Summary
Boswellia sacra Flueck (family Burseraceae) tree is wounded to produce frankincense. We report its de novo assembled genome (667.8 Mb) comprising 18,564 high-confidence protein-encoding genes. Comparing conserved single-copy genes across eudicots suggest >97% gene space assembly of B. sacra genome. Evolutionary history shows B. sacra gene-duplications derived from recent paralogous events and retained from ancient hexaploidy shared with other eudicots. The genome indicated a major expansion of Gypsy retroelements in last 2 million years. The B. sacra genetic diversity showed four clades intermixed with a primary genotype—dominating most resin-productive trees. Further, the stem transcriptome revealed that wounding concurrently activates phytohormones signaling, cell wall fortification, and resin terpenoid biosynthesis pathways leading to the synthesis of boswellic acid—a key chemotaxonomic marker of Boswellia. The sequence datasets reported here will serve as a foundation to investigate the genetic determinants of frankincense and other resin-producing species in Burseraceae.
Subject areas: Evolutionary biology, Genomics, Plant biology
Graphical abstract

Highlights
-
•
Assembly and architecture of frankincense producing Boswellia sacra Flueck
-
•
Comparative genomics and evolutionary history of frankincense tree within orders
-
•
Transcriptome of stem part and gene expression patterns of wounding to the tree
-
•
Resin biosynthesis pathway and related CYP450 enzymes and gene families
Evolutionary biology; Genomics; Plant biology
Introduction
Family Burseraceae comprises resin-producing species and is widely known for myrrh (Commiphora) and frankincense (Boswellia). Resin products from these two genera are essential for medicinal, commercial, cultural, and religious purposes (Ernst, 2008). The genus Boswellia consists of 24 species that have somatic chromosome numbers of 2n = 22. Of these species, Boswellia sacra Flueck is one of the most essential members: a small-to-medium-sized tree endemic to Oman and Yemen (Coppi et al., 2010; Daly et al., 2010; Ernst, 2008; Langenheim, 2003; Mengistu et al., 2013; Tadesse et al., 2004). Its habitats range from arid subtropical to extreme desert conditions (Coppi et al., 2010; Ernst, 2008). The trees are self-pollinated and bisexual (Daly et al., 2010) (Figure S1). B. sacra is confronted with extreme heat, drought, and low soil fertility during its growth. The tree is famous for producing high-quality oleo-gum resin, known as frankincense or Luban (in Arabic). Resin is obtained by scalping the tree’s outer bark during the dry season (Tadesse et al., 2004). Depending on its size, age, and location, the tree can be tapped for resin four to eight times per year. The tree responds to wounding by discharging a milky viscous liquid (Langenheim, 2003; Mengistu et al., 2013). The strong winds and high temperature of the surrounding environment evaporate the moisture to leave behind a crystalline material (Rijkers et al., 2006). The annual production of frankincense obtained from different species of Boswellia is ∼5,000 to 6,000 tons (Bongers et al., 2019). Tree mortality due to tapping and slow regeneration rates has declined in the existing populations. A possible decrease of >70% over the next 25 years has been predicted (Muys, 2019). Most current harvesting is from trees that are wild or were planted in ancient times. There are efforts to establish regenerating populations of some Boswellia species, but these will require a decade to reach adulthood (Bongers et al., 2019). Hence, promoting sustainable frankincense extraction, propagating in-situ conservation practices, and understanding physiology will drastically improve population growth and resin production.
The chemical profile of frankincense essential oil consists of more than 300 identified mono-, di-, and triterpenes. In addition, >130 phytochemicals belonging to different classes (sterols, di, tri, and sesqui-terpenoids) have been isolated and identified from different species of Boswellia. These include the superior anti-inflammatory agents—boswellic acids (Al-Harrasi et al., 2018b; Halliwell and Gutteridge, 2015) known as chemotaxonomic markers of genus Boswellia (Miyamoto et al., 2014; Pospíšil et al., 2014). Numerous studies (Al-Harrasi et al., 2018b; Gaylord et al., 2007; Knebel et al., 2008; Rehman et al., 2018) have demonstrated that boswellic acids and their derivatives are potent metabolites against different human diseases (Al-Harrasi et al., 2021; Halliwell and Gutteridge, 2015; Mannino et al., 2016; Shah et al., 2009). These compounds, precursors, and intermediates are synthesized through methylerythritol 4-phosphate or mevalonic acid pathways using isopentenyl diphosphate (IPP) dimethylallyl diphosphate (DMAPP) as the backbone to form significant resin constituent’s synthesis. The resin is transported by specialized resin canals (axial or radial) into the outer epidermal region of the stem (Al-Harrasi et al., 2018b; Halliwell and Gutteridge, 2015; Langenheim, 2003). The biosynthesis of resin diterpenoids in conifers has been well characterized recently. However, the triterpenoid-based resin synthesis in Boswellia and Commiphora has not yet been studied because of an absence of gene content data (Al-Harrasi et al., 2018b; Khan et al., 2018a, 2018b). Characterization of B. sacra genes will assist the development of new strategies for improving resin biosynthesis and encouraging the resilient growth of tapped trees.
Population studies on Boswellia are needed as a foundation for future efforts toward improvement, use, and preservation, particularly for discovering sources of useful genetic variation. A few studies have reported using microsatellite markers to understand the genetic diversity of the frankincense population in Africa (Addisalem et al., 2016; Eshete et al., 2012; Mengistu et al., 2013; Tolera et al., 2013). However, little is known about B. sacra or other species of the incense tree family (Burseraceae) at the genomic level. This family comprises 18 genera with ∼540 species and has no genomic datasets. The absence of a reference genome for any member of the Burseraceae hinders the elucidation of mechanisms underlying resin production, wounding stress tolerance, or adaptation to their stressful endemic environments. Thus, the current study will provide insights into the genomic structure, evolutionary history, population dynamics, and responses to wounding stress in B. sacra. We generated a B. sacra genome sequence of 667.8 Megabase pairs (Mb), evidence for ancient polyploidy, and a wealth of identified genes with expression levels influenced by the frankincense harvest process.
Results
Genome sequencing and assembly of B. sacra
An improved high-molecular-weight DNA extraction method from leaf parts was developed to avoid the interference of resin (Methods S1). To generate an initial assembly and determine genome size, de novo sequencing strategies were utilized on Illumina platforms with different library sizes (pair-ended, 300 bp and 550 bp). This approach yielded ∼250.5 Gb of data (Figure S2), equivalent to 357x sequence coverage of the genome. This was followed by additional sequencing of Illumina mate-pair libraries (3Kb and 8Kb) to improve assembly size and help fill gaps. There is no genome sequence data reported for genus Boswellia or family Burseraceae. The results of k-mer analysis revealed approximately ∼597 Mb with ∼137x peak depth (Figure S3). To further improve the assembly, single-molecule real-time (SMRT [PacBio]) sequencing was performed to produce 49.7 Gb of data (average insert size of 15–20 kb). This provided another 70x coverage of the genome. For improved assembly, both Illumina paired end (PE) reads and PacBio long reads were combined de novo to create a polished hybrid assembly (Figure S4; Table S1).
The final hybrid assembly contained 16,350 scaffolds with a total scaffold length of 667,847,569 (Table 1). About 2,560 scaffolds comprised 78% of genome. The longest scaffold was 5.4 Mb (Table 1). Furthermore, scaffolds of >50 kb size covered ∼79% of the entire assembly. BUSCO (Simão et al., 2015) analysis based on a plant-specific database (2,121 genes) revealed 2070 BUSCO genes (97.5%) in the B. sacra assembly. Of these, 1889 (89%) were single-copy genes and 181 (8.5%) were duplicated genes (Figure S5). The mitochondrial genomic sequences were separated from the genomic datasets. Its analysis showed a mitochondrial genome size of 470,365 bp with 43.8% GC. The size of this genome is more significant than that of Arabidopsis thaliana (367,808 bp), as that of Carica papaya (476,890 bp) but smaller than that of Citrus sinensis (640,906 bp; Figures S6 and S7; Table S2).
Table 1.
Genome assembly features of Boswellia sacra
| Length of genome assembly (bp) | 667,841,942 |
| Total length of scaffolds (bp) | 667,847,569 |
| Number of scaffolds | 16,363 |
| N50 of scaffolds (bp) | 209,191 |
| L50 of scaffolds | 653 |
| Total length of contigs (bp) | 755,139,700 |
| Number of contigs | 22,349 |
| N50 of contigs (bp) | 161,367 |
| GC content (%) | 35.15 |
| Longest scaffold | 5,497,760 |
| Longest contig | 5,452,056 |
| Fraction of genome in >50 kb scaffolds | 78.78% |
| Total length of retrotransposons (Class I, bp) | 214,185 |
| Total length of retrotransposons (Class II, bp) | 95,061 |
| Total length of genes (bp) | 63,644,645 |
| Number of high confidence genes | 18,564 |
| Average gene length (bp) | 3,428.39 |
| Number of mRNA | 20,890 |
| Number of exons | 131,908 |
| Average exon length (bp) | 204.11 |
| Average exon number | 6.31 |
| Number of annotated genes | 15,527 |
Genome annotation and transposable element discovery in B. sacra genome
The B. sacra genome was annotated to help understand its basic structural and functional features. Because there is no reference genome available for any close relative, even at the family or tribe levels, we relied on de novo methods as a first step. The GC content of B. sacra was 35.2% (Figure 1). We constructed a customized digital transposable elements (TEs) library through ab initio, homology, and structure-based TEs predicting tools. This was also applied to the current dataset to annotate the genome (Table S3). We identified 311.1 Mb of repeats that comprised ∼46.6% of the genome (Tables S4 and S5). This level of repeat content is relatively standard for genomes in the 600–800 Mb range. This overall 46.6% repeat content includes 278.3 Mb (41.6%) of retroelements and 24.6 Mb (3.7%) of DNA transposons (Table S4). Long terminal repeat retrotransposons (LTR-RTs) comprised the most significant repeats (276 Mb, 41.4% of the genome). Class I and Class II total length of retrotransposons were 214,185 bp and 95,061 bp, respectively (Table 1; Figure 1).
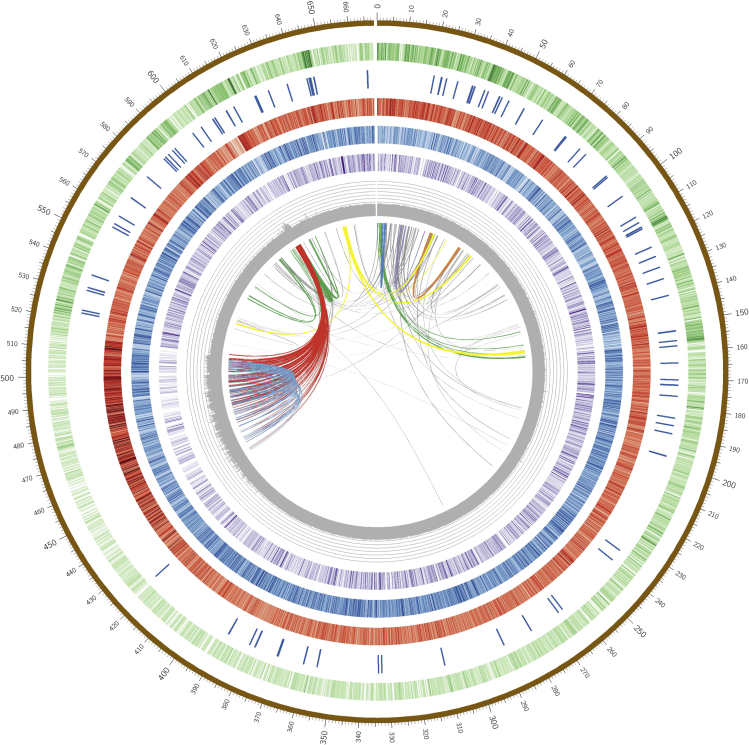
Figure 1.
Genomic landscape of B. sacra based on final de novo assembly
Concentric rings present: all scaffolds (brown) as a concatenated singular molecule, gene density (green), location and presence of genes involved in terpenoid biosynthesis (dark blue), simple repeat density (red), transposable elements distribution (blue), LTR-RT density (purple), GC content (gray), and inner circle show the major collinear gene blocks across different scaffolds.
