Summary
Autism spectrum disorder (ASD) is a neurodevelopmental disorder with high genetic heterogeneity, affecting one in 44 children in the United States. Recent genomic sequencing studies from autistic human individuals indicate that PARK2, a gene that has long been considered in the pathogenesis of Parkinson's disease, is involved in ASD. Here, we report that Prkn knockout (KO) mice demonstrate autistic-like behaviors including impaired social interaction, elevated repetitive behaviors, and deficits in communication. In addition, Prkn KO mice show reduced neuronal activity in the context of sociability in the prelimbic cortex. Cell morphological examination of layer 5 prelimbic cortical neurons shows a reduction in dendritic arborization and spine number. Furthermore, biochemistry and immunocytochemistry analyses reveal alterations in synapse density and the molecular composition of synapses. These findings indicate that Prkn is implicated in brain development and suggest the potential use of the Prkn KO mouse as a model for autism research.
Subject areas: Behavioral neuroscience, Molecular neuroscience, Developmental neuroscience, Model organism
Graphical abstract

Highlights
-
•
Prkn KO mice show autistic-like behaviors
-
•
Prkn KO mice show reduced sociability-related neuronal activity in the prelimbic cortex
-
•
Prkn loss leads to a reduction in dendritic arborization
-
•
Prkn loss alters synaptic proteins and reduces synapse density
Behavioral neuroscience; Molecular neuroscience; Developmental neuroscience; Model organism
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder with a rising prevalence in the population, estimated to affect one in 44 children by far in the United States (Maenner et al., 2021). ASD individuals are diagnosed behaviorally by three core characteristics including impaired social interaction, repetitive and stereotypical behaviors, and deficits in communication (Landa, 2008). In addition, ASD demonstrates a variety of comorbidities including intellectual disability (Srivastava and Schwartz, 2014), anxiety (Zaboski and Storch, 2018), attention-deficit hyperactivity disorder (Rao and Landa, 2014), and seizures (El Achkar and Spence, 2015; Viscidi et al., 2013). Despite the shared common behavioral traits, the genetic etiological factors of ASD are highly heterogeneous. Studies from twins and families of ASD individuals initially revealed the strong genetic heritability of ASD (Colvert et al., 2015; Morrow et al., 2008; Sandin et al., 2017; Tick et al., 2016). Recent studies from large-scale genetic sequencing of ASD populations have revealed high occurrence (∼10% of cases) of genomic DNA copy number variations (CNVs) in ASD, in either heritable or de novo forms (Shishido et al., 2014). Moreover, sequence analysis of CNVs from ASD individuals mapped a wide variety of ASD-related genes (Glessner et al., 2009; He et al., 2013; Levy et al., 2011; Morrow et al., 2008; Pinto et al., 2010; Sebat et al., 2007; Stein et al., 2013). In addition to dosage changes due to CNVs, ASD-related genes can also result from missense mutations, nonsense mutations, insertions, or deletions (Anney et al., 2010; Satterstrom et al., 2020).
Two categories of functionally related genes with high occurrence in CNVs are recognized for their preferential linkage to ASD: neuronal cell-adhesion molecule genes such as CNTNAP2 and NLGN3, and the ubiquitin-proteasome system genes, such as UBE3A (Glessner et al., 2009). Genes from both categories are implicated in synapse formation and synaptic transmission, consistent with the commonly found cellular phenotype of synaptopathy in ASD. Interestingly, both duplication and deletion of the chromosome locus 6q23, a region only harboring the gene PRKN within the human genome, have been identified in ASD individuals (Girirajan et al., 2013; Glessner et al., 2009; He et al., 2013; Levy et al., 2011; Morrow et al., 2008; Pinto et al., 2010; Prasad et al., 2012; Scheuerle and Wilson, 2011; Yin et al., 2016), implicating the involvement of the PRKN gene in the neurobiology of ASD. Notedly, PRKN deletions are the dominant form by case numbers (Glessner et al., 2009; Scheuerle and Wilson, 2011). Also, in duplication cases, the recurrent duplicated region with certain exons might cause early termination of translation by frameshift (Girirajan et al., 2013). These observations suggest that a lack of PRKN gene product leads to neurodevelopmental defects in ASD.
Parkin, encoded by the PRKN (PARK2) gene, is an E3 ubiquitin-protein ligase and has a well-established role in early-onset Parkinson's disease (Fearnley and Lees, 1991; Kitada et al., 1998; Lucking et al., 2000). As a RING-between-RING E3 ligase, Parkin either directly catalyzes the transfer of ubiquitin during ubiquitination, or functions as a scaffold protein to bring specific substrates to other E3 ligases, therefore enabling the regulation of a broader spectrum of substrates and biological processes in the cell (Seirafi et al., 2015). Interestingly, a recent study reported that there is a high frequency of parkinsonism (32%) among ASD individuals older than 39 years, implying a mechanistic convergence between Parkinson's disease and ASD (Starkstein et al., 2015). Accumulating evidence suggests that Parkin may play a role in regulating synapse number, synaptic composition, and transmission in neurons. It has been shown that knockdown of Parkin alters glutamatergic synaptic transmission by reducing surface levels of AMPA receptors (AMPARs) and NMDA receptors (NMDARs) (Cortese et al., 2016; Zhu et al., 2018), which is in line with the common features of synaptopathy in ASD where synaptic transmission, synapse number, and levels of neurotransmitter receptor expression are altered. It has been also shown that over-expression of Parkin promotes excitatory synapse pruning, whereas knockdown of Parkin blocks the pruning effect in cultured hippocampal neurons (Helton et al., 2008), implying the divergent mechanisms underlying the duplication or deletion of the Prkn gene in the etiology of ASD. However, there is no direct evidence linking the change in PRKN gene dosage to autistic-like behaviors. Here, we report that the loss of the Prkn gene in mice results in autistic-like behaviors, accompanied with altered neuronal activity, abnormalities in synapse formation and synaptic molecular composition. These findings indicate a direct involvement of Parkin in the etiology of autism, suggesting the potential of using Prkn knockout (KO) mouse as a novel animal model of ASD.
Results
Prkn KO mice show deficits in social interaction
To examine whether the Prkn KO mice demonstrate autistic-like phenotypes, we compared Prkn KO mice and wild-type (WT) mice on their behavioral characteristics using classic behavioral paradigms during postnatal development (Figure 1A). Impaired social interaction is one of the most recognizable manifestations of autistic behaviors in human individuals with ASD (Battle, 2013). Therefore, we performed three-chamber tests to assess their sociability and interest in social novelty (Crawley, 2007) (Figure 1B). Initially, a subject mouse was placed in the middle chamber and allowed to explore all three chambers to become acquainted with the environment. To examine sociability, an age- and gender-matched WT stranger mouse (stranger 1) was introduced into a wired cage at one side chamber, and the subject mouse was allowed to freely navigate in the three-chamber apparatus for 5 min. The time spent in each chamber and in closeness, proximity with the side cages was quantified. WT mice spent more time in the chamber interacting with stranger 1 (Figures 1C and 1D), as compared to the time spent in the empty chamber. Surprisingly, rather than social preference to stranger 1, Prkn KO mice showed a reversed preference by spending significantly more time in the empty chamber (Figures 1C and 1D) (genotype factor F (1, 23) = 0.08, p = 0.78; chamber factor F (2, 46) = 19.24, p < 0.0001; interaction F (2, 46) = 8.20, p = 0.0009; NWT = 10, NPrkn2 KO = 15, two-way ANOVA). To further evaluate the preference toward stranger 1, we calculated the preference index (see STAR Methods). While a preference toward stranger 1 was found in WT mice, Prkn KO mice showed a reverse preference toward the empty chamber in terms of time spent in the designated chamber (Figure 1E). We further measured the time spent in close proximity to each cage (area within 3 cm from the rim of the wired cage). Consistent with the result based on the time spent in the chambers, WT mice spent more time in close proximity to the cage containing stranger 1 (Figure 1H). On the contrary, Prkn KO mice spent a significantly longer time in close proximity to the empty side (Figure 1E; genotype factor F (1, 23) = 2.92, p = 0.10; chamber factor F (1, 23) = 1.26, p = 0.27; interaction F (1, 23) = 17.77, p = 0.0003; NWT = 10, NPrkn KO = 15, two-way ANOVA). Preference indexes showed that WT mice had a preference toward stranger 1 in terms of the time spent in proximity to the stranger 1 cage, whereas Prkn KO mice showed a reverse preference toward the empty side (Figure 1I).
Figure 1.
Prkn KO mice show impaired social interaction and abnormal response to social novelty
(A) Behavioral tests timeline during mouse developmental stages.
(B) Experimental paradigm for three-chamber social interaction tests.
(C) Representative traces of mouse movement from sociability test (left) and social novelty test (right) from Prkn KO and WT mice.
(D) In the social interaction test, Prkn KO mice spent a significantly longer time in the chamber with the empty cage, whereas WT mice spent more time in the chamber with the “stranger 1” mouse.
(E) Quantification of preference index (defined in STAR Methods) showed a reversal in preference for stranger 1 in Prkn KO mice as compared to WT mice.
(F) In the social novelty test, Prkn KO mice did not show a preference between “stranger 1” and the newly introduced “stranger 2” mouse, whereas WT mice spent significantly more time in the chamber of stranger 2 mouse.
(G) Quantification of preference index showed no preference between stranger 2 and stranger 1 in Prkn KO mice as compared to the preference toward stranger 2 in WT mice.
(H and J) Time spent in close proximity (within 3 cm) of the “stranger 1” versus “empty” cage (H) or the “stranger 1” versus “stranger 2” (J).
(I) Quantification of preference index showed a reversal in preference for stranger 1 in Prkn KO mice as compared to WT mice.
Error bars represent SEM. ∗p < 0.05, ∗∗p < 0.01, n. s., not significant; in (D), (F), (H), and (J) repeated measures two-way ANOVA with Bonferroni post hoc test; in (E), (G), (I), and (K) two-tailed Student’s t test.
Next, we placed a novel WT mouse (stranger 2) into the cage at the initially empty side chamber (Figures 1B and 1C). As expected, WT mice spent more time in the chamber with stranger 2, demonstrating a preference for social novelty. In contrast, Prkn KO mice displayed no preference toward the novel social target by spending an equal amount of time in either of the side chambers containing stranger 1 and stranger 2 (Figure 1F; genotype factor F (1, 23) = 2.91, p = 0.10; chamber factor F (2, 46) = 19.53, p < 0.0001; interaction F (2, 46) = 7.87, p = 0.0012; NWT = 10, NPrkn KO = 15, two-way ANOVA). Consistently, the preference index in terms of the time spent in the chamber indicates that WT mice had a preference toward stranger 2, whereas Prkn KO mice had no clear preference toward either side (Figure 1G). Next, the time spent in closeness, proximity to the cages with stranger 2 or stranger 1 was quantified. Consistent with results from the time spent in the chambers, WT mice spent more time in close proximity to the cage with stranger 2, suggesting a preference toward stranger 2 (Figure 1F), whereas Prkn KO mice showed no preference between the two side chambers by spending a relatively equal amount of time in close proximity to the cages with stranger 1 and stranger 2, respectively (Figure 1J; genotype factor F (1, 23) = 8.63, p = 0.007; chamber factor F (1, 23) = 6.19, p = 0.021; interaction F (1, 23) = 4.53, p = 0.044; NWT = 10, NPrkn KO = 15, two-way ANOVA). These results show that Prkn KO mice have impairments in social interaction and reduced preference for social novelty.
