Abstract
The potential of a bacterial toxin-antitoxin gene system for use in containment control in eukaryotes was explored. The Escherichia coli relE and relB genes were expressed in the yeast Saccharomyces cerevisiae. Expression of the relE gene was highly toxic to yeast cells. However, expression of the relB gene counteracted the effect of relE to some extent, suggesting that toxin-antitoxin interaction also occurs in S. cerevisiae. Thus, bacterial toxin-antitoxin gene systems also have potential applications in the control of cell proliferation in eukaryotic cells, especially in those industrial fermentation processes in which the escape of genetically modified cells would be considered highly risky.
Genetically modified microorganisms (GMMs) used in the biotechnological industry are normally kept physically closed off from their surroundings. The strains used are attenuated and will not survive very long if they escape into the environment. In recent years there has been a growing interest in the deliberate release of GMMs into the environment. GMMs suitable for release could, for example, be used for bioremediation of polluted soils, for biocontrol of fungicidal and insecticidal pests in agriculture, or as live vaccines in biomedicine. Upon release, such strains must be able to proliferate and compete with the indigenous strains present. However, to ensure safety, uncontrolled spread of GMMs into the environment must be prevented. Safety can be achieved through biological containment if the GMMs self-destruct by expression of killing genes after fulfilling their jobs. Several bacterial toxins are good candidates for use in bacterial containment systems, including membrane-destabilizing or pore-forming proteins (4, 15) and enzymes attacking the genetic material of the cell (1, 2, 7). The design and applications of active biological containment systems are reviewed and discussed in several publications (8, 9, 12, 13).
Recently, relBE, members of a new toxin-antitoxin gene family, have been found in Escherichia coli (5). To date, relBE homologues have been identified in a broad range of both gram-negative and gram-positive bacteria and in archaea (5, 6). The relE gene encodes a small (11-kDa) protein that is extremely toxic to bacterial cells, and the relB gene encodes an antitoxin of similar size that counteracts the cell killing activity of the RelE toxin (5, 6). The specific molecular targets of the RelE protein, as well as the physiological role of the RelE-RelB toxin-antitoxin system in bacteria, is still speculative (3, 5). So far, no relBE homologues have been found in eukaryotes.
In the study described here, we analyzed whether this toxin-antitoxin gene system also could be used to control proliferation of eukaryotic cells. For this purpose, we used Saccharomyces cerevisiae as a general model for eukaryotes, specifically fungi. We showed that expression of relE strongly inhibits the growth of yeast cells and that the products of relE and relB interact.
Strains and media.
E. coli TOP10 (Invitrogen) was routinely used during vector constructions. The bacteria were maintained and grown in Luria-Bertani medium (14) supplemented with ampicillin (100 μg/ml). The yeast strain used was S. cerevisiae 281288DIV-36 (MAT a his4-15 ura3-52 trp1; Y493 from our laboratory collection). Yeast strains were transformed as described previously (11). Transformed yeast cells were grown in liquid or solid SC−ura (synthetic complete medium without uracil and with 2% glucose), SC−ura+gal (synthetic complete medium without uracil and with 2% galactose), SC−ura−met (synthetic complete medium without uracil and methionine and with 2% glucose), or SC−ura−met+gal (synthetic complete medium without uracil and methionine and with 2% galactose) (11). When necessary, the media were solidified by addition of agar to 2% (wt/vol).
Construction of a vector for expression of the RelE toxin in S. cerevisiae.
The DNA manipulations were performed according to standard methods (14). All PCR amplifications (20 cycles consisting of 40 s of denaturation, 40 s of annealing, and 1 min of extension) were performed with Vent DNA polymerase (New England Biolabs) using a PTC-100 thermocycler (MJ Research Inc.). After agarose gel electrophoresis, amplified fragments were isolated using a DNA purification kit from Qiagen.
The coding region of the relE gene from E. coli was PCR amplified from pMG223 (5) by using the sense primer 5′-TAGGTACCATGGCGTATTTTCTGG-3′ and the antisense primer 5′-TGAATTCCTCGACTCAGAG-3′. KpnI and EcoRI restriction enzyme recognition sites, which were added at the 5′ ends of the sense and antisense primers, respectively, are underlined. The PCR product was inserted into the KpnI-EcoRI site of the polylinker of the yeast expression vector pYES2 (Invitrogen) to yield the plasmid pPK727.