Among the LTR-RTs, Gypsy accounted for 119.1 Mb and Copia comprised 57.1 Mb of the B. sacra genome. Of the DNA TEs, terminal inverted repeat (TIR) elements contribute ∼20.5 Mb (3.1% of the genome) with exceptionally high numbers of Tc1 and hAT superfamily members (Table S4). The other DNA TEs, helitrons, account for only 0.6% of the genome (Table S5). Because Gypsy retrotransposons are particularly abundant, we analyzed their distribution and insertion date across scaffolds (Figure S8). Estimating insertion dates using LTR divergence (SanMiguel et al., 1998), we observed that >80% of the intact elements were amplified within the last 1.5 million years, and ∼10% inserted <60,000 years ago (Figure S9). Similar findings were observed in the genome of sweet orange (Xu et al., 2013). Moreover, this result agrees with analyses of most angiosperm genomes, where LTR-RT insertion dates occurred predominantly within the last few million years. We found 292,812 simple sequence repeats (SSRs) in the B. sacra genome (Tables S6 and S7), where the majority of SSRs consisted of mononucleotide (191,132; 65.2%) followed by dinucleotide repeats (70,875; 24.2%; Figure S10; Figure 1).
Gene discovery
Given the absence of close relatives in genome databases, our gene discovery process combined ab initio and homology-based searches (Table S3). After ab initio gene prediction with GeneMark-ET (Hoff et al., 2016), we selected previously reported genomes from the order Sapindales as gene-homology sources. These genomes were cashew (Anacardium occidentale), papaya (C. papaya), citrus (Citrus x sinensis), and grape (Vitis vinifera). In addition, we also used 13 other Rosid genomes to predict gene models through a homology search-based approach (Table S3). A total of 363,734 protein sequences of cashew (82,170), papaya (22,914), citrus (76,250), and grape (182,400), in addition to RNA-Seq data (∼88Gb) generated for this study (Table S8) and RNA-Seq data (ERR2040466) from the 1000 Plant (1KP) Transcriptomes (Initiative, 2019) project yielding 12,486,410 total read pairs (∼2.2 Gb of data), were used in gene structure prediction. The gene prediction yielded 121,043 gene models. About 30% of these candidate genes (38,913) were probable transposable element genes or genes for non-coding RNAs (Table S9). The remaining gene models were categorized as high confidence if they were supported by RNA-Seq data (≥50% of coding region covered and ≥5x mean coverage) or protein sequences of cashew, papaya, citrus, and grape, and low confidence for all other genes and for those single-exon genes that had a coding region of fewer than 300 bases (100 aa).
We were left with 18,564 high-confidence protein-coding gene models based on these criteria. The combined length of genes was 63.6 Mb (average length 3,428 bp), or ∼9.5% of the genome (Table S10). About 83% (15,527) of genes were assigned with proposed functional annotation. Including both the high-confidence and all other genome models, the transcripts, exons/CDS, introns, and single-exon genes were 20,890, 131,908, 111,018, and 3077, respectively (Table 1 and S10). BUSCO gene set assessment showed that 95.4% of eudicot orthologs were found in the resulting gene models (Figure S5).
Comparisons of gene content and collinearity in species closely related to B. sacra
To investigate the evolutionary characteristics of B. sacra, we compared the genome collinearity within and between the B. sacra and related species of C. sinensis, A. occidentale in the order Sapindales, while the genomes of V. vinifera and C. papaya only affected by core eudicot hexaploidy event and are selected for outgroups. The inter-genomic dotplot for B. sacra showed higher internal synteny with taxonomically related species C. sinensis than A. occidentale (Figure S11). The colinear genes within B. sacra were assessed and compared to reference genome sequences using ColinearScan (Wang et al., 2006). These results indicate a low level of retained gene content across B. sacra generated by polyploidy—an expected ancientness of duplications (Figure 1).
Their shared ancient polyploidy makes the comparison of collinearity across species even more complicated. This complexity can be resolved by undertaking within-species comparisons for each homologous genome. During analysis of blocks with four or more collinear genes, we found that the highest duplicated genes (13,314) were in A. occidentale (10,494 pairs) and the fewest (1094) were in B. sacra (631 pairs). These two species are closely related in phylogeny (Figures 2A and 2B), but the number of retained homoeologous genes tends to be variable (Tables S11 and S12). Furthermore, we found similar conclusions for blocks with 20 or more collinear genes. The recent polyploidization is unique to the A. occidentale lineage, which may be a reason for the abundance of orthologous duplications.
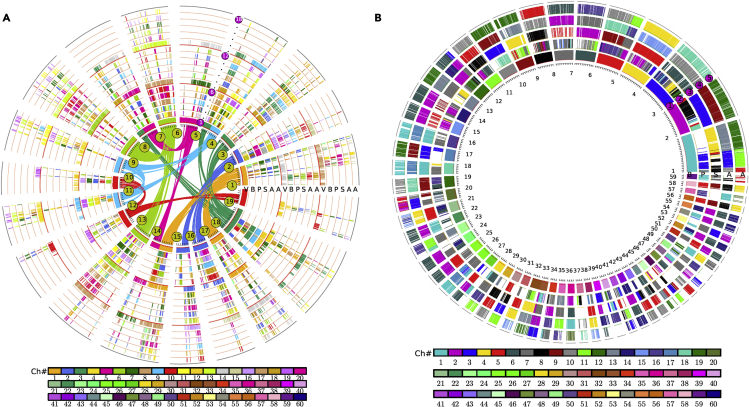
Figure 2.
Genome alignments and comparisons
Each circle represents chromosomes or chromosomal segments from compared genomes, represented by collinear genes. Each gene is represented by a short line to construct the chromosomes or chromosomal regions.
(A) Multiple alignment between five selected genomes (V: V. vinifera, (B) B. sacra, P: C. papaya, S: C. sinensis, (A) A. occidentale) with V. vinifera (grape) as the reference genome. The inner circle shows 19 chromosomes within the grape genome comprising tripled homeologs of seven assumed ancestral chromosomes, and colored lines connect the homoeologous genes. The 15 circles can be divided into three groups according to the major eudicot-common hexaploidy (ECH), in which the first group, the inner five circles, represents the orthologs between them, and the other two groups (the intermediate and outer five circles) are paralogs comparing to the first group generated from the ECH event.
(B) Circles show the multiple alignment between the compared four genomes (B: B. sacra, P: C. papaya, S: C. sinensis, (A) A. occidentalie) with Boswellia sacra as reference. The five circles show their orthologous relationship between the compared genomes, with A. occidentalie comprising two circles to show an extra genome doubling in its genome, in contrast to the other three genomes. The colored boxes indicate chromosome number in their respective source plant as shown in the color scheme at bottom of (A) and (B).
We also assessed the inter-genomic collinearity for 10,555–17,529 collinear genes between two species. The number of collinear genes between B. sacra and A. occidentale was higher (17,529), but the longest collinear blocks (282 genes) were shorter than those between B. sacra and C. sinensis (624 genes). For blocks with >50 colinear genes, we found 47 blocks when B. sacra was compared to A. occidentale (averaging 93 genes in length), while there were 42 blocks in B. sacra compared to C. sinensis (averaging 97 genes in length). Hence, the conserved collinearity between B. sacra and C. sinensis is longer than between B. sacra and A. occidentale (Table S11).
With the V. vinifera genome as a reference and collinear gene IDs, we constructed a hierarchical and event-related multiple genome alignment of V. vinifera, B. sacra, C. papaya, C. sinensis, and A. occidentale to investigate the collinearity of orthologous/homologous genes. The information on collinear genes (Table S13) was translated into a circular map to show the alignment of the five genomes (Figures 2A and 2B). The homoeologous collinearity was used to reveal inter and intragenomic homology and identify polyploidization events. To accommodate genes specific to B. sacra but not available in the grape genome or represented by the earlier alignments, we also constructed genome homology within B. sacra as reference.
Local multiple alignments with related species
Using V. vinifera chromosomes 3, 18, and 4 produced by eudicot-common hexaploidy (Zhuang et al., 2019) (ECH) as a reference, we observed the linear alignment of 1.72–1.82 Mb on chromosome 3 with its corresponding regions from B. sacra and several other genomes. Analysis revealed that this region of grape chromosome 3 was orthologous to those regions from 0.55 to 0.76 Mb on scaffold 30 of B. sacra, with nine collinear genes. This region also shares orthology with the region of 12.14–12.23 Mb on chromosome 5 of C. papaya, the region from 0.92 to 1.16 Mb on chromosome 8 of C. sinensis, and the corresponding regions of chromosomes 3 and 11 of A. occidentale (Figure S12). Also, using B. sacra as a reference, the 0.08 to 0.25 Mb regions on scaffold two and the corresponding regions from other genomes were aligned (Figure S13).
Gene duplication
Our interspecies and intraspecies genomic comparisons have helped to unravel the genome structure of B. sacra. The analysis also allowed distinguishing between the duplicated genes derived from the ECH events versus independently derived paralogs. A total of 9,637 duplicated genes from five species were analyzed. The ECH event showed 3,866 genes (40%) in V. vinifera, 627 genes (6.2%) in B. sacra, and comparatively 285 genes (2.8%) in C. papaya genome (Table S14). While in C. sinensis, there were 1,755 (17%) paralogous genes. A. occidentale genome, on the other hand, went through a unique duplication after ECH events with 4,067 genes (∼40%), the most in selected genomes. A. occidentale special paleo-tetraploidization duplication produced 10,127 genes, which is 2-fold higher than ECH-related genes (Table S14).
The distribution and clustering of orthologous genes (OrthoVenn2 (Wang et al., 2015)) in B. sacra with four species (V. vinifera, C. sinensis, C. climentina, A. occidentale) showed that the protein-coding genes formed 34,868 clusters, 33,129 orthologous clusters (with genes from at least two species), and 1,739 single-copy gene clusters in B. sacra (Figures S14 and S15). The category of the highest count was observed in transcript regulation (26%) and defense-related responses (7.6%), suggesting its significant role in Boswellia (Table S15). Similarly, a total of 26,424 orthogroups (gene families) were identified using genes from B. sacra, V. vinifera, A. occidentale, C. clementina, C. sinensis, and C. papaya (Table S16), of which 7,574 groups contained at least one copy of a gene from each of these species. These orthogroups contained 234,162 genes (90.4% of all compared species genes) distributed well in the six species. Phylogeny of discrete gene duplication events within the 26,424 ortho-groups was shown by constructing alignments of multiple sequences, tree dispositions, and gene-based tree reconciliation. The analysis revealed 114,986 gene-duplication events in the tree (Figure S16). A total of 1,694 gene duplications occurred in the ancestor of these species at nodes 0 and 374 in the ancestor of the C. papaya and the other four species. In total, 107,298 gene duplication events occurred on terminal branches of the species tree. In contrast, 135 gene duplications were mapped at node 3 to differentiate the B. sacra and A. occidentale lineages. Similarly, 1,758 and 49,175 gene duplication events occurred since the divergence of B. sacra and A. occidentale, respectively. The considerable difference between these two is primarily the extra polyploidy in the A. occidentale lineage.
Evolutionary divergence of B. sacra
Analysis of organellar genomes provided a consistent phylogenetic tree for B. sacra and related Rosids, but it was worthwhile to determine whether the nuclear genome followed the same evolutionary pattern. To understand the evolutionary divergence of the nuclear genome, we depicted synonymous nucleotide substitutions on synonymous substitution sites (Ks) between the collinear genes (Zhang et al., 2002). It was noted that the Ks of ECH-produced duplications should be pretty similar, and their clustering might obscure other Ks values for paralogous duplications (Wang et al., 2018b). Normal distributions were used to represent the complex Ks distribution, and the principal one was used to describe the corresponding evolutionary events (Wang et al., 2018b). Considering the different but ECH-related Ks peaks of five species, we adopted the widely accepted concept that the grape has the slowest standard and corrected the evolutionary rates of the compared five genomes (Wang et al., 2018b). Hence, the maximum likelihood estimate from inferred Ks means of ECH-produced duplicated genes was aligned to have the same value as grape (Figure 3A; Table S17). Based on the Ks values, we deduced the polyploidizations and divergence events in the V. vinifera, B. sacra, C. papaya, C. sinensis, and A. occidentale lineages (Figure 3B; Table S17). The results indicated that the genomes of B. sacra, C. sinensis, and C. papaya are similar to the V. vinifera genome that underwent no further polyploidization after ECH. At the same time, the A. occidentale experienced extra whole genome duplication after ECH. Because of the different evolutionary histories of the compared genomes, we corrected all the related genomes using V. vinifera as a rate reference to date each evolutionary event. After correction, we estimated the shared ECH event at ∼115–130 mya (million years ago) and another polyploidization event (AST) in the A. occidentale lineage at ∼29–33 mya (Figure 3C; Table S18). We also used these data to date the divergencies of B. sacra and the other compared four genomes. The divergence of the B. sacra lineages from the shared V. vinifera and C. papaya lineage was dated to ∼80–91 mya. Comparing the B. sacra, C. sinensis, and A. occidentale, we found that B. sacra similarly adheres to the branches of C. sinensis and A. occidentale about 48–54 mya and 50–56 mya, respectively. Furthermore, considering that there may be saturation of synonymous nucleotide variation for tens of millions of years, for caution, we also conducted a comparative analysis of nonsynonymous rate (Ka) in parallel (Figure S17; Tables S19 and S20). We found that the time of polyploidization events and species divergence was close to that estimated by Ks analysis, except for the B. sacra diverged from V. vinifera and C. papaya date to ∼68–77 mya, which is younger than the estimation from Ks analysis.