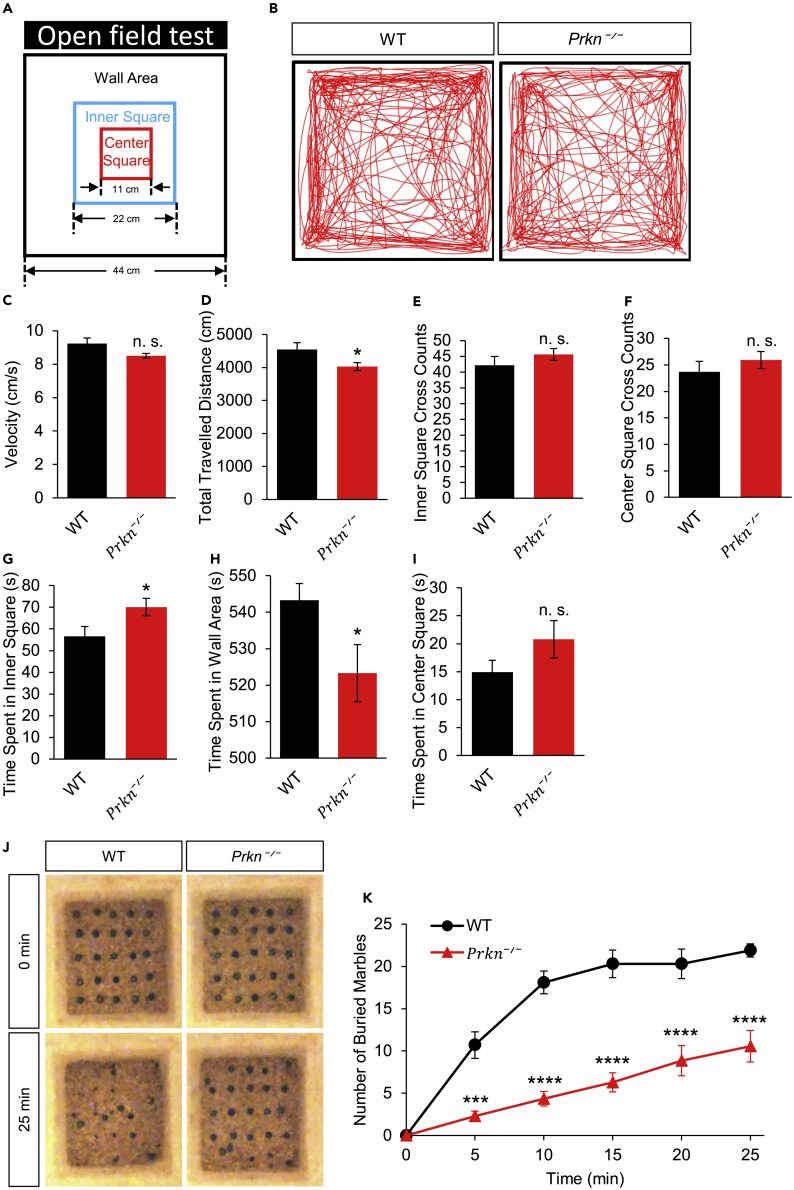
Prkn KO mice show abnormalities in open field and marble burying tests
Mouse models of ASD often demonstrate comorbidities including deficits in locomotion and altered level in anxiety. Therefore, we performed the open field test (Figure 2A), which is indicative of locomotion and anxiety, and the marble burying test, which is indicative of anxiety (Angoa-Perez et al., 2013; Seibenhener and Wooten, 2015; Treit et al., 1981). In the open field test, Prkn KO mice showed no difference in the average travel velocity compared to that of the WT mice (Figures 2B and 2C; WT: 9.23 ± 0.33 cm/s, N = 10; Prkn KO: 8.50 ± 0.13 cm/s, N = 14; p = 0.06), suggesting normal motor functions. A slight reduction in the total distance traveled was detected, which was caused by more frequent pausing and rearing (Figure 2D; WT: 4541.46 ± 207.86 cm, N = 10; Prkn KO: 4026.44 ± 115.73 cm, N = 14; p = 0.04). Compared to the WT mice, Prkn KO mice showed comparable numbers in inner square crossings (Figure 2E; WT: 42.20 ± 2.74, N = 10; Prkn KO: 45.57 ± 1.85, N = 14; p = 0.32) and center square crossings (Figure 2F; WT: 23.70 ± 1.99, N = 10; Prkn KO: 25.93 ± 1.61, N = 14; p = 0.39). However, Prkn KO mice spent a longer time in the inner square area (Figure 2G; WT: 56.60 ± 4.52 Sec, N = 10; Prkn KO: 70.14 ± 4.03 Sec, N = 14; p = 0.03) and less time near the wall area (Figure 2H; WT: 543.30 ± 4.56 Sec, N = 10; Prkn KO: 523.30 ± 7.83 Sec, N = 14; p = 0.03). These findings indicate that Prkn KO mice have normal locomotion with probably a reduced level of anxiety.
Figure 2.
Reduced anxiety-like behavior and normal locomotion in Prkn KO mice
(A) Illustration and dimensions of the arena for the open field test.
(B) Representative traces of mouse movement in open field tests of WT and Prkn KO mice.
(C–I) Assessments of mouse locomotion and anxiety levels from open field tests of both WT (N = 10) and Prkn KO mice (N = 14). (C and D) Prkn KO mice traveled significantly less distance (D) with no significant difference in average travel velocity (C) as compared to WT mice. (E and F) Prkn KO mice showed no difference in the counts of inner square crossings as well as center square crossings. (G–I) Time spent in the inner square area, wall area, and center square area. Prkn KO mice spent significantly more time in the inner square and less time in the wall area as compared to WT mice (G and H). However, Prkn KO mice showed no significant difference in the time spent in the center square as compared to WT mice (I).
(J and K) Marble burying test. The quantity of buried marbles in the cage of Prkn KO mice was significantly reduced as compared to that of WT mice during the 25 min time course. WT, N = 10; Prkn KO, N = 14.
All graphs depict mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, n. s., not significant; in (C to I) two-tailed Student’s t test; in (K), repeated measures two-way ANOVA with Bonferroni post hoc test.
In the marble burying test, WT or Prkn KO mice were individually placed into a cage with 25 marble balls arrayed in a five multiplied by five formation on the bedding (Figure 2J). When the buried marbles were counted 25 min later, we found that the Prkn KO mice buried significantly fewer marble balls across the 25-min period, suggesting a reduction in anxiety and/or interest in new environment (Greco et al., 2013) (Figures 2J and 2K; genotype factor F (1, 22) = 46.52, p < 0.0001; time factor F (5, 110) = 95.13, p < 0.0001; interaction F (5, 110) = 17.62, p < 0.0001; NWT = 10, NPrkn KO = 14, two-way ANOVA).
Increased repetitive behaviors in Prkn KO mice
An increase in stereotyped repetitive behaviors is a hallmark of ASD. In rodents, self-grooming is an innate behavior, which has been widely used to assess repetitive behavior (Peca et al., 2011; Rothwell et al., 2014; Silverman et al., 2010; Smith et al., 2011). To evaluate grooming behavior, WT or Prkn KO mice were individually placed in a holding cage, with a thin layer of bedding to prevent digging behavior. After a 5-min acclimation period, animal activities were recorded for 10 min to quantify the number of grooming episodes and the total duration of grooming. We found that Prkn KO mice displayed a higher level of groom behavior in terms of both the number of grooming episodes (Figure 3A; WT: 10.77 ± 1.20, N = 13; Prkn KO: 20.80 ± 3.12, N = 10; p = 0.01) and the total time of grooming (Figure 3B; WT: 67.92 ± 8.26 Sec, N = 13; Prkn KO: 101.30 ± 11.91 Sec, N = 10; p = 0.03). In addition to grooming behavior, rearing, an exploratory behavior during which animals stand up on their hind paws, has also been used to represent repetitive behavior and anxiety status (Pogorelov et al., 2005; Sturman et al., 2018; Wohr and Scattoni, 2013). Consistent with the grooming test, we found that the frequency of rearing behavior from the open field test was significantly increased in Prkn KO mice as compared to WT mice (Figure 3C; WT: 91.60 ± 5.09, N = 10; Prkn KO: 120.80 ± 3.74, N = 14; p = 0.0072). The rearing could be either standing up freely without support or standing against the wall. Interestingly, this increased rearing resulted mainly from standing up freely in the middle of the arena floor (Figure 3D; WT: 37.50 ± 3.33, N = 10; Prkn KO: 62.21 ± 3.29, N = 14; p = 0.016), rather than standing against the wall (Figure 3E; WT: 54.16 ± 3.00, N = 10; Prkn KO: 58.57 ± 2.97, N = 14; p = 0.22). These results suggest that Prkn KO mice have elevated levels of repetitive behaviors. Frequent rearing in the central area is consistent with the notion of a reduced level in anxiety in Prkn KO mice.
Figure 3.
Excessive self-grooming and rearing behaviors in Prkn KO mice
(A and B) Prkn KO mice showed an elevated number of self-grooming episodes (A) with significantly more time spent self-grooming during the 10-min session (B), as compared to WT mice.
(C–E) In the open field test, Prkn KO mice displayed elevated repetitive rearing behavior during the arena exploration as compared to WT mice. During the test, Prkn KO mice showed an increase in rearing behavior (C), mainly the unsupported rearing behavior within the arena (D). Prkn KO mice showed no difference in supported rearing against the wall (E). For the self-grooming test, WT, N = 13; Prkn KO, N = 10.
For rearing test, WT, N = 10; Prkn KO, N = 14. All bar graphs and data depict mean ± SEM. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, n. s., not significant; two-tailed Student’s t test.
Vocalization recordings show altered communication in Prkn KO mouse pups
Impaired communication is another core characteristic for ASD individuals. In rodents, communication is achieved through ultrasonic vocalizations. To determine possible impairments in communication in Prkn KO mice, we recorded and analyzed pup vocalization during early postnatal development at the ages of P5 (postnatal day 5), P7, P9, P11, and P13. To induce the calling response, each individual pup was separated from the mother mouse, and isolation induced vocalization from the pup was recorded for 5 min (Figure 4A). Analysis of the vocalization recordings showed that both the WT and Prkn KO pups had a similar developmental trend in call rate, with an early increase from P5 to P7 followed by a decrease until the last recording time point at P13 (Figure 4B). However, the absolute call rate in Prkn KO pups was significantly reduced at P7, P9, and P11 compared to that in the WT pups (Figure 4B; P5: NWT = 14, NPrkn KO = 11, p > 0.99; P7-P13: NWT = 25, NPrkn KO = 21; P7: p = 0.03; P9: p = 0.03; P11: p = 0.0024; P13: p > 0.99; genotype factor F (1, 199) = 19.30, p < 0.0001; time factor F (4, 199) = 10.47, p < 0.001; interaction F (4, 199) = 1.31, p = 0.27). Prkn KO pups also demonstrated a longer call duration (Figure 4C; P5: NWT = 14, NPrkn KO = 11, p = 0.0087; genotype factor F (1, 199) = 1.88, p = 0.17; time factor F (4, 199) = 35.90, p < 0.0001; interaction F (4,199) = 2.80, p = 0.027) and a higher peak frequency at P5 (Figure 4F; P5: NWT = 14, NPrkn KO = 11, p = 0.01; genotype factor F (1, 199) = 3.62, p = 0.06; time factor F (4, 199) = 8.62, p < 0.0001; interaction F (4,199) = 2.92, p = 0.0225), but not at other time points. There was a significant reduction in the total call time for Prkn KO pups at P9 and P11 (Figure 4D; P9 & P9: NWT = 25, NPrkn KO = 21; P9: p = 0.02; P11: p = 0.04; genotype factor F (1, 199) = 7.13, p = 0.0082; time factor F (4, 199) = 24.12, p < 0.0001; interaction F (4,199) = 1.86, p = 0.1199). The maximum peak frequency from Prkn KO pups at P7 was significantly higher than that from WT pups (Figure 4G; P7: NWT = 25, NPrkn KO = 21; P7: p = 0.02; genotype factor F (1, 199) = 6.37, p = 0.01; time factor F (4, 199) = 33.40, p < 0.0001; interaction F (4,199) = 1.24, p = 0. 2943). The peak amplitude was comparable between WT and Prkn KO pups (Figure 4E; genotype factor F (1, 199) = 0.96, p = 0.3298; time factor F (4, 199) = 51.99, p < 0.0001; interaction F (4,199) = 1.66, p = 0.16). When the recordings were further analyzed by a smaller time unit, we found that the calls were evenly distributed for both the WT and Prkn KO pups (Figures S1A–S1E). At P5 (Figure S1A) and P13 (Figure S1E), there was no significant difference in call number during each minute between WT and Prkn KO pups (P5: genotype factor F (1, 23) = 0.27, p = 0.61; time factor F (4, 92) = 3.15, p = 0.018; interaction F (4, 92) = 1.10, p = 0.36; NWT = 14, NPrkn KO = 14; P13: genotype factor F (1, 44) = 0.99, p = 0.32; time factor F (4, 176) = 2.74, p = 0.03; interaction F (4, 176) = 0.97, p = 0.42; NWT = 25, NPrkn KO = 21). At P7, the difference was significant at the third and fifth minutes (Figure S1B; P7: third minute: p = 0.02; fifth minute: p = 0.01; genotype factor F (1, 44) = 5.66, p = 0.021; time factor F (4, 176) = 1.21, p = 0.31; interaction F (4, 176) = 2.697, p = 0.032; NWT = 25, NPrkn KO = 21). At P9, Prkn KO pups showed a reduction in call number at the second and fourth minutes compared to WT pups (Figure S1C; P9: second minute: p = 0.02; fourth minute: p = 0.02; fifth minute: p = 0.62; genotype factor F (1, 44) = 8.25, p = 0.006; time factor F (4, 176) = 3.04, p = 0.018; interaction F (4, 176) = 0.91, p = 0.46; NWT = 25, NPrkn KO = 21). At P11, Prkn KO pups showed a reduction in call number at the first, fourth, and fifth minutes during the recording session as compared to WT pups (Figure S1D; P11: first minute: p = 0.02; fourth minute: p = 0.01; fifth minute: p = 0.0026; genotype factor F (1, 44) = 12.75, p = 0.0009; time factor F (4, 176) = 7.60, p < 0.0001; interaction F (4, 176) = 0.74, p = 0.56; NWT = 25, NPrkn KO = 21). These results demonstrate abnormalities in communication during early development in Prkn KO mice as compared to WT mice.
