subunits. S1subunit is involved in the attachment of the virus in the host's range and cellular orientation, while S2 subunits check the membrane fusion of the viral to the host cell membrane [11].The Membrane glycoprotein is a triple-spanning membrane protein that drives the assembly of the SARS-CoV-2 by binding to the nucleocapsid [12]. The Envelope protein is an integral membrane protein highly involved in viral pathogenesis.
The Nucleocapsid glycoprotein stabilizes the viral RNA structure via two different domains. These proteins play a crucial role in the virus entry, fusion, and survival in the host cells; hence, we can be quoted as the potent drug target [13]. Numerous single mutations have been realized in the SARS-CoV-2 genomes since the virus first erupted in 2019, which have induced the emergence of risky variants. Although genetic changes have also been noted from other genomic regions, such as ORFs 7b, 8, 10, and nsp3, several SARS-CoV-2 genes, including S, N, and NSP12 (RdRP), have evidenced substantial mutability. As in ORF10 protein, Hassan et al. found 128 single and 35 co-occurring mutations directly influencing pathogenicity. Additionally, they noted that missense mutations had drastically reduced ORF10's stability [14]. With the incidence of highly frequent mutations in ORF7b and ORF8, demographically relying upon mutations and a cyclical emergence of dominant mutations have been reported in various countries as variants of concern [15]. High viral transmission and immune escape have been linked to genetic alterations R203K and G204R in the N protein and Q27stop in ORF8 [16]. To identify and develop a potential target for the SARS CoV-2, it is crucial to be well versed in the intricate mechanism of viral replication along with the knowledge of host-virus interaction and its regulation inside the host's body.
3.2. Lifecycle and pathogenesis of SARS CoV-2 virus
Intending to discover a novel strategy to combat the fatal SARS-CoV-2 virus, it is crucial to be aware of the molecular pathways governing the pathogenesis of the virus. A comprehensive study performed by Bosch et al. classified the life cycle of SARS-CoV-2 with the host tissues into five stages: Attachment, Viral entry, Viral replication, and Protein synthesis, Maturation, and Release [17]. The SARS-CoV-2 virus employs the virion to invade the host cell to complete its life cycle by the mode of replication within the host cell to increase the copy number of the virus particles. This ultimately leads to cellular infection as well as increased transmissibility. The pathway by which the COVID-19 virus mediates its lifecycle and induces viral pathogenesis requires generous efforts to resolve. Nevertheless, as per the published studies so far, the SARS-CoV-2 replication cycle mainly initiates with-.
Attachment of SARS-CoV-2 viral spike protein to the host cell receptor (ACE-2) where the spike protein facilitates cell tropism by directing the S protein to penetrate specific host tissues consisting of host cell receptors angiotensin-converting enzyme 2 (ACE) for viral entry. Angiotensin-converting enzyme 2 (ACE-2) is a cellular receptor encoded mainly in the lung, heart, kidneys, intestine, and arteries. S glycoprotein undergoes cleavage into the S1 and S2 subunits by the presence of an extracellular protease Transmembrane protease serine2 (TMPRESS2). The S1 subunit of the S protein attaches itself to the ACE-2 receptor while S2 underwent further cleavage with the help of TMPRESS2 for membrane fusion [16]. The viral entry is mediated via any one of the two processes. First, fusion of viral and host cell membrane to allow the viral genome admittance within-host cytosol. Second, endocytosis of the ACE-2 receptor-bound virus enclosed in a vesicle to grant entry into the host cytoplasm and subsequently viral RNA genome released in the host cytoplasm. Following that, the viral genome uncoats to set the viral RNA free. Host cell machinery for the translation is hijacked for viral RNA translation of polyproteins (pp1a and pp1ab) and critical viral proteases. The polyproteins are further processed to 16 nonstructural proteins with the assistance of 3 CLpro and PLpro forms complex. Formation of Clpro and PLpro complex with RNA-dependent RNA polymerase to synthesize a negative strand of RNA as a template. This serves as a template for positive-stranded RNA viral genome, replication of the complete RNA genome and generation of subgenomic mRNA for the process of translation of viral structural and accessory proteins. This newly formed structural (e.g., S, E, M, and N) and nonstructural accessory protein assemble at the transitional zones to trafficked between ER and Golgi body in a membrane-enclosed vesicle. Both the cellular organelle are found associated with post-transcriptional regulation, protein synthesis, packaging of protein, and transport [17, 18]. Lastly, with the maturation of SARS-CoV-2 virion, they are translocated encased in a vesicle towards the host cell membrane to undergo exocytosis to discharge into the extracellular milieu. This release of the virion doesn't rip the host cell apart hence referred as non-lytic exocytosis. This invasion contributes to the virulence of the protein which is an infective mediator in the community transmission of respiratory droplets. The infected cells undergo apoptosis/necrosis that activates the host's inflammatory response. This can be marked by the assembly of pro-inflammatory cytokines and the production of T helper cells or macrophages [18]. This turns out with a dramatized immune reaction of uncontrolled cytokines and chemokines levels circulating into the blood, thus embarking on a condition known as a cytokine storm. This condition drives the progression of the systemic inflammatory response, which brings about a state of acute respiratory distress syndrome (ARDS) and a series of tissue/organ damage [19]. This makes a critically diseased condition and boosts COVID-19-linked co-morbidities like coagulopathy, thrombosis, inflammation, and multiple organ failure in severely ill COVID-19 patients [20].
4. MiRNA-biogenesis and mechanism of action
An organism's genome comprising DNA/RNA directs the regulation of cellular processes, cell to cell communication, and homeostasis [21]. The regulation of these complex processes is carried out with the help of protein encoded by the nucleic acids. Surprisingly, only 1–2 % of the human RNA is transcribed by DNA and could be categorized under the coding region, while the rest of the lot is derisively known as junk or noncoding RNA. These noncoding RNAs (ncRNA) are emerging as the major players in evolution and are known to mediate chunks of signaling pathways essential for an organism's survival. These ncRNA could be in the form of circular RNA (circRNA), long ncRNA (lncRNA), small nucleolar RNA (snoRNA), or Micro RNA (miRNA) [22].
miRNAs are short noncoding RNAs having an approximate length of 22 nucleotides. Most of the miRNA are synthesized as a result of DNA transcription, which at the outset forms primary miRNAs and is later processed to precursor miRNAs [23]. Most miRNAs target the 3′UTR of the specific mRNA, but in the case of virus miRNA, it has been found to bind at 5′UTR to repress gene expression. Current literature has mentioned that miRNA ferries among the different subcellular localization to direct the transcription and translation pathways [24]. Any strange behavior mediated by these miRNAs is a strong indicator of any host infection. miRNAs are mainly reported in two forms: extracellular or circulating miRNA found in the extracellular fluids, whereas cellular miRNA is present inside the cell [25], [26]. miRNA begins its biogenesis with post – or co-transcriptional modification of RNA polymerase II/III transcripts. Generally, miRNAs are either intragenic, processed mainly from noncoding genes or introns and few from exons of the mRNA transcripts or intergenic transcribed autonomously by their promoter with any host gene involvement. miRNAs also encode a family of clusters having comparable seed regions. Broadly miRNA biosynthesis can take place in one of the two pathways, viz. Canonical or Noncanonical [27]. The Canonical pathway of miRNA biogenesis, also known as the classical pathway, initiates the formation of a >70 nucleotides long pri-miRNA transcript from miRNA genes. After that, the pri-miRNA is subjected to a microprocessor complex consisting of Drosha and DGCR8, which metabolizes the former to a ~65nucleotide length pre-miRNA [28]. This pre miRNA travels from the nucleus to the cytoplasm with the help of Ran-GTP depending on exportin-5 and modifies itself to miRNA duplex ~22 nucleotides. This step is facilitated by another complex of Dicer and RNA binding protein (TRBP/PACT) [29]. One strand of the duplex RNA is degraded while the other remains intact and associates with Argonaute (AGO) to form a miRNA-RISC complex. Contrarily the noncanonical pathway began with the cleavage of small hairpin RNA (shRNA) by the microprocessor complex and exported to the cytoplasm associated with Exportin-5/Ran GTP. From here on, these can progress by Dicer independent or Dicer dependent pathways. In the former pathway, the pre-miRNA is processed in AGO-2 dependent manner [28]. The latter pathway could be observed in the case of Mirtrons and (m7G) 7-methylguanine capped pre-miRNA, which complete their maturation in a Dicer-dependent manner. Mirtrons differ from m7Gcap in nucleocytoplasmic shuttling, where Exportin exports mirtrons-5/RanGTP and m7Gcap by Exportin 1, ultimately leading to the miRISC complex [30]. This new complex complements the 3′ or 5′UTR (completely/partially) of the target mRNA where the RISC enforces a family of GW182 proteins to attach with polyAbinding proteins (PABPs) employing CCR4-NOT deadenylase complex [31]. This complex destabilizes it, resulting in degradation via exoribonuclease. It is worth mentioning that complete complementary interaction induces degradation of target mRNA while partial complementary interaction inhibits translation. Fascinatingly miRNA merely embodies 1 % of the total human gene; however, it could umpire the expression of about 60 % of the protein-encoding gene (Fig. 1 ) [32].
Fig. 1.
Schematic mode of the interplay between human miRNA and SARS CoV-2 viral miRNA in COVID-19 infected individuals.
(A) Conventional mechanism of eukaryotic host miRNA biogenesis and regulation of host gene expression. (B) The life cycle of SARS-CoV-2 virus with possible means of SARS-CoV-2 genome integration in the host cell and contribution of SARS-CoV-2 virus miRNAs to the manifestation of viral infection 1) virus attachment to host cell receptors (ACE-2) via its spike glycoprotein to penetrate the host cell. 2) Viral replication and host miRNA response alternation, as well as the maintenance of miRNA targeting sites in the viral genome 3) synthesis and discharge of new viruses to invade unaffected host tissues. 4) Ultimately, triggering the immunosuppression followed by a hyperinflammatory response in the host body.
5. miRNAs as therapeutic biomarker
The miRNAs have been acknowledged as potential biomarkers in case of a broad range of infections from tuberculosis to HIV to globally encountered current acute respiratory syndrome, SARS-CoV-2. These noncoding RNA is associated with modulation of gene expression and provides a perfect means to evaluate alternation in the miRNA expression, uniquely discriminating a complicated case from an uncomplicated one. These small molecules meet almost all the requisite criteria to take the position of an ideal biomarker; thus, they are highly specific, accessible, marked sensitivity, extended long half-life in sera, rapid, and clinically economical (https://www.fda.gov/drugs). The detection of miRNAs is performed using standard means involving qPCR, northern blotting, microarrays, mass spectroscopy, and Next-generation sequencing (NGS). Recent trends reveal that researchers favor the use of NGS and qPCR more frequently due to the accuracy of these approaches in terms of enhanced sensitivity, specificity, and stability with vast input of total RNA. Initial symptoms prevailing in most viral diseases are often vague viz. pain, headache, fever, sore throat, etc., giving negligible information about the exact cause of infection and its causative agent. A study on Herpes simplex virus-1 miRNAs has shown satisfactory specificity, differentiating an active infection stage with miRH1 from a latent one with miR-H2-6. Thus, it proves that miRNA profiling can give an upper hand during the treatment of patients when the regular diagnostics treatments could not succeed. Oncologists were the first to utilize this approach, and it is only recently that it could be applied for SARS CoV-2 [33], [34].
Furthermore, these miRNAs could be easily detected in the blood, serum, plasma, saliva, cerebrospinal fluid (CSF), urine, and histological biopsies and can escape the blood-brain barrier [35], [36].
Noncoding transcripts data are accumulated in specified computational databases, and many registered miRNAs are on the verge of getting FDA approved. A majority of 90 % of miRNAs occur in association with proteins, while the rest, 10 %, are encapsulated in exosomes and microbubbles, giving protection under adverse extracellular conditions. Recent findings have proposed ample host and virus-specific biomarkers accountable for SARS CoV-2 infection [35]. Respiratory illnesses often share a similar lung morphological aspect but differ in pathophysiological aspects. Grasselli et al. described the pathological conditions of SARS CoV-2 as a severe endothelial damage adjunct with high D-dimer, disordered plasma membrane, and strikingly amplified mortality rate. Examination of COVID-19 patients undoubtedly could permit the early detection of disease severances and subsequent follow-ups [37], [38].
5.1. SARS CoV-2 virus encoded miRNA as potential biomarker
With the encounter of the deadly virus, the host can trigger an extraordinary immune response which can set about differential expression of host and viral miRNAs as elaborated formerly. Viral miRNA mainly functions by modulating different life cycle phases and inducing/inhibiting the expression of host defense mediated pathways. Viral miRNAs encoded by the SARS-CoV-2 genome guard their replication machinery and escape the host's immunity by targeting various host genes, finally attenuating it. Identification of viral miRNA could be accomplished by two main approaches, i.e., in silico analysis and sequencing of small cloned RNA [39]. Recent studies have predicted 3377 human target genes denoting it to be a unique target for 170 SARS-CoV-2 encoded matured miRNAs [40]. Earlier, a study was conducted by Usme-Ciro et al. on miRNA, which predicted that the biogenesis pathway followed by RNA virus encoding miRNA follows the non-canonical miRNA biogenesis pathway. This pathway proceeded in a Dicer- or Drosha-independent manner [41]. Lately, Drosha-dependent miRNA processing in cytoplasmic RNA viruses has also come to light. But in the case of SARS CoV-2, researchers are still not able to explain miRNA biogenesis and hence pose a hot topic of interest for different scientists [42].
Viral miRNAs have been widely focused on in recent years to understand their mechanism of action to develop therapeutics. Mostly viral miRNAs have shown their role in overall pathogenesis by regulating host gene promoters. The findings of an in silico study have shown an analogy between the SARS CoV2 genome and regulatory host miRNA such as miR8066, miR5197, miR3611, miR3934-3p, miR1307-3p, miR3691-3p, miR1468-5p [40]. Few of the viral miRNAs exhibiting their action along different life cycle stages and pathogenesis are compiled below.
The works of Dr. Xingyin Liu et al. elaborated on the role of virus-encoded MR 147-3p in priming TMPRSS2 gene expression in host gastrointestinal cells [40]. This causes a viral entry and could be the possible explanation for COVID-19 mediated gastrointestinal symptoms. TMPRSS2-and endothelial cell surface protein, known to activate viral S protein, attaches to the ACE2 receptor aiding viral evasion. The virus entails the transcriptional and translational apparatus of the host for carrying out its replication, transcription, and protein synthesis. Hence SARS CoV-2 employs viral miRNAs to endorse their needs by protecting self mRNA from host degradation [43]. They do so by repressing the attachment of RNA polymerase II to the host promoter during transcriptional initiation by targeting transcriptional factors viz. (TAF4, TAF5, TAF7L, MAFG, TCF4, TFDP2, TRPS1, BRF1, SOX11) and human mediator complexes like MED1, MED9, MED12L, and MED19 and inhibiting few mRNA deadenylases complexes like CCR4-NOT (CNOT4, CNOT10, and CNOT6L) [44]. The SARS-CoV-2 can invade other tissues by releasing and breaking off from the host cell. The viral MR359-5p upregulates the expression of MYH9 (non-muscle myosin heavy chain 9), which promotes attachment, invasion, migration, and release within the host cell. The architecture of virus particles is subjected to viral uncoating, which is facilitated by the overexpression of MYH9. Studies have shown that MR359-5p also enhances the expression of Integrin β-5 protein which is associated with cytoskeleton-related and adherence functions driving viral surfing, trafficking, and internalization. Additionally, Fu et al. study suggested the presence of viral miRNA miR-nsp3-3p in the host sera, which might be another potential biomarker option [45], [46]. The pathogenesis of the viral infection is emanated by inflammation, arresting of host cell apoptosis, immunosuppression, and other pathological damages [47], [48].
Khan et al. have demonstrated the contribution of viral miRNAs in the modulation of a variety of signaling pathways such as insulin signaling, Wnt, T cell-mediated immunity, autophagy, FGF receptor binding, Type I Interferon signaling pathways, NF-κB, PI3K/Akt, TNF-alpha, ErbB, mTOR, VEGF, TGF-beta and MAPK signaling pathways, and the Notch signaling pathway which affect heart and brain development [40]. SARS CoV-2 employs viral miRNA, which dysregulates host miRNAs crucial for fatty acid, metabolism, insulin resistance, renal and myocardial processes, and peroxisomes proliferator-activated signaling (PPAR) wrap up with COVID-19 infection followed by an added overcomplicated comorbidity [40].
The SARS-CoV-2 viral miRNA exhibits hybridization of vital human proteins like beta and gamma-globin 2 in the hemoglobin subunit, IFN 1, and olfactory receptor proteins. This concludes in oxygen fluctuations in vital organs and dysosomia, as in the case of COVID 19 [49]. Another study on the virus-derived miRNA MR345-5p by Arsian et al. and team described the 5′UTR target of the TGF-Beta receptor 3 (TGFBR3) gene, which has an activating effect that provokes the host's innate and adaptive immune response, generates regulatory T cells, and thereafter encourages Th1 differentiation, and maybe the cause of pathological trauma. Chemokine-mediated cytokine storm occurs due to activation of critical host immune response and chemokine signaling pathway targeted by viral miRNAs (miR-1-5p,miR-2-5p, miR-3-5p, miR-4-5p, miR-5-5p, and miR-6-5p [50].
