Summary
Precision remodeling of glycans in their native environments is pivotal for understanding glycan-mediated biological events and has important biotechnological implications in fields of clinical diagnosis, glyco-immune checkpoint therapy, and so forth. However, the influence of aglycone-steric diversity on the selectivity of glycan remodeling has been largely overlooked, limiting the application in complex biological scenarios. Here, we report the achievement of aglycone sterics-selective enzymatic glycan remodeling by controlled grafting of functional polymers from glycoenzyme. Through tuning polymer length, a series of enzyme-polymer composites with varying substrate permeability are prepared, which afford an activity pattern-based differentiation strategy for aglycone sterics. This leads to the implementation of glycolipid’s partner screening, and aglycone sterics-selective glycan remodeling in a complex biological environment. We further orchestrate the polymer length adjustment with external cues to regulate aglycone-steric selectivity in a multi-faceted fashion, resulting in an unexpected enhancement of glycolipid remodeling, and temporal control of glycan remodeling on live cells.
Subject areas: Biological sciences, Biochemistry, Glycobiology, Enzyme engineering
Graphical abstract

Highlights
-
•
Functional polymer is used to tune the glycan remodeling activity of a glycoenzyme
-
•
Glycoenzyme-polymer composites display an aglycone sterics-dependent activity
-
•
The composites enable aglycone sterics-selective glycan remodeling in complex media
-
•
Temporal and two-dimensional regulation of glycan remodeling is achieved, respectively
Biological sciences; Biochemistry; Glycobiology; Enzyme engineering
Introduction
Glycosylation is the most abundant and diverse modification type for biomolecules (Reily et al., 2019; Schjoldager et al., 2020). Through sequential concerted steps in the endoplasmic reticulum and the Golgi apparatus, complex glycan structures can be modified on proteins or lipids with varying compositions, sequences, linkages, and lengths (Reily et al., 2019; Schjoldager et al., 2020). Recently, glycans have also been found to be modified on RNA (Flynn et al., 2021). These sugars not only extend the chemical repertoire of biomolecules but also profoundly affect the intrinsic properties and the corresponding biological functions (Varki, 2017). In animals, glycans are abundantly present in many forms, from freely diffused saccharides and secreted glycoconjugates (e.g., glycoproteins, glycolipids, proteoglycans) in the extracellular matrix, to whole-cell glycocalyx (i.e., membrane-bound glycoconjugates). To facilitate discussion, we define the parent structure block (including saccharides, proteins, lipids, cells) that links to the target glycosyl group as “aglycone” in this work.
The diversity of both glycans and aglycones makes the understanding of glycan-mediated biological processes very challenging. Various biology and chemical biology investigation tools (Cheng et al., 2021), such as affinity-based probes (Pilobello et al., 2007), direct chemical modification (Zeng et al., 2009), metabolic glycan labeling (Mahal et al., 1997), chemoenzymatic glycan editing (Zheng et al., 2011; Chaubard et al., 2012; Hong et al., 2019), have been developed in the past decades to advance our abilities to probe and manipulate glycans. Among those, guiding glycoenzyme-aglycone binding by modifying glycoenzymes with antibodies (Gray et al., 2020; Belardi et al., 2013), aptamers (Hui et al., 2017; Guo et al., 2020; Li et al., 2021), or nanobodies (Ge et al., 2021), holds a prominent position to endow glycan remodeling with cell (or protein) specificity. However, the robust physical connection between glycoenzyme and aglycone deprives glycan remodeling processes of (internal) tunability and (external) responsiveness, which leads to the incompetence in differentiating aglycone sterics.
The extremely wide range of aglycone sterics of substrates is a unique feature of glycoenzyme-mediated glycan remodeling. For example, when galactose oxidase (GO), an enzyme that catalyzes the oxidation of D-galactose/N-acetyl-D-galactosamine (Gal/GalNAc) at the C-6 position (Rannes et al., 2011), is added into biological environments, Gal, GalNAc, galactosylated/GalNAcylated polysaccharides, proteins, lipids, in freely diffused or membrane-bound states, all have the potential to be processed because of the external restriction-free accessibility of GO’s activity center. We thus envisioned that endowing glycan remodeling with aglycone-steric selectivity by fine-tuning glycoenzyme’s accessibility would add a new dimension to the spatial control of glycan remodeling. This would allow one to differentiate glycoproteins and glycolipids carrying the same target glycan, or selectively manipulate the glycoforms of secreted glycoconjugates without affecting epithelial cell glycocalyx (e.g., gastrointestinal tract mucus labeling, exosomal glycan labeling). Note that aglycone-steric differentiation cannot be achieved by other types of glycan labeling strategies, such as direct chemical modification and metabolic glycan labeling, because of the working principles. Besides the installation of a modulation component on glycoenzyme to allow for tuning of enzyme’s accessibility, further introduction of external-stimuli responsiveness may enable multiple regulations (or switch) of aglycone-steric selectivity, thus contributing to the realization of novel functions, for example, temporal control. Such a modulation platform will facilitate the dissection and utilization of glycosylation functions in many biological contexts.
With these criteria in mind, we herein devise a multi-faceted modulation platform for glycoenzyme accessibility, by means of polymer-based, controlled single-enzyme caging, to achieve and regulate the aglycone-steric selectivity of glycan remodeling. Owing to the feature of tailored synthesis and well-defined properties of synthetic polymers (Liu and Gao, 2021; Lu et al., 2020; Kaupbayeva and Russell, 2020), wrapping up protein with a polymer shell has been extensively used to alter protein’s bioactivity and stability (Heredia et al., 2005; Lele et al., 2005; Cummings et al., 2013; Murata et al., 2013; Kovaliov et al., 2018), tune solubility (Baker et al., 2019), improve circulating half-life (Harris and Chess, 2003), decrease immunogenicity (Liu et al., 2014), and adjust substrate/inhibitor affinity (Liu et al., 2013; Murata et al., 2014; Kaupbayeva et al., 2019). More importantly, different regulation mechanisms can be introduced to proteins by conjugation with stimuli-responsive polymers (Heredia et al., 2005; Cummings et al., 2013; Murata et al., 2013; Chen and Hoffman, 1993; Stayton et al., 1995; Shimoboji et al., 2002, 2003; Boyer et al., 2007; De et al., 2008; Mackenzie and Francis, 2013; Gobbo et al., 2018). The pioneering works of Maynard’s group (Heredia et al., 2005; Bontempo and Maynard, 2005) and Russell’s group (Lu et al., 2020) have proved that atom transfer radical polymerization (ATRP)-based grafting-from techniques are attractive routes for the preparation of protein-polymer composites in a controlled fashion (Boyer et al., 2016; Matyjaszewski, 2018). We thus utilized this method to graft temperature-responsive polymers with various lengths from GO, as a model glycoenzyme, through a cleavable initiator containing a disulfide group (Scheme 1A). Owing to the permeability hindrance effect of polymer, the catalytic activities of the enzyme-polymer composites display an aglycone-steric dependence (Scheme 1B). Via polymer length adjustment (Shimoboji et al., 2003), we acquire an activity pattern-based differentiation principle for galactosylated/GalNAcylated aglycones. This leads to the implementation of immobilization-free and label-free screening of protein partners for a glycolipid, and aglycone sterics-selective glycan remodeling in the complex biological environment. Moreover, by orchestrating thermal or chemical stimuli with length adjustment, multi-faceted modulation of aglycone-steric selectivity is achieved (Scheme 1C), resulting in the implementation of enhancement of glycolipid modification and temporal control of glycan remodeling on live cells. Thus, this work provides a unique, aglycone sterics-based perspective for the modulation of glycan remodeling, which will contribute to the development of precision glycan manipulation tools for fields of clinical diagnosis, glyco-immune checkpoint therapy, tissue engineering, bottom-up synthetic biology, and so forth.
Scheme 1.
Scheme of endowing glycoenzyme with aglycone-steric selectivity by grafting a tunable functional polymer shell
(A) Scheme of the ATRP polymerization of poly(N-isopropylacrylamide) (PN) from GO, which can catalyze the oxidation of Gal/GalNAc.
(B) Schematic illustration of aglycone-steric selectivity of the GO-PN composite.
(C) Scheme of the multi-faceted modulation of PN shell.
Results
Synthesis and characterization
To demonstrate our strategy, we first tagged bis[2-(2′-bromoisobutyryloxy)ethyl]disulfide (BiBOEDS) onto the amine groups of GO via disulfide exchange using N-succinimidyl3-(2-pyridyldithio)propionate (SPDP)-based linkage chemistry (Scheme S1) (Wu et al., 2019), and yielded GO-initiator conjugate (GO-Br, I0) with a BiBOEDS-to-GO molar ratio of approximately 4:1 (Figure S1). We then chose N-isopropylacrylamide (NIPAm, M0) as the model monomer, and grafted it from GO-Br using N,N,N′,N”,N″-pentamethyldiethylenetriamine (PMDETA) as the ligand, and L-ascorbic acid (Vc) as the reducing agent, to generate CuI, the catalyst of ATRP, from CuII (Scheme 1A) (Wu et al., 2019). By adjusting the feed molar ratio and polymerization time, a set of GO-poly(N-isopropylacrylamide) (GO-PN) composites, GO-PN (1∼15), with different degrees of polymerization, were successfully prepared (Tables S1 and S2), as evidenced by the phase and color change during the polymerization reaction. To verify the grafting of PNs from GO and investigate whether GO-PNs possess length adjustability (Theodorou et al., 2020), thermo sensitivity (Heredia et al., 2005; Cummings et al., 2013; Chen and Hoffman, 1993; Stayton et al., 1995; Boyer et al., 2007; De et al., 2008; Mackenzie and Francis, 2013; Wu et al., 2019), and chemical responsiveness (Boyer et al., 2007; Wu et al., 2019; Liu et al., 2007), we chose GO-PN (1∼3) (M0/I0/PMDETA/CuII/Vc = 15000/1/59/59/59, 1-, 2-, 3-h polymerization) as the model enzyme-polymer composites.
