Abstract
The manifestation of intra- and inter-tumor heterogeneity hinders the development of ubiquitous cancer treatments, thus requiring a tailored therapy for each cancer type. Specifically, the reprogramming of cellular metabolism has been identified as a source of potential drug targets. Drug discovery is a long and resource-demanding process aiming at identifying and testing compounds early in the drug development pipeline. While drug repurposing efforts (i.e., inspecting readily available approved drugs) can be supported by a mechanistic rationale, strategies to further reduce and prioritize the list of potential candidates are still needed to facilitate feasible studies. Although a variety of ‘omics’ data are widely gathered, a standard integration method with modeling approaches is lacking. For instance, flux balance analysis is a metabolic modeling technique that mainly relies on the stoichiometry of the metabolic network. However, exploring the network’s topology typically neglects biologically relevant information. Here we introduce Transcriptomics-Informed Stoichiometric Modelling And Network analysis (TISMAN) in a recombinant innovation manner, allowing identification and validation of genes as targets for drug repurposing using glioblastoma as an exemplar.
Subject terms: Cancer, Computational biology and bioinformatics, Drug discovery
Introduction
Cancer is the first or second cause of premature death in 70% of countries1. Tumors are characterized by multiple biological capabilities conferring cellular proliferation and metastasis (dissemination to other tissues and colonization), where metabolic reprogramming is crucial to fulfil biosynthetic requirements underlying these phenotypes2. The disparity of oxygen and nutrient availability within the surrounding tumor microenvironment promotes different metabolic states amongst cells, which show a continuum in gene expression within sub-clonal populations3. Consequently, metabolic dependencies and vulnerabilities are shaped by the cell lineage and the environment, rather than being universal4. Isocitrate dehydrogenase-1 wild-type Glioblastoma (GBM) is the most aggressive brain tumor in adults5, characterized by a high degree of invasiveness through brain parenchyma6. Different subtypes and transcriptional subpopulations may co-exist within the same tumor, where a quiescent subpopulation of GBM stem cells (GSC) can facilitate the development of radio- or chemo-resistance, leading to inevitable disease relapse7–9. Therefore, novel therapeutic approaches should aim to target all cell lineages10, block GSC differentiation11, or combine both strategies9. In particular, recent studies have shown the suitability of chemicals disrupting energy metabolism in GBM12 or blocking de novo pyrimidine synthesis in GSC13. Conversely, the application of immunotherapy and many drug compounds is hindered by the impermeability of the blood–brain–barrier14.
Phenotypic screening of compound libraries can help identify putative drug candidates, albeit being a costly process and requiring further target deconvolution steps15. Although gene essentiality is used as a proxy for potential drug targeting, its context-dependent and evolvable nature hinders its usage, for example: (i) genetic ablation affects both the catalytic activity (which may not be essential) and any other function (e.g. essential structural); (ii) incomplete knockdown or off-target effects; (iii) genetic divergence of cell lines from the original tumor; (iv) metabolic response of cells to non-physiological growth conditions16–19.
Whilst simulating ligand–target interactions (molecular docking) provides a shortlist of compounds to be tested, the scarcity of structural data limits its application20. In addition, this approach prevents the identification of compounds indirectly affecting metabolism, for example by altering the expression levels of multiple genes21. Therefore, information-rich computational strategies to generate manageable lists of drug candidates should be considered as a viable complement to the drug discovery phase. The set of metabolic fluxes (i.e., metabolic or transport reaction rates) can be either estimated via 13C metabolic flux analysis (13C-MFA) for the central carbon metabolism or modelled by applying flux balance analysis (FBA) to a genome-scale model (GSM).
A GSM consists of a stoichiometric matrix connecting metabolites (substrates and products) with reactions, where the latter are associated with genes following the gene-protein-reaction formalism22. FBA predicts a flux distribution upon the optimization for a biological task (normally the biomass formation), represented by a reaction in the GSM23. Whilst 13C-MFA mainly informs about the central carbon metabolism4,24, transcriptomics data can be applied to obtain cell-specific models from GSMs and can predict in silico gene essentiality25–27. Other strategies based on network analysis allow identification of highly connected reactions or reactions which exclusively consume/produce a metabolite (choke points) that if inhibited, would potentially disrupt metabolism28.
