Abstract
Long-read sequencing (LRS) approaches shed new light on the complexity of viral (Kakuk et al., 2021 [1]; Boldogkői et al., 2019 [2]; Depledge et a., 2019 [3]), bacterial (Yan et al., 2018 [4]) and eukaryotic (Tilgner et al., 2014 [5]) transcriptomes. Emerging RNA viruses are zoonotic (Woolhouse et al., 2016 [6]) and create public health problems, e.g. influenza pandemic caused by H1N1 virus in (Fraser et al., 2009 [7]), as well as the current SARS-CoV-2 pandemic (Kim et al., 2020 [8]). In this study, we carried out nanopore sequencing for generating transcriptomic data valuable for structural and kinetic profiling of six important human pathogen RNA viruses, the H1N1 subtype of Influenza A virus (IVA), the Zika virus (ZIKV), the West Nile virus (WNV), the Crimean-Congo hemorrhagic fever virus (CCHFV), the Coxsackievirus [group B serotype 5 (CVB5)] and the Vesicular stomatitis Indiana virus (VSIV), and the response of host cells upon viral infection. The raw sequencing data were filtered during basecalling and only high quality reads (Qscore ≥ 7) were mapped to the appropriate viral and host genomes. Length distribution of sequencing reads were assessed and statistics of data were plotted by the ReadStat.4 python script. The datasets can be used to profile the transcriptomic landscape of RNA viruses, provide information for novel gene annotations, can serve as resource for studying the virus-host interactions, and for the analysis of RNA base modifications. These datasets can be used to compare the different sequencing techniques, library preparation approaches, bioinformatics pipelines, and to analyze the RNA profiles of viruses with small RNA genomes.
Keywords: Zika virus, Crimean-Congo hemorrhagic fever virus, West Nile virus, Coxsackievirus, Influenza a virus, Vesicular stomatitis Indiana virus, Transcriptome profiling, Third-generation sequencing
Specifications Table
| Subject | Biological sciences |
| Specific subject area | Omics: Transcriptomics Virology |
| Type of data | Raw BAM files Figure Table |
| How the data were acquired | Sequencing – Oxford Nanopore MinION R9.4 SpotOn and Flongle Flow Cells Basecalling – Guppy 3.6 Statistics – in house script (https://github.com/moldovannorbert/seqtools) |
| Data format | filtered data: after basecalling, reads were filtered based on quality score (Qscore ≥ 7) and passed mapped reads are stored in BAM files |
| Description of data collection | Various cell cultures were infected with six human pathogen RNA viruses. Total RNA was isolated from the infected cells at different time points after viral infection. Libraries were generated and then sequencing reactions were carried out on a MinION (Oxford Nanopore Technologies) device. Guppy 3.6 was used for basecalling and minimap2 for aligning the raw reads to the viral and host genomes. |
| Data source location | Coxsackievirus (CVB5)
|
Vesicular stomatitis Indiana virus
|
|
| Data accessibility | The available BAM files containing the reads aligned to the reference genomes are available at ENA and can be used without restrictions. Repository name: European Nucleotide Archive (ENA) Data identification numbers: PRJEB46600 (H1N1), PRJEB46591 (CCHFV, WNV and ZIKV), PRJEB46598 (CVB5), PRJEB46127 (VSIV) Direct links to the dataset: https://www.ebi.ac.uk/ena/browser/view/PRJEB46600 https://www.ebi.ac.uk/ena/browser/view/PRJEB46591 https://www.ebi.ac.uk/ena/browser/view/PRJEB46598 https://www.ebi.ac.uk/ena/browser/view/PRJEB46127 Supplementary files are available at figshare: doi:10.6084/m9.figshare.19228416 |
Value of the Data
-
•
Regularly recurring epidemics and pandemics underlie the importance of the molecular analysis of RNA viruses. Understanding the molecular genetic mechanisms of these pathogens is essential for developing a defense strategy against them.