Figure 4.
Abnormalities in ultrasonic vocalization in Prkn KO mice
(A) Representative traces of ultrasonic vocalization recordings from WT and Prkn KO pups at P5 and P7. Color scale encodes the voice amplitude.
(B) Call rate (number of calls per minute) profiling of ultrasonic vocalizations from isolated pups during postnatal development (every 2 days from P5 to P13) in WT and Prkn KO mice. Prkn KO mice showed a significant reduction in call rate at P7, P9, and P11 as compared to WT mice (WT, N = 25; Prkn KO, N = 21), but not at P5 (WT, N = 14; Prkn KO, N = 11) or P13 (WT, N = 25; Prkn KO, N = 21).
(C–G) Measurement of other parameters of ultrasonic vocalization from isolated pups during postnatal development. At P5, Prkn KO mice showed a longer call duration (C) and a higher average peak frequency (F) as compared to those in WT mice, but no difference was detected at other time points. At P9 and P11, Prkn KO mice showed a reduction in total call time as compared to WT mice (D). At P7, Prkn KO mice displayed a higher peak maximum frequency as compared to WT mice (G). No difference in peak amplitude was detected throughout postnatal development (E).
Data in (A) is presented as individual data points and mean ± SEM The rest graphs depict mean ± SEM. ∗p < 0.05, ∗∗p < 0.01; two-way ANOVA with Bonferroni post hoc test.
Prkn KO mice show normal spatial learning and memory capacity
Alterations in cognitive functions including learning and memory are common comorbidities of ASD (Blundell et al., 2010; Gilbert et al., 2020; Moretti et al., 2006; Silva et al., 1997). Previous studies have revealed that synaptic plasticity is impaired in the hippocampus of Prkn KO mice (Hanson et al., 2010; Itier et al., 2003; Kitada et al., 2009), suggesting the possibility of alterations in learning and memory formation. We therefore performed Barnes maze tests to examine hippocampus-dependent spatial memory. In brief, each test subejct mouse was placed into a brightly lit and noise-exposed circular platform with 20 holes evenly distributed along the edge of the maze. During the 3 days of training, mice were allowed to freely explore the maze for 5 min and memorize the location of the single escape hole among the 20 holes. One or six days after the training sessions, animals were placed back into the maze to be tested individually for memory retention. To assess the learning capacity of Prkn KO mice, we first quantified the primary latency (the time spent to reach the escape hole) (Figure 5A; acquisition day 1 to 6-day post-test: p > 0.99; genotype factor F (1, 27) = 1.15, p = 0.29; time factor F (4, 108) = 5.05, p = 0.0009; interaction F (4, 108) = 0.24, p = 0.92; NWT = 12, NPrkn KO = 17) and the primary hole distance (the distance covered to reach the escape hole) during acquisitions and in memory retention tests (Figure 5B; acquisition day 1 to 6-day post-test: p > 0.99; genotype factor F (1, 27) = 1.15, p = 0.29; time factor F (4, 108) = 5.05, p = 0.0009; interaction F (4, 108) = 0.24, p = 0.92; NWT = 12, NPrkn KO = 17). WT and Prkn KO mice showed similar measurements, suggesting a normal learning capacity of the KO mice. During the 24-hour post-test (Figure 5C) and 6-day post-test (Figure 5D), the escape hole was sealed just like the other holes, and the test subject searched the platform using spatial memory, attempting to locate the escape hole. When the time spent on each quadrant was quantified, we found no significant difference at either the 24-hour (Figure 5E; target quadrant: WT: p > 0.99; Left quadrant: p = 0.39; Right quadrant: WT: p > 0.99; opposite quadrant: WT: p > 0.99; genotype factor F (1, 108) = 0, p > 0.99; quadrant factor F (3, 108) = 38.16, p < 0.0001; interaction F (3, 108) = 1.50, p = 0.22; NWT = 12, NPrkn KO = 17) or the 6-day post-test (Figure 5F; all quadrants: p > 0.99; genotype factor F (1, 108) = 0, p > 0.99; quadrant factor F (3, 108) = 63, p < 0.0001; interaction F (3, 108) = 0.39, p = 0.76; NWT = 12, NPrkn KO = 17), suggesting comparable memory retention in Prkn KO and WT mice. Furthermore, we assessed the nose probe counts at each hole during the test sessions (Figures 5C and 5D). Consistent with a lack of memory deficits, we found that the probe numbers for the target region holes (hole “-1”, “0” and “1”) were comparable between the WT and Prkn KO mice during the 24-hour post-test (Figure 5G; for all holes: p > 0.99; genotype factor F (1, 27) = 0.21, p = 0.65; hole position factor F (19, 513) = 23.36, p < 0.0001; interaction F (19, 513) = 0.68, p = 0.84; NWT = 12, NPrkn KO = 17) and the 6-day post-test (Figure 5H; “10”-“-1” and “-4”-“-7” and “-9”: p > 0.99; “-2”: p = 0.055; “-3”: p = 0.086; “-8”: p = 0.94; genotype factor F (1, 27) = 4.053, p = 0.0542; hole position factor F (19, 513) = 22.88, p < 0.0001; interaction F (19, 513) = 0.85, p = 0.64; NWT = 12, NPrkn KO = 17). These findings suggest that hippocampus-dependent spatial learning and memory remain intact in Prkn KO mouse.
Figure 5.
Prkn KO mice show normal spatial learning and memory in Barnes maze test
(A) Plots of primary latency throughout the training phase and testing phase. Prkn KO and WT mice spent comparable amounts of time locating the escape hole in the acquisition and testing sessions.
(B) Plots of distance traveled before reaching the target hole (primary hole distance) throughout the training phase with an open escape hole and the testing phase with a closed escape hole. Prkn KO and WT mice traveled comparable distances before locating the target hole through the acquisition and testing sessions.
(C and D) Representative movement traces of 24-h and 6-day post-acquisition test sessions.
(E and F) Prkn KO mice showed no significant difference from WT mice in the time spent in each of the four quadrants during the 24-h and 6-day post-acquisition test sessions.
(G and H) Prkn KO mice showed no significant difference from WT mice in the number of nose probes at the target (hole “0”) and its neighboring holes (hole “1”, “-1”) during the 24-h and 6-day post-acquisition test sessions.
Data (A to B, and E to H) depict mean ± SEM. WT, N = 12; Prkn KO, N = 17. Repeated measures two-way ANOVA with Bonferroni post hoc test.
Prkn KO mice show abnormalities in neuronal activity, dendritic arborization, and spine morphology
Accumulating evidence suggests that abnormalities in neuronal activity, synaptic connectivity, and an altered excitatory-inhibitory (E/I) ratio are shared cellular features underlying ASD. E/I imbalance in ASD often results from altered glutamatergic and GABAergic neurotransmission in related brain regions and neural circuits (Gilbert and Man, 2017; Won et al., 2013). Among the brain regions, the medial prefrontal cortex (mPFC) has been shown to be critically implicated in social behaviors in ASD (Franklin et al., 2017; Ibrahim et al., 2021; Ko, 2017; Sacai et al., 2020). To test whether medial PFC neuronal activity is altered in Prkn KO mice in the context of sociability, we examined c-fos expression as an indicator of neuronal activity in mouse brains following the sociability test. To this end, WT and Prkn KO mice at the age of eight weeks were divided into naive and sociability groups. Naive mice of both WT (naive; n = 7) and Prkn KO (naive; n = 4) were sacrificed without experiencing the three-chamber sociability test. As for the sociability group, both WT (sociability; n = 5) and Prkn KO mice (sociability; n = 5) were subjected to the three-chamber sociability test and then kept in isolation for 90 min before sacrificing to allow sufficient expression of the behavioral-relevant neuronal activity-induced c-fos protein. Mouse brain slices were immunostained with antibodies against c-fos for detection of recently activated neurons (Figure 6A). c-fos positive cells were quantified in the prelimbic (PrL) cortex (Figure 6B), as well as in specific cortical layers (Figures 6C, 6D, and S2A–S2D). In both WT and Prkn KO mouse brains, the sociability test resulted in significantly more c-fos positive cells in the PrL cortex as compared to that of naive mice (Figure 6B), suggesting the involvement of the PrL region in sociability. Within either the naive or sociability group, the number of c-fos positive cells was reduced in Prkn KO mice, suggesting reduced neuronal activity in this brain region (Figure 1B; genotype factor F (1, 17) = 24.32, p = 0.0001; behavior factor F (1, 17) = 29.6, p < 0.0001; interaction F (1, 17) = 0.50, p = 0.49; NWT-naïve = 7, NWT-sociability = 5, NPrkn KO-naïve = 4, NPrkn KO-sociability = 5, two-way ANOVA). Furthermore, we quantified the number of c-fos positive cells in layers 1, 2, 3, 5, 6a, and 6b of the PrL cortex. Surprisingly, c-fos positive cell number in layer 5 showed a similar trend as that from the whole PrL cortex area (Figure 6C; genotype factor F (1, 17) = 20.27, p = 0.0003; behavior factor F (1, 17) = 24.17, p = 0.0001; interaction F (1, 17) = 0.30, p = 0.59; NWT-naïve = 7, NWT-sociability = 5, NPrkn KO-naïve = 4, NPrkn KO-sociability = 5, two-way ANOVA), whereas no significant difference was detected in other layers (Figures 6D, 6E, and S2A–S2D). We also counted the number of NeuN positive cells and found no difference in total neuron numbers in the PrL and IL regions between WT and Prkn KO mouse brains (Figures S3A–S3C), indicating that the change in c-fos positive cells was not due to a change in total number of neurons.
Figure 6.
Reduced neuronal activity during the sociability test and aberrance in dendritic arborization and spines in Prkn KO mice
(A) Representative coronal brain slice images at approximately Bregma 1.93 mm from the prelimbic (PrL) and infralimbic (IL) region of the prefrontal cortex immunostained by neuronal activity marker c-fos from behaviorally naive mice (WT mouse brains, N = 7; Prkn KO mouse brains, N = 5) and mice that were subject to the sociability test (WT mouse brains, N = 4; Prkn KO mouse brains, N = 5). Nuclei were stained by Hochest. Naive mice remained in home cages, whereas other mice were subjected to the three-chamber sociability test before brain slice preparation for c-fos immunostaining. Scale bar, 500 μm.
(B–E) Quantification of c-fos positive neurons in the PrL (B), layer 5 of the PrL (C), layer 2 of the PrL(D), and the IL (E), respectively. (B) Quantification showed that social interaction caused an increase in c-fos positive neurons in the PrL in both WT and Prkn KO mice; however, the increase in c-fos positive neurons in KO mice was significantly smaller than that in WT mice. In naive mice, c-fos positive neuron number was reduced in Prkn KO mice as compared to WT mice. (C) In layer 5 of the PrL, a change similar to that in the PrL overall was observed. No significant change was observed in layer 2 (D) of the PrL, as well as in the IL (E).
(F) Representative tracing images from layer 5 pyramidal neurons of the PrL of WT and Prkn KO mouse brains. Scale bar, 20 μm.
(G) Sholl analysis on the basal dendritic arborization of layer 5 pyramidal neurons in the PrL (N = 18 cells from three brains, for both WT and Prkn KO mice).
(H–K) Quantitative analysis of layer 5 pyramidal neurons of the PrL on total dendrite length (G), dendrite tip number (H), longest dendrite length (I) and average dendrite length (J).
(L) Representative images of Golgi staining of basal dendritic spines of layer 5 pyramidal neurons of the PrL. Scale bar, 2 μm.
(M) Analysis of spine subtypes and filopodia (WT, N = 18 cells from three brains; Prkn KO, N = 16 cells from three brains).
(N–P) Quantification of spine density (N), spine length (O) and spine head width (P) of layer 5 pyramidal neurons in the PrL.
Data in (B to E), (G to K) and (M to P) are presented as Mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; in (B to E), two-way ANOVA with Turkey post hoc test; in (G), repeated measures two-way ANOVA with Bonferroni post hoc test; in (H to K and M to P), two tailed Student’s t test.