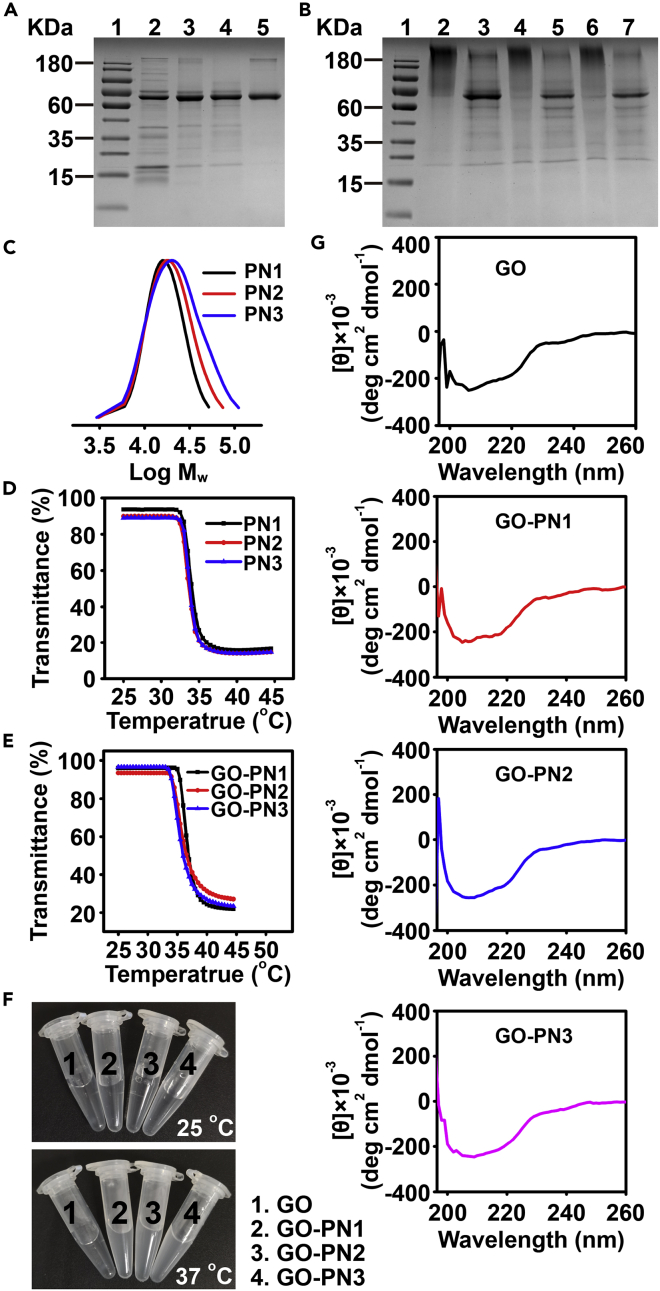
The sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (Theodorou et al., 2020) showed that the as-prepared GO-PN1 (before removal of uncoated GO) smeared on the gel with higher molecular weight compared with native GO, GO-SPDP, and GO-Br (Figure 1A, Lane 6), indicating the formation of polymer on GO. Considering that GO is a copper metalloenzyme (Rannes et al., 2011), the possibility of performing ATRP in the absence of exogenous CuII was excluded. No composite could be observed even after a 12-h reaction (Figure S2). Meanwhile, we also performed control experiments to demonstrate the dependence of polymerization on the presence of Vc and PMDETA (Figure S2) The proportion of coated GO was estimated to be more than 90% using a densitometry semi-quantification analysis (Theodorou et al., 2020) (Figure 1A, Lane 6). The uncoated GO could be readily removed by ultrafiltration with a molecular-weight cutoff of 100 kDa, as manifested by the diminishment of the native GO band (Figure 1B, Lane 2). This washing step also removed other polymerization reagents. For the following experiments, GO-PNs after separation from free GO were used. With increasing polymerization time, the smear of GO-PN (1∼3) ran slower (Figure 1B) (Boyer et al., 2007), suggesting increasing molecular weight.
Figure 1.
Characterization of GO-PNs
(A) SDS-PAGE analysis of species from GO-PN preparation after ultrafiltration with a molecular weight cutoff of 30 kDa. Lane 1: Marker; lane 2: GO; lane 3: GO-SPDP; lane 4: GO-SPDP + TCEP (5 mM, added after ultrafiltration); lane 5: GO-Br; lane 6: GO-PN1.
(B) SDS-PAGE analysis of GO-PN (1∼3) after ultrafiltration using a molecular-weight cutoff of 100 kDa with and without subsequent TECP treatment (5 mM). Lane 1: Marker; lane 2: GO-PN1; lane 3: GO-PN1+TCEP; lane 4: GO-PN2; lane 5: GO-PN2+TCEP; lane 6: GO-PN3; lane 7: GO-PN3+TCEP. Samples with an equivalent GO amount of 5 μg were separated on 10% SDS-PAGE gel, stained with Coomassie blue, and imaged.
(C) GPC analysis of PN (1∼3) cleaved from GO-PN (1∼3) (Table S1).
(D and E) LCST curves of (D) PN (1∼3) and (E) GO-PN (1∼3).
(F) Representative photographs of GO and GO-PN (1∼3) at 25 and 37 oC.
(G) CD spectra of GO and GO-PN (1∼3).
Cleavage of PNs from GO was implemented using 5 mM tris(2-carboxyethyl)phosphine (TCEP) (Sun et al., 2019b) to reduce the disulfide bond (Boyer et al., 2007; Wu et al., 2019; Liu et al., 2007), which was verified by the reappearance of GO single band at 68 kDa (Figure 1B, Lanes 3, 5, 7). We proceeded to characterize the cleaved PN (1∼3) by gel permeation chromatography (GPC, Figure 1C), which showed monomodal traces. An increase in the molecular weight of PNs was observed with elongating polymerization time (Theodorou et al., 2020), and relatively low polydispersity indices (PDI) of 1.24 (weight average molecular weight (Mw) = 18915), 1.36 (Mw = 22531), and 1.54 (Mw = 27783) were achieved for PN1, PN2, and PN3 (Table S1), respectively.
PN is miscible below lower critical solution temperature (LCST) but precipitates above LCST as a result of releasing water around the isopropyl side groups (Boyer et al., 2007). Turbidity measurements of cleaved PN (1∼3) revealed that the LCST of PNs was 33.8 °C (Figure 1D). The phase transition property was inherited by GO-PNs as proven by observable phase separation and the LCST curve measurement. GO was tested under the same conditions as a negative control (Figures 1E, 1F, and S3). The LCST values of GO-PN (1∼3) were higher than those of PNs, owing to the protein increased the hydrophilicity of the hybrid structures (Boyer et al., 2007). The LCST of the composites decreased from GO-PN1 to GO-PN3, which was in agreement with a previous report (Boyer et al., 2007). The reason may lie in that along with increasing the molecular weight, the influence from the hydrophilic protein becomes less profound while the effect of polymer becomes dominant (Boyer et al., 2007).
We also verified the existence of protein in GO-PN (1∼3) by the Protein A280 method (Figure S4), and the retention of protein conformation by circular dichroism (CD) spectra (Figures 1G and S5) (Theodorou et al., 2020). The CD spectra of both GO and GO-PN composites showed a continuous change between 200 and 230 nm, consistent with the GO crystal structure, which contains mostly beta-sheets and random coil/loops (Rogers et al., 2007). These spectra were basically identical to the one published in the literature (Paukner et al., 2014). The minimal differences between the spectra of GO and GO-PNs indicate insignificant structural change after polymer modification.
We next sought to confirm that the polymer grafting would preserve the inherent enzymatic activity of GO using GO-PN1 (with relatively shorter polymer length, thus lower shielding effect, vide infra) as the model. We quantified H2O2, the enzymatic catalytic product of substrate Gal, using a GO activity assay kit (Figure S6). We noted that either the initiator anchoring procedure or the ATRP polymerization displayed negligible influence on GO activity (Figure S7A). The glycan remodeling specificity for GO-PN1 remained intact, as revealed by the indiscernible signal when we replaced Gal or GalNAc with other types of monosaccharides (Figure S7B). By contrast, minimal oxidation activity was observed if GO was omitted from the system (Figure S7C). Therefore, GO-PN composites are necessary for catalytic activity to occur. These data collectively illustrate that GO can survive the polymerization conditions.
Cell sterics-dependent activity inhibition of GO-PNs
Typically, attaching polymer chains to enzymes imposes permeability hindrance (Chen and Hoffman, 1993; Liu et al., 2007), which introduces interactions that compete with enzyme-substrate binding (Keefe and Jiang, 2012). The polymer layer can be tuned to allow substrates below a certain size threshold to diffuse to the active site of enzyme (Liu et al., 2013, 2014; Kaupbayeva et al., 2019, 2021). We thus wondered whether we could utilize the shielding effect of polymer to endow glycoenzyme with differentiation capability towards distinct aglycone s terics. Note that almost all current studies avoid discussing enzymatic activity when live cells act as the substrates; however, this can not be bypassed for glycan remodeling, because of the complexity of glyco-substrates in in vivo scenarios. In this context, we systematically tested the effect of polymer length on GO-PN remodeling activity towards live cell-carried Gal/GalNAc by adjusting reactant composition and polymerization time. The cytocompatibility of GO, PN1, and GO-PN1 was firstly validated and no toxicity was observed at the concentration tested (Figure S8).