-
•
Only a few LRS transcriptomic data of the RNA viruses have been generated so far, however these studies focus on various protocols and techniques developed by the authors’ laboratories rather than the viruses itself [9,10]. Although the genomes of these viruses have previously been reported, their transcriptomic architectures have not been well-characterized. They were also examined by short-read sequencing [11], or by qRT-PCR [12]. The primary aim of the generation of our datasets is its use for the characterization of the transcriptomic architecture of H1N1, CCHFV, ZIKV, WNV, VSIV and CVB5 viruses.
-
•
This dataset can be used to profile the virus-host interactions and the global changes of host gene expression. These data can also be included in meta-analyses to characterize the host cell response to infection using various viruses. The uploaded BAM files can be further analyzed, or LRS bioinformatics pipelines can be tested on it (most of them can be found at LONG-READ-TOOLS [13]).
-
•
This dataset can be used to profile the virus-host interactions and the global changes of host gene expression.
-
•
Our data can be reused or reanalyzed and it can be compared with other datasets with the aim to obtain a better reannotation of viral transcriptomes. Such type of integrative approach has already been applied in Herpesvirus research [14]. In addition to the canonical RNAs, we expect to obtain novel transcripts encoding unknown ORFs with fusion, deletion or frameshift or novel RNA isoforms, by following meta-analytic approach.
-
•
Combining various short-read RNA-Seq and LRS techniques allow fine detailing of viral transcriptome annotation [15] therefore this dataset is also useful for the gene expression dynamics profiling of H1N1, CCHFV, ZIKV, WNV, VSIV and CVB5 viruses.
1. Data Description
Here we report the transcriptome datasets of six, important human pathogen RNA viruses, the H1N1 subtype of Influenza A virus (IVA), the Zika virus (ZIKV), the West Nile virus (WNV), the Crimean-Congo hemorrhagic fever virus (CCHFV), the Coxsackievirus [group B serotype 5 (CVB5)] and the Vesicular stomatitis Indiana virus (VSIV), and their host cell's transcriptome in different time points of infection, obtained using long read sequencings (Fig. 1, Table 1A, Supplementary Information). Table 1. summarizes the characteristics of the examined viruses, parameters of the experimental design and the mapped read counts of each viral and host cell transcriptomes.
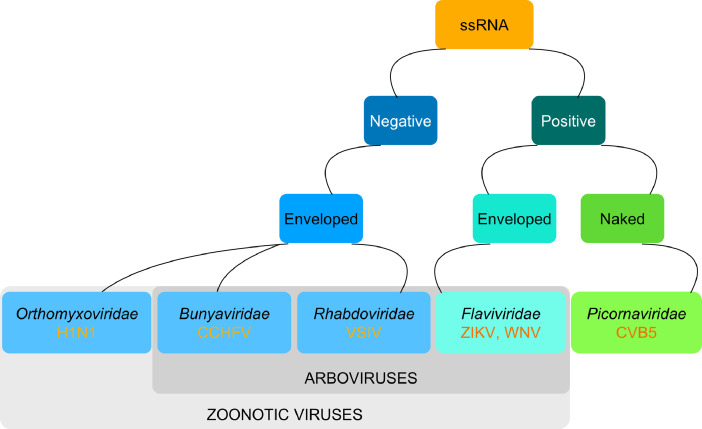
Fig. 1.
Classification of viruses examined in this study.
Table 1.
Details of the viruses, the experimental setup, the raw data statistics and ENA accession numbers A. Basic information of the examined viruses. B. Experimental conditions and summary statistics of the obtained dataset. Abbreviations: kb: kilobase, ss: single-stranded, MDCK: Madin-Darby canine kidney, Vero: kidney epithelial cells extracted from an African green monkey (Chlorocebus sabaeus), sp: species, MOI: multiplicity of infection C. European Nucleotide Archive (ENA) project accession numbers. D. GenBank IDs of viral and host reference genomes used for this study.