To this end, we developed Transcriptomics-Informed Stoichiometric Modelling And Network analysis (TISMAN), a recombinant innovation introducing the concept of “extended choke point” and exploiting biologically-rich information aiming at developing a workflow providing a stringent search of potential drug targets and compounds to be tested for GBM as an exemplar disease type. Briefly, published data were exploited to build condition specific GBM metabolic models, which were analyzed to identify and rank the reactions of interest according to different indicative criteria. Several chemical-gene interaction databases were used to identify and prioritize approved drugs or experimental compounds either directly targeting proteins or indirectly affecting the corresponding gene expression levels. Chemicals with a proven impact against other cancer types but which have not been tested for GBM, were prioritized for experimental validation using patient-derived GBM in vitro models.
Materials and methods
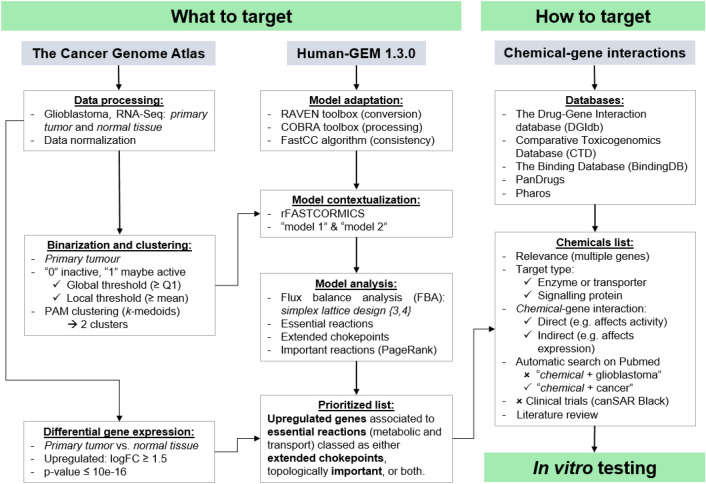
The workflow developed in this study comprised three main parts (Fig. 1): (i) “what to target”: identification of either metabolic or transport (e.g., between cellular compartments) reactions of interest; (ii) “how to target”: identification of chemicals (readily available approved drugs or experimental compounds); (iii) in vitro validation of the selected compounds. Briefly, transcriptomics data from GBM tumors was used to: (a) contextualize a generic human GSM, yielding two GBM-specific models; (b) identify up-regulated genes. Next, the models were subjected to FBA to identify essential reactions, and the network topology was inspected to pinpoint certain features (choke points and centrality). Overall, the workflow aimed to identify proteins that if targeted, would potentially hinder cellular metabolism, impairing tumor metabolic viability and/or invasion. Chemicals targeting metabolism either directly (i.e., affecting the activity) or indirectly (i.e., affecting gene expression) were ranked, with five candidates prioritized for in vitro testing. Files and scripts for MATLAB and R can be found in Github (https://github.com/CTA-code/TISMAN).
Figure 1.
TISMAN (Transcriptomics-Informed Stoichiometric Modelling And Network analysis), the workflow developed in this study. It comprised three main parts (what to target, how to target and in vitro testing), where both MATLAB and R scripts were used.