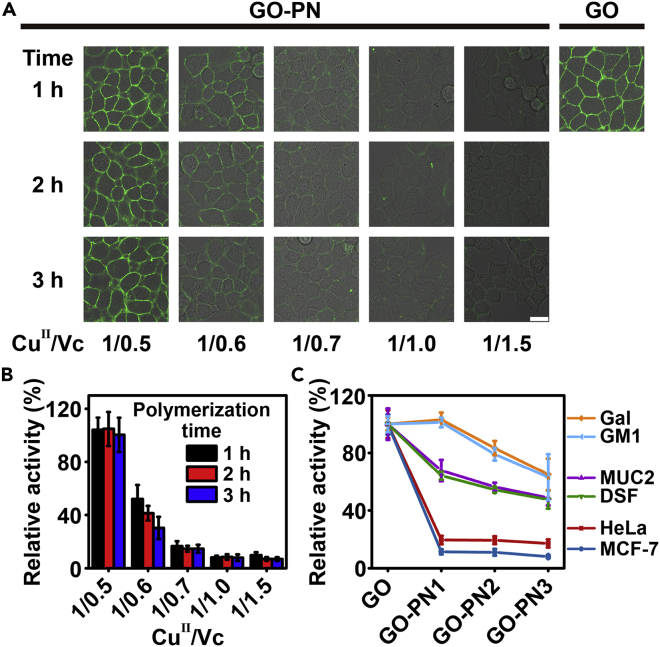
We treated MCF-7 cells with GO-PN (1∼15) at 4°C for 1 h, respectively. The hydroxyl group at the C6 position of terminal Gal/GalNAc on the cell surface can be oxidized to the aldehyde group (Rannes et al., 2011), and then labeled with fluorescein-5-thiosemicarbazide (FTZ) to form a stable hydrazone linkage (Zhang et al., 2019). The mean fluorescence intensity (MFI) of FTZ on GO-PN (1∼15) treated cells was normalized to MFI of equimolar GO-treated cells, to obtain a relative activity (%) for each type of GO-PNs. From the confocal laser scanning microscopy (CLSM) data, we noticed a distinct labeling pattern (Figures 2A, 2B, and S9): (1) an increase of Vc loading provides accelerated polymerization kinetics and a higher molecular weight (this was also verified by ATRP grafting PN from bovine serum albumin, Figure S10) (Chen et al., 2011; Averick et al., 2012), hence increasing the shielding effect of PNs. For example, the group with CuII/Vc = 1:1.0 (GO-PN (1∼3)) exhibited an average relative activity of only 10%. Considering that PN fabrication did not alter GO intrinsic activity while PNs alone only resulted in background activity signals (Figures S7A and S7C), the reduced apparent GO activity towards cell surface Gal/GalNAc is mainly attributed to the PN shielding effect around GO active site (Liu et al., 2013; Keefe and Jiang, 2012). (2) Elongation of polymerization time gradually decreases GO-PN activity, as witnessed by the FTZ signal changes such as for the group with CuII/Vc = 1/0.6 (Figures 2A, 2B, and S9). These phenomena imply the possibility of facile modulation of GO remodeling activity towards cell surface glycoconjugates by adjusting the polymerization time to tune substrate permeability to the GO active site (Liu et al., 2007). This was also supported by a parallel cell-based GO-PN activity test using the composite series (CuII/Vc = 1/1.0) with shorter polymerization time (Figure S11).
Figure 2.
Study of the activity of GO-PNs towards substrates with distinct aglycone sterics
(A) CLSM images of MCF-7 cells treated with GO-PN (1∼15) (five different reactant ratios, three different polymerization times, Tables S1 and S2) or GO (0.05 mg/mL of equivalent GO concentration) for 60 min. The treated cells were washed and fluorescently labeled by FTZ. All fluorescence images were taken using the same instrumental setting. Scale bar: 20 μm.
(B) Quantification of GO-PN activity for remodeling Gal/GalNAc on live cells. Relative activity: MFI at cellular periphery for GO-PN treated cells divided by MFI for GO treated cells.
(C) Relative activity of GO-PN (1∼3) towards various substrates. Activity was normalized by setting naked GO as 100% for each type of substrates. Data represent the mean and standard deviation of triplicate experiments.
Activity pattern-based discrimination of aglycones with distinct sterics
Having demonstrated the blockage of cellular glycan remodeling by the PN shell, we hypothesized that galactosylated/GalNAcylated substrates with smaller aglycones would cross this cell-prohibiting polymer shell, thus enabling an aglycone sterics-selective glycan remodeling. To test this hypothesis, we chose GO-PN (1∼3) in the following experiments because they could largely prohibit the permeation of MCF-7 and HeLa cells (Figure 2A, column 4, and S12), while GO-PN1 exhibited comparable activity with native GO towards Gal (Figure S7A). Four substrates with different molecular weights, including mucin 2 (MUC2, 640 kDa) (Hayakawa et al., 2020), desialylated fetuin (DSF, 48 kDa) (Pal et al., 2012), monosialotetrahexosylganglioside (GM1, 1546 Da) (Komura et al., 2016) and galactose (Gal, 180 Da), were carefully scrutinized. We treated each substrate with GO-PN (1∼3) for 1 h and measured the normalized absorption value (setting native GO activity of equal enzyme amount as 100% for each type of substrates) of the generated H2O2 to obviate the influence from Gal/GalNAc expression level or substrate concentration on activity comparison. We observed similar activity trends for glycoproteins MUC2 and DSF: the relative activity gradually decreased from GO-PN1 (68% for MUC2 and 64% for DSF) to GO-PN3 (49% for MUC2 and 47% for DSF) (Figure 2C). These values were substantially higher than GO-PN (1∼3) activities towards cells (∼10% for MCF-7 and ∼19% for HeLa), thus indicating that freely diffused glycoproteins are more accessible to PN-shielded glycoenzymes. Consistent with the hypothesis, the shielding effect of PNs towards Gal and GM1 was even weaker (Figure 2C): the relative activities were constantly higher than those for glycoproteins and cells.
To provide mechanistic insights into the different behaviors of GO-PNs towards distinct aglycone sterics, we measured the kinetic parameters using Gal, GM1, and MUC2 as substrates (Table 1, Figure S13). The Michaelis-Menten constant (Km) of GO-PN (1∼3) was lower than that of GO for each substrate assessed, suggesting higher affinity of GO-PNs for substrates than that of native GO (Murata et al., 2013; Keefe and Jiang, 2012). This might be attributed to either allosteric conformation changes in the GO active site or a more favorable microenvironment for substrate partitioning (Chen and Hoffman, 1993). With polymer elongating, both Gal and GM1 displayed higher enzyme-substrate affinity (smaller values of Km) with the enzyme. As to MUC2, insignificant difference could be observed for GO-PN (1∼3), which might be owing to the relative large size of MUC2 compared with the variation between PN shells. We also observed a decrease in kcat for GO-PN (1∼3) compared to that for GO when using Gal or MUC2 as substrates, suggesting that the maximum number of Gal/GalNAc that could be oxidized at a given GO concentration per minute was reduced (Murata et al., 2013). Interestingly, as to GM1, the kcat for GO-PN1 was even larger than that for GO, probably owing to the hydrophobic interaction between PN1 and the aglycone ceramide (Hunter et al., 2018; Hannun and Obeid, 2018). With regard to kcat/Km (the rate of the reaction) (Murata et al., 2013), the variation trends for substrate Gal and MUC2 were in good agreement with the data in Figure 2C. In toto, we conclude that PN (1∼3) can, indeed, regulate glyco-substrate permeability to afford an activity pattern-based discrimination strategy for substrates with distinct aglycone sterics.
Table 1.
Michaelis-Menton analysis of the catalytic activity of GO and GO-PN (1∼3) towards substrate Gal, GM1 and MUC2 (25°C)
| Substrate | Sample | Km (μM) | kcat × 103 (min−1) | kcat/Km × 103 (μM−1 min−1) |
|---|---|---|---|---|
| Gal | GO | 930 ± 24 | 600 ± 92 | 0.643 ± 0.082 |
| GO-PN1 | 787 ± 6 | 519 ± 93 | 0.659 ± 0.114 | |
| GO-PN2 | 642 ± 121 | 318 ± 146 | 0.469 ± 0.139 | |
| GO-PN3 | 519 ± 24 | 212 ± 56 | 0.405 ± 0.090 | |
| GM1 | GO | 666 ± 66 | 97.1 ± 18.6 | 0.149 ± 0.042 |
| GO-PN1 | 612 ± 87 | 106 ± 15 | 0.175 ± 0.036 | |
| GO-PN2 | 527 ± 78 | 73.7 ± 0.8 | 0.142 ± 0.019 | |
| GO-PN3 | 423 ± 51 | 61.5 ± 8.4 | 0.147 ± 0.025 | |
| MUC2 | GO | 0.819 ± 0.103 | 164 ± 22 | 205 ± 53 |
| GO-PN1 | 0.620 ± 0.090 | 106 ± 12 | 175 ± 43 | |
| GO-PN2 | 0.636 ± 0.112 | 101 ± 8 | 163 ± 38 | |
| GO-PN3 | 0.619 ± 0.097 | 96.5 ± 9.2 | 159 ± 23 |
Screening of protein partner for glycolipid
To demonstrate the application of the aglycone-steric differentiation, we next exploited the activity pattern to screen protein partners for small-sized, galactosylated/GalNAcylated substrate. The basic assumption was that efficient substrate-protein interaction might lead to a substantial aglycone-steric increase (or even Gal site occupation), which would reduce the enzyme accessibility to different extents for different GO-PNs. We chose GM1 as the model substrate, and assessed two types of candidate proteins: 1) cholera toxin B subunit (CTxB), which can bind to the pentasaccharide of GM1 (Merritt et al., 1994), and 2) bovine serum albumin (BSA). We, respectively, added GO, GO-PN (1∼3) to GM1 in the presence or absence of each candidate protein (Figure 3A). After background subtraction, the absorption readout via a Gal oxidase assay was normalized to that for the corresponding control (without the addition of candidate protein), to obtain an AGM1+X/AGM1 ratio (X represents candidate protein, Figure 3B). With polymer length increasing, the AGM1+X/AGM1 for CTxB gradually decreased, whereas BSA caused little change. This phenomenon can be mainly attributed to the steric change in GM1 on CTxB binding. The Gal site occupation by CTxB on GM1 is not a limiting factor in the system, as, for native GO, CTxB-mixed sample still preserved 87% of the AGM1+X/AGM1 ratio for BSA-mixed one. These data demonstrate the application of the activity pattern to differentiate glycolipid and its protein complex, providing a simple approach for Gal/GalNAc-binding protein (e.g., galectins) profiling in solution, which features immobilization-free and label-free operation.
Figure 3.
Screening of protein partners for GM1 using GO-PNs
(A) Schematic illustration. The binding between GM1 and CTxB leads to an obvious increase of aglycone sterics, thus reducing the enzymatic activity to different extents for different GO-PNs.
(B) Quantitative analysis of GO or GO-PN (1∼3) activity (0.0025 mg/mL of equivalent GO concentration) on the addition of CTxB (0.8 μM, red column) or BSA (4 μM, blue column) as relative to that without protein added (represented by AGM1+X/AGM1, X = CTxB or BSA) using 100 μM GM1 as the substrate. Data represent the mean and standard deviation of triplicate experiments. Statistical analysis: t-test (∗∗p < 0.005; ∗∗∗p < 0.0005; NS, not significant).