| A | Viruses | IVA | ZIKV | WNV | CCHFV | CVB5 | VSIV | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Genome length | 13.5 kb | 10.8 kb | 11.0 kb | 19.2 kb | 7.3 kb | 11 kb | ||||
| Genome type | (-)ssRNA | (+)ssRNA | (+)ssRNA | (-)ssRNA | (+)ssRNA | (-)ssRNA | ||||
| Number of segments | 8 | - | - | 3 | - | - | ||||
| Number of genes | 11 | 1 | 1 | 3 | 1 | 5 | ||||
| PolyA-tailed mRNA | ✓ | - | - | - | ✓ | ✓ | ||||
| 5’-Cap | ✓ | ✓ | ✓ | ✓ | - | ✓ | ||||
| Vectors | - | Aedes mosquitoes | Culex mosquitoes | Ixodid (hard) ticks (Hyalomma sp) | - | Black flies (Simulium sp) Sand flies (Lutzomyia sp) |
||||
| Reservoirs | Wild birds | Monkeys, Human | Wild birds | Hard ticks | Human | Horse, cattle, pig | ||||
| B | Viral strain | H1N1 | MR766 | Own isolate, Serbia, 2014 | Kosova Hoti | B5 | ||||
| Host cell(s) | MDCK | Vero | Vero | Vero | Vero | Vero |
T98G |
|||
| Examined time points p.i. | 1 h, 4 h, 7 h | 24 h, 72 h | 24 h, 72 h | 24 h, 72 h | 1h, 6h, 15h, 24h | 1 h, 6 h, 15 h, 24 h |
||||
| V1 | V2 | G1 | G2 | |||||||
| MOI | 10 | low | low | low | low | 5 | 5 | 5 | 5 | |
| Transcript read counts | 10916 | 592 | 21790 | 528 | 1508 | 193820 | 273882 | 289807 | 255610 | |
| Average read lengths | 812 | 634 | 491 | 683 | 630 | 805 | 954 | 1021 | 1122 | |
| Maximum mapped read length | 2.28 kb | 2.59 kb | 2.60 kb | 1.86 kb | 6.99 kb | 4.45 kb | 6.38 kb | 5.83 kb | 6.24 kb | |
| C | Project ID (ENA) | PRJEB46600 | PRJEB46591 | PRJEB46598 | PRJEB46127 | |||||
| D | Virus reference genome ID | GCF_001343785.1 | NC_012532.1 | NC_001563.2 | NC_005300.2 | AF114383 |
NC_001560.1 |
|||
| Host reference genome ID | GCA_000002285.4 | GCF_000409795.2 | GCF_000409795.2 | GCF_000409795.2 | GCF_000409795.2 | GCA_000409795.2 | GCA_000001405.28 | |||
This article reports the dataset of transcriptome analysis of canine kidney (MDCK) cell line infected with H1N1 influenza viruses, the African green monkey kidney cell line (Vero) infected with ZIKV, WNV, CCHFV and CVB5 viruses. VSIV transcriptome analysis carried out on Vero and T98G human glioblastoma cell lines (Table 1B). Mock infected cells were also sequenced, as controls. Barcode sequences, used to distinguish between the sequencing libraries of various samples are listed in Supplementary Table 1.