Pawlica et al. and their team conducted an in-vitro experiment on lung-derived cell lines and nasal swabs, which led them to discover a viral miRNA-like small RNA viz. CoV2-miR-O7a (SARS-CoV-2 miRNA-like ORF7a-derived small RNA). They demonstrated its association with Argonaute proteins, a core component of RNAi gene silencing. They also managed to identify Basic Leucine Zipper ATF-Like Transcription Factor 2 (BATF2) as its putative target, which takes part in interferon signaling [51]. Another in vitro study was done by Meng et al. where they isolated viral miRNA from COVID-19 infected cell lines like Vero E6 and Calu-3. They sorted three significant viral miRNAs (v-miRNA-N-28612, v-miRNA-N-29094, and v-miRNA-N-29443) extracted from the N gene of the viral genome. Further RT-qPCR analysis denoted the suppression of host transcripts viz. ACO1, BCAS1, BNIP3L, CLDN10, DMBX1, and SNCA by virtue of upregulated v-miRNA-N. They also demonstrated a few additional binding targets for viral miRNA like IL-1, NLRP3, and caspase 1 in the host inflammasome [52].
In-silico study of SARS-CoV-2 encoding miRNA targets the Ca2+ signaling pathway, which acts as a key activator that subsequently affects other signaling pathways branching it. A variety of prediction tools are used to compare the resemblance of small noncoding miRNA of the host to viral miRNAs to identify an efficient target to effectively build a human-friendly therapeutic vaccine [39]. One of the computational works of Tie Ying Yu et al. has identified three miRNA precursor sequences with the help of BLAST analysis of novel SARS-CoV-2 and then functionally annotated mature and precursor sequences [53]. Aydemir et al. figured out that 40 SARS-CoV-2 encoded miRNA and their respective targets using computational tools. Those targets were significantly involved in various gene signaling pathways, including TGFB, JAK/STAT (STAT1 and STAT5B), NFKB; tumor-suppressing- AKT1, PTEN, RB1, epigenetic factors like Histone deacetylases and JARIDs, and transcription factors. Liu et al. conducted a computational project aiming to check the role of SARS CoV-2 encoded miRNAs in the lung tissues. Their findings suggested that viral miRNAs were associated with amplifying chemokine signaling pathways by inducing proinflammatory cytokines, mainly CXC chemokine ligand 16 (CXCL16) and arrestin beta [39]. These cytokines were predicted to be the binding targets of MR147-5p. Similarly, they also revealed MR66-3p to target the TNF-alpha transcription enhancer in the spleen, which operates as a chief cytokine during inflammation [54].
The viral genome encoded miRNA also has a permissive role in the immunosuppressive activity. They practice this by dwindling the IFN-I signaling pathways and autophagy. Liu et al. have elaborated on the role of viral encoded MR328-5p in the over-expression of the Retinoid X receptor alpha (RXRα) gene, whose upregulation hampers host immunity by repressing IFN1 machinery. The STAT family also downregulates the IFN response. The COVID virus has also been reported to hijack host miRNAs in the form of proviral factors such as hsa miR-146b and hsa-miR-939, which have a key role in host immunity. The findings of Saini S et al. have given away that the viral miRNA MD241-3P misbalances vascular pulmonary homeostasis by curbing the BMPR2 gene in the bone, which undergoes TGF-β signaling. Therefore, these decrease the antiviral response in the host body and ultimately lead to lung diseases [44], [55].
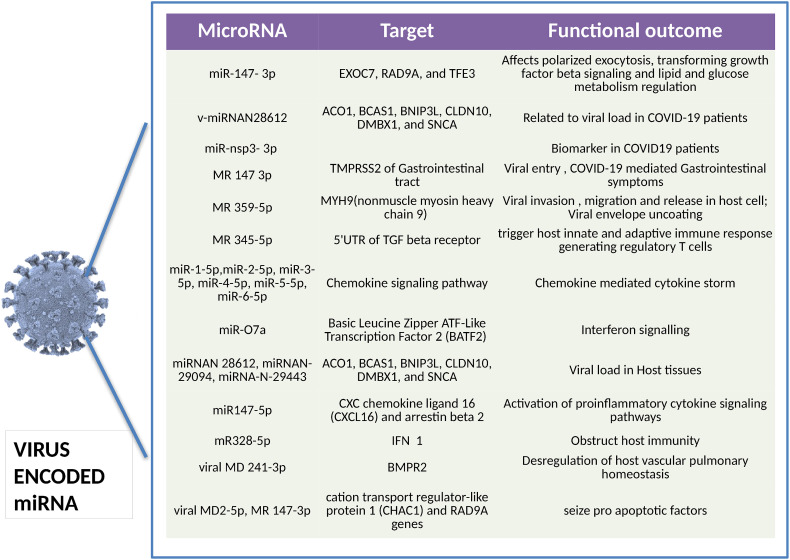
Viral miRNAs like MD2-5p and MR147-3p, which are predicted to stop the activity of pro-apoptotic components such as cation transport regulator-like protein 1 (CHAC1) and RAD9A genes, respectively, impede the host's apoptotic defense mechanism. By interfering with host miR-376b-3p, which is primarily coupled to increase rapamycin expression, SARS-CoV-2 virus also suppresses autophagy [56]. The scientific community has acknowledged these viral miRNAs as a budding space of research that has already taken its pace leading to the breakthrough of novel viral miRNAs. (Fig. 2 ).
Fig. 2.
SARS-CoV-2 encoded miRNAs present in human serum during COVID-19 infection [14], [15], [16], [54], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76].
5.2. Host encoded miRNAs as potential biomarker
The dialogue between the human host and the pathogen has shed light on the underlying host-pathogen noncoding miRNAs. miRNA promotes the regulation of about one-third of the entire human protein encoded gene set [77]. Multiple studies on miRNA have shown its crucial role in the pathogenesis of various human diseases such as cancer, neurological, cardiovascular, and many infectious diseases. The interaction of the host and the pathogen arises under miRNA's function in intercellular communication [78]. Multiple studies have highlighted the role of human miRNAs in fighting viral infections. Since the RNA of single-stranded RNA viruses (ssRNA viruses) and host mRNA are structurally similar, the binding of miRNA is a simple process for the host. According to studies, the miRNA typically associates with a group of proteins to form a RISC (RNA-induced silencing complex), which specifically targets the 3′UTR of target mRNAs and suppresses or destroys the protein synthesis pathway. However, it is discovered that host miRNAs bind to the 5′UTR or gene promoters of viral RNA and then cause translational repression. One miRNA from an infected host cell has been shown to perform a variety of roles, ranging from activating important immune pathways to acting as a proviral factor by interfering with immune pathways. By weakening the host's immune surveillance pathways, the host miRNA can act as a proviral. Host miRNAs are sometimes referred to as bipartite because they promote viral replication, target some of the critical host immune responses to aid viral immune evasion, or persistently alter the number of free miRNAs in a cell [79]. SARS CoV-2 enters the host cell via the ACE-2 receptors expressed on Type 2 pneumocyte cells in particular. Finally, these miRNAs suppress Toll-Like Receptors (TLRs), which are known to generate antiviral responses by stimulating the production of multiple inflammatory cytokines (IL-1, IL-6, and IL-8) as well as endothelial function governing inherent receptors. Centa et al. reported suppressed expression of miR-26a, miR-29b, and miR-34a in tissue samples from COVID-infected patients' lungs. Surprisingly, they discovered a reciprocal correlation between the levels of Interleukins and the miRNAs mentioned above. With the increased expression of IL-4, the level of miR-29b-3p shrinks, and likewise, upregulated IL-6 and ICAM-1 lead to low levels of miR-26a-5p while IL-8 caused an increased level of miR-29b-3p.In the case of miR-34a, its downregulation in CD4+ and CD8+ T cells lifts the level of kappa-light chain enhancer of activated B cells; consequently, the Interleukins show the host miRNA expression during inflammation [80].
As mentioned in the literature of Tang et al., miR-146a, miR-21, and miR-142 in COVID19 affected induce the inflammatory machinery by promoting MAPK and NF-Ƙb signaling [81]. It was identified that restricting miR-21 or miR-142 would promote multiple cascading reactions and activation of the JAK-STAT cell signaling circuit, resulting in proinflammatory status advancements. It is necessary to mention the virus particles' tactics in targeting the host cell mitochondria. Mitochondria, also known as the cell's powerhouse, mediate critical cellular processes like cell cycle, signaling, metabolism, differentiation, immunity, and cell death. For SARS CoV-2 infection, the primary basis of mitochondrial trauma is the production of reactive oxygen species ROS and the stimulation of inflammatory mediators. The virus can modulate this function by directing miRNAs to crash the mitochondrial defense mechanism against ROS; for example, miR-2392 works together with mitochondrial DNA (mtDNA) to efficiently shut the genes encoding for proteins such as methionine-R-sulfoxide reductase, COX 6B1 and COX-10, NADH: Ubiquinone Oxidoreductase Subunit S5 (NDUFS5), CKMT1A (mitochondrial creatine kinase), Mitochondrial Ribosomal Protein L34 (MRPL34), and adenylate kinase 4 (AK4). Besides this, host miRNA (miR-302b and miR-372) targets the mitochondrial antiviral signaling protein (MAVS), which occurs in the outer mitochondrial membrane and is responsible for innate antiviral immunity. Targeting these proteins leads to the silencing of type 1 interferon signaling via inhibition of Interferon regulatory factors 1 and 2 [82]. The most obvious finding that emerged from the analysis of the host-mediated miRNA is that one of the host miRNAs, i.e., miR-2392, is known to promote glycolysis by specifically thrusting hexokinase two pyruvate kinase enzymes which play a key role in sustaining the viral spike protein glycosylation and their replication [82].
Host miRNA plays a distinguished role in the life cycle of SARS CoV2 as well. Prior studies have noted the importance of host miRNAs in blocking the attachment and entrance of the SARS CoV-2 by virtue of its effect of distinct miRNA families on ACE2 and TMPRSS receptor genes. Matarese A et al.'s work on human lung microvascular endothelial cells and human umbilical vein endothelial cells reveal that hsa-miR-98-5p binds the 3′UTR of TMPRSS2 transcript. hsa-miR-125a-5p miRNA with miR-200 family are known to target 3′UTR of ACE2 mRNA in contrast to hsa-let-7e-5p, which binds to the 3′UTR of TMPRSS2 [83]. Moreover, the ACE 2 receptor is also hindered by hsa-miR-23b-5p and hsa miR-769-5p in its 3′UTR [84]. Liu et al.'s manuscript has mentioned another miRNA has-miR-4661-3p which reportedly binds and represses the 3′UTR of the S gene [85]. Plenty of previous research has been conducted keeping the S protein gene at the center, citing a few of them, including- hsa-miR-510-3p, hsa-miR-624-5p, and hsamiR-497-5p target S glycoprotein RNA; hsa-miR-23b and hsa-miR-98 is specifically targeting S protein by cleaving Interferon-beta genes and VP1 genes; hsa-miR-622, hsa-miR-761, miR-A3r, has-miR-15b-5p, miR-A2r, and hsa-miR-196a-5p also target the S protein gene while miR-338-3p, miR-4661-3p, miR-4761-5p, hsa-miR-4464, hsa-miR-1234-3p, hsa-miR-7107-5p, and hsa-miR-885-5p is unique to the RBD of S gene [86]. Tang h et al. carried out a study on the S glycoprotein associated CLEC4M gene, which was investigated to get suppressed by hsa-miR-4462 and hsa-miR-5187-5p. Additionally, hosts encoding hsa-miR-3934-3p are involved in the biosynthesis of extracellular matrix proteins such as Heparan Sulfate ProteoGlycan. In contrast, hsa-miR-8066 is involved in the synthesis of mucin-type O-glycan synthesis, a kind of post-translational modification [87].
Much of earlier research has been carried out to know the targets of host miRNA to restrain viral replication, transcription, and translation. It includes hsa-miR-203b-3p, which targets ORF1ab and ORF3a of influenza; hsa-miR-497-5p, hsa-miR-21-3p, and hsa-miR-195-5p are specific to nonstructural protein synthesis; lung epithelial tissue expressing hsa-miR-497-5p, hsa-miR-21-3p, and hsa-miR-195-5p also inhibit replication by nucleotide deletion in viral ssRNA coding strands [88], [89], [90].
Complimentary miRNAs (cc-miRNA) such as cc-miR1c, cc-miR2c, cc-miR3c, cc-miR4c, cc-miR5c, cc-miR6c, and cc-miR7c bred by host miRNAs are thought to be responsible for post transcriptional silencing of the SARS-CoV-2. Previous research has shown that these miRNAs have a high affinity for the viral genome, either directly, as in 5p-miRNA and miR-5197-3p, or indirectly, as in a few miRNAs such as miR-1273a, miR-1273d, miR-1272, miR-1292-5p, miR-3143, miR-1226-5p, and miR-7161-3p. Few works in literature like Chen et al. reported that miR-1307-3p and miR-3613-5p regulate the TGF signaling beta signaling by binding to the 3′UTR, where the upregulation of the former activates semaphorin signaling. In contrast, the latter's suppression reduces the level of FGF 2 and VCAM1, thus leading to pulmonary pathogenesis [91]. The ORF9 gene fragment encodes N glycoprotein, which is an essential component of the SARS CoV-2 virus. A cocktail of antisense miRNAs binds to the viral 5′UTR, 3′UTR, and ORF9, preventing viral protein synthesis and assembly. Similarly, hsa-miR-126 and hsa-miR-378 inhibit N protein production by preventing translation and cleaving the IFN- gene in that order [89].
In a study that set out to determine the miRNA's engagement in inflammation, apoptosis, and immune response, Khan M-A-A-K et al. found the defensive role of hsa-miR-17-5p, hsa-miR-20b-5p, and hsa-miR-323a-5p host miRNA in the process [87]. Few host miRNAs(hsa-miR-516b-3p, hsa-miR-3529-3p, and hsa-miR-6749-3p) act reversibly by the RELA gene, thus encouraging viral infection. The host also assigns a small number of miRNAs (miR-21, miR-125b-5p, miR-199a, miR-211, miR-138, miR-211, miR-146a, and miR-146) as antagomiRs to reduce the lung epithelium trauma by virtue of the cytokine storm [92].
Owing their biregulatory role, host miRNAs are known to downregulate various signaling pathways associated with host immune suppression and censored proinflammatory cytokines, including IFN-, TGF-, interleukin (IL), IGF1 (insulin-like growth factor 1), tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), and toll-like receptors (TLRs). miRNAs may also be used in defense mechanisms by reducing host cell apoptosis [93]. P53 modulated miRNAs like miR-101, miR-100, miR-99a/b, miR-7, miR-107, let-7, miR-199, miR-200, miR-15/16, miR-223, miR-143/145, and miR-17 function either by binding to mTOR mRNA directly downregulating it or silencing their downstream target molecule [94].
Fulzele et al. conducted in-vitro experiments, and their findings show that the miRNome of this SARS CoV-2 infected lung tissue had altered miRNAs expression levels by using miR-15b-5p, miR-15a-5p, miR-548c-5p, miR-548d-3p, miR-409-3p, miR-30b-5p, and miR-505, and this will be applicable for the rest of the human coronavirus [95]. Exosomal miRNAs are an excellent delivery system for miRNAs (detailed information in the preceding sections). Patients in the high dimer fragment group had lower levels of miRNA-103a, miR-145, and miR-885, while miR-424 was upregulated [95]. Demirci MDS et al. did a detailed in-silico analysis in which they attempted to predict the miRNA binders for different targets of the SARS CoV-2 virus. They concluded that, aside from ORF6, which has miR-190a-5p as a target site, and the envelope target (E) protein, which has miR-3672 as a target site, the rest of the viral component had a plethora of miRNA binders. MiR-325, miR-34a-5p, miR-6820-5p, miR-1252-5p, miR-1262, miR-2355-3p, miR-382-5p, miR-215-3p, miR-5047, miR-6779-5p miRNAs were discovered to target the viral membrane, and miR-8066, miR-1911-3p, miR-4259, miR-6838-3p, miR-208a-5p.
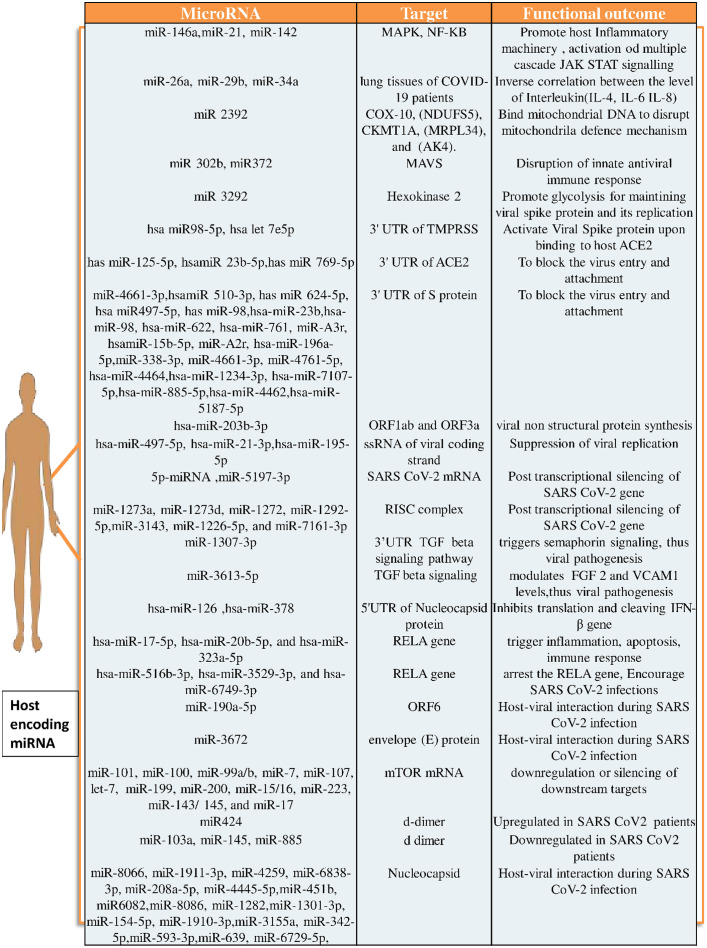
These findings unequivocally demonstrate that SARS-CoV-2 miRNAs, in addition to host miRNAs, are involved in the pathogenesis process. Despite being a cost- and time-efficient method, this theoretical foundation needs experimental validation. For this, numerous techniques are used to validate in-silico results, including microarray profiling, RNA sequencing, miRNA profiling, RT-PCR, Northern, Western, and ELISA blotting as well as immunocytochemistry [39] (Fig. 3 ).