We then examined neuronal morphology by Golgi staining. We measured the basal dendritic arborization from layer 5 pyramidal neurons in the PrL cortex from both WT and Prkn KO mice (Figure 6F). The analysis showed a reduction in the complexity of basal dendritic arborization in Prkn KO mice relative to the WT mice (Figure 6G; distance F (48, 1274) = 141.20, p < 0.0001; genetic F (1, 1274) = 195.10, p < 0.0001; interaction F (48, 1274) = 2.84, p < 0.0001; NWT = 18 from three brains, NPrkn KO = 18 from three brains). We observed decreases in total dendritic length (Figure 6H), dendritic tip number (Figure 6I), and the length of the longest dendritic branch (Figure 6J; total dendritic length: WT: 2198.35 ± 111.10 μm, N = 18 cells from three brains; Prkn KO: 1624.93 ± 86.99 μm, N = 18 cells from three brains, p = 0.0003; dendritic tip number: WT: 26.94 ± 0.99, N = 18 cells from three brains; Prkn KO: 22.17 ± 1.14, N = 18 cells from three brains, p = 0.0033; longest dendritic branch WT: 209.65 ± 13.65 μm, N = 18 cells from three brains; Prkn KO: 163.92 ± 7.10 μm, N = 18 cells from three brains, p = 0.0064). In contrast, the average dendrite length in the Prkn KO mice remained the same as in WT mice (Figure 6K; WT: 81.97 ± 3.64 μm, N = 18 cells from three brains; Prkn KO: 74.12 ± 2.72 μm, N = 18 cells from three brains, p = 0.09). Next, we measured the spine and filopodia density. Prkn KO mice showed a significant reduction in spine density (Figure 6L). Among different types of spines, the densities of mushroom and thin spines, as well as filopodia, were reduced in Prkn KO mice (Figures 6M and 6N; mushroom spine density: WT: 8.37 ± 0.35 per 10 μm; Prkn KO: 4.77 ± 0.28 per 10 μm p < 0.0001; thin spine density: WT: 3.68 ± 0.23 per 10 μm; Prkn KO: 2.70 ± 0.25 per 10 μm p = 0.0078; stubby spine density: WT: 1.64 ± 0.18 per 10 μm; Prkn KO: 1.38 ± 0.22 per 10 μm p = 0.36; filipodia density: WT: 1.66 ± 0.14 per 10 μm; Prkn KO: 0.89 ± 0.14 per 10 μm p = 0.0007; spine density: WT: 13.70 ± 0.34 per 10 μm; Prkn KO: 8.86 ± 0.44 per 10 μm p < 0.0001; NWT = 18 from three brains; NPrkn KO = 16 cells from three brains), whereas no significant differences were found between WT and Prkn KO mice in spine length (Figure 6O; WT: 1.15 ± 0.03 μm; Prkn KO: 1.13 ± 0.03 μm, p = 0.56; NWT = 18 from three brains; NPrkn KO = 16 cells from three brains) and spine head width (Figure 6P) (WT: 0.88 ± 0.01 μm; Prkn KO: 0.90 ± 0.02 μm p = 0.52; NWT = 18 from three brains; NPrkn KO = 16 cells from three brains). We then compared the spine density at apical and basal dendrites of hippocampal CA1 pyramidal neurons. Interestingly, at both locations, no significant difference in spine density was detected (Figures S4A–S4E). Thus, at the PrL PFC, Prkn KO leads to defects in spine formation and maturation, as well as a reduction in basal and sociability-related neuronal activity.
Alterations of synaptic components in Prkn KO mice
Given the observed changes in neuronal activity and spine morphology in Prkn KO mice, we further assessed the expression of synaptic proteins from the brain lysates of Prkn KO and WT mice. Western blotting results revealed region-specific changes. From frontal cortical lysate, ubiquitin signals were reduced in Prkn KO mouse brains, consistent with the lack of the E3 ligase Parkin (Figures 7A and 7C). Also, the glutamatergic receptors GluA1, GluA2, GluA3, and GluN1 were down-regulated. Interestingly, we also observed a reduction in the potassium channel Kir2.1, and the inhibitory synapse components Gephyrin and GABAergic receptor (GABAR) subunit α1 (Ubiquitin: p = 0.0027; Synaptophysin: p = 0.48; Gephyrin: p = 0.047; PSD-95: p = 0.52; PICK 1: p = 0.83; GluA1: p = 0.01; GluA2: p = 0.025; GluA3: p = 0.01; GluN1: p = 0.0017; GluN2A: p = 0.73; GABAR α1: p = 0.97; Kir2.1: p = 0.01; NSF: p = 0.67; NWT = 6, NPrkn KO = 8). In contrast, ubiquitin signals in the hippocampus remained unchanged across WT and Prkn KO mouse brains (Figures 7B and 7D). GluA2 level was increased in this region, while the inhibitory synapse scaffold protein Gephyrin and GABARα1 were both down-regulated (Ubiquitin: p = 0.5344; Synaptophysin: p = 0.09; Gephyrin: p = 0.0083; PSD-95: p = 0.82; PICK 1: p = 0.23; GluA1: p = 0.071; GluA2: p = 0.038; GluA3: p = 0.30; GluN1: p = 0.78; GluN2A: p = 0.36; GABAR α1: p = 0.049; Kir2.1: p = 0.14; NSF: p = 0.086; NWT = 6, NPrkn KO = 8).
Figure 7.
Synaptic component analysis in Prkn KO mouse brains
(A and B) Western blotting using frontal cortical area (A) and hippocampal lysates (B) of P100 WT and Prkn KO mice (WT, N = 6; Prkn KO, N = 8).
(C and D) Quantification of western blot immuno-intensities.
(E) Western blotting analysis of synaptosome preparations from frontal cortical area of P100 Prkn KO and WT mice.
(F–I) Quantification of the synaptic protein levels for GluA1 (F), GluA2 (G), GluN1 (H), and PSD-95 (I); N = 4 for both WT and Prkn KO mice. GAPDH was probed as a loading control.
(J) Representative coronal brain slice images of layer 5 from the PrL immunostained with the pre-synaptic markers VGlut and VGAT. Scale bar, 5 μm.
(K and L) Quantification of the pre-synaptic puncta per unit area for excitatory pre-synaptic marker VGlut(K) and inhibitory pre-synaptic marker VGAT (L); N = 9 brains for both WT and Prkn KO mice.
All bar graphs are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01. Two-tailed Student’s t test.
To compare the biochemical composition of synapses between KO and WT mouse brains, we prepared synaptosome fractions from the whole frontal cortical region of both WT and Prkn KO mouse brains. Western blotting analysis showed that the levels of excitatory post-synaptic receptors, including GluA1, GluA2, and GluN1, were significantly reduced in Prkn KO mice (Figures 7E–7H). In contrast, the amount of post-synaptic scaffolding protein PSD-95 in the synaptosome remained the unaltered between the two strains (Figure 7I) (GluA1: p = 0.028; GluA2: p = 0.0011; GluN1: p = 0.0086; PSD-95: p = 0.51; NWT = 4, NPrkn KO = 4). Changes in protein amount could result from regulation of gene transcription, mRNA translation, or protein turnover. To evaluate the effect at the transcriptional level, we performed qPCR to measure the mRNA levels of certain post-synaptic proteins. Using tissue from the frontal cortical region, we found that the mRNA abundance of GluA1, GluA2, Kir2.1, and GluN1 in the Prkn KO mice were not changed as compared to that of the WT mice (Figures S5A–S5D) (Gria1: p = 0.75; Gria2: p = 0.51; Kcnj2: p = 0.78; Grin1: p = 0.21; NWT = 6, NPrkn KO = 6). In the hippocampus, the same results were observed (Figures S5E–S5H) (Gria1: p = 0.30; Gria2: p = 0.57; Kcnj2: p = 0.089; Grin1: p = 0.14; NWT = 6, NPrkn KO = 6). These results suggest that changes in the synaptic proteins in Prkn KO mice probably resulted from dysregulation in protein synthesis or stability, rather than altered gene regulation. Given the changes in ubiquitination intensity in the KO mice, ubiquitination, proteasome-mediated degradation, and ubiquitin-dependent autophagy may be responsible for the observed changes in synaptic proteins including glutamate receptors (Huo et al., 2015; Jarzylo and Man, 2012; Lin et al., 2011; Wang et al., 2017).
To determine whether changes also occurred at the pre-synaptic sites in Prkn KO mice, we stained brain slices from WT and Prkn KO mice for pre-synaptic markers of inhibitory or excitatory synapses. We chose VGlut1 as the pre-synaptic markers of excitatory synapses, whereas VGAT was chosen as the pre-synaptic markers of inhibitory synapses. Immunosignals were quantified in layers 5 and 2 of the PrL cortex, as well as in the CA1 hippocampal region. In layer 5, the puncta density of VGAT was not changed in Prkn KO mouse brains (Figure 7L), but puncta densities of the excitatory pre-synaptic marker VGlut1 were reduced as compared to that in WT mouse brains (Figure 7K; VGlut1: WT: 3.05 × 105 ± 13,772 per 1 mm2, N = 9; Prkn KO: 2.52 × 105 ± 8,508 per 1 mm2, N = 9, p = 0.0063). In layer 2 of the PrL cortex, the puncta densities of VGlut1 and VGAT were both decreased in Prkn KO mice (Figures S6B and S6C; VGlut1: WT: 3.20 × 105 ± 6,397 per 1 mm2, N = 9; Prkn KO: 2.60 × 105 ± 10,339 per 1 mm2, N = 9, p = 0.00024; VGAT: WT: 1.35 × 105 ± 3,453 per 1 mm2, N = 9; Prkn KO: 1.13 × 105 ± 2,940, N = 9, p = 0.00016). In the hippocampal CA1 region, the puncta density of the inhibitory synaptic markers VGAT was reduced at the apical dendrite region (Figure S6F; VGAT: WT: 9.87 × 104 ± 2,218 per 1 mm2, N = 9; Prkn KO:7.28 × 104 ± 3,299 per 1 mm2, N = 9, p < 0.0001). However, the puncta density of the excitatory synaptic markers VGlut1 was not altered (Figure S6E). These data together suggest that the synapse connection is altered in Prkn KO mice, and the changes are specific to the brain region and the synapse type.
Discussion
ASD is a neurodevelopmental disorder with high heterogeneity. Recent genetic sequencing studies from ASD individuals revealed a high occurrence of CNVs in their genome, suggesting that genomic CNVs could be a cause of ASD and highlighting the genes within the CNV genomic fragments for their relevance to the etiology of ASD (Glessner et al., 2009; He et al., 2013; Levy et al., 2011; Morrow et al., 2008; Pinto et al., 2010; Yin et al., 2016). PRKN is a gene at chromosome locus 6q23 within the CNV region identified from multiple ASD cases (Girirajan et al., 2013; Prasad et al., 2012; Scheuerle and Wilson, 2011; Yin et al., 2016). Interestingly, both duplication and deletion of the PRKN gene are identified in ASD individuals with a higher occurrence rate in gene deletion. The PRKN gene product Parkin is an E3 ligase that has been intensively studied in early-onset Parkinson's disease, suggesting its important role in the nervous system. Notedly, a recent study indicated the high frequency of parkinsonism (32%) within ASD individuals older than 39 years (Starkstein et al., 2015). It is intriguing to speculate that the gene dosage change of PRKN due to the CNVs could act as an etiological factor of ASD.
Prkn KO mouse models have been generated for the study of Parkinson's disease (Harvey et al., 2008; Stephenson et al., 2012). It is worth noting that the Prkn KO mouse model seems unable to fully recapitulate the behavioral characteristics of Parkinons's disease in human, nor the cell death of dopaminergic neurons in the substantia nigra in mice of 6–18 months or 18–22 months old (Goldberg et al., 2003; Perez and Palmiter, 2005), suggesting that the Prkn KO mouse may have features of other neurological disorders. Indeed, we found that the Prkn KO mouse demonstrates autistic-like behaviors, including impaired sociability, elevated repetitive behaviors, and alterations in communication. In the region of the PrL cortex, immunohistology analysis revealed that neuronal activity is reduced in Prkn KO mice in the context of sociability. Morphological analysis based on Golgi staining showed a reduction in the complexity of the dendritic arborization and dendritic spine number in layer 5 pyramidal neurons, and biochemistry and immunostaining analyses indicated altered synaptic protein composition and synapse number.
In this study, several autistic-like behaviors were detected in the Prkn KO mice. First, using the three-chamber sociability and social novelty test, we found that the Prkn KO mice have impairments in sociability and social novelty. Notably, Prkn KO mice showed a reversed or a lack of preference in the sociability test and social novelty test, strongly indicating aberrant sociability. Second, our profiling results from pup isolation-induced ultrasonic vocalization recordings showed a significant reduction in call rate from Prkn KO pups. Differences were detected at certain developmental time points in other measurements including call duration, call amplitude, and peak frequency. Interestingly, the WT and Prkn KO mice showed similar trends along early postnatal development, suggesting a developmental delay in Prkn KO mice instead of a complete disruption of neural developmental processes. Third, self-grooming test revealed an increase in the number of grooming episodes, as well as time spent grooming in Prkn KO mice. In addition, open field tests indicated an elevation in rearing behavior. The findings on grooming and rearing behaviors demonstrate an elevated repetitive behavior in Prkn KO mice.