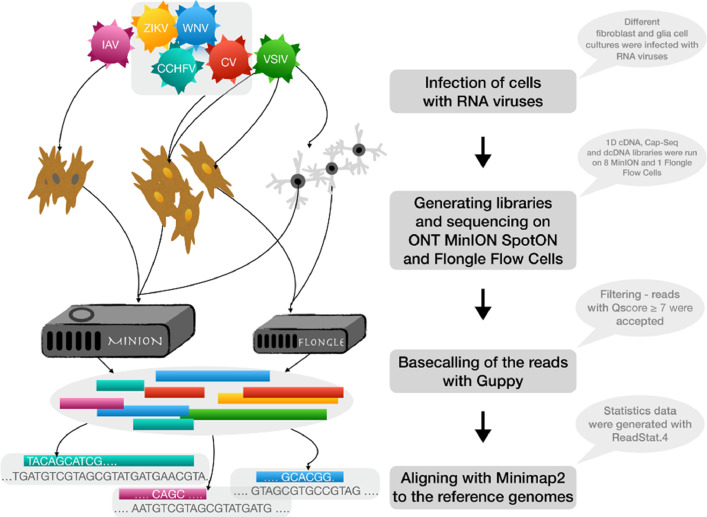
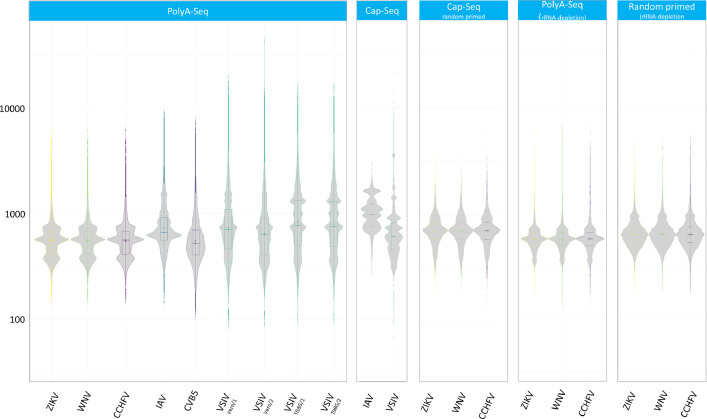
The MinION R9.4 type and Flongle flow cells from the Oxford Nanopore Technologies were used for long read sequencing. The bioinformatic pipeline included the basecalling and de-barcoding of FAST5 raw files into FASTQ files, then mapping the reads to the appropriate genomes and the generation of read distribution statistics by the ReadStat.4 tool. The experimental design, the bioinformatic pipeline is depicted on Fig. 2. Read length distributions in each sequencing libraries (PolyA-seq, random primed, Cap-Seq, PolyA-Seq and random primed combined with ribodepletion and Cap-selection combined with ribodepletion) were calculated and depicted on Fig. 3.
Fig. 2.
Schematic overview of the study and the bioinformatic pipeline.
Fig. 3.
Violin plots of read length distribution of the sequencing data.
Sequencing data were deposited in compressed BAM files in the European Nucleotide Archive (ENA) and are publicly available under the accession numbers enlisted in Table 1C.
Altogether the experiments yielded 23,254,878 sequencing reads, of which 4,689,145 mapped to the various viral and 18,565,733 to the host genomes (Table 1D). The average length for the ONT 1D cDNA sequencing ranged between 566 and 1,369 bps, whereas the Cap-Seq library preparation approach generated an average length of 725 – 1,092 bp. More statistical details can be found in Supplementary Table 2.
Supplementary Table 1. Details of library preparation. Experimental design, type of library preparations, time point of infection, sequence of barcodes and amplification count
Supplementary Table 2. Summary of mapped read statistics in each sequencing library sample after quality filtering. Read count, mean read length, minimum, maximum read length, median read length, and mapped read count, mapped mean read length, mapped minimum and maximum read length and standard deviations (stdev) are listed in the table.
Supplementary Information. More information on the viruses analyzed in this study.
2. Experimental Design, Materials and Methods
The data presented here have been obtained from cell cultures infected by ZIKV, CCHFV, CVB5, VSIV, WNV and H1N1. The wet and dry lab workflows have been illustrated in Fig. 2.
2.1. Viruses and Cell Lines
The Coxsackievirus was provided by Dr. Andrea Kátai (Public Health and Food Chain Safety Service of Government Office for Csongrád County, Laboratory Department, Szeged, Hungary). The WNV was isolated in Vojvodina, Serbia in 2013 [16]. The mouse-adapted influenza type A virus of subtype H1N1 (A/Puerto Rico/8/1934) was kindly provided by the National Center for Epidemiology (Budapest, Hungary). The VSIV was obtained from the collection of the Department of Medical Microbiology and Immunobiology, University of Szeged (Szeged, Hungary). The Kosova Hoti strain of CCHFV used in our experiments was isolated at the Balkan Peninsula [17]. The African ZIKV, strain MR766 was obtained from the European Virus Archive.