Figure 3.
Evolutionary comparative assessment of B. sacra
(A) Phylogenetic tree of B. sacra (Bs), A. occidentale (Ao), C. papaya (Cp), C. sinensis (Cs), and V. vinifera (Vv). The core eudicot common-hexaploidy (ECH) is blue, and A. occidentale special tetraploidization (AST) is demonstrated by red colors flash. Gene phylogeny of the three paralogous genes in the Vv, Cp, Bs, Cs, and Ao are denoted by Vv1, Vv2, and Vv3, and Cp1, Cp2, and Cp3, and Cs1, Cs2, and Cs3, and Bs1, Bs2, and BS3, respectively, which ECH produces, and each has 2 orthologs and 4 out paralogs in Ao genome (e.g., V1 has 2 orthologs, Ao11 and Ao12, and 4 out paralogs, Ao21, Ao22, Ao31, and Ao32 in Ao genome). The tree was based on the presence of homologous genes.
(B and C) Ks dating before and after evolutionary rate corrections. Anonymous nucleotide substitution rate (Ks) distribution between the selected genomes (V: V. vinifera, (B) B. sacra, P: C. papaya, S: C. sinensis, (A) A. occidentale). The solid curve lines show the events of polyploidization that appeared in genomes, while the dash lines represent the divergent events between the two compared genomes. The x axes represent the Ks values, and the y axes represent the density of the compared homologous genes. The C shows the correction comparing with the B (Tables S17–S20).
Genetic diversity and population structure of frankincense tree
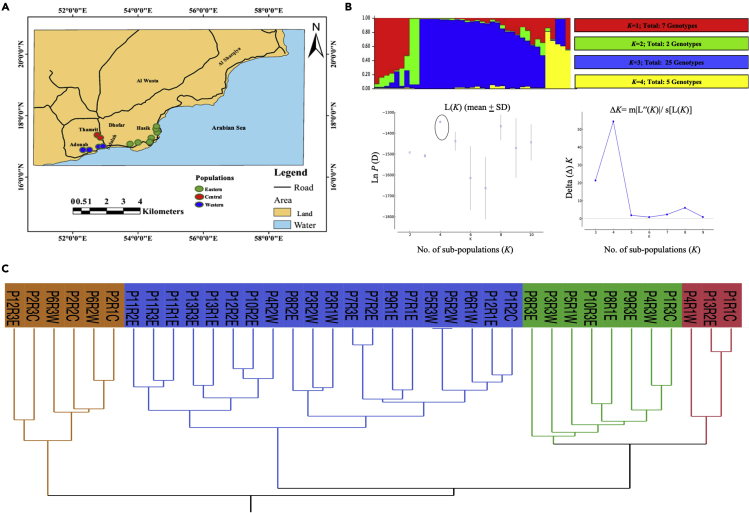
Boswellia is faced with population collapse because of unsustainable resin harvest, which exposes the trees to attack by mites, beetles, and fungal pathogenesis (Groenendijk et al., 2012) (Figure S18). Most frankincense trees grow across the Dhofar region of Oman. Thirteen populations (P) have been categorized as distinctive to three different areas in the Dhofar (C-central, W-western, and E-eastern; Figure 4A). Chloroplast DNA was sequenced at a low depth (112,936,195 bp, 703x chloroplast genome size) and then scanned for the sequences of trnG-psbA, rbcl, ycf, and rpoC1. These genes were explicitly designed for B. sacra. The genomic DNA was extracted from the leaves of twenty trees, each representing three distinctive regions (13 populations), to understand the B. sacra population structure. Sequenced data was analyzed through STRUCTURE (Pritchard et al., 2000b), resulting in 1,775 single nucleotide polymorphisms (Figure 4B). The peak delta (Δ) K was observed at K = 4, suggesting the presence of four main population clusters. This resulted in four distinctive clades, and more than 64.1% of the genotypes were clustered in the main clade comprising the eastern and western populations. These are all found along the coast of the Arabian Sea. The other three clades were geographically dispersed (Figure 4C).
Figure 4.
Population genetic diversity and structure of B. sacra
(A) The tree populations are widely distributed across Dhofar region of Oman, with characteristic 13 populations in different areas. Most of these populations are confluence between sub-tropical arid and desert conditions.
(B) Based on the diversity-related barcodes derived from the chloroplast genome of B. sacra, the population structure was determined, where the sequence data revealed four major subpopulations. The criterion for the choice of model used to detect the most probable value of K was ΔK, which is an ad hoc quantity related to the second-order change in the log probability of data concerning the number of clusters inferred by STRUCTURE (Evanno et al., 2005).
(C) A hierarchal clustering was generated for all four barcodes of the sequence-based data from 13 populations using JMP Pro 14v. “P” is for the unique population sampled, “W, C, or E” shows western, central, and eastern populations, R stands for replication of each genotype.
Wounding responses in B. sacra associated with frankincense production
We exposed several selected trees to wounding stress (30 min, 3, 6, 12, 24 h, and 3 days) and compared their endogenous salicylic acid (SA) and jasmonic acid (JA) levels to unwounded control trees. We found that JA level was significantly increased from 3- to 12-fold during wounding compared with control, while SA showed a similar trend, but the concentrations were exponentially lesser than JA (Figure 5A). To further explore wounding response, we extracted RNA from wounded and unwounded stem tissues to perform RNA-seq (Methods S2). The heatmap depicts 1,451 upregulated differentially expressed genes (DEGs) with significant (p < 0.05) differential expression based on the criterion of q-values (which are adjusted p-values based on Benjamin-Hochberg correction to reduce false positives; Figure 5B). Among these, 118 were annotated as previously known wounding response-related genes in other plants. These genes exhibited 2- to 7-fold higher expressions in the wounded stems (Table S21). The GO categories of these highly expressed DEGs were related to functional processes such as cell-wall biology (biosynthesis, remodeling, or transport), defense mechanisms, carbohydrate metabolism, and oxidative stress responses (Figure 5C; Table S22). Downregulated DEGs were 386, primarily including hypothetical proteins and elongation factors. To help validate the transcript profiling results, some major DEGs had their transcript levels assayed by quantitative real-time PCR. The results revealed that the DEGs involved in terpenoid synthesis, cell wall function, and development were significantly (2- to 8-fold; p < 0.05) upregulated during wounding (Figure 5D). Exogenous application of phytohormones has been shown to regulate plant response during abiotic stresses. We applied JA at 30 min after wounding to see whether it alters the transcript profile in B. sacra. The results indicated that exogenous JA strongly activates cell wall-associated hydrolase and xyloglucan endotransglucosylase/hydrolase protein five genes (Figure 5E) that play an essential role in cell wall fortification. Overall, the calcium and cascade of phytohormones homeostasis (e.g., auxin, ethylene, gibberellic acid, and JA; Tables S21 and S22) are a significant partner in wounding responses that have triggered long-distance signaling during tissue damage.
Figure 5.
Wound-induced changes in transcript factors of B. sacra
(A) After varying time intervals, responses to wounding were assessed by quantifying the endogenous jasmonic acid (JA) and salicylic acid (SA). Gas chromatograph-mass spectroscopy coupled with selected ion monitoring and high-performance liquid chromatography was used. Samples were supplemented with a deuterated internal standard during the extraction phase (Kim et al., 2012) to correct for losses due to extraction. The experiment was repeated three times and compared with a healthy control stem section.
(B) Digital gene expressions profile heatmap was generated from the transcriptomic data of B. sacra tree wounded after 30 min and compared with non-wounded control stem part. The color scale represents the expression fold change. Three biological replicates were used for the RNA-seq.
(C) Different functional classes of significantly regulated transcripts during wounding to stem section. The pie graph shows the number of upregulated genes falling into specific functional categories. This classification is derived from the BLAST2GO analysis of the transcriptome.
(D) Quantitative real-time PCR analysis of significantly expressed genes from the wounded stem section. These genes were selected based on expression patterns in the transcriptomic data. These were divided into four categories depending upon their function BLAST2GO analysis. Gene expression patterns were validated for xyloglucan endotransglucosylase (XGL), cell wall-associated hydrolase (CAH), arabinogalactan (AGN), flavonoid 3′,5′-hydroxylase (FDHA), pectin methyltransferase (PMT), cellulose synthase A (CSA1), amino acid transporter (AT), beta amyrin (BAM), squalene epoxidase (SQE), germacrene D-synthase (GSD), cytochrome p450 (p450), stress enhanced protein (SEPA), and farnesyl diphosphate synthase (FDS). Elongation factor alpha was used as an internal control and quantitative real-time PCR data were calculated using 2-ΔΔCt. Each experiment was replicated three times.
(E) mRNA gene expression related to cell wall-hydrolase (CAH) and xyloglucan endotranglucolase (XGL) regulation during wounding stress to B. sacra. qRT – PCR results show exogenous jasmonic acid’s effect on the wounded stems from B. sacra. The asterisk shows the mean values that were significantly different (p < 0 .05).
Genes involved in resin synthesis in Boswellia
Resin from Boswellia has been viewed as one of the significant defense responses to wounding and pathogenic attacks (Cabrita, 2018; Trapp and Croteau, 2001). Resin, synthesized via methylerythritol 4-phosphate (MEP) or mevalonic acid (MVA) pathways (Bai et al., 2019; Mao et al., 2019), is transported to wounded sites through resin canals (Khan et al., 2018b; Mengistu et al., 2013). The resin is composed of more than 300 chemical constituents (Al-Harrasi et al., 2019a) identified as monoterpenes (KEGG pathway: map00902) and diterpenes (KEGG pathway: map00904), and triterpenes (KEGG pathway: map00909; map01062). The pathways of resin biosynthesis converge either through MEP (monoterpenes and diterpenes) or MVA pathways at farnesyl diphosphate through farnesyl diphosphate synthase (FPPS), forming squalene or oxidosqualene via squalene synthase or epoxidase (SQS/SE). This has been noted as a backbone of triterpenoid biosynthesis (KEGG pathway: map00909) in several angiosperm species, including C. sinensis and V. vinifera.
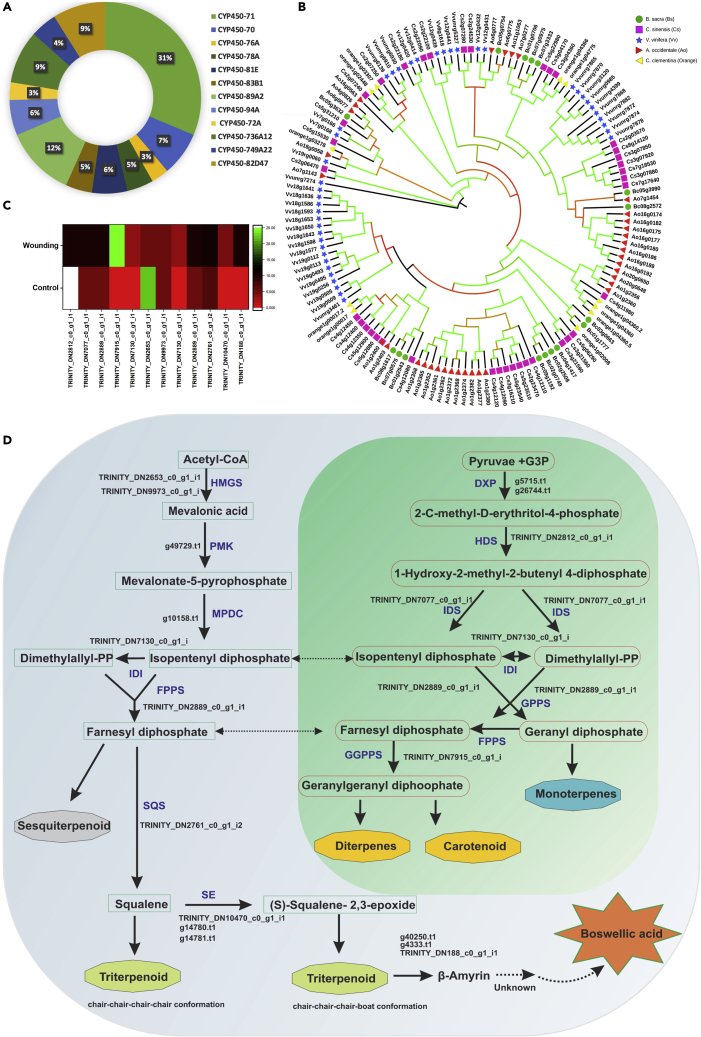
The Cytochrome p450 family is involved in various modifications during the late steps in the biosynthesis of terpene-based resin constituents (Keeling and Bohlmann, 2006). More than 70 genes related to the CYP450 family in B. sacra, with CYP70, CYP89A2, and CYP71 subfamily particularly abundant (Figure 6A; Table S23). The genomic dataset reveals genes from subfamily CYP71, specifically CYP72A and CYP716B1, that are known to play a crucial role downstream (Nelson et al., 2008) in triterpenoid biosynthesis in the resin acid formation field (Banerjee and Hamberger, 2018). DEGs related to CYP71 (especially D9) were upregulated. Also, CYP71 has been noted for its role in geraniol and nerol epoxidase activity and sesquiterpene synthesis (Diaz-Chavez et al., 2013), a vital component of the frankincense yield (Niebler et al., 2016) resin but of largely unknown function. The DEGs related to geraniol through geranyl diphosphate synthase were significantly upregulated in the wounding process (Table S24). Interestingly, we also found genes related to subfamily CYP76, previously found to oxidize (Höfer et al., 2014), geraniol (Figure 6A), which are an active ingredients of fragrance in essential oils (Chen and Viljoen, 2010) and frankincense (Mertens et al., 2009).