Fig. 3.
Expression of human derived miRNAs during SARS CoV-2 infection [14], [15], [16], [59], [60], [61], [62], [63], [64], [65], [66], [67], [96], [97], [98], [99], [100].
5.3. COVID-19 co-morbidities associated biomarkers
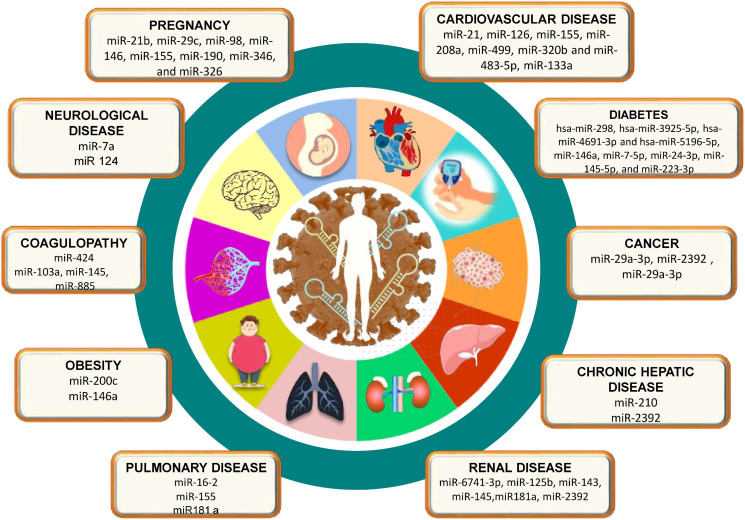
Multiple comorbidities, namely hypertension, cardiovascular and cerebrovascular conditions, diabetes, lung, liver, and kidney disease; cancer patients on chemotherapy; smokers; transplant recipients; and patients taking steroids chronically have been associated with an increased risk of SARS CoV-2 infection and enhanced disease severity [101]. Considering the documented studies on differential expression of miRNAs in tumorigenic tissues or even expression of viral miRNAs to hijack the host immune system [102], miRNAs have been explored widely as a potential biomarker as a potential alternative to prevalent therapeutics in recent times. Here, we have reviewed a few studies that documented the possible avenues of miRNA applications in the diagnostic and therapeutic of SARS-CoV-2 comorbidities (Fig. 4 ).
Fig. 4.
Differential miRNA as biomarkers in COVID-19 infected patients with various comorbidities.
5.3.1. Diabetes
In an extensive meta-analysis conducted by Kumar et al. [103], including 33 studies (16,003 patients) to explore the SARS CoV-2 progression in diabetic patients, a two-fold increase in mortality and severity of COVID-19 were documented, demonstrating the significant correlation between the two [103]. In another subsequent study, diabetes was associated with a significant increase in the risk of mortality when studied against hospitalized patients [104]. Investigating the role of human pancreas miRNAs in the interplay between COVID-19 and diabetes, Pathak and Mishra [105] reported that the upregulation of diabetic biomarkers post SARS-CoV-2 infection can influence the targeting of SARS-CoV-2 genome by hsa-miR-298, hsa-miR-3925-5p, hsa-miR-4691-3p, and hsa-miR-5196-5p miRNAs preferentially over cell's Diabetes-associated messenger RNAs (mRNAs) in the human pancreas leading to diabetic as well as SARS-CoV-2 complications [105]. In another study, wherein the relation between the diabetic and COVID-19 affecting cardiovascular conditions was studied, the expression of two miRNAs, miR-15b-5p and miR-30e-3p, was predicted to target the SARS-CoV-2 genome and monitoring their expression could prove to be potential biomarkers [106]. Simultaneously, upregulation of miRNA miR-133a was also predicted to be associated with a number of roles, including regulation of lipotoxicity and ACE-2 receptor function in congestive heart failure conditions, having basically a protective role against impaired cardiovascular contractility and therefore a promising candidate for investigation in heart failure associated with both diabetic and COVID-19 complications [106]. miR-133a, when studied in relation to arrhythmia, which has also been associated with SARS-CoV-2, also showed to be expressed and involved along with the miRNAs miR-1, miR-208, miR-328, miR-21, miR-212, and miR-590 which thereby demonstrates a potential to be a futuristic biomarker and therapeutic targets [106]. Subsequent studies on the analysis of circulating serum exosomal miRNAs in elderly diabetic patients demonstrate that the four miRNAs, miR-7-5p, miR-24-3p, miR-145-5p, and miR-223-3p has the potential to directly inhibit the SARS-CoV-2 replication and S protein expression and their reduced expression in elderly and diabetic persons has been correlated with decreased inhibition of SARS-CoV-2 replication, promising them to be significant diagnosis and therapeutic targets [107]. Analyzing host miRNAs for their anti-viral effects in further studies conducted by Roganović [108] has also led to the identification of miR-146a, which has been demonstrated to target the SARS-CoV-2 genome as well as downregulation of miR-146a in patients with diabetics, obesity, and hypertension has been found to be associated with the systematic effects of SARS-CoV-2 including enhanced inflammation and fibrosis thereby marking miR-146a to be a potential future biomarker [108].
5.3.2. Kidney diseases, cancer
Considering the expression of the SARS-CoV-2 receptor, ACE-2 in the renal tubular cells, patients affected by associated kidney diseases like nephropathy, chronic kidney diseases, and glomerulonephritis are at a higher risk of SARS-CoV-2 infections as well, and their early diagnosis is certainly crucial. Studies have reported that downregulation of ACE-2 pathways in SARS-CoV-2 affected individuals may lead to podocyte injuries and cause acute kidney injuries [109]. miRNAs are essential for kidney functions, and as a result, they have presented an interesting site for diagnosis and therapeutic targets over the years against kidney diseases. Widiasta, Ahmedz et al., in their review, reports a number of ACE-2 associated gene enhancers and gene silencer miRNAs, of which the miR-29, a fibrotic gene silencer and reported fibrotic-protectant, is proposed for further studies as a potential biomarker [109]. In-silico studies analyzing the role of miRNAs as pathological biomarkers have also demonstrated a potential correlation between differential expression of miR-6741-3p during kidney diseases and their susceptibility to SARS-CoV-2 infection, thereby demanding a further in-depth analysis of miR-6741-3p as a potential target [110]. Srivastava, Swayam Prakash et al., in their paper, suggested that the miRNAs that have been reported to alter the ACE-2 expression level in COVID-induced diabetic kidney diseases, including nephropathy and glomerulosclerosis, could be analyzed for their diagnostic roles and reports a number of miRNAs including, miR-125b, miR-143, miR-145, miR181a, which could be targeted for further studies [111]. Arisan et al. studied the correlation between the SARS-CoV-2 virus and the comorbidities associated with it, reporting seven miRNAs, miRs 8066, 5197, 3611, 3934-3p, 1307-3p, 3691-3p, 1468-5p linking them to viral pathogenicity and host immune responses and could be potential biomarker targets. In another one of the studies, miR-2392 was analyzed for its role as a potential therapeutic target and was demonstrated to be associated with a number of SARS-CoV-2 associated comorbidities including cardiovascular diseases, hyperesthesia, CNS failure and kidney failure, with results observing an enhanced miR-2392 expression in SARS-CoV-2 infected cells and predicting their utilization as a positive biomarker for SARS-CoV-2 infection [50].
The multi-systemic SARS-CoV-2 infection observed in critically immunocompromised individuals increases the risk of SARS-CoV-2 complications in cancer patients as well and essentializes early diagnosis. miR-2392, which has been studied in kidney diseases as a potential COVID-19 biomarker, was also extensively studied in cancer tissues, where it is suggested to be a dominant factor in cellular invasion and metastasis and suggests miR-2392 targeting could decrease SARS-CoV-2 infection [112]. In silico analysis of mature miRNAs from the SARS-CoV-2 genome identifies nCoV-MD3-3P that targets p53 and subverts-reduces its antiviral responses resulting in diseases manifestation, proposes it to be a potential biomarker [74]. miR-29a-3p, which has been reported to be a suppressor miRNA inhibiting cancer cell proliferation and invasion, when studied against SARS-CoV-2, depicts interaction with possible inhibition of viral translation and replication and demands further analysis of their potential anti-SARS-CoV-2 effects [113].
5.3.3. Cardiovascular diseases
Underlying cardiovascular diseases or myocardial injuries observed during SARS-CoV-2 infection have been linked to increased COVID-19 severity. As a result, studying cardiovascular diseases linked to miRNAs could lead to antiviral therapies against SARS-CoV-2. The increased expression of circulating miRNAs miR-21, miR-126, miR-155, miR-208a, and miR-499 in serum of critically ill-COVID-19 patients has been reported, with possible differentiation between severely ill, mechanically ventilated Influenza-ARDS and COVID-19 patients, demonstrating their significance as SARS-CoV-2 biomarkers [114]. miR-320, which has been reported to be involved in the regulation of cardiac injury and dysfunction via antithetical regulation of Hsp20 in ischemic heart diseases, was observed to have a significantly hyper-expression in deceased COVID-19 patients, with reports marking a seven-fold increase of both miR-320b and miR-483-5p expressed patients in dying during in-hospital stay for COVID-19, marking them as possible COVID-19 stratification biomarkers [115]. miR-29, which has been previously associated with kidney diseases, is reported to promote pathologic hypertrophy of cardiac myocytes and their serum expression analysis in hospitalized COVID-19 patients observes a reduced expression with an elevation of their mRNA targets correlating with the disease severity, possibly aiding in futuristic SARS-CoV-2 diagnostic targets [116]. In another study, analyzing the cardiometabolic miRNAs associations with SARS-CoV-2 severity, myocyte-derived (myomiR) miR-133a along with liver-derived miR-122 were reported to be associated with inflammation-induced myocyte damage and hepatic acute phase responses respectively, which upon further analysis reported to be possible markers of SARS-CoV-2 severity and mortality [117].
5.3.4. Pregnancy and SARS-CoV-2
Pregnant women are quite a susceptible group with a higher risk of SARS-CoV-2 infection probability, with predictions even suggesting possible vertical transmissions and can be either asymptomatic or symptomatic [118]. miRNA profiled from the human placenta of infected and uninfected control subjects reported the isolation of a number of miRNAs including miR-21b, miR-29c, miR-98, miR-146, miR-155, miR-190, miR-346, and miR-326 which acts in viral replication through direct-indirect mechanisms and could be potential biomarkers [119]. Complications in pregnancy, including in HIV-associated preeclampsia interlinking SARS-CoV-2 and HIV infection, report identification of potential host miRNAs with anti-SARS-CoV-2 infection properties and differential expression including miR-28, miR-125b, miR-150, miR-223, and miR-382 by the studies of Huang, Jialing et al., while Nersisyan et al., studies report miR-21, miR-195-5p, miR-16-5p, miR-3065-5p, miR-424-5p, and miR-421 miRNAs which can be further studied for their HIV-associated preeclampsia and COVID-19 associations [84], [120], [121]. Although it is worth mentioning that the lack of studies correlating SARS-CoV-2 infection and its association with miRNA expression profiles in pregnant women significantly constricts the data and therefore warrants further studies in this field in the future.
5.3.5. Coagulopathy
The coagulopathy caused by COVID-19 is nearly similar to coagulopathies that are encountered in other critical infections [122]. In a study on 26 COVID-19 infected patients, Gambardella et al., reported the significantly higher level of miR-424, while miR-103a, miR-145, and miR-885 showed low expression levels in patients, having higher D-dimer compared to a lower one [123]. In the same study, it has also been found that miR-145 directly targets the tissue factor, whereas von Willebrand Factor is a direct target of miR-885. Notably, miR-424 is related to hypercoagulability, while low expression of miR-103a is associated with deep vein thrombosis. Even though the procedure for both miRNAs have not been currently exposed. miR-424 can alone be a predictor of pulmonary embolism and acute myocardial infarction in COVID-19 patients, but miR-103a was found to modulate D-dimer levels individually [123]. In COVID-19 infection, upregulation of miR-9, dysregulates the EP300 transcription factor, resulting in increased plasminogen activator inhibitor 1 (PAI-1). This PAI-1 might be an inhibitor of both tissue plasminogen activator (tPA) and urokinase plasminogen activator (uPA) [124], [125]. The two most common plasminogen-activating serine proteases are tPA and uPA. Plasmin is a breakdown fibrin polymer at certain places, causes the breakdown of blood clots, and the inactive precursors of plasmin are plasminogen [126], [127].
5.3.6. Pulmonary diseases
ARDS, which indicates severe acute lung injury, is the most common concern of SARS-CoV-2 infection, and nearly 20 % of infected people will reach this stage [128]. Under normal settings, miR-16-2 reduces both productions as well as expression of TNF-α and IL-6 while improving anti-inflammatory IL-10 in the lung's macrophages. Nevertheless, in COVID-19 infected patients, the production of pro-inflammatory cytokines increases IL-6, MCP-1, TNF-α, and IL-1, whereas it lowers the IL-10 production and TGF-β [129]. During the acute phase, patients infected with COVID-19, showed higher miR-1246 expression levels, which increases the expression of ACE2 in the lungs, along with that also increases vascular permeability and heightens apoptosis, causing injury in the lung epithelial [130]. miR-155 is located in lymphoid cells of the lungs, shown higher in COVID-19 patients. IL-1β, TNF-α, PAMPs, and DAMPs are all examples of inflammatory stimuli by this miRNA [131]. Studies show that miR-181 expression was lowered in older people, activating innate and adaptive immune systems [132]. Recent studies reported that expression of miR-181a miRNA is solitary downregulated in SARS-CoV-2 infected patients, increasing viral clearance. Furthermore, low levels of miR-181a boost viral entry by increases the transcription of TMPRSS2 and ECA2 mRNA. This variation exacerbates lung injury by lowering the Bcl-2 molecule that controls cell apoptosis [130], [133]. In bronchoalveolar stem cells, expression of miR-223 (BASCs) downregulated activates numerous inflammatory pathways by increasing ACE2 receptors, potentiating viral replication. This leads to the lung surface's destruction and also loss of repair capacity [134]. Lastly, miR-211 has been linked with enhanced inflammatory response and increased attraction for actual target sites in COVID-19 [135].
5.3.7. Neurological disease
Coronavirus arrives in CNS through ACE2 receptor present in choroid plexus and neurons that are inhibitory or excitatory in nature. Receptors of astrocytes and oligodendrocytes may also act as an entry point for SARS-CoV-2 into the CNS [136]. Kim et al. reported that high levels of miR-7a in COVID-19 patients suppress the transcription of numerous proteins, like α-synuclein. The suppression of proteins transcription may cause neurodegenerative diseases like oxidative stress, fragmentation of mitochondria, and autophagy, increasing the risk of neuronal death and maximizing harm in the brain and cognitive dysfunction after CVD occurrence [137]. The miRNA miR-24 can regulate the expression of NRP1, which is a transmembrane receptor that helps in virus entry; however, in COVID-19, this miRNA is downregulated, increasing the production of NRP1 and, as a result, the neurological damage increases [138]. Keikha et al. reported that in severe COVID-19 infection, miR-124 expression decreases in CNS [139].
5.3.8. Obesity
Papannarao et al. reported higher expression of miR-200c might escalate vulnerability to COVID-19 in obese people. As miRNAs are the initial molecules that regulate the cell cycle, circulating miR-200c might be a biomarker to recognize individuals at risk of developing severe COVID-19 infection [140]. Overexpression of miR-155 in obese mice's ATM causes insulin resistance. A study on human adipocytes treated with TNF-α, resulted in elevated miR-155 expression and inflammation, and underlined the importance of miR-155 [141]. miR-146a downregulation has been discovered in diabetes, obesity, and hypertension, resulting in increased inflammation and fibrosis and systemic consequences associated with severe COVID-19 infection [108]. Adipocyte and adipose tissue function anomalies have been linked to fat deposition in obesity. Downregulation of miR-126 increases hypercytokinemia and ARDS vulnerability, resulting in severe COVID-19 infection in younger obese patients as compared to older obese patients [142], [143].
5.3.9. Chronic liver disease
Organ-specific miRNAs with the most considerable changes in severe COVID-19 were myomiR miR-133a and liver-derived miR-122, and both were linked to 28-day ICU mortality. Another essential characteristic of miR-122 is its remarkable tissue selectivity, which provides information about the underlying pathology [144], [145]. Liver-derived miRNA miR-192-5p was a significant predictor of mortality during an ICU stay, whereas hypoMSCs have much greater levels of miR-210 than norMSCs. miR-210 also decreased the hepatocyte apoptosis rate [146]. RNA-seq data from hepatic, lymph node, heart, kidney, and lung got throughout dissections of SARS-CoV-2 infected patients with low or high viral loads were used to correlate miR-2392 to SARS-CoV-2 infections. During the associating link between gene fold change values, there was an important and positive link to liver tissue, but no link was found when comparing C2 curated gene sets [147]. Hence, it may be concluded that miRNAs play a crucial role in COVID-19 multi-organ problems.