All laboratory manipulations associated with ZKV, WNV and CCHFV were performed in a BSL-4 suite laboratory, at the Szentágothai Research Centre, University of Pécs. The CVB5, the VSIV experiments, as well as the H1N1 laboratory works were carried out at BSL-2 laboratories at University of Szeged and at the National Food Chain Safety Office, respectively.
For the propagation of H1N1 virus, IVA stocks were prepared in 10-day-old embryonated eggs, and the titer was determined by plaque assay on MDCK cells. Cells were infected with 1 ml influenza virus suspension using a multiplicity of infection (MOI)=10 and the infection was arrested 1, 4 and 7h p.i. VSIV was propagated on Vero and human glioblastoma cell line (T98G) using MOI=5. The infection was terminated at 1h, 6h, 15h and 24h p.i. CVB5 was cultured on Vero cell line and the infection was arrested 1h, 6h, 15h, 24h p.i. ZIKV, WNV and CCHFV were grown to high titers on Vero cells. Supernatants were aliquoted and then frozen at −80°C. Cells were infected with low titer (not determined) of viruses for this study. Cells were incubated for 24 and 72 h. Incubations were carried out at 37°C in a humidified atmosphere containing 5% CO2.
2.2. RNA Extraction
For the purification of total RNAs from the viral infected cells, the NucleoSpin® RNA kit (Macherey-Nagel) was applied. Cells were collected by low-speed centrifugation, then lysis was carried out with the addition of a chaotropic ion containing buffer solution and ß-Mercapthoethanol in order to inactivate the RNase enzymes. The lysates were filtered by using the NucleoSpin® Filter (Macherey-Nagel kit) and centrifuged at 11,000 x g for 1 min at room temperature (RT). Filters were discarded and ethanol (70%) was used to wash the samples. The lysates were loaded onto the NucleoSpin® RNA Column (Macherey-Nagel kit) and centrifuged at 11,000 x g for 30 s, which enables RNA to bind to the RNA Column. Membrane desalting buffer and then rDNase (both from the Macherey-Nagel kit) were loaded onto the membrane and incubated at RT for 15 min to remove residual DNA. Samples were treated with wash buffer (Macherey-Nagel RNA kit) and then the RNAs were eluted from the membrane of the RNA Column in RNase-free water (Ambion). Total RNA samples were then treated with Ambion® TURBO DNA-free™ Kit (Ambion) to remove the potential DNA contamination. The protocols were carried out according manufacturer's instructions.
2.3. Library Preparation
Our aim was to obtain full-length sequencing reads, therefore we used oligo(dT) primers for reverse transcription (RT) following the ONT's 1D Strand switching cDNA by ligation method. A combination of a 5’-Cap selection protocol with the ONT's 1D library preparation) was also used to obtain the exact 5’-end of the RNAs, [18]. Random oligonucleotide primers were also used for the RT reactions in order to detect potential non-polyA-tailed transcripts.
2.4. PolyA-Selection
The polyA(+) fraction of RNA was purified from half of the total RNA samples by using the Oligotex mRNA Mini Kit (Qiagen), following the “Spin Columns” method of the kit.
2.5. Ribodepletion
Ribosomal (r)RNAs were removed from the other half of total RNAs by using the Ribo-Zero Magnetic Kit H/M/R (Epicentre/Illumina) according to the kit's manual.
2.6. Polyadenylation
Because some or all of the mRNAs of ZIKV, CCHFV and WNV are not polyadenylated (Table 1A), polyA-tails were attached to the rRNA-depleted samples using Escherichia coli Poly(A) Tailing Kit (Invitrogen; as described by the manual of the kit) in order to capture the 3’-ends of RNAs.