Figure 6.
Boswellic acid biosynthesis-related enzymes and gene families in Boswellia sacra
(A) Cyp450 gene families in the genome datasets of B. sacra.
(B) The identification of Terpene synthase (TPS)-related gene families in B. sacra and its comparison with four species. The phylogenetic tree was constructed with a maximum likelihood (ML) approach using RAxML(8.1.16) with 1000 bootstrap replicates. The green color branches show high bootstrap values.
(C) The significantly high DEGs related to terpenoid biosynthesis pathway in control versus wounding stress.
(D) Proposed biosynthesis of resin synthesis in Boswellia based on genomic and transcriptomic datasets.
Previous studies show that terpenoids synthesis was significantly activated upon wounding, including increased terpene synthase (TPS) activity, which can serve as a marker for resin acid synthesis (Chen et al., 2018). Hence, we assessed the TPS gene family in B. sacra and compared it with related species in this study. Identification of TPS gene families of five species and the construction of phylogenetic trees were used to determine B. sacra TPS genes (Figure 6B). The TPS gene family contains key enzymes in the regulatory network of terpenoid biosynthesis and is derived from ancestral triterpene synthase and prenyltransferase enzymes that play essential roles in synthesizing various terpenoids, including triterpenoids (Karunanithi and Zerbe, 2019; Külheim et al., 2015; Martin et al., 2010; Smit et al., 2019; Tholl, 2006). A thorough analysis identified 160 candidate TPS genes in five species, comprising 16 in B. sacra (Bs), 35 in A. occidentale (Ao), eight in C. papaya (Cp), 53 in V. vinifera (Vv), and 48 in C. sinensis (Cs). The analysis revealed gene family expansions in some lineages but relatively low TPS gene numbers in papaya and frankincense trees.
In various tree (e.g., eucalyptus) (Kanagendran et al., 2018), the terpenoids biosynthesis pathway has been associated with a potential role in defense signaling. We found that 87 genes involved in terpenoid synthesis during the wound-induced defense mechanism were significantly upregulated (Figure 6C). The dataset revealed that some significant DEGs are acetyl-CoA C-acetyltransferase, glyceraldehyde-3-phosphate dehydrogenase, isopentenyl phosphate kinase, farnesyl diphosphate synthase, geranylgeranyl diphosphate synthase, squalene synthase, and amyrin synthase. These genes contribute to the synthesis and/or modification of boswellic acid (Al-Harrasi et al., 2018a), a pentacyclic terpenoid.
We found gene sets related to SQS/SE (g82460.t1; Table S25; Figure 6D) while their related DEGs were significantly upregulated during wounding. Amyrin is another proposed precursor of boswellic acid—a critical taxonomic marker for the genus Boswellia (Al-Harrasi et al., 2018a, 2018b, 2019b; Bongers et al., 2019; Muys, 2019) and one of the significant constituents of frankincense. A total of 17 genes involved in amyrin synthesis were annotated in the B. sacra genome (Table S26; Figure 6D), which belonged to beta-amyrin synthase (AmyS) and oxidase. We also noticed that DEGs related to amyrin showed significant upregulation in wounding from the transcriptome data.
Furthermore, we examined paralogous and orthologous genes related to wounding stress and cell-wall biogenesis in B. sacra (Figure S19). The results showed no orthologous genes for xyloglucan endotransglucosylases (Bc09g0214 and Bc09g1941) compared to two resin-producing confers Pinus taeda, Picea abies, and numerous angiosperms, including A. thaliana. Two genes, expansins like A2 (Bc07g2837 (AbuQamar, 2014); cell wall remodeling and stability) and methyltransferase PMT13 (Bc09g1982; part of the vacuolar membrane), had the following: orthologous gene present in B. sacra and A. thaliana but was not found in P. taeda and P. abies. Surprisingly, we detected 197 paralogous genes for Lol (Bc07g2837; related to pollen allergy in maize) and five for O-fucosyltransferase 20 (Bc02g1551; cell wall development-related) in B. sacra. The DEGs related to O-fucosyltransferase 20 and cell wall hydrolases were also upregulated during wounding stress.
Discussion
The unsustainable harvest poses a significant threat to this genus (Muys, 2019). It may result in death for more than 70% of trees in the next 25 years. Hence, the genome sequence of B. sacra is pivotal to understanding the entire family Burseraceae (comprising ∼540 species), especially the Boswellia genus. Genomics and transcriptomics will provide a foundation for understanding frankincense and myrrh genetics. In addition, this work will improve the availabilities of sequence datasets in Sapindales that comprise more than nine families and 7,500 (Bachelier and Endress, 2009) species, where only 16 sequenced genomes are available. A de novo sequence assembly and analysis approaches were used to uncover a haploid genome of 667.8 Mb for B. sacra—nearly 2-fold higher than the genomes of fruit-crop C. sinensis (Rutaceae) (Xu et al., 2013) and medicinal plant Azadirachta indica (Meliaceae) (Krishnan et al., 2016). The sequence quality of current assembly is the highest reported for this genus and family compared to some initial attempts on Boswellia papyrifera (Marçais and Kingsford, 2011). Furthermore, we extracted the chloroplast DNA with a specific protocol (Shi et al., 2012). We sequenced it to identify a complete chloroplast genome (160,543 bp) compared with taxonomically related species in order Sapindales (Khan et al., 2017). The phylogenomic analysis of mitochondrial and chloroplast genomes of B. sacra formed a close relationship with A. indica and C. sinensis (Khan et al., 2017).
Among the most closely related genomes that have been sequenced so far at the tribe level, B. sacra’s genome is almost 45.1% larger than that of sweet orange (367 Mb) (Xu et al., 2013), 28.8% than grape (475Mb) (Jaillon et al., 2007), 44.2% than papaya (372Mb) (Ming et al., 2008), and 33.6% than cashew nut (443.2Mb) (Zimin et al., 2013). However, it was 5.5% lesser than B. papyrifera (Addisalem et al., 2015) (705Mb) without fully sequenced. Adopting several close species annotated genes and ab initio gene modeling, high-confidence protein-encoding genes (18,564; ∼9.5% of the genome) were lesser than citrus (Xu et al., 2013), grape (Jaillon et al., 2007), and in papaya (Ming et al., 2008). B. sacra has 42%, 39.1%, and 24.9% fewer predicted genes than citrus (Xu et al., 2013), grape (Jaillon et al., 2007), and papaya (Ming et al., 2008), respectively. This has been attributed well in the literature due to gene duplication, the difference in ancestral ploidy, and variation in the transposon amplification (Bennetzen et al., 2005). The current study showed 46.6% repeats in the genome, which is slightly higher than V. vinifera (Jaillon et al., 2007) and 2-fold more than C. sinensis (Xu et al., 2013), while the LTR-RTs shared the most significant proportion (41.4%). Similarly, the ∼730 Mb sorghum (Paterson et al., 2009) is ∼61% repeats, and the ∼640 Mb Eucalyptus genome is ∼50% repeats (Myburg et al., 2014). Their relationships maybe become more relative because the extra polyploidization event in A. occidentale accelerated its evolutionary rate. This is consistent with the most recent WGD event in B. sacra being the ancient hexaploidy shared by all eudicots and inconsistent with a new WGD (whether tetraploidy or hexaploidy) occurring in the lineage after the most recent common ancestor of Boswellia and Anacardium. Previously, eudicot-common hexaploidy (ECH) was found in Arabidopsis (Initiative, 2000), grape (Jaillon et al., 2007), and citrus (Xu et al., 2013), helping to unravel their common ancestral genome (Zheng et al., 2009). Due to ancient hexaploidy, intraspecies analysis of similarity in gene content and gene order revealed that the ancestral genome structure was conserved during genome evolution (Wang et al., 2018b). The results showed that ECH occurred 115–130 mya (Jiao et al., 2011; Vekemans et al., 2012), as reported in previous publications (Jaillon et al., 2007; Paterson et al., 2012), and we inferred AST events that happened in 29–33 mya. B. sacra splits 49–55 mya from the nearest ancestors in taxonomy C. sinensis whereas from A. occidentale ancestors 46–52 mya. This is intriguing those ancestral woody plants that might circumvent further ploidies while improving their ability to confront biotic or abiotic environmental challenges to augment adaptation.
Frankincense production is a defense response to wounding (Muys, 2019). Excessive wounding practices are responsible for the decline in B. sacra tree populations; a better understanding of the wounding response may be vital to improving the stem regeneration process (Al-Harrasi et al., 2018b). In all studied vascular plants, jasmonic acid (JA) and salicylic acid (SA) are critical regulators of the wounding response (Ding et al., 2018; Zhou and Memelink, 2016). Boswellia resists the intensities of high heat and wounding by activating defense responses. Specifically, during wounding, JA and SA are potent regulators (Ding et al., 2018; Zhou and Memelink, 2016) that were significantly activated, while a variation in concentration of SA compared to JA also showed a dominating role of JA during the defense. This was in synergy with other wild trees especially crops (Caarls et al., 2015; Xu et al., 2019). We applied JA, which activated cell-wall synthesis-related genes, suggesting that exogenous JA treatment of wounds may improve the tree’s wound healing and defense responses without halting resin production. This approach has been widely used for other crops (Balusamy et al., 2015; Villarreal-García et al., 2016), but the application of JA in combination with other phytohormones such as gibberellins to wild tree-like B. sacra still needs to be tested.
Interestingly, some cell-wall synthesis DEGs were expressed, suggesting that the tree also triggers tissue regeneration after wounding during resin secretion. From the DEGs analysis, we found that the responses of B. sacra to wounding were quite similar to that seen in other plants (Li et al., 2019) (Kazan, 2015; Koo, 2018). However, resin production to cover damaged tissues by activating terpenoids synthesis is prominent and essential to studying Boswellia. The resin is produced either constitutively (flow of stored resin) or induced (synthesis in response) flow to the wounding site via interconnected canals (axial or radial). The upregulation of several genes involved in MEP and MVA pathways leads to increased SQE and AmyS, which are recently proposed precursors of boswellic acids. This was also attributed to the regulation of CYP76, and CYP71 genes that are considerably involved in late triterpene structural modifications (Thimmappa et al., 2014) and functionalization. Further characterization through synthetic biology approaches could essentially lead to understanding and transforming the resin synthesis to enhance conservation efforts in resin extraction from trees (Al-Harrasi et al., 2018a).
Increased resin extraction practices from the Boswellia population pose a more significant threat to the existence of this resource. In contrast, little is known about the population dynamics that are often affected by a combination of factors, including ancient planting locations, current harvesting-induced stresses, possible consumer preferences for particular ecotypes, and differential growth of genotypes in different environments/locations (Addisalem et al., 2015, 2016; Halliwell and Gutteridge, 2015; Langenheim, 2003; Tadesse et al., 2004). Our analysis indicated four distinctive genotypes, with more significant variation between western/eastern and center populations. Although climate and the overall growth ecology can impact the genotype (Gray et al., 2016), the anthropogenic intrusion of unsustainable harvest methods can also markedly affect tree growth and tolerance over time (Aitken et al., 2008). Our data indicate the rare germplasm classes that are most in need of protection. The predominant germplasms seem to be the most productive, partly because they are in the highest rainfall zones. It would be interesting to see what additional diversity is present in the Yemen endogenous regions and whether transplantation of different germplasms into each climactic zone affects resin quality, quantity, or tree sustainability. Crosses between different germplasms can now be planned to optimize diversity for segregating traits that can be mapped as a step toward B. sacra preservation and improvement.