6. Potential MiRNA-based antiviral strategies targeting SARS CoV-2
Following the development of an FDA-approved mRNA vaccine, researchers have focused on the regulatory role of miRNA in disease pathogenesis. It has painted the possibility of progress in developing a novel antiviral strategy. Aside from RNA vaccines, many new promising RNA-targeted strategies have been explored. The ability of miRNA to manipulate SARS-CoV-2 - host interaction by umpiring RNA degradation or inhibition of protein synthesis is being investigated in recent studies. As a result, we attempt to highlight the potential therapeutic approaches based on miRNA that could alter the landscape of conventional mRNA vaccines for COVID-19.
6.1. miRNA-based-restoration therapy: MiRNA mimics and precursors
The main motto behind miRNA restoration therapy is to increase miRNA levels that are downregulated in the diseased cell [148], [149]. It is executed by using synthetic miRNA mimics or Precursors to replace the miRNA molecule loss. miRNA mimics have sequences identical to the sequences of endogenous matured miRNA[150]. Since these mimics don't contain new sequences, this approach does not have the backdrop of vector-mediated toxicity while conducting a synthesis. Inculcating vector-based restoration of miRNA using expression vectors like the retrovirus, lentivirus or adenovirus is extensively practiced to achieve a long-term expression of miRNAs and is administered intravenously [151]. They do so by inserting precursor miRNA sequences to promoters involving downstream RNA polymerase and subsequent upregulation of the desired miRNA. For instance, MRG-201 and MRX34 mimic the action of miR-29 and miR-34a, respectively [152], [153]. Another strategy in a similar line is conducted by using miRNA precursors. This also can enhance the matured miRNA levels, but they are employed in the intermediate state, which further gets matured. There are findings that these precursor miRNAs have a modulatory role in recognizing the target [154], [155]. A recent work conducted by Jafari et al. showed evidently how the effects dissipated by pre-miRNA differ from that of miRNA from an anticancer perspective [156]. Enormous research is conducted to induce the upregulation of the desired miRNA. A fluoroquinolone antibiotic, enoxacin is a must mention antibiotic which enhances the overexpression of target miRNAs, i.e., miR-125a, miR-139, miR-199b, and miR-23b [157]. Although these strategies are quite nonspecific, this area still grabs the minds of researchers. Young et al. worked on small organic molecules and recognized a few molecules such as 2-(2-(Dimethylamino)ethyl)-5-amino-1H-benzo[de]isoquinoline-1,3(2H)-dione responsible for the expression of miR-122 [158].
6.2. miRNA based- inhibition therapy: MiRNA inhibitors and sponges
Lately, considerable literature has grown up around the theme of RNA-based therapies such as antisense oligonucleotides (ASOs) technology which has proposed the use of multiple classes of compounds to serve the purpose of miRNA inhibitors [159]. ASOs can be described as small single-stranded nucleic acid that exclusively targets cognate RNA. In the following section, we systematically confer the mechanism, chemical modifications, and the optimized delivery strategy governing around ASOs. Existing literature indicates that it carries out its function utilizing two prime mechanisms depending on the nature of the chemical backbone. (A) RNase H independent ASOs act as steric blockers subjecting to disrupting the RNA secondary structure, splicing machinery, and RNA-Protein interactions. (B) RNase-dependent ASOs also referred to as GapmeRs, employ and catalyze specified RNA's RNase-mediated cleavage. This leads to suppressed expression of RNA and protein. The chemical modifications form the basis of differences in the ASO mechanism [160]. These chemical modifications increase the efficacy by boosting its potency and pharmacokinetics while lowering its toxicity. One of the most familiar modifications is the substitution of one of the non-bridging oxygen with a sulfur atom in the phosphodiester bond, which leads to the formation of a first-generation ASO. Phosphorothioate (PS). Second-generation ASOs comprises a modification in the 2′ position of the sugar moiety, which embraces the druggable properties of ASO. Adding to the fact, the development of the Third generation ASO involves many modifications, including Locked Nucleic Acid(LNAs), Phosphorodiamidate morpholino oligomers (PMOs), and Peptide nucleic acids(PNAs), which increase the affinity to target miRNA. These modifications based ASOs, upon binding to the target entity, carry out RNase H1 cleavage [161], [162], [163]. Modifications such as LNA and PS have a nonspecific nature while dealing with the proteins resulting in diminished urinary clearance rate and an enhanced half-life. While Peptide-nucleic acid (PNA)-based ASOs and methylcytosinenucleobase-modified ASOs have better binding affinity and don't activate the immune response PNAs are less hydrophilic, and have a quick urinary clearance rate; hence are not a good choice for carrying out inhibition [164]. Fazi et al. work fascinatingly suggest the plus point of using ASOs is that it does not require any vector to travel to the target cells; they could internalize themselves by micropinocytosis or gymnosis. This approach was first practiced against SARS-CoV, considering it to have the closest resemblance to SARS CoV-2 and targeted ORF 1 a of the virus and also the 5′UTR regulatory sequence of the positive RNA strand [165]. Yuchen nan et al. mentioned a promising ability of ASO to have antiviral properties. They modified the chemical sugar moiety mainly with a morpholine ring (PMOs [antisense morpholinooligomers] and P-PMOs [peptide-conjugated antisense morpholinooligomers]) [166]. To develop ASO specific to SARS CoV-2, it is important to realize that the virus harbors 14 ORFs, which in specific combination encode for the spike, nucleocapsid, membrane, envelope, or accessory proteins. Hence Zhu et al. pointed out the significance of these proteins in designing accurate ASO targeting specific proteins [167].
Aside from that, the concept of a miRNA sponge differs greatly from that of an inhibitor in that the sponge transgene is mostly carried by a virus vector, and the transcribed mRNA contains multiple binding sites for the target miRNA. This method outperforms the existing methods of miRNA knockout or miRNA in loss of function for inhibition therapy. The use of miRNA sponges carrying transgenes is prevalent in a wide range of cells that would not be possible using ASOs [168]. To achieve continuous miRNA inhibition, ASOs would require repeated transfection, whereas a miR sponge can perform uninterrupted inhibition. These miRNA sponges are members of the RNA class, which contains numerous artificial miRNA binding sites that compete for miRNA binding with miRNA targets. Furthermore, these miRNA sponges are now designed using a simple enzymatic ligation method to customize circular RNA. This sponge has the ability to inhibit matured miRNA exogenously, making it a potentially useful therapeutic option. Surprisingly, these circular RNAs have more than one binding site in the circular sequence and are easily carried by viral vectors, allowing them to regulate multiple miRNAs [169].
6.3. miRNA biogenesis interference therapy
Drosha and Dicer are two important enzymes that regulate the miRNA synthesis machinery. Attempting to weaken or delete these enzymes would result in an overall imbalance of miRNA levels in the body. A recent ACE-2 study discovered that a small number of miRs are responsible for downregulating ACE-2 expression in cardiomyocytes by binding to ACE-2 mRNA and protein. Aside from cardiac remodeling, cardiomyopathy, and heart failure, deletion of dicer enzymes in cardiomyocyte-specific cells would make them more susceptible to viral infection. In an orthotopic mouse model, a DGCR8 gene responsible for microprocessor complex unit formation was deleted. In a rat model, it was discovered that it alternated miRNA biogenesis and cognitive and behavioral impairment. Our body consists of thousands of miRNA bodies dictating gene expression. In case of inhibition of miRNA processing enzyme would have a redundant outcome, which is not favorable for therapeutic designing [170], [171]. The upregulation of miR-200c represses the expression of ACE-2 in both human and rat cardiomyocytes. This fact would pave the way to design miR-200c based therapy by targeting the viral RNAs and miRNAs to lessen the treatment window and disarm the SARS CoV-2 pathogenesis potential. Existing literature also highlights the effect of Dicer mutation in animal models, which exhibits reduced cellular miRNA biogenesis and ultimately lowers the inflammatory activation, which increases the host conformity to murine cytomegalovirus infection [172]. It has also been predicted that varied Dicer expression in different hosts would result in distinct miRNA expression and, therefore, susceptibility to viral pathogens, including SARS CoV-2 [173]. This Dicer regulated protein expression could be utilized to bind those protein targets which otherwise could not be accessible with conventional drug compounds. e.g., Dicerna Pharmaceuticals formulated the use of DICER substrate siRNA (DsiRNA), which could silence oncogene protein myc, which could condense the tumor volume in mouse models [174]. Kaur et al. recognized miR-214, miR-98, and miR-32 to target TMPRSS2 and ultimately silence it. Deletion of miRNA regulatory protein could change the high binding affinity of these miRNAs with this SARS CoV-2 receptor which could help in devising an antiviral drug lead [175], [176].
6.4. miRNA-based target site blocker therapy
Target site blocker (TSB) is single-stranded RNA molecule that usually acts as a barrier between miRNA and its target site to prevent the action of the target gene [177]. It functions by blocking the binding site of miRNA at the 3′UTR region of the target mRNA gene. Usually, it targets a single site while the remaining target gene continues to be regulated. The main objective behind subjecting this TSB to miRNA therapy is to check whether the miRNA interaction with a target gene is responsible for the desired effect. For instance, a CAV1 TSB phenotypically resembles a miR-199a-5p inhibitor and exhibits the way miR-199a-5p plays its role as a crucial factor of TGFβ signaling in lung endothelial tissues. A commonly employed Target site blocker miRCURY LNA miRNA mainly determines the pathways associated with miRNA inhibition detected based on phenotypic analysis. A study has also shown the use of TSM to validate the communication of miR-155 with PAD4 throughout the neutrophil action in the extracellular space during inflammatory infections [178].
6.5. Prospective miRNA-mediated helicase suppression therapy
Wang et al. described a new class of host enzymes known as DEAD-box helicase acting as a double-edged sword in viral replication. The precise mechanism of their involvement in virus replication is still a mystery, but in general, DEAD-box helicase has been portrayed as an ATP-dependent chaperone that manipulates the RNA by disordering its secondary and tertiary structure RNA and protein configuration [179], [180]. It has been shown to have a remarkable response during transcription, post-translational modification, miRNA biosynthesis, translation, and RNA degradation in the host. Also, it has been revealed that many such helicases like DDX1, DDX3, DDX5, and DDX17 have a role in host cell–virus dialogue which benefits the virus in their maintenance and survival [180]. Considering these facts, research is required to provide therapeutic involvement at the stage of miRNA biosynthesis, which could be administered as a substitute to existing approaches. This approach has not seen any parallel investigation specifically for SARS-CoV-2; hence more information on this area would help shape a greater degree of accuracy [181].
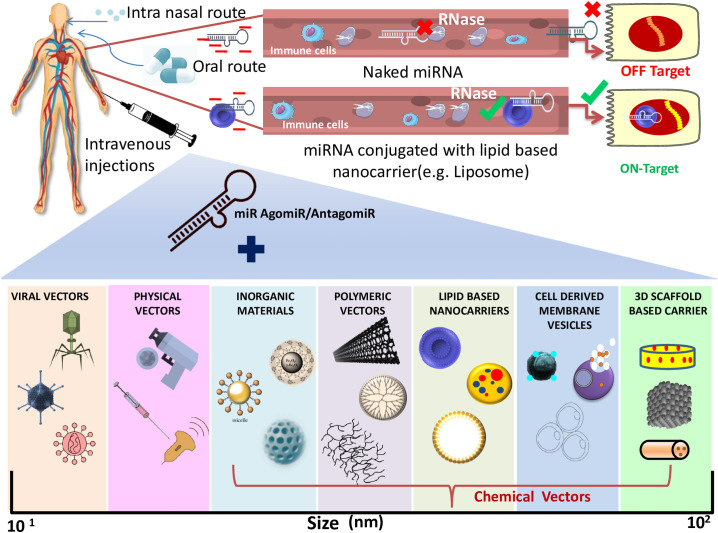
7. Recent miRNA-mediated delivery system
One of the major shortcomings encountered while designing miRNA-based therapeutics is the ease of delivering these miRNAs to the target tissues. The conventional delivery vehicle faces a critical issue of toxicity owing to its negative charge, short half-life, and restricted stability [182]. Consequently, these short non-coding naked miRNAs fall into the trap of redundant off- or on-target effects due to their charge, get rejected by the host immunity, or undergo disruption rapidly by the nucleases occurring in the bloodstream. Keeping all these in mind, the scientific community came up with the idea of formulating a vehicle conjugated with miRNA to effectively fix all the inadequacies. In this framework, recently, Viral and Nonviral vectors carriers have been utilized in vivo, including bacteriophages, exosomes, liposomes, micelles, nanoparticles, intravenous injection, and oral delivery systems in the preclinical and clinical interventions. These are manipulated at a few genomic sites to inhibit their replication machinery, making them safer.
Virus-mediated carrier system includes the exploitation of adenovirus or adeno-associated viruses (AVVs), lentiviruses, and retroviruses; Mowa et al. mentioned the use of Adenoviridae family derived Hepatitis B adenoviral vectors [183]. These vectors carried pri-miR-122/5, pri-miR-31/5, and primiR-31/5-8-9, which hindered viral reproduction in the human hepatocytes. Another study on Adeno associated viral vectors has elicited persistent gene expression in the delivery of specific miRNA Spinal and bulbar muscular atrophy (SBMA) [184]. Working on a similar line, another scientist, Ramanujam et al. [185] recognized retrovirus as an effective means of miRNA carrier. This RNA virus can invade the host cell, undergo reverse transcription, and finally integrate with the host genome to induce sustained gene expression [185]. It can create a space of up to 8 kb to incorporate a generous amount of genetic materials. Likewise, lentivirus too integrates into the host DNA, subjecting to persistent expression. Theis et al. experiment has discovered a marked recovery in spinal cord injured mice using lentivirus as an excellent vehicle for miR-133b. The key advantage of using these vectors is that it accounts for high transfection competency and remarkable gene expression of miRNA agomirs or antagomirs [186], [187]. However, this viral vector restricts their usage in the clinical setup since they solicit strong host immune response, mutations due to inserted sequences, and cytotoxicity in a few cases, making their use quite delicate [188], [189].
Considering the above limitations, alternatively, the use of nonviral miRNA vectors is the major focus of the researchers. The nonviral vehicle system involves both physical and chemical-based approaches. Physical-based approaches mainly deal with increasing the plasma membrane permeability momentarily to allow gene delivery at ease. Hence the host cell is subjected to hydrodynamics, electroporation, gene/biolistic guns, ultrasound, or laser-based technique. In contrast to that, the chemical-based approaches utilize the use of nanocarriers. These nanocarriers can be further sectioned into four classes.
-
(1)
The first class includes Inorganic material-based nanocarriers comprising gold nanoparticles (AuNPs), Fe3O4-based nanoparticles, silica-based, and magnetic nanoparticles. These are tailored by attaching amino or thiol groups onto their surface to allow the transmission of miRNA to target cells/tissues. The physicochemical properties like shape, surface functionalization, surface area, biocompatibility, and amphiphilicity offer advantages over the viral vector. Despite this, these bodies also have clearance and toxicity issues.
-
(2)
Second class of nanosystem includes Polymeric material-based nanocarrier, which could be either natural or synthetic. Polysaccharides, proteins, or peptides are natural polymers offering easy uptake, have low toxicity, and can imitate the natural environment. Unfortunately, the host immune system and sera pose a significant disadvantage to these nanocarriers. Instead, synthetic polymer-based nanocarriers are the biomaterials that generate less cellular damage and lower cytotoxicity. Examples include polyethyleneimine (PEI), poly (lactic-co-glycolic acid) (PLGA), and dendrimers. Unlikely these polymers are known to have low transfection ability and are nonbiodegradable. miRNA-mediated therapy could have the upper hand over other therapies in the context of COVID-19 since it can be easily administered in the upper and lower respiratory tract by mere intranasal inhalation. Polymer-based Oligonucleotides are hydrophilic, and recent technologies have an aerosolized miR-93 amalgamated viral vaccine to confer protection by fluently getting absorbed in the lungs. Another approach by Dyawanapelly et al., which thrived in phase 2 clinical trials, demonstrated the use of siRNA against the nucleocapsid protein of respiratory syncytial virus (RSV) [190], [191], [192]. Moreover, antagomiRs are a new class of oligonucleotides that acts as miRNA silencer has shown their application in severe COVID-19 patients who have encountered cytokine storm [193]. Their administration in polymer-based nanocarriers enables remarkable results in the inflammatory landscape.
-
(3)
The third class of nanovectors is lipid-based nanocarriers, one of the most studied vectors. This takes into account that lipid complexes known as liposomes/lipoplex complexed or encapsulated with desired miRNA act as miRNA carriers and are of three types divided based on charge- cationic, anionic, and neutral liposomes. These are simple to synthesize, stable, elevated loading efficiency, functionalize, and less cytotoxic. These nano delivery systems have gained ample popularity due to their broad range of druggable targets, small dosage, treatable side effects, and could be manufactured on a large scale. A single-step manufacturing procedure can synthesize a huge amount of small lipid nanoparticles within a short period. Recently reported commercial nanocarrier formulations such as Lipofectamine®, SiPORT, and DharmaFECT are cationic lipoplexes which have generously given positive results in in-vitro and in-vivo setup and are amphiphilic apt for oligonucleotides delivery [191]. The most crucial con behind this liposome delivery system is the stability of these vectors in the host sera owing to its nonspecific binding protein targets. Thus to address this issue and enhance the circulatory half-life, they are first conjugated with polyethylene glycol (PEG) to enhance the stability, efficiency, and sustained RNAi delivery by receptor-mediated endocytosis.