2.7. Cap-Selection
The Lexogen's TeloPrime Full-Length cDNA Amplification Kit was used to generate cDNAs from the capped mRNAs (Table 1A). This method is based on specific double-stranded adapters, which ligates only to the first strand cDNAs if the inverted Gs of the cap structure are present. The cDNAs were produced from total RNAs using oligo(dT) primers and RT enzyme following the recommendations of the kit's manual. The ligation step was carried out at 25°C, overnight, then it was followed by the generation of the second cDNA strand using the forward PCR primer and the enzyme mix (both from the Lexogen kit). The following program was used: 1 cycle of 90 s at 95.8°C, 60 s at 62°C, 5 min at 72°C. Finally, the double-stranded cDNAs were amplified by PCR using PCR mix, both the forward and reverse PCR primers and enzyme mix (from the Lexogen kit). Sixteen PCR cycles were performed according to the followings: 1 cycle of 95.8°C for 30 s, 50°C for 45 s, 72°C for 20 min, then 15 cycles of 95.8°C for 30 s, 62°C for 30 s, 72°C for 20 min, and a final extension step at 72°C for 20 min.
2.8. ONT 1D Library Preparation
The strand switching cDNA by ligation library preparation approach was used to generate libraries by using the ONT's Ligation Sequencing kit (SQK-LSK108). In brief, the first cDNA strand was synthesed using SuperScript IV Reverse Transcriptase enzyme (Thermo Fisher Scientific) and an anchored oligo(dT) [(VN)T20] or random primers. A 5′ strand-switching adapter (containing three O-methyl-guanine RNA bases) was added to the mixture. The PCR reactions were performed with Kapa HiFi DNA polymerase (Kapa Biosystems)/LongAmp Taq 2X Master Mix (New England Biolabs), the end repair was carried out with NEBNext End repair/dA-tailing Module, then the adapter ligation with NEB Blunt/TA Ligase Master Mix (New England Biolabs). The cDNAs were cleaned up between each step using Agencourt AMPure XP magnetic beads (Beckman Coulter). Cap-Seq libraries were generated from the Cap-selected cDNAs with the addition of ONT adapters using NEB Blunt/TA Ligase Master Mix.
2.9. ONT Direct cDNA Sequencing
Libraries were prepared from the VSIV samples (1, 6, 15 and 24 h pi) using the direct cDNA (dcDNA) Sequencing Kit (SQK-DCS109, ONT) according to the manufacturer's instructions. In short, Poly(A)+ RNA samples, the VN oligo(dT) primer from the ONT kit and dNTP were mixed and incubated at 65°C for 5 min. After this denaturation step, RT Buffer, RNaseOUT (Thermo Fisher Scientific), and Strand-Switching Primer (from the ONT Kit) were added. The samples were incubated at 42°C for 2 min. Maxima H Minus Reverse Transcriptase enzyme was added to the samples. RT and strand-switching reactions were performed at 42°C for 90 min. It was followed by heat inactivation step (85°C, 5 min). RNase Cocktail Enzyme Mix (Thermo Fisher Scientific) was added to remove the residual RNAs (37°C,10 min). The purified (AMPure XP), and amplified (LongAmp Taq Master Mix) cDNAs were end-repaired using the NEBNext Ultra II End repair enzyme and buffer. The Adapter Mix was ligated to the end-prepped samples by using the Blunt/TA Ligation Master Mix (NEB).
2.10. Barcoding
1D and Cap-Seq libraries were barcoded using PCR Barcoding Expansion 1-96 (EXP-PBC096) kit, while the dcDNA samples with the Native Barcoding (12) Kit (ONT) following the manufacturer's recommendations (Supplementary Table 1). Mock-infected samples, and samples from the earlier time points (1 and 2 h p.i.) were sequenced separately from the later time point samples in order to avoid potential ‘barcode hopping’ issue.
2.11. Sequencing
Library concentration was detected by using Qubit 2.0 Fluorometer and Qubit (ds)DNA HS Assay Kit (Thermo Fisher Scientific). Samples were loaded and sequenced on ONT MinION R9.4 SpotOn or Flongle Flow Cells (Supplementary Table 1).
2.12. Data Processing and Analysis
Base calling was performed using Guppy 3.6, thereafter the high quality reads (Qscore ≥ 7) of FASTQ files were aligned to the appropriate viral and host reference genomes using minimap2 (Table 1D). Statistical analysis of mapped reads was carried out by the readstat.py program suite (https://github.com/moldovannorbert/seqtools). Mapped SAM files have been compressed by samtools and all BAM files have been deposited in the European Nucleotide Archive (ENA) under the ENA project and sample accession numbers listed in Table 1C.
