Abstract
Cancer has been found as a heterogeneous disease with various subtypes and aims to destroy the body's normal cells abruptly. As a result, it is essential to detect and prognosis the distinct type of cancer since they may help cancer survivors with treatment in the early stage. It must also divide cancer patients into high- and low-risk groups. While realizing efficient detection of cancer is frequently a time-taking and exhausting task with the high possibility of pathologist errors and previous studies employed data mining and machine learning (ML) techniques to identify cancer, these strategies rely on handcrafted feature extraction techniques that result in incorrect classification. On the contrary, deep learning (DL) is robust in feature extraction and has recently been widely used for classification and detection purposes. This research implemented a novel hybrid AlexNet-gated recurrent unit (AlexNet-GRU) model for the lymph node (LN) breast cancer detection and classification. We have used a well-known Kaggle (PCam) data set to classify LN cancer samples. This study is tested and compared among three models: convolutional neural network GRU (CNN-GRU), CNN long short-term memory (CNN-LSTM), and the proposed AlexNet-GRU. The experimental results indicated that the performance metrics accuracy, precision, sensitivity, and specificity (99.50%, 98.10%, 98.90%, and 97.50) of the proposed model can reduce the pathologist errors that occur during the diagnosis process of incorrect classification and significantly better performance than CNN-GRU and CNN-LSTM models. The proposed model is compared with other recent ML/DL algorithms to analyze the model's efficiency, which reveals that the proposed AlexNet-GRU model is computationally efficient. Also, the proposed model presents its superiority over state-of-the-art methods for LN breast cancer detection and classification.
1. Introduction
Cancer is a group of uncontrolled development cells in the body, which may spread to any organ abruptly [1, 2]. There are many distinct kinds of cancer, but lung cancer, breast cancer (BC), and skin cancer are the most prevalent. According to the World Health Organization (WHO) reports, the cancer death ratio is up to 9.2 million in lung cancer and 1.7 million in skin cancer, while breast cancer has caused 627,000 deaths [3, 4]. Breast cancer survival is strongly tied to the size of the tumor at the time of diagnosis; patients who have a tumor that is even less than 10 mm in size have a 98% chance of survival. When the tumor is 30 mm in size, 70% of breast cancer cases are diagnosed [5]. Therefore, the size of a breast tumor has a big impact on the detection and survival of a patient. There are so many imaging techniques used for the detection and classification of BC, such as X-ray [6], ultrasound [7], and CT scan [8]. Scientists use several methods to determine different kinds of cancer and present with symptoms, such as early-phase screening (EPS). In addition, they have developed unique approaches for the early identification of the prognosis in cancer treatment. Because of the invention of new technologies in medicine, vast volumes of cancer data have been collected and available for bioinformatics and the scientific community for evaluation and testing. However, the prediction of breast cancer disease is among the most fascinating and demanding challenges in healthcare, including incorrect classification by using these investigated diagnosis techniques.
Furthermore, a breast cancer diagnosis in clinics relies on pathologists' visual investigation of patients, who are highly qualified cancer specialists. However, this process is manual, which is a time-consuming and exhausting task that is very susceptible to human mistakes because most cells are usually a portion of irregular, arbitrary, and uncontrolled visual angles. It is important to determine whether a tumor is malignant or benign. The objective is to distinguish between the two classes. Since malignant tumors need early treatment because it is cancerous cell and spread abruptly. To limit and avoid future issues from occurring, the problem is a binary classification task to recognize malignant and benign issues that can be addressed using various machine learning and deep learning (ML/DL) algorithms [9–18]. The use of machine learning approaches to decrease the risk of developing cancer, recurrence, and survival prediction might increase the accuracy by 20% to 25% than last year [18]. However, handling high-dimensional and unbalanced data sets are challenging for ML methods.
To handle the above informative challenges, we have introduced a hybrid deep learning AlexNet gated recurrent unit (AlexNet-GRU) model to outperform a pathologist in terms of accuracy, precision sensitivity, and specificity of lymph node (LN) breast cancer detection. The following are the major contributions of this research:
We have proposed a novel hybrid deep learning AlexNet-GRU model that automatically extracts features from the PCam data set and accurately identifies metastatic cancer
The AlexNet-GRU classifier is used for successful metastatic cancer detection in clinical and biomedical research
We have compared the performance of the proposed AlexNet-GRU model, the detection accuracy, and other performance metrics (precision, sensitivity, specificity, and time complexity) with the convolutional neural network GRU (CNN-GRU), CNN long short-term memory (CNN-LSTM) models, and other current ML/DL algorithms including state-of-the-art classification methods on the same data set to check the efficiency of the proposed model
The paper is organized as follows. Section 2 provides the literature survey related to metastatic cancer detection. A framework for lymph node breast cancer detection, including explaining the experimental data set and data preprocessing, is presented in Section 3. A full discussion of the proposed model is given in Section 4. The experimental study of the hybrid model, including ML/DL methods with performance evaluation and discussion, is given in Section 5. Finally, the conclusion and future scope of the research are shown in Section 6.
2. Related Work
Various research has been done in metastatic cancer image classification, primarily to improve the evaluation performance of LN cancer detection. Previously, the classification of metastatic cancer images was investigated using several ML/DL approaches, including K-nearest neighbors (K-NN), support vector machines (SVM), convolutional neural networks (CNN), and so on. Sun et al. [19] used a graph-based method to classify breast cancer accurately and got an accuracy of 88.90% but had high time complexity (sec.), and they also compared SVM and K-NN algorithms. The comparative study indicated that SVM has high accuracy than K-NN in classifying LN breast cancer detection. Machhale et al. [20] introduced the CNNs model and achieved 89% accuracy in classifying malignant or benign cancer. Arunava [21] investigated the accuracy and error rate of K-NN and Naïve Bayesian methods of breast cancer classification and found that K-NN had better accuracy (85.90%) and lower error rate than Nave Bayesian approaches. Nawaz et al. [22] investigated CNN's approach to breast cancer detection. The method considered various types of breast cancer, and the results showed 88.4% accuracy.