Aglycone sterics-selective glycan remodeling in the complex cellular system
The ability to achieve aglycone sterics-selective glycan remodeling in the complex physiological environment should enable numerous studies of glycobiology. One of the interesting applications is to discriminate between membrane-anchored and freely diffused glycoconjugates. This is particularly significant for in vivo glycan-targeting diagnosis and therapy, because secreted glycoconjugates, such as most mucus components, should be distinguished from those anchored on the epithelial cell layer. For proof of concept, we designed a model system by mixing mucin 2 (MUC2) protein with MCF-7 cells (Figure 4A). Treatment by GO-PN1 resulted in glycan remodeling and subsequent FTZ labeling on MUC2 rather than MCF-7 cells, while on the contrary, indiscriminate labeling was found for the two substrates when GO was used (Figures 4B, 4C, and S14). These data suggest that the polymer shell on GO can efficiently block cells but allow easily diffused glycoproteins to access GO. The reduced FTZ fluorescence on MUC2 for the GO-PN1 group compared with that for the GO group may be owing to the decrease of catalysis activity by PN1 masking (Figures 4B and 4C). These results demonstrate the feasibility to perform aglycone sterics-selective glycan remodeling in complex, cell-involved biological environments.
Figure 4.
Selective remodeling of MUC2-bound Gal/GalNAc in the presence of MCF-7 cells by GO-PN1
(A) Schematic illustration.
(B) MCF-7/MUC2 (640 μg/mL) mixture was treated with GO or GO-PN1 (0.05 mg/mL of equivalent GO concentration) for 1 h. The cells were then washed, fluorescently labeled with FTZ, and imaged using CLSM (left). The washed buffer containing MUC2 was collected, purified, fluorescently labeled with FTZ, and measured at an excitation wavelength of 488 nm (Right). Scale bar: 20 μm.
(C) Quantitative analysis of the relative activity of GO and GO-PN1 in (B). The activity of GO-PN1 was normalized to the corresponding GO activity as 100% for MCF-7 cells and MUC2, respectively. Data represent the mean and standard deviation of triplicate experiments.
Enhancement of glycolipid remodeling by multi-faceted modulation
Having demonstrated the modulation of aglycone-steric selectivity by polymer length adjustment, we then proceeded to integrate the external-stimuli responsiveness of polymer shell to achieve superimposed regulation of GO activity. Given the thermal sensitivity of PNs (Heredia et al., 2005; Chen and Hoffman, 1993; Stayton et al., 1995; Boyer et al., 2007; De et al., 2008; Mackenzie and Francis, 2013; Gobbo et al., 2018; Wu et al., 2019), we firstly investigated the combined effect of polymer shielding and temperature elevation on the catalytic activity of GO and GO-PN (1∼3) towards GM1 or Gal at 25 or 37°C. For native GO, the elevation of temperature only led to a minimal increase of activity, regardless of substrates used (Figures 5A and 5B). As to GO-PNs, we initially hypothesized that above LCST, the polymer conformation would change from an extended state into a collapsed state, resulting in the formation of a hydrophobic polymer shell and thus stronger blocking (Cummings et al., 2013). Surprisingly, temperature increase (from 25 to 37°C) resulted in an enhancement of catalytic activity towards GM1 for GO-PN (1∼3) (Figure 5A), while negligible influence was observed when using Gal as the substrate (Figure 5B). Most notably, the activity of GO-PN1 towards GM1 was 1.4 times as that of native GO at 37°C. Though GO-PN (1∼3) displayed an inverse correlation between enzymatic activity and PN length towards GM1 owing to polymeric shielding hindrance, the activity of GO-PN3 towards GM1 was still comparable with that of GO at 37°C. One hypothesis for the enhanced GM1 catalytic activity is the lateral hydrophobic-hydrophobic interactions between the collapsed polymer chain and the ceramide tail of GM1, which would enhance the affinity of GM1 in the polymer shell and thus increase the effective substrate concentration in the close proximity to the enzyme (Figure 5C) (Kovaliov et al., 2018). The superimposed modulation of temperature and polymer length represents a 2D mass transportation regulation mechanism for the encapsulated glycoenzyme, resulting in different activity patterns for substrates Gal and glycolipid at high temperatures.
Figure 5.
Enhancement of glycolipid modification by elevating temperature
(A and B) The absorbance for the oxidation product of (A) GM1 and (B) Gal (0.1 mM) under catalysis by GO, GO-Br, and GO-PN (1∼3) (0.0025 mg/mL of equivalent GO concentration) at 25 or 37°C, respectively.
(C) Schematic illustration for the enhancement of GO activity towards GM1 by PN grafting and temperature elevation. Data represent the mean and standard deviation of triplicate experiments.
Temporal control of glycan remodeling on live cells
Finally, we tested the feasibility of switching aglycone-steric selectivity by orchestrating an external chemical stimulus (Figure 6A). Installation of disulfide bonds between PN and GO enables controllable cleavage of the PN shell from GO using a reducing reagent. We thus designed an off-on switch experiment for live-cell glycan remodeling. We chose TCEP of 5 mM to cleave the S–S bond because at this concentration TCEP exerted indiscernible influence on GO (or GO-PN1) activity (Figure S15) and monosaccharide specificity (Figure S16), and displayed minimized cytotoxicity (Figure S17) (Sun et al., 2019b). The cleavage facilitates the release of PN1 and thus the access of cell-surface Gal/GalNAc to the GO catalytic site. Galactoaldehydes were then visualized by FTZ labeling and CLSM. As expected, simultaneous administration of GO-PN1 and TCEP resulted in robust fluorescence labeling at cell periphery, in a time-dependent manner (Figures 6B, 6C, S18, and S19). In addition, we excluded the undesired influence from extracellular glutathione (GSH) (Figure S20), which also demonstrated the stability of GO-PN1 in a complex environment (Cheng et al., 2011). These data demonstrate the feasibility of using the proposed multi-faceted modulation platform to accomplish temporal control of cellular glycan remodeling.
Figure 6.
TCEP-triggered Gal/GalNAc remodeling and labeling on live cells
(A) Schematic illustration.
(B) CLSM images of MCF-7 cells treated with GO-PN1 (0.05 mg/mL of equivalent GO concentration) in the presence and absence of TCEP (5 mM) for 30, 40, 50, and 60 min. The treated cells were washed and fluorescently labeled by FTZ. Scale bar: 20 μm.
(C) Quantitative analysis of GO-PN1 activity with or without TCEP. Relative activity: MFI of cellular periphery from (B) divided by MFI for GO treated cells (Figure S19). Data represent the mean and standard deviation of triplicate experiments.
Discussion
In summary, we have endowed glycoenzyme with aglycone-steric selectivity by wrapping up enzyme with a polymer-based functional shell. We proved that the polymer shell acts as a tunable and switchable permeation hinder on the GO surface. By adjusting polymer length, we found different activity patterns for distinct aglycone sterics using a series of GO-PNs and GO, thus affording a label-free strategy for the differentiation of aglycone sterics for the first time. We demonstrated the utility of this activity pattern by differentiating glycolipid-protein complex from glycolipid itself and selective remodeling and then labeling of freely diffused MUC2 in the presence of live cells. The multi-functional (thermal-sensitive and cleavable) polymer shell also enabled us to perform multiple regulation of aglycone-steric selectivity, resulting in enhanced glycolipid remodeling activity and temporal control of glycan remodeling on live cells. This proof-of-principle study would serve as an experimental paradigm to guide the selectivity regulation of other types of glycan-modifying enzymes, such as glycosyltransfereases and glycosidases. Also, by virtue of the advancements in responsive polymer design and synthesis (Sun et al., 2019a), the strategy can be readily adapted to other stimuli-responsive versions, such as using pH, light, or electromagnetic field as the trigger, for diverse application scenarios (Kang et al., 2022; Xie et al., 2022). Notably, the switchable glycan remodeling on the cell surface would be further combined with targeted delivery techniques (Li et al., 2021) to achieve tumor-specific, in vivo glycan remodeling, which may obviate the side effects from glycan remodeling on non-targeted cells/tissues. We thus believe this work provides an elegant yet powerful strategy for the modulation of glycan remodeling and will contribute to the development of glycan-targeting analytical, synthesis, and research tools as well as therapeutic techniques.