2.13. Technical Validation
The concentration of total RNA, polyadenylated RNA samples as well as the sequencing ready libraries were quantified by Qubit 2 Fluorometer and Qubit RNA Broad-Range, High Sensitivity RNA, and High Sensitivity dsDNA Assay Kits, respectively. The quality of the nucleic acids was checked by using the Agilent TapeStation 4150. RNA samples with RIN ≥ 9.0 were used for library preparation. Three or two replicates were used for sequencing the VSIV and CV samples, respectively. Various library preparation methods were used in order to provide independent methods for the validation of novel transcripts. To monitor the effect of VSIV infection on host cell transcriptome, mock-infected cells were also sequenced.
Ethics Statements
Not applicable.
CRediT authorship contribution statement
István Prazsák: Investigation, Formal analysis, Validation, Visualization. Zsolt Csabai: Investigation, Validation. Gábor Torma: Formal analysis, Visualization. Henrietta Papp: Investigation. Fanni Földes: Investigation. Gábor Kemenesi: Investigation. Ferenc Jakab: Resources, Supervision. Gábor Gulyás: Data curation, Visualization. Ádám Fülöp: Data curation. Klára Megyeri: Investigation, Resources. Béla Dénes: Investigation, Resources. Zsolt Boldogkői: Investigation, Conceptualization, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition. Dóra Tombácz: Investigation, Methodology, Resources, Writing – original draft, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Funding: This work was supported by the National Research, Development, and Innovation Office grants FK 128252 to DT and K 128247 to ZB. The project was also funded by the Lendület (Momentum) I Program of the Hungarian Academy of Sciences LP-2020/8 to DT.
ZC was supported by the UNKP-21-4-SZTE-126 New National Excellence Program of the Ministry for Innovation and Technology from the source of the National Research, Development and Innovation Fund. GG was supported by the UNKP-21-3-SZTE-51 New National Excellence Program of the Ministry for Innovation and Technology from the source of the National Research, Development and Innovation Fund
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2022.108386.
Contributor Information
Zsolt Boldogkői, Email: boldogkoi.zsolt@med.u-szeged.hu.
Dóra Tombácz, Email: tombacz.dora@med.u-szeged.hu.
Appendix. Supplementary materials
Data Availability
References
- 1.Kakuk B., Kiss A.A., Torma G., Csabai Z., Prazsák I., Mizik M., Megyeri K., Tombácz D., Boldogkői Z. Nanopore assay reveals cell-type-dependent gene expression of vesicular stomatitis indiana virus and differential host cell response. Pathogens. 2021;10:1196. doi: 10.3390/pathogens10091196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boldogkői Z., Moldován N., Balázs Z., Snyder M., Tombácz D. Long-read sequencing - a powerful tool in viral transcriptome research. Trends Microbiol. 2019;27:578–592. doi: 10.1016/j.tim.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Depledge D.P., Srinivas K.P., Sadaoka T., Bready D., Mori Y., Placantonakis D.G., Mohr I., Wilson AC. Direct RNA sequencing on nanopore arrays redefines the transcriptional complexity of a viral pathogen. Nat. Commun. 2019;10:754. doi: 10.1038/s41467-019-08734-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan B., Boitano M., Clark T.A., Ettwiller L. SMRT-cappable-Seq reveals complex operon variants in bacteria. Nat. Commun. 2018;9:3676. doi: 10.1038/s41467-018-05997-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tilgner H., Grubert F., Sharon D., Snyder M.P. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc. Natl. Acad. Sci. USA. 2014;111:9869–9874. doi: 10.1073/pnas.1400447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woolhouse M.E., Brierley L., McCaffery C., Lycett S S. Assessing the epidemic potential of RNA and DNA viruses. Emerg. Infect. Dis. 2016;22:2037–2044. doi: 10.3201/eid2212.160123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser C., Donnelly C.A., Cauchemez S., Hanage W.P., Van M.D. Kerkhove T.D.H, Griffin J., Baggaley R.F., Jenkins H.E., Lyons E.J., Jombart T., Hinsley W.R., Grassly N.C., Balloux F., Ghani A.C., Ferguson N.M., Rambaut A., Pybus O.G., Lopez-Gatell H., Alpuche-Aranda C.M., Chapela I.B., Zavala EP E.P., Guevara D.M., Checchi F., Garcia E., Hugonnet S., Roth C. WHO rapid pandemic assessment collaboration. pandemic potential of a strain of influenza A (H1N1): early findings. Science. 2009;324:1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wongsurawat T., Jenjaroenpun P., Taylor M.K., Lee J., Tolardo A.L., Parvathareddy J., Kandel S., Wadley T.D., Kaewnapan B., Athipanyasilp N., Skidmore A., Chung D., Chaimayo C., Whitt M., Kantakamalakul W., Sutthent R., Horthongkham N., Ussery D.W., Jonsson C.B, Nookaew I. Rapid sequencing of multiple RNA viruses in their native form. Front. Microbiol. 2019;10:260. doi: 10.3389/fmicb.2019.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan C., Maurer-Stroh S., Wan Y., Sessions O.M., de Sessions P.F. A novel method for the capture-based purification of whole viral native RNA genomes. AMB Express. 2019;9:45. doi: 10.1186/s13568-019-0772-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozak R.A., Fraser R.S., Biondi M.J., Majer A., Medina S.J., Griffin B.D., Kobasa D., Stapleton P.J., Urfano C., Babuadze G., Antonation K., Fernando L., Booth S., Lillie B.N., Kobinger G.P. Dual RNA-Seq characterization of host and pathogen gene expression in liver cells infected with Crimean-Congo hemorrhagic fever virus. PLoS Negl. Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seong R.K., Lee J.K., Cho G.J., Kumar M., Shin O.S. mRNA and miRNA profiling of Zika virus-infected human umbilical cord mesenchymal stem cells identifies miR-142-5p as an antiviral factor. Emerg. Microbes Infect. 2020;9:2061–2075. doi: 10.1080/22221751.2020.1821581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amarasinghe S.L., Ritchie M.E., Gouil Q. long-read-tools.org: an interactive catalogue of analysis methods for long-read sequencing data. Gigascience. 2021;10:giab003. doi: 10.1093/gigascience/giab003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tombácz D., Torma G., Gulyás G., Moldován N., Snyder M., Boldogkői Z. Meta-analytic approach for transcriptome profiling of herpes simplex virus type 1. Sci. Data. 2020;7:223. doi: 10.1038/s41597-020-0558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Depledge D.P, Mohr I., Wilson A.C. Going the Distance: optimizing RNA-Seq strategies for transcriptomic analysis of complex viral genomes. J. Virol. 2018;93 doi: 10.1128/JVI.01342-18. e01342-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zana B., Kemenesi G., Herczeg R., Dallos B., Oldal M., Marton S., Krtinic B., Gellért Á., Bányai K., Jakab F. Genomic characterization of West Nile virus strains derived from mosquito samples obtained during 2013 Serbian outbreak. J. Vector Borne Dis. 2016;53:379–383. [PubMed] [Google Scholar]
- 17.Duh D., Nichol S.T., Khristova M.L., Saksida A., Hafner-Bratkovič I., Petrovec M., Dedushaj S Ahmeti I., Avšič-Županc T. The complete genome sequence of a Crimean-Congo Hemorrhagic fever virus isolated from an endemic region in Kosovo. Virol. J. 2008;5:7. doi: 10.1186/1743-422X-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tombácz D., Prazsák I., Csabai Z., Moldován N., Dénes B., Snyder M., Boldogkői Z. Long-read assays shed new light on the transcriptome complexity of a viral pathogen. Sci. Rep. 2020;10:13822. doi: 10.1038/s41598-020-70794-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.