-
(4)
The last class of nanocarriers is Cell-derived membrane vesicles, commonly referred to as Extracellular vesicles. These are usually associated with cell-to-cell communication and occur in different forms based on their sizes, such as Exosomes (40–120 nm), Microvesicles (100–1000 nm), or Apoptotic bodies (50–5000 nm) [190], [192]. Wang et al. have mentioned that EVs are advantageous over other delivery systems as they have negligible antigenicity, high delivery to the target tissue, null cytotoxicity, and potentially superior delivery option in miRNA-based therapeutics. Red Blood cells (RBCs) have a central role in pulmonary respiration and transport of oxygen and carbon dioxide hence conferring uninterrupted circulation. In agreement to a study, RBC consists of a small number of mRNAs and miRNAs. This RBC is associated with nanoparticles to succor ten times the advantage of nanoparticles alone [107]. Apart from that, immune cells such as T cells, Macrophages, Dendritic cells, and Natural cells are optimized in an encapsulated manner to transport miR/miR antagomir [194], [195], [196], [197], [198], [199], [200], [201], [202], [203], [204], [205], [206], [207], [208]. Fascinatingly Parayath et al.'s study revealed the injectable mode of administration of the T cells to selectively carry these small molecules to the target site. The specificity could also be enhanced by temporarily altering the circulating T cells to disease-specific antigens in an ex vivo manner [209]. Amusingly plant-derived exosomes vesicles are widely implicated in the current scenario with the name of edible nanoparticles (ENP). These ENPs could be extracted from plants like ginger and grapefruit. The SARS-CoV-2 miRNA of interest could be administered with this ENP, and potent therapy could be deduced in the interest of human mankind. The miR-530b-5p is known to specifically target the SARS CoV-2 genome by binding to the ribosome slippage between ORF1a and ORF1b. Using these ENPs, these miRNA could be accumulated in the inner linings of lung tissue in various biological entities [210]. In the face of the current pandemic, researchers have been trying hard to utilize novel delivery strategies to apply to miRNA. Zhang et al. work; introduced an innovative technique using nanobody, a miniature form of the engineered antibody. They utilized a miRNA crosslinked nanobody functionalized nanogel popularly known as Three-dimensional (3D) scaffold-based carriers. This nanogel paves a new avenue in the miRNA-mediated therapeutics world. It is important to note that these nanoparticles safeguard the miRNA from getting depleted so that they can passage all over the body effortlessly. Using these nano carrier-based carriers can combat COVID-19 by developing effective RNAi-based therapy subjecting to any of the above-mentioned delivery machinery [211].
A plant-derived miRNA -MIR2911 in a honeysuckle decoction (HD) has been notably known to inhibit viral replication and stimulate the negative conversion of moderately ill COVID-19 patients. Conversely, HD-MIR2911 failed to get absorbed in the gastrointestinal tract (GI) in patients with SID1 transmembrane family member 1 polymorphism. Thus, this miRNA requires scientific intervention focusing on the ways to deliver this small ncRNA in the host tissues suggesting potential miRNA-based COVID-19 therapeutics (Fig. 5 ) [212], [213], [214].
Fig. 5.
Currently available delivery vehicle for the effective administration of miRNA-based therapeutics. It could be categorized broadly into viral and non-viral vectors. 1) Viral vector includes Lentiviruses, Bacteriophages, and Retroviruses. 2) Non-viral vector is further categorized into physical and chemical vectors. (i) Physical vector (biolistic gun, electroporation, ultrasound, etc.) (ii) chemical vector could be (a) inorganic material like gold nanoparticles (AuNPs), Fe3O4-based nanoparticles, silica-based nanoparticles, or magnetic nanoparticles; (b) polymeric vector such as polyethyleneimine (PEI), poly (lactic-co-glycolic acid) (PLGA), and dendrimers; (c) lipid-based nanocarriers like cationic, anionic, and neutral liposomes; (d) cell-derived membrane vesicles viz. Exosomes, microvesicles, or apoptotic bodies; lastly, (e) 3D scaffold based carrier comprises porous, fibrous scaffolds, etc. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
8. Challenges in the development of miRNA-based therapy
Scientists around the globe are extensively engaged in formulating a therapeutic cure against the lethal SARS-CoV-2 viral infection. Despite advancements in molecular therapies, it is still a difficult hurdle to get down the virus spread. Scrutinizing over every possible molecular strategy, miRNA-mediated therapies are gaining enormous attention in scientific society. Despite miRNAs distinctive nature and favorable characteristics being in talk, it is still challenging to release an FDA-approved miRNA vaccine into the market. Statistical studies suggest that fewer than twenty miRNAs of therapeutic value have passed the initial phase of the clinical trial, but none could make it to phase III clinical trials. Zhang et al. mentioned that around 60 siRNA drugs had entered the clinical trials, among which two of them have been approved by the authorities. The study compared both miRNA and siRNA therapeutics that revealed a higher fraction of rejected/terminated clinical studies of miRNA therapeutics than successful siRNA therapeutics [208]. This implies several unexplainable obstacles on the road to miR therapeutic candidates. There could be many possible reasons, including fewer vaccine nominees, multiple gene targets, less complementarity, etc. siRNA displays 100 % complementarity with the target gene and has 1–3 gene targets, making it a better choice. Multiple targets reduce the efficacy of the therapeutic approach. This miRNA usually functions by either inhibiting translation or downregulating the stability of the target gene. A miRNA-based therapeutics, i.e., miR-34a mimic altered two host immune pathways but failed to enter phase II trials due to immune toxicity issues [208]. Another study denoted a mode of action of a miR-122 molecule which revealed only a few genes participated in modulating the jaundice disease pathway [208]. Nonetheless, the experiments conducted could not show any results in the phase-I subjects but appeared in the phase-II trial cases. These instances exhibit the ill effects of multiple targets, which fail the process of developing a miR therapeutic. Either chemical modifications or targeted delivery could address these limitations by using vectors. Moreover, the application of plant-derived miRNA (e.g., aly-miR396a-5p) affects the pathogenic genome but escapes the host gene and ultimately doesn't disturb the body's immune response. This novel approach will make a miR and efficient therapeutics option exclusively targeting the specified tissue/organ using a competent miRNA modulator, thus minimizing nonspecific interactions. The complex physiochemical nature of miRNA and miRNA inhibitors entails severe limitations in developing miRNA-based therapeutics.
A large number and growing body of literature have focused on the nature of DNA or noncoding RNA-based miRNAs. Richard S Geary et al. suggested that these nucleic acid bodies carry a negative charge which endorses nonspecific attachment with blood serum proteins, restrained passive diffusion for cellular entry eventually declining urinary clearance [215]. It is found that ASOs like PNAs or unmodified siRNA don't hold any charge; they have a low affinity to plasma proteins. Hence, they are subjected to rapid clearance from the body by the route of metabolism or excretion from the body [216]. Accumulation of such nonspecific ASOs could ultimately lead to sequence complementarity to nonspecific target tissues. The main concerns while dealing with miRNA mimics includes: Restrained efficacy, activated complement cascade pathway, triggered partial thromboplastin time, and mounting up in nonspecific tissues is clinically insignificant for a good therapeutic candidate [217], [218], [219].
The prime requirement of designing a drug involves a drug target specific to a disease, i.e., SARS CoV-2 in the present case. A single miRNA has the property of regulating multiple targets; hence extracting a single mRNA is a demanding task [220]. In addition to that, various biological factors such as miRNA expression, accessibility to AGO2, availability of ample gene of interest, their expression, and successive off-target effects further restrict even the movement of therapeutic miRNA. For a miR therapeutic to display its effect, escaping the ribonucleases enzyme while circulating in the host blood is crucial [221]. For a naked miRNA, an unaltered 2′OH of ribose sugar is prone to quick degradation by the nuclease in the biological system. Additionally, chemical modifications such as LNA, PS, o-methyl, O-(2-Methoxyethyl), or Fluro boost the binding stability and affinity, improving the chances of miRNA attachment to the target genes [222], [223].
It has been noted that chemical modifications in the 5′ end of the antisense strand are more vulnerable to getting ruined; hence biologists have thought to modify phosphorothioate at the sense strand to avoid such molecular asymmetrical glitches [224], [225]. The endosomal system facilitates the trafficking of miRNA, which shows the transition from the early endosomal state to the late endosomal stage and further lysosomal state, which surrounds an acidic milieu showing active nuclease degradation. Consecutively for a miRNA mimic to show its effect, it is fundamental to interact with the miRNA RISC complex. While formulating miRNA-based mimics or inhibitors, it is essential to check whether the miR escapes the endosomal system or not. This constitutes a significant challenge that can be sorted by alternative delivery systems like photosensitive polyplexes or lipoplexes sensitive to pH facilitating separation from endosomes [226], [227]. Li et al. identified concerns arising due to toxicity mediated by chemical modifications in the complement cascade, blood coagulation, decreased WBC counts, activation of immune cells, or sequence independence [228]. These side effects occur as a sign of toxicity accompanying chemical modifications. Another inadequacy related to miRNA-based inhibitors is poor absorption in a few regions, such as skeletal tissues or the blood-brain barrier [225], [228]. For efficient uptake of these miRNA molecules, the size of the nanoparticles also establishes an important characteristic for penetration to leaky vessels. Hence, optimizing chemical conditions such as pH and hydrophilicity, which decide the size and urinary clearance, determines the warranted generation of biocompatible miRNA therapeutic options [229], [230].
9. Future headways for the development of MiR-therapeutics
Central to clinical research, the study of miRNA has been a booming interest over the past two decades. The lack of targeted vaccines has existed as a loop hole for these COVID-19 pandemic years. With the technological advancements in the field of vaccinology, it has somehow been ineffective against the current rising Variants of concern (VOC). Therefore there is an urgent need to address novel anti-SARS-CoV-2 strategies and biomarkers or diagnostic approaches in the coming times. In order to eliminate the spread of COVID-19 infection, plasma from SARS-CoV-2 infection recovering patients are utilized and treated against severely infected patients attributed to the presence of antibodies and miRNAs. The presence of miRNA has a role in regulating the cytokine storm by blocking host receptors like ACE-2 and TMPRSS2. Previously published studies strongly suggest the importance of knowing the host cell response and virus metabolism to draw similarities with the nearest resembling organism [231]. Amidst the limited treatment choice, we can suppose that an RNA-based vaccine is a protagonist for long-term resistance against the SARS CoV-2 disease. Studies established by investigators such as Hum et al. and Demongeot et al. have examined the influence of miRNA in the viral genome, modulation of the host microenvironment, and the innate immune system of the host biological system [49], [232]. A considerable amount of literature has been published on miRNA targeted therapy, mostly drawing the usage of miRNA antagonists or inhibitors viz. Cobomarsen- miR-155 inhibitor [233]. Also, miRNA mimics such as miR-29 mimic – Remlarsen [233]; Okuyan and Begen [234] attempts have introduced a Santaris Pharma A/S manufactured Miravirsen-a miR-122 antagonist and also the first miRNA-based drug passing Phase II clinical trials that suppresses the levels of viral RNA. With respect to the miRNA SARS-CoV-2 interaction, it can undergo both suppression and amplification of viral gene expression. It is important to bear in mind the above-mentioned limitations as a note of caution, which opens doors for new interventions in the field of miRNA-based therapy [235]. Identifying plant-based miR inhibitors, target-specific delivery, more efficient competent miR inhibitors, or advancement of microvesicle-mediated targeted delivery are budding opportunities in the advancing miR therapeutics era. The field of miR-based therapy got introduced to plant-derived miRs, which get easily taken up by the gut/intestinal microbiota. Publish studies have mentioned the use of ginger exosome-like nanoparticles for the purpose of better miR (e.g., Plant miR2911) stability and its better absorption in GI tract than synthetic miRs (miR2911). Similarly, ginger-derived miRNAs (aly-miR396a-5p and rlcv-miR-rL1-28-3p) facilitates specific inhibition of expression of nonstructural and spike protein. These natural miRNAs have high scalability and are on the verge of getting their validation, which opens a novel opportunity to design new drugs that could be administered orally [236]. A very interesting observation was made by a study conducted by Mangukia et al. and group, where they identified dietary plant derived miRNAs from fruits such as Citrus sinensis (Orange), Prunus persica (Peaches), Vitis vinifera (Grapes) and Malus domestica (Apple) to target SARS-CoV-2 viral genomes. They highlighted the role of miRNA csi miR169–3p extracted from Citrus sinensis to specifically target the 3′UTR region with a conserved sequence of “CTGCCT” to target all collocated 772 Omicron variants globally [69].
miR-therapeutic modulators, for instance, synthetic RNA/DNA mimics like Peptide Nucleic Acids (PNAs) are neutral charged and poorly soluble in water. This limitation is resolved by using gamma PNAs which cut the shortcomings of regular PNAs by having a hydrophilic nature and neutral charges that prevent nonspecific accumulation or adherence on the surface of nontarget tissues. It is the sole known molecule with the ability to invade DNA/RNA duplex with CG content as high as 100 % by undergoing tail clamp modifications [237]. It has been employed in various fields ranging from biological to clinical applications, CRISPR Cas-9 gene editing, nanotechnology, and gene targeting. It is quite rampant to have immune system-related toxicity; hence, the investigators are switching to cell-derived microvesicles to effectively deliver miRNA [238]. This view is highly supported and applied to reduce the inflammation of cells by using platelet-derived microvesicles as a delivery vehicle enclosing miRNAs like miR-302b, miR-142-3p, and miR-223. The miRNAs miR-302b and miR-372 regulate mitochondrial metabolism via the SLC25A12 transporter, which controls MAVS-mediated antiviral innate immunity [239].
Nanoformulations-based methods are also becoming handy and can be administered in the form of nasal sprays. Scientists have deduced the relevance of miRNA in COVID-19-mediated comorbidities, like cardiac [240], pulmonary [241], or neurological diseases [242]. Viral miRNAs control the various signaling pathways mentioned above, creating a hypoxic microenvironment leading to severe consequences. Recently, considerable literature has grown up around the theme of COVID-19 animal models, which has boosted the preclinical trials of candidate vaccines [243]. A mRNA vaccine mRNA-1273 displayed quick protection to both upper and lower respiratory tract, for which Corbett et al. used a nonhuman primate model [244]. An investigation conducted by Rezk et al. evaluated the effectiveness of ivermectin in reducing the fatality percentage by influencing the expression profile of miR-2029, miR-233-3p, on a group of 300 COVID-19-infected hospitalized patients. This shows the marked correlation of miRNA with the COVID-19 existing therapy [73]. Studies have related the positive results from the COVID-19 RT-PCR test to changes in the miRNA profiles in nasal swabs, which could potentially classify the case self-reliantly [65]. Investigations are expanded with larger groups of infected people, including pre-symptomatic, asymptomatic, and infection caused by different variants. Additionally, to see whether this notion endures during the COVID-19 incubation period (on average 6–7 days) or if this could be incorporated on real grounds for efficient detection and prognosis. miRNA biomarkers may be useful in providing infection evidence to doctors since miRNA responses reflect the host's response to infection.
10. Conclusion
As the COVID-19 pandemic unfolded, the world has seen diverse versions of mutants that restrict the disease to die out completely, escape the effectiveness of current therapeutics, and ability to sustain pre- and asymptomatic human-to-human transmission. In such a scenario, there is always a possibility of outbreak clusters; hence the focus should be on identifying the early signs of COVID-19 infection irrespective of the arising variants of concern exhibiting patterns of genomic mutations. The present study was undertaken to comprehensively account for small noncoding MiRNAs as a critical player in the viral illness game. Even though the class of miRNA has grabbed attention recently, they have been successfully calculated as the biomarker for a range of diseases over more than a decade. Researchers have already detected changes in miRNA expression levels in body fluids, including saliva, urine, breast milk, plasma samples, and serum in cardiovascular disorders, cancer, and viral infections. The miRNA-derived biomarkers offer a potent therapeutic option, being highly stable, specific, and resistant to several RNase III, making them an eligible biomarker for early disease detection. Computational and in vitro studies have manifested dynamics of miRNA expression contesting against the virus as a double-edged sword where it could potentially refrain and function as a proviral molecule during the disease onset. In addition, we addressed the virus-encoded miRNA's ability to persist in the host body by triggering pro-inflammatory cytokines or altering host signaling pathways, mishandling the host immune system and resulting in immune suppression and pathogenesis. Next-generation miRNAs therapy is advancing with the concept of personalized medicine, which could be incorporated by conjugating miRNA with a viral or nonviral delivery vehicle. In addition to that, the comorbidities contained with the SARS-CoV-2 disorient the existing clinical practice. Apart from that is high time to switch our focus on the cross-kingdom RNA molecules (plant miRNAs) which could turn the perspective of modulating this chronic contagious disease to the next stage. The acknowledgment of small noncoding miRNAs as a promising curative agent has shed light on the path of gene therapy, thus ruling out lethal viral infections.
CRediT authorship contribution statement
Mamta Panda: Conceptualization, Data curation, Software analysis and Validation, Writing original draft.
Elora Kalita: Conceptualization, Data curation, Software analysis and Validation, Writing original draft.
Satyendra Singh: Conceptualization, Data curation, Software analysis and Validation, Writing original draft.
Ketan Kumar: Drafting the revised manuscript.
Abhishek Rao: Drafting the revised manuscript.
Vijay Kumar Prajapati: Conceptualization, Supervision, Writing original draft and finalizing draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
MP and EK are thankful to Central University for lab facilities. SS is thankful to Central University of Rajasthan for fellowship. VKP is thankful to the Central University of Rajasthan for providing lab facility.