Limitations of the study
In this study, the prepared GO-PNs smeared on the SDS-PAGE gel, which suggested that the grafting of PN onto GO did not proceed uniformly. This might be attributed to the random conjugation of different amounts of the initiator to the amine groups of GO. The inability to obtain composites with definite conjugation sites and thus polymer-to-enzyme ratio hinders the precise regulation of enzyme accessibility and activity to substrates of different aglycone sterics. Thus, the main limitation of this study is the lack of modification site specificity. In future research, we aim to combine the site-specific protein labeling techniques with click chemistry to precisely install the initiator on a specific site of the enzyme and investigate the influence of the modification site as well as polymer composition and length on the distinguishing capability of glycoenzymes toward aglycone sterics.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Bis[2-(2′-bromoisobutyryloxy)ethyl]disulfide (BiBOEDS) | Sigma-Aldrich | Cat# 723169 |
| Dimethyl sulfoxide | Sigma-Aldrich | Cat# D4540 |
| Tris(2-carboxyethyl)phosphine hydrochloride | Sigma-Aldrich | Cat# 646547 |
| Sinapic acid (SA) | Sigma-Aldrich | Cat# D7927 |
| D-(+)-galactose (Gal) | Sigma-Aldrich | Cat# G0750 |
| N-acetyl-D-(+)-galactosamine (GalNAc) | Sigma-Aldrich | Cat# A2795 |
| D-(+)-glucose (Glc) | Sigma-Aldrich | Cat# G8270 |
| D-(+)-mannose (Man) | Sigma-Aldrich | Cat# M2069 |
| N-acetyl-neuraminic acid (Sia) | Sigma-Aldrich | Cat# A0812 |
| Sucrose (Suc) | Sigma-Aldrich | Cat# V900116 |
| D-(−)-ribose (Rib) | Sigma-Aldrich | Cat# V900389 |
| D-(+)-xylose (Xyl) | Sigma-Aldrich | Cat# X1500 |
| D-(−)-fructose (Fru) | Sigma-Aldrich | Cat# F0127 |
| Fluorescein-5-thiosemicarbazide (FTZ) | Sigma-Aldrich | Cat# 46985 |
| Mucin 2 (MUC2) | Sigma-Aldrich | Cat# M2378 |
| Bovine serum albumin (BSA) | Sigma-Aldrich | Cat# V900933 |
| Fetuin | Sigma-Aldrich | Cat# MB3121 |
| N-Succinimidyl3-(2-pyridyldithio)propionate (SPDP) | Aladdin | Cat# N159714 |
| Methanol (MeOH) | Aladdin | Cat# M116118 |
| N-isopropylacrylamide (NIPAm) | Aladdin | Cat# I106818 |
| N,N,N′,N″,N″-pentamethyldiethylenetriamine (PMDETA) | Aladdin | Cat# P106715 |
| Glutathione (GSH) | Aladdin | Cat# G105427 |
| Tetrahydrofuran (THF) | Aladdin | Cat# T103263 |
| Acetonitrile | Aladdin | Cat# A104440 |
| NuPAGETM LDS sample buffer (4×) | Thermo Fisher Scientific | Cat# NP0008 |
| Fetal bovine serum (FBS) | Thermo Fisher Scientific | Cat# 10099141 |
| CuCl2⋅2H2O | Rhawn Reagent | Cat# R019783 |
| Potassium ferrocyanide (K4[Fe(CN)6]) | Macklin | Cat# P871953 |
| Monosialotetrahexosylganglioside (GM1) | Macklin | Cat# G873919 |
| Cholera toxin B subunit (CTxB) | Absin Bioscience | Cat# abs80001 |
| Trifluoroacetic acid | TCI (Shanghai) Development | Cat# A5711 |
| α2-3,6,8,9 Neuraminidase A (NEU) | New England Biolabs | Cat# P0722L |
| Sodium hydroxide (NaOH) | Nanjing Chemical Reagent | Cat# 82001 |
| L-ascorbic acid (Vc) | Sangon Biotech | Cat# A610021 |
| Tris(hydroxymethyl)aminomethane hydrochloride (Tris-HCl) | Sangon Biotech | Cat# B548122 |
| Tris-glycine | Sangon Biotech | Cat# B040129 |
| Galactose oxidase (GO) | Sangon Biotech | Cat# A004520 |
| Sodium dodecyl sulfate (SDS) | Sangon Biotech | Cat# C520001 |
| Coomassie brilliant blue | Sangon Biotech | Cat# A100472 |
| Color-prestained protein Mw marker | Sangon Biotech | Cat# B300002 |
| Goat serum | Sangon Biotech | Cat# E510009 |
| Phosphate buffer saline (PBS, pH 7.4) | KeyGen Biotech | Cat# KGB5001 |
| RPMI-1640 medium | KeyGen Biotech | Cat# KGM31800 |
| Dulbecco’s modified Eagle’s medium (DMEM) | KeyGen Biotech | Cat# KGM12800 |
| Trypsin | KeyGen Biotech | Cat# KGF023 |
| Critical commercial assays | ||
| Bicin-choninic acid (BCA) Protein Assay Kit | Cowin Biotech | Cat# CW0014S |
| AmplexTM Red Galactose/Galactose Oxidase Assay Kit | Thermo Fisher Scientific | Cat# A22179 |
| 10% TGX Stain-FreeTM FastCastTM Acrylamide Kit | Bio-Rad | Cat# 1610183 |
| Cell Counting Kit-8 (CCK8) | KeyGen Biotech | Cat# KGA317 |
| Experimental models: Cell lines | ||
| Human: MCF-7 cells | KeyGen Biotech | Cat# KG031 |
| Human: HeLa cells | KeyGen Biotech | Cat# KG042 |
| Software and algorithms | ||
| Leica Application Suite Advanced Fluorescent | Leica | https://www.leica-microsystems.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lin Ding (dinglin@nju.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model
Cell lines and cell culture
MCF-7 cells were cultured in RPMI-1640 medium supplemented with streptomycin (0.1 mg/mL), penicillin (0.1 mg/mL) and 10% FBS (v/v). HeLa cells were cultured in DMEM supplemented with streptomycin (0.1 mg/mL), penicillin (0.1 mg/mL) and 10% FBS (v/v). Cells were seeded in a four-well confocal dish (1×104 per well) and cultured overnight in an incubator at 37°C containing 5% CO2, and the cell number was measured using a Countess II Automated Cell Counter (Thermo Fisher).
Method details
Preparation of GO-PNIPAm composites (GO-PNs)
GO (2 mg/mL) dissolved in PBS (pH 8.0), and SPDP (100 mM) dissolved in DMSO were prepared respectively. After adding 20 μL of SPDP solution into 2 mL GO solution, the mixture was gently shaken for 2 h at room temperature, followed by purification with an ultrafiltration tube (30 kDa MWCO, Merck Millipore). The as-prepared GO-SPDP was diluted with PBS (pH 8.0) to 1.8 mg/mL (of equivalent GO concentration), and allowed to react with TCEP solution (60 mM) for 30 min to generate thiol-modified GO (GO-SH). After ultrafiltration with PBS (pH 8.0), 400 μL BiBOEDS (10 mM) was added dropwise to GO-SH (1 mL, 2.7 mg/mL) and gently shaken for 2 h. The crude product was dialyzed against PBS for 24 h at 4°C using a dialysis tube (3500 Da MWCO, Yuanye Biotech) to obtain GO-Br.
NIPAm (200 mg), PMDETA (1.2 mg), CuCl2⋅2H2O (1.2 mg) were dissolved with H2O (2 mL) and methanol (MeOH) (1 mL) (Wu et al., 2019) to obtain a blue monomer solution in a Schlenk flask at NIPAm/PMDETA/CuCl2 = 255/1/1 (molar ratio). The flask was sealed carefully, frozen with liquid nitrogen, and deoxygenated with nitrogen for 15 min 200 μL Vc (3.1 mg/mL for Entries 4∼6; 3.7 mg/mL for Entries 7∼9; 4.3 mg/mL for Entries 10∼12; 6.2 mg/mL for Entries 1∼3 and 16∼18; 9.3 mg/mL for Entries 13∼15, Tables S1 and S2) was then added into the thawed monomer solution under nitrogen atmosphere, deoxygenated for 15 min, and mixed with 1 mL deoxygenated GO-Br solution (2 mg/mL). After another freeze-deoxygenation-thaw cycle, the Schlenk flask was placed on a magnetic stirrer at room temperature for different periods of time (solution color changed into light green). The reaction was quenched by exposing to the atmosphere (solution color turned blue). GO-PN (1∼3) (Entries 1∼3, Table S1) were prepared using Vc (6.2 mg/mL), with polymerization time for 1, 2, 3 h, respectively. The crude products were then ultrafiltrated (100 kDa MWCO, Merck Millipore) with PBS at 4°C to generate GO-PNs. To demonstrate the dependence of polymerization on the presence of exogenous CuII, Vc and PMDETA, several control synthesis experiments were performed with polymerization time for 3 and 12 h, followed by ultrafiltration (30 kDa MWCO).
GO activity calibration curve
According to AmplexTM Red Galactose/Galactose Oxidase Assay Kit protocol, GO catalyzes the oxidation of Gal at the C6 position to produce hydrogen peroxide (H2O2). H2O2 reacts with Amplex Red reagent under catalysis by horseradish peroxidase (HRP) to produce resorufin, a red fluorescent oxidation product. Stock solutions including 200 U/mL GO, 20 mM Gal, 10 mM Amplex Red reagent, and 100 U/mL HRP were firstly prepared. 50 μL of Amplex Red, 10 μL of HRP, 50 μL of Gal and 4.89 mL of 1× reaction buffer (AmplexTM Red Galactose/Galactose Oxidase Assay Kit, Thermo Fisher Scientific Inc.) were mixed to prepare the 2× working solution buffer. GO solutions with various concentrations in 1× reaction buffer were added to a 96-well plate (50 μL per well), followed by addition of 50 μL of 2× working solution buffer. The mixture was incubated at room temperature for 1 h, and then analyzed on a multiskan FC microplate photometer (Thermo Fisher Scientific). The GO activity calibration curve was obtained by recording the absorbance value at 560 nm.
Activity determination of GO and GO-PNs
To investigate the effects from PN preparation process on GO activity, GO, GO-SPDP, GO-SPDP + TCEP (This refers to 0.005 mg/mL GO-SPDP solution after co-incubation with 5 mM TCEP for 1 h and then removal of TCEP, hereafter), GO-Br, GO-PN1 and GO-PN1+TCEP (This refers to 0.005 mg/mL GO-PN1 solution after co-incubation with 5 mM TCEP for 1 h and then removal of TCEP, hereafter) were diluted to 0.005 mg/mL of equivalent GO concentration using 1× reaction buffer, respectively. 50 μL of each sample was transferred into 96-well plate, mixed with 50 μL of 2× working solution buffer. After reaction at room temperature for 1 h, the absorbance value was recorded.
To determine the activity of GO-PN1 towards different monosaccharides, different 2× working solution buffer counterparts were prepared by mixing 50 μL of Amplex Red reagent, 10 μL of HRP, 4.89 mL of 1× reaction buffer with 50 μL of different monosaccharides (Gal, GalNAc, Glc, Man, Sia, Suc, Rib, Xyl and Fru, 20 mM). Then 50 μL of GO-PN1 (0.005 mg/mL of equivalent GO concentration) with and without TCEP (5 mM TCEP) in 1× reaction buffer were transferred into 96-well plate, followed by addition of 50 μL of different 2× working solution buffer counterparts. After reaction at room temperature for 1 h, the absorbance value was recorded.
To evaluate the activity of PN (1∼3) towards Gal, 50 μL of GO, PN1, PN2 or PN3 in 1× reaction buffer (0.005 mg/mL) was transferred into 96-well plate respectively, and then mixed with 50 μL of 2× working solution buffer. After reaction at room temperature for 1 h, the absorbance value was recorded.
To obtain time course of absorbance for Gal oxidation under catalysis by GO or GO-PN (1∼3), 50 μL of GO or GO-PN (1∼3) (0.005 mg/mL of equivalent GO concentration) in 1× reaction buffer was added into 96-well plate respectively, and then mixed with 50 μL of 2× working solution buffer. The absorbance value was recorded for 1 h at 10 min intervals.
Cellular viability assay
According to the protocol of Cell Counting Kit-8, MCF-7 cells (1×104, 100 μL per well) were cultured in 96-well plates for 24 h. After washing and blocking, the cells were subjected to incubation with 100 μL of GO, PN1 (0.05 mg/mL), GO-PN1, and the mixture of GO-PN1 and TCEP (GO or GO-PN1: 0.05 mg/mL of equivalent GO concentration; TCEP: 5 mM), respectively, at 4°C for 1 h. After washing, the cells were allowed to incubate with RPMI-1640 medium containing 10% FBS and 10% CCK8 at 37°C for 4 h. Then the absorption at 450 nm was measured, and the cell viability (%) was calculated based on the equation: cell viability (%) = (average OD value of treated cells/average OD value of control cells) × 100%.