What to target
Transcriptomics data analysis
Data processing: RNA-Seq data for GBM and healthy counterpart astrocytic tissue (herein referred to as ‘primary tumor’ and ‘normal tissue’) was obtained from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) and processed using the R package TCGAbiolinks29. This information was used for two independent steps, binarization and clustering. Expression data from primary tumor was binarized by considering two simultaneous thresholds: (i) global (≥ Q1) across all genes and samples, and (ii) local (≥ mean), which was gene-specific across samples. Genes were flagged as either maybe active (“1”) or inactive (“0”) if fulfilling both criteria or not, respectively, so that each sample was codified by a binary vector25. Next, R’s PAM function (Partitioning Around Medoids, or k-medoids) was used to cluster the data into two groups (optimal number of clusters according to the Silhouette method), yielding a representative binary vector (or medoid) for each cluster30. Focusing on upregulated genes in cancerous tissue compared to normal tissue is a widely used strategy to identify potential therapeutic genes31. Therefore, both primary tumor and normal tissue datasets were used to determine the differential gene expression: genes with logFC ≥ 1.5 and p-value ≤ 10–16 were deemed upregulated in the tumor32.
Stoichiometric modeling and network analysis
Model adaptation: the latest version of a human GSM (Human-GEM 1.3.0, comprising 13,417 reactions, 10,135 metabolites and 3,628 genes33) was converted and adapted to a COBRA toolbox paradigm by using the RAVEN toolbox34 and the COBRA toolbox22. Next, a consistent model was generated by FastCC prior to the contextualization step, so that all reactions were active (i.e., nonzero flux) for at least one feasible flux distribution35.
Model contextualization: the two medoids were used with rFASTCORMICS27 to build two cancer-specific models (COBRA toolbox v. 2, with IBM CPLEX 12.8 in MATLAB R2016b run in Windows 10 to ensure solver compatibility).
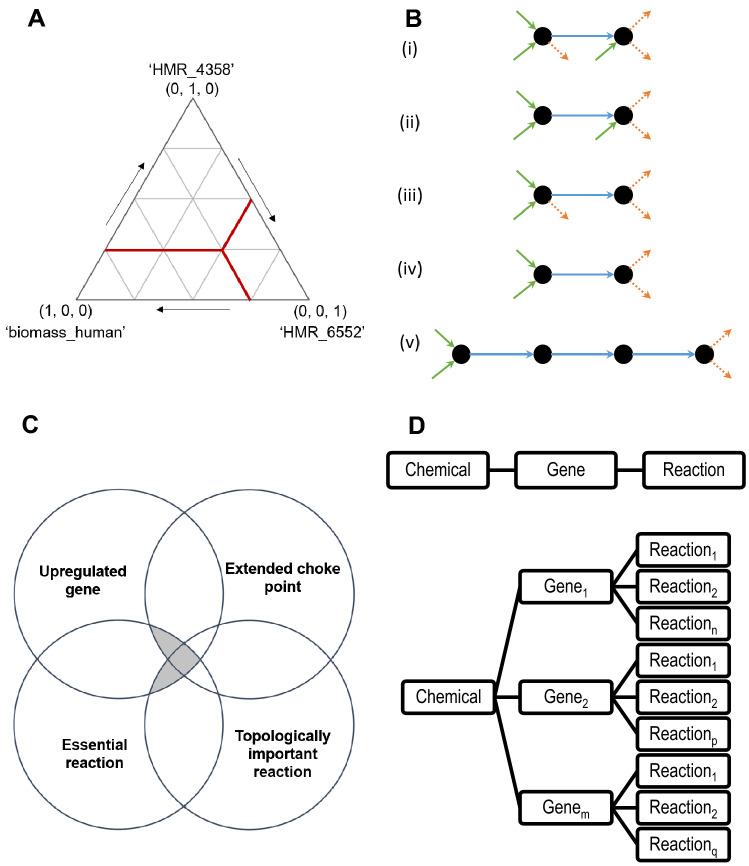
Model analysis: an upper bound of 2 mmol/gDW-h was set for the oxygen uptake rate, as suggested for previous cancer models36,37. For the sake of modelling a variety of metabolic states, several objective functions were considered: maximization of the biomass yield (‘biomass_human’), maximization of the ATP yield (‘HMR_4358’), and maximization of 1-phosphatidyl-1D-myo-inositol-4-phosphate production (‘HMR_6552’), a lipid which induces curvature in the lipid membrane38. The latter reaction is catalyzed by synaptojanin 2 (SYNJ2) a polyphosphoinositide phosphatase related to GBM invasion39 and a mediator of metastasis in breast cancer40. Instead solving a multi-objective linear problem41, the weighted global criterion method was used to reformulate it as a single objective problem directly solvable by FBA23,42. Therefore, a simplex lattice design for FBA was designed by varying the weight of the three reactions, allowing to study the aforementioned three objective functions and mixtures amongst them in 15 single objective problems (Fig. 2A). In addition, an extra case for the minimization of the sum of fluxes was considered, yielding 16 final tests. Since certain optimal solutions were the same, the number of non-redundant solutions for each model was calculated. Reactions of interest fulfilled several criteria (Fig. 2C): related to an upregulated gene in the tumor; essential reaction; extended choke point and/or topologically important. Considering the full GSM and the flux through the