Limitations of the study
In the current study, we investigated the genome architecture and evolutionary history of B.csacra. We have performed a significant amount of genomics, transcriptomics, molecular, and biochemical-related work to understand the tree physiology. We have identified several key enzymes that can play a role in boswellic acid synthesis pathway; however, further experimentation on biocatalysts and gene cloning-based approaches may usher in-depth functionality of these enzymes. Furthermore, new method and technologies for detailed chromosomal structure can help to improve the assembly, annotation, and findings of the missing links in resin biosynthetic pathways.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Boswellia sacra | This paper | NA |
| Chemicals, peptides, and recombinant proteins | ||
| SPRI magnetic beads | Beckman Coulter | Cat#B23317 |
| RNAse A | Qiagen | Cat#19101; RRID:SCR_006680 |
| Qubit dsDNA BR Assay | Invitrogen | Cat#12102 |
| Taq polymerase | New England Biolabs | Cat#M0267S |
| PBS | Beckman Coulter | Cat#6603369 |
| BSA | Vector Laboratories | Cat#H1200 |
| CTAB | Sigma | Cat#H6269 |
| EDTA | Sigma | Cat#E6758 |
| Tris | Sigma | Cat#V900312 |
| 2-Mercaptoethanol | ThermoFisher | Cat#21985023 |
| Acetone | ThermoFisher | Cat#T_702A060015 |
| anhydrous ethanol | ThermoFisher | Cat#E/0550DF/15 |
| Critical commercial assays | ||
| Femto Pulse | Agilent Technologies | Cat#M5330AA |
| Qubit fluorimeter | Thermofisher | Cat#Q33216 |
| NEBNext Ultra II DNA library preparation Kit | New Englands Biolabs | Cat#E7103 |
| Qubit dsDNA HS reagent assay kit | Thermofisher | Cat#Q32851 |
| PacBio SMRT Cells v2.0 | Pacific Biosciences | Cat#101-389-001 |
| Deposited data | ||
| RNA-Seq | This paper | GSE131217 |
| Whole genome sequence data | This paper | SNVD00000000.1 |
| BioProject | This paper | PRJNA379064 |
| Bio samples | This paper | SAMN06578933 |
| Software and algorithms | ||
| Clustal Omega | Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega Sievers., F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins D Molecular Systems Biology 7 Article number: 539 http://10.1038/msb.2011.75 | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| Structure | Inference of Population Structure Using Multilocus Genotype Data Pritchard et al. (2000a) |
https://web.stanford.edu/group/pritchardlab/structure.html |
| JMP ver.12 | SAS Institute Inc., Cary, NC | https://www.jmp.com/en_us/home.html |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| FastQC | FastQC (2016). FastQC: a quality control tool for high throughput sequence data | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| MaSuRCA (Maryland Super Read Cabog Assembler) v3.3.1 | (Zimin et al., 2013) | https://github.com/alekseyzimin/masurca |
| Canu | Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Research. (2017). http://10.1101/gr.215087.116 | https://github.com/marbl/canu |
| JELLYFISH (v1.1.11) | (Marçais and Kingsford, 2011) | http://www.cbcb.umd.edu/software/jellyfish/ |
| Chromosomer v0.1.4a | (Tamazian et al., 2016) | https://academic.oup.com/gigascience/article/5/1/s13742-016-0141-6/2737417?login=false |
| BRAKER v2.1.4 | (Hoff et al., 2016, 2019) | https://github.com/Gaius-Augustus/BRAKER/releases |
| RepeatMasker v4.0 | (Smit et al., 2015, 2017) | https://www.repeatmasker.org/ |
| STAR v2.7 | (Dobin et al., 2013) | https://physiology.med.cornell.edu/faculty/skrabanek/lab/angsd/lecture_notes/STARmanual.pdf |
| GeneMark-ET v4.46 | (Hoff et al., 2016) | http://topaz.gatech.edu/GeneMark/ |
| TransposonPSI | Haas, B. (2007) | (http://transposonpsi.sourceforge.net/) |
| Infernal v1.1.3 | (Nawrocki and Eddy, 2013) | http://eddylab.org/infernal/ |
| ColinearScan | (Wang et al., 2006) | https://github.com/SunPengChuan/wgdi-doc/blob/master/source/collinearity.rst |
| Circos software | (Krzywinski et al., 2009) | http://circos.ca/software/ |
| RepeatMasker v4.0 | (Smit et al., 2013, 2015, 2017) | https://www.repeatmasker.org/ |
| TopHat | (Trapnell et al., 2009) | https://github.com/infphilo/tophat |
| Blast2GO v.2.6.0 | (Conesa et al., 2005) | https://github.com/blast2go-apps |
Resource availability
All the resources are available with this manuscript.
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ahmed Al-Harrasi (aharrasi@unizwa.edu.om).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Plant material
Leaf samples of Boswellia sacra were collected from a preserved tree population at the Museum of Frankincense Land (Salalah, Oman; 17°0ʹ32.58ʹʹ N, 54°08ʹ8.56ʹʹ E, 11 m altitude). Leaf samples from healthy trees were collected and transported in liquid nitrogen. High molecular weight (HMW) genomic DNA was extracted from leaves using a unique method designed to avoid resin interference-based degradation and contamination of DNA (Methods S1). For population genetic analysis, samples were collected from different geographical locations, as described previously in Khan et al. (2018b). For the transcriptome analysis, wounds were induced by scalping the outer bark with a sterilized knife in the middle of the stem of healthy trees (Khan et al., 2017, 2018a).
Method details
DNA extraction, library preparation, and sequencing
Mitochondrial DNA was extracted according to a previously published protocol (Khan et al., 2018a; Strehle et al., 2018). HMW DNA (5μg) was processed for library (300bp and 550bp) preparation using the manufacturer’s protocol (Illumina NovaSeq 6000 and MiSeq) for a de novo sequencing (each 100bp PE) strategy (Figure S2). To improve the initial assembly, long-read sequencing data were generated. Fresh HMW DNA (10μg; Methods S1) was processed according to the manufacturer’s protocol for PacBio Sequel (Library 20Kb; 70x coverage of genome size on SMRT cell). Sequencing was performed at the Center for Genomics and Bioinformatics, Texas A&M University. FastQC (v0.11.6) (FastQC, 2016) was used to check raw read quality. To reduce biases in the analysis, an in-house script was used to filter out reads where less than 90% of the bases were below Q20. Trimmomatic (v0.36) (Bolger et al., 2014) was used to remove adapter sequences.
Genome survey and size estimation
For the de novo assembly obtained through analysis of shotgun sequence data, a genome survey was performed on the Illumina data with JELLYFISH (v1.1.11) (Marçais and Kingsford, 2011) first to estimate B. sacra genome size. In this step, 17, 21, and 25 k-mers were used for calculating genome size. The relative genome size was analyzed with a Cyflow Ploidy Analyser (Partec, Germany) with excitation for PI at 532 nm as described in Dolezel et al. (Doležel and Bartoš, 2005).
Sequence assembly
After the genome sequence data generation with Illumina technology, de novo assembly was performed using Platanus (v1.2.4) (Kajitani et al., 2014). In this step, paired-end reads and mate-pair reads of various insert sizes were used. Scaffolding and gap-filling processes were performed on the same assembler with mate-pair reads. Platanus assembles fragments of DNA into contiguous sequences and constructs scaffolds out of contigs based on information in the paired ends. The complicated graphs resulting from heterozygosity are simplified during the scaffolding and contig assembly. The workflow included (i) Platanus v.1.2.4 (Kajitani et al., 2014) assembly adapted for Illumina with default settings and using trimmed 357× PE reads; (ii) first PacBio assembly using Canu v.1.5 (Koren et al., 2017), based on default parameters and keeping an expected genome size of ∼750 Mb; (iii) Falcon (Chin et al., 2016) assembler for the second assembly of PacBio reads corrected by Canu; and (iv) hybrid assembly using MaSuRCA (Maryland Super Read Cabog Assembler) v3.3.1 (Zimin et al., 2013), using the option of CABOG assembler based on both PacBio and Illumina PE libraries (Figure S4; Table S1). The MaSuRCA-polish tool indicated that the consensus quality of the assembly was 99.9%, which was computed by mapping Illumina PE reads to the assembly.
For the final high-quality assembly, the pipeline used for genome assembly is shown in Figures S2 and S4. Genome assembly was first done with PacBio reads using two long read assemblers, Canu (Koren et al., 2017) and Falcon (Chin et al., 2016), followed by polishing with Arrow (https://github.com/pacificbiosciences/genomicconsensus/) and Pilon (Walker et al., 2014) but the assemblies generated were not as good as one generated using MaSurCA v3.3.1 (Zimin et al., 2013) utilizing both PacBio and Illumina data. A hybrid assembly using MaSurCA was briefly implemented with both PacBio and Illumina reads. QuorUM (Marçais et al., 2015) error correction was used on the Illumina paired-end reads, and these were first extended into super-reads. Super-reads were then used to create mega-reads combined with the PacBio reads (Zimin et al., 2017). These reads and linking pairs, which preserved the information if two mega-reads come from the same PacBio read in the presence of a gap, were provided to a modified version of the CABOG assembler (Miller et al., 2008) for generating contigs and scaffolds. Scaffolds in the resulting assembly were then linked using a similar technique as the MaSurCA super-reads algorithm. The draft assembly was then polished using the MaSuRCA-polish (Garrison and Marth, 2012; Jain et al., 2018; Li and Durbin, 2009), an assembly consensus quality evaluation, and polishing tool that comes with MaSuRCA v3.3.4. The completeness of the final, polished assembly was assessed with BUSCO v3.0 (Simão et al., 2015; Waterhouse et al., 2017) using the Eudicot ortholog dataset and the statistics of the assembly were generated using the BBMap Stats toolkit v38.06 (Bushnell, 2018).
B. sacra genome assembly into pseudo-molecules
Chromosomer v0.1.4a (Tamazian et al., 2016) maximized assembly length. The alignment score ratio threshold (Ratio) distinguished anchored and unplaced fragments. The insertion size is recommended to be equal to or greater than the sequencing library size: gap size = 5000; lens>=1000).
Gene prediction
We used an integrated approach to predict genes. This approach included ab initio and homology searching. Homology searches employed protein sequences of cashew (Anacardium occidentale), sweet orange (Citrus sinensis), grape (Vitis vinifera), and papaya (Carica papaya) and RNA-Seq data of B. sacra generated in this study and those (ERR2040466) from the 1000 Plant (1KP) Transcriptome’s project (PRJEB21674). Gene structure prediction was made with BRAKER v2.1.4 (Hoff et al., 2016, 2019), using the option of proteins of close homology and RNA-Seq data (RNA-Seq and protein supported training) (Barnett et al., 2011; Gremme et al., 2005; Li and Durbin, 2009). The assembly was soft-masked for simple repeats and low complexity sequences using RepeatMasker v4.0 (Smit et al., 2015, 2017). RNA-Seq data were first aligned to the assembly using STAR v2.7 (Dobin et al., 2013). The alignment was used to train GeneMark-ET v4.46 (Hoff et al., 2016), from which predicted gene structures were used to train Augustus v3.3.3 (Hoff et al., 2016) (Stanke et al., 2008) together with the spliced alignment of proteins. The training parameters and extrinsic evidence generated were used for gene prediction by Augustus. The completeness of the gene prediction was assessed with BUSCO v3.0 (Simão et al., 2015; Waterhouse et al., 2017) using the Eudicot ortholog dataset. Predicted proteins that showed sequence homology to known transposable element (TE)-encoded proteins were identified with TransposonPSI (http://transposonpsi.sourceforge.net/). Identification of non-coding RNA was made with Infernal v1.1.3 (Nawrocki and Eddy, 2013), together with Rfam (Kalvari et al., 2017, 2018) and tRNAscan-SE v2.0.3 (Chan and Lowe, 2019). Functional annotation of the protein sequences was performed by BLASTp against the NCBI non-redundant protein database (nr, Oct 2019) and an in-house script (available upon request) that uses the most frequent longest-common substrings of subject sequences as an initial annotation.
Colinearity block analysis
We inferred colinear genes within B. sacra and reference genomes, and between them, using ColinearScan (Wang et al., 2006) provide a function to evaluate the statistical significance of blocks of colinear genes. ColinearScan was used for inferring colinear genes keeping maximum gap length (50 genes, often assumed in previous inferences) in collinearity between neighboring genes in a chromosomal region. Further analysis was performed for the homologous blocks with colinear gene pairs (Wang et al., 2018a). Collinearity significance was checked statistically through CollinearScan.
Homologous gene dot plot and construction of the multiple genome alignments
The Multiple Genome Colinearity Scan (MCSCAN) toolkit from the database “Plant Genome Duplication Database (PGDD)” is available online (http://www.plantgenome.uga.edu/pgdd/). It was implemented to produce dot plots of the comparative gene content and order between B. sacra and related genomes. All grape genes were arranged in the first column to use the grape genome as a reference for constructing a collinearity table. Although the grape genome is diploid, it is derived from an ancient hexaploid. There are often three orthologous copies for any genes, so the gene information is included in three columns in the table. Each specific gene ID was put in the respective column whenever an expected location of a corresponding colinear gene was in the table. However, when the gene was missing at that location due to translocation or gene loss, the corresponding cell was filled with a dot. Without extra duplications, we assigned one column for B. sacra, C. papaya, and C. sinensis genomes. We assigned two columns to this species because of the A. occidentale special tetraploidization (AST) event. Consequently, 18 columns formed the table, derived mostly from diploids, representing 3-fold and 4-fold homology in the polyploids.
Visualization
A high-level data visualization of the B. sacra genome (Figure JJ-1) has been generated utilizing the Circos software (Krzywinski et al., 2009) from a concatenated assembly of all scaffolds in no particular order, separated by a tiny 5-N stretch of nucleotides. Protein-coding genes (green), terpenoid-related genes (blue), and transposable elements (blue range) were extracted from the gene prediction file generated by BRAKER v2.1.4 (Hoff et al., 2016, 2019). RepeatMasker v4.0 (Smit et al., 2013, 2015, 2017)was used to determine simple repeats (red). LTRretriever (Ou and Jiang, 2018), LTRharvest (Ellinghaus et al., 2008), and LTR_FINDER (Xu and Wang, 2007) detected Long Terminal Repeat-Retrotransposons (LTR-RTs) on the genome (purple). GC content deviations from the mean (35.15) were calculated in 50,000 bp non-overlapping windows and displayed with higher-than-average GC% toward the outer, and less than average GC% toward the inner of the circle. Collinear blocks of more than five proteins were determined by BLASTp v2.9 (Gish and States, 1993) of all proteins against self, and collinearity positions were calculated by MCScanX (Wang et al., 2012) and displayed as arcs on the inner ring. In-house Perl scripts were used to remap data from original locations (on scaffolds) to the new pseudo map (available on request).
Ks calculation, distribution fitting, and correction
The Nei-Gojobori approach adopted the Bioperl Statistical module to estimate the synonymous substitution of nucleotides (Ks). Considering the sequenced species different but core-eudicot-common hexaploidy (ECH)related Ks peaks, we adopted the widely accepted concept that grape is evolving the most slowly (Wang et al., 2019). Hence, the grape comparison corrected the evolutionary rates of compared genomes. The estimated likelihood inferred from Ks represents the means of duplicated genes produced by ECH. Following the slowly evolving grape genome, these were aligned to have the same value. The gene pair duplicated variation was obtained as discussed in detail by Wang et al. (2019). Orthologous gene groups (orthogroups) were inferred using OrthoFinder (Emms and Kelly, 2015). Phyldog (Boussau et al., 2013) was used to simultaneously infer the gene trees for each orthogroup and reconcile them with the species' phylogeny. To identify the phylogenetic position of gene duplication events, the multiple sequence alignments for the amino acid sequences of each orthogroup were inferred using mafft-linsi (Katoh and Standley, 2013) and the alignments subject to gene-tree to species-tree reconciliation using Phyldog. To identify retained gene duplication event, the following two filtration criteria as reported by Emms et al. (2016), were applied: 1) A gene duplication event must have supporting evidence for its occurrence from at least two species and 2) all genes resulting from a gene duplication event must be retained in the genomes of all species that diverged after the duplication event. Thus, all gene duplication events analyzed in this work are fully retained in all descendant species that evolved after the gene duplication event. The gene trees for all the gene duplication events are available.
Transcriptomic analysis of the wounded B. sacra stem
Five-year-old healthy tree’s outer bark of stems (secondary branches at least 3–4 inches in diameter and a meter above ground) were wounded (4 × 6 cm wide; approximately 3 mm deep) with a sharp sterilized knife and collected immediately liquid nitrogen after 30min. Similarly, healthy trees with no prior history of wounding were selected as control. Six trees with twelve secondary branches were selected for experiment, followed by pooling samples into three biological replicates for control and wounded stem parts. The stem part was ground into fine powder with liquid nitrogen. Using the Pure Link Plant RNA reagent kit (Life Technologies, USA) with some modifications to the extraction method, total HMW RNA was extracted from the 1.0g of stem part (Methods S2). The Quality, integrity and quantity of RNA were checked using gel electrophoresis and a Qubit 3.0 fluorophotometer and Bioanalyzer (Agilent Technologies, Santa Clara, CA). The RiboMinus Plant Kit for RNA-Seq (Invitrogen) was used to remove ribosomal RNA (rRNA), followed by a quality check on the Bioanalyzer. The cDNA libraries were constructed using the Ion Torrent RNA-seq kit V2. External RNA Controls Consortium (ERCC) RNA spike-in control mixes were added according to the manufacturer’s protocol (Life Technologies USA). The mRNA-enriched template was loaded onto Ion 540 Chips for the transcriptome sequencing on an Ion Torrent S5. A total of 3 independent runs were sequenced. The generated sequence data were subjected to standard methods of quality control, filtering and adopter removal using FastQC (FastQC, 2016) Trimmomatic 0.33 (Bolger et al., 2014), annotation of rRNA as putative proteins (Tripp et al., 2011) and Sort-MeRNA version 2.0 (Kopylova et al., 2012). Trinity version 2.0.6 (Grabherr et al., 2011; Haas et al., 2013) was used for conducting transcript assembly. To evaluate assembly completeness, Bench-marking Universal Single-Copy Orthologs (BUSCO) (Simão et al., 2015) version 1.1 was used as a database of expected single-copy genes. The BUSCO score was reduced by CD-Hit (Li and Godzik, 2006) and the raw assembly was filtered by RSEM-EVAL’s contig impact score (Li et al., 2014). The Trinity differential expression module (Kopylova et al., 2012) was used for sample-specific expression analysis. Using TopHat (Trapnell et al., 2009), the original reads were mapped back against the reference transcriptome using default values. For the wounded and non-wounded stem tissue, gene and transcript expression levels were calculated and tested for the significant difference using Cuffdiff v.2.2.156 to identify differentially expressed genes (DE-Gs). Those genes were selected with a considerable difference based on Q < 0.05. Annotation of the de novo assembled transcripts of B. sacra was performed by BLAST search with Blast2GO v.2.6.0 (Conesa et al., 2005) software.
Quantification and statistical analysis
Differentially regulated genes by qRT-PCR
Total RNA was extracted just before and 30 min after wounding the stem to validate candidate DE-Gs. Genes associated with biochemical and stress responses, cell wall synthesis and terpenoid biosynthesis were analyzed. The sequences retrieved from the annotated data were used to design sequence-specific primers through Primer3Plus (Untergasser et al., 2007). The synthesized cDNA was used for amplification in a Power Up “SYBR” green Master Mix on Quant Studio 5 (Applied Biosystems Life Technologies) PCR reaction. Reactions were performed at 94○C for 10 min in stage 1, 35 cycles at 94○C for 45 s followed by 65 ○C for 45 s and 72○C for 1 min, and finally, the extension temperature was set at 72°C for 2 min. A threshold level of 0.1 was selected for gene amplification. Gene expressions results were analyzed using the delta CT method in Microsoft excel. For fold change gene expression analysis, the following formula was used: ; where
according to Schmittgen and Kenneth, (2008). Graph-pad prism (v7.0 San Diago, CA, USA) was used for graphical analysis.
Endogenous phytohormone analysis
McCloud and Baldwin (1997) described the quantification of jasmonic acid. Freeze-dried tissue of about 100mg was extracted in 70:30 (v/v) acetone and acetic acid (50 mM), with ∼20 ng [9,10-2H2]-9,10-dihydro-JA (∼20 ng) as an internal standard. The fragment ion was carefully observed at m/z = 83 amu, corresponding to the base peaks of JA and internal standards for the quantification of JA. All the experiments were performed in triplicate. About 200 mg of freeze-dried stem tissue was used to quantify salicylic acid (SA), as Seskar et al. (1998) described. The experiment was performed in triplicate.
Population structure analysis
Leaf samples from 13 different populations of B. sacra were used to extract DNA, amplified via PCR by selecting chloroplast genome-based genetic loci and sequenced on Sanger platform. Population structure and identification of admixed individuals were performed using the model-based software program STRUCTURE 2.2 (Pritchard et al., 2000b). Groupings were assigned to individuals with > 70% of genome fraction value. Genetic differences were then illustrated for the likelihood ratio (LR) to describe genetic differences among the subgroups. Then, the best partition models for all individuals were described by maximizing LR and, finally, for the best subpopulation count (K), the ad-hoc approach is proposed.
Acknowledgments
The authors wish to thank Adil Khan, Muhammad Numan, and Usadel Bjorn’s technical suggestions during this work.
Funding Source: The authors A.L.K. and A.A-H. thank The Research Council (ORG/BES/16/007; Ministry of Higher Education, Research & Innovation – Oman) and University of Nizwa (Internal Research Grant) for their financial support to this work. Also, the author A.L.K. thank financial support of University of Houston-National University Research Funds (NURF-R0507404).
Author contributions
A.L.K. and A.A-H., designed the project and arranged funding, S.A., J.J.R., and C-S. L., performed the genome assembly and annotation, T.S., population diversity analysis, A.A-R., D.S., and I-J.L., edited the manuscript, J.B. and J.W., performed the TE analysis, X.S., C.L., J.Y., Z.Z., F.M., J.Y., C.W., H.G., and X.W., performed evolutionary history analysis and prepared graphs, A.L.K. and A.A-H., wrote the manuscript.
Declaration of interests
All the authors declare no conflict of interest.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104574.
Contributor Information
Ahmed Al-Harrasi, Email: aharrasi@unizwa.edu.com.
Xi-Yin Wang, Email: wang.xiyin@hotmail.com.
Supplemental information
Data and code availability
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. No code is produced in this paper.