To investigate the effect of TCEP on cellular viability, according to the protocol of Cell Counting Kit-8, MCF-7 cells (1×104, 100 μL per well) were cultured in 96-well plates for 24 h. After washing with PBS, the cells were incubated with 100 μL TCEP at different concentrations (1, 2.5, 5, 7.5, 10 mM) at 4°C for 1 h, then rinsed with PBS again. RPMI-1640 medium containing 10% FBS and 10% CCK8 was then added into each well and allowed to incubate at 37°C for 4 h. The absorption at 450 nm was measured, and the cell viability (%) was calculated as mentioned above. The control cells were those without TCEP incubation.
GO-PN remodeling activity assay
To modulate GO remodeling activity on live cells, a series of GO-PN composites were synthesized with different conditions, including five feed molar ratios and various polymerization time. MCF-7 or HeLa cells were washed with PBS for three times, and blocked using PBS containing 10% goat serum at 37°C for 30 min. The cells were then washed three times with PBS, and incubated with GO or different GO-PNs (0.05 mg/mL of equivalent GO concentration) at 4°C for 1 h. After gentle washing with PBS for three times, the cells were subjected to reaction with 100 μL PBS containing 10 mM aniline, 100 μM FTZ and 5% fetal bovine serum (FBS) at 4°C for 1 h, followed by PBS washing for three times. Then the cells were imaged with an SP8 STED confocal laser scanning microscopy (Leica). All images were analyzed by Leica Application Suite Advanced Fluorescent (LAS AF) software.
For freely diffused substrates, different 2× working solution buffers (5 mL) were prepared by mixing 50 μL of Amplex Red reagent, 10 μL of HRP, and different substrates (50 μL 20 mM Gal, 100 μL 10 mM GM1, 160 μL 20 mg/mL MUC2, or 160 μL 20 mg/mL desialylated fetuin) in 1× reaction buffer (AmplexTM Red Galactose/Galactose Oxidase Assay Kit, Thermo Fisher Scientific Inc.). 50 μL of GO or GO-PN (1∼3) (0.005 mg/mL of equivalent GO concentration) in 1× reaction buffer was transferred into 96-well plate, followed by addition of 50 μL of different 2× working solution buffers. After reaction at room temperature for 1 h, the absorbance value was recorded.
Determination of michaelis-menten parameters
Different 2× working solution buffer counterparts (1 mL) were prepared by mixing 10 μL of Amplex Red reagent, 2 μL of HRP, and different samples (GO or GO-PN (1∼3), 100 μL, 0.05 mg/mL of equivalent GO concentration) in 1× reaction buffer. To each substrate solution (Gal, GM1 or MUC2, 50 μL) with a given concentration, 50 μL of different 2× working solution buffer counterparts were respectively added. The concentration series for substrates were as follows: Gal (μM), 100, 200, 300, 400, 500; GM1 (μM), 400, 500, 600, 700, 800; MUC2 (μg/mL), 400, 500, 600, 700, 800. After reaction at room temperature for 30 min, the absorbance value was recorded.
Glycolipid binding partner screening
Different 2× working solution buffer counterparts (1 mL) were prepared by mixing 10 μL of Amplex Red reagent, 2 μL of HRP, and different samples (GO or GO-PN (1∼3), 100 μL, 0.05 mg/mL of equivalent GO concentration) in 1× reaction buffer. PBS containing GM1 (200 μM) and CTxB (1.6 μM) was incubated at room temperature for 1 h. A control sample was also prepared by incubating GM1 (200 μM) and BSA (8 μM) in PBS at room temperature for 1 h. The above-mentioned samples (50 μL) were individually added into 96-well plate, and then mixed with 50 μL of different 2× working solution buffer counterparts. After reaction at room temperature for 1 h, the absorbance value was recorded. A five-fold concentration of BSA compared to CTxB was added to the system due to the assumption that each CTxB has five subunits for binding with GM1.
Selective glycan remodeling in a mixture of MUC2 and cells
After washing and blocking as mentioned above, the MCF-7 cells were subjected to incubation with the mixture of GO (or GO-PN1, 0.05 mg/mL of equivalent GO concentration) and MUC2 (640 μg/mL) at 4°C for 1 h. The supernatants and cells were both collected. The supernatants were centrifuged at 4°C at 10,000 rpm for 6 min to remove suspended cells, and were subjected to reaction with PBS containing 10 mM aniline, 100 μM FTZ, and 100 mM K4[Fe(CN)6] (to preclude further Gal/GalNAc remodeling during this labeling step) at 4°C for 1 h. After ultrafiltration (100 kDa MWCO) to remove small molecules, samples were dialyzed against PBS for 24 h at 4°C using a dialysis tube (300 kDa MWCO) to remove GO (or GO-PN1). The obtained MUC2 was diluted to 0.1 mg/mL and subjected to fluorescence detection (F-7100 fluorescence spectrophotometer, Hitachi) under excitation wavelength at 488 nm. The cells were gently washed with PBS for three times, and subjected to reaction with 100 μL PBS containing 10 mM aniline, 100 μM FTZ, and 5% FBS at 4 oC for 1 h. After washing with PBS for three times, the cells were imaged with an SP8 CLSM.
Enhancement of GO-PN activity towards GM1 by raising temperature
GO, GO-Br, GO-PN1, GO-PN2 and GO-PN3 (0.005 mg/mL of equivalent GO concentration) were diluted in 1× reaction buffer, respectively. Each sample of 50 μL was added into 96-well plate, and then mixed with 50 μL of different 2× working solution buffer containing 100 μM Amplex Red reagent, 200 mU/mL HRP, and 200 μM Gal (or GM1). After reaction at 25 or 37°C for 1 h, the absorbance value was recorded.
Investigation of the effect of TCEP cleavage time on cell surface glycan remodeling
After washing and blocking, MCF-7 cells were subjected to incubation with GO (0.05 mg/mL), GO-PN1 (0.05 mg/mL of equivalent GO concentration), or the mixture of GO-PN1 (0.05 mg/mL of equivalent GO concentration) and TCEP (5 mM) at 4°C for different periods of time (30, 40, 50, 60 min). After washing with PBS for three times, the cells were subjected to reaction with 100 μL PBS containing 10 mM aniline, 100 μM FTZ, and 5% FBS at 4°C for 1 h. After washing with PBS for three times, the cells were imaged with an SP8 CLSM.
TCEP-triggered glycan remodeling on live cells
After washing and blocking, MCF-7 cells were subjected to incubation with GO (0.05 mg/mL) in the presence of TCEP at different concentrations (1, 5, 10, 15, 20, 25 mM) at 4°C for 1 h. After washing with PBS for three times, the cells were subjected to reaction with 100 μL PBS containing 10 mM aniline, 100 μM FTZ, and 5% FBS at 4°C for 1 h. After washing with PBS for three times, the cells were imaged with an SP8 CLSM.
To exclude the influence of extracellular GSH on glycan remodeling on live cells, after washing and blocking, MCF-7 cells were respectively subjected to incubation with GO, the mixture of GO-PN1 and TCEP, and the mixture of GO-PN1 and GSH (GO or GO-PN1: 0.05 mg/mL of equivalent GO concentration; TCEP: 5 mM; GSH: 20 μM) at 4°C for 1 h. After washing with PBS for three times, the cells were subjected to reaction with 100 μL PBS containing 10 mM aniline, 100 μM FTZ, and 5% FBS at 4°C for 1 h. After washing with PBS for three times, the cells were imaged with an SP8 CLSM.
Characterization techniques
MALDI-TOF MS characterization
GO, GO-SPDP and GO-Br were firstly desalted by ultrafiltration with water (30 kDa MWCO). The matrix, saturated SA solution, was prepared using a solvent containing 50% ultrapure water (Millipore Simplicity System, Bedford), 50% acetonitrile and 0.1% trifluoroacetic acid. 1 μL of matrix solution and 1 μL of sample solution (5.6 mg/mL for GO, 6.5 mg/mL for GO-SPDP, and 4.6 mg/mL for GO-Br) were successively applied onto the MALDI target plate. After solvent evaporation, another 1 μL of matrix solution was spotted onto the plate and allowed to dry at 37°C. Samples were then analyzed by MALDI-TOF mass spectrometer (MALDI-7090 MALDI TOF mass spectrometer, Shimadzu).
SDS-PAGE characterization
Samples dissolved in PBS with equivalent GO concentration of 0.67 mg/mL were shaken at room temperature for 1 h, respectively. Then each sample solution was diluted with LDS sample buffer to 0.5 mg/mL of equivalent GO concentration. 10 μL of the mixture was loaded into hand casting 10% TGX Stain-Free polyacrylamide gels for electrophoresis separation using a standard Tris-glycine running buffer system (Electrophoresis Analyzer, Bio-Rad) at 200 V for 40 min. Gels were stained with Coomassie brilliant blue at room temperature for 30 min, decolorized with decolorizing solutions, and imaged on a Bio-Rad ChemDoc XRS facility.
GPC characterization
The prepared GO-PN (1∼3) were further purified by heating at 40°C for 5 min, and centrifuged at 40°C at 10,000 rpm for 6 min. The resulting GO-PN pellet was resuspended with water to wash, and then heated and recentrifuged as above for three times. Each type of GO-PNs (0.9 mg/mL of equivalent GO concentration) was mixed with TCEP (60 mM), and shaken at room temperature for 1 h to cleave the PNs. The mixture was incubated at 40°C for 5 min and centrifuged at 40°C at 10,000 rpm for 6 min to remove the GO in the supernatant. The resulting PN pellet was resuspended with water, heated and recentrifuged as above for three times, and then lyophilized. The PNs were redissolved in THF (10 mg/mL), filtered with 0.22 μm filter, and the Mn, Mw, and PDI were measured on Waters gel permeation chromatography (GPC) system at a flow rate of 1.0 mL/min. The GPC is equipped with a Waters 1515 HPLC pump, a Waters 2414 refractive index detector, and a tandem column system with Waters Styragel HR 2, Styragel HR 4 and Styragel HR 5 columns.
LCST curve measurement
GO (0.1 mg/mL), PN (1∼3) (1 mg/mL) and GO-PN (1∼3) (of equivalent GO concentration of 0.1 mg/mL) were dissolved in PBS, respectively. LCST curves were measured at 490 nm by a UV-VIS spectrophotometer (Cary 3500, Agilent). Samples were heated from 25 to 45°C at 1°C/min. The LCST was defined as the temperature at which the transmittance of the solution was 50% of the maximum.