biomass reaction when maximizing for it in FBA, a reaction was deemed essential for a test (same network, different objective function(s)) when shutting it down (no flux through allowed) caused a ≥ 5% reduction of said metabolic flux. Conversely, topological aspects were studied by exploring reduced metabolic networks. A condition-specific model was generated for each unique flux distribution produced by FBA, so that the stoichiometric matrix only comprised active elements (reactions and metabolites). Single choke points were identified as reactions associated exclusively with producing or consuming a metabolite. Double choke points were then defined as reactions associated exclusively with producing and consuming metabolites. To ensure a more stringent selection criterion, the concept of extended choke points was introduced, defined as double choke points surrounded by single choke points (consuming and producing) (Fig. 2B). Alternatively, topologically important reactions were identified by MATLAB’s in-built PageRank function43. This analysis was applied to a directed graph (i.e., considering the flux direction) derived from an adjacency matrix, where the centrality of the nodes (reactions) was determined. Directly calculating the adjacency matrix from the stoichiometric matrix would yield a metabolite-centric graph (metabolites as nodes)44. Therefore, an intermediate step converting the stoichiometric matrix into its conjugate transpose (reactions as nodes) was included prior to generating the adjacency matrix.
Figure 2.
Strategies to identify gene targets for drug repurposing. (A) Simplex lattice design for objective function weight values distribution. Each outer vertex represents a case where a single objective function is selected: maximization of the biomass yield (‘biomass_human’), maximization of the ATP yield (‘HMR_4358’), and maximization of 1-phosphatidyl-1D-myo-inositol-4-phosphate production (‘HMR_6552’). For illustrative purposes, the test comprising weight values (0.25, 0.25, 0.50) is labeled in red. (B) Examples of metabolites-reactions relationships (dots and arrows, respectively). The blue arrows represent the production of the metabolite on the right from the metabolite on the left. Inward green arrows refer to production reactions from metabolites not drawn, while outward orange dotted arrows represent consumption reactions. (i) Not a choke point, (ii) single choke point on the consumption side, (iii) single choke point on the production side, (iv) double choke point, and (v) extended choke point. (C) Venn diagram showing the subsets of reactions of interest. In all cases, reactions (metabolic or of transport) were essential and carried out by proteins related to upregulated genes, as well as identified as extended choke point and/or topologically important reactions. (D) (Top) Direct chemical–gene interaction, where a compound affects the activity of a single protein (e.g., an inhibitor). (D) (Bottom) Indirect chemical–gene interaction, where a single compound targets an element such as a signaling protein, that leads to changes in the expression levels of multiple genes, thus affecting several reactions in a pleiotropic manner.
Prioritized list: reactions of interest for each medoids were identified by independently selecting those identified as extended choke points in at least half of the non-redundant optimal solutions (occurrence), and those within the top 10% of central reactions according to PageRank. Finally, results from both medoids were combined by summation of occurrence or centrality scores, respectively.