It is known that ASD individuals often demonstrate comorbidities such as locomotion abnormalities, alterations in anxiety, and cognitive defects. However, our results from the open filed test and Barnes maze test did not reveal notable differences between Prkn KO and WT mice, which are consistent with previous behavioral measurements in Prkn KO mice (Goldberg et al., 2003; Perez and Palmiter, 2005). Our assessment of the Prkn KO mouse model suggests a reduction in anxiety level and normal spatial memory. In line with our findings, a case study of two ASD children with PRKN CNVs reported that the child with a microdeletion of the PRKN gene showed reluctance for social interactions and a calm demeanor, but was cognitively normal, whereas the other child with a micro-duplication of the PRKN gene was cognitively impaired (Scheuerle and Wilson, 2011). In addition, previous electrophysiological studies reported normal hippocampal LTP in Prkn KO mice (Hanson et al., 2010; Itier et al., 2003), consistent with our findings of normal spatial learning and memory in Prkn KO mice.
An accumulating amount of evidence indicates that cortical excitation/inhibition imbalance is a convergent mechanism in ASD (Nelson and Valakh, 2015). It has been shown that in the PrL cortex, a shift in the E/I balance toward excitation led to autistic social dysfunction (Yizhar et al., 2011). When neuronal activity in the PrL PFC was examined, we found a brain region (PrL)- and layer (layer 5)-specific reduction in the number of c-fos positive neurons by immunostaining in Prkn KO mice following sociability tests. This finding implies that a shift in the E/I balance toward inhibition may also contribute to sociability deficits. In line with our findings, previous research revealed that many risk genes for autism and schizophrenia are highly expressed in fetal and infant L5/6 PFC projection neurons (Bakken et al., 2016; Willsey et al., 2013). Mechanistic studies in mice further show that functional disruption of subcortically projecting L5/6 mPFC neurons is responsible for abnormalities in sociability either in the autism model (Brumback et al., 2018) or in the juvenile social isolation model (Yamamuro et al., 2020). In the juvenile social isolation model, the activation of the mPFC to posterior paraventricular thalamus projection promotes sociability (Yamamuro et al., 2020). Interestingly, a recent single-cell sequencing study from the dorsolateral PFC (but not from the mPFC) of postmortem ASD brains revealed that synaptic signaling of upper-layer excitatory neurons (layers ⅔ and 4) are preferentially affected in autism, underlying the importance of layer ⅔ excitatory neurons in the dorsolateral PFC in the ASD individuals (Velmeshev et al., 2019). Taken together, these lines of evidence suggest the PFC subdivision and layer-specific involvement in ASD. Our data indicate that in Prkn KO mice, activity of the PrL layer 5 neurons may be more relevant to the impairment in sociability. Further electrophysiological studies and functional cell labeling assays will help to determine the identity of the neurons in the PrL cortex that contribute to the defects in sociability in this model.
At the structural level, our examination revealed a reduction in the complexity of dendritic arborization and spine density in the PFC of Prkn KO mouse. Aberrant neuronal morphology has been shown as a common cellular phenotype in many ASD mouse models, such as Fmr1 KO (Mineur et al., 2006; Spencer et al., 2005, 2008), Shank3 KO (Peca et al., 2011), Ube3a two × Tg (Khatri et al., 2018; Krishnan et al., 2017; Smith et al., 2011), and our Nexmif KO mouse models (Gilbert and Man, 2016; Gilbert et al., 2020). In addition, we examined synaptic protein composition and found a down-regulation in excitatory glutamatergic receptors including GluA1, GluA2, GluA3, and GluN1 in the PFC. In contrast, a reduction in inhibitory synaptic proteins including gephyrin and GABARα1 was observed in the hippocampus. Consistently, our analysis using synaptosomal fractionation showed a reduction in glutamatergic receptors including GluA1, GluA2, and GluN1, suggesting functional alterations in synaptic transmission and neuronal activity in the brains of Prkn KO mice. In addition, immunostaining of excitatory and inhibitory synaptic marker proteins revealed synaptic puncta change in a layer- and brain region-specific manner in Prkn KO mouse. In line with these findings, studies using cultured hippocampal neurons revealed that acute knockdown of Parkin results in a reduction in surface AMPAR levels (Cortese et al., 2016; Zhu et al., 2018). Together, these findings support that abnormalities in synaptic maturation and function are key components of the neuropathological events underlying ASD.
Ubiquitin-proteasome pathway has been shown to be one of the most common molecular processes implicated in ASD pathogenesis (Louros and Osterweil, 2016). As an E3 ligase, Parkin is involved in the proteasomal degradation of ubiquitinated proteins and mitophagy, a special form of autophagy specifically degrading damaged mitochondria (Bingol et al., 2014; Lazarou et al., 2015). One recent study reveals that Parkin plays a role in synaptic vesicle autophagy in Bassoon-deficient mice (Hoffmann-Conaway et al., 2020), suggesting that Parkin can participate in more forms of autophagy. In addition, both up- and down-regulation of autophagy have been associated with ASD. For example, autophagic activity has been found to be down-regulated in the brains of ASD, leading to excessive spine formation, which suggests the importance of autophagy in the early development of neuronal connectivity and likely also in the balanced functionality of neural circuits in the brain (Tang et al., 2014). In contrast, in the Cc2d1a mouse model of autism, it is shown that the mRNA levels of key autophagy molecules LC3 and Beclin-1 are elevated (Dana et al., 2020). Notably, knockdown of Parkin could increase LC3-II level in vitro in both HEK cells and neuroblast-like SH-SY5Y cells (Chen et al., 2010), and in vivo in the liver (Williams et al., 2015), a sign of elevated autophagy, implying that the loss-of-function Prkn KO mouse brain might have elevated autophagy. Related to these findings, we found a significant reduction in the complexity of dendritic arborization and spine density in Prkn KO mice, accompanied by alterations in neurotransmitter receptors and other synaptic proteins. Thus, autophagic processes may potentially contribute to the synaptopathy of Prkn KO mice.
In summary, this study demonstrates that the Prkn KO mouse manifests autistic-like behavioral characteristics and provides new insights into the cellular and synaptic aberrance resulting from the loss of the Prkn gene. These findings suggest the potential value of the Prkn KO mouse as an animal model of ASD.
Limitations of the study
Our study shows that Prkn gene KO in mice results in autistic-like behaviors recapitulating the phenotypes in ASD individuals with PRKN gene deletion. However, there is usually only one affected allele in ASD individuals with PRKN gene deletion, whereas in the mouse model both alleles are deleted. Further exploration on the haploinsufficiency of Prkn gene in mice would be informative to the understanding of Prkn-related ASD. In addition, the activity of L5 neurons in PrL cortex was indicated by c-fos staining, and the nature of the activity change remains unclear. Future studies by electrophysiological methods will clarify whether the reduction in neuronal activity is due to weakened synaptic inputs and/or a reduction in excitability.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-GluA1 (1:2000) | EMD Milipore | Cat. #MAB2263; RRID:AB_11212678 |
| Mouse anti-Parkin (1:4000) | Cell Signaling Technology | Cat. #4211S; RRID:AB_2159920 |
| Mouse anti-Ubiquitin (1:1000) | Cell Signaling Technology | Cat. #3936S; RRID:AB_331292 |
| Rabbit anti-Synaptophysin (1:2000) | Abcam | Cat. #ab32127; RRID:AB_2286949 |
| Mouse anti-Gephryin (1:1000) | Neuromab | Cat. #75-443; RRID: AB_2636851 |
| Mouse anti-PSD-95 (1:5000) | Synaptic Systems | Cat. #124011; RRID:AB_10804286 |
| Mouse anti-PICK1 (1:1000) | Santa Cruz | Cat. #sc-166654; RRID:AB_2018022 |
| Mouse anti-GluA2 (1:2000) | EMD Millipore | Cat. #MAB397; RRID:AB_2113875 |
| Rabbit anti-GluA3 (1:2000) | Cell Signaling Technology | Cat. #5117S; RRID:AB_10544796 |
| Mouse anti-NR1(1:1000) | NeuroMab | Cat. #75-272; RRID:AB_2877408 |
| Mouse anti-NR2A (1:1000) | EMD Millipore | Cat. #07-632; RRID:AB_310837 |
| Mouse anti-GABAR α1 (1:1000) | NeuroMab | Cat. #75-136; RRID:AB_2877288 |
| Rabbit anti-Kir2.1 (1:1000) | Abcam | Cat. #ab65796; RRID:AB_1140953 |
| Rabbit anti-NSF (1:2000) | Cell Signaling Technology | Cat. #2145S; RRID:AB_2155696 |
| Mouse anti- GAPDH (1:5000) | EMD Millipore | Cat. #MAB374; RRID:AB_2107445 |
| Rabbit anti-c-Fos (1:100) | Cell Signaling Technology | Cat. #2250; RRID:AB_2247211 |
| Mouse anti-NeuN (1:500) | EMD Millipore | Cat. #MAB377; RRID:AB_2298772 |
| Mouse anti-VGAT (1:400) | NeuroMab | Cat. #75-457; RRID:AB_2877230 |
| Mouse anti-VGlut1 (1:400) | NeuroMab | Cat. # 75-066; RRID:AB_2877383 |
| Anti-rabbit IgG-HRP (1:5000) | Bio-Rad | Cat. #170-6515; RRID:AB_11125142 |
| Anti-mouse IgG-HRP (1:5000) | Bio-Rad | Cat. #170-6516; RRID:AB_11125547 |
| Anti-mouse Alexa Fluor 488 (1:500) | Invitrogen | Cat. #A21121; RRID:AB_2535764 |
| Anti-rabbit Alexa Fluor 555 (1:500) | Invitrogen | Cat. #A21428; RRID:AB_2535849 |
| Experimental models: Organisms/strains | ||
| Prkn KO mice (B6.129S4-Prkntm1Shn/J) | THE JACKSON LABORATORY | 006582; RRID:IMSR_JAX:006582 |
| C57BL/6J | THE JACKSON LABORATORY | 000664; RRID:IMSR_JAX:000664 |
| Software and algorithms | ||
| ImageJ | N/A | https://wsr.imagej.net/distros/win/ij153-win-java8.zip |
| Prism 7 | GraphPad Software | N/A |
Resource availability
Lead contact
Hengye Man (hman@bu.edu).
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact.
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
All animals were maintained, handled, and processed in accordance with the policies of the Institutional Animal Care and Use Committee (IACUC) at Boston University.
Prkn KO mice (B6.129S4-Prkntm1Shn/J; Stock# 006582) and wild-type (WT) mice (C57BL/6J; Stock#000664) were purchased from the Jackson Laboratory (Bar Harbor, Maine). Mouse colonies were housed in the Laboratory Animal Care Facility (LACF) at Boston University (Charles River Campus) with a 12-hour light/12-hour dark cycle, at approximately 25°C, and ad libitum access to food and water. To obtain homozygous wild type and knockout mice, heterozygous males and heterozygous females were used for breeding. The genotypes of the littermates were determined by PCR with the following primers: 5′-GCAGAATTACAGCAGTTACCTGG-3′ (Common); 5′-CCTACACAGAACTGTGACCTGG-3′ (Wild type reverse); 5′-ATGTTGCCGTCCTCCTTGAAGTCG-3′ (Mutant reverse).
Method details
Behavioral tests
Behavioral tests were performed on litter mates from the age of P5 to P90. The sequential order and the specific times of behavioral tests are: ultrasonic vocalization recording (P5-P13), Three-chamber test (at the age of week 8), open field test (at the age of week 9), Marble burying (at the age of week 10), Self-grooming test (at the age of week 11) and Barnes maze test (at the age of week 12). Male and female homozygous mice at the age between P5 to P13 were used for the ultrasonic vocalization tests, and males at the age between 8 to 12 weeks were used for all other behavior experiments. During the battery of behavioral tests, one aggressor male was removed from the home cage due to the animal welfare policy and therefore excluded from the behavior data analysis for open field and marble burying tasks. All behavioral tests were performed between 9 am and 4 pm during the light cycle.
Ultrasonic vocalization recording
Ultrasonic vocalization recording of pup isolation calls was performed as described in our previous work (Gilbert et al., 2020). On the test day, individual pups were removed randomly from the litter and placed at the bottom center of a sound-proof cylindrical plastic container at constant 25°C. Recording was then performed with UltraSoundGate 116Hb (basic) with a condenser ultrasound microphone CM16 from Avisoft Bioacoustics (Germany) to capture the ultrasonic vocalization from isolated pups. The acoustic data were recorded with a sampling rate of 300 kHz in 16-bit format. After recording, the tested pup was labeled with a marker and placed immediately back to its litter. Between trials, the container was thoroughly wiped with 70% ethanol and 2% Virkon to remove odors and air-dried before the next trial. All pups were individually recorded every two days from postnatal day 5 (P5) until postnatal day 13 (P13). All recorded data were transferred to Avisoft SASLab Pro (Avisoft Bioacoustics) and filtered to eliminate amplitudes under 30 kHz before further processing. All the call traces were extracted blindly using an automatic detection function with a proper threshold and manually checked. Call numbers, duration, total call time, entire peak frequency, peak maximum frequency, and peak amplitude were analyzed from the labeled call traces.