Construction of a vector for expression of the RelE-RelB toxin-antitoxin in S. cerevisiae.
A modified version of the pYES2 expression vector was constructed by removing the GAL1 promoter and inserting the methionine (MET25) promoter from S. cerevisiae. PMET25 was amplified from pYC012 (10) by PCR using the forward primer 5′-AGACTAGTCCCGGGCTTAATTAAATAATATAC-3′ and the reverse primer 5′-AGACTAGTGGATCCTGTATGGATGGGGGTA-3′. SpeI restriction enzyme recognition sites added at the 5′ ends of both primers are underlined, and the BamHI site added in the reverse primer is in italics. PGAL1 was removed from pYES2 as an SpeI fragment, and the SpeI-digested PCR fragment was inserted into the SpeI site of this vector, yielding plasmid pPK908. Twenty-three nucleotides at the 5′ end of the PCR product were removed because of the presence of an SpeI restriction site in PMET25. This deletion did not appear to have any significant effect on the promoter activity. The correct orientation of the inserted promoter was verified by restriction enzyme digestions.
The region of pPK727 including the relE gene under the control of the GAL1 promoter and the CYC1 transcriptional terminator was amplified by PCR with the following set of forward and reverse primers to introduce ClaI restriction sites (underlined): 5′-AGATCGATTACAGGGCGCGTGGGGATGATC-3′ and 5′-AGATCGATAATACGCAAACCGCCTCTCCCC-3′. The amplified PCR product was inserted into the unique ClaI site of pPK908 to yield plasmid pPK988.
The coding region of the relB gene from E. coli was amplified from plasmid pBD2430 by PCR (5) using the sense primer 5′-TAGGTACCATGGGTAGCATTAACC-3′ and the antisense primer 5′-ATGGATCCTCAGAGTTCATCCAGC-3′. Each of these primers possesses a BamHI site at its 5′ end (underlined). The amplified relB coding region was inserted into the BamHI site of pPK988 downstream of the MET25 promoter, yielding plasmid pPK1006. The orientation of relB was verified by restriction enzyme digestions, and the DNA sequence of the PCR product of relB was verified by DNA sequence analysis.
Expression of relE in S. cerevisiae.
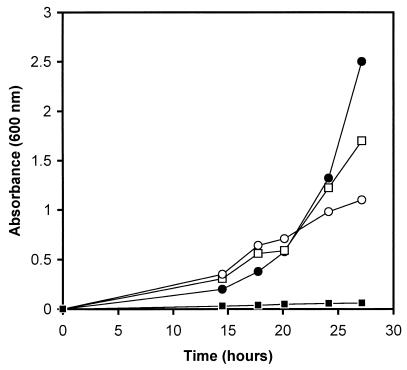
Yeast cells transformed with either plasmid pPK727 (containing relE under the control of the GAL1 promoter) or the pYES2 vector were grown overnight in SC−ura, then diluted 1:100 in SC−ura or SC−ura+gal. Growth experiments were performed in 250-ml bottles with 20-ml cultures at 28°C with heavy agitation. The presence of glucose repressed the relE gene. Expression of relE from the GAL1 promoter in pPK727 was induced by galactose when the SC−ura was replaced by SC−ura+gal. A clear inhibitory effect on cell growth in SC−ura+gal was observed for yeast cells containing the pPK727 plasmid, whereas no such inhibition was observed for yeast cells containing the empty pYES2 vector (Fig. 1). The same inhibitory effect was also observed when pPK727 was introduced into other yeast strains (data not shown).
FIG. 1.
Effect of relE expression, under the regulation of the GAL1 promoter, on cell growth of S. cerevisiae. Cells harboring pPK727 were grown in SC−ura (□) or SC−ura+gal (■), and cells harboring pYES2 as a control were grown in SC−ura (○) or SC−ura+gal (●).