In the research [23], Montazeri et al. created a rule-based method; they presented the classification of breast cancer and got accuracy up to 89% in the experimental study. Hossain and Rahaman [24] proposed a fuzzy logic method for categorizing bone cancers with 90.75% accuracy. A computer-aided approach [24] is also used to classify lung cancer that achieved an accuracy rate of 75.1%. Skin cancer classification was carried out using the visual geometry group NET (VGGNET) model, which was shown to be accurate at 78.66% [25]. The authors proposed a rough set technique for feature extraction and an SVM classifier for the breast cancer classification in women. They used a rough reduction technique to eliminate identical characteristics from the model to enhance accuracy. The authors in [25] proposed a rough set approach to extract features and an SVM classifier to classify breast cancer. They evaluated their proposed model's accuracy, sensitivity, and specificity by categorizing cancer images and comparing the results. Their findings showed that their method had the highest accuracy value of 92% and was more efficient than recent ML/DL models on the same data set. Aylin [26] presented a feature extraction method with a rough set method for breast cancer classification and an SVM classifier. They also implemented a rough reduction method to reduce unnecessary features from the model to improve accuracy and precision. In addition, the techniques mentioned above offer greater accuracy as compared to other investigated models. In addition, SVMs, artificial neural networks (ANNs), the K-NN, and various additional algorithms were used in breast cancer classification [26].
Saber et al. [27] proposed a deep learning model for automatic BC detection using the transfer learning method and six performance elevation matrices. For feature extraction and classification, they utilized five pretrained DL models, that is, Inception-V3, ResNet50, VGG19, VGG16, and Inception-V2 ResNet. The VGG16 model was shown to be effective for BC detection, with an accuracy of 98.96%. Togaçar et al. [28] presented a novel deep learning model for BC classification based on the concept of CNN. The model comprises two main parts. The hyper-column method attention component and the residual block are the two aspects of the model. When analyzing the BreakHis data set, the model achieves 98.80% accuracy. Ting et al. [29] proposed a new framework for BC classification named CNN improvement for BC classification (CNNI-BCC), using a supervised deep learning neural network to classify breast cancer. Experimental results showed that CNNI-BCC outperformed existing studies and attained an accuracy of 89.47%.
Eroglu et al. [30] proposed a CNN-based hybrid model for BC classification. The extracted features from breast cancer ultrasound images using AlexNet, ResNet50, MobileNetv2, and ResNet50. The extracted features are concatenated with the help of the mRMR method to increase accuracy. Finally, these features are classified with the SVM. The proposed hybrid model achieved 95.6% classification accuracy. Masud et al. [31] presented their own custom CNN model for BC detection. Moreover, they used eight pretrain, deep learning models for two BC data sets classification using a transfer learning strategy. The ResNet50 with Adam optimizer obtained the best accuracy of 92.4%, and VGG16 achieved the maximum AUC 0.97 score.
Another study [31] used AlexNet CNN's structure to detect mitosis images in breast histology and found 87% accuracy, and they compared performance metrics (sens, spec, and sens). The networks were capable of arranging the image in a pixel-by-pixel fashion. Gecer et al. [32] employed a DL technique to automatically identify and investigate the invasive ductal carcinoma (IDC) tissue zones and accurately detect cancer. Jason [33] demonstrated context-aware stacked CNNs for the classification of breast whole slide images (WSIs) into simple, DCIS (ductal carcinoma in situ), and IDC (in ductal carcinoma). The WSI classification achieved an area under the curve of 0.852 for classifying malignant and nonmalignant slides. In addition, the AlexNet CNN structure cannot store the memory of previous time-series patterns, where it is difficult to directly learn the most important and representative features of breast cancer that are given in the form of time series [34]. A GRU model is used with the AlexNet CNN structure to solve the above challenges. To improve the accuracy and other performance metrics (pres (%), sens (%), spec (%), and time complexity (sec) of classifications) [21, 35–42]. The literature related to this research is presented in Table 1.
Table 1.
The literature employed ML/DL models for breast cancer detection.
| Publication | Algorithms | Data set | Detection accuracy (%) | Time complexity (ms) |
|---|---|---|---|---|
| [35] | RF, K-NN | UCI-cancer | 87%, 88% | N/A |
| [36] | DNN | BCW (breast cancer Wisconsin) | 75% | High |
| [37] | DCNN, CNN | BreakHis | 90%, 90.1% | High |
| [21] | CNN | MIAS | 90% | High |
| [38] | RF, SVM, K-NN | UCI-cancer WBCD11, WBCD32 | 76%, 75%, 74% | N/A |
| [39] | DNN | BCW (breast cancer Wisconsin) | 79.01% | High |
| [40] | SVM, RF, K-NN | BCW (breast cancer Wisconsin) | 77%, 74%, 75% | High |
| [41] | GRU, RNN | UCI-cancer | 78.90% | N/A |
| [42] | CNN | BCW (breast cancer Wisconsin) | 70.50% | High |
3. A Framework for LN Breast Cancer Detection
This section proposes a new hybrid deep learning (AlexNet-GRU) model for metastatic (tumor) cancer detection and presents its framework, architecture, and classification performance evaluation. Besides, a conceptual representation of the entire classification scheme adopted in this study is shown in Figure 1 that consists of data collection, preprocessing, class labeling, model building, and evaluation for LN breast cancer detection.
Figure 1.

The basic block diagram of the AlexNet-GRU model for lymph node (LN) breast cancer detection.