Three chamber sociability and social novelty test
Sociability and social novelty tests were performed as described in our previous study (Gilbert et al., 2020). Each chamber measures 47 cm (length) × 23 cm (width) × 30 cm (height). Briefly, male mice aged 8 weeks were habituated to three empty chambers for 10 mins each day for three days prior to the social test. Target subjects (Stranger 1 and Stranger 2) were age- and gender-matched WT mice. On the test day for the sociability test, a test subject was allowed to freely explore the whole space without target subjects for 5 mins, followed by introducing an unfamiliar mouse (Stranger 1) in one side chamber. The test animals were allowed to explore the full space again and were video recorded for 5 mins. In the following social novelty test, the additional second unfamiliar mouse (Stranger 2) was placed into the opposite side chamber without Stranger 1 and again the test animal was set free to explore the full space and was video recorded for 5 mins. The footage was then converted to a series of images at 0.5 sec intervals and traced with an ImageJ plug-in (MTrackJ) manually to quantify the time spent in each chamber and the proximity to the cages. The test animals were always released from the middle chamber for all trials. The position of the empty cage and the cage of target subjects (Stranger 1 and Stranger 2) was assigned randomly between different test animals to avoid the impact of environmental factors. Preference Index in the sociability test is defined as following: (TimeStranger1-Timeempty)/(Timestranger1+Timeempty); preference index in social novelty test is defined as following: (TimeStranger2-Timestranger1)/(Timestranger2+Timestranger1). All behavior videos were captured by a Logitech c920 webcam and analyzed blindly to the genotypes.
Open field test
The open field test was performed as described in previous protocols with minor modifications (Seibenhener and Wooten, 2015). During the test, test subject mice were allowed to explore the arena (L × W × H, 44 cm × 44 cm × 35 cm) individually and were video recorded for 10 mins. Animal movement was traced and analyzed blindly. Rearing behavior was counted blindly as well.
Marble burying test
The marble burying test was performed as described in our previous study (Gilbert et al., 2020). Mice were individually placed into a cage (L × W × H, 25.5 cm × 23 cm × 16.5 cm) with 25 (5 × 5 array) evenly distributed marble balls (1 cm, diameter) on top of ∼ 5 cm thick wood chip beddings and were video recorded continuously for 25 mins. Marble balls exposing less than or equal to 1/3 of their original area (at 0-minute time point) were considered buried and the number of buried marble balls were blindly counted at 0, 5, 10, 15, 20- and 25-minute time points from the footage blindly to genotype.
Self-grooming behavior
The self-grooming behavior test was performed as described in our previous work (Gilbert et al., 2020). Mice were placed in a regular housing cage with a fresh thin layer (∼0.5 cm) of wood chip beddings and were allowed to acclimate for 5 mins. The mice were then video recorded for 10 mins from the side view of the cage by a camera. The total time spent grooming and the number of grooming episodes were measured blindly.
Barnes Maze test
The protocol of the Barnes Maze test is adopted and modified from previous studies (Wang et al., 2017). The circular maze measures 122 cm in diameter with twenty holes, 5 cm in diameter, equidistantly drilled around the perimeter of the maze. The escape cage was placed beneath the escaping hole under the maze. Fluorescent lights and a loud buzzer served as the aversive stimuli during the test. Between trials, the odor was removed by wiping with 70% ethanol and 2% Virkon. During the test, the whole maze was isolated by a white curtain with distinct pictures as visual cues for spatial directions. All mice were habituated to the maze on day 1 to be guided to the escaping hole (Adaptation). From day 2 to day 4, all animals went through a similar procedure for 5 mins to acquire the location of the escape hole without guidance (Acquisition) and were video recorded for learning assessment. Memory retention was tested 24 hours and 6 days following all acquisition sessions. During the memory retention tests, mice were allowed to freely explore the maze with the sealed escaping hole and were video recorded for 4 mins. Mouse movement was traced blindly for proper measurement of the primary latency (time spent to find the escaping hole for the first time), primary hole distance (distance travelled before reaching the escaping hole for the first time), duration of stays in each quadrant, and the number of nose probes at each hole.
Immunohistochemistry
Ninety minutes after the sociability behavioral test, mice were anesthetized in a CO2 chamber and transcranially perfused with ice-cold 4% paraformaldehyde in PBS. The brains were then removed from the skull and placed in fixation solution (4% paraformaldehyde in PBS) at 4°C overnight, followed by incubation in a 30% sucrose PBS solution at 4°C for 3 days until sinking to the bottom. The dehydrated brains were placed in embedding molds (Fisher Scientific, catalog #12-20), submerged in optimal cutting temperature-embedded medium (Tissue-Tek, catalog #25608-930), and quickly frozen by placing the molds with brains in a dry-ice bath with ethanol. Frozen brains were sliced in 50 μm sections on a Leica CM 1850 cryostat (Leica Biosystems) at −20°C. Slices were immediately transferred onto Superforst PLUS adhesion microscope slides (Fisher Scientific, catalog #12-550-15). Brain slices on slides were rehydrated in PBS for 1 hour, followed by permeabilization in 1% Triton X-100 in PBS overnight at 4°C and blocking in 20% bovine serum albumin with 0.1% Triton X-100 in PBS for 1 hour at room temperature. Slices were then incubated with properly diluted primary antibodies made in 2% goat serum with 0.1% Triton X-100 in PBS for 1 day at 4°C, washed three times with PBS, and incubated with properly diluted Alexa Fluor conjugated secondary antibodies in 1% goat serum with 0.1% Triton X-100 for 3 hours at room temperature. Nuclei were labeled with Hoechst, followed by three washes with PBS. Brain slices were soaked in Prolong Gold anti-fade mountant (ThermoFisher Scientific, catalog #P36930) and covered with cover-glass (Fisher Scientific, catalog #12-544-18). Nail posh was applied to the edges of the cover-glass to secure the sealing of the brain slices. Brain sliced were then stored at 4°C in dark before imaging.
Golgi impregnation staining
As described in our previous studies (Gilbert et al., 2020; Khatri et al., 2018), whole brains collected from male Prkn KO and WT mice at P100 were subjected to Golgi neuron staining according to the manufacturer’s instructions (FD Neurotechnologies). Briefly, fresh brains were immersed in an impregnation solution at room temperature for 2 weeks in the dark. Brains were then transferred to a cryo-protectant solution for 3 days in the dark at room temperature before slicing. Following this, brains were sliced coronally in 100 μm thick slices for the optimal condition to observe the morphology and mounted on gelatin-coated microscopy slides. Brain slices were then dehydrated in a series of gradient ethanol solutions. Finally, the slices were immersed in xylene and mounted with Permount mounting medium (Thermo Fisher Scientific) and coverglass. Brain slices were then subjected to imaging. Images obtained from Golgi-stained slices were measured and traced using ImageJ for spines and NeuronJ for dendrites.
Sholl analysis
Layer 5 pyramidal neurons from the Prelimbic cortical region of the prefrontal cortex were analyzed. Layer 5 pyramidal neurons were identified based on the distance of the soma and pia (around 400 μm) as well as the pyramidal shape of soma with a relatively larger soma area in reference to the Allen Brain Atlas and mouse brain atlas (Gabbott et al., 2005; Mitric et al., 2019; Paxinos and Franklin, 2019; Van Eden and Uylings, 1985). Dendritic arborization was traced against original stack Golgi staining images using the NeuronJ plugin. The center of the soma was used as a reference point and 49 concentric circles were made on the tracings: the starting radius was at 10 μm and the ending radius was at 250 μm. The number of intersections at each concentric circle was measured and plotted.
Brain lysate and crude synaptosome preparation
For brain lysate, mouse brains were removed and dissected to separate the prefrontal cortex and the hippocampus. Tissue was minced and homogenized in ice-cold modified Radio-immunoprecipitation assay buffer (RIPA) lysis buffer [50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP40, 1% Sodium deoxycholate (SDOC) and 1% sodium dodecyl sulfate (SDS)] containing mini cOmplete protease inhibitors (Roche). Proteins were further solubilized by sonication and a 30-min incubation on a rotor at 4°C, followed by centrifugation for 10 mins at 13,000 g. The supernatant was collected, and the protein concentration was determined using the PierceTM BCA protein assay kit (ThermoFisher Scientific Inc). All samples were processed and boiled in Laemmli buffer at 95°C with the same protein concentration.
For crude synaptosome fraction purification, prefrontal cortex tissue from P100 Prkn KO and WT mice was minced and homogenized in an ice-cold solution (0.32 M sucrose, 1 mM NaHCO3, 1 mM MgCl2, 0.5 mM CaCl2) by 12 strokes in the homogenizer. Samples were centrifuged at 1400 g for 10 mins to harvest the supernatant (S1). The supernatant (S1) was then transferred to a new tube and centrifuged at 13,800 g for 10 mins to obtain the pellet (P2). The P2 fraction containing the synaptosome was re-suspended in RIPA lysis buffer. Protein concentration was measured using the PierceTM BCA protein assay kit (ThermoFisher Scientific Inc.) and samples of lysates, the S2 and the P2 fraction were diluted to the same protein concentration with RIPA lysis buffer and denatured in Laemmli buffer at 95°C for 10 mins for further Western blotting analysis.
Western blotting
Proteins from brain lysates were separated by SDS-PAGE, transferred to PVDF membranes, and incubated with the desired primary antibodies diluted in TBST (0.05% Tween 20) against certain target proteins, followed by incubation with secondary antibodies conjugated to horseradish peroxidase (HRP) (in Key resource table with proper dilution ratio). The bands were visualized by the ECL substrate (ThermoFisher Scientific Cat. #32106). The intensity of the bands was measured and quantified by ImageJ. All values were normalized to corresponding GAPDH intensities and then normalized to controls where appropriate before statistical analysis.
mRNA analysis
For RT-PCR, total RNA from frontal cortical area or hippocampus was extracted using the RNeasy® Mini Kit (Qiagen) according to the manufacturer’s instructions. cDNA was reverse transcribed from 500 ng of total RNA for each sample using SuperScript® III First-Strand kit (Invitrogen) in accordance with the manufacture’s procedure. cDNA samples were then used as a template for further mRNA level analysis.
For qPCR, 1 μL of the reverse transcribed cDNA templates was used as a template and mixed with primer pairs for Gria1 (5′-GTCCGCCCTGAGAAATCCAG-3′, 5′-CTCGCCCTTGTCGTACCAC-3′), Gria2 (5′-TTCTCCTGTTTTATGGGGACTGA-3′, 5′-CTACCCGAAATGCACTGTATTCT-3′), Grin1 (5′-GATGGAGGTCTTCAAATCTGCC-3′, 5′-GGCCCTTCTGAAAAACCCCT-3′) and Kcnj2 (5′-ATGGGCAGTGTGAGAACCAAC-3′, 5′-TGGACTTTACTCTTGCCATTCC-3′). 10 μL reactions were prepared with Power SYBR® Green PCR Master Mix kit (Applied Biosystems) according to the manufacturer’s protocol and placed in Applied Biosystems 7900HT Fast Real-Time PCR system for real-time monitoring. Primer pairs were from the PrimerBank database (https://pga.mgh.harvard.edu/primerbank/) (Spandidos et al., 2008, 2010; Wang and Seed, 2003).
Imaging and analysis
Golgi staining brain slices were imaged with an inverted phase contrast microscope at either 10× with an air objective for dendritic morphology or at 63× with an oil immersion objective for spine morphology (Zeiss Axiovert 200M, Carl Zeiss, Carl-Zeiss-Strasse, Oberkochen, Germany). Images for dendritic morphology were taken along the z stack at a step of 10 μm and images for spines were acquired at a step of 1 μm. A segment of secondary dendrite from a designated area was selected for spine morphological analysis. Spine length, spine head width, and spine base width were measured using ImageJ. Mushroom spines are defined as spine length ≤ 2 μm and spine head width >2 × spine base width; thin spines are defined as spine head width ≤ 2 × spine base width; stubby spines are defined as spine length ≤ 2 μm and spine head width = 0.6 to 1.5 × spine base width; filopodia are defined as spine length >3 μm.
Brain slice immunostaining images were acquired with Leica DM6 confocal microscopy by Leica Application Suite X (LAS X) software (Leica, Wetzlar, Germany). Exposure time was fixed throughout the imaging process with a confocal fluorescence microscope at 10× with an air objective or 63× with oil immersion objective. The whole brain slices were scanned at 5 μm step along the z-Axis. For c-fos positive cell analysis, all raw images were processed within ImageJ with Enhanced Contrast with saturated = 1, Apply LUT, Auto Thresholding with Yen method and despeckle. The processed image was used for c-fos positive cell counting. As for synaptic puncta analysis, all raw images were processed within ImageJ with brightness and contrast adjustment, followed by thresholding with the same value to select the puncta for counting.