Thus, the product of the relE gene is also active in S. cerevisiae. The relE gene could be maintained in yeast cells and induced when necessary, leading to cell death. However, the inhibition was not complete, despite the very peculiar appearance of the yeast cells, and it seemed that some of the cells were still growing, although extremely slowly (Fig. 1).
Expression of relE-relB in S. cerevisiae.
Yeast cells transformed with plasmid pPK1006 (containing relE under the control of the GAL1 promoter and relB under the control of the MET25 promoter) were studied to see if there were interactions between the two proteins. The overnight culture was diluted 1:100 in four different growth media, SC−ura, SC−ura−met, SC−ura+gal, and SC−ura−met+gal. Growth experiments were performed in 250-ml bottles with 20-ml cultures at 28°C with heavy agitation. Expression of relE was induced by galactose (in SC−ura+gal and SC−ura−met+gal) but repressed by the presence of glucose. Expression of relB was induced in the absence of methionine (in SC−ura−met and SC−ura−met+gal) but repressed in the presence of methionine. Cells grown in SC−ura−met+gal showed higher growth rates than cells grown in the corresponding medium supplemented with methionine (Table 1). These results indicate that expression of relB counteracts the growth-inhibiting effect of the relE gene to some extent. A difference in doubling time was also observed for cells grown in SC−ura or SC−ura−met and can be explained by the fact that small amounts of the RelE toxin are synthesized also in the absence of galactose, due to a leaky activity of the uninduced GAL1 promoter. This growth-inhibiting effect was counteracted by the action of the RelB protein in SC−ura−met (Table 1). The relatively small difference in doubling time between yeast cells grown in SC−ura−met+gal and those grown in SC−ura+gal suggested that either the level of relB expression was too low to fully counteract the toxic effect of the induced relE expression or the RelB-RelE interaction in the yeast cell was very weak.
TABLE 1.
Effect of expression of relE and relB on the growth rate of S. cerevisiae harboring plasmid pPK1006a
| Growth medium | Induced gene(s) | Doubling time (h) |
|---|---|---|
| SC−ura−met | PMET25-relB | 3.5 |
| SC−ura | None | 5.5 |
| SC−ura−met+gal | PMET25-relB, PGAL1-relE | 13.0 |
| SC−ura+gal | PGAL1-relE | 20.0 |
pPK1006 carries the relE gene, induced in the presence of galactose, and the relB gene, induced in the absence of methionine.
Conclusions.
The data presented here clearly demonstrate that expression of the bacterial relE gene is also toxic to S. cerevisiae cells. The fact that the growth rates of relB-expressing cells are higher than those of non-relB-expressing cells suggests that both gene products function in S. cerevisiae. Thus, bacterial toxin-antitoxin systems can also be applied in eukaryotic cells. It is likely, even if so far it cannot be demonstrated directly, that the actions of the two genes occur at the protein level. The absence of a complete counteraction of the activity of relE by relB in SC−ura−met+gal could be partially reversed if a stronger promoter controls the relB gene, leading to overproduction of antitoxin compared to toxin. So far, it is not known whether several RelB molecules are required to counteract a single RelE molecule. Further experiments, in which the expression level of RelB is substantially higher than that of RelE, might demonstrate a total counteraction of RelE by RelB. Such data would support the hypothesis that the molecular targets for the RelE and RelB proteins in bacteria and S. cerevisiae are the same.
The relE-relB toxin-antitoxin genes, as well as similar toxin-antitoxin pairs, could be used as part of a containment system in genetically modified yeasts as well as other fungi. For example, the relE gene under the control of the glucose-repressed promoter could be used as a containment control in those industrial fermentation processes in which the escape of genetically modified yeast cells would be considered highly risky. Under fermentation conditions, the relE gene is suppressed by high levels of glucose, and if yeast cells escape from the fermentation tank they will self-destruct upon derepression of relE because of the extremely low levels of glucose in the environment. Because of the leakiness of the GAL1 promoter, the relB gene could be constitutively expressed at a low level to ensure optimal growth under repressed conditions. Interestingly, it was previously demonstrated that expression of the relE gene in a mammalian cell line also led to inhibition of cell proliferation (K. Gerdes, M. Gotfredsen, H. Grøndlund, K. Pedersen, and P. Kristoffersen, U.S. patent application USSN 60/085067). Experiments analyzing the applications of the RelE-RelB toxin-antitoxin gene system for gene and cancer therapy are also in progress.