How to target
Identification of relevant drugs and chemicals
Five databases were used, either programmatically or manually: The Drug-Gene Interaction database (DGIdb) Nov 2020, v4.2.045; Comparative Toxicogenomics Database (CTD), Oct 202046; The Binding Database (BindingDB), Nov 202047; PanDrugs, version: 2018.11.748; and Pharos, version 3.2.049. To exploit published chemical–gene interactions, a gene-centric list of potential candidates was generated from the reactions of interest. Since a gene may participate in several reactions, occurrence and centrality scores for each gene were normalized and separately summated. Similarly, as a chemical may target (directly or indirectly) one or several genes (Fig. 2D), drug-centric lists were created, and a final relevance score was calculated as the sum of occurrences and centrality scores. Automatic searches on Pubmed were performed to identify which drugs have been tested for cancers (except for GBM), followed by a literature review and search for clinical trials in canSAR50 to finally select five compounds for in vitro testing.
In vitro testing
Cell culture
Glioblastoma INvasive margin (GIN28, GIN31) and Glioblastoma Contrast-Enhanced core (GCE28, GCE31) cell lines have been previously derived at Queen’s Medical Centre, Nottingham, authenticated using short tandem repeat genotyping (Supplementary Table S1) and used at passages 21–29, 27–36, 15–23 and 28–37, respectively51,52. Cell lines were cultured using adherent T75 flasks (Corning) in Dulbecco’s Modified Eagle Medium (Gibco) supplemented with 10% FBS (Sigma-Aldrich) and incubated under humidified growth conditions at 37 °C and 5% CO2.
Metabolic viability assay
All cell lines were seeded in 96 well plates (Costar) at 5 × 103 cells/well and incubated for 24 h prior to treatment. Cells were treated with afuresertib (Selleckchem), isorhamnetin (Merck), formononetin (Merck), pyrogallol (Merck) and taxifolin (Merck) in a 16-point dose response (n = 6) over 72 h, with concentrations ranging from 1 to 500 μM. Appropriate vehicle (0.1% DMSO) and positive cell death controls (10% DMSO) were used. Viability was assessed by adding 1:10 PrestoBlue (ThermoFisher) over 45 min and measuring the resulting fluorescence at 544/590 nm (excitation/emission) on a FLUOstar Omega microplate reader (BMG LABTECH). Fluorescence intensity was normalized against that of control wells to determine percentage viability of drug-treated cells.
Invasion assay
The ability of the compounds to inhibit invasion was investigated for all cell lines using a Transwell collagen barrier assay. 24 well ThinCert™ 8 µm cell culture inserts (Greiner) were coated with 10 µg of mouse collagen IV (Cultrex, Trevigen) and incubated at 37 °C overnight. Cells were seeded at 13,500–15,000 cells/insert in 0% FBS DMEM with added treatment and allowed to invade towards 10% FBS DMEM over 24 h at 37 °C and 5% CO2. Invasion of untreated cells towards 10% and 0% FBS was used as controls. Following incubation, non-invading cells were removed from the top chamber using a cotton swab and media was replaced in the bottom chamber with 1:10 PrestoBlue in 0% DMEM and incubated for 45 min with fluorescence measured at 544/590 nm. Number of invading cells was extrapolated from a standard curve comparing fluorescence intensity to cell seeding density (serially diluted from 20,000 to 234 cells/well). Finally, percentage invasion was calculated as the ratio of the number of invading cells to number of seeded cells.
Statistical analysis
Statistical data analysis and curve fitting was performed using GraphPad Prism (v8.4.3). Statistical significance was analyzed via one-way ANOVA with p < 0.05 considered significant. Inhibitory concentration values (IC50 and IC25 for afuresertib and taxifolin, respectively) were generated from sigmoidal dose–response curves with upper and lower 95% confidence intervals.
Results and discussion
This study revealed three main outcomes: a widely applicable workflow based on metabolic modelling and network analysis (TISMAN), a list of potential drug targets for further investigation, and repurposing of drug compounds exerting inhibitory effects on GBM metabolic viability and invasion. Accessible high throughput sequencing of biopsies transformed the usage of RNA-sequencing in neuro-oncology practice53. Therefore, transcriptomics data was used to generate two contextualized models from a generic human GSM25–27, and to identify upregulated genes in cancerous cells32. Specifically, GBMs comprise pro-tumorigenic subpopulations of stem cells able to develop resistance to therapy7–9,54. While using FBA to obtain in silico gene essentiality is not a novel idea23, defining an appropriate objective function for normal and cancerous tissues to elucidate differences cannot be easily done2,55. Therefore, our analyses focused on cancer-specific models under an array of metabolic states and considering only reactions related to genes upregulated in GBM versus normal tissue. Conversely, recent metabolic modelling studies for GBM solely focused on the maximization of biomass yield, but generated c. 140 patient-derived GSMs56.