Quantification and statistical analysis
Statistics
For all behavioral test, N represents mouse subject used in the test. For the three-chamber social test (WT, N = 10 mice; Prkn KO, N = 15 mice), marble burying test (WT, N = 10 mice; Prkn KO, N = 14 mice), ultrasonic vocalization recordings (P5: WT, N = 14 mice; Prkn KO, N = 11 mice; P7, P9, P11, and P13: WT, N = 25 mice; Prkn KO, N = 21 mice), Barnes maze test (WT, N = 12; Prkn KO, N = 17), sholl analysis (N = 18 cells from 3 brains for both WT and Prkn KO mice), and c-fos positive neurons number comparison (behaviorally naïve mice, WT: N = 7 brains; Prkn KO: N = 5 brains; sociability mice, WT: N = 4 brains; Prkn KO: N = 5 brains) a two-way ANOVA was applied to determine differences between multiple comparisons with two variables, followed by the Bonferroni or Turkey post hoc test where appropriate. For the open field test (WT, N = 10 mice; Prkn KO, N = 14 mice), grooming test (WT, N = 13 mice; Prkn KO, N = 10 mice), rearing test (WT, N = 10 mice; Prkn KO, N = 14 mice), neuronal morphological analysis (in prefrontal cortex, WT, N = 18 cells from 3 brains; Prkn KO, N = 16 cells from 3 brains; in hippocampus, WT, N = 18 cells from 3 brains; Prkn KO, N = 13 cells from 3 brains), synaptic puncta number analysis and NeuN positive cell number count (N = 9 brains for both WT and Prkn KO mice), western blotting (tissue lysate from frontal cortical area and hippocampus: WT, N = 6 brains; Prkn KO, N = 8 brains; synaptosomal fraction: WT, N = 4 brains; Prkn KO, N = 4 brains) , and RT-PCR analysis (for both frontal cortical area and hippocampus, WT, N = 6 brains; Prkn KO, N = 6 brains) a two-tailed student’s t-test was applied as appropriate.
Prism 7 (GraphPad Software) was used for the graphics and statistical analysis; p < 0.05 was considered statistically significant. All values are presented as mean ± SEM.
Acknowledgments
We would like to thank the Man Lab members for helpful discussion, and Kathryn Ann Odamah for assistance in manuscript preparation. This work was supported by NIH grant R01 MH079407 (H.Y.M.).
Author contributions
H.Y.M. directed and supervised the study. Y.H. performed majority of the experiments and the data analysis. W.L. performed partial westerns. Q.H. performed qPCR experiments. Y.T. participated in mouse breeding, genotyping, and brain tissue collection. Y.H. and H.Y.M. wrote the manuscript.
Declaration of interests
The authors declare that there is no potential conflict of interest.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104573.
Supplemental information
Data and code availability
-
•
All raw behavioral test footages, original western blot images and microscopy data reported in this paper will be shared by the Lead contact upon request.
-
•
No original code has been generated in this study.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.
References
- Angoa-Perez M., Kane M.J., Briggs D.I., Francescutti D.M., Kuhn D.M. Marble burying and nestlet shredding as tests of repetitive, compulsive-like behaviors in mice. JoVE. 2013;82:50978. doi: 10.3791/50978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anney R., Klei L., Pinto D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S., Sykes N., Pagnamenta A.T., et al. A genome-wide scan for common alleles affecting risk for autism. Hum. Mol. Genet. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken T.E., Miller J.A., Ding S.L., Sunkin S.M., Smith K.A., Ng L., Szafer A., Dalley R.A., Royall J.J., Lemon T., et al. A comprehensive transcriptional map of primate brain development. Nature. 2016;535:367–375. doi: 10.1038/nature18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle D.E. Diagnostic and statistical manual of mental disorders (DSM) Coda. 2013;25:191–192. doi: 10.1590/s2317-17822013000200017. [DOI] [PubMed] [Google Scholar]
- Bingol B., Tea J.S., Phu L., Reichelt M., Bakalarski C.E., Song Q., Foreman O., Kirkpatrick D.S., Sheng M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510:370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- Blundell J., Blaiss C.A., Etherton M.R., Espinosa F., Tabuchi K., Walz C., Bolliger M.F., Sudhof T.C., Powell C.M. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J. Neurosci. 2010;30:2115–2129. doi: 10.1523/jneurosci.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumback A.C., Ellwood I.T., Kjaerby C., Iafrati J., Robinson S., Lee A.T., Patel T., Nagaraj S., Davatolhagh F., Sohal V.S. Identifying specific prefrontal neurons that contribute to autism-associated abnormalities in physiology and social behavior. Mol. Psychiatr. 2018;23:2078–2089. doi: 10.1038/mp.2017.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Gao F., Li B., Wang H., Xu Y., Zhu C., Wang G. Parkin mono-ubiquitinates Bcl-2 and regulates autophagy. J. Biol. Chem. 2010;285:38214–38223. doi: 10.1074/jbc.m110.101469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvert E., Tick B., McEwen F., Stewart C., Curran S.R., Woodhouse E., Gillan N., Hallett V., Lietz S., Garnett T., et al. Heritability of autism spectrum disorder in a UK population-based twin sample. JAMA Psychiatr. 2015;72:415. doi: 10.1001/jamapsychiatry.2014.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese G.P., Zhu M., Williams D., Heath S., Waites C.L. Parkin deficiency reduces hippocampal glutamatergic neurotransmission by impairing AMPA receptor endocytosis. J. Neurosci. 2016;36:12243–12258. doi: 10.1523/jneurosci.1473-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley J.N. Mouse behavioral assays relevant to the symptoms of autism. Brain Pathol. 2007;17:448–459. doi: 10.1111/j.1750-3639.2007.00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H., Bayramov K.K., Delibaşı N., Tahtasakal R., Bayramov R., Hamurcu Z., Sener E.F. Disregulation of autophagy in the transgenerational Cc2d1a mouse model of autism. Neuromolecular Med. 2020;22:239–249. doi: 10.1007/s12017-019-08579-x. [DOI] [PubMed] [Google Scholar]
- El Achkar C.M., Spence S.J. Clinical characteristics of children and young adults with co-occurring autism spectrum disorder and epilepsy. Epilepsy Behav. 2015;47:183–190. doi: 10.1016/j.yebeh.2014.12.022. [DOI] [PubMed] [Google Scholar]
- Fearnley J.M., Lees A.J. Ageing and Parkinson's disease: substantia nigra regional selectivity. Brain. 1991;114(Pt 5):2283–2301. doi: 10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- Franklin T.B., Silva B.A., Perova Z., Marrone L., Masferrer M.E., Zhan Y., Kaplan A., Greetham L., Verrechia V., Halman A., et al. Prefrontal cortical control of a brainstem social behavior circuit. Nat. Neurosci. 2017;20:260–270. doi: 10.1038/nn.4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbott P.L., Warner T.A., Jays P.R., Salway P., Busby S.J. Prefrontal cortex in the rat: projections to subcortical autonomic, motor, and limbic centers. J. Comp. Neurol. 2005;492:145–177. doi: 10.1002/cne.20738. [DOI] [PubMed] [Google Scholar]
- Gilbert J., Man H.Y. The X-linked autism protein KIAA2022/KIDLIA regulates neurite outgrowth via N-cadherin and delta-catenin signaling. eNeuro. 2016;3 doi: 10.1523/eneuro.0238-16.2016. ENEURO.0238-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J., Man H.Y. Fundamental elements in autism: from neurogenesis and neurite growth to synaptic plasticity. Front. Cell. Neurosci. 2017;11:359. doi: 10.3389/fncel.2017.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J., O'Connor M., Templet S., Moghaddam M., Di Via Ioschpe A., Sinclair A., Zhu L.Q., Xu W., Man H.Y. NEXMIF/KIDLIA knock-out mouse demonstrates autism-like behaviors, memory deficits, and impairments in synapse formation and function. J. Neurosci. 2020;40:237–254. doi: 10.1523/jneurosci.0222-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S., Dennis M.Y., Baker C., Malig M., Coe B.P., Campbell C.D., Mark K., Vu T.H., Alkan C., Cheng Z., et al. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am. J. Hum. Genet. 2013;92:221–237. doi: 10.1016/j.ajhg.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner J.T., Wang K., Cai G., Korvatska O., Kim C.E., Wood S., Zhang H., Estes A., Brune C.W., Bradfield J.P., et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M.S., Fleming S.M., Palacino J.J., Cepeda C., Lam H.A., Bhatnagar A., Meloni E.G., Wu N., Ackerson L.C., Klapstein G.J., et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J. Biol. Chem. 2003;278:43628–43635. doi: 10.1074/jbc.m308947200. [DOI] [PubMed] [Google Scholar]
- Greco B., Managò F., Tucci V., Kao H.T., Valtorta F., Benfenati F. Autism-related behavioral abnormalities in synapsin knockout mice. Behav. Brain Res. 2013;251:65–74. doi: 10.1016/j.bbr.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J.E., Orr A.L., Madison D.V. Altered hippocampal synaptic physiology in aged parkin-deficient mice. Neuromolecular Med. 2010;12:270–276. doi: 10.1007/s12017-010-8113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey B.K., Wang Y., Hoffer B.J. Transgenic rodent models of Parkinson's disease. Acta Neurochir. Suppl. 2008;101:89–92. doi: 10.1007/978-3-211-78205-7_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Sanders S.J., Liu L., De Rubeis S., Lim E.T., Sutcliffe J.S., Schellenberg G.D., Gibbs R.A., Daly M.J., Buxbaum J.D., et al. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9:e1003671. doi: 10.1371/journal.pgen.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helton T.D., Otsuka T., Lee M.C., Mu Y., Ehlers M.D. Pruning and loss of excitatory synapses by the parkin ubiquitin ligase. Proc. Natl. Acad. Sci. USA. 2008;105:19492–19497. doi: 10.1073/pnas.0802280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann-Conaway S., Brockmann M.M., Schneider K., Annamneedi A., Rahman K.A., Bruns C., Textoris-Taube K., Trimbuch T., Smalla K.H., Rosenmund C., et al. Parkin contributes to synaptic vesicle autophagy in Bassoon-deficient mice. Elife. 2020;9:e56590. doi: 10.7554/elife.56590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y., Khatri N., Hou Q., Gilbert J., Wang G., Man H.Y. The deubiquitinating enzyme USP46 regulates AMPA receptor ubiquitination and trafficking. J. Neurochem. 2015;134:1067–1080. doi: 10.1111/jnc.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim K., Soorya L.V., Halpern D.B., Gorenstein M., Siper P.M., Wang A.T. Social cognitive skills groups increase medial prefrontal cortex activity in children with autism spectrum disorder. Autism Res. 2021;14:2495–2511. doi: 10.1002/aur.2603. [DOI] [PubMed] [Google Scholar]
- Itier J.M., Ibanez P., Mena M.A., Abbas N., Cohen-Salmon C., Bohme G.A., Laville M., Pratt J., Corti O., Pradier L., et al. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum. Mol. Genet. 2003;12:2277–2291. doi: 10.1093/hmg/ddg239. [DOI] [PubMed] [Google Scholar]
- Jarzylo L.A., Man H.Y. Parasynaptic NMDA receptor signaling couples neuronal glutamate transporter function to AMPA receptor synaptic distribution and stability. J. Neurosci. 2012;32:2552–2563. doi: 10.1523/jneurosci.3237-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri N., Gilbert J.P., Huo Y., Sharaflari R., Nee M., Qiao H., Man H.Y. The autism protein Ube3A/E6AP remodels neuronal dendritic arborization via caspase-dependent microtubule destabilization. J. Neurosci. 2018;38:363–378. doi: 10.1523/jneurosci.1511-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Kitada T., Pisani A., Karouani M., Haburcak M., Martella G., Tscherter A., Platania P., Wu B., Pothos E.N., Shen J. Impaired dopamine release and synaptic plasticity in the striatum of parkin-/- mice. J. Neurochem. 2009;110:613–621. doi: 10.1111/j.1471-4159.2009.06152.x. [DOI] [PubMed] [Google Scholar]
- Ko J. Neuroanatomical substrates of rodent social behavior: the medial prefrontal cortex and its projection patterns. Front. Neural Circ. 2017;11:41. doi: 10.3389/fncir.2017.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V., Stoppel D.C., Nong Y., Johnson M.A., Nadler M.J.S., Ozkaynak E., Teng B.L., Nagakura I., Mohammad F., Silva M.A., et al. Autism gene Ube3a and seizures impair sociability by repressing VTA Cbln1. Nature. 2017;543:507–512. doi: 10.1038/nature21678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa R.J. Diagnosis of autism spectrum disorders in the first 3 years of life. Nat. Clin. Pract. Neurol. 2008;4:138–147. doi: 10.1038/ncpneuro0731. [DOI] [PubMed] [Google Scholar]
- Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., Sideris D.P., Fogel A.I., Youle R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D., Ronemus M., Yamrom B., Lee Y.H., Leotta A., Kendall J., Marks S., Lakshmi B., Pai D., Ye K., et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Lin A., Hou Q., Jarzylo L., Amato S., Gilbert J., Shang F., Man H.Y. Nedd4-mediated AMPA receptor ubiquitination regulates receptor turnover and trafficking. J. Neurochem. 2011;119:27–39. doi: 10.1111/j.1471-4159.2011.07221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louros S.R., Osterweil E.K. Perturbed proteostasis in autism spectrum disorders. J. Neurochem. 2016;139:1081–1092. doi: 10.1111/jnc.13723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücking C.B., Dürr A., Bonifati V., Vaughan J., De Michele G., Gasser T., Harhangi B.S., Meco G., Denèfle P., Wood N.W., et al. Association between early-onset Parkinson's disease and mutations in the parkin gene. N. Engl. J. Med. 2000;342:1560–1567. doi: 10.1056/nejm200005253422103. [DOI] [PubMed] [Google Scholar]
- Maenner M.J., Shaw K.A., Bakian A.V., Bilder D.A., Durkin M.S., Esler A., Furnier S.M., Hallas L., Hall-Lande J., Hudson A., et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 Years - autism and developmental disabilities monitoring network, 11 sites, United States, 2018. MMWR Surveill. Summ. 2021;70:1–16. doi: 10.15585/mmwr.ss7011a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineur Y.S., Huynh L.X., Crusio W.E. Social behavior deficits in the Fmr1 mutant mouse. Behav. Brain Res. 2006;168:172–175. doi: 10.1016/j.bbr.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Mitrić M., Seewald A., Moschetti G., Sacerdote P., Ferraguti F., Kummer K.K., Kress M. Layer- and subregion-specific electrophysiological and morphological changes of the medial prefrontal cortex in a mouse model of neuropathic pain. Sci. Rep. 2019;9:9479. doi: 10.1038/s41598-019-45677-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P., Levenson J.M., Battaglia F., Atkinson R., Teague R., Antalffy B., Armstrong D., Arancio O., Sweatt J.D., Zoghbi H.Y. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J. Neurosci. 2006;26:319–327. doi: 10.1523/jneurosci.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow E.M., Yoo S.Y., Flavell S.W., Kim T.K., Lin Y., Hill R.S., Mukaddes N.M., Balkhy S., Gascon G., Hashmi A., et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson S.B., Valakh V. Excitatory/inhibitory balance and circuit homeostasis in autism spectrum disorders. Neuron. 2015;87:684–698. doi: 10.1016/j.neuron.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G., Franklin K.B. Paxinos and Franklin's the Mouse Brain in Stereotaxic Coordinates, Compact: The Coronal Plates and Diagrams. Fifth Edition. Academic Press; 2019. [Google Scholar]
- Peça J., Feliciano C., Ting J.T., Wang W., Wells M.F., Venkatraman T.N., Lascola C.D., Fu Z., Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F.A., Palmiter R.D. Parkin-deficient mice are not a robust model of parkinsonism. Proc. Natl. Acad. Sci. USA. 2005;102:2174–2179. doi: 10.1073/pnas.0409598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S., et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorelov V.M., Rodriguiz R.M., Insco M.L., Caron M.G., Wetsel W.C. Novelty seeking and stereotypic activation of behavior in mice with disruption of the Dat1 gene. Neuropsychopharmacology. 2005;30:1818–1831. doi: 10.1038/sj.npp.1300724. [DOI] [PubMed] [Google Scholar]
- Prasad A., Merico D., Thiruvahindrapuram B., Wei J., Lionel A.C., Sato D., Rickaby J., Lu C., Szatmari P., Roberts W., et al. A discovery resource of rare copy number variations in individuals with autism spectrum disorder. G3 (Bethesda) 2012;2:1665–1685. doi: 10.1534/g3.112.004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P.A., Landa R.J. Association between severity of behavioral phenotype and comorbid attention deficit hyperactivity disorder symptoms in children with autism spectrum disorders. Autism. 2014;18:272–280. doi: 10.1177/1362361312470494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell P.E., Fuccillo M.V., Maxeiner S., Hayton S.J., Gokce O., Lim B.K., Fowler S.C., Malenka R.C., Südhof T., Sudhof T.C. Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell. 2014;158:198–212. doi: 10.1016/j.cell.2014.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacai H., Sakoori K., Konno K., Nagahama K., Suzuki H., Watanabe T., Watanabe M., Uesaka N., Kano M. Autism spectrum disorder-like behavior caused by reduced excitatory synaptic transmission in pyramidal neurons of mouse prefrontal cortex. Nat. Commun. 2020;11:5140. doi: 10.1038/s41467-020-18861-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandin S., Lichtenstein P., Kuja-Halkola R., Hultman C., Larsson H., Reichenberg A. The heritability of autism spectrum disorder. J. Am. Med. Assoc. 2017;318:1182. doi: 10.1001/jama.2017.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterstrom F.K., Kosmicki J.A., Wang J., Breen M.S., De Rubeis S., An J.Y., Peng M., Collins R., Grove J., Klei L., et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–584. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuerle A., Wilson K. PARK2 copy number aberrations in two children presenting with autism spectrum disorder: further support of an association and possible evidence for a new microdeletion/microduplication syndrome. Am J Med Genet B Neuropsychiatr. Genet. 2011;156:413–420. doi: 10.1002/ajmg.b.31176. [DOI] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J., et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener M.L., Wooten M.C. Use of the Open Field Maze to measure locomotor and anxiety-like behavior in mice. JoVE. 2015;96:e52434. doi: 10.3791/52434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seirafi M., Kozlov G., Gehring K. Parkin structure and function. FEBS J. 2015;282:2076–2088. doi: 10.1111/febs.13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido E., Aleksic B., Ozaki N. Copy-number variation in the pathogenesis of autism spectrum disorder. Psychiatr. Clin. Neurosci. 2014;68:85–95. doi: 10.1111/pcn.12128. [DOI] [PubMed] [Google Scholar]
- Silva A.J., Frankland P.W., Marowitz Z., Friedman E., Lazlo G., Cioffi D., Jacks T., Bourtchuladze R. A mouse model for the learning and memory deficits associated with neurofibromatosis type I. Nat. Genet. 1997;15:281–284. doi: 10.1038/ng0397-281. [DOI] [PubMed] [Google Scholar]
- Silverman J.L., Tolu S.S., Barkan C.L., Crawley J.N. Repetitive Self-Grooming Behavior in the BTBR Mouse Model of Autism Is Blocked by the mGluR5 Antagonist MPEP. Neuropsychopharmacology. 2010;35:976–989. doi: 10.1038/npp.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.E.P., Zhou Y.D., Zhang G., Jin Z., Stoppel D.C., Anderson M.P. Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci. Transl. Med. 2011;3:103ra97. doi: 10.1126/scitranslmed.3002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A., Wang X., Wang H., Dragnev S., Thurber T., Seed B. A comprehensive collection of experimentally validated primers for Polymerase Chain Reaction quantitation of murine transcript abundance. BMC Genom. 2008;9:633. doi: 10.1186/1471-2164-9-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A., Wang X., Wang H., Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792–D799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer C.M., Alekseyenko O., Serysheva E., Yuva-Paylor L.A., Paylor R. Altered anxiety-related and social behaviors in the Fmr1 knockout mouse model of fragile X syndrome. Gene Brain Behav. 2005;4:420–430. doi: 10.1111/j.1601-183x.2005.00123.x. [DOI] [PubMed] [Google Scholar]
- Spencer C.M., Graham D.F., Yuva-Paylor L.A., Nelson D.L., Paylor R. Social behavior in Fmr1 knockout mice carrying a human FMR1 transgene. Behav. Neurosci. 2008;122:710–715. doi: 10.1037/0735-7044.122.3.710. [DOI] [PubMed] [Google Scholar]
- Srivastava A.K., Schwartz C.E. Intellectual disability and autism spectrum disorders: causal genes and molecular mechanisms. Neurosci. Biobehav. Rev. 2014;46:161–174. doi: 10.1016/j.neubiorev.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkstein S., Gellar S., Parlier M., Payne L., Piven J. High rates of parkinsonism in adults with autism. J. Neurodev. Disord. 2015;7:29. doi: 10.1186/s11689-015-9125-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein J.L., Parikshak N.N., Geschwind D.H. Rare inherited variation in autism: beginning to see the forest and a few trees. Neuron. 2013;77:209–211. doi: 10.1016/j.neuron.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson S., Taylor J., Lockhart P. In: Mechanisms in Parkinson’s Disease-Models and Treatments. Dushanova J., editor. InTech; 2012. Parkinson’s disease and parkin: insights from Park2 knockout mice. [Google Scholar]
- Sturman O., Germain P.L., Bohacek J. Exploratory rearing: a context- and stress-sensitive behavior recorded in the open-field test. Stress. 2018;21:443–452. doi: 10.1080/10253890.2018.1438405. [DOI] [PubMed] [Google Scholar]
- Tang G., Gudsnuk K., Kuo S.H., Cotrina M.L., Rosoklija G., Sosunov A., Sonders M.S., Kanter E., Castagna C., Yamamoto A., et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83:1482. doi: 10.1016/j.neuron.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tick B., Bolton P., Happé F., Rutter M., Rijsdijk F. Heritability of autism spectrum disorders: a meta-analysis of twin studies. J. Child Psychol. Psychiatr. 2016;57:585–595. doi: 10.1111/jcpp.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treit D., Pinel J.P., Fibiger H.C. Conditioned defensive burying: a new paradigm for the study of anxiolytic agents. Pharmacol. Biochem. Behav. 1981;15:619–626. doi: 10.1016/0091-3057(81)90219-7. [DOI] [PubMed] [Google Scholar]
- Van Eden C.G., Uylings H.B.M. Postnatal volumetric development of the prefrontal cortex in the rat. J. Comp. Neurol. 1985;241:268–274. doi: 10.1002/cne.902410303. [DOI] [PubMed] [Google Scholar]
- Velmeshev D., Schirmer L., Jung D., Haeussler M., Perez Y., Mayer S., Bhaduri A., Goyal N., Rowitch D.H., Kriegstein A.R. Single-cell genomics identifies cell type-specific molecular changes in autism. Science. 2019;364:685–689. doi: 10.1126/science.aav8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscidi E.W., Triche E.W., Pescosolido M.F., McLean R.L., Joseph R.M., Spence S.J., Morrow E.M. Clinical characteristics of children with autism spectrum disorder and co-occurring epilepsy. PLoS One. 2013;8:e67797. doi: 10.1371/journal.pone.0067797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Li S., Gilbert J., Gritton H.J., Wang Z., Li Z., Han X., Selkoe D.J., Man H.Y. Crucial roles for SIRT2 and AMPA receptor acetylation in synaptic plasticity and memory. Cell Rep. 2017;20:1335–1347. doi: 10.1016/j.celrep.2017.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.A., Ni H.M., Haynes A., Manley S., Li Y., Jaeschke H., Ding W.X. Chronic deletion and acute knockdown of parkin have differential responses to acetaminophen-induced mitophagy and liver injury in mice. J. Biol. Chem. 2015;290:10934–10946. doi: 10.1074/jbc.m114.602284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey A.J., Sanders S.J., Li M., Dong S., Tebbenkamp A.T., Muhle R.A., Reilly S.K., Lin L., Fertuzinhos S., Miller J.A., et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wöhr M., Scattoni M.L. Behavioural methods used in rodent models of autism spectrum disorders: current standards and new developments. Behav. Brain Res. 2013;251:5–17. doi: 10.1016/j.bbr.2013.05.047. [DOI] [PubMed] [Google Scholar]
- Won H., Mah W., Kim E. Autism spectrum disorder causes, mechanisms, and treatments: focus on neuronal synapses. Front. Mol. Neurosci. 2013;6:19. doi: 10.3389/fnmol.2013.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro K., Bicks L.K., Leventhal M.B., Kato D., Im S., Flanigan M.E., Garkun Y., Norman K.J., Caro K., Sadahiro M., et al. A prefrontal-paraventricular thalamus circuit requires juvenile social experience to regulate adult sociability in mice. Nat. Neurosci. 2020;23:1240–1252. doi: 10.1038/s41593-020-0695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C.L., Chen H.I., Li L.H., Chien Y.L., Liao H.M., Chou M.C., Chou W.J., Tsai W.C., Chiu Y.N., Wu Y.Y., et al. Genome-wide analysis of copy number variations identifies PARK2 as a candidate gene for autism spectrum disorder. Mol. Autism. 2016;7:23. doi: 10.1186/s13229-016-0087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O., Fenno L.E., Prigge M., Schneider F., Davidson T.J., O'Shea D.J., Sohal V.S., Goshen I., Finkelstein J., Paz J.T., et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaboski B.A., Storch E.A. Comorbid autism spectrum disorder and anxiety disorders: a brief review. Future Neurol. 2018;13:31–37. doi: 10.2217/fnl-2017-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Cortese G.P., Waites C.L. Parkinson's disease-linked Parkin mutations impair glutamatergic signaling in hippocampal neurons. BMC Biol. 2018;16:100. doi: 10.1186/s12915-018-0567-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All raw behavioral test footages, original western blot images and microscopy data reported in this paper will be shared by the Lead contact upon request.
-
•
No original code has been generated in this study.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.