Acknowledgments
We thank Jørgen Hansen from Carlsberg Research Laboratory for providing the plasmid pYC012 containing the MET25 promoter.
REFERENCES
- 1.Ball T K, Saurugger P N, Benedik M J. The extracellular nuclease gene of Serratia marcescens and its secretion from Escherichia coli. Gene. 1987;57:183–192. doi: 10.1016/0378-1119(87)90121-1. [DOI] [PubMed] [Google Scholar]
- 2.Diaz E, Munthali M, de Lorenzo V, Timmis K N. Universal barrier to lateral spread of specific genes among microorganisms. Mol Microbiol. 1994;5:855–861. doi: 10.1111/j.1365-2958.1994.tb00477.x. [DOI] [PubMed] [Google Scholar]
- 3.Gerdes K. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacteriol. 2000;182:561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerdes K, Poulsen L K, Thisted T, Nielsen A K, Martinussen J, Andreasen P H. The hok killer gene family in gram-negative bacteria. New Biol. 1990;11:946–956. [PubMed] [Google Scholar]
- 5.Gotfredsen M, Gerdes K. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol. 1998;29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- 6.Grøndlund H, Gerdes K. Toxin-antitoxin systems homologous with relBE of Escherichia coli plasmid P307 are ubiquitous in prokaryotes. J Mol Biol. 1999;285:1401–1415. doi: 10.1006/jmbi.1998.2416. [DOI] [PubMed] [Google Scholar]
- 7.Molin, S., M. Givskov, C. S. Kristensen, A. K. Bej, and L. Eberl. October 1995. Method of limiting the survival of genetically engineered microorganisms in their environment. U.S. patent 5,834,233.
- 8.Molin S, Boe L, Jensen L B, Kristensen C S, Givskov M, Ramos J L, Bej A K. Suicidal genetic elements and their use in biological containment of bacteria. Annu Rev Microbiol. 1993;47:139–166. doi: 10.1146/annurev.mi.47.100193.001035. [DOI] [PubMed] [Google Scholar]
- 9.Molina L, Ramos C, Ronchel M-C, Molin S, Ramos J L. Construction of an efficient biologically contained Pseudomonas putida strain and its survival in outdoor assays. Appl Environ Microbiol. 1998;64:2072–2078. doi: 10.1128/aem.64.6.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olesen K, Johannesen P F, Hoffmann L, Sørensen S B, Gjermansen C, Hansen J. The pYC plasmids, a series of cassette-based yeast plasmid vectors providing means of counter-selection. Yeast. 2000;16:1035–1045. doi: 10.1002/1097-0061(200008)16:11<1035::AID-YEA606>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 11.Piskur J, Kielland-Brandt M C. Folding and secretion of Saccharomyces cerevisiae carboxypeptidase Y are influenced by fusion with short heterologous peptides. Biotechnol Appl Biochem. 1993;18:239–257. [PubMed] [Google Scholar]
- 12.Ramos J L, Andersen P, Jensen L B, Ramos C, Ronchel M C, Diaz E, Timmis K N, Molin S. Suicide microbes on the loose. Bio/Technology. 1995;1:35–37. doi: 10.1038/nbt0195-35. [DOI] [PubMed] [Google Scholar]
- 13.Ronchel M C, Ramos C, Jensen L B, Molin S, Ramos J L. Construction and behavior of biologically contained bacteria for environmental applications in bioremediation. Appl Environ Microbiol. 1995;61:2990–2994. doi: 10.1128/aem.61.8.2990-2994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 15.Witte A, Reisinger G R, Sackl W, Wanner G, Lubitz W. Characterization of Escherichia coli lysis using a family of chimeric E-L genes. FEMS Microbiol Lett. 1998;164:159–167. doi: 10.1111/j.1574-6968.1998.tb13081.x. [DOI] [PubMed] [Google Scholar]