3.1. Data Collection and Analysis
The cancer data set is obtained from the Kaggle [43]. The data set is an updated version of the original PCam data set, and there is no duplication within the data set. The collection contains 220,901 lymph node cancer images, with patches taken from 400 slide scan images from 162 women were LN breast cancer diagnosed and tested at University Medical Center, Netherlands. The first experimental training data set includes 170 WSIs samples of lymph node, and the second training data set consists of 100 WSIs. The data set contains high-resolution images (2,040 × 1,536 pixels). All the slides were scanned at 0.25 micro/pr using the same scanner to ensure consistency. Reduced-resolution images were sampled to a smaller region of pixel 50 × 50. It is organized into two main categories: metastatic tumors (cancer) and nonmetastatic (no-cancer) tumors, including 89,118 samples of LN breast cancer and 130,893 images of nonmetastatic tumors. The Kaggle data set is split into training data set 80% and testing 20% to avoid overfitting. Empirical studies show that the best results are obtained if we use 20–30% of the data for testing and the remaining 70–80% of the data for testing. The images are in the RGB color space and the PNG format.
Figure 2 presents the ratio of the cancerous photos and noncancer images. The training distribution of noncancerous/cancer was approximately 60/40.
Figure 2.

The ratio of malignant (cancerous images) and benign (noncancer images).
3.2. Data Preprocessing
Preprocessing is very important for accurate classification performance. It is usually done on data before categorization. On the “cancer data set,” preprocessing methods are essential to enhance the model's classification accuracy. The most frequently utilized preprocessing methods are class labeling, picture scaling, data augmentation, random cropping, and sliding with the crop in breast cancer detection [44].
3.2.1. Class Labeling
The cancer data set is subjected to the preprocessing method of class labeling that is conducted on the data set. The data set presentation of the class labeling is as follows: the label 0 for the image represents the patients who have no cancer (nonmetastatic cancer), while 1 represents the patient's “cancer” (metastatic cancer), as shown in Figure 3.
Figure 3.
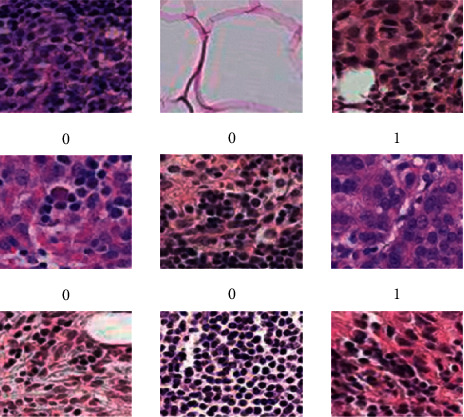
Breast images of metastatic cancer and non-metastatic.
3.2.2. Resizing
Resizing is another preprocessing method used in this study, as shown in Figure 4. The proposed model needs the same image size to obtain the desired results in the training process, so all the images were resized into identical dimensions.
Figure 4.

Resizing technique for breast cancer image data set.
3.2.3. Data Augmentation
A data augmentation process was used in the cancer data set. Typically, a convolution neural network's structure performs better when working with big data, yet collecting vast amounts of data is challenging. Because of the limited quantity of data available, the convolution neural network suffers from an overfitting issue that reduces the performance evaluation of the models. The data augmentation approach is the most effective method of overcoming the underlying problem. The data augmentation of Kaggle cancer data sets is graphically shown in Figure 5.
Figure 5.

Data augmentation for breast cancer data set.
3.2.4. Random Cropping
An additional preprocessing method, namely random cropping with convolution neural networks (CNNs), was used to process the cancer data set. In the process of randomly cropping various sections large dimensional of images, the quantity of data available for the model needs significant data for training purposes. Figure 6 presents the random cropping employed in the current study.
Figure 6.

Random cropping of the breast cancer data set.
3.2.5. Sliding with Crop
In sliding with the crop process, initially, the images of the PCam data set were cropped. To increase the number of images, they should be arranged from left to right and top to bottom to handle the overfitting issue. Figure 7 shows sliding with the crop process.
Figure 7.

Sliding with a crop of the breast cancer image data set.
3.2.6. Row Major Order
Multi-dimensional arrays of images are stored in one dimension or a single row when they are stored in row-major order. Images may be in greyscale or RGB format, depending on their size. Greyscale images are composed of two channels, including white and black. RGB pictures are three channels: green, red, and blue. RGB images are more complex than greyscale pictures. The row-major order is shown in Figure 8.
Figure 8.

Row major order of the breast cancer image data set.
4. Proposed System Model
After preprocessing steps, the next section is to feed data into the proposed model (AlexNet-GRU). This section introduces the AlexNet and GRU model, including training parameters for lymph node (LN) breast cancer detection and classification.
4.1. AlexNet Model
The AlexNet is the submodel of CN that has made significant contributions, particularly in applying objection detection, image classification, and so on. It had won the Image Net LSVRC-2012 competition by a considerable margin (15.3% error rates vs. 26.2% error rates in second place). The network's design was very similar to that of the LeNet network. However, it was deeper, had more filters per layer, and featured convolutional layers. It was composed of convolutions, max pooling, ReLU activations, and dropout layers. Figure 9 describes the basic structure of the AlexNet model. The following steps present the mathematical formula of the AlexNet model as follows.
Figure 9.

Structure of the AlexNet model.
4.1.1. Convolution Layers
The convolution layer is the main building block of the AlexNet model. It consists of different learnable filters that extract the input data features. The feature map assumes that the cth of layer d is represented by ycd and that the Jth at layer d – 1 is represented by ycd−1. The value of ycd is calculated as follows:
| (1) |
where |b| is the number of mapping features at layer d, acd is a biased term distributed throughout all connections to the c feature map, and bc is a subset of feature maps in layer d − 1 related to units c, at layer d.
4.1.2. Activation Function (ReLU_Func)
The activation d(y) of ReLU_Func can be expressed as follows:
| (2) |
where the range value is from 0 to y.
4.1.3. Max_Pooling
The max_pooling can be mathematically represented as follows:
| (3) |
where
| (4) |
A triplet (cd, Jd, dd) finding one component in the input xd and another triplet (cd+1, J1+1, dd+1) to determine the location of one component inside y are needed. The pooling output yc(d+1)jd+1, d comes from xcl,jd,ddd with the given condition is true. The (cd, jd) − th distinct entry belongs to the (cd+1, jd+1)th subregion.