Identification of genes of interest
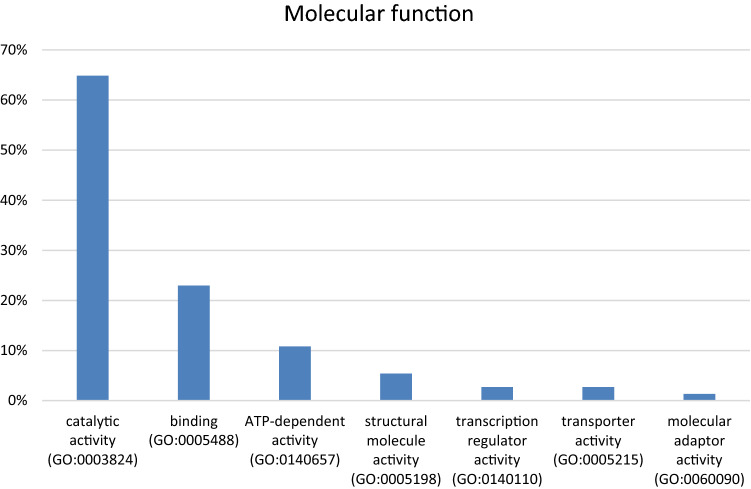
A final list of 168 upregulated genes associated with essential reactions classed as extended chokepoints and/or topologically important, was generated. Specifically, 74 genes contained hits in the chemical-gene interaction databases (Fig. 3). The remaining 94/168 genes had no reported chemical–gene interactions, so they were deemed as potential drug targets yet to be explored (Dataset S1)58.
Figure 3.
Functional classification of the 74 genes of interest. Using the PANTHER Classification System (17.0)58 showed that 65% of the 74 aforementioned genes have a catalytic function, which relates to the underlying metabolic modelling to TISMAN. A functional enrichment analysis performed in STRING v1157 identified oxidase and ligase activity in lipid metabolism to be particularly prevalent in this gene list.
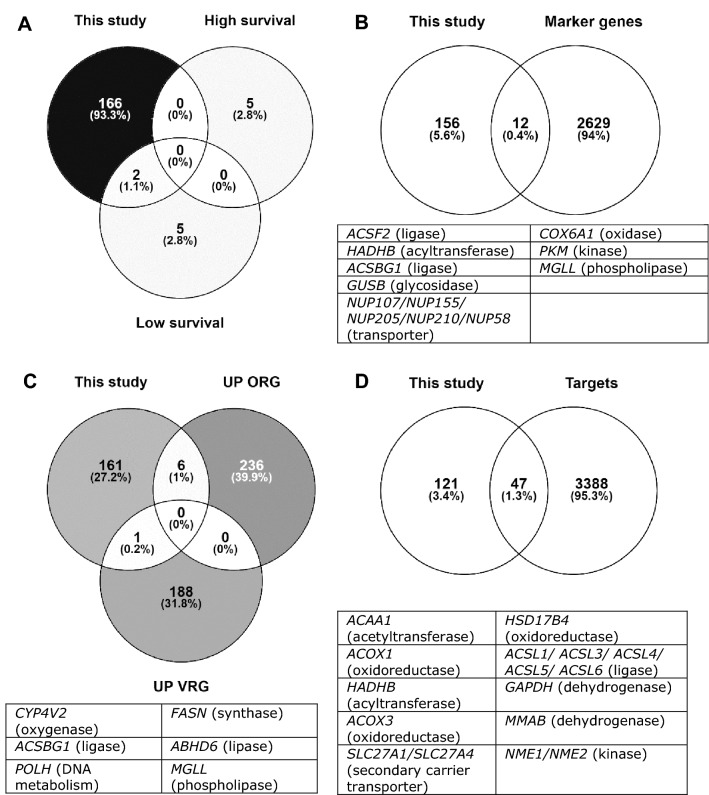
Given the genetic heterogeneity which manifests within GBM7–9,54, predicted genes of interest were compared against modelling and experimental approaches applied to cancerous tissue or specific subpopulations (Fig. 4). In Larsson et al. (Fig. 4A), patient-derived GSMs for GBM were generated from transcriptomics data, and gene essentiality predicted by FBA56. Two of the genes pinpointed for the low survival models agreed with our results: ACOT8 (acyl-CoA thioesterase 8) and GPAT4 (glycerol-3-phosphate acyltransferase 4), both related to lipid metabolism. The former is targeted by three drugs with antineoplastic activity which decrease its expression: doxorubicin52,59, orlistat60,61, and paclitaxel60,62. GPAT4 is the only gene of the GPAT family expressed in the brain, and it has recently been identified as a potential drug target for treating obesity-associated depression63.
Figure 4.
Comparison of genes of interest identified in this study against published works: (A) Larsson et al.56, which highlighted essential genes for models for high and low survival, (B) Richards et al.9, where the list of marker genes comprises the top 50 genes for each one of the 86 intra-GSC clusters, (C) Wang et al.54, where upregulated genes for outer RG-like and ventricular RG-like (UP ORG and UP VRG, respectively) were considered, and (D) Liu et al.67, where “targets” comprise both genes and miRNAs. Diagrams were made using Venny68. Panels B, C and D include tables informing about the protein type (most of them are enzymes).