UV-vis characterization
PN (1∼3), GO, GO-SPDP, GO-Br, GO-PN (1∼3) and mixtures of GO-PN (1∼3) and TCEP were shaken at room temperature for 1 h, respectively. GO and its conjugates were dissolved with an equivalent GO concentration of 0.2 mg/mL, and the concentrations for PN (1∼3) and TCEP were 0.2 mg/mL and 5 mM, respectively. The absorbance of each sample was measured from 200 to 600 nm by a UV-VIS-NIR spectrophotometer (UV-3600, Shimadzu).
CD characterization
GO, GO-SPDP, GO-Br and GO-PN (1∼3) (of equivalent GO concentration of 0.5 mg/mL) were respectively dissolved in 20 mM Tris-HCl for CD measurement from 180 to 260 nm using Applied Photophysics Circular Dichroism (Applied Photophysics).
Quantification and statistical analysis
All data were representative results from at least three independent experiments and presented as mean ± standard deviation. The Student’s t test was used for statistical analysis. ∗∗p < 0.005, and ∗∗∗p < 0.0005 were considered statistically significant, while p ≥ 0.05 was considered statistically insignificant (NS).
Acknowledgments
We gratefully acknowledge support from the National Natural Science Foundation of China (21974067, 21708019, 2207070006), the National Key Research and Development Program of China (2018YFC1004704), Fundamental Research Funds for the Central Universities (021414380502), and Programs for High-Level Entrepreneurial and Innovative Talents Introduction of Jiangsu Province (Individual and Group Program).
Author contributions
L.D. and A.M. conceived the project. L.D., R.X., H.J., A.M., and X.W. designed the experiments. A.M., Y.Z., G.W., T.Z., X.C., and X.W. conducted the experiments and data analysis. L.D., A.M., R.X., and H.W. co-wrote the article.
Declaration of interests
The authors declare no competing interests.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104578.
Contributor Information
Ran Xie, Email: ranxie@nju.edu.cn.
Lin Ding, Email: dinglin@nju.edu.cn.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Averick S., Simakova A., Park S., Konkolewicz D., Magenau A.J.D., Mehl R.A., Matyjaszewski K. ATRP under biologically relevant conditions: grafting from a protein. ACS Macro Lett. 2012;1:6–10. doi: 10.1021/mz200020c. [DOI] [PubMed] [Google Scholar]
- Baker S.L., Munasinghe A., Kaupbayeva B., Rebecca Kang N., Certiat M., Murata H., Matyjaszewski K., Lin P., Colina C.M., Russell A.J. Transforming protein-polymer conjugate purification by tuning protein solubility. Nat. Commun. 2019;10:4718. doi: 10.1038/s41467-019-12612-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belardi B., de la Zerda A., Spiciarich D.R., Maund S.L., Peehl D.M., Bertozzi C.R. Imaging the glycosylation state of cell surface glycoproteins by two-photon fluorescence lifetime imaging microscopy. Angew. Chem. Int. Ed. 2013;52:14045–14049. doi: 10.1002/anie.201307512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontempo D., Maynard H.D. Streptavidin as a macroinitiator for polymerization: in situ Protein−Polymer conjugate formation. J. Am. Chem. Soc. 2005;127:6508–6509. doi: 10.1021/ja042230+. [DOI] [PubMed] [Google Scholar]
- Boyer C., Bulmus V., Liu J., Davis T.P., Stenzel M.H., Barner-Kowollik C. Well-defined protein-polymer conjugates via in situ RAFT polymerization. J. Am. Chem. Soc. 2007;129:7145–7154. doi: 10.1021/ja070956a. [DOI] [PubMed] [Google Scholar]
- Boyer C., Corrigan N.A., Jung K., Nguyen D., Nguyen T.K., Adnan N.N.M., Oliver S., Shanmugam S., Yeow J. Copper-mediated living radical polymerization (atom transfer radical polymerization and copper(0) mediated polymerization): from fundamentals to bioapplications. Chem. Rev. 2016;116:1803–1949. doi: 10.1021/acs.chemrev.5b00396. [DOI] [PubMed] [Google Scholar]
- Chaubard J.L., Krishnamurthy C., Yi W., Smith D.F., Hsieh-Wilson L.C. Chemoenzymatic probes for detecting and imaging fucose-α(1-2)-galactose glycan biomarkers. J. Am. Chem. Soc. 2012;134:4489–4492. doi: 10.1021/ja211312u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Liu D., Ji N., Tan Z., Zong G., Qu R., Wang C. Samarium(III)-based AGET ATRP of acrylonitrile using ascorbic acid as reducing agent. J. Macromol. Sci. 2011;48:284–290. doi: 10.1080/10601325.2011.552345. [DOI] [Google Scholar]
- Chen G., Hoffman A.S. Preparation and properties of thermoreversible, phase-separating enzyme-oligo(N-isopropylacrylamide) conjugates. Bioconjug. Chem. 1993;4:509–514. doi: 10.1021/bc00024a013. [DOI] [PubMed] [Google Scholar]
- Cheng B., Tang Q., Zhang C., Chen X. Glycan labeling and analysis in cells and in vivo. Annu. Rev. Anal. Chem. 2021;14:363–387. doi: 10.1146/annurev-anchem-091620-091314. [DOI] [PubMed] [Google Scholar]
- Cheng R., Feng F., Meng F., Deng C., Feijen J., Zhong Z. Glutathione-responsive nano-vehicles as a promising platform for targeted intracellular drug and gene delivery. J. Control. Release. 2011;152:2–12. doi: 10.1016/j.jconrel.2011.01.030. [DOI] [PubMed] [Google Scholar]
- Cummings C., Murata H., Koepsel R., Russell A.J. Tailoring enzyme activity and stability using polymer-based protein engineering. Biomaterials. 2013;34:7437–7443. doi: 10.1016/j.biomaterials.2013.06.027. [DOI] [PubMed] [Google Scholar]
- De P., Li M., Gondi S.R., Sumerlin B.S. Temperature-regulated activity of responsive polymer-protein conjugates prepared by grafting-from via RAFT polymerization. J. Am. Chem. Soc. 2008;130:11288–11289. doi: 10.1021/ja804495v. [DOI] [PubMed] [Google Scholar]
- Flynn R.A., Pedram K., Malaker S.A., Batista P.J., Smith B.A.H., Johnson A.G., George B.M., Majzoub K., Villalta P.W., Carette J.E., Bertozzi C.R. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell. 2021;184:3109–3124.e22. doi: 10.1016/j.cell.2021.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Ramirez D.H., Yang B., D'Souza A.K., Aonbangkhen C., Wong S., Woo C.M. Target protein deglycosylation in living cells by a nanobody-fused split O-GlcNAcase. Nat. Chem. Biol. 2021;17:593–600. doi: 10.1038/s41589-021-00757-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbo P., Patil A.J., Li M., Harniman R., Briscoe W.H., Mann S. Programmed assembly of synthetic protocells into thermoresponsive prototissues. Nat. Mater. 2018;17:1145–1153. doi: 10.1038/s41563-018-0183-5. [DOI] [PubMed] [Google Scholar]
- Gray M.A., Stanczak M.A., Mantuano N.R., Xiao H., Pijnenborg J.F.A., Malaker S.A., Miller C.L., Weidenbacher P.A., Tanzo J.T., Ahn G., et al. Targeted glycan degradation potentiates the anticancer immune response in vivo. Nat. Chem. Biol. 2020;16:1376–1384. doi: 10.1038/s41589-020-0622-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Tao J., Li Y., Feng Y., Ju H., Wang Z., Ding L. Quantitative localized analysis reveals distinct exosomal protein-specific glycosignatures: implications in cancer cell subtyping, exosome biogenesis, and function. J. Am. Chem. Soc. 2020;142:7404–7412. doi: 10.1021/jacs.9b12182. [DOI] [PubMed] [Google Scholar]
- Hannun Y.A., Obeid L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018;19:175–191. doi: 10.1038/nrm.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J.M., Chess R.B. Effect of pegylation on pharmaceuticals. Nat. Rev. Drug Discov. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- Hayakawa S., Matsushita T., Yokoi Y., Wakui H., Garcia-Martin F., Hinou H., Matsuoka K., Nouso K., Kamiyama T., Taketomi A., Nishimura S.I. Impaired O-glycosylation at consecutive threonine TTX motifs in mucins generates conformationally restricted cancer neoepitopes. Biochemistry. 2020;59:1221–1241. doi: 10.1021/acs.biochem.0c00007. [DOI] [PubMed] [Google Scholar]
- Heredia K.L., Bontempo D., Ly T., Byers J.T., Halstenberg S., Maynard H.D. In situ preparation of Protein−"Smart" polymer conjugates with retention of bioactivity. J. Am. Chem. Soc. 2005;127:16955–16960. doi: 10.1021/ja054482w. [DOI] [PubMed] [Google Scholar]
- Hong S., Shi Y., Wu N.C., Grande G., Douthit L., Wang H., Zhou W., Sharpless K.B., Wilson I.A., Xie J., Wu P. Bacterial glycosyltransferase-mediated cell-surface chemoenzymatic glycan modification. Nat. Commun. 2019;10:1799. doi: 10.1038/s41467-019-09608-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui J., Bao L., Li S., Zhang Y., Feng Y., Ding L., Ju H. Localized chemical remodeling for live cell imaging of protein-specific glycoform. Angew. Chem. Int. Ed. 2017;56:8139–8143. doi: 10.1002/anie.201703406. [DOI] [PubMed] [Google Scholar]
- Hunter C.D., Guo T.L., Daskhan G., Richards M.R., Cairo C.W. Synthetic strategies for modified glycosphingolipids and their design as probes. Chem. Rev. 2018;118:8188–8241. doi: 10.1021/acs.chemrev.8b00070. [DOI] [PubMed] [Google Scholar]
- Kang Z., Wang C., Zhang Z., Liu Q., Zheng Y., Zhao Y., Pan Z., Li Q., Shi L., Liu Y. Spatial distribution control of antimicrobial peptides through a novel polymeric carrier for safe and efficient cancer treatment. Adv. Mater. 2022;34:e2201945. doi: 10.1002/adma.202201945. [DOI] [PubMed] [Google Scholar]
- Kaupbayeva B., Murata H., Wilson A.L., Matyjaszewski K., Minden J.S., Russell A.J. Molecular sieving on the surface of a nano-armored protein. Biomacromolecules. 2019;20:1235–1245. doi: 10.1021/acs.biomac.8b01651. [DOI] [PubMed] [Google Scholar]