4.1.4. Forward Pass of CNN
The forward pass of a CNN may be expressed mathematically as follows:
| (5) |
The forward pass of the CNN layer is shown in equation (5). Here, c1, cK−1, and cK are the CNNs, c1, dK−1, dK show the layer processing, and E expresses the cost function. Let us assume that j presents the actual value, while cK represents the cost function values can be mathematically expressed as follows:
| (6) |
4.2. Gated Recurrent Unit Network
The GRU model was most often employed in recurrent neural networks (RNN) to address the vanishing gradient issue, as shown in Figure 10. In comparison to LSTM, GRU has three major gates and an internal cell state, making it more efficient. The information is stored in a secret state inside the GRU. The backward and forward information is provided with the update gate (z), whereas the previous knowledge is presented in the reset gate (r). The current memory gate uses the reset gate to preserve and keep the required information from the previous state of the computer. With the input modulation gate, it is possible to introduce nonlinearity into the input while also providing it with the characteristics of a zero-mean. According to the following definition, the mathematical description of the fundamental GRU of rest and updated gates is
| (7) |
where Wrx and Wxz are weight parameters and br and bz represent biased.
Figure 10.

The basic structure of the GRU model.
4.3. AlexNet-GRU Model
In this research, we propose the AlexNet-GRU model for breast cancer classification. The proposed model is composed of seven convolutions, four max-pooling, three completely linked layers, and ReLU as an activation function. The proposed AlexNet-GRU model's structure and employed parameters are shown in Figure 11 and Table 2, respectively.
Figure 11.

The general structure of the proposed AlexNet-GRU model.
Table 2.
Training parameters of the proposed AlexNet-GRU model.
| Layers | K_Size | Input (shape) | Act_Func | Output (shape) |
|---|---|---|---|---|
| Con_Layer_1 | 3 × 3 | (60, 60, 3) | ReLU_Func | (58, 58, 128) |
| B-Norm_1 | — | (58, 58, 128) | — | (58, 58, 128) |
| Max_pooling_1 | 2 × 2 | (58, 58, 128) | — | (57, 57, 128) |
| Drop_out = 0.9 | — | (57, 57, 128) | — | (57, 57, 128) |
| Con_Layer_2 | 2 × 3 | (57, 57, 128) | ReLU_Func | (55, 55, 256) |
| B-Norm_2 | — | (57, 57, 128) | — | (55, 55, 256) |
| Max_pooling_2 | 2 × 2 | (55, 55, 256) | — | (54, 54, 256) |
| Drop_out = 0.9 | — | (54, 54, 256) | — | (54, 54, 256) |
| Con_Layer_3 | 2 × 3 | (54, 54, 256) | ReLU_Func | (52, 52, 256) |
| B-Norm_3 | — | (52, 52, 256) | — | (52, 52, 256) |
| Max_pooling_3 | 2 × 2 | (52, 52, 256) | — | (51, 51, 256) |
| Dropout = 0.5 | — | (51, 51, 256) | — | (51, 51, 256) |
| Con_Layer_4 | 2 × 3 | (51, 51, 256) | ReLU_Func | (49, 49, 256) |
| B-Norm_4 | — | (49, 49, 256) | — | (49, 49, 256) |
| Dropout = 0.9 | — | (49, 49, 256) | — | (49, 49, 256) |
| Con_Layer_5 | 2 × 3 | (49, 49, 256) | ReLU_Func | (47, 47, 256) |
| B-Norm_5 | — | (47, 47,256) | — | (47, 47, 256) |
| Dropout = 0.9 | — | (47, 47, 256) | — | (47, 47, 256) |
| Con_Layer_6 | 2 × 3 | (47, 47, 256) | ReLU_Func | (45, 45, 256) |
| B-Norm_6 | — | (45, 45, 256) | — | (45, 45, 256) |
| Dropout = 0.9 | — | (45, 45, 256) | — | (45, 45, 256) |
| Con_Layer_7 | 2 × 3 | (45, 45, 256) | ReLU_Func | (45, 45, 512) |
| B-Norm_7 | — | (43, 43, 512) | — | (43, 43, 512) |
| Max_pooling_4 | 3 × 2 | (43, 43, 512) | — | (42, 42, 512) |
| Dropout = 0.5 | — | (42, 42, 512) | — | (42, 42, 512) |
| Flatten | — | (42, 42, 512) | — | (903,168) |
| Dense1 | — | (903,168) | — | (1,024) |
| B-Norm_8 | — | (1,024) | — | (1,024) |
| Drop_out = 0.3 | — | (1,024) | — | (1,024) |
| Dense2 | — | (1,024) | — | (2,000) |
| B-Norm_9 | — | (2,000) | — | (2,000) |
| GRU | (1,024) | — | — | |
| Dense3 | — | (2,000) | — | (2,000) |
Initially, the input shape of the image was (60, 60, 3) size, where the image height was 60 pixels and width 60 pixels with RGB, and the number of channels was 3.
To extract the features from the input shape, it passes through the first convolutional layer of the proposed model, where the output shape of the feature map was 128. Additionally, the stride and the kernel size of the convolutional layer were (3 × 3) and 1. While padding was the same for all of the layers of the proposed model and rectified linear units (ReLU) were used as activation function to decrease the nonlinearity dimension problem, followed by conventional layer 1. After the convolutional layer 1, the output shape contained (60, 60) size and 128 feature maps. The pooling layer decreases the training parameter up to (58, 58) size that speeds up the process of the proposed model. After the pooling layer, the training parameter (58, 58, 128) were passed through drop_out to prevent the model from overfitting issues. An initial dropout of 0.9 was used in the convolution layer to avoid the overfitting problem. After each conventional and max-pooling layer, the training parameter was substantially decreased, followed by activation function (ReLU) and drop out. After the training process of the traditional and max-pooling layers, the data must be composed into an ID array to use as input for a fully connected layer implemented by flatten with training parameters (42, 42) size and features map (512) had been produced. After completing all the convolutional layers process, the dropout was used that made 1,024 feature maps. A GRU model was employed with a fully connected layer composed of 1,024 neurons to solve the vanishing gradient problem. After the GRU model, two fully connected layers were also used. Finally, SoftMax operations were implemented with the final linked layer.