A subpopulation of GSC can facilitate developing resistance to therapy, promoting disease relapse7–9. In Richards et al.9 certain marker genes for intra-GSC clusters were identified, some of which were also highlighted in this work as potential drug targets (Fig. 4B): ACSF2 (acyl-CoA Synthetase Family Member 2), MGLL (monoglyceride lipase), and PKM (pyruvate kinase M1/2). Although ACSF2 is functionally associated with fatty acid metabolism, ACSF2 did not affect cell anchorage-dependent growth in U87 GBM cells64. MGLL is highly expressed in the brain and in aggressive cancers and its role in self-renewal and tumorigenic behavior has been demonstrated in murine models for GBM, where pharmacological inhibition of the translated protein showed promising results65. Finally, overexpression of pyruvate kinase M1/2 is a trait occurring in brain tumors but not in other cancers, where it is involved in metabolic and signaling functions that promote cell proliferation, migration and chemoresistance. Knocking down PKM alters central carbon metabolism and decreases the activation of oncogenes, thus making it a viable therapy target66.
Other genes of interest also identified by both Richards et al.9 and this study, currently lack (i) extensive evidence of therapeutic relevance to treating cancer and/or (ii) cognate ligands or compounds affecting their expression levels (or outdated chemical–gene interactions in databases): ACSBG1 (acyl-CoA synthetase bubblegum family member 1), COX6A1 (cytochrome c oxidase subunit 6A1), GUSB (glucuronidase beta), HADHB (acetyl-CoA acyltransferase), as well as multiple nucleoporins (NUP58, NUP107, NUP155, NUP205 and NUP201). A recent study in renal cell carcinoma cells showed that ACSBG1 induces ferroptosis, an iron-dependent regulated cell death proposed as a therapeutic approach for cancer treatment69. This mechanism plays a pro-tumorigenic effect in GBM by promoting necrosis, which is a sign of poor prognosis70. Interestingly, depleting ACSL4 (acyl-CoA synthetase long chain family member 4), a gene highlighted by the workflow, also inhibited tumor necrosis70. COX6A1, has been reported as an anti-apoptotic gene in U373MG GBM cells, involved in mitochondrial energy metabolism71. HADHB is involved in fