Abstract
The atmosphere connects habitats across multiple spatial scales via airborne dispersal of microbial cells, propagules and biomolecules. Atmospheric microorganisms have been implicated in a variety of biochemical and biophysical transformations. Here, we review ecological aspects of airborne microorganisms with respect to their dispersal, activity and contribution to climatic processes. Latest studies utilizing metagenomic approaches demonstrate that airborne microbial communities exhibit pronounced biogeography, driven by a combination of biotic and abiotic factors. We quantify distributions and fluxes of microbial cells between surface habitats and the atmosphere and place special emphasis on long-range pathogen dispersal. Recent advances have established that these processes may be relevant for macroecological outcomes in terrestrial and marine habitats. We evaluate the potential biological transformation of atmospheric volatile organic compounds and other substrates by airborne microorganisms and discuss clouds as hotspots of microbial metabolic activity in the atmosphere. Furthermore, we emphasize the role of microorganisms as ice nucleating particles and their relevance for the water cycle via formation of clouds and precipitation. Finally, potential impacts of anthropogenic forcing on the natural atmospheric microbiota via emission of particulate matter, greenhouse gases and microorganisms are discussed.
Keywords: aeromicrobiology, bioaerosols, microbial biogeography, microbial dispersal, microbial ice nucleation, One Health
This review identifies ecological drivers of microbial distribution in the atmosphere, evidence for their involvement in biochemical and biophysical transformations, and risks from anthropogenic forcing.
Introduction
An implicit assumption in microbial ecology is that extensive atmospheric transport between habitats occurs due to the aerosolization and long-range transport of microorganisms (Womack et al. 2010, Zhou and Ning 2017). At any time, the atmosphere supports vast and highly variable numbers of microbial cells, pollen, propagules, cell fragments, viruses and biomolecules (Després et al. 2012), which account for primary biological aerosols (bioaerosols) estimated to represent 25% of all particles larger than 200 nm (Jaenicke 2005). Most cells in the atmosphere are metabolically inactive during transport between source and sink destinations and experience a high turnover (Burrows et al. 2009). The fate of viable bioaerosols is important to maintenance of diversity and resilience in terrestrial and marine ecosystems globally (Womack et al. 2010, Hanson et al. 2012, Barberan et al. 2014). Bioaerosols also have important health-related impacts via the dispersal of animal (Lai et al. 2009) and plant (Brown and Hovmøller 2002) pathogens, gene flow of antibiotic resistance genes (Pal et al. 2016) and microbially derived allergens (Woo et al. 2013). In addition, the recognition that some microorganisms mediate biochemical (Deguillaume et al. 2008) and biophysical transformations (Morris et al. 2008) during brief residence periods in the atmosphere offers the prospect of an active microbial contribution to atmospheric processes.
In this review, the unique complexity of the atmospheric environment and its microbiota is identified and the extent and limitations to current understanding of its ecological relevance are considered (Fig. 1). The scope encompasses microorganisms that occur in outdoor atmospheric air for extended periods or travel across regional or inter-continental distances. The person-to-person transmission of human pathogens facilitated by exhaled respiratory droplets and bioaerosols in air, e.g. SARS-CoV-2 (Kutter et al. 2018, Leung 2021), and human-associated microorganisms suspended in dust within the indoor built environment (Hospodsky et al. 2012) are excluded. This is because they involve brief residence times for microorganisms in air and defining the nature, extent and importance of aerosol transmission for these scenarios is still under active discussion (Leung 2021). Specific focus is given to how insights from molecular ecological studies have identified atmospheric microbiology as key to understanding the interdependence of ecosystems via flux of diverse taxa between atmosphere and surface habitats, and critical examination of the genetic and physiological evidence for microbial activity in the atmosphere. Furthermore, because the atmosphere is facing profound change due to anthropogenic emissions, consideration is given to how this may impact its microbiology in future.
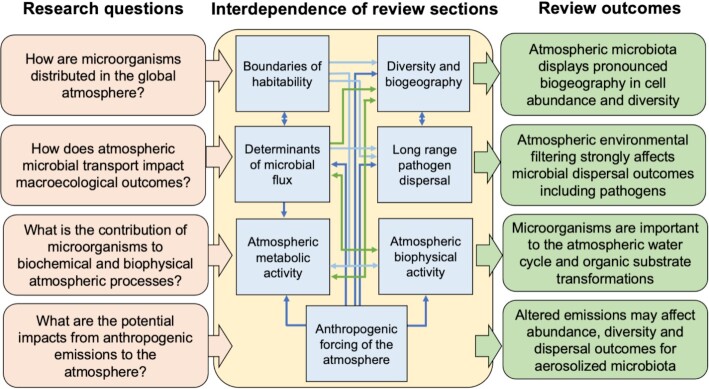
Figure 1.
Structure of this review. Schematic of the research questions and outcomes of this review, with the interdependence of topics within the review indicated by arrows. Arrow colours are for clarity of viewing and do not reflect different processes.
Boundaries for habitability in the atmosphere
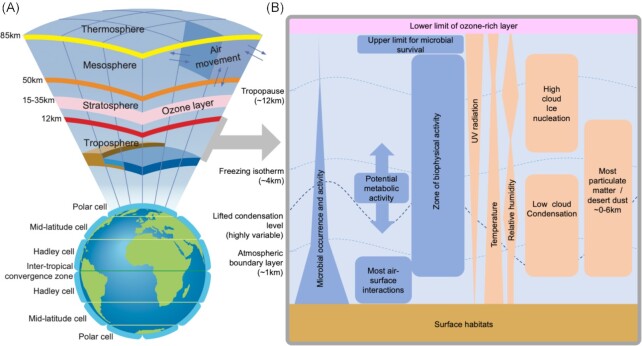
The atmosphere comprises a series of vertically delineated concentric layers whose altitude and depth are primarily defined by thermal properties (Fig. 2A). The troposphere is the lowermost atmospheric layer that contains 75% of its molecular and gaseous mass as well as most water vapour and atmospheric particles (Barry and Chorley 2010), as well as the majority of microbial cells (Fig. 2B). This is where water cycles through clouds, precipitation and surface environments. It includes the atmospheric boundary layer (a.k.a. planetary boundary layer) that is in direct contact with the surface, and the overlaying free troposphere that is of great relevance for regional and global dispersal due to long-range movement of air masses. Vertical mixing above the troposphere is limited due to thermal inversion. Above the troposphere, the bulk of atmospheric ozone is located in the lower stratosphere and this is likely to represent the altitudinal limit to survival for the majority of microbial cells. This is due to lethal UV exposure at higher altitudes as a function of reduced attenuation by ozone and extended residence time for suspended biological particles at higher altitudes (Bryan et al. 2019).
Figure 2.
Defining the atmospheric ecological niche. (A) The atmosphere comprises a series of vertically delineated concentric layers that are primarily defined by thermal properties. The troposphere contains 75% of the molecular and gaseous mass of the atmosphere and almost all of the water vapour, clouds and aerosolized particles including microorganisms. (B) Four boundaries that occur at varying altitude within the troposphere are important in terms of atmospheric microbial ecology: The atmospheric boundary layer delineates the region of air closest to the surface where the bulk of surface–atmosphere interactions occur, and this includes exchange of microorganisms with terrestrial and marine ecosystems. The lifted condensation level (LCL) delineates the layer above which cloud formation occurs. It is variable with temperature and humidity and delineates the thermal limit for condensation of saturated water vapour on particulate surfaces. The freezing isotherm delineates the layer above which freezing occurs and is also the limit for active non-psychrophilic microbial metabolism. We note that the relationship between the atmospheric boundary layer, the LCL and the freezing isotherm, which are highly dynamic, is significantly more complex than presented in the figure, e.g. both the LCL and the freezing isotherm can sit at ground level well within the boundary layer. The boundary between the LCL and freezing isotherm represents the theoretical range for mesophilic microbial metabolic activity, with potential for psychrophilic and xerophilic activity to extend beyond this zone. The tropopause marks the upper limit of the troposphere and mixing with upper layers is limited by the thermal inversion in the stratosphere. The bulk of atmospheric ozone is located in the lower stratosphere and this likely delineates the altitudinal limit of survival for many atmospheric microorganisms. Blue shapes denote biotic attributes and orange boxes denote physical attributes.
The troposphere presents an extremely physico-chemically challenging but nonetheless potentially habitable environment for microorganisms. The presence of sufficient liquid water in cells is a fundamental prerequisite for any microbial activity and is subject to atmospheric relative humidity that is determined by physical conditions and evapotranspiration. In the troposphere, relative humidity is generally bimodal, with higher values in the lower and upper troposphere and a drier mid-troposphere (Fig. 2B) (Gettelman et al. 2006). The LCL is the altitude at which vapour pressure of an air parcel becomes saturated and above the LCL, at slight supersaturations, the water vapour will condense on aerosol particles and form clouds. The LCL varies widely depending on the temperature and water vapour content, which fluctuate with latitude, location and time, determining the cloud patterns (Fig. 2B). Microbial exploitation of low moisture in xeric environments (Pointing and Belnap 2012, Lebre et al. 2017) suggests that sub-saturated relative humidity levels throughout the troposphere are not inhibitory to adapted microbial taxa although the lower limit of relative humidity that supports active metabolism has not been determined. In contrast, only regions of high relative humidity and clouds are likely to support most microorganisms not specifically adapted to low water activity. Cloud droplets provide aqueous microenvironments (Hill et al. 2007, Šantl-Temkiv et al. 2013a, Joly et al. 2015), also the thin layer of liquid water that occurs at the surface of bioaerosols under subsaturated conditions may be sufficient to sustain microbial activity (Stevenson et al. 2015a).
Photo-oxidative and thermal stress are among other major challenges to microbial survival and proliferation in the troposphere (Fig. 2B). A strong altitude-dependent radiation gradient occurs in the troposphere due to the attenuation by ozone, oxygen, water vapour and particulates, and overall UV-A and UV-B radiation increases ∼10–20% with every km in altitude (Barry and Chorley 2010). Whilst UV radiation exposure per se is a strong environmental filter to microbial survival and influences diurnal bacterial occurrence in air (Tong and Lighthart 2006), it is unlikely to be a limiting factor to radiation-tolerant taxa. Air temperature decreases with altitude in the troposphere at the rate of ∼6.5°C per km from ambient near-surface temperatures to −50°C or lower at the tropopause (NOAA, NASA and US Air Force 1976) in the absence of inversions. The freezing isotherm occurs at variable altitudes between ground level and ∼5 km according to season, latitude, terrain and atmospheric conditions (Fig. 2B). This delineates the mechanism of precipitation formation and the altitudinal limit for non-psychrophilic microbial metabolism. Microbial metabolism has been demonstrated at temperatures below those encountered even in the uppermost region of the troposphere (Amato and Christner 2009), and so whilst the freezing isotherm undoubtedly represents a physiological boundary, temperature per se is unlikely to inhibit potential microbial metabolism or survival in the troposphere.
A peculiarity of the atmosphere contrasting with other environments such as soils and oceans is its highly dynamic nature with high cell turnover rates (Burrows et al. 2009). Temperature, humidity and liquid water content can change within minutes, exposing cells to repeated thermal and osmotic shock, and freeze–thaw cycles during residence in air. Additionally, as air density decreases with altitude so does the spatial proximity of cells and their potential substrates. The limited availability of surfaces in the atmosphere is also a challenge to microbial community development as it limits conventional biofilm formation and the possibility for microbial interactions, e.g. via quorum sensing (Monier and Lindow 2003). Overall, the atmosphere has been described as possessing qualities of a chaotic system in terms of its physicochemical complexity (Hochman et al. 2019), and this underlines its heterogeneity as a microbial environment.
Diversity and biogeography of atmospheric microorganisms
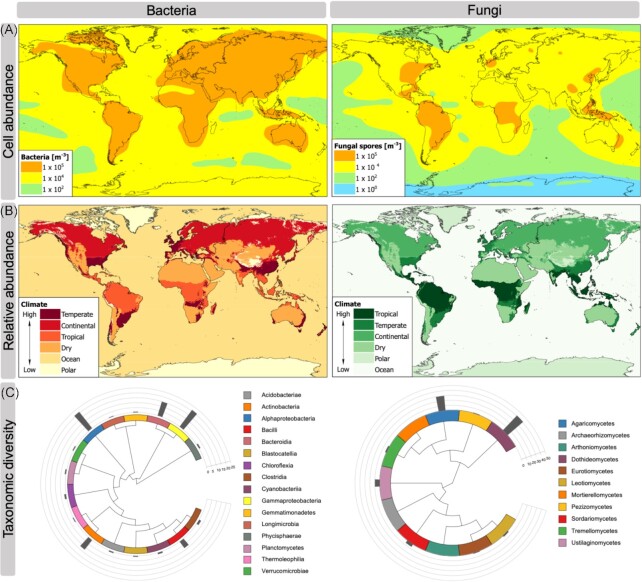
The physical volume of the troposphere dwarfs that of the equivalent microbially inhabited layers of the ocean (i.e. the photic zone) or land (i.e. topsoil) although microbial abundance is several orders of magnitude lower (Whitman et al. 1998, Bar-On et al. 2018). On a global scale, modelled atmospheric cell concentrations fall within the range of 100–105 per m3 for bacteria and fungi (Burrows et al. 2009, Hoose et al. 2010, Spracklen and Heald 2014), and observed geographic patterns in relative abundance of cells are broadly congruent with modelled estimates of their biogeography (Mayol et al. 2017, Tignat-Perrier et al. 2019, Archer et al. 2022) (Fig. 3A and B). Generally, the major delineation in global microbial abundance occurs between air masses above terrestrial and marine ecosystems, with concentration of bacterial and fungal cells highest above terrestrial surfaces and decreasing markedly with distance from land (Mayol et al. 2017). Direct cell counting (Mayol et al. 2017), estimates from sequencing of environmental DNA (Bryan et al. 2019, Gusareva et al. 2019, Tignat-Perrier et al. 2019, Archer et al. 2022) and historical cultivation approaches (Després et al. 2012) collectively indicate that the most abundant cellular microorganisms within the atmospheric boundary layer are bacteria and fungi. Other microbial groups such as archaea and protists appear to comprise a relatively minor component in air (Cáliz et al. 2018, Gusareva et al. 2019, Archer et al. 2022). Estimates from fresh snowfall and from cloud water suggest 4 × 102–4 × 103 bacterial cells per m3 and 4 × 100–4 × 102 eukaryotic cells per m3 assuming condensed water content of 0.4 g per m3 (Christner et al. 2008b, Tesson et al. 2016, Amato et al. 2017, Hu et al. 2018, Šantl-Temkiv et al. 2019). Cells associated with aerosolized desert dust may reach 107 per m3 (Maki et al. 2016). Aggregations of cells that may be free-floating or attached to the surface of particles may create localized elevated concentrations of biomass. In this regard microorganisms within clouds and particulate dust plumes are likely to be significant. The global total mass of clouds has been estimated at 194 000 Tg (Pruppacher and Jaenicke 1995), and the fraction of the atmosphere under cloud cover has been estimated at 67% globally (King et al. 2013). Up to 2 billion tonnes of desert dust is transported in the atmosphere annually (Shepherd et al. 2016), and a comparable mass of soil dust particulates are also emitted to the atmosphere by arable lands every year (Borrelli et al. 2017). Attempts to extrapolate the various estimates in order to estimate global atmospheric cell abundance have a high level of uncertainty and have not been able to adequately encompass variability between boundary layer, troposphere, stratosphere, cloud and dust-free regions, clouds and dust. Nonetheless, a global scale meta-analysis of bacterial abundance across biomes indicated ∼5 × 1019 cells may occur globally in the atmospheric boundary layer, compared with ∼1.2 × 1029 in oceans and 2.6 × 1029 in soils globally (Whitman et al. 1998). This estimate was several orders of magnitude lower than those for ocean, soil and subsurface biomes, and reflects the extremely low density of cells in the atmosphere. The number of viruses in the atmosphere is uncertain but may be significant (Reche et al. 2018). Cell-free microbial metabolites and cell fragments also constitute a significant portion of suspended particulate organic matter in air, above both terrestrial and ocean surfaces (Wilson et al. 2015, Tobo et al. 2019).
Figure 3.
Patterns of microbial diversity and abundance in the atmosphere. (A) Simulated concentration of bacterial cells and fungal spores in the near surface atmospheric boundary layer, adapted from Hoose, Kristjánsson and Burrows (2010). Models assumed a uniform bacterial cell diameter of 1 µm and fungal spore diameter of 5 µm (Burrows et al. 2009), and emission rates were estimated from observed and modelled data (Burrows et al. 2009, Heald and Spracklen 2009). (B) Predicted global relative abundance of bacteria and fungi in the near-surface atmospheric boundary layer from observed quantitative PCR of rRNA genes in air above locations within different climatically defined biomes (Archer et al. 2022). The use of quantitative PCR data is cautiously interpreted here as relative abundance and is not used to infer cell numbers. These data are also domain specific and cannot be directly compared between bacteria and fungi. (C)Taxonomic composition by class of bacteria and fungi in the near-surface atmospheric boundary layer inferred from rigorously decontaminated environmental rRNA gene sequence data from globally distributed locations, adapted from Archer et al. (2022). Coloured boxes indicate classes encountered at a relative abundance of ≥0.1%. Grey bars indicate percentage relative abundance for each class.
Microbiological diversity estimations for the atmosphere have been strongly influenced by methodological biases (Šantl-Temkiv et al. 2020). Direct comparisons of cultivation versus molecular ecological approaches have indicated very few atmospheric microorganisms are cultivable (Temkiv et al. 2012), and this is supported by order of magnitude higher diversity estimates from molecular ecology surveys (Cáliz et al. 2018, Archer et al. 2019, 2020, 2022, Tignat-Perrier et al. 2019, Uetake et al. 2020). The recent emergence of portable high-volume air sampling devices has greatly helped in recovery of sufficient biomass for diversity estimation with sample sizes of several hundred cubic metres of air now possible during short timeframes (Šantl-Temkiv et al. 2020). A persistent issue for diversity estimation has been in distinguishing genuine microbial signatures from the unavoidable contaminants that occur from equipment and reagents when interrogating ultra-low biomass samples. Assessment of contamination during sequence-based diversity estimation indicates that at the very low sample DNA template concentrations encountered in air, reagent-derived contaminants may form a significant component of sequenced diversity (Salter et al. 2014). For this reason a rigorous approach to mitigation is required during experimental and bioinformatic workflows (Eisenhofer et al. 2019). Therefore, it is also necessary to strongly nuance findings where decontamination effort and outcome were not fully reported.
Nonetheless, a growing body of recent research indicates a consensus in the understanding of microorganisms in bulk air (i.e. including free cells, aggregates and particulate-associated cells). Taxa adapted to survival under high UV irradiance, moisture limitation and other challenges are over-represented in atmospheric communities, which is indicative of strong environmental filtering. This is supported by evidence that atmospheric microbial communities exhibit distinct phylogenetic structuring and elevated abundance of stress-tolerant taxa within the boundary layer and at higher altitudes (Tignat-Perrier et al. 2019, Archer et al. 2022). Thus theoretical frameworks in ecology that have made implicit assumptions about ubiquitous microbial distribution in air are being validated and revised (Dumbrell et al. 2010, Lowe and McPeek 2014, Zhou and Ning 2017).
Across broad geographic, altitudinal and temporal scales in the troposphere taxonomic diversity broadly reflects recruitment from underlying surface source habitats, and commonly encountered bacteria affiliate with the classes Alphaproteobacteria, Gammaproteobacteria, Bacteroidia, Actinobacteria and Bacilli (Fig. 3C). Spore-forming bacteria are particularly prevalent and also taxa adapted to environmental stresses encountered in the atmosphere, e.g. desiccation, oxidative stress (Aalismail et al. 2019, Archer et al. 2022). The commonly encountered fungal classes in air largely comprise Dothideomycetes and Agaricomycetes (Cáliz et al. 2018, Gusareva et al. 2019, Tignat-Perrier et al. 2019, Els et al. 2019b, Archer et al. 2022) (Fig. 3C). These are highly diverse fungal groups mostly encountered in the air as spores rather than hyphae. The estimation of atmospheric viral diversity is an emerging topic with few studies to date: Across a range of urban, coastal and forest land use types, the most abundant DNA viruses recovered belonged to the ssDNA Gemini-,Circo-,Nano- and Microviridae (Whon et al. 2012). The study recovered lower abundance of dsDNA viruses but acknowledged that methodological limitations may have affected recovery for this group (Whon et al. 2012). In another study Geminiviruses associated with the phyllosphere and plant diseases were the most abundant group (García-Arenal and Zerbini 2019). A laboratory simulation of virus aerosolization from marine sea spray concluded that the Polydnoviridae and Alloherpesviridae viruses were enriched in aerosols (Michaud et al. 2018). The RNA viruses were not targeted in these ecological studies and so uncertainties remain about their distribution in free air.
Biogeographic regions for atmospheric microorganisms have been proposed based upon global air circulation patterns of latitudinally defined air cells (Fig. 2A) (Womack et al. 2010). This is supported by recent evidence for regionalization (Uetake et al. 2020) and biogeographic patterns in atmospheric bacterial and fungal diversity on a global scale (Archer et al. 2022). Furthermore, these studies point towards a highly complex biogeography that also reflects surface habitats and climatic factors. A high microbial species richness within a given air mass in the atmospheric boundary layer reflects recruitment from a variety of terrestrial and marine sources, and source tracking has indicated combined influence of local and distantly recruited taxa to observed diversity (Cáliz et al. 2018, Archer et al. 2022). Diversity estimates for near-ground air within the atmospheric boundary layer at various locales have indicated bacterial and fungal communities that were correlated with local abiotic variables such as temperature, precipitation and humidity or land use/surface cover, reflecting the role of local cell emissions (Bowers et al. 2011, Fröhlich-Nowoisky et al. 2012, Tignat-Perrier et al. 2019, Archer et al. 2022, Spring et al. 2021). Thus, diversity of fungi is relatively higher above regions with a well-developed phyllosphere (Lymperopoulou et al. 2016, Archer et al. 2022, Zhou et al. 2021), and lowest above oceans (Mayol et al. 2017, Archer et al. 2020, Uetake et al. 2020) and low productivity terrestrial systems such as drylands and polar/alpine regions where underlying surface communities are less diverse and microbially dominated by bacteria (Archer et al. 2019, 2022). Several studies have also related diversity to factors associated with dispersal such as history of air masses and UV radiation, suggesting combined influence of predominant remote sources and conditions during transit (Bowers et al. 2012, Cáliz et al. 2018, Archer et al. 2019, Uetake et al. 2019, Els et al. 2019b, 2020, Tignat-Perrier et al. 2020).
Temporal patterns in local atmospheric microbiota at a specific location reflect seasonal changes in underlying surface cover and the trajectory of incoming air masses. Inter-seasonal variation in the atmospheric boundary layer was found to be absent (Gusareva et al. 2019), weak (Tignat-Perrier et al. 2020), pronounced for some taxa (Bowers et al. 2012) or stochastic (Els et al. 2019b), depending on the location. Still, a large number of studies have identified differences in bacterial and/or fungal communities in summer versus winter at temperate and sub-tropical terrestrial locations (Bowers et al. 2012, 2013, Woo et al. 2013, Barberán et al. 2014, Cáliz et al. 2018, Uetake et al. 2019, Els et al. 2019b). Two of the scarce continuous long-term studies demonstrated a clear and well-delineated seasonal pattern in bacterial and fungal communities at near-ground locations (Woo et al. 2013) and above the atmospheric boundary layer (Cáliz et al. 2018). At polar latitudes elevated cell abundance in air corresponded to summer with snow-free terrestrial surfaces (Šantl-Temkiv et al. 2019). Patterns may also occur over shorter timeframes, with a pronounced diel variation of specific bacterial and fungal groups observed for the equatorial tropics (Gusareva et al. 2019). Together these studies strengthen the view that the underlying phyllosphere is a major determinant of atmospheric microbial diversity as they are related to patterns of plant ontogeny and growing season. As the elevation above ground increases, the source area of influence widens (Schmid 2002, Hsieh and Katul 2009), and the small-scale spatial and temporal heterogeneities observed near the ground blur (Archer et al. 2019, Els et al. 2019a, 2022). Above the atmospheric boundary layer long-range air movements progressively decouple atmospheric diversity from its emission sources (Burrows et al. 2009) (Smith et al. 2018).
Whilst most research has focused on the troposphere and specifically the atmospheric boundary layer, understanding the biogeographic limit of microbial occurrence and survival at higher altitudes is also of interest. It has been postulated that limited microbial survival and transport may occur in the stratosphere (DasSarma and DasSarma 2018), but the role of the stratosphere in microbial dispersal may be limited due to the combined effects of extreme environmental stress and extended residence times (Bryan et al. 2019). Several studies have employed balloon and aircraft platforms to recover stratospheric aerosol samples and they have described a limited diversity and viability of bacterial and fungal taxa (Wainwright et al. 2003, Smith et al. 2010, 2011, 2018, Bryan et al. 2019), with taxa not markedly differentiated from those occurring in the troposphere below (Smith et al. 2018). Even though estimates of abundance vary greatly a recent study suggested that bacterial abundance in the troposphere and lower stratosphere may be comparable (Bryan et al. 2019). However, there is currently insufficient evidence to draw robust conclusions about the occurrence, transport or viability of microorganisms in the stratosphere. The discipline of astrobiology has also considered the potential role of the stratosphere and upper atmosphere in the potential interplanetary dispersal of microorganisms as free cells or via mineral vectors (Yang et al. 2009) but this will require major investigative effort to be further considered.