5. Performance Evaluation and Discussion
This section presents the classification performance (accuracy, precision, sensitivity, and specificity), including experimental setup and enhanced comparative study of the proposed model (AlexNet-GRU) with CNN-GRU, CNN-LSTM, and the recent ML/DL models for lymph node breast cancer detection.
5.1. Experimental Setup
We have utilized an Intel Core i6 processor and a graphics-processing unit (GPU) from NVIDIA for this experiment. The proposed model has also been trained by merging Keras with the Python 3.8 programming environment. Detailed descriptions of the employed software and hardware characteristics are presented in Table 3.
Table 3.
The experimental setup.
| GPU | 1,060 6 GB, Nvidia GeForce |
| CPU | Core-i6, 6th Generation, and 2.80 GHz processor |
| RAM | 16 GB |
| Operating system | Windows 64 bit |
| Libraries | Pandas, NumPy, Scikit-learn, TensorFlow, and Keras |
| Languages | Python 3.7 |
5.2. Performance Metrics
The performance metrics are taken into consideration to evaluate the proposed model's viability and to distinguish between breast cancer (tumor) and normal tissue properly, such as the following:
TP: The cancer samples were positive
TN: It refers to negative (noncancer) samples
FP: It presents negative but is predictable to be positive
FN: It presents positive samples predictable to be negative by the model
The mathematical representations of the accuracy, precision, sensitivity, and specificity that are used as performance metrics to identify cancer in most cases are as follows:
| (8) |
5.3. Accuracy, Precision, Sensitivity, and Specificity
This study introduced three models, CNN-GRU, CNN-LSTM, and the proposed AlexNet-GRU model, to compare cancer image classification. Every model has two classes, such as 0 and 1. The result indicates that the proposed model exhibits the best accuracy, precision, sensitivity, and specificity, while CNN-GRU shows the lowest performance in terms of performance metrics, as shown in Figure 12. Table 4 shows the experimental results of the employed models' accuracy, precision, sensitivity, and specificity.
Figure 12.

The classification performance of the proposed AlexNet-GRU, CNN-GRU, and CNN-LSTM models for binary recognition task of BC detection.
Table 4.
The experimental results of the proposed AlexNet-GRU, CNN-GRU, and CNN-LSTM models for BC image classification.
| Performancemetrics (%) | Classification | CNN-GRU (%) | CNN-LSTM (%) | Proposedmodel (%) | Data set | Other parameters |
|---|---|---|---|---|---|---|
| Accuracy (%) | Binary class | 97.10 | 97.90 | 99.10 | Kaggle | Batch size = 100,epoch = 100, optimizer = Adam |
| Precision (%) | 96.90 | 97.70 | 98.70 | |||
| Sensitivity (%) | 96.87 | 97.60 | 98.71 | |||
| Specificity (%) | 96.59 | 97.10 | 98.51 |
5.4. Receiver-Operating Characteristic (ROC) Analysis
The ROC is an experimental tool to express the diagnostic tests' evaluation performance or the ratio of sensitivity and 1-specificity. The ROC of the proposed AlexNet-GRU model, CNN-LSTM, and CNN-GRU of the binary class recognition task of breast cancer detection is shown in Figure 13. The ROC indicates that the proposed model accurately classifies metastatic cancer (tumor or cancer) and nonmetastatic cancer (no cancer) of lymph nodes compared to CNN-LSTM and CNN-GRU models.
Figure 13.

The ROC performance of the given models: (a) proposed AlexNet-GRU, (b) CNN-GRU, and (c) LSTM-GRU.
5.5. Time Complexity (ms)
This section represents the time complexity of the models based on the testing. Because the training is performed mostly offline and not considered in the evaluation process. Furthermore, the testing technique is viewed as a critical statistic since it reveals the efficiency and general performance, important performance indicators. On the breast cancer detection task, the proposed AlexNet-GRU model is time-efficient for binary class recognition tasks compared to CNN-GRU and CNN-LSTM models, as shown in Figure 14.
Figure 14.

The time complexity (ms) of the binary BC detection.
Performance metrics are compared with existing deep learning algorithms, such as CNN, deep neural network (DNN), LSTM, GRU, and BiLSTM for binary tasks to demonstrate the efficiency of the proposed AlexNet-GRU model for binary tasks. As shown in Figure 15, the findings reveal that all of the current deep learning algorithms are less accurate (%) than the proposed model, while CNN and LSTM are the least accurate models among all of the models. In Table 5, the proposed model's performance is compared to the state-of-the-art ML/DL algorithms for the BC classification task.
Figure 15.

The classification performance comparison of the proposed model with other existing DL models in BC detection.
Table 5.
Comparative study of the proposed model with recent ML/DL models.
| Publication | Cancer type | Models | Classification type | Data set | Accuracy (%) |
|---|---|---|---|---|---|
| [45] | Breast cancer | ANN | Binary class | SNPs | 69% |
| [46] | Colon carcinomatosis | BN | Binary class | Clinical, pathologic | 71% |
| [47] | Multiple myeloma | SVM, DCNN | Multi-class | DDSM (CBIS-DDSM) | 71%, 93% |
| [48] | Breast cancer | DCNNs | Bi-multi-class | Kaggle | 90%, 93% |
| [49] | Breast cancer | DCNNs | Multi-class | Wisconsin diagnostic breast cancer (WDBC) | 88% |
| [50] | Breast cancer | ResNet | Multi-class | Kaggle | 85% |
| [51] | Breast cancer | Pretrained CNN | Binary class | Kaggle | 90% |
| Proposed model | Breast cancer | AlexNet-GRU | Binary class | Kaggle | 99.50% |
The proposed model shows better performance accuracy (%). However, the proposed model has some disadvantages. The AlexNet-GRU method has high time complexity (ms) and requires high computational power and specialized hardware such as a good GPU to expedite the training process.
Recently, there also exist some other artificial intelligence technologies that can be used in next-generation healthcare systems, that is, federated learning algorithms [52], deep learning algorithms [53], reinforcement, and deep reinforcement learning algorithms [54–58]. In addition, next-generation industries, that is, Industry 4.0 and Industry 5.0, would play a crucial role in developing advanced equipment used in health treatment [59].
6. Conclusions
This research intended to classify lymph node breast cancer detection using the proposed Alex net-GRU model. To train the model, the preprocessing step was done by using various types of techniques to achieve the best classification performance. Furthermore, the experimental study was performed with CNN-GRU, CNN-LSTM, and recent ML/DL models. We used a well-known Kaggle (PCam) data set to classify lymph node cancer samples. The experimental results indicated that the performance metrics accuracy, precision, sensitivity, and specificity (99.50%, 98.10%, 98.90%, and 97.50%) of the proposed model can reduce the pathologist errors made during the diagnosis process of incorrect classification and significantly better performance than CNN-GRU and CNN-LSTM models. To analyze the model efficiency, the proposed model was compared with recent ML/DL algorithms that revealed that the proposed AlexNet-GRU model is computationally efficient and outperformed. Also, the proposed model presented its superiority over state-of-the-art methods for breast cancer detection and classification. The proposed AlexNet-GRU model showed about 3% best accuracy and other performance metrics. Therefore, the presented AlexNet-GRU model is a promising technique for categorizing lymph node breast cancer detection and classification. This research can be further expanded by adding one or more layers with the AlexNet-GRU model and will be tested on multi-classification breast cancer detection and not limited to binary recognition tasks.
Data Availability
The data are publicly available at https://www.kaggle.com/c/histopathologic-cancer-detection/data.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
References
- 1.Ou F. S., Michiels S., Shyr Y., Adjei A. A., Oberg A. L. Biomarker discovery and validation: statistical considerations. Journal of Thoracic Oncology . 2021;16(4):537–545. doi: 10.1016/j.jtho.2021.01.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chtihrakkannan R., Kavitha P., Mangayarkarasi T., Karthikeyan R. Breast cancer detection using machine learning. International Journal of Innovative Technology and Exploring Engineering . 2019;8(11):3123–3126. [Google Scholar]
- 3.Chaurasia V., Pal S., Tiwari B. Prediction of benign and malignant breast cancer using data mining techniques. Journal of Algorithms & Computational Technology . 2018;12(2):119–126. doi: 10.1177/1748301818756225. [DOI] [Google Scholar]
- 4.Solanki Y. S., Chakrabarti P., Jasinski M., et al. A hybrid supervised machine learning classifier system for breast cancer prognosis using feature selection and data imbalance handling approaches. Electronics . 2021;10(6):p. 699. doi: 10.3390/electronics10060699. [DOI] [Google Scholar]
- 5.Roslidar R., Rahman A., Muharar R., et al. A review on recent progress in thermal imaging and deep learning approaches for breast cancer detection. IEEE Access . 2020;8:116176–116194. doi: 10.1109/access.2020.3004056. [DOI] [Google Scholar]
- 6.Singh S. P., Urooj S. A. Breast cancer detection using pcpcet and adewnn: a geometric invariant approach to medical X-ray image sensors. IEEE Sensors Journal . 2016;16(12):4847–4855. doi: 10.1109/jsen.2016.2533440. [DOI] [Google Scholar]
- 7.Guo R., Lu G., Qin B., Fei B. Ultrasound imaging technologies for breast cancer detection and management: a review. Ultrasound in Medicine and Biology . 2018;44(1):37–70. doi: 10.1016/j.ultrasmedbio.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nover A. B., Jagtap S., Anjum W., et al. Modern breast cancer detection: a technological review. International Journal of Biomedical Imaging . 2009;2009:1–14. doi: 10.1155/2009/902326.902326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang J., Rangayyan R. M., Xu J., Naqa I. E., Yang Y. Computer-aided detection and diagnosis of breast cancer with mammography: recent advances. IEEE Transactions on Information Technology in Biomedicine . 2009;13(2):236–251. doi: 10.1109/titb.2008.2009441. [DOI] [PubMed] [Google Scholar]
- 10.Ahmad I., Ullah I., Khan W. U., et al. Efficient algorithms for E-healthcare to solve multiobject fuse detection problem. Journal of Healthcare Engineering . 2021;2021:1–16. doi: 10.1155/2021/9500304.9500304 [DOI] [Google Scholar]
- 11.Raza A., Ayub H., Khan J. A., et al. A hybrid deep learning-based approach for brain tumor classification. Electronics . 2022;11(7):p. 1146. doi: 10.3390/electronics11071146. [DOI] [Google Scholar]
- 12.Ahmad I., Liu Y., Danish J., Shahab A. A decision-making technique for solving order allocation problem using a genetic algorithm. IOP Conference Series: Materials Science and Engineering . 2020;853(1) doi: 10.1088/1757-899x/853/1/012054.012054 [DOI] [Google Scholar]
- 13.Ahmad I., Liu Y., Danish J., Nadia S., Danish S., Shahab A. A review of artificial intelligence techniques for selection & evaluation. IOP Conference Series: Materials Science and Engineering . 2020;853(1) doi: 10.1088/1757-899x/853/1/012055.012055 [DOI] [Google Scholar]
- 14.Tufail A. B., Ullah I., Tufail A. B., et al. Diagnosis of diabetic retinopathy through retinal fundus images and 3D convolutional neural networks with limited number of samples. Wireless Communications and Mobile Computing . 2021;2021:1–15. doi: 10.1155/2021/6013448.6013448 [DOI] [Google Scholar]
- 15.Koscielny S. Why most gene expression signatures of tumors have not been useful in the clinic. Science Translational Medicine . 2010;2:p. 14. doi: 10.1126/scitranslmed.3000313. [DOI] [PubMed] [Google Scholar]
- 16.Amrane M., Oukid S., Gagaoua I., Ensarİ T. Breast cancer classification using machine learning. Proceedings of the Electric Electronics, Computer Science, Biomedical Engineerings’ Meeting (EBBT); April 2018; Istanbul, Turkey. IEEE; pp. 1–4. [DOI] [Google Scholar]
- 17.Michiels S., Koscielny S., Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet (London, England) . 2005;365:488–492. doi: 10.1016/s0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- 18.Kourou K., Exarchos T. P., Konstantinos P. E., Michalis V. K., Dimitrios I. F. Machine learning applications in cancer prognosis and prediction. Computational and Structural Biotechnology Journal . 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun W., Zheng B., Qian W. Computer-aided lung cancer diagnosis with deep learning algorithms. Proceedings of the Medical imaging computer-aided diagnosis International Society for Optics and Photonics; March 2016; Bellingham, WA, U.S.A. [DOI] [Google Scholar]
- 20.Machhale K., Nandpuru H. B., Kapur V., Kosta L. MRI brain cancer classification using hybrid classifier (SVM-KNN). Proceedings of the International Conference on Industrial Instrumentation and Control (ICIC); May 2015; Pune, India. IEEE; pp. 60–65. [DOI] [Google Scholar]
- 21.Arunava A. Convolutional Neural network. Proceedings of the International Conference On Applied Sciences And Engineering; June 2018; Pattaya, Thailand. pp. 62–72. [Google Scholar]
- 22.Nawaz W., Ahmed S., Tahir A., Khan H. A. Classification of breast cancer histology images using alexnet. Proceedings of the International Conference Image Analysis and Recognition; June 2018; Berlin, Germany. Springer; pp. 869–876. [DOI] [Google Scholar]
- 23.Montazeri M., Montazeri M., Montazeri M., Beigzadeh A. Machine learning models in breast cancer survival prediction. Technology and Health Care . 2016;24(1):31–42. doi: 10.3233/thc-151071. [DOI] [PubMed] [Google Scholar]
- 24.Hossain E., Rahaman M. A. Bone cancer detection classification using fuzzy clustering neuro-fuzzy classifier. Proceedings of the 2018 4th International Conference on Electrical Engineering and Information & Communication Technology (iCEEiCT); September 2018; Dhaka, Bangladesh. IEEE; pp. 541–546. [Google Scholar]
- 25.Lakshmanaprabu S. k., Mohanty S. N., Shankar K., Arunkumar N., Ramirez G. Optimal deep learning model for classification of lung cancer on CT images. Future Generation Computer Systems . 2019;92:374–382. doi: 10.1016/j.future.2018.10.009. [DOI] [Google Scholar]
- 26.Aylin C. layers of convolutional neural network. Fundamentals Of Convolution Neural Network . 2017;46:139–144. [Google Scholar]
- 27.Saber A., Sakr M., Seida O. M. A., Keshk A., Chen H. A novel deep-learning model for automatic detection and classification of breast cancer using the transfer-learning technique. IEEE Access . 2021;9:71194–71209. doi: 10.1109/access.2021.3079204. [DOI] [Google Scholar]
- 28.Toğaçar M., Özkurt K. B., Ergen B., Cömert Z. BreastNet: a novel convolutional neural network model through histopathological images for the diagnosis of breast cancer. Physica A: Statistical Mechanics and Its Applications . 2020;545 doi: 10.1016/j.physa.2019.123592.123592 [DOI] [Google Scholar]
- 29.Ting F. F., Tan Y. J., Sim K. S. Convolutional neural network improvement for breast cancer classification. Expert Systems with Applications . 2019;120:103–115. doi: 10.1016/j.eswa.2018.11.008. [DOI] [Google Scholar]
- 30.Eroğlu Y., Yildirim M., Çinar A. Convolutional Neural Networks based classification of breast ultrasonography images by hybrid method with respect to benign, malignant, and normal using mRMR. Computers in Biology and Medicine . 2021;133 doi: 10.1016/j.compbiomed.2021.104407.104407 [DOI] [PubMed] [Google Scholar]
- 31.Masud M., Eldin Rashed A. E., Hossain M. S. Convolutional neural network-based models for diagnosis of breast cancer. Neural Computing & Applications . 2020:1–12. doi: 10.1007/s00521-020-05394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gecer B., Aksoy S., Mercan E., Shapiro L. G., Weaver D. L., Elmore J. G. Detection and classification of cancer in whole slide breast histopathology images using deep convolutional networks. Pattern Recognition . 2018;84:345–356. doi: 10.1016/j.patcog.2018.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jason B. A Gentle Introduction to cross-entropy for machine learning. Machine learning mastery . 2019 [Google Scholar]
- 34.Karthik S., Srinivasa Perumal R., Chandra Mouli P. V. S. S. R. Knowledge Computing and its Applications . Singapore: Springer; 2018. Breast cancer classification using deep neural networks; pp. 227–241. [DOI] [Google Scholar]
- 35.Lopez A. R., Nieto X. G., Burdick J., Marques O. Skin lesion classification from dermoscopic images using deep learning techniques. Proceedings of the International Conference on Biomedical Engineering (BioMed); February 2017; Innsbruck, Austria. IEEE; pp. 49–54. [DOI] [Google Scholar]
- 36.Mohsen H., Dahshan E. S. A., Horbaty E. S. M., Salem A. B. M. Classification using deep learning neural networks for brain tumors. Future Computing and Informatics Journal . 2018;3(1):68–71. doi: 10.1016/j.fcij.2017.12.001. [DOI] [Google Scholar]
- 37.Javeed D., Gao T., Khan M. T., Ahmad I. A hybrid deep learning-driven SDN enabled mechanism for secure communication in internet of things (IoT) Sensors . 2021;21(14):p. 4884. doi: 10.3390/s21144884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rizhevsky A., Sutskever I., Hinton G. E. Imagine classification with deep convolutional neural networks. Advances in Neural Information Processing Systems . 2012;25:1097–1105. [Google Scholar]
- 39.Spanhol F. A., Oliveira L. S., Petitjean C., Heutte L. A dataset for breast cancer histopathological image classification. IEEE Transactions on Biomedical Engineering . 2016;63(7):1455–1462. doi: 10.1109/tbme.2015.2496264. [DOI] [PubMed] [Google Scholar]
- 40.Sharma S. Activation functions in neural network. Expert Systems with Applications . 2017;47:132–244. [Google Scholar]
- 41.Sanjeevi M. Different types of Machine learning and their types. Machine Learning and Their Applications . 2017;90:360–362. [Google Scholar]
- 42.Vesal S., Ravikumar N., Davari A., Ellmann S., Maier A. Classification of breast cancer histology images using transfer learning. Proceedings of the International Conference Image Analysis and Recognition; January 2018; Islamabad, Pakistan. Springer; pp. 812–819. [DOI] [Google Scholar]
- 43.kaggle. Histopathologic Cancer Detection. 2017. https://www.kaggle.com/c/histopathologic-cancer-detection .
- 44.Waddell M., Page D., Shaughnessy J. Predicting cancer susceptibility from single-nucleotide polymorphism data: a case study in multiple myeloma. Proceedings of the 5th International Workshop on Bioinformatics; August 2005; New York, NY, U.S.A. [Google Scholar]
- 45.Ayer T., Alagoz O., Chhatwal J., Shavlik J. W., Kahn C. J., Burnside E. S. Breast cancer risk estimation with artificial neural networks revisited. Cancer . 2010;116(14):3310–3321. doi: 10.1002/cncr.25081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Listgarten J., Damaraju S., Poulin B., et al. Predictive models for breast cancer susceptibility from multiple single nucleotide polymorphisms. Clinical Cancer Research . 2004;10(8):2725–2737. doi: 10.1158/1078-0432.ccr-1115-03. [DOI] [PubMed] [Google Scholar]
- 47.Saffar A. A. M., Tao H., Talab M. A. Review of deep convolution neural network in image classification. Proceedings of the International Conference on Radar, Antenna, Microwave, Electronics, and Telecommunications (ICRAMET); October 2017; Jakarta, Indonesia. IEEE; pp. 26–31. [DOI] [Google Scholar]
- 48.Ragab D. A., Sharkas M. S. J. Breast cancer detection using deep convolutional neural networks and support vector machines. PeerJ . 2019;7 doi: 10.7717/peerj.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nawaz M., Sewissy A. A., Hassan T. Multi-class breast cancer classification using deep learning convolutional neural network. International Journal of Advanced Computer Science and Applications . 2018;9(6):316–322. doi: 10.14569/ijacsa.2018.090645. [DOI] [Google Scholar]
- 50.Joseph A., Abdullahi M., Sahalu B. J., Hayatu H. I., Haruna C. Improved multi-classification of breast cancer histopathological images using handcrafted features and deep neural network (dense layer) Intelligent Systems with Applications . 2022;14 doi: 10.1016/j.iswa.2022.200066.200066 [DOI] [Google Scholar]
- 51.Khan M. H. M, Jahangeer N. B., Dullull W., et al. Multi- class classification of breast cancer abnormalities using Deep Convolutional Neural Network (CNN) PLoS One . 2021;16(8) doi: 10.1371/journal.pone.0256500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vesal S., Ravikumar N., Davari A., Ellmann S., Maier A. Classification of breast cancer histology images using transfer learning. Proceedings of the International Conference Image Analysis and Recognition; June 2018; Berlin, Germany. Springer; [DOI] [Google Scholar]
- 53.Shome D., Waqar O., Khan W. U. Federated Learning and Next-Generation Wireless Communications: A Survey on Bidirectional Relationship. Transactions on Emerging Telecommunications Technologies . 2022 doi: 10.1002/ett.4458. [DOI] [Google Scholar]
- 54.Khan W. U., Nguyen T. N., Jameel F., et al. Leaning-based Resource Allocation for Backscatter-Aided Vehicular Networks. IEEE Transactions on Intelligent Transportation Systems . 2022 doi: 10.1109/TITS.2021.3126766. [DOI] [Google Scholar]
- 55.Liu J., Ahmed M., Mirza M. A., et al. RL/DRL Meets Vehicular Task Offloading Using Edge and Vehicular Cloudlet: A Survey. IEEE Internet of Things Journal . 2022;9(11) doi: 10.1109/JIOT.2022.3155667. [DOI] [Google Scholar]
- 56.Khan W. U. Opportunities for Physical Layer Security in UAV Communication Enhanced with Intelligent Reflective Surfaces. 2022. https://arxiv.org/abs/2203.16907 .
- 57.Hasan T., Malik J., Bibi I., et al. Securing industrial internet of things against botnet attacks using hybrid deep learning approach. IEEE Transactions on Network Science and Engineering . 2022 doi: 10.1109/tnse.2022.3168533. [DOI] [Google Scholar]
- 58.Khan W. U., Ihsan A., Nguyen T. N., Muhammad A. J., Ali Z. NOMA-enabled backscatter communications for green transportation in automotive-industry 5.0. IEEE Transactions on Industrial Informatics . 2022:p. 1. doi: 10.1109/tii.2022.3161029. [DOI] [Google Scholar]
- 59.Zaher A. M., Eldeib A. M. Breast cancer classification using deep belief networks. Expert Systems with Applications . 2016;46:139–144. doi: 10.1016/j.eswa.2015.10.015. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data are publicly available at https://www.kaggle.com/c/histopathologic-cancer-detection/data.