References
- 1.Gaunt E.R., Hardie A., Claas E.C., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Callaway E. The coronavirus is mutating - does it matter? Nature. 2020;585:174–177. doi: 10.1038/d41586-020-02544-6. [DOI] [PubMed] [Google Scholar]
- 3.Seyedpour S., Khodaei B., Loghman A.H., Seyedpour N., Kisomi M.F., Balibegloo M., et al. Targeted therapy strategies against SARS-CoV-2 cell entry mechanisms: a systematic review of in vitro and in vivo studies. J. Cell. Physiol. 2021;236:2364–2392. doi: 10.1002/jcp.30032. [DOI] [PubMed] [Google Scholar]
- 4.Lu T.X., Rothenberg M.E. MicroRNA. J. Allergy Clin. Immunol. 2018;141:1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demongeot J., Seligmann H. SARS-CoV-2 and miRNA-like inhibition power. Med. Hypotheses. 2020;144 doi: 10.1016/j.mehy.2020.110245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow J.T.-S., Salmena L. Prediction and analysis of SARS-CoV-2-targeting MicroRNA in human lung epithelium. Genes. 2020;11:1002. doi: 10.3390/genes11091002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO, n.d.WHO. WHO Weekly Report (Situation Report).12.n.d.
- 8.Pontecorvi G., Bellenghi M., Ortona E., Carè A. microRNAs as new possible actors in gender disparities of COVID-19 pandemic. Acta Physiol (Oxf). 2020;230 doi: 10.1111/apha.13538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182 doi: 10.1016/j.cell.2020.06.043. 812-27.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tahir M. Coronavirus genomic nsp14-ExoN, structure, role, mechanism, and potential application as a drug target. J. Med. Virol. 2021;93:4258–4264. doi: 10.1002/jmv.27009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chams N., Chams S., Badran R., Shams A., Araji A., Raad M., et al. Vol. 383. 2020. COVID-19: a multidisciplinary review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 13.Chams N., Chams S., Badran R., Shams A., Araji A., Raad M., et al. COVID-19: a multidisciplinary review. Front. Public Health. 2020;8:383. doi: 10.3389/fpubh.2020.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassan S.S., Lundstrom K., Serrano-Aroca Á., Adadi P., Aljabali A.A., Redwan E.M., et al. Emergence of unique SARS-CoV-2 ORF10 variants and their impact on protein structure and function. Int. J. Biol. Macromol. 2022;194:128–143. doi: 10.1016/j.ijbiomac.2021.11.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirabara S.M., Serdan T.D., Gorjao R., Masi L.N., Pithon-Curi T.C., Covas D.T., et al. SARS-COV-2 variants: differences and potential of immune evasion. Front. Cell. Infect. Microbiol. 2022:1401. doi: 10.3389/fcimb.2021.781429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hulswit R.J., de Haan C.A., Bosch B.J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashemian S.M., Shafigh N., Afzal G., Jamaati H., Tabarsi P., Marjani M., et al. Plasmapheresis reduces cytokine and immune cell levels in COVID-19 patients with acute respiratory distress syndrome (ARDS) Pulmonology. 2021;27:486–492. doi: 10.1016/j.pulmoe.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 22.Ratti M., Lampis A., Ghidini M., Salati M., Mirchev M.B., Valeri N., et al. MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as new tools for cancer therapy: first steps from bench to bedside. Target. Oncol. 2020;15:261–278. doi: 10.1007/s11523-020-00717-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., et al. Molecular basis for the recognition of primary microRNAs by the drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro J.S., Langlois R.A., Pham A.M., Tenoever B.R. Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA. 2012;18:1338–1346. doi: 10.1261/rna.032268.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Havens M.A., Reich A.A., Duelli D.M., Hastings M.L. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012;40:4626–4640. doi: 10.1093/nar/gks026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verma P., Pandey R.K., Prajapati P., Prajapati V.K. Vol. 7. 2016. Circulating microRNAs: potential and emerging biomarkers for diagnosis of human infectious diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B., Li J., Cairns M.J. Identifying miRNAs, targets and functions. Brief. Bioinform. 2014;15:1–19. doi: 10.1093/bib/bbs075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girardi E., López P., Pfeffer S. On the importance of host microRNAs during viral infection. Front. Genet. 2018;9:439. doi: 10.3389/fgene.2018.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabekkodu S.P., Shukla V., Varghese V.K., J D.S., Chakrabarty S., Satyamoorthy K. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. Camb. Philos. Soc. 2018;93:1955–1986. doi: 10.1111/brv.12428. [DOI] [PubMed] [Google Scholar]
- 30.Titov I.I., Vorozheykin P.S. Comparing miRNA structure of mirtrons and non-mirtrons. BMC Genomics. 2018;19:114. doi: 10.1186/s12864-018-4473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saliminejad K., Khorram Khorshid H.R., Soleymani Fard S., Ghaffari S.H. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019;234:5451–5465. doi: 10.1002/jcp.27486. [DOI] [PubMed] [Google Scholar]
- 32.Chauhan N., Jaggi M., Chauhan S.C., Yallapu M.M. COVID-19: fighting the invisible enemy with microRNAs. Expert Rev. Anti-Infect. Ther. 2021;19:137–145. doi: 10.1080/14787210.2020.1812385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chugh P., Dittmer D.P. Potential pitfalls in microRNA profiling. Vol. 3. Wiley Interdisciplinary Reviews: RNA; 2012. pp. 601–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.HQ McLean, Belongia E.A., Kieke B.A., Meece J.K., Fry A.M. Open Forum Infectious Diseases. Oxford University Press; 2015. Impact of late oseltamivir treatment on influenza symptoms in the outpatient setting: results of a randomized trial. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bala S., Petrasek J., Mundkur S., Catalano D., Levin I., Ward J., et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Vol. 56. 2012. pp. 1946–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turchinovich A., Samatov T.R., Tonevitsky A.G., Burwinkel BJFig. Circulating miRNAs: cell–cell communication function? Vol. 4. 2013. p. 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grasselli G., Cattaneo E., Florio G., Ippolito M., Zanella A., Cortegiani A., et al. Mechanical ventilation parameters in critically ill COVID-19 patients: a scoping review. Crit. Care. 2021;25:115. doi: 10.1186/s13054-021-03536-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ojha R., Nandani R., Chatterjee N., Prajapati V.K.J.C.R. Emerging role of circular RNAs as potential biomarkers for the diagnosis of human diseases. 2018. pp. 141–157. [DOI] [PubMed] [Google Scholar]
- 39.Bautista-Becerril B., Pérez-Dimas G., Sommerhalder-Nava P.C., Hanono A., Martínez-Cisneros J.A., Zarate-Maldonado B., et al. miRNAs, from evolutionary junk to possible prognostic markers and therapeutic targets in COVID-19. Vol. 14. 2021. p. 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khan M., Sany M., Us R., Islam M., Islam ABMMJFig. Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. 2020. p. 765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Usme-Ciro J.A., Campillo-Pedroza N., Almazán F., Gallego-Gomez JCJVj. Cytoplasmic RNA viruses as potential vehicles for the delivery of therapeutic small RNAs. Vol. 10. 2013. pp. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Havens M.A., Reich A.A., Duelli D.M., Hastings MLJNar. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Vol. 40. 2012. pp. 4626–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh D., Mathews M.B., Mohr IJCSHpib. Vol. 5. 2013. Tinkering with translation: protein synthesis in virus-infected cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Demirci M.D.S., Adan A.J.P. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. Vol. 8. 2020. p. e9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao J., Xiao S., Xiao Y., Wang X., Zhang C., Zhao Q., et al. MYH9 is an essential factor for porcine reproductive and respiratory syndrome virus infection. Vol. 6. 2016. pp. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Z., Wang J., Xu Y., Guo M., Mi K., Xu R., et al. 2020. Implications of the virus-encoded miRNA and host miRNA in the pathogenicity of SARS-CoV-2. [Google Scholar]
- 47.Benvenuto D., Angeletti S., Giovanetti M., Bianchi M., Pascarella S., Cauda R., et al. Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. Vol. 81. 2020. pp. e24–e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martines R.B., Ritter J.M., Matkovic E., Gary J., Bollweg B.C., Bullock H., et al. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease,United States. Vol. 26. 2020. p. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demongeot J., Seligmann H.J.M.H. Vol. 144. 2020. SARS-CoV-2 and miRNA-like inhibition power. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arisan E.D., Dart A., Grant G.H., Arisan S., Cuhadaroglu S., Lange S., et al. The prediction of miRNAs in SARS-CoV-2 genomes: hsa-miR databases identify 7 key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Vol. 12. 2020. p. 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pawlica P., Yario T.A., White S., Wang J., Moss W.N., Hui P., et al. Vol. 118. 2021. SARS-CoV-2 expresses a microRNA-like small RNA able to selectively repress host genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meng F., Siu G.K.-H., Mok B.W.-Y., Sun J., Fung K.S., Lam J.Y.-W., et al. Viral microRNAs encoded by nucleocapsid gene of SARS-CoV-2 are detected during infection, and targeting metabolic pathways in host cells. Vol. 10. 2021. p. 1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu T.-Y., Chen M., Wang C.-DJJoES. Vol. 19. 2021. Technology. Annotation of miRNAs in COVID-19 coronavirus. [Google Scholar]
- 54.Raman C., Ren C., Boas S., Sestero C.M., Chellappan R., Kesterson R.A., et al. TCR signaling strength dependent regulation of T cell proliferation, survival and Th differentiation by TGF-βR3 (betaglycn) Am. Assoc. Immnol. 2017:201–208. [Google Scholar]
- 55.Saini S., Saini A., Thakur C.J., Kumar V., Gupta R.D., Sharma JKJMbrc. Genome-wide computational prediction of miRNAs in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) revealed target genes involved in pulmonary vasculature and antiviral innate immunity. Vol. 9. 2020. p. 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao D., Mikosz A.M., Ringsby A.J., Anderson K.C., Beatman E.L., Koike K., et al. MicroRNA-126-3p inhibits angiogenic function of human lung microvascular endothelial cells via LAT1 (L-type amino acid transporter 1)-mediated mTOR (mammalian target of rapamycin) signaling. Vol. 40. 2020. pp. 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arisan E.D., Dart A., Grant G.H., Arisan S., Cuhadaroglu S., Lange S., et al. The prediction of miRNAs in SARS-CoV-2 genomes: hsa-miR databases identify 7 key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses. 2020;12:614. doi: 10.3390/v12060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benvenuto D., Angeletti S., Giovanetti M., Bianchi M., Pascarella S., Cauda R., et al. Evolutionary analysis of SARS-CoV-2: how mutation of non-structural protein 6 (NSP6) could affect viral autophagy. J. Infect. 2020;81:e24–e27. doi: 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhavya B., Pathak E., Mishra R. 2022. Endogenous pancreatic microRNAs differentially target the Delta, Omicron, and Wuhan SARS-CoV-2 genomes to upregulate the diabetes-associated genes. bioRxiv. [Google Scholar]
- 60.Cao D., Mikosz A.M., Ringsby A.J., Anderson K.C., Beatman E.L., Koike K., et al. MicroRNA-126-3p inhibits angiogenic function of human lung microvascular endothelial cells via LAT1 (L-type amino acid transporter 1)-mediated mTOR (mammalian target of rapamycin) signaling. Arterioscler. Thromb. Vasc. Biol. 2020;40:1195–1206. doi: 10.1161/ATVBAHA.119.313800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Condrat C.E., Thompson D.C., Barbu M.G., Bugnar O.L., Boboc A., Cretoiu D., et al. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells. 2020;9:276. doi: 10.3390/cells9020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Demirci M.D.S., Adan A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ. 2020;8 doi: 10.7717/peerj.9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Demongeot J., Seligmann H. SARS-CoV-2 and miRNA-like inhibition power. Med. Hypotheses. 2020;144 doi: 10.1016/j.mehy.2020.110245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fan Z., Zhang Q., Chen H., He P., Li Y., Si M., et al. Circulating microRNAs as a biomarker to predict therapy efficacy in hepatitis C patients with different genotypes. Microb. Pathog. 2017;112:320–326. doi: 10.1016/j.micpath.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 65.Farr R.J., Rootes C.L., Stenos J., Foo C.H., Cowled C., Stewart C.R. Detection of SARS-CoV-2 infection by microRNA profiling of the upper respiratory tract. Plos one. 2022;17 doi: 10.1371/journal.pone.0265670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao J., Xiao S., Xiao Y., Wang X., Zhang C., Zhao Q., et al. MYH9 is an essential factor for porcine reproductive and respiratory syndrome virus infection. Sci. Rep. 2016;6:1–13. doi: 10.1038/srep25120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Havens M.A., Reich A.A., Duelli D.M., Hastings M.L. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012;40:4626–4640. doi: 10.1093/nar/gks026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Z., Wang J., Xu Y., Guo M., Mi K., Xu R., et al. 2020. Implications of the virus-encoded miRNA and host miRNA in the pathogenicity of SARS-CoV-2. arXiv preprint arXiv:200404874. [Google Scholar]
- 69.Mangukia N., Rao P., Patel K., Pandya H., Rawal R.M. Unveiling the nature's fruit basket to computationally identify Citrus sinensis csi-mir169–3p as a probable plant miRNA against reference and omicron SARS-CoV-2 genome. Comput. Biol. Med. 2022;105502 doi: 10.1016/j.compbiomed.2022.105502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martines R.B., Ritter J.M., Matkovic E., Gary J., Bollweg B.C., Bullock H., et al. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease,United States. Emerg. Infect. Dis. 2020;26:2005. doi: 10.3201/eid2609.202095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meng F., Siu G.K.-H., Mok B.W.-Y., Sun J., Fung K.S., Lam J.Y.-W., et al. Viral microRNAs encoded by nucleocapsid gene of SARS-CoV-2 are detected during infection, and targeting metabolic pathways in host cells. Cells. 2021;10:1762. doi: 10.3390/cells10071762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pawlica P., Yario T.A., White S., Wang J., Moss W.N., Hui P., et al. SARS-CoV-2 expresses a microRNA-like small RNA able to selectively repress host genes. Proc. Natl. Acad. Sci. 2021;118 doi: 10.1073/pnas.2116668118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rezk N., Elsayed Sileem A., Gad D., Khalil A.O. miRNA-223-3p, miRNA-2909 and cytokines expression in COVID-19 patients treated with ivermectin. Zagazig Univ.Med.J. 2022;28:573–582. [Google Scholar]
- 74.Saini S., Saini A., Thakur C.J., Kumar V., Gupta R.D., Sharma J.K. Genome-wide computational prediction of miRNAs in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) revealed target genes involved in pulmonary vasculature and antiviral innate immunity. Mol.Biol.Res.Commun. 2020;9:83. doi: 10.22099/mbrc.2020.36507.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Usme-Ciro J.A., Campillo-Pedroza N., Almazán F., Gallego-Gomez J.C. Cytoplasmic RNA viruses as potential vehicles for the delivery of therapeutic small RNAs. Virol. J. 2013;10:1–10. doi: 10.1186/1743-422X-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walsh D., Mathews M.B., Mohr I. Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ramchandran R., Chaluvally-Raghavan P. miRNA-mediated RNA activation in mammalian cells. Adv. Exp. Med. Biol. 2017;983:81–89. doi: 10.1007/978-981-10-4310-9_6. [DOI] [PubMed] [Google Scholar]