References
- AbuQamar S. Expansins: cell wall remodeling proteins with a potential function in plant defense. J. Plant Biochem. Physiol. 2014;02:1000e118. doi: 10.4172/2329-9029.1000e118. [DOI] [Google Scholar]
- Addisalem A., Bongers F., Kassahun T., Smulders M. Genetic diversity and differentiation of the frankincense tree (Boswellia papyrifera (Del.) Hochst) across Ethiopia and implications for its conservation. For. Ecol. Manag. 2016;360:253–260. doi: 10.1016/j.foreco.2015.10.038. [DOI] [Google Scholar]
- Addisalem A.B., Esselink G.D., Bongers F., Smulders M.J.M. Genomic sequencing and microsatellite marker development for Boswellia papyrifera, an economically important but threatened tree native to dry tropical forests. AoB Plants. 2015;7:plu086. doi: 10.1093/aobpla/plu086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken S.N., Yeaman S., Holliday J.A., Wang T., Curtis-McLane S. Adaptation, migration or extirpation: climate change outcomes for tree populations. Evol. Appl. 2008;1:95–111. doi: 10.1111/j.1752-4571.2007.00013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Harrasi A., Csuk R., Khan A., Hussain J. Distribution of the anti-inflammatory and anti-depressant compounds: incensole and incensole acetate in genus Boswellia. Phytochemistry. 2019;161:28–40. doi: 10.1016/j.phytochem.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Al-Harrasi A., Hussain H., Csuk R., Khan H.Y. Elsevier; 2018. Chemistry and Bioactivity of Boswellic Acids and Other Terpenoids of the Genus Boswellia. [Google Scholar]
- Al-Harrasi A., Khan A.L., Asaf S., Al-Rawahi A. Biology of Genus Boswellia. Springer; 2019. Frankincense tree physiology and its responses to wounding stress; pp. 53–70. [Google Scholar]
- Al-Harrasi A., Khan A.L., Rehman N.U., Csuk R. Biosynthetic diversity in triterpene cyclization within the Boswellia genus. Phytochemistry. 2021;184:112660. doi: 10.1016/j.phytochem.2021.112660. [DOI] [PubMed] [Google Scholar]
- Al-Harrasi A., Rehman N.U., Khan A.L., Al-Broumi M., Al-Amri I., Hussain J., Hussain H., Csuk R. Chemical, molecular and structural studies of Boswellia species: β-Boswellic Aldehyde and 3-epi-11β-Dihydroxy BA as precursors in biosynthesis of boswellic acids. PLoS One. 2018;13:e0198666. doi: 10.1371/journal.pone.0198666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachelier J.B., Endress P.K. Comparative floral morphology and anatomy of Anacardiaceae and Burseraceae (Sapindales), with a special focus on gynoecium structure and evolution. Bot. J. Linn. Soc. 2009;159:499–571. doi: 10.1111/j.1095-8339.2009.00959.x. [DOI] [Google Scholar]
- Bai Q., He B., Cai Y., Lian H., Zhang Q. Transcriptomic and metabolomic analyses reveal several critical metabolic pathways and candidate genes involved in resin biosynthesis in Pinus massoniana. Mol. Genet. Genomics. 2019;295:327–341. doi: 10.1007/s00438-019-01624-1. [DOI] [PubMed] [Google Scholar]
- Balusamy S.R.D., Rahimi S., Sukweenadhi J., Kim Y.-J., Yang D.-C. Exogenous methyl jasmonate prevents necrosis caused by mechanical wounding and increases terpenoid biosynthesis in Panax ginseng. Plant Cell, Tissue Organ. Cult. 2015;123:341–348. doi: 10.1007/s11240-015-0838-8. [DOI] [Google Scholar]
- Banerjee A., Hamberger B. P450s controlling metabolic bifurcations in plant terpene specialized metabolism. Phytochem. Rev. 2018;17:81–111. doi: 10.1007/s11101-017-9530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett D.W., Garrison E.K., Quinlan A.R., Strömberg M.P., Marth G.T. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J.L., Ma J., Devos K.M. Mechanisms of recent genome size variation in flowering plants. Ann. Bot. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers F., Groenendijk P., Bekele T., Birhane E., Damtew A., Decuyper M., Eshete A., Gezahgne A., Girma A., Khamis M.A., Lemenih M., Mengistu T., Ogbazghi W., Sass-Klaassen U., Tadesse W., Teshome M., Tolera M., Sterck F.J., Zuidema P.A. Frankincense in peril. Nat. Sustain. 2019;2:602–610. doi: 10.1038/s41893-019-0322-2. [DOI] [Google Scholar]
- Boussau B., Szöllősi G.J., Duret L., Gouy M., Tannier E., Daubin V. Genome-scale coestimation of species and gene trees. Genome Res. 2013;23:323–330. doi: 10.1101/gr.141978.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B. University of California; 2018. BBMap Short-Read Aligner, and Other Bioinformatics Tools. 2015. [Google Scholar]
- Caarls L., Pieterse C.M.J., Van Wees S.C.M. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 2015;6:170. doi: 10.3389/fpls.2015.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrita P. Resin flow in conifers. J. Theor. Biol. 2018;453:48–57. doi: 10.1016/j.jtbi.2018.05.020. [DOI] [PubMed] [Google Scholar]
- Chan P.P., Lowe T.M. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 2019;1962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., He X., Chen J., Gu T., Liu P., Xu T., Teale S.A., Hao D. Traumatic resin duct development, terpenoid formation, and related synthase gene expression in Pinus massoniana under feeding pressure of monochamus alternatus. J. Plant Growth Regul. 2018;38:897–908. doi: 10.1007/s00344-018-9900-1. [DOI] [Google Scholar]
- Chen W., Viljoen A. Geraniol—a review of a commercially important fragrance material. South Afr. J. Bot. 2010;76:643–651. doi: 10.1016/j.sajb.2010.05.008. [DOI] [Google Scholar]
- Chin C.-S., Peluso P., Sedlazeck F.J., Nattestad M., Concepcion G.T., Clum A., Dunn C., O'Malley R., Figueroa-Balderas R., Morales-Cruz A., et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Coppi A., Cecchi L., Selvi F., Raffaelli M. The Frankincense tree (Boswellia sacra, Burseraceae) from Oman: ITS and ISSR analyses of genetic diversity and implications for conservation. Genet. Resour. Crop Evol. 2010;57:1041–1052. doi: 10.1007/s10722-010-9546-8. [DOI] [Google Scholar]
- Daly D., Harley M., Martínez-Habibe M., Weeks A. In: THE FAMILIES AND GENERA OF VASCULAR PLANTS. Kubitzki, editor. Springer; 2010. Burseraceae; pp. 76–104. [Google Scholar]
- Diaz-Chavez M.L., Moniodis J., Madilao L.L., Jancsik S., Keeling C.I., Barbour E.L., Ghisalberti E.L., Plummer J.A., Jones C.G., Bohlmann J. Biosynthesis of sandalwood oil: santalum album CYP76F cytochromes P450 produce santalols and bergamotol. PLoS One. 2013;8:e75053. doi: 10.1371/journal.pone.0075053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Sun T., Ao K., Peng Y., Zhang Y., Li X., Zhang Y. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell. 2018;173:1454–1467.e15. doi: 10.1016/j.cell.2018.03.044. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J., Bartoš J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005;95:99–110. doi: 10.1093/aob/mci005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Kurtz S., Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 2008;9:18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D.M., Covshoff S., Hibberd J.M., Kelly S. Independent and parallel evolution of new genes by gene duplication in two origins of C4 photosynthesis provides new insight into the mechanism of phloem loading in C4 species. Mol. Biol. Evol. 2016;33:1796–1806. doi: 10.1093/molbev/msw057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst E. Frankincense: systematic review. Bmj. 2008;337:a2813. doi: 10.1136/bmj.a2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshete A., Sterck F.J., Bongers F. Frankincense production is determined by tree size and tapping frequency and intensity. For. Ecol. Manag. 2012;274:136–142. doi: 10.1016/j.foreco.2012.02.024. [DOI] [Google Scholar]
- Evanno G., Regnaut S., Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294x.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- FastQC 2016. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 2012 doi: 10.48550/arXiv.1207.3907. Preprint at. [DOI] [Google Scholar]
- Gibson Gaylord T., Barrows F.T., Teague A.M., Johansen K.A., Overturf K.E., Shepherd B. Supplementation of taurine and methionine to all-plant protein diets for rainbow trout (Oncorhynchus mykiss) Aquaculture. 2007;269:514–524. doi: 10.1016/j.aquaculture.2007.04.011. [DOI] [Google Scholar]
- Gish W., States D.J. Identification of protein coding regions by database similarity search. Nat. Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray L.K., Hamann A., John S., Rweyongeza D., Barnhardt L., Thomas B.R. Climate change risk management in tree improvement programs: selection and movement of genotypes. Tree Genet. Genomes. 2016;12:23. doi: 10.1007/s11295-016-0983-1. [DOI] [Google Scholar]
- Gremme G., Brendel V., Sparks M.E., Kurtz S. Vol. 47. Information and Software Technology; 2005. Engineering a software tool for gene structure prediction in higher organisms. pp. 965–978. [Google Scholar]
- Groenendijk P., Eshete A., Sterck F.J., Zuidema P.A., Bongers F. Limitations to sustainable frankincense production: blocked regeneration, high adult mortality and declining populations. J. Appl. Ecol. 2012;49:164–173. doi: 10.1111/j.1365-2664.2011.02078.x. [DOI] [Google Scholar]
- Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B., Gutteridge J.M. Oxford University Press; 2015. Free Radicals in Biology and Medicine. [Google Scholar]
- Höfer R., Boachon B., Renault H., Gavira C., Miesch L., Iglesias J., Ginglinger J.-F., Allouche L., Miesch M., Grec S., et al. Dual function of the cytochrome P450 CYP76 family from Arabidopsis thaliana in the metabolism of monoterpenols and phenylurea herbicides. Plant Physiol. 2014;166:1149–1161. doi: 10.1104/pp.114.244814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff K.J., Lange S., Lomsadze A., Borodovsky M., Stanke M. BRAKER1: unsupervised RNA-seq-based genome annotation with GeneMark-ET and AUGUSTUS: table 1. Bioinformatics. 2016;32:767–769. doi: 10.1093/bioinformatics/btv661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff K.J., Lomsadze A., Borodovsky M., Stanke M. Gene Prediction. Springer; 2019. Whole-genome annotation with BRAKER; pp. 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Initiative A.G. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Initiative O.T.P.T. One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019;574:679–685. doi: 10.1038/s41586-019-1693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O., Aury J.-M., Noel B., Policriti A., Clepet C., Casagrande A., Choisne N., Aubourg S., Vitulo N., Jubin C., et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., Tyson J.R., Beggs A.D., Dilthey A.T., Fiddes I.T., et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018;36:338–345. doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wickett N.J., Ayyampalayam S., Chanderbali A.S., Landherr L., Ralph P.E., Tomsho L.P., Hu Y., Liang H., Soltis P.S., et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Kajitani R., Toshimoto K., Noguchi H., Toyoda A., Ogura Y., Okuno M., Yabana M., Harada M., Nagayasu E., Maruyama H., et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24:1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I., Argasinska J., Quinones-Olvera N., Nawrocki E.P., Rivas E., Eddy S.R., Bateman A., Finn R.D., Petrov A.I. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2017;46:D335–D342. doi: 10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I., Nawrocki E.P., Argasinska J., Quinones-Olvera N., Finn R.D., Bateman A., Petrov A.I. Non-coding RNA analysis using the Rfam database. Curr. Protoc. Bioinformatics. 2018;62:e51. doi: 10.1002/cpbi.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagendran A., Pazouki L., Bichele R., Külheim C., Niinemets Ü. Temporal regulation of terpene synthase gene expression in Eucalyptus globulus leaves upon ozone and wounding stresses: relationships with stomatal ozone uptake and emission responses. Environ. Exp. Bot. 2018;155:552–565. doi: 10.1016/j.envexpbot.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunanithi P.S., Zerbe P. Terpene synthases as metabolic gatekeepers in the evolution of plant terpenoid chemical diversity. Front. Plant Sci. 2019;10:1166. doi: 10.3389/fpls.2019.01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015;20:219–229. doi: 10.1016/j.tplants.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Keeling C.I., Bohlmann J. Diterpene resin acids in conifers. Phytochemistry. 2006;67:2415–2423. doi: 10.1016/j.phytochem.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Khan A.L., Al-Harrasi A., Asaf S., Park C.E., Park G.-S., Khan A.R., Lee I.-J., Al-Rawahi A., Shin J.-H. The first chloroplast genome sequence of Boswellia sacra, a resin-producing plant in Oman. PLoS One. 2017;12:e0169794. doi: 10.1371/journal.pone.0169794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.L., Al-Harrasi A., Shahzad R., Imran Q.M., Yun B.-W., Kim Y.-H., Kang S.-M., Al-Rawahi A., Lee I.-J. Regulation of endogenous phytohormones and essential metabolites in frankincense-producing Boswellia sacra under wounding stress. Acta Physiol. Plant. 2018;40:113. doi: 10.1007/s11738-018-2688-6. [DOI] [Google Scholar]
- Khan A.L., Mabood F., Akber F., Ali A., Shahzad R., Al-Harrasi A., Al-Rawahi A., Shinwari Z.K., Lee I.-J. Endogenous phytohormones of frankincense producing Boswellia sacra tree populations. PLoS One. 2018;13:e0207910. doi: 10.1371/journal.pone.0207910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.W., Kim J.B., Cho S.M., Chung M.N., Lee Y.M., Chu S.M., Che J.H., Kim S.N., Kim S.Y., Cho Y.S., et al. Anthocyanin changes in the Korean purple-fleshed sweet potato, Shinzami, as affected by steaming and baking. Food Chem. 2012;130:966–972. doi: 10.1016/j.foodchem.2011.08.031. [DOI] [Google Scholar]
- Knebel L., Robison D.J., Wentworth T.R., Klepzig K.D. Resin flow responses to fertilization, wounding and fungal inoculation in loblolly pine (Pinus taeda) in North Carolina. Tree Physiol. 2008;28:847–853. doi: 10.1093/treephys/28.6.847. [DOI] [PubMed] [Google Scholar]
- Koo A.J. Metabolism of the plant hormone jasmonate: a sentinel for tissue damage and master regulator of stress response. Phytochem. Rev. 2018;17:51–80. doi: 10.1007/s11101-017-9510-8. [DOI] [Google Scholar]
- Kopylova E., Noé L., Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N.M., Jain P., Gupta S., Hariharan A.K., Panda B. An improved genome assembly of Azadirachta indica A. Juss. G3. 2016;6:1835–1840. doi: 10.1534/g3.116.030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol İ., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Külheim C., Padovan A., Hefer C., Krause S.T., Köllner T.G., Myburg A.A., Degenhardt J., Foley W.J. The Eucalyptus terpene synthase gene family. BMC Genomics. 2015;16:450. doi: 10.1186/s12864-015-1598-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenheim J.H. Timber Press; 2003. Plant Resins: Chemistry, Evolution, Ecology, and Ethnobotany. [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Fillmore N., Bai Y., Collins M., Thomson J.A., Stewart R., Dewey C.N. Evaluation of de novo transcriptome assemblies from RNA-Seq data. Genome Biol. 2014;15:553. doi: 10.1186/s13059-014-0553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Bob H., Alec W., Tim F., Jue R., Nils H., Gabor M., Goncalo A., Richard D. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Yan A., Bhatia N., Altinok A., Afik E., Durand-Smet P., Tarr P.T., Schroeder J.I., Heisler M.G., Meyerowitz E.M. Calcium signals are necessary to establish auxin transporter polarity in a plant stem cell niche. Nat. Commun. 2019;10:726. doi: 10.1038/s41467-019-08575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Mannino G., Occhipinti A., Maffei M. Quantitative determination of 3-O-Acetyl-11-Keto-βBoswellic acid (AKBA) and other boswellic acids in boswellia sacra Flueck (syn. B. Carteri birdw) and boswellia serrata roxb. Molecules. 2016;21:1329. doi: 10.3390/molecules21101329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J., He Z., Hao J., Liu T., Chen J., Huang S. Identification, expression, and phylogenetic analyses of terpenoid biosynthesis-related genes in secondary xylem of loblolly pine (Pinus taeda L.) based on transcriptome analyses. PeerJ. 2019;7:e6124. doi: 10.7717/peerj.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Yorke J.A., Zimin A. QuorUM: an error corrector for Illumina reads. PLoS One. 2015;10:e0130821. doi: 10.1371/journal.pone.0130821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.M., Aubourg S., Schouwey M.B., Daviet L., Schalk M., Toub O., Lund S.T., Bohlmann J. Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 2010;10:226. doi: 10.1186/1471-2229-10-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloud E.S., Baldwin I.T. Herbivory and caterpillar regurgitants amplify the wound-induced increases in jasmonic acid but not nicotine in Nicotiana sylvestris. Planta. 1997;203:430–435. doi: 10.1007/s004250050210. [DOI] [Google Scholar]
- Mengistu T., Sterck F.J., Fetene M., Bongers F. Frankincense tapping reduces the carbohydrate storage of Boswellia trees. Tree Physiol. 2013;33:601–608. doi: 10.1093/treephys/tpt035. [DOI] [PubMed] [Google Scholar]
- Mertens M., Buettner A., Kirchhoff E. The volatile constituents of frankincense–a review. Flavour Fragr. J. 2009;24:279–300. doi: 10.1002/ffj.1942. [DOI] [Google Scholar]
- Miller J.R., Delcher A.L., Koren S., Venter E., Walenz B.P., Brownley A., Johnson J., Li K., Mobarry C., Sutton G. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics. 2008;24:2818–2824. doi: 10.1093/bioinformatics/btn548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R., Hou S., Feng Y., Yu Q., Dionne-Laporte A., Saw J.H., Senin P., Wang W., Ly B.V., Lewis K.L.T., et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S., Martinez G.R., Medeiros M.H., Di Mascio P. Singlet molecular oxygen generated by biological hydroperoxides. J. Photochem. Photobiol. B. 2014;139:24–33. doi: 10.1016/j.jphotobiol.2014.03.028. [DOI] [PubMed] [Google Scholar]
- Muys B. Frankincense facing extinction. Nat. Sustainability. 2019;2:665–666. doi: 10.1038/s41893-019-0355-6. [DOI] [Google Scholar]
- Myburg A.A., Grattapaglia D., Tuskan G.A., Hellsten U., Hayes R.D., Grimwood J., Jenkins J., Lindquist E., Tice H., Bauer D., et al. The genome of Eucalyptus grandis. Nature. 2014;510:356–362. doi: 10.1038/nature13308. [DOI] [PubMed] [Google Scholar]
- Nawrocki E.P., Eddy S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.R., Ming R., Alam M., Schuler M.A. Comparison of cytochrome P450 genes from six plant genomes. Trop. Plant Biol. 2008;1:216–235. doi: 10.1007/s12042-008-9022-1. [DOI] [Google Scholar]
- Niebler J., Zhuravlova K., Minceva M., Buettner A. Fragrant sesquiterpene ketones as trace constituents in frankincense volatile oil of Boswellia sacra. J. Nat. Prod. 2016;79:1160–1164. doi: 10.1021/acs.jnatprod.5b00836. [DOI] [PubMed] [Google Scholar]
- Ou S., Jiang N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2018;176:1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., Bowers J.E., Bruggmann R., Dubchak I., Grimwood J., Gundlach H., Haberer G., Hellsten U., Mitros T., Poliakov A., et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Paterson A.H., Wendel J.F., Gundlach H., Guo H., Jenkins J., Jin D., Llewellyn D., Showmaker K.C., Shu S., Udall J., et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492:423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- Pospíšil P., Prasad A., Rác M. Role of reactive oxygen species in ultra-weak photon emission in biological systems. J. Photochem. Photobiol. B. 2014;139:11–23. doi: 10.1016/j.jphotobiol.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Pritchard J.K., Matthew S., Peter Donnelly. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman N.U., Ali L., Al-Harrasi A., Mabood F., Al-Broumi M., Khan A.L., Hussain H., Hussain J., Csuk R. Quantification of AKBA in boswellia sacra using NIRS coupled with PLSR as an alternative method and crossvalidation by HPLC. Phytochem. Anal. 2018;29:137–143. doi: 10.1002/pca.2721. [DOI] [PubMed] [Google Scholar]
- Rijkers T., Ogbazghi W., Wessel M., Bongers F. The effect of tapping for frankincense on sexual reproduction in Boswellia papyrifera. J. Appl. Ecol. 2006;43:1188–1195. doi: 10.1111/j.1365-2664.2006.01215.x. [DOI] [Google Scholar]
- SanMiguel P., Gaut B.S., Tikhonov A., Nakajima Y., Bennetzen J.L. The paleontology of intergene retrotransposons of maize. Nat. Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Kenneth J.L. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Seskar M., Shulaev V., Raskin I. Endogenous methyl salicylate in pathogen-inoculated tobacco Plants1. Plant Physiol. 1998;116:387–392. doi: 10.1104/pp.116.1.387. [DOI] [Google Scholar]
- Shah B.A., Qazi G.N., Taneja S.C. Boswellic acids: a group of medicinally important compounds. Nat. Prod. Rep. 2009;26:72–89. doi: 10.1039/b809437n. [DOI] [PubMed] [Google Scholar]
- Shi C., Hu N., Huang H., Gao J., Zhao Y.-J., Gao L.-Z. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLoS One. 2012;7:e31468. doi: 10.1371/journal.pone.0031468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Smit A., Hubley R., Green P. 2013. RepeatMasker Open-4.0. 2013–2015.http://www.repeatmasker.org [Google Scholar]
- Smit A., Hubley R., Green P. 2015. RepeatMasker Open-4.0. 2013–2015.http://www.repeatmasker.org [Google Scholar]
- Smit A., Hubley R., Green P. 2017. RepeatMasker Open-4.0 (2013-2015)http://www.repeatmasker.org 2015. [Google Scholar]
- Smit S.J., Vivier M.A., Young P.R. Linking terpene synthases to sesquiterpene metabolism in grapevine flowers. Front. Plant Sci. 2019;10:177. doi: 10.3389/fpls.2019.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Diekhans M., Baertsch R., Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Strehle M.M., Purfeerst E., Christensen A.C. A rapid and efficient method for enriching mitochondrial DNA from plants. Mitochondrial DNA B. 2018;3:239–242. doi: 10.1080/23802359.2018.1438856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadesse W., Feleke S., Eshete T. Comparative study of traditional and new tapping methods on frankincense yield of boswellia papyirifera. Ethiopian J. Nat. Resour. 2004;6:287–299. [Google Scholar]
- Tamazian G., Dobrynin P., Krasheninnikova K., Komissarov A., Koepfli K.P., O'Brien S.J. Chromosomer: a reference-based genome arrangement tool for producing draft chromosome sequences. Gigascience. 2016;5:38. doi: 10.1186/s13742-016-0141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmappa R., Geisler K., Louveau T., O'Maille P., Osbourn A. Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 2014;65:225–257. doi: 10.1146/annurev-arplant-050312-120229. [DOI] [PubMed] [Google Scholar]
- Tholl D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 2006;9:297–304. doi: 10.1016/j.pbi.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Tolera M., Sass-Klaassen U., Eshete A., Bongers F., Sterck F.J. Frankincense tree recruitment failed over the past half century. For. Ecol. Manag. 2013;304:65–72. doi: 10.1016/j.foreco.2013.04.036. [DOI] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp S., Croteau R. Defensive resin biosynthesis in conifers. Annu. Rev. Plant Biol. 2001;52:689–724. doi: 10.1146/annurev.arplant.52.1.689. [DOI] [PubMed] [Google Scholar]
- Tripp H.J., Hewson I., Boyarsky S., Stuart J.M., Zehr J.P. Misannotations of rRNA can now generate 90% false positive protein matches in metatranscriptomic studies. Nucleic Acids Res. 2011;39:8792–8802. doi: 10.1093/nar/gkr576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekemans D., Proost S., Vanneste K., Coenen H., Viaene T., Ruelens P., Maere S., Van de Peer Y., Geuten K. Gamma paleohexaploidy in the stem lineage of core eudicots: significance for MADS-box gene and species diversification. Mol. Biol. Evol. 2012;29:3793–3806. doi: 10.1093/molbev/mss183. [DOI] [PubMed] [Google Scholar]
- Villarreal-García D., Nair V., Cisneros-Zevallos L., Jacobo-Velázquez D.A. Plants as biofactories: postharvest stress-induced accumulation of phenolic compounds and glucosinolates in broccoli subjected to wounding stress and exogenous phytohormones. Front. Plant Sci. 2016;7:45. doi: 10.3389/fpls.2016.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K., et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Sun P., Li Y., Liu Y., Yang N., Yu J., Ma X., Sun S., Xia R., Liu X., et al. An overlooked paleotetraploidization in cucurbitaceae. Mol. Biol. Evol. 2018;35:16–26. doi: 10.1093/molbev/msx242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Yuan J., Yu J., Meng F., Sun P., Li Y., Yang N., Wang Z., Pan Y., Ge W., et al. Recursive paleohexaploidization shaped the durian genome. Plant Physiol. 2019;179:209–219. doi: 10.1104/pp.18.00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-P., Yu J.-G., Li J., Sun P.-C., Wang L., Yuan J.-Q., Meng F.-B., Sun S.-R., Li Y.-X., Lei T.-Y., et al. Two likely auto-tetraploidization events shaped kiwifruit genome and contributed to establishment of the Actinidiaceae family. iScience. 2018;7:230–240. doi: 10.1016/j.isci.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Shi X., Li Z., Zhu Q., Kong L., Tang W., Ge S., Luo J. Statistical inference of chromosomal homology based on gene colinearity and applications to Arabidopsis and rice. BMC Bioinformatics. 2006;7:447. doi: 10.1186/1471-2105-7-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Coleman-Derr D., Chen G., Gu Y.Q. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2015;43:W78–W84. doi: 10.1093/nar/gkv487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.-h., Jin H., Marler B., Guo H., et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse R.M., Seppey M., Simão F.A., Manni M., Ioannidis P., Klioutchnikov G., Kriventseva E.V., Zdobnov E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2017;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.-X., Qian L.-X., Wang X.-W., Shao R.-X., Hong Y., Liu S.-S., Wang X.-W. A salivary effector enables whitefly to feed on host plants by eliciting salicylic acid-signaling pathway. Proc. Nat. Acad. Sci. USA. 2019;116:490–495. doi: 10.1073/pnas.1714990116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Chen L.-L., Ruan X., Chen D., Zhu A., Chen C., Bertrand D., Jiao W.-B., Hao B.-H., Lyon M.P., et al. The draft genome of sweet orange (Citrus sinensis) Nat. Genet. 2013;45:59–66. doi: 10.1038/ng.2472. [DOI] [PubMed] [Google Scholar]
- Xu Z., Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Vision T.J., Gaut B.S. Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Mol. Biol. Evol. 2002;19:1464–1473. doi: 10.1093/oxfordjournals.molbev.a004209. [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Albert V.A., Leebens-Mack J., Bell C.D., Paterson A.H., Zheng C., Sankoff D., de Pamphilis C.W., Wall P.K., Soltis P.S. Polyploidy and angiosperm diversification. Am. J. Bot. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Zhou M., Memelink J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 2016;34:441–449. doi: 10.1016/j.biotechadv.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Zhuang W., Chen H., Yang M., Wang J., Pandey M.K., Zhang C., Chang W.-C., Zhang L., Zhang X., Tang R., et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019;51:865–876. doi: 10.1038/s41588-019-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A.V., Marçais G., Puiu D., Roberts M., Salzberg S.L., Yorke J.A. The MaSuRCA genome assembler. Bioinformatics. 2013;29:2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A.V., Puiu D., Luo M.-C., Zhu T., Koren S., Marçais G., Yorke J.A., Dvořák J., Salzberg S.L. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res. 2017;27:787–792. doi: 10.1101/gr.213405.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. No code is produced in this paper.