- Kaupbayeva B., Russell A.J. Polymer-enhanced biomacromolecules. Prog. Polym. Sci. 2020;101:101194. doi: 10.1016/j.progpolymsci.2019.101194. [DOI] [Google Scholar]
- Kaupbayeva B., Boye S., Munasinghe A., Murata H., Matyjaszewski K., Lederer A., Colina C.M., Russell A.J. Molecular dynamics-guided design of a functional protein-ATRP conjugate that eliminates protein-protein interactions. Bioconjug. Chem. 2021;32:821–832. doi: 10.1021/acs.bioconjchem.1c00098. [DOI] [PubMed] [Google Scholar]
- Keefe A.J., Jiang S. Poly(zwitterionic)protein conjugates offer increased stability without sacrificing binding affinity or bioactivity. Nat. Chem. 2012;4:59–63. doi: 10.1038/nchem.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komura N., Suzuki K.G.N., Ando H., Konishi M., Koikeda M., Imamura A., Chadda R., Fujiwara T.K., Tsuboi H., Sheng R., et al. Raft-based interactions of gangliosides with a GPI-anchored receptor. Nat. Chem. Biol. 2016;12:402–410. doi: 10.1038/nchembio.2059. [DOI] [PubMed] [Google Scholar]
- Kovaliov M., Allegrezza M.L., Richter B., Konkolewicz D., Averick S. Synthesis of lipase polymer hybrids with retained or enhanced activity using the grafting-from strategy. Polymer. 2018;137:338–345. doi: 10.1016/j.polymer.2018.01.026. [DOI] [Google Scholar]
- Lele B.S., Murata H., Matyjaszewski K., Russell A.J. Synthesis of uniform Protein−Polymer conjugates. Biomacromolecules. 2005;6:3380–3387. doi: 10.1021/bm050428w. [DOI] [PubMed] [Google Scholar]
- Li S., Mao A., Huo F., Wang X., Guo Y., Liu L., Yan C., Ding L., Ju H. A localized glyco-editing probe for revelation of protein-specific glycan function. Mater. Today. 2021;49:85–96. doi: 10.1016/j.mattod.2021.04.015. [DOI] [Google Scholar]
- Liu J., Bulmus V., Herlambang D.L., Barner-Kowollik C., Stenzel M.H., Davis T.P. In situ formation of protein-polymer conjugates through reversible addition fragmentation chain transfer polymerization. Angew. Chem. Int. Ed. 2007;46:3099–3103. doi: 10.1002/anie.200604922. [DOI] [PubMed] [Google Scholar]
- Liu M., Johansen P., Zabel F., Leroux J.-C., Gauthier M.A. Semi-permeable coatings fabricated from comb-polymers efficiently protect proteins in vivo. Nat. Commun. 2014;5:5526. doi: 10.1038/ncomms6526. [DOI] [PubMed] [Google Scholar]
- Liu M., Tirino P., Radivojevic M., Phillips D.J., Gibson M.I., Leroux J.-C., Gauthier M.A. Molecular sieving on the surface of a protein provides protection without loss of activity. Adv. Funct. Mater. 2013;23:2007–2015. doi: 10.1002/adfm.201202227. [DOI] [Google Scholar]
- Liu X., Gao W. Precision conjugation: an emerging tool for generating protein-polymer conjugates. Angew. Chem. Int. Ed. 2021;60:11024–11035. doi: 10.1002/anie.202003708. [DOI] [PubMed] [Google Scholar]
- Lu J., Wang H., Tian Z., Hou Y., Lu H. Cryopolymerization of 1, 2-dithiolanes for the facile and reversible grafting-from synthesis of protein-polydisulfide conjugates. J. Am. Chem. Soc. 2020;142:1217–1221. doi: 10.1021/jacs.9b12937. [DOI] [PubMed] [Google Scholar]
- Mahal L.K., Yarema K.J., Bertozzi C.R. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- Mackenzie K.J., Francis M.B. Recyclable thermoresponsive polymer–cellulase bioconjugates for biomass depolymerization. J. Am. Chem. Soc. 2013;135:293–300. doi: 10.1021/ja309277v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyjaszewski K. Advanced materials by atom transfer radical polymerization. Adv. Mater. 2018;30:1706441. doi: 10.1002/adma.201706441. [DOI] [PubMed] [Google Scholar]
- Merritt E.A., Sarfaty S., Akker F.V.D., L'Hoir C., Martial J.A., Hol W.G.J. Crystal structure of cholera toxin B-pentamer bound to receptor GM1 pentasaccharide. Protein Sci. 1994;3:166–175. doi: 10.1002/pro.5560030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata H., Cummings C.S., Koepsel R.R., Russell A.J. Polymer-based protein engineering can rationally tune enzyme activity, pH-dependence, and stability. Biomacromolecules. 2013;14:1919–1926. doi: 10.1021/bm4002816. [DOI] [PubMed] [Google Scholar]
- Murata H., Cummings C.S., Koepsel R.R., Russell A.J. Rational tailoring of substrate and inhibitor affinity via ATRP polymer-based protein engineering. Biomacromolecules. 2014;15:2817–2823. doi: 10.1021/bm5008629. [DOI] [PubMed] [Google Scholar]
- Pal D., Dasgupta S., Kundu R., Maitra S., Das G., Mukhopadhyay S., Ray S., Majumdar S.S., Bhattacharya S. Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nat. Med. 2012;18:1279–1285. doi: 10.1038/nm.2851. [DOI] [PubMed] [Google Scholar]
- Paukner R., Staudigl P., Choosri W., Sygmund C., Halada P., Haltrich D., Leitner C. Galactose oxidase from Fusarium oxysporum--expression in E. coli and P. pastoris and biochemical characterization. PLoS One. 2014;9:e100116. doi: 10.1371/journal.pone.0100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilobello K.T., Slawek D.E., Mahal L.K. A ratiometric lectin microarray approach to analysis of the dynamic mammalian glycome. Proc. Nati. Acad. Sci. U S A. 2007;104:11534–11539. doi: 10.1073/pnas.0704954104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannes J.B., Ioannou A., Willies S.C., Grogan G., Behrens C., Flitsch S.L., Turner N.J. Glycoprotein labeling using engineered variants of galactose oxidase obtained by directed evolution. J. Am. Chem. Soc. 2011;133:8436–8439. doi: 10.1021/ja2018477. [DOI] [PubMed] [Google Scholar]
- Reily C., Stewart T.J., Renfrow M.B., Novak J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019;15:346–366. doi: 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M.S., Tyler E.M., Akyumani N., Kurtis C.R., Spooner R.K., Deacon S.E., Tamber S., Firbank S.J., Mahmoud K., Knowles P.F., et al. The stacking tryptophan of galactose oxidase: a second-coordination sphere residue that has profound effects on tyrosyl radical behavior and enzyme catalysis. Biochemistry. 2007;46:4606–4618. doi: 10.1021/bi062139d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager K.T., Narimatsu Y., Joshi H.J., Clausen H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020;21:729–749. doi: 10.1038/s41580-020-00294-x. [DOI] [PubMed] [Google Scholar]
- Shimoboji T., Larenas E., Fowler T., Kulkarni S., Hoffman A.S., Stayton P.S. Photoresponsive polymer–enzyme switches. Proc. Natl. Acad. Sci. U S A. 2002;99:16592–16596. doi: 10.1073/pnas.262427799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoboji T., Larenas E., Fowler T., Hoffman A.S., Stayton P.S. Temperature-Induced switching of enzyme activity with smart Polymer−Enzyme conjugates. Bioconjug. Chem. 2003;14:517–525. doi: 10.1021/bc025615v. [DOI] [PubMed] [Google Scholar]
- Stayton P.S., Shimoboji T., Long C., Chilkoti A., Ghen G., Harris J.M., Hoffman A.S. Control of protein–ligand recognition using a stimuli-responsive polymer. Nature. 1995;378:472–474. doi: 10.1038/378472a0. [DOI] [PubMed] [Google Scholar]
- Sun H., Kabb C.P., Sims M.B., Sumerlin B.S. Architecture-transformable polymers: reshaping the future of stimuli-responsive polymers. Prog. Polym. Sci. 2019;89:61–75. doi: 10.1016/j.progpolymsci.2018.09.006. [DOI] [Google Scholar]
- Sun M., Wong J.Y., Nugraha B., Ananthanarayanan A., Liu Z., Lee F., Gupta K., Fong E.L.S., Huang X., Yu H. Cleavable cellulosic sponge for functional hepatic cell culture and retrieval. Biomaterials. 2019;201:16–32. doi: 10.1016/j.biomaterials.2019.01.046. [DOI] [PubMed] [Google Scholar]
- Theodorou A., Liarou E., Haddleton D.M., Stavrakaki I.G., Skordalidis P., Whitfield R., Anastasaki A., Velonia K. Protein-polymer bioconjugates via a versatile oxygen tolerant photoinduced controlled radical polymerization approach. Nat. Commun. 2020;11:1486. doi: 10.1038/s41467-020-15259-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. Biological roles of glycans. Glycobiology. 2017;27:3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wu S., Ma S., Yan F., Weng Z. Cytocompatible modification of thermoresponsive polymers on living cells for membrane proteomic isolation and analysis. Anal. Chem. 2019;91:3187–3194. doi: 10.1021/acs.analchem.8b04201. [DOI] [PubMed] [Google Scholar]
- Xie R., Wang X., Wang Y., Ye M., Zhao Y., Yandell B.S., Gong S. pH-responsive polymer nanoparticles for efficient delivery of Cas9 ribonucleoprotein with or without donor DNA. Adv. Mater. 2022;34:2110618. doi: 10.1002/adma.202110618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Li Y., Yu X., Ju H., Ding L. Switchable enzymatic accessibility for precision cell-selective surface glycan remodeling. Chem. Eur J. 2019;25:10505–10510. doi: 10.1002/chem.201902113. [DOI] [PubMed] [Google Scholar]
- Zeng Y., Ramya T.N.C., Dirksen A., Dawson P.E., Paulson J.C. High-efficiency labeling of sialylated glycoproteins on living cells. Nat. Methods. 2009;6:207–209. doi: 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T., Jiang H., Gros M., Soriano del Amo D., Sundaram S., Lauvau G., Marlow F., Liu Y., Stanley P., Wu P. Tracking N-acetyllactosamine on cell-surface glycans in vivo. Angew. Chem. Int. Ed. 2011;50:4113–4118. doi: 10.1002/anie.201100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.