- 78.Khan M.A., Sany M.R.U., Islam M.S., Islam A. Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front. Genet. 2020;11:765. doi: 10.3389/fgene.2020.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trobaugh D.W., Klimstra W.B. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol. Med. 2017;23:80–93. doi: 10.1016/j.molmed.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Centa A., Fonseca A.S., Ferreira S., Azevedo M.L.V., Vaz de Paula C.B., Nagashima S., et al. Deregulated miRNA expression is associated with endothelial dysfunction in post-mortem lung biopsies of COVID-19 patients. Am. J. Physiol. Lung Cell Mol. Physiol. 2020;320:L405–L412. doi: 10.1152/ajplung.00457.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang H., Gao Y., Li Z., Miao Y., Huang Z., Liu X., et al. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin. Transl.Med. 2020;10 doi: 10.1002/ctm2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McDonald J.T., Enguita F.J., Taylor D., Griffin R.J., Priebe W., Emmett M.R., et al. Role of miR-2392 in driving SARS-CoV-2 infection. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matarese A., Gambardella J., Sardu C., Santulli G. miR-98 regulates TMPRSS2 expression in human endothelial cells: key implications for COVID-19. Biomedicines. 2020;8:462. doi: 10.3390/biomedicines8110462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nersisyan S., Engibaryan N., Gorbonos A., Kirdey K., Makhonin A., Tonevitsky A. Potential role of cellular miRNAs in coronavirus-host interplay. PeerJ. 2020;8 doi: 10.7717/peerj.9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Arghiani N., Nissan T., Matin M.M. Role of microRNAs in COVID-19 with implications for therapeutics. Biomed. Pharmacother. 2021;144 doi: 10.1016/j.biopha.2021.112247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sardar R., Satish D., Birla S., Gupta D. Integrative analyses of SARS-CoV-2 genomes from different geographical locations reveal unique features potentially consequential to host-virus interaction, pathogenesis and clues for novel therapies. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e04658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Khan M., Sany M., Us R., Islam M., Islam A.B.M.M. Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front. Genet. 2020;765 doi: 10.3389/fgene.2020.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen L., Zhong L. Genomics functional analysis and drug screening of SARS-CoV-2. Genes Dis. 2020;7:542–550. doi: 10.1016/j.gendis.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cong Y., Ulasli M., Schepers H., Mauthe M., V'Kovski P., Kriegenburg F., et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 2020;94 doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cullen B.R. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 2011;25:1881–1894. doi: 10.1101/gad.17352611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen L., Zhong L.J.G. diseases. Genomics functional analysis and drug screening of SARS-CoV-2. Vol. 7. 2020. pp. 542–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mirzaei R., Mahdavi F., Badrzadeh F., Hosseini-Fard S.R., Heidary M., Jeda A.S., et al. The emerging role of microRNAs in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Int. Immunopharmacol. 2021;90 doi: 10.1016/j.intimp.2020.107204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ivanisenko N.V., Seyrek K., Kolchanov N.A., Ivanisenko V.A., Lavrik I.N. The role of death domain proteins in host response upon SARS-CoV-2 infection: modulation of programmed cell death and translational applications. Cell Death Discov. 2020;6:101. doi: 10.1038/s41420-020-00331-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ramaiah M.J. mTOR inhibition and p53 activation, microRNAs: the possible therapy against pandemic COVID-19. Gene Rep. 2020;20 doi: 10.1016/j.genrep.2020.100765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fulzele S., Sahay B., Yusufu I., Lee T.J., Sharma A., Kolhe R., et al. COVID-19 virulence in aged patients might be impacted by the host cellular microRNAs abundance/profile. Aging Dis. 2020;11:509–522. doi: 10.14336/AD.2020.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Centa A., Fonseca A.S., da Silva Ferreira S.G., Azevedo M.L.V., De Paula C.B.V., Nagashima S., et al. Deregulated miRNA expression is associated with endothelial dysfunction in post-mortem lung biopsies of COVID-19 patients. Am. J. Phys. Lung Cell. Mol. Phys. 2021;320:L405–L412. doi: 10.1152/ajplung.00457.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen L., Zhong L. Genomics functional analysis and drug screening of SARS-CoV-2. GenesDis. 2020;7:542–550. doi: 10.1016/j.gendis.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cong Y., Ulasli M., Schepers H., Mauthe M., V'kovski P., Kriegenburg F., et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J.Virol. 2020;94 doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cullen B.R. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 2011;25:1881–1894. doi: 10.1101/gad.17352611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ivanisenko N.V., Seyrek K., Kolchanov N.A., Ivanisenko V.A., Lavrik I.N. The role of death domain proteins in host response upon SARS-CoV-2 infection: modulation of programmed cell death and translational applications. Cell Death Discov. 2020;6:1–10. doi: 10.1038/s41420-020-00331-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sanyaolu A., Okorie C., Marinkovic A., Patidar R., Younis K., Desai P., et al. Comorbidity and its impact on patients with COVID-19. SN Compr.Clin.Med. 2020;2:1069–1076. doi: 10.1007/s42399-020-00363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Giza D.E., Vasilescu C., Calin G.A. Key principles of miRNA involvement in human diseases. Discoveries. 2014;2 doi: 10.15190/d.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kumar A., Arora A., Sharma P., Anikhindi S.A., Bansal N., Singla V., et al. Is diabetes mellitus associated with mortality and severity of COVID-19? A meta-analysis. Diabetes Metab. Syndr. Clin. Res. Rev. 2020;14:535–545. doi: 10.1016/j.dsx.2020.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Corona G., Pizzocaro A., Vena W., Rastrelli G., Semeraro F., Isidori A.M., et al. Diabetes is most important cause for mortality in COVID-19 hospitalized patients: systematic review and meta-analysis. Rev.Endocr.Metab.Disord. 2021;22:275–296. doi: 10.1007/s11154-021-09630-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pathak E., Mishra R. Deciphering the link between diabetes mellitus and SARS-CoV-2 infection through differential targeting of microRNAs in the human pancreas. J. Endocrinol. Investig. 2022;45:537–550. doi: 10.1007/s40618-021-01693-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mishra P.K., Tandon R., Byrareddy S.N. Diabetes and COVID-19 risk: an miRNA perspective. Am. J. Phys. Heart Circ. Phys. 2020;319:H604–H609. doi: 10.1152/ajpheart.00489.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang Y., Zhu X., Jiang X.M., Guo J., Fu Z., Zhou Z., et al. Decreased inhibition of exosomal miRNAs on SARS-CoV-2 replication underlies poor outcomes in elderly people and diabetic patients. Signal Transduct. Target. Ther. 2021;6:300. doi: 10.1038/s41392-021-00716-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roganović J. Downregulation of microRNA-146a in diabetes, obesity and hypertension may contribute to severe COVID-19. Med. Hypotheses. 2021;146 doi: 10.1016/j.mehy.2020.110448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Widiasta A., Sribudiani Y., Nugrahapraja H., Hilmanto D., Sekarwana N., Rachmadi D. Potential role of ACE2-related microRNAs in COVID-19-associated nephropathy. Non-coding RNA Res. 2020;5:153–166. doi: 10.1016/j.ncrna.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Safdar M., Khan M.S., Karim A.Y., Omar S.A., Smail S.W., Saeed M., et al. SNPs at 3′ UTR of APOL1 and miR-6741-3p target sites associated with kidney diseases more susceptible to SARS-COV-2 infection: in silico and in vitro studies. Mamm. Genome. 2021;32:389–400. doi: 10.1007/s00335-021-09880-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Srivastava S.P., Srivastava R., Chand S., Goodwin J.E. Coronavirus disease (COVID)-19 and diabetic kidney disease. Pharmaceuticals. 2021;14:751. doi: 10.3390/ph14080751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McDonald J.T., Enguita F.J., Taylor D., Griffin R.J., Priebe W., Emmett M.R., et al. Role of miR-2392 in driving SARS-CoV-2 infection. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Siniscalchi C., Di Palo A., Russo A., Potenza N. Human microRNAs interacting with SARS-CoV-2 RNA sequences: computational analysis and experimental target validation. Front. Genet. 2021;12:760. doi: 10.3389/fgene.2021.678994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Garg A., Seeliger B., Derda A.A., Xiao K., Gietz A., Scherf K., et al. Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Eur. J. Heart Fail. 2021;23:468–475. doi: 10.1002/ejhf.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Giuliani A., Matacchione G., Ramini D., Di Rosa M., Bonfigli A.R., Sabbatinelli J., et al. Circulating miR-320b and miR-483-5p levels are associated with COVID-19 in-hospital mortality. Mech. Ageing Dev. 2022;202 doi: 10.1016/j.mad.2022.111636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Keikha R., Hashemi-Shahri S.M., Jebali A. The relative expression of miR-31, miR-29, miR-126, and miR-17 and their mRNA targets in the serum of COVID-19 patients with different grades during hospitalization. Eur. J. Med. Res. 2021;26:1–8. doi: 10.1186/s40001-021-00544-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gutmann C., Khamina K., Theofilatos K., Diendorfer A.B., Burnap S.A., Nabeebaccus A., et al. Association of cardiometabolic microRNAs with COVID-19 severity and mortality. Cardiovasc. Res. 2022;118:461–474. doi: 10.1093/cvr/cvab338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wastnedge E.A., Reynolds R.M., Van Boeckel S.R., Stock S.J., Denison F.C., Maybin J.A., et al. Pregnancy and COVID-19. Physiol. Rev. 2021;101:303–318. doi: 10.1152/physrev.00024.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Saulle I., Garziano M., Fenizia C., Cappelletti G., Parisi F., Clerici M., et al. MiRNA profiling in plasma and placenta of SARS-CoV-2-infected pregnant women. Cells. 2021;10:1788. doi: 10.3390/cells10071788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Abel T., Moodley J., Naicker T. The involvement of microRNAs in SARS-CoV-2 infection comorbid with HIV-associated preeclampsia. Curr. Hypertens. Rep. 2021;23:1–13. doi: 10.1007/s11906-021-01138-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang J., Wang F., Argyris E., Chen K., Liang Z., Tian H., et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 122.Levi M., Thachil J., Iba T., Levy J.H. Coagulation abnormalities and thrombosis in patients withCOVID-19. Vol. 7. The Lancet Haematology; 2020. pp. e438–e440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gambardella J., Sardu C., Morelli M.B., Messina V., Marfella R., Maggi P., et al. Exosomal microRNAs drive tromboembolism in COVID-19. Vol. 142. 2020. p. A221-A. [Google Scholar]
- 124.Idell S., Zwieb C., Boggaram J., Holiday D., Johnson A.R., Raghu GJAJoP-L.C., et al. Mechanisms of fibrin formation and lysis by human lung fibroblasts: influence of TGF-beta and TNF-alpha. Vol. 263. 1992. pp. L487–L494. [DOI] [PubMed] [Google Scholar]
- 125.Khurana P., Gupta A., Sugadev R., Sharma Y., Varshney R., Ganju L., et al. nSARS-Cov-2, pulmonary edema and thrombosis: possible molecular insights using miRNA-gene circuits in regulatory networks. Vol. 2. 2020. pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bautista-Becerril B., Pérez-Dimas G., Sommerhalder-Nava P.C., Hanono A., Martínez-Cisneros J.A., Zarate-Maldonado B., et al. miRNAs, from evolutionary junk to possible prognostic markers and therapeutic targets in COVID-19. Viruses. 2021;14 doi: 10.3390/v14010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Whyte C.S., Mutch NJJh. uPA-mediated plasminogen activation is enhanced by polyphosphate. Vol. 106. 2021. p. 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Parasher AJPmj. COVID-19: current understanding of its pathophysiology, clinical presentation and treatment. Vol. 97. 2021. pp. 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chow J.T., Salmena L. Prediction and analysis of SARS-CoV-2-targeting MicroRNA in human lung epithelium. Genes. 2020;11 doi: 10.3390/genes11091002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ferruelo A., Peñuelas Ó., Lorente JaJAotm. Vol. 6. 2018. MicroRNAs as biomarkers of acute lung injury. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mahesh G., Biswas RJJoI, Research C. MicroRNA-155: a master regulator of inflammation. Vol. 39. 2019. pp. 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Knarr M., Avelar R.A., Sekhar S.C., Kwiatkowski L.J., Dziubinski M.L., McAnulty J., et al. miR-181a initiates and perpetuates oncogenic transformation through the regulation of innate immune signaling. Vol. 11. 2020. pp. 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pierce J.B., Simion V., Icli B., Pérez-Cremades D., Cheng H.S., Feinberg M.W.J.G. Computational analysis of targeting SARS-CoV-2, viral entry proteins ACE2 and TMPRSS2, and interferon genes by host microRNAs. Vol. 11. 2020. p. 1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mirzaei R., Mahdavi F., Badrzadeh F., Hosseini-Fard S.R., Heidary M., Jeda A.S., et al. Vol. 90. 2021. The emerging role of microRNAs in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang S., Li Z., Chen Q., Wang L., Zheng J., Lin Z., et al. NF-κB-induced microRNA-211 inhibits interleukin-10 in macrophages of rats with lipopolysaccharide-induced acute respiratory distress syndrome. Vol. 45. 2018. pp. 332–342. [DOI] [PubMed] [Google Scholar]
- 136.Desforges M., Le Coupanec A., Dubeau P., Bourgouin A., Lajoie L., Dubé M., et al. Human coronaviruses and other respiratory viruses: underestimated opportunistic pathogens of the central nervous system? Vol. 12. 2019. p. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kim T., Mehta S.L., Morris-Blanco K.C., Chokkalla A.K., Chelluboina B., Lopez M., et al. The microRNA miR-7a-5p ameliorates ischemic brain damage by repressing α-synuclein. 2018;11 doi: 10.1126/scisignal.aat4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mone P., Gambardella J., Wang X., Jankauskas S.S., Matarese A., Santulli G.J.R.S. miR-24 targets SARS-CoV-2 co-factor Neuropilin-1 in human brain microvascular endothelial cells: Insights for COVID-19 neurological manifestations. 2021. pp. rs. 3–rs-192099. [Google Scholar]
- 139.Keikha R., Hashemi-Shahri S., Jebali A.J.N. 2021. The miRNA neuroinflammatory biomarkers in COVID-19 patients with different severity of illness. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Papannarao J.B., Schwenke D., Manning P.J., Katare RJm. 2021. Upregulated miR-200c may increase the risk of obese individuals to severe COVID-19. [Google Scholar]
- 141.Karkeni E., Astier J., Tourniaire F., El Abed M., Romier B., Gouranton E., et al. Obesity-associated inflammation induces microRNA-155 expression in adipocytes and adipose tissue: outcome on adipocyte function. Vol. 101. 2016. pp. 1615–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kokai L.E., Marra K., Rubin J.P.J.T.R. Adipose stem cells: biology and clinical applications for tissue repair and regeneration. Vol. 163. 2014. pp. 399–408. [DOI] [PubMed] [Google Scholar]
- 143.Mormile RJAomr. 2021. Obesity and severe COVID-19 in the young: is downregulation of miR-126 a piece of the SARS-COV2 pathogenicity puzzle? In memory of my Dad Sossio Mormile. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gutmann C., Khamina K., Theofilatos K., Diendorfer A.B., Burnap S.A., Nabeebaccus A., et al. Association of cardiometabolic microRNAs with COVID-19 severity and mortality. Cardiovasc. Res. 2021;118:461–474. doi: 10.1093/cvr/cvab338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Willeit P., Skroblin P., Kiechl S., Fernandez-Hernando C., Mayr MJEhj. Liver microRNAs: potential mediators and biomarkers for metabolic and cardiovascular disease? Vol. 37. 2016. pp. 3260–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kojima Y., Tsuchiya A., Ogawa M., Nojiri S., Takeuchi S., Watanabe T., et al. Mesenchymal stem cells cultured under hypoxic conditions had a greater therapeutic effect on mice with liver cirrhosis compared to those cultured under normal oxygen conditions. Regen.Ther. 2019;11:269–281. doi: 10.1016/j.reth.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McDonald J.T., Enguita F.J., Taylor D., Griffin R.J., Priebe W., Emmett M.R., et al. Role of miR-2392 in driving SARS-CoV-2 infection. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gaddam R.R., Jacobsen V.P., Kim Y.-R., Gabani M., Jacobs J.S., Dhuri K., et al. Microbiota-governed microRNA-204 impairs endothelial function and blood pressure decline during inactivity in db/db mice. Vol. 10. 2020. pp. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vikram A., Kim Y.-R., Kumar S., Li Q., Kassan M., Jacobs J.S., et al. Vascular microRNA-204 is remotely governed by the microbiome and impairs endothelium-dependent vasorelaxation by downregulating Sirtuin1. Nat. Commun. 2016;7:1–9. doi: 10.1038/ncomms12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Austin TbM. First microRNA mimic enters clinic. Nat. Biotechnol. 2013;31:577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- 151.Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 152.Gallant-Behm C.L., Piper J., Lynch J.M., Seto A.G., Hong S.J., Mustoe T.A., et al. A microRNA-29 mimic (Remlarsen) represses extracellular matrix expression and fibroplasia in the skin. J. Investig. Dermatol. 2019;139:1073–1081. doi: 10.1016/j.jid.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 153.Hong D.S., Kang Y.-K., Borad M., Sachdev J., Ejadi S., Lim H.Y., et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer. 2020;122:1630–1637. doi: 10.1038/s41416-020-0802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu G., Min H., Yue S., Chen C.-Z. Pre-miRNA loop nucleotides control the distinct activities of mir-181a-1 and mir-181c in early T cell development. PloS one. 2008;3 doi: 10.1371/journal.pone.0003592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Trujillo R.D., Yue S.B., Tang Y., O'Gorman W.E., Chen C.Z. The potential functions of primary microRNAs in target recognition and repression. EMBO J. 2010;29:3272–3285. doi: 10.1038/emboj.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jafari N., Abediankenari S., Hossein-Nataj H. miR-34a mimic or pre-mir-34a, which is the better option for cancer therapy? KatoIII as a model to study miRNA action in human gastric cancer cells. Cancer Cell Int. 2021;21:1–15. doi: 10.1186/s12935-021-01872-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shan G., Li Y., Zhang J., Li W., Szulwach K.E., Duan R., et al. A small molecule enhances RNA interference and promotes microRNA processing. Nat. Biotechnol. 2008;26:933–940. doi: 10.1038/nbt.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Young D.D., Connelly C.M., Grohmann C., Deiters A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J. Am. Chem. Soc. 2010;132:7976–7981. doi: 10.1021/ja910275u. [DOI] [PubMed] [Google Scholar]
- 159.Dhuri K., Bechtold C., Quijano E., Pham H., Gupta A., Vikram A., et al. Antisense oligonucleotides: an emerging area in drug discovery and development. J. Clin. Med. 2020;9:2004. doi: 10.3390/jcm9062004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Verma N.K., Fazil M.H.U.T., Duggan S.P., Kelleher D. Combination therapy using inhalable GapmeR and recombinant ACE2 for COVID-19. Front. Mol. Biosci. 2020;7:197. doi: 10.3389/fmolb.2020.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dragulescu-Andrasi A., Rapireddy S., Frezza B.M., Gayathri C., Gil R.R., Ly D.H. A simple γ-backbone modification preorganizes peptide nucleic acid into a helical structure. J. Am. Chem. Soc. 2006;128:10258–10267. doi: 10.1021/ja0625576. [DOI] [PubMed] [Google Scholar]
- 162.Swenson C.S., Heemstra J.M. Peptide nucleic acids harness dual information codes in a single molecule. Chem. Commun. 2020;56:1926–1935. doi: 10.1039/c9cc09905k. [DOI] [PubMed] [Google Scholar]
- 163.Zhou P., Wang M., Du L., Fisher G.W., Waggoner A., Ly D.H. Novel binding and efficient cellular uptake of guanidine-based peptide nucleic acids (GPNA) J. Am. Chem. Soc. 2003;125:6878–6879. doi: 10.1021/ja029665m. [DOI] [PubMed] [Google Scholar]
- 164.Swayze E.E., Siwkowski A.M., Wancewicz E.V., Migawa M.T., Wyrzykiewicz T.K., Hung G., et al. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fazi F., Racanicchi S., Zardo G., Starnes L.M., Mancini M., Travaglini L., et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 166.Nan Y., Zhang Y.J. Antisense phosphorodiamidate morpholino oligomers as novel antiviral compounds. Front. Microbiol. 2018;9:750. doi: 10.3389/fmicb.2018.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhu C., Lee J.Y., Woo J.Z., Xu L., Nguyenla X., Yamashiro L.H., et al. 2021. An intranasal ASO therapeutic targeting SARS-CoV-2. bioRxiv. 2021.05.17.444397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ebert M.S., Sharp P.A. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 170.Liu Q., Du J., Yu X., Xu J., Huang F., Li X., et al. miRNA-200c-3p is crucial in acute respiratory distress syndrome. Cell Discov. 2017;3:17021. doi: 10.1038/celldisc.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Otsuka M., Jing Q., Georgel P., New L., Chen J., Mols J., et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 172.Ostermann E., Tuddenham L., Macquin C., Alsaleh G., Schreiber-Becker J., Tanguy M., et al. Deregulation of type I IFN-dependent genes correlates with increased susceptibility to cytomegalovirus acute infection of dicer mutant mice. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang M., Gu B., Chen X., Wang Y., Li P., Wang K. The function and therapeutic potential of Epstein-Barr virus-encoded microRNAs in cancer. Mol. Ther. Nucleic Acids. 2019;17:657–668. doi: 10.1016/j.omtn.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dicerna Pharmaceuticals I . 2014. Phase Ib/2, multicenter, dose escalation study of DCR-MYC in patients with hepatocellular carcinoma. December 10. [Google Scholar]
- 175.Barbu M.G., Condrat C.E., Thompson D.C., Bugnar O.L., Cretoiu D., Toader O.D., et al. MicroRNA involvement in signaling pathways during viral infection. Front. Cell Dev. Biol. 2020;8:143. doi: 10.3389/fcell.2020.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kaur T., Kapila S., Kapila R., Kumar S., Upadhyay D., Kaur M., et al. Tmprss2 specific miRNAs as promising regulators for SARS-CoV-2 entry checkpoint. Virus Res. 2021;294 doi: 10.1016/j.virusres.2020.198275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Baumann V., Winkler J. miRNA-based therapies: strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med. Chem. 2014;6:1967–1984. doi: 10.4155/fmc.14.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.De Santi C., Fernández Fernández E., Gaul R., Vencken S., Glasgow A., Oglesby I.K., et al. Precise targeting of miRNA sites restores CFTR activity in CF bronchial epithelial cells. Mol. Ther. 2020;28:1190–1199. doi: 10.1016/j.ymthe.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Taschuk F., Cherry S. DEAD-box helicases: sensors, regulators, and effectors for antiviral defense. Viruses. 2020;12 doi: 10.3390/v12020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang X., Wang R., Luo M., Li C., Wang H.X., Huan C.C., et al. (DEAD)-box RNA helicase 3 modulates NF-κB signal pathway by controlling the phosphorylation of PP2A-C subunit. Oncotarget. 2017;8:33197–33213. doi: 10.18632/oncotarget.16593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tanner N.K., Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 182.Di Martino M.T., Leone E., Amodio N., Foresta U., Lionetti M., Pitari M.R., et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin. Cancer Res. 2012;18:6260–6270. doi: 10.1158/1078-0432.CCR-12-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mowa M.B., Crowther C., Arbuthnot P. Therapeutic potential of adenoviral vectors for delivery of expressed RNAi activators. Expert Opin. Drug Deliv. 2010;7:1373–1385. doi: 10.1517/17425247.2010.533655. [DOI] [PubMed] [Google Scholar]
- 184.Miyazaki Y., Adachi H., Katsuno M., Minamiyama M., Jiang Y.M., Huang Z., et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat. Med. 2012;18:1136–1141. doi: 10.1038/nm.2791. [DOI] [PubMed] [Google Scholar]
- 185.Ramanujam D., Sassi Y., Laggerbauer B., Engelhardt S. Viral vector-based targeting of miR-21 in cardiac nonmyocyte cells reduces pathologic remodeling of the heart. Mol. Ther. 2016;24:1939–1948. doi: 10.1038/mt.2016.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Koonin E.V., Senkevich T.G., Dolja V.V. The ancient virus world and evolution of cells. Biol. Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Xie J., Ameres S.L., Friedline R., Hung J.H., Zhang Y., Xie Q., et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat. Methods. 2012;9:403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sadelain M., Papapetrou E.P., Bushman F.D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer. 2011;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 189.Ylä-Herttuala S. Endgame: glybera finally recommended for approval as the first gene therapy drug in the European Union. Mol. Ther. 2012;20:1831–1832. doi: 10.1038/mt.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bai Z., Wei J., Yu C., Han X., Qin X., Zhang C., et al. Non-viral nanocarriers for intracellular delivery of microRNA therapeutics. J. Mater. Chem. B. 2019;7:1209–1225. doi: 10.1039/c8tb02946f. [DOI] [PubMed] [Google Scholar]
- 191.Boca S., Gulei D., Zimta A.A., Onaciu A., Magdo L., Tigu A.B., et al. Nanoscale delivery systems for microRNAs in cancer therapy. Cell. Mol. Life Sci. 2020;77:1059–1086. doi: 10.1007/s00018-019-03317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yang N. An overview of viral and nonviral delivery systems for microRNA. Int. J. Pharm. Investig. 2015;5:179–181. doi: 10.4103/2230-973X.167646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Guterres A., de Azeredo Lima C.H., Miranda R.L., Gadelha M.R. What is the potential function of microRNAs as biomarkers and therapeutic targets in COVID-19? Infect. Genet. Evol. 2020;85 doi: 10.1016/j.meegid.2020.104417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Akao Y., Iio A., Itoh T., Noguchi S., Itoh Y., Ohtsuki Y., et al. Microvesicle-mediated RNA molecule delivery system using monocytes/macrophages. Mol. Ther. 2011;19:395–399. doi: 10.1038/mt.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Badeau B.A., Comerford M.P., Arakawa C.K., Shadish J.A., DeForest C.A. Engineered modular biomaterial logic gates for environmentally triggered therapeutic delivery. Nat. Chem. 2018;10:251–258. doi: 10.1038/nchem.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Gonzalez H., Hagerling C., Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 2018;32:1267–1284. doi: 10.1101/gad.314617.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Muzykantov V.R. Drug delivery by red blood cells: vascular carriers designed by mother nature. Expert Opin. Drug Deliv. 2010;7:403–427. doi: 10.1517/17425241003610633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Neviani P., Wise P.M., Murtadha M., Liu C.W., Wu C.H., Jong A.Y., et al. Natural killer-derived exosomal miR-186 inhibits neuroblastoma growth and immune escape mechanisms. Cancer Res. 2019;79:1151–1164. doi: 10.1158/0008-5472.CAN-18-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Patente T.A., Pinho M.P., Oliveira A.A., Evangelista G.C.M., Bergami-Santos P.C., Barbuto J.A.M. Human dendritic cells: their heterogeneity and clinical application potential in cancer immunotherapy. Front. Immunol. 2018;9:3176. doi: 10.3389/fimmu.2018.03176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Soundara Rajan T., Gugliandolo A., Bramanti P., Mazzon E. In vitro-transcribed mRNA chimeric antigen receptor T cell (IVT mRNA CAR T) therapy in hematologic and solid tumor management: a preclinical update. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21186514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sun H., Shi K., Qi K., Kong H., Zhang J., Dai S., et al. Natural killer cell-derived exosomal miR-3607-3p inhibits pancreatic cancer progression by targeting IL-26. Front. Immunol. 2019;10:2819. doi: 10.3389/fimmu.2019.02819. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 202.Sun L., Fan F., Li R., Niu B., Zhu L., Yu S., et al. Different erythrocyte microRNA profiles in low- and high-altitude individuals. Front. Physiol. 2018;9:1099. doi: 10.3389/fphys.2018.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Surman M., Drożdż A., Stępień E., Przybyło M. Extracellular vesicles as drug delivery systems - methods of production and potential therapeutic applications. Curr. Pharm. Des. 2019;25:132–154. doi: 10.2174/1381612825666190306153318. [DOI] [PubMed] [Google Scholar]
- 204.Wang Y., Chen X., Tian B., Liu J., Yang L., Zeng L., et al. Nucleolin-targeted extracellular vesicles as a versatile platform for biologics delivery to breast cancer. Theranostics. 2017;7:1360–1372. doi: 10.7150/thno.16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wayne E.C., Long C., Haney M.J., Batrakova E.V., Leisner T.M., Parise L.V., et al. Targeted delivery of siRNA lipoplexes to cancer cells using macrophage transient horizontal gene transfer. Adv. Sci. (Weinh.) 2019;6 doi: 10.1002/advs.201900582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wculek S.K., Cueto F.J., Mujal A.M., Melero I., Krummel M.F., Sancho D. Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2020;20:7–24. doi: 10.1038/s41577-019-0210-z. [DOI] [PubMed] [Google Scholar]
- 207.Weber E.W., Maus M.V., Mackall C.L. The emerging landscape of immune cell therapies. Cell. 2020;181:46–62. doi: 10.1016/j.cell.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zhang S., Cheng Z., Wang Y., Han T. The risks of miRNA therapeutics: in a drug target perspective. Drug Des. Dev. Ther. 2021;15:721–733. doi: 10.2147/DDDT.S288859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Parayath N.N., Stephan S.B., Koehne A.L., Nelson P.S., Stephan M.T. In vitro-transcribed antigen receptor mRNA nanocarriers for transient expression in circulating T cells in vivo. Nat. Commun. 2020;11:6080. doi: 10.1038/s41467-020-19486-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Budak H., Bulut R., Kantar M., Alptekin B. MicroRNA nomenclature and the need for a revised naming prescription. Brief. Funct. Genomics. 2016;15:65–71. doi: 10.1093/bfgp/elv026. [DOI] [PubMed] [Google Scholar]
- 211.Zhang Q., Ding F., Liu X., Shen J., Su Y., Qian J., et al. Nanobody-guided targeted delivery of microRNA via nucleic acid nanogel to inhibit the tumor growth. J. Control. Release. 2020;328:425–434. doi: 10.1016/j.jconrel.2020.08.058. [DOI] [PubMed] [Google Scholar]
- 212.Budak H., Bulut R., Kantar M., Alptekin B. MicroRNA nomenclature and the need for a revised naming prescription. Brief.Funct.Genomics. 2016;15:65–71. doi: 10.1093/bfgp/elv026. [DOI] [PubMed] [Google Scholar]
- 213.Chin A.R., Fong M.Y., Somlo G., Wu J., Swiderski P., Wu X., et al. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016;26:217–228. doi: 10.1038/cr.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang K., Li H., Yuan Y., Etheridge A., Zhou Y., Huang D., et al. The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PloS one. 2012;7 doi: 10.1371/journal.pone.0051009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 216.McMahon B.M., Mays D., Lipsky J., Stewart J.A., Fauq A., Richelson E. Pharmacokinetics and tissue distribution of a peptide nucleic acid after intravenous administration. Antisense Nucleic Acid Drug Dev. 2002;12:65–70. doi: 10.1089/108729002760070803. [DOI] [PubMed] [Google Scholar]
- 217.Advani R., Lum B.L., Fisher G.A., Halsey J., Geary R.S., Holmlund J.T., et al. A phase I trial of aprinocarsen (ISIS 3521/LY900003), an antisense inhibitor of protein kinase C-alpha administered as a 24-hour weekly infusion schedule in patients with advanced cancer. Investig. New Drugs. 2005;23:467–477. doi: 10.1007/s10637-005-2906-0. [DOI] [PubMed] [Google Scholar]
- 218.Desai A.A., Schilsky R.L., Young A., Janisch L., Stadler W.M., Vogelzang N.J., et al. A phase I study of antisense oligonucleotide GTI-2040 given by continuous intravenous infusion in patients with advanced solid tumors. Ann. Oncol. 2005;16:958–965. doi: 10.1093/annonc/mdi178. [DOI] [PubMed] [Google Scholar]
- 219.Kahan B.D., Stepkowski S., Kilic M., Katz S.M., Van Buren C.T., Welsh M.S., et al. Phase I and phase II safety and efficacy trial of intercellular adhesion molecule-1 antisense oligodeoxynucleotide (ISIS 2302) for the prevention of acute allograft rejection. Transplantation. 2004;78:858–863. doi: 10.1097/01.tp.0000128857.77893.d2. [DOI] [PubMed] [Google Scholar]
- 220.Iannitti T., Morales-Medina J.C., Palmieri B. Phosphorothioate oligonucleotides: effectiveness and toxicity. Curr. Drug Targets. 2014;15:663–673. doi: 10.2174/1389450115666140321100304. [DOI] [PubMed] [Google Scholar]
- 221.Zhang Z., Qin Y.W., Brewer G., Jing Q. MicroRNA degradation and turnover: regulating the regulators. Wiley Interdiscip. Rev. RNA. 2012;3:593–600. doi: 10.1002/wrna.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Lennox K.A., Behlke M.A. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011;18:1111–1120. doi: 10.1038/gt.2011.100. [DOI] [PubMed] [Google Scholar]
- 223.Segal M., Biscans A., Gilles M.E., Anastasiadou E., De Luca R., Lim J., et al. Hydrophobically modified let-7b miRNA enhances biodistribution to NSCLC and downregulates HMGA2 in vivo. Mol. Ther. Nucleic Acids. 2020;19:267–277. doi: 10.1016/j.omtn.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Huang Y., Ma Y., Guo Y., Zou L., Jin H., Zhong L., et al. Exploring directional invasion of serum nuclease into siRNA duplexes by asymmetrical terminal modifications. ChemMedChem. 2014;9:2111–2119. doi: 10.1002/cmdc.201402115. [DOI] [PubMed] [Google Scholar]
- 225.Segal M., Slack F.J. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin Drug Discov. 2020;15:987–992. doi: 10.1080/17460441.2020.1765770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Christie R.J., Nishiyama N., Kataoka K. Delivering the code: polyplex carriers for deoxyribonucleic acid and ribonucleic acid interference therapies. Endocrinology. 2010;151:466–473. doi: 10.1210/en.2009-1045. [DOI] [PubMed] [Google Scholar]
- 227.Linsley C.S., Wu B.M. Recent advances in light-responsive on-demand drug-delivery systems. Ther. Deliv. 2017;8:89–107. doi: 10.4155/tde-2016-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Li Z., Rana T.M. Therapeutic targeting of microRNAs: current status and future challenges. Nat. Rev. Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 229.Masarudin M.J., Cutts S.M., Evison B.J., Phillips D.R., Pigram P.J. Factors determining the stability, size distribution, and cellular accumulation of small, monodisperse chitosan nanoparticles as candidate vectors for anticancer drug delivery: application to the passive encapsulation of [(14)C]-doxorubicin. Nanotechnol. Sci. Appl. 2015;8:67–80. doi: 10.2147/NSA.S91785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zein R., Sharrouf W., Selting K. Physical properties of nanoparticles that result in improved cancer targeting. J. Oncol. 2020;2020 doi: 10.1155/2020/5194780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rizkita L.D., Astuti IJJoPA. The potential of miRNA-based therapeutics in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: a review. Vol. 11. 2021. pp. 265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Hum C., Loiselle J., Ahmed N., Shaw T.A., Toudic C., Pezacki J.P.J.D. MicroRNA mimics or inhibitors as antiviral therapeutic approaches against COVID-19. Vol. 81. 2021. pp. 517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Gallant-Behm C.L., Piper J., Lynch J.M., Seto A.G., Hong S.J., Mustoe T.A., et al. A microRNA-29 mimic (Remlarsen) represses extracellular matrix expression and fibroplasia in the skin. Vol. 139. 2019. pp. 1073–1081. [DOI] [PubMed] [Google Scholar]
- 234.Okuyan H.M., Begen M.A.J.M.R. miRNAs as attractive diagnostic and therapeutic targets for Familial MediterraneanFever. Vol. 31. 2021. pp. 949–959. [DOI] [PubMed] [Google Scholar]
- 235.El-Nabi S.H., Elhiti M., El-Sheekh MJMh. Vol. 143. 2020. A new approach for COVID-19 treatment by micro-RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kalarikkal S.P., Sundaram G.M.J.T., Pharmacology A. Vol. 414. 2021. Edible plant-derived exosomal microRNAs: exploiting a cross-kingdom regulatory mechanism for targeting SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.He G., Rapireddy S., Bahal R., Sahu B., Ly DHJJotACS. Strand invasion of extended, mixed-sequence B-DNA by γPNAs. Vol. 131. 2009. pp. 12088–12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Masarudin M.J., Cutts S.M., Evison B.J., Phillips D.R., Pigram P.J.J.N. science, applications. Factors determining the stability, size distribution, and cellular accumulation of small, monodisperse chitosan nanoparticles as candidate vectors for anticancer drug delivery: application to the passive encapsulation of [14C]-doxorubicin. Vol. 8. 2015. p. 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Yasukawa K., Kinoshita D., Yaku K., Nakagawa T., Koshiba TJJoBC. The microRNAs miR-302b and miR-372 regulate mitochondrial metabolism via the SLC25A12 transporter, which controls MAVS-mediated antiviral innate immunity. Vol. 295. 2020. pp. 444–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Garg A., Seeliger B., Derda A.A., Xiao K., Gietz A., Scherf K., et al. Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Vol. 23. 2021. pp. 468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Jiang C., Guo Y., Yu H., Lu S., Meng L.J.A. Asthma, Immunology C. Pleiotropic microRNA-21 in pulmonary remodeling: novel insights for molecular mechanism and present advancements. Vol. 15. 2019. pp. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Prasad K., AlOmar S.Y., Alqahtani S.A.M., Malik M., Kumar VjMn. Brain disease network analysis to elucidate the neurological manifestations of COVID-19. Vol. 58. 2021. pp. 1875–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Muñoz-Fontela C., Dowling W.E., Funnell S.G., Gsell P.-S., Riveros-Balta A.X., Albrecht R.A., et al. Animal models for COVID-19. Vol. 586. 2020. pp. 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Vol. 383. 2020. pp. 1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]