What are the determinants of microbial flux between the atmosphere and surface habitats?
The atmosphere has long been identified as a major conduit for microbial transport between surface habitats, e.g. Womack et al. (2010), and this has a profound impact on the function and resilience of terrestrial and marine surface communities due to its contribution to gene flow, community assembly and dispersal of pathogens (Zhou and Ning 2017). Recent developments in ecological theory have begun to challenge the long-held view that microbial transport is a neutral process (Lowe and McPeek 2014), and recent evidence from large-scale biogeographic (Mayol et al. 2017, Archer et al. 2022) and temporal (Cáliz et al. 2018, Tignat-Perrier et al. 2020) studies of atmospheric microbiota increasingly support this view. Deterministic influence due to environmental filtering and microbial stress tolerance both play a significant role in shaping atmospheric microbiota (Archer et al. 2019, 2022), and hence the composition of assemblages that are exchanged between atmosphere and surface habitats.
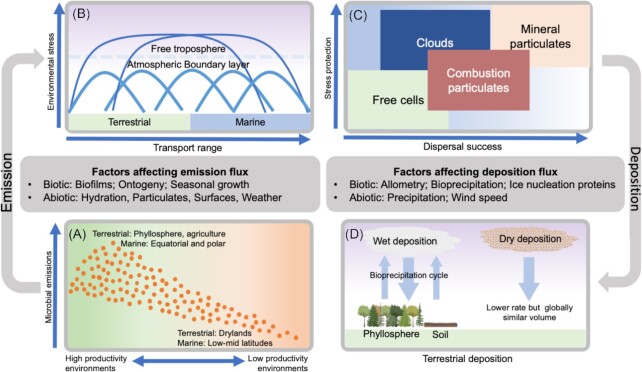
The major determinants of microbial fluxes in the atmosphere between source and sink habitats are emission, survival during transport and deposition (Fig. 4). Emission from the surface, known as aerosolization, is dependent upon the substrate and a major factor is the nature of the interface with air, such as the composition of plant cover (Zhou et al. 2021), cohesiveness of soil (Joung et al. 2017, Archer et al. 2022) or chemistry of the water-surface microlayer (Michaud et al. 2018) (Fig. 4A). Emissions from terrestrial and marine environments are higher when productivity of the underlying surface environment is greater (Burrows et al. 2009) (Fig. 4A). The processes affecting microbial emissions are highly variable and subject to stochastic influence from events such as dust storms (González-Toril et al. 2020) and wildfires (Moore et al. 2021). Biotic microbial traits that affect aerosolization are also important, including cell hydrophobicity and membrane composition (Burger and Bennett 1985, Michaud et al. 2018), extracellular polymeric substance (EPS) production (Morris and Monier 2003), spore production and release (Lagomarsino Oneto et al. 2020), and allometry (Norros et al. 2014). For viruses the possession of a hydrophobic envelope has been linked with a tendency to aerosolize and to desiccation tolerance (Michaud et al. 2018). Modelling emissions for particulates 1–3 µm diameter as a proxy for single and particle-associated bacterial cells revealed that the total annual emissions were in the range of 7.6 × 1023–3.5 × 1024 cells largely originating from terrestrial environments, i.e. crops, grasslands and shrubs (Burrows et al. 2009, 2013). This equated to an estimated annual emission of 470–1100 Gg bacterial biomass (Burrows et al. 2013). Fungal emissions occur predominantly as free spores and were estimated at 9.5 × 1023 total spores from terrestrial surfaces per year, which amounts to ∼50 Tg of fungal biomass annually (Elbert et al. 2007). The emission rate of viral particles from surface habitats is currently uncertain.
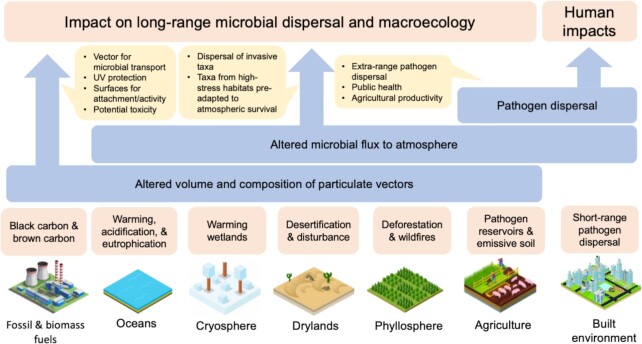
Figure 4.
Factors affecting atmospheric microbial transport and macroecological outcomes. (A) Emission and recruitment of microorganisms to the atmosphere are strongly dependent on the underlying surface habitats. This includes major delineations between terrestrial and marine locations, and a strong influence of productivity in underlying habitats. Biotic traits may also be important to emission variations between taxa. (B) Above terrestrial and marine surfaces at local scales transport occurs largely within the atmospheric boundary layer over relatively short distances. At larger spatial scales microbial transport involves transit above the boundary layer and this results in longer residence times and greater environmental filtering.(C) The association of cells with clouds or particulates directly impacts survival and transport. Combustion particulates may be less efficient vectors than mineral particles from desert dust and soil because they co-aerosolize with lower numbers of microorganisms, and they are associated with toxic combustion products. (D) Deposition of cells from the atmosphere occurs via wet deposition as rainfall, hail and snow, and via dry deposition that relies on sedimentation of cells due to gravity and occurs at lower velocities than wet deposition. Wet deposition over continents is typically induced by ice nucleation, which leads to preferential deposition of ice nucleation active cells and contributes to bioprecipitation and other feedbacks.
The major marine aerosolization process is initiated by breaking waves that produce small air bubbles that scavenge microorganisms and organic compounds from the sea column and transport them to the sea surface (Carlucci and Williams 1965, Aller et al. 2005), where they burst to eject small droplets containing salts, biogenic material and microorganisms into the atmosphere (Blanchard and Syzdek 1970, Blanchard 1989, Afeti and Resch 1990). Even though oceans are thought to represent weak sources of airborne microorganisms on a global per surface area basis, bioaerosol emissions include bacteria and viruses (Aller et al. 2005, Wilson et al. 2015, Mescioglu et al. 2019, Uetake et al. 2020) and selective transfer and enrichment of organic compounds in marine aerosols has been shown experimentally (Rastelli et al. 2017). In addition, large spatial and temporal variability in marine emissions and effects on atmospheric processes may occur due to marine biological processes and meteorological factors (Wang et al. 2015, Wilson et al. 2015, DeMott et al. 2016).
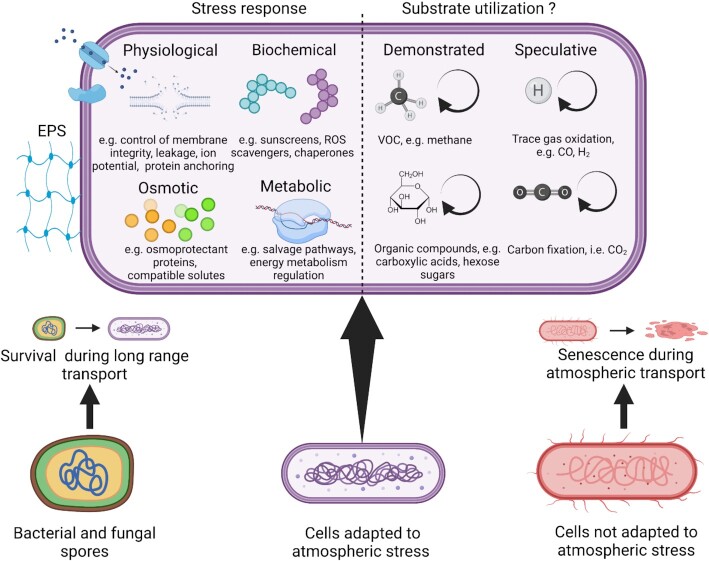
Microbial transport fluxes are affected by the altitude that the cells reach, which is a deterministic factor related to stress exposure (Fig. 4B). Survival during transit in air is influenced by biotic traits involved in stress tolerance, and particularly with regard to mitigating the effects of low water activity (Stevenson et al. 2015b), desiccation (Potts 1994), photo-oxidative UV (Daly 2009, Sharma et al. 2017), general oxidative stress (Ziegelhoffer and Donohue 2009, Ezraty et al. 2017) and low temperatures/freezing (Amato 2013) (Fig. 5). Dormancy and sporulation are strategies that provide protection against major stressors and functional metagenomics of air has indicated genes associated with these traits are over-represented in atmospheric communities compared with underlying surface habitats (Aalismail et al. 2019, Archer et al. 2022). Many spores as well as dormant and active cells possess pigment compounds such as melanins, mycosporines and scytonemins that protect against oxidative and desiccation stress largely due to scavenging of reactive species (Gao and Garcia-Pichel 2011). A number of biodiversity surveys in air have revealed taxa known to produce protective pigments are over-represented in air compared with underlying terrestrial and marine habitats (Fröhlich-Nowoisky et al. 2012, Tignat-Perrier et al. 2019, Archer et al. 2022), which implies a survival advantage. Additional strategies to counter stress may include the secretion of EPSs to mitigating both low water activity via hygroscopic carbohydrate polymers as well as provide extracellular protection from oxidative stress (Matulová et al. 2014). Aerosolization of cells as aggregates, biofilms or attached to particulates may also confer survival advantages during dispersal (Monier and Lindow 2003, Morris and Monier 2003) (Fig. 4), but may also reduce residence time in air due to increased size and density (Burrows et al. 2009).
Figure 5.
Stress response and potential substrate utilization by atmospheric microorganisms. Cells that possess adaptation to atmospheric environmental stressors are potentially capable of metabolic activity during atmospheric transport. A range of intracellular protective and repair strategies to reactive oxygen species (ROS) and other stressors may be complemented by extracellular protection due to EPSs that also confer protection against xeric, thermal and ultraviolet radiation (UVR) stress. Atmospheric bacteria oxidize volatile organic compounds (VOC) such as methane and a range of small organic compound carboxylic acids, pentose and hexose sugars, amino acids and phenolic compounds. Oxidation of other atmospheric trace gases and carbon fixation may also be potential pathways for activity in the atmosphere although supporting data are currently lacking.
Metagenomic profiling of atmospheric communities has begun to reveal prevalence of genes encoding cold shock, oxidative stress and UV repair enzymes (Aalismail et al. 2019, Archer et al. 2022), and metatranscriptomics of cloud water has revealed active metabolic responses to counter oxidative, osmotic and thermal stresses (Amato et al. 2019). Physiological responses against simulated atmospheric stress have also been observed for taxa isolated from the atmosphere. Strains of Pseudomonas syringae isolated from rain and cloud water withstood aerosolization, freezing, osmotic shock, short-term exposure to UVA and UVB radiation (Alsved et al. 2018, de Araujo et al. 2019, Ling et al. 2021), and oxidative stress imposed by hydrogen peroxide (H2O2) (Wirgot et al. 2019). Metabolomic studies on P. syringae also revealed a suite of responses to cold shock under simulated atmospheric conditions (Jousse et al. 2017), including production of cryoprotectants, antioxidants, alkaloids and metabolites involved in energy metabolism. The persistence of viruses in aerosols and traits that facilitate survival is not well constrained at this time (Leung 2021). However, a laboratory simulation has shown that a number of ssDNA, dsDNA and RNA viruses can remain viable in an aerosolized form across a range of temperature, humidity and UV exposures (Verreault et al. 2015). The infectivity of airborne viruses towards microorganisms suspended in the atmosphere is unknown and further investigation is required to determine if such interactions may impact survival during transport.
A very large fraction (97%) of depositing bacterial cells have been found to be particle associated (Reche et al. 2018), and with a large mean diameter (∼9 μm) (Woo and Yamamoto 2020), thus supporting the idea that the mixing state of cells in air, i.e. free cells, aggregates or cells vectored on particulate matter, has a significant effect on cell residence time (Fig. 4C). The deposition of microorganisms occurs through a combination of wet and dry deposition (Després et al. 2012). The wet deposition is affected by a combination of in-cloud partitioning during cloud droplet/ice particle formation and below-cloud aerosol scavenging processes, which can both contribute to segregation of taxa between the wet and dry phases (Bauer et al. 2003, Moore et al. 2020, Woo and Yamamoto 2020). In particular, cloud condensation ability was modelled to shorten the average bacterial residence time in air (and hence dispersal distance) from ∼8 to 3 days (Burrows et al. 2009) and ice nucleating microbial cells were shown to be efficiently precipitated (Amato et al. 2015, Stopelli et al. 2015). Wet deposition rates of 107–5 × 109 bacteria cells m−2 d–1 have been reported that exceed dry deposition (Reche et al. 2018, Woo and Yamamoto 2020). Dry deposition velocity is largely determined by allometric considerations, such as biological particle size, shape and density, as well as meteorology (Fig. 4D). Globally the volume of wet and dry deposition may be similar due to the frequency of precipitation events versus dry periods (Reche et al. 2018, Woo and Yamamoto 2020). Patterns in deposition of fungal spores and pollen are thought to follow a similar pattern although taxon-specific variation is likely due to spore characteristics (Aylor 2017, Woo and Yamamoto 2020). Deposition of viruses from the free troposphere has been estimated at 0.26 × 109 to >7 × 109 viral particles/day and was positively correlated with fine organic aerosols (<0.7 μm) but displayed no significant variation between wet and dry deposition (Reche et al. 2018).
A pertinent question for atmospheric microbial communities is the extent to which the complex variation in emission and deposition fluxes, together with environmental filtering during transit, influence the assembly of terrestrial (including freshwater) and marine surface communities. The flux of biomass containing diverse taxa from air to land or oceans is likely smaller by magnitudes than that of standing communities but given the ability of microorganisms to rapidly exploit favourable ecological niches, atmospheric flux should be viewed as a critical control point in aspects of connectivity and ecological resilience across biomes (Dumbrell et al. 2010, Lowe and McPeek 2014, Zhou and Ning 2017). This occurs by facilitating microbial dispersal between spatially separated natural systems (Kellogg and Griffin 2006), contributing to maintenance of diversity via recruitment (Jones and Lennon 2010), promoting dispersal of dormant propagules that may act as microbial seed banks (Lennon and Jones 2011) and dispersal of phages that exert critical control on microbial population turnover (Sandra et al. 2004).
What is the evidence for long distance atmospheric pathogen dispersal?
The person to person transmission of pathogens over short distances in air is well documented and covered in excellent recent reviews (Kutter et al. 2018, Leung 2021). Human respiratory pathogens and particularly viruses are transmitted by a variety of means that include respiratory droplets (≥0.1 mm diameter) that are rapidly deposited from air, and respiratory aerosols (≤0.1 mm diameter) that may remain suspended for longer periods and thus disperse over longer distances up to several metres (Leung 2021). Current knowledge on the extent and relative importance of this short-range person to person aerosol transmission is currently not well constrained, e.g. Kutter et al. (2018) and Leung (2021). Airborne genetic signatures of human bacterial and viral pathogens have also been detected in aerosols of confined built environments with point source emissions such as livestock facilities, e.g. Nehme et al. (2008) and Zhao et al. (2014), and indoor wastewater treatment plants, e.g. Yang et al. (2019). Such bioaerosols appear to be highly localized and evidence for significant transport in free atmospheric air beyond point sources is lacking. These scenarios are significant for public health but there is currently a lack of compelling evidence that they pose risk associated with long-range atmospheric transport.
Long-range atmospheric dispersal of pathogens (i.e. over hundreds to thousands of kilometres) typically requires protection from atmospheric environmental stress as evidenced by the taxonomy and physiological state of recovered taxa, e.g. Mayol et al. (2017), Cáliz et al. (2018) and Šantl-Temkiv et al. (2018). Despite this, some evidence exists for regional dispersal of pathogens up to hundreds of kilometres without any obvious adaptation to environmental stress during atmospheric transport, e.g. foot and mouth diseases virus (Sorensen et al. 2000, Björnham et al. 2020). More typically long-range pathogen dispersal involves preadaptation due to the formation of spores or other resting structures (Fröhlich-Nowoisky et al. 2009), possession of stress tolerance traits such as oxidative stress avoidance and repair pathways (Archer et al. 2022), and/or association with particulate vectors (Maki et al. 2019). Vectoring on biological surfaces also provides a protected means of long-distance dispersal: Wind-borne transport of Anopheles mosquitoes carrying the Plasmodium parasite was recorded over several hundred kilometres in the Sahel region of Africa (Huestis et al. 2019), and pollen has been demonstrated as a vector for bacteria and bacterial allergens (Oteros et al. 2019). Correlations have been made between regional trans-continental scale atmospheric dust vectoring and pathogen dispersal (Griffin 2007, Tobias et al. 2019). Consistent inter-annual dynamics for specific taxa may exist that make them foreseeable over time with dominance of plant pathogens over those of animals and humans (Triadó-Margarit et al. 2022).
Fungal spores are particularly well suited to long-range dispersal and are problematic as a threat to agriculture and global food security (Brown and Hovmøller 2002, Savary et al. 2019). Many plant pathogens have historically displayed distinct regionalization but globalization of agriculture has exacerbated range shifts (Bebber et al. 2014). Airborne fungal pathogens have been detected above land with agricultural land use (Nicolaisen et al. 2017, Archer et al. 2022), and some fungal pathogens are able to disperse across inter-continental distances and reestablish infections even when host plants are seasonally absent (Brown and Hovmøller 2002). A striking example is the reemergence of wheat stem rust as a threat to global food supply. The disease caused by the basidiomycete fungus Puccinia graminis f. sp. tritici is dispersed readily in air by basidiospores. It was virtually eradicated in the twentieth century but a highly pathogenic new strain, Ug99 and its variants, emerged in the last decade and have threatened to devastate global wheat cultivation (Singh et al. 2011). Combination of disease surveillance and dispersal modelling has resulted in strong evidence for atmospheric transmission as a major determinant of wheat rust epidemiological zones (Meyer et al. 2017).
Evidence for long-range atmospheric dispersal of human pathogens also exists. Seasonal outbreaks of meningococcal meningitis in sub-Saharan Africa have been correlated with bacterial vectoring on dust (Jusot et al. 2017), and this also appears to occur for fungal diseases such as Valley Fever caused by Coccidioides spores transmitted on desert dust across the southwestern United States and northern Mexico (Kollath et al. 2019), and across the Atlantic Ocean to Europe (Triadó-Margarit et al. 2022). In addition to human pathogens, atmospheric transport of microbially derived allergens (Woo et al. 2013) and antibiotic resistance genes (Pal et al. 2016, Li et al. 2018, Caliz et al. 2022) may pose health risks to human, agricultural and natural populations (Bai et al. 2022). Atmospheric microbiology is therefore relevant to the One Health conceptual framework that seeks to highlight how the wellbeing of humans, animals, plants and the environment are inter-connected (CDC 2020). Emission, transport and deposition of microorganisms via the atmosphere at various spatial scales are central to some relationships identified by this framework. It is envisaged as important to improved understanding of the transboundary aspect of how pathogens, antibiotic resistance and invasive taxa traverse regions and exert negative outcomes.
What is the evidence for metabolically active atmospheric microorganisms?
The majority of cells in the atmosphere are unlikely to be metabolically active but are simply undergoing transport between source and sink destinations with a high turnover (Burrows et al. 2009). Given that most fungi in air occur as dormant spores it is likely that the active fraction of atmospheric microbiota is comprised almost exclusively of bacteria that possess adaptation to atmospheric environmental stress (Archer et al. 2022) (Fig. 5). Putative generation times for bacteria in cloud water have been estimated at 3.6–19.5 days (Sattler et al. 2001), which fits within their modelled airborne residence times (Burrows et al. 2009). However, whether metabolic transformations in situ under the highly fluctuating atmospheric conditions are sufficient to sustain only quasi-dormancy, or if cell homeostasis and reproduction are energetically feasible within the short timeframe of favourable conditions during atmospheric transport remains unexplored.
Evidence from environmental samples has demonstrated that atmospheric microorganisms possess the potential to transform substrates encountered in the atmosphere (Fig. 5). Molecular ecological surveys of cloud water (Amato et al. 2019), free air (Gusareva et al. 2019, Archer et al. 2022) and airborne desert dust plumes (Aalismail et al. 2019) indicate diverse autotrophic and heterotrophic metabolic pathways in atmospheric assemblages that broadly reflect source environments. Photosynthesis may be limited in atmospheric microorganisms due to light inhibition (Barber and Andersson 1992), but clouds and desert dust provide shading against damaging levels of direct UV radiation, and diverse viable and stress-tolerant microalgae and cyanobacteria have been reported from clouds (Tesson and Šantl-Temkiv 2018). A range of pathways indicating atmospheric trace gas metabolism including hydrogen, methane and carbon monoxide were widespread among atmospheric metagenomes (Archer et al. 2022) and methane oxidation at atmospheric concentrations has been demonstrated by Methylocystis and Methylosinus enriched from the atmosphere (Šantl-Temkiv et al. 2013b), although it is unclear if this is energetically feasible under atmospheric conditions. Most research, however, has focused on the potential for heterotrophic metabolism of simple organic compounds in clouds as these are envisaged as ‘hotspots’ for atmospheric microbial activity.
Microbial assemblages in cloud water have revealed general markers of metabolic activity under simulated atmospheric conditions, including ATP, high ribosomal content, transcribed RNA, and enzymatic and respiratory activities (Klein et al. 2016, Šantl-Temkiv et al. 2018, Amato et al. 2019). Under conditions of liquid water and substrate availability, favourable temperature and reduced stress exposure, atmospheric microorganisms appear capable of active heterotrophic metabolism as evidenced by chemical speciation in recovered cloud water (Vaitilingom et al. 2013, Bianco et al. 2019). A comparative metagenomic and metatranscriptomic study of cloud water assemblages highlighted that active metabolism in clouds is closely coupled with satisfying the energetic demands of active stress response metabolism and particularly with regard to mitigating oxidative stress (Amato et al. 2019). A complex metabolism was envisaged that also included synthesis of cryoprotectant and EPSs implicated in a protective role. The study identified C1 and C2 compounds as important substrates and ammonium as the major nitrogen source (Amato et al. 2019). Another combined metatranscriptomic and metabolic study of cloud water provided strong evidence for phenol metabolism by bacteria, and this was envisaged as a possible atmospheric substrate derived from anthropogenic emissions (Lallement et al. 2018).
Several studies have demonstrated the ability of atmospheric isolates to metabolize compounds encountered in clouds: A screening of 60 atmospheric isolates including the bacterial genera Bacillus, Pseudomonas, Micrococcus and Sphingomonas as well as fungal yeasts from cloud water revealed that under laboratory conditions all were capable of degrading diverse carboxylic acids and volatile organic compounds (Amato et al. 2007). Atmospheric bacterial isolates of Bacillus sp., Frigoribacterium sp., Pseudomonas sp. and the methanotrophic genera Methylocystis sp. and Methylosinus sp. have been demonstrated to oxidize common atmospheric volatile organic compounds including formaldehyde and methanol (Husárová et al. 2011, Šantl-Temkiv et al. 2013b). Rhodococcus enclensis isolated from cloud water was shown to metabolize catechol and phenol under simulated atmospheric conditions at rates comparable to that of chemical transformations in the atmosphere (Jaber et al. 2020). Mono- and disaccharides may also serve as substrates, as demonstrated for a Bacillus strain isolated from cloud water (Matulová et al. 2014).
Based on bulk cloud water measurements, which were unable to account for the multiphase structure and highly dynamic nature of clouds, it has been proposed that heterotrophic microbial activity could be the main driver for the oxidation of common atmospheric organic compounds in air during night and could compete with photochemistry during the day (Vaïtilingom et al. 2010). Modelling has indicated that given the small fraction (∼10–4) of spatially separated microdroplets that contain microbial cells, the quantitative contribution of microbial activity to the transformation of atmospheric compounds is likely to be low (Fankhauser et al. 2019), although this needs to be verified with experimental approaches simulating in situ atmospheric conditions. More detailed modelling, accounting for the in situ cloud structure, has suggested that even though the biodegradation of highly water-soluble organic gasses, such as dicarboxylic acids, may have been overestimated, the biodegradation of organic gases with intermediate solubility, such as acetate and formate, is more efficient and may represent a significant sink for these compounds (Khaled et al. 2021).
What is the microbial role in atmospheric biophysical processes?
Microbial involvement in biophysical transformations occurs independently from the metabolic activity that is thought to be rare in the atmosphere. Biological ice nucleation contributes to cloud formation and precipitation, and more generally to climate and the hydrological cycle. It also impacts microbial flux between atmospheric and surface habitats, and recent advances have shed new insight on the mechanistic basis for this. Diverse microorganisms and biomolecules, including bacteria (particularly Gammaproteobacteria) (Joly et al. 2013), fungi (Fröhlich-Nowoisky et al. 2014), microalgae (Tesson and Šantl-Temkiv 2018) and microbially derived cell-free ice nucleating proteins (INpro) (O′Sullivan et al. 2015) can nucleate ice in super-cooled cloud droplets at temperatures much warmer than other atmospheric particles (between −13 and −1°C) and have been implicated in cloud processes. This property has been linked with specific surface- or excreted proteins and is essentially a biophysical process that occurs independently of cell viability or association (Fig. 6). Whilst terrestrial sources are dominant on a global level, marine sources associated with INpro excreted by marine microalgae (Wilson et al. 2015) may be important in remote regions unaffected by continents (e.g. Southern Ocean) (DeMott et al. 2016).
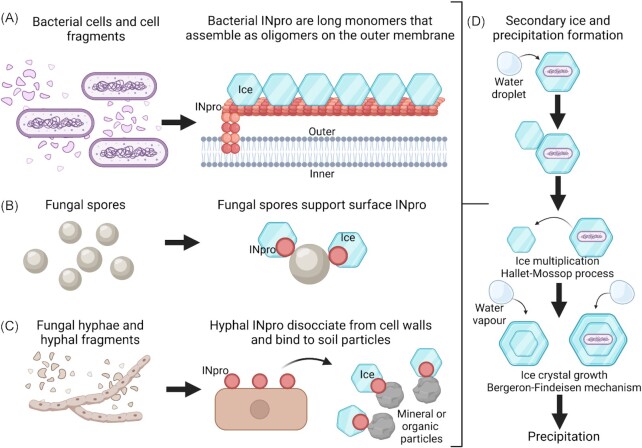
Figure 6.
Biophysical role of atmospheric microorganisms in ice nucleation. An important biophysical interaction of atmospheric microorganisms with cloud formation is ice nucleation by specific microbial proteins that contributes to precipitation and the hydrological cycle, as well as impacting microbial deposition from the atmosphere to surface habitats. (A)Bacterial ice nucleating proteins (INpro) are long repetitive proteins that are anchored on the surface of the outer membrane and nucleate ice through a yet to be fully resolved mechanism. They may be cell associated or retain activity in cell fragments. Monomers or small oligomers nucleate ice at temperatures from −7 to −10°C and higher order oligomers of INpro form at well-defined sizes that nucleate ice between −2 and −5°C. Fungal spores and hyphae are associated with INpro that exhibit different molecular structure and properties than those in bacteria, but they are yet to be fully described. (B) Fungal spores are the dominant state for fungi in the atmosphere and support surface-associated INpro and ice nucleating activity. (C) Fungal hyphae occur very rarely in the atmosphere and so the major contribution arises from fungal INpro that are easily dissociated from cell walls and adhere to soil particulates that subsequently become aerosolized. (D) Low concentrations of atmospheric INpro nucleate cloud ice and may induce subsequent secondary ice formation via the Hallet–Mossop process. This process involves freezing of supercooled cloud droplets that encounter primary ice particles (riming) and subsequent ejection of frozen droplets that form independent ice particles (splintering). Primary and secondary ice particles form precipitation through the Bergeron–Findeisen mechanism where ice growth occurs at the expense of surrounding water droplets. Images in panels (A–D) are not to scale: in panels (A–C) hexagonal shapes represent ice embryos forming on INpro and in panel (D) hexagonal shapes represent ice particles in clouds.
While ice nucleating bacteria are genetically diverse and have recently been proposed to employ different mechanisms for ice nucleation (Failor et al. 2017), among identified biological ice nucleation active molecules, the best understood are bacterial INpro. Recent advances have shed new insight on the mechanistic basis of their nucleation. These are large repetitive membrane-associated proteins (>120 kDa), which nucleate ice through a yet to be fully resolved mechanism (Pandey et al. 2016, Hudait et al. 2018, Lukas et al. 2020). They were recently found to structure interfacial water molecules at low temperature promoting ice formation (Roeters et al. 2021) (Fig. 6). Monomers or small oligomers nucleate ice at temperatures from −7 to −10°C and are known as Type C INpro; and higher order oligomers of INpro nucleate ice between −2 and −5°C and are known as Type A INpro (Wex et al. 2015, Ling et al. 2018). Laboratory studies using cloud simulation chambers have shown that the activity of bacterial INpro is independent of cell viability (Hartmann et al. 2013, Amato et al. 2015). Fungal spores and hyphae also carry INpro that exhibit different molecular structure and properties to those in bacteria and they are yet to be fully characterized. Fungal spores are the dominant state for fungi in the atmosphere and support surface-associated INpro and ice nucleating activity (Pouleur et al. 1992, Morris et al. 2013) (Fig. 6). Fungal hyphae occur very rarely in the atmosphere and so the major contribution arises from dissociation of fungal INpro from cell walls and adhesion to soil particulates that subsequently become aerosolized (O′Sullivan et al. 2015) (Fig. 6). The ubiquitous soil fungal genera Fusarium and Mortierella have been demonstrated to excrete INpro into the environment and soil particles with associated INpro were capable of ice nucleation (Fröhlich-Nowoisky et al. 2014, O′Sullivan et al. 2015).
Once they induce freezing, microbial INpro can also contribute to the formation of secondary ice particles in clouds that enhances the impact of low abundance biological INPs through ice multiplication, particularly at high sub-zero temperatures (Korolev and Leisner 2020) (Fig. 6). Rain that is produced via the ice phase through the Hallett–Mossop process strongly dominates over continents and is common over mid-latitude oceans (Mülmenstädt et al. 2015). This process occurs in mixed phase clouds, where some of the supercooled cloud droplets freeze due to the presence of ice nucleating particles. Water vapour is preferentially deposited on thus-formed ice particles and allows them to grow to precipitation sizes whilst being replenished by evaporating cloud droplets (Fig. 6). Ice nucleation active bacteria that are aerosolized from the phyllosphere were suggested to drive the bioprecipitation cycle by inducing rain formation. Rain enhances the growth of vegetation and epiphytic microorganisms, which in turn leads to enhanced emissions of ice nucleation active bacteria resulting in a positive feedback (Fig. 4D) (Morris et al. 2014). Bioprecipitation may be important above regions with a well-developed phyllosphere where emissions of bacterial INpro are highest, and accompanied by relatively low levels of abiotic INPs (Huffman et al. 2013, Healy et al. 2014). Other precipitation-related feedbacks that increase bioaerosol generation and thus enhance the atmospheric INpro include immediate and delayed interactions of rain with soil and plants (Huffman et al. 2013, Joung et al. 2017). These positive feedbacks occur alongside nonbiological ice nucleation and precipitation and the significance of these processes for meteorology and climate is yet to be quantified.
How might anthropogenic activities affect atmospheric microbiota?
The atmosphere is a primary sink for anthropogenic emissions of greenhouse gases, chemical pollutants and particulates from fossil fuel combustion (Archer and Pointing 2020). Additional particulate emissions occur as a result of biomass burning due to land clearance and wildfires, soil destabilization due to land use change and expansion of intensive agriculture, and climate change induced desertification (Archer and Pointing 2020). Most of these emissions are concentrated in the troposphere where long-range microbial transport, metabolism and biophysical transformations occur. Atmospheric forcing due to emissions has been acute in the industrial age and this has the potential to create positive and negative impacts on the dispersal outcomes for microorganisms transported in the atmosphere (Archer and Pointing 2020) (Fig. 7).
Figure 7.
Potential anthropogenic impacts on atmospheric microbiology. Direct and indirect anthropogenic forcing of the atmosphere will potentially impact the emission and deposition flux, atmospheric burden, biodiversity, biophysical and metabolic activity of microorganisms in the atmosphere. We identify three main outcomes of anthropogenic forcing that are of potential concern, although these are likely inter-linked and overlap. They are altered atmospheric transport arising from changes to particulate matter (PM) burden in the atmosphere; shifts in atmospheric microbial emission and deposition flux due to change in surface microbial communities; and impacts on human systems due to agricultural and human pathogen dispersal. We advocate for future research focus to address these impacts.
Gaseous emissions from fossil fuel combustion including SOx and NOx have been shown to inhibit cell function and survival of Pseudomonas strains isolated from clouds (Kondakova et al. 2016). Greenhouse gaseous, e.g. methane (Šantl-Temkiv et al. 2013b), can be metabolized by atmospheric bacteria. However, as in situ microbial activity and its influence on atmospheric chemistry is still not well constrained, it also remains unclear if elevated substrate levels in the atmosphere have the potential to significantly affect microbial activity and its outcomes (Archer and Pointing 2020). The ice nucleation activity of bioaerosols may also become more important to precipitation patterns in a warming world because they are more efficient at warmer temperatures from −10°C to 0°C, compared with mineral particulates (Christner et al. 2008a), and INpro emissions are likely enhanced by biomass burning and intensive agriculture (Moore et al. 2021). The potential impacts from direct and indirect anthropogenic particulate emissions are more apparent and widespread. Anthropogenic particulates including brown carbon from biomass combustion and black carbon from fossil fuel combustion account for ∼20% of global emissions (Chen et al. 2018). Mineral dust and soil emissions due to climate change impact on desertification (Pointing and Belnap 2014) and agricultural emissions (Salawu-Rotimi et al. 2021). Elevated concentrations of anthropogenic and desert dust particulates have been shown to influence atmospheric microbial diversity (Maki et al. 2019), and this may have implications for dispersal outcomes at deposition sites. The transport of microorganisms in plumes of airborne particulates is envisaged to provide additional protection from UV damage during atmospheric transport (González-Toril et al. 2020). Particulates arising from combustion are more hygroscopic than natural mineral particles and whilst this may be viewed as potentially facilitating greater survival and microbial activity, it must be balanced against the often toxic levels of PAH associated with these particulates (Hayakawa et al. 2018). Anthropogenic forcing through particulate emissions may also affect ice nucleation patterns in clouds and thus impact microbial flux and deposition in the atmosphere more generally through changes in precipitation.
A recent modelling study suggests that overall microbial diversity in the atmosphere may decline under future climate scenarios and this may severely impact ecosystem connectivity and function (Ontiveros et al. 2021). Conversely, the magnitude of emissions may increase due to the effects of climate-change forcing or pollution, e.g. due to marine phytoplankton blooms (Lewis et al. 2020) or desertification (Pointing and Belnap 2014). Strengthened emissions from point sources are anticipated for pathogenic microorganisms, antibiotic resistance genes and allergens, e.g. wastewater treatment plants (Han et al. 2020), livestock facilities (Zhao et al. 2014) and urban centres (Brodie et al. 2007, Woo et al. 2013). These emissions have direct public health relevance although dispersal appears to be localized. However, recent long-term studies of microbiota in rain and snow deposited at high altitude above the atmospheric boundary layer identified up to 3% of the total airborne microbiota as potential pathogens (Triadó-Margarit et al. 2022) and observed long-range transport of antibiotic resistance genes indicative of diffuse distribution related to agricultural sources (Caliz et al. 2022). Assessing the extent of anthropogenic impacts remains somewhat speculative as more dedicated studies are needed both to provide a better constrained understanding of current processes and to investigate impacts of highly complex anthropogenic changes to the atmospheric microbiota.
Concluding remarks
The discipline of aeromicrobiology has benefitted enormously from recent advances in methodology that overcome the limitations associated with the study of ultra-low biomass habitats, and we view this is beneficial in a wider sense to improved ecological understanding of how the atmosphere links to surface terrestrial and marine biomes. Specifically, the ability to accurately estimate microbial diversity in air through rigorous decontamination of environmental sequence data has revealed that the atmospheric boundary layer supports a highly diverse but nonrandomly assembled microbiota. Atmospheric microbial assemblages are taxonomically structured reflecting strong environmental filtering, as well as distinct spatio-temporal biogeographic signals indicating a complex interplay of local and regional recruitment to the atmosphere. The long-held ecological view of atmospheric transport as neutral to dispersal outcomes is therefore no longer valid. Thus, the assumptions about how atmospheric microbial transport impacts community assembly and biogeography need to be revised. Knowledge gaps in this regard include high-quality quantitative data on fluxes between surface environments and the atmosphere, as well as improved understanding of postdepositional processes. Methodological improvements that directly address the problem of contamination in ultra-low biomass air samples from higher altitudes will also help resolve the vertical distribution of microorganisms in the atmosphere.
In the absence of evidence for widespread metabolic activity that influences biogeochemical outcomes, lack of microbial interactions and food webs or evidence for cell proliferation, we conclude that microbial biochemical transformations make a very limited contribution to overall atmospheric chemistry although they may be important in localized habitable areas such as clouds or plumes of hydrated particles. The notion of a metabolically active atmospheric microbiome must therefore be contemplated with a high degree of caution. The molecular mechanisms underpinning biogenic ice nucleation are becoming better resolved for bacterial INpro but are still lacking for other microbial groups. Given that the temperature range at which biogenic ice nucleation occurs is relevant in the context of a warming climate, we urge greater focus on microbial ice nucleation to predict its future impacts on the water cycle. Development of improved laboratory simulations for atmospheric conditions and in situ measurements will assist with further understanding of atmospheric microbial metabolic and biophysical activity and its boundaries in air.
In a broader sense, estimates of the pan-global microbiota and its ecological relevance require greater consideration of biomass in the gas-phase atmospheric microbial environment alongside solid-phase terrestrial and liquid-phase aquatic habitats. The increasing threats to environmental, organismal and human health due to changes in atmospheric transport of microorganisms because of anthropogenic forcing will be more clearly understood when empirical considerations of the atmospheric microbiota are incorporated into analytical frameworks, including the One Health and Planetary Health concepts. We therefore urge expansion of atmospheric microbiological monitoring and inclusion as part of global-scale atmospheric data networks.
Contributor Information
Tina Šantl-Temkiv, Department of Biology, Aarhus University, DK-8000 Aarhus, Denmark; Stellar Astrophysics Centre, Department of Physics and Astronomy, Aarhus University, DK-8000 Aarhus, Denmark.
Pierre Amato, Institut de Chimie de Clermont-Ferrand, SIGMA Clermont, CNRS, Université Clermont Auvergne, 63178, Clermont-Ferrand, France.
Emilio O Casamayor, Centre for Advanced Studies of Blanes, Spanish Council for Research (CSIC), 17300, Blanes, Spain.
Patrick K H Lee, School of Energy and Environment, City University of Hong Kong, Hong Kong, China; State Key Laboratory of Marine Pollution, City University of Hong Kong, Hong Kong, China.
Stephen B Pointing, Yale-NUS College, National University of Singapore, Singapore 138527; Department of Biological Sciences, National University of Singapore, Singapore 117558.
Funding
This work was supported by the following research grants: TS-T was supported by the Danish National Research Foundation [grant number DNRF106], the Aarhus University Research Foundation Nova programme [grant number AUFF-E-2015-FLS-9-10], the Villum Foundation [grant numbers 23175 and 37435], the NOVO Interdisciplinary Synergy Programme [grant number NNF19OC0056963] and the Independent Research Fund Denmark [grant number 9145-00001B]. PA was supported by the French National Research Agency Make Our Planet Great Again (MOPGA) programme [grant number ANR-17-MOPGA-0013]. EOC was supported by the Spanish State Research Agency [grant number AEI- MICINN] and European Regional Development Fund [grant number INTERACTOMA RTI2018-101205-B-I00]. PKHL was supported by the Environment Conservation Fund of the Government of Hong Kong [grant number 2019–12]. SBP was supported by the Ministry of Education - Singapore and Yale-NUS College [grant number R-607-265-331-121].
Conflict of interest
The authors declare no conflict of interest.
References
- Aalismail NA, Ngugi DK, Díaz-Rúa Ret al. . Functional metagenomic analysis of dust-associated microbiomes above the Red Sea. Sci Rep. 2019;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afeti GM, Resch FJ. Distribution of the liquid aerosol produced from bursting bubbles in sea and distilled water. Tellus B Chem Phys Meteorol. 1990;42:378–84. [Google Scholar]
- Aller JY, Kuznetsova MR, Jahns CJet al. . The sea surface microlayer as a source of viral and bacterial enrichment in marine aerosols. J Aerosol Sci. 2005;36:801–12. [Google Scholar]
- Alsved M, Holm S, Christiansen Set al. . Effect of aerosolization and drying on the viability of Pseudomonas syringae cells. Front Microbiol. 2018;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato P, Besaury L, Joly Met al. . Metatranscriptomic exploration of microbial functioning in clouds. Sci Rep. 2019;9:4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato P, Christner BC. Energy metabolism response to low-temperature and frozen conditions in Psychrobacter cryohalolentis. Appl Environ Microbiol. 2009;75:711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato P, Demeer F, Melaouhi Aet al. . A fate for organic acids, formaldehyde and methanol in cloud water: their biotransformation by micro-organisms. Atmos Chem Phys. 2007;7:4159–69. [Google Scholar]
- Amato P, Joly M, Besaury Let al. . Active microorganisms thrive among extremely diverse communities in cloud water. PLoS One. 2017;12:e0182869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato P, Joly M, Schaupp Cet al. . Survival and ice nucleation activity of bacteria as aerosols in a cloud simulation chamber. Atmos Chem Phys. 2015;15:6455–65. [Google Scholar]
- Amato P. Energy metabolism in low temperature and frozen conditions. In: Yumoto I (ed). Cold-Adapted Microorganisms. 1st edn.Poole: Caister Academic Press, 2013. [Google Scholar]
- Archer S, Lee K, Caruso Tet al. . Global biogeography of atmospheric microorganisms reflects diverse recruitment and environmental filtering. Res Sq. 2022. https://www.researchsquare.com/article/rs-244923/v4(14 February 2022, date last accessed). [Google Scholar]
- Archer SDJ, Lee KC, Caruso Tet al. . Air mass source determines airborne microbial diversity at the ocean–atmosphere interface of the Great Barrier Reef marine ecosystem. ISME J. 2020;14:871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SDJ, Lee KC, Caruso Tet al. . Airborne microbial transport limitation to isolated antarctic soil habitats. Nat Microbiol. 2019;4:925–32. [DOI] [PubMed] [Google Scholar]
- Archer SDJJ, Pointing SB. Anthropogenic impact on the atmospheric microbiome. Nat Microbiol. 2020;5:229–31. [DOI] [PubMed] [Google Scholar]
- Aylor D. Aerial Dispersal of Pollen and Spores. St Paul: APS Press, 2017. [Google Scholar]
- Bai H, He L-Y, Wu D-Let al. . Spread of airborne antibiotic resistance from animal farms to the environment: dispersal pattern and exposure risk. Environ Int. 2022;158:106927. [DOI] [PubMed] [Google Scholar]
- Bar-On YM, Phillips R, Milo R. The biomass distribution on earth. Proc Natl Acad Sci USA. 2018;115:6506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber J, Andersson B. Too much of a good thing: light can be bad for photosynthesis. Trends Biochem Sci. 1992;17:61–6. [DOI] [PubMed] [Google Scholar]
- Barberan A, Casamayor EO, Fierer N. The microbial contribution to macroecology. Front Microbiol. 2014;5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberán A, Henley J, Fierer Net al. . Structure, inter-annual recurrence, and global-scale connectivity of airborne microbial communities. Sci Total Environ. 2014;487:187–95. [DOI] [PubMed] [Google Scholar]
- Barry RG, Chorley RJ. Atmosphere, Weather and Climate. 9th edn. New York: Routledge, 2010. [Google Scholar]
- Bauer H, Giebl H, Hitzenberger Ret al. . Airborne bacteria as cloud condensation nuclei. J Geophys Res Atmos. 2003;108:4658. [Google Scholar]
- Bebber DP, Holmes T, Gurr SJ. The global spread of crop pests and pathogens. Glob Ecol Biogeogr. 2014;23:1398–407. [Google Scholar]
- Bianco A, Deguillaume L, Chaumerliac Net al. . Effect of endogenous microbiota on the molecular composition of cloud water: a study by Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR MS). Sci Rep. 2019;9:7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björnham O, Sigg R, Burman J. Multilevel model for airborne transmission of foot-and-mouth disease applied to Swedish livestock. PLoS One. 2020;15:1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard DC, Syzdek L. Mechanism for the water-to-air transfer and concentration of bacteria. Science (80-). 1970;170:626–8. [DOI] [PubMed] [Google Scholar]
- Blanchard DC. The ejection of drops from the sea and their enrichment with bacteria and other materials: a review. Estuaries. 1989;12:127. [Google Scholar]
- Borrelli P, Lugato E, Montanarella Let al. . A new assessment of soil loss due to wind erosion in European agricultural soils using a quantitative spatially distributed modelling approach. L Degrad Dev. 2017;28:335–44. [Google Scholar]
- Bowers RM, Clements N, Emerson JBet al. . Seasonal variability in bacterial and fungal diversity of the near-surface atmosphere. Environ Sci Technol. 2013;47:12097–106. [DOI] [PubMed] [Google Scholar]
- Bowers RM, McCubbin IB, Hallar AGet al. . Seasonal variability in airborne bacterial communities at a high-elevation site. Atmos Environ. 2012;50:41–9. [Google Scholar]
- Bowers RM, McLetchie S, Knight Ret al. . Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J. 2011;5:601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie EL, DeSantis TZ, Moberg Parker JPet al. . Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci USA. 2007;104:299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JKM, Hovmøller MS. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science (80-). 2002;297:537–41. [DOI] [PubMed] [Google Scholar]
- Bryan NC, Christner BC, Guzik TGet al. . Abundance and survival of microbial aerosols in the troposphere and stratosphere. ISME J. 2019;13:2789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger SR, Bennett JW. Droplet enrichment factors of pigmented and nonpigmented Serratia marcescens: possible selective function for prodigiosin. Appl Environ Microbiol. 1985;50:487–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows SM, Butler T, Jöckel Pet al. . Bacteria in the global atmosphere - Part 2: modeling of emissions and transport between different ecosystems. Atmos Chem Phys. 2009;9:9281–97. [Google Scholar]
- Burrows SM, Rayner PJ, Butler Tet al. . Estimating bacteria emissions from inversion of atmospheric transport: sensitivity to modelled particle characteristics. Atmos Chem Phys. 2013;13:5473–88. [Google Scholar]
- Caliz J, Subirats J, Triado-Margarit Xet al. . Global dispersal and temporal dynamics in atmospheric remote depositions of antibiotic resistance genes. Environ Int. 2022;160:107077. [DOI] [PubMed] [Google Scholar]
- Cáliz J, Triadó-Margarit X, Camarero Let al. . A long-term survey unveils strong seasonal patterns in the airborne microbiome coupled to general and regional atmospheric circulations. Proc Natl Acad Sci USA. 2018;115:12229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlucci AF, Williams PM. Concentration of bacteria from sea water by bubble scavenging. ICES J Mar Sci. 1965;30:28–33. [Google Scholar]
- CDC . One Health. 2020. https://www.cdc.gov/onehealth/basics/index.html(14 February 2022, date last accessed). [Google Scholar]
- Chen S, Jiang N, Huang Jet al. . Quantifying contributions of natural and anthropogenic dust emission from different climatic regions. Atmos Environ. 2018;191:94–104. [Google Scholar]
- Christner BC, Cai R, Morris CEet al. . Geographic, seasonal, and precipitation chemistry influence on the abundance and activity of biological ice nucleators in rain and snow. Proc Natl Acad Sci USA. 2008a;105:18854–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christner BC, Morris CE, Foreman CMet al. . Ubiquity of biological ice nucleators in snowfall. Science (80-). 2008b;319:1214. [DOI] [PubMed] [Google Scholar]
- Daly MJ. A new perspective on radiation resistance based on Deinococcus radiodurans. Nat Rev Micro. 2009;7:237–45. [DOI] [PubMed] [Google Scholar]
- DasSarma P, DasSarma S. Survival of microbes in Earth's stratosphere. Curr Opin Microbiol. 2018;43:24–30. [DOI] [PubMed] [Google Scholar]
- de Araujo GG, Rodrigues F, Gonçalves FLTet al. . Survival and ice nucleation activity of Pseudomonas syringae strains exposed to simulated high-altitude atmospheric conditions. Sci Rep. 2019;9:7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguillaume L, Leriche M, Amato Pet al. . Microbiology and atmospheric processes: chemical interactions of primary biological aerosols. Biogeosciences. 2008;5:1073–84. [Google Scholar]
- DeMott PJ, Hill TCJ, McCluskey CSet al. . Sea spray aerosol as a unique source of ice nucleating particles. Proc Natl Acad Sci USA. 2016;113:5797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després VR, Alex Huffman J, Burrows SMet al. . Primary biological aerosol particles in the atmosphere: a review. Tellus Ser B Chem Phys Meteorol. 2012;64:15598. [Google Scholar]
- Dumbrell AJ, Nelson M, Helgason Tet al. . Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 2010;4:337–45. [DOI] [PubMed] [Google Scholar]
- Eisenhofer R, Minich JJ, Marotz Cet al. . Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. 2019;27:105–17. [DOI] [PubMed] [Google Scholar]
- Elbert W, Taylor PE, Andreae MOet al. . Contribution of fungi to primary biogenic aerosols in the atmosphere: wet and dry discharged spores, carbohydrates, and inorganic ions. Atmos Chem Phys. 2007;7:4569–88. [Google Scholar]
- Els N, Baumann-Stanzer K, Larose Cet al. . Beyond the planetary boundary layer: bacterial and fungal vertical biogeography at Mount Sonnblick, Austria. Geo Geogr Environ. 2019a;6:e00069. [Google Scholar]
- Els N, Larose C, Baumann-Stanzer Ket al. . Microbial composition in seasonal time series of free tropospheric air and precipitation reveals community separation. Aerobiologia (Bologna). 2019b;35:671–701. [Google Scholar]
- Ezraty B, Gennaris A, Barras Fet al. . Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol. 2017;15:385–96. [DOI] [PubMed] [Google Scholar]
- Failor KC, Schmale DG, Vinatzer BAet al. . Ice nucleation active bacteria in precipitation are genetically diverse and nucleate ice by employing different mechanisms. ISME J. 2017;11:2740–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser AM, Antonio DD, Krell Aet al. . Constraining the impact of bacteria on the aqueous atmospheric chemistry of small organic compounds. ACS Earth Sp Chem. 2019;3:1485–91. [Google Scholar]
- Fröhlich-Nowoisky J, Burrows SM, Xie Zet al. . Biogeography in the air: fungal diversity over land and oceans. Biogeosciences. 2012;9:1125–36. [Google Scholar]
- Fröhlich-Nowoisky J, Hill TCJ, Pummer BGet al. . Ice nucleation activity in the widespread soil fungus mortierella alpina. Biogeosciences Discuss. 2014;11:12697–731. [Google Scholar]
- Fröhlich-Nowoisky J, Pickersgill D A, Després VRet al. . High diversity of fungi in air particulate matter. Proc Natl Acad Sci USA. 2009;106:12814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Garcia-Pichel F. Microbial ultraviolet sunscreen. Nat Rev Microbiol. 2011;9:791–802. [DOI] [PubMed] [Google Scholar]
- García-Arenal F, Zerbini FM. Life on the edge: geminiviruses at the interface between crops and wild plant hosts. Annu Rev Virol. 2019;6:411–33. [DOI] [PubMed] [Google Scholar]
- Gettelman A, Collins WD, Fetzer EJet al. . Climatology of upper-tropospheric relative humidity from the atmospheric infrared sounder and implications for climate. J Clim. 2006;19:6104–21. [Google Scholar]
- González-Toril E, Osuna S, Viúdez-Moreiras Det al. . Impacts of Saharan dust intrusions on bacterial communities of the low troposphere. Sci Rep. 2020;10:6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DW. Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin Microbiol Rev. 2007;20:459–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusareva ES, Acerbi E, Lau KJXet al. . Microbial communities in the tropical air ecosystem follow a precise diel cycle. Proc Natl Acad Sci USA. 2019;116:23299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Li L, Wang Yet al. . Composition, dispersion, and health risks of bioaerosols in wastewater treatment plants: a review. Front Environ Sci Eng. 2020;15:38. [Google Scholar]
- Hanson CA, Fuhrman JA, Horner-Devine MCet al. . Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol. 2012;10:497–506. [DOI] [PubMed] [Google Scholar]
- Hartmann S, Augustin S, Clauss Tet al. . Immersion freezing of ice nucleation active protein complexes. Atmos Chem Phys. 2013;13:5751–66. [Google Scholar]
- Hayakawa K, Tang N, Nagato EGet al. . Long term trends in atmospheric concentrations of polycyclic aromatic hydrocarbons and nitropolycyclic aromatic hydrocarbons: a study of Japanese cities from 1997 to 2014. Environ Pollut. 2018;233:474–82. [DOI] [PubMed] [Google Scholar]
- Heald CL, Spracklen DV. Atmospheric budget of primary biological aerosol particles from fungal spores. Geophys Res Lett. 2009;36:L09806. [Google Scholar]
- Healy DA, Huffman JA, O'Connor DJet al. . Ambient measurements of biological aerosol particles near Killarney, Ireland: a comparison between real-time fluorescence and microscopy techniques. Atmos Chem Phys. 2014;14:8055–69. [Google Scholar]
- Hill KA, Shepson PB, Galbavy ESet al. . Processing of atmospheric nitrogen by clouds above a forest environment. J Geophys Res Atmos. 2007;112:D11301. [Google Scholar]
- Hochman A, Alpert P, Harpaz Tet al. . A new dynamical systems perspective on atmospheric predictability: eastern mediterranean weather regimes as a case study. Sci Adv. 2019;5:eaau0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoose C, Kristjánsson JE, Burrows SM. How important is biological ice nucleation in clouds on a global scale?. Environ Res Lett. 2010;5:24009. [Google Scholar]
- Hospodsky D, Qian J, Nazaroff WWet al. . Human occupancy as a source of indoor airborne bacteria. PLoS One. 2012;7:e34867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CI, Katul G. The lagrangian stochastic model for estimating footprint and water vapor fluxes over inhomogeneous surfaces. Int J Biometeorol. 2009;53:87–100. [DOI] [PubMed] [Google Scholar]
- Hu W, Niu H, Murata Ket al. . Bacteria in atmospheric waters: detection, characteristics and implications. Atmos Environ. 2018;179:201–21. [Google Scholar]
- Hudait A, Odendahl N, Qiu Yet al. . Ice-nucleating and antifreeze proteins recognize ice through a diversity of anchored clathrate and ice-like motifs. J Am Chem Soc. 2018;140:4905–12. [DOI] [PubMed] [Google Scholar]
- Huestis DL, Dao A, Diallo Met al. . Windborne long-distance migration of malaria mosquitoes in the Sahel. Nature. 2019;574:404–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman JA, Prenni AJ, DeMott PJet al. . High concentrations of biological aerosol particles and ice nuclei during and after rain. Atmos Chem Phys. 2013;13:6151–64. [Google Scholar]
- Husárová S, Vaïtilingom M, Deguillaume Let al. . Biotransformation of methanol and formaldehyde by bacteria isolated from clouds. Comparison with radical chemistry. Atmos Environ. 2011;45:6093–102. [Google Scholar]
- Jaber S, Lallement A, Sancelme Met al. . Biodegradation of phenol and catechol in cloud water: comparison to chemical oxidation in the atmospheric multiphase system. Atmos Chem Phys. 2020;20:4987–97. [Google Scholar]
- Jaenicke R. Abundance of cellular material and proteins in the atmosphere. Science (80-). 2005;308:73. [DOI] [PubMed] [Google Scholar]
- Joly M, Amato P, Sancelme Met al. . Survival of microbial isolates from clouds toward simulated atmospheric stress factors. Atmos Environ. 2015;117:92–8. [Google Scholar]
- Joly M, Attard E, Sancelme Met al. . Ice nucleation activity of bacteria isolated from cloud water. Atmos Environ. 2013;70:392–400. [Google Scholar]
- Jones SE, Lennon JT. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA. 2010;107:5881–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung YS, Ge Z, Buie CR. Bioaerosol generation by raindrops on soil. Nat Commun. 2017;8:14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousse C, Dalle C, Canet Iet al. . Metabolomic study of the response to cold shock in a strain of Pseudomonas syringae isolated from cloud water. Metabolomics. 2017;14:11. [DOI] [PubMed] [Google Scholar]
- Jusot J-F, Neill DR, Waters EMet al. . Airborne dust and high temperatures are risk factors for invasive bacterial disease. J Allergy Clin Immunol. 2017;139:977–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg CA, Griffin DW. Aerobiology and the global transport of desert dust. Trends Ecol Evol. 2006;21:638–44. [DOI] [PubMed] [Google Scholar]
- Khaled A, Zhang M, Amato Pet al. . Biodegradation by bacteria in clouds: an underestimated sink for some organics in the atmospheric multiphase system. Atmos Chem Phys. 2021;21:3123–41. [Google Scholar]
- King MD, Platnick S, Menzel WPet al. . Spatial and temporal distribution of clouds observed by MODIS onboard the Terra and Aqua satellites. IEEE Trans Geosci Remote Sens. 2013;51:3826–52. [Google Scholar]
- Klein AM, Bohannan BJM, Jaffe DAet al. . Molecular evidence for metabolically active bacteria in the atmosphere. Front Microbiol. 2016;7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollath DR, Miller KJ, Barker BM. The mysterious desert dwellers: Coccidioides immitisand Coccidioides posadasii, causative fungal agents of coccidioidomycosis. Virulence. 2019;10:222–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondakova T, Catovic C, Barreau Met al. . Response to gaseous NO2 air pollutant of P. fluorescens airborne strain MFAF76a and clinical strain MFN1032. Front Microbiol. 2016;7:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev A, Leisner T. Review of experimental studies of secondary ice production. Atmos Chem Phys. 2020;20:11767–97. [Google Scholar]
- Kutter JS, Spronken MI, Fraaij PLet al. . Transmission routes of respiratory viruses among humans. Curr Opin Virol. 2018;28:142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagomarsino Oneto D, Golan J, Mazzino Aet al. . Timing of fungal spore release dictates survival during atmospheric transport. Proc Natl Acad Sci USA. 2020;117:5134–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai KM, Emberlin J, Colbeck I. Outdoor environments and human pathogens in air. Environ Heal A Glob Access Sci Source. 2009;8:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallement A, Besaury L, Tixier Eet al. . Potential for phenol biodegradation in cloud waters. Biogeosciences. 2018;15:5733–44. [Google Scholar]
- Lebre PH, De Maayer P, Cowan DA. Xerotolerant bacteria: surviving through a dry spell. Nat Rev Microbiol. 2017;15:285–96. [DOI] [PubMed] [Google Scholar]
- Lennon JT, Jones SE. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol. 2011;9:119–30. [DOI] [PubMed] [Google Scholar]
- Leung NHL. Transmissibility and transmission of respiratory viruses. Nat Rev Microbiol. 2021;19:528–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KM, van Dijken GL, Arrigo KR. Changes in phytoplankton concentration now drive increased arctic ocean primary production. Science (80-). 2020;369:198–202. [DOI] [PubMed] [Google Scholar]
- Li J, Cao J, Zhu Yet al. . Global survey of antibiotic resistance genes in air. Environ Sci Technol. 2018;52:10975–84. [DOI] [PubMed] [Google Scholar]
- Ling M, Marshall IPG, Rosati Bet al. . Properties relevant to atmospheric dispersal of the ice-nucleation active Pseudomonas syringae strain R10.79 isolated from rain water. Aerobiologia (Bologna). 2021;37:225–41. [Google Scholar]
- Ling ML, Wex H, Grawe Set al. . Effects of ice nucleation protein repeat number and oligomerization level on ice nucleation activity. J Geophys Res Atmos. 2018;123:1802–10. [Google Scholar]
- Lowe WH, McPeek MA. Is dispersal neutral?. Trends Ecol Evol. 2014;29:444–50. [DOI] [PubMed] [Google Scholar]
- Lukas M, Schwidetzky R, Kunert ATet al. . Electrostatic interactions control the functionality of bacterial ice nucleators. J Am Chem Soc. 2020;142:6842–6. [DOI] [PubMed] [Google Scholar]
- Lymperopoulou DS, Adams RI, Lindow SE. Contribution of vegetation to the microbial composition of nearby outdoor air. Appl Environ Microbiol. 2016;82:3822–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki T, Kurosaki Y, Onishi Ket al. . Variations in the structure of airborne bacterial communities in tsogt-ovoo of Gobi Desert area during dust events. Air Qual Atmos Heal. 2016:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki T, Lee KC, Kawai Ket al. . Aeolian dispersal of bacteria associated with desert dust and anthropogenic particles over continental and oceanic surfaces. J Geophys Res Atmos. 2019;124:5579–88. [Google Scholar]
- Matulová M, Husárová S, Capek Pet al. . Biotransformation of various saccharides and production of exopolymeric substances by cloud-borne Bacillus sp. 3B6. Environ Sci Technol. 2014;48:14238–47. [DOI] [PubMed] [Google Scholar]
- Mayol E, Arrieta JM, Jiménez MAet al. . Long-range transport of airborne microbes over the global tropical and subtropical ocean. Nat Commun. 2017;8:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescioglu E, Rahav E, Belkin Net al. . Aerosol microbiome over the mediterranean sea diversity and abundance. Atmosphere (Basel). 2019;10. DOI: 10.3390/atmos10080440. [Google Scholar]
- Meyer M, Cox JA, Hitchings MDTet al. . Quantifying airborne dispersal routes of pathogens over continents to safeguard global wheat supply. Nat Plants. 2017;3:780–6. [DOI] [PubMed] [Google Scholar]
- Michaud JM, Thompson LR, Kaul Det al. . Taxon-specific aerosolization of bacteria and viruses in an experimental ocean-atmosphere mesocosm. Nat Commun. 2018;9:2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier J-M, Lindow SE. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc Natl Acad Sci USA. 2003;100:15977–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RA, Bomar C, Kobziar LNet al. . Wildland fire as an atmospheric source of viable microbial aerosols and biological ice nucleating particles. ISME J. 2021;15:461–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RA, Hanlon R, Powers Cet al. . Scavenging of sub-micron to micron-sized microbial aerosols during simulated rainfall. Atmosphere (Basel). 2020;11:80. [Google Scholar]
- Morris CE, Conen F, Alex Huffman Jet al. . Bioprecipitation: a feedback cycle linking earth history, ecosystem dynamics and land use through biological ice nucleators in the atmosphere. Glob Chang Biol. 2014;20:341–51. [DOI] [PubMed] [Google Scholar]
- Morris CE, Monier J-M. The ecological significance of biofilm formation by plant-associated bacteria. Annu Rev Phytopathol. 2003;41:429–53. [DOI] [PubMed] [Google Scholar]
- Morris CE, Sands DC, Bardin Met al. . Microbiology and atmospheric processes: an upcoming era of research on bio-meteorology. Biogeosciences Discuss. 2008;5:191–212. [Google Scholar]
- Morris CE, Sands DC, Glaux Cet al. . Urediospores of rust fungi are ice nucleation active at >−10°C and harbor ice nucleation active bacteria. Atmos Chem Phys. 2013;13:4223–33. [Google Scholar]
- Mülmenstädt J, Sourdeval O, Delanoë Jet al. . Frequency of occurrence of rain from liquid-, mixed-, and ice-phase clouds derived from A-Train satellite retrievals. Geophys Res Lett. 2015;42:6502–9. [Google Scholar]
- Nehme B, Létourneau V, Forster RJet al. . Culture-independent approach of the bacterial bioaerosol diversity in the standard swine confinement buildings, and assessment of the seasonal effect. Environ Microbiol. 2008;10:665–75. [DOI] [PubMed] [Google Scholar]
- Nicolaisen M, West JS, Sapkota Ret al. . Fungal communities including plant pathogens in near surface air are similar across northwestern Europe. Front Microbiol. 2017;8:1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOAA, NASA, US Air Force . U.S. Standard Atmosphere. Washington D.C., 1976. [Google Scholar]
- Norros V, Rannik Ü, Hussein Tet al. . Do small spores disperse further than large spores?. Ecology. 2014;95:1612–21. [DOI] [PubMed] [Google Scholar]
- O′Sullivan D, Murray BJ, Ross JFet al. . The relevance of nanoscale biological fragments for ice nucleation in clouds. Sci Rep. 2015;5:8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontiveros VJ, Cáliz J, Triadó-Margarit Xet al. . General decline in the diversity of the airborne microbiota under future climatic scenarios. Sci Rep. 2021;11:20223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oteros J, Bartusel E, Alessandrini Fet al. . Artemisia pollen is the main vector for airborne endotoxin. J Allergy Clin Immunol. 2019;143:369–77. [DOI] [PubMed] [Google Scholar]
- Pal C, Bengtsson-Palme J, Kristiansson Eet al. . The structure and diversity of human, animal and environmental resistomes. Microbiome. 2016;4:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Usui K, Livingstone RAet al. . Ice-nucleating bacteria control the order and dynamics of interfacial water. Sci Adv. 2016;2:e1501630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointing SBSB, Belnap J. Disturbance to desert soil ecosystems contributes to dust-mediated impacts at regional scales. Biodivers Conserv. 2014;23:1659–67. [Google Scholar]
- Pointing SBSB, Belnap J. Microbial colonization and controls in dryland systems. Nat Rev Microbiol. 2012;10:551–62. [DOI] [PubMed] [Google Scholar]
- Potts M. Desiccation tolerance of prokaryotes. Microbiol Mol Biol Rev. 1994;58:755–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouleur S, Richard C, Martin Jet al. . Ice nucleation activity in Fusarium acuminatum and Fusarium avenaceum. Appl Environ Microbiol. 1992;58:2960–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruppacher HR, Jaenicke R. The processing of water vapor and aerosols by atmospheric clouds, a global estimate. Atmos Res. 1995;38:283–95. [Google Scholar]
- Rastelli E, Corinaldesi C, Dell'anno Aet al. . Transfer of labile organic matter and microbes from the ocean surface to the marine aerosol: an experimental approach. Sci Rep. 2017;7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reche I, D'Orta G, Mladenov Net al. . Deposition rates of viruses and bacteria above the atmospheric boundary layer. ISME J. 2018;12:1154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeters SJ, Golbek TW, Bregnhøj Met al. . Ice-nucleating proteins are activated by low temperatures to control the structure of interfacial water. Nat Commun. 2021;12:1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salawu-Rotimi A, Lebre PH, Vos HCet al. . Gone with the wind: microbial communities associated with dust from emissive farmlands. Microb Ecol. 2021;82:859–69. [DOI] [PubMed] [Google Scholar]
- Salter SJ, Cox MJ, Turek EMet al. . Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandra C-C, Anne B, Marie-Lise Det al. . Phage–host interaction: an ecological perspective. J Bacteriol. 2004;186:3677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šantl-Temkiv T, Finster K, Dittmar Tet al. . Hailstones: a window into the microbial and chemical inventory of a storm cloud. PLoS One. 2013a;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šantl-Temkiv T, Finster K, Hansen BMet al. . Viable methanotrophic bacteria enriched from air and rain can oxidize methane at cloud-like conditions. Aerobiologia (Bologna). 2013b;29:373–84. [Google Scholar]
- Šantl-Temkiv T, Gosewinkel U, Starnawski Pet al. . Aeolian dispersal of bacteria in southwest greenland: their sources, abundance, diversity and physiological states. FEMS Microbiol Ecol. 2018;94:fiy031. [DOI] [PubMed] [Google Scholar]
- Šantl-Temkiv T, Lange R, Beddows Det al. . Biogenic sources of ice nucleating particles at the high Arctic site villum research station. Environ Sci Technol. 2019;53:10580–90. [DOI] [PubMed] [Google Scholar]
- Šantl-Temkiv T, Sikoparija B, Maki Tet al. . Bioaerosol field measurements: challenges and perspectives in outdoor studies. Aerosol Sci Technol. 2020;54:520–46. [Google Scholar]
- Sattler B, Puxbaum H, Psenner R. Bacterial growth in supercooled cloud droplets. Geophys Res Lett. 2001;28:239–42. [Google Scholar]
- Savary S, Willocquet L, Pethybridge SJet al. . The global burden of pathogens and pests on major food crops. Nat Ecol Evol. 2019;3:430–9. [DOI] [PubMed] [Google Scholar]
- Schmid HP. Footprint modeling for vegetation atmosphere exchange studies: a review and perspective. Agric For Meteorol. 2002;113:159–83. [Google Scholar]
- Sharma A, Gaidamakova EK, Grichenko Oet al. . Across the tree of life, radiation resistance is governed by antioxidant Mn2+, gauged by paramagnetic resonance. Proc Natl Acad Sci USA. 2017;114:9253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd G, Terradellas E, Baklanov Aet al. . Global assessment of sand and dust storms. United Nations Environment Program (UNEP). 2016. [Google Scholar]
- Singh RP, Hodson DP, Huerta-Espino Jet al. . The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol. 2011;49:465–81. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Griffin DW, McPeters RDet al. . Microbial survival in the stratosphere and implications for global dispersal. Aerobiologia (Bologna). 2011;27:319–32. [Google Scholar]
- Smith DJ, Griffin DW, Schuerger AC. Stratospheric microbiology at 20 km over the pacific ocean. Aerobiologia (Bologna). 2010;26:35–46. [Google Scholar]
- Smith DJ, Ravichandar JD, Jain Set al. . Airborne bacteria in earth's lower stratosphere resemble taxa detected in the troposphere: results from a new NASA aircraft bioaerosol collector (ABC). Front Microbiol. 2018;9:1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen JH, Mackay DKJ, Jensen Cet al. . An integrated model to predict the atmospheric spread of foot-and-mouth disease virus. Epidemiol Infect. 2000;124:577–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklen D V., Heald CL. The contribution of fungal spores and bacteria to regional and global aerosol number and ice nucleation immersion freezing rates. Atmos Chem Phys. 2014;14:9051–9. [Google Scholar]
- Spring AM, Domingue KD, Kerber T Vet al. . Land use effects on airborne bacterial communities are evident in both near-surface and higher-altitude air. Diversity. 2021;13. DOI: 10.3390/d13020085. [Google Scholar]
- Stevenson A, Burkhardt J, Cockell CSet al. . Multiplication of microbes below 0.690 water activity: implications for terrestrial and extraterrestrial life. Environ Microbiol. 2015a;17:257–77. [DOI] [PubMed] [Google Scholar]
- Stevenson A, Cray JA, Williams JPet al. . Is there a common water-activity limit for the three domains of life?. ISME J. 2015b;9:1333–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopelli E, Conen F, Morris CEet al. . Ice nucleation active particles are efficiently removed by precipitating clouds. Sci Rep. 2015;5:16433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temkiv TŠ, Finster K, Hansen BMet al. . The microbial diversity of a storm cloud as assessed by hailstones. FEMS Microbiol Ecol. 2012. DOI: 10.1111/j.1574-6941.2012.01402.x. [DOI] [PubMed] [Google Scholar]
- Tesson SVM, Šantl-Temkiv T. Ice nucleation activity and aeolian dispersal success in airborne and aquatic microalgae. 2018;9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson SVM, Skjøth A, Šantl-temkiv T. Airborne microalgae: insights, opportunities, and challenges. Appl Environ Microbiol. 2016;82:1978–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tignat-Perrier R, Dommergue A, Thollot Aet al. . Global airborne microbial communities controlled by surrounding landscapes and wind conditions. Sci Rep. 2019;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tignat-Perrier R, Dommergue A, Thollot Aet al. . Seasonal shift in airborne microbial communities. Sci Total Environ. 2020;716:137129. [DOI] [PubMed] [Google Scholar]
- Tobias A, Karanasiou A, Amato Fet al. . Health effects of desert dust and sand storms: a systematic review and meta-analysis. Environ Epidemiol. 2019;3. DOI: 10.1097/01.EE9.0000610424.75648.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobo Y, Adachi K, DeMott PJet al. . Glacially sourced dust as a potentially significant source of ice nucleating particles. Nat Geosci. 2019;12:253–8. [Google Scholar]
- Tong Y, Lighthart B. Effect of simulated solar radiation on mixed outdoor atmospheric bacterial populations. FEMS Microbiol Ecol. 2006;26:311–6. [Google Scholar]
- Triadó-Margarit X, Cáliz J, Casamayor E. A long-term atmospheric baseline for intercontinental exchange of airborne pathogens. Environ Int. 2022;158:106916. [DOI] [PubMed] [Google Scholar]
- Uetake J, Hill TCJ, Moore KAet al. . Airborne bacteria confirm the pristine nature of the southern ocean boundary layer. Proc Natl Acad Sci USA. 2020;117:13275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake J, Tobo Y, Uji Yet al. . Seasonal changes of airborne bacterial communities over tokyo and influence of local meteorology. Front Microbiol. 2019;10:1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaïtilingom M, Amato P, Sancelme Met al. . Contribution of microbial activity to carbon chemistry in clouds. Appl Environ Microbiol. 2010;76:23–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaitilingom M, Deguillaume L, Vinatier Vet al. . Potential impact of microbial activity on the oxidant capacity and organic carbon budget in clouds. Proc Natl Acad Sci USA. 2013;110:559–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault D, Marcoux-Voiselle M, Turgeon Net al. . Resistance of aerosolized bacterial viruses to relative humidity and temperature. Appl Environ Microbiol. 2015;81:7305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright M, Wickramasinghe NC, Narlikar J Vet al. . Microorganisms cultured from stratospheric air samples obtained at 41 km. FEMS Microbiol Lett. 2003;218:161–5. [DOI] [PubMed] [Google Scholar]
- Wang X, Sultana CM, Trueblood Jet al. . Microbial control of sea spray aerosol composition: a tale of two blooms. ACS Cent Sci. 2015;1:124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wex H, Augustin-Bauditz S, Boose Yet al. . Intercomparing different devices for the investigation of ice nucleating particles using Snomax® as test substance. Atmos Chem Phys. 2015;15:1463–85. [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whon TW, Kim M-S, Roh SWet al. . Metagenomic characterization of airborne viral DNA diversity in the near-surface atmosphere. J Virol. 2012;86:8221–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TW, Ladino LA, Alpert PAet al. . A marine biogenic source of atmospheric ice-nucleating particles. Nature. 2015;525:234–8. [DOI] [PubMed] [Google Scholar]
- Wirgot N, Lagrée M, Traïkia Met al. . Metabolic modulations of Pseudomonas graminis in response to H2O2 in cloud water. Sci Rep. 2019;9. DOI: 10.1038/s41598-019-49319-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womack AM, Bohannan BJM, Green JL. Biodiversity and biogeography of the atmosphere. Philos Trans R Soc Lond B Biol Sci. 2010;365:3645–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo AC, Brar MS, Chan Yet al. . Temporal variation in airborne microbial populations and microbially-derived allergens in a tropical urban landscape. Atmos Environ. 2013;74:291–300. [Google Scholar]
- Woo C, Yamamoto N. Falling bacterial communities from the atmosphere. Environ Microbiome. 2020;15:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Li L, Wang Yet al. . Airborne bacteria in a wastewater treatment plant: emission characterization, source analysis and health risk assessment. Water Res. 2019;149:596–606. [DOI] [PubMed] [Google Scholar]
- Yang Y, Yokobori S, Yamagishi A. Assessing panspermia hypothesis by microorganisms collected from the high altitude atmosphere. Biol Sci Sp. 2009;23:151–63. [Google Scholar]
- Zhao Y, Aarnink A, De Jong Met al. . Airborne microorganisms from livestock production systems and their relation to dust. Crit Rev Environ Sci Technol. 2014;44:1071–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Ning D.. Stochastic community assembly: does it matter in microbial ecology?. Microbiol Mol Biol Rev. 2017;81:e00002–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S-Y-D, Li H, Giles Met al. . microbial flow within an air-phyllosphere-soil continuum. Front Microbiol. 2021;11:3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelhoffer EC, Donohue TJ. Bacterial responses to photo-oxidative stress. Nat Rev Microbiol. 2009;7:856–63. [DOI] [PMC free article] [PubMed] [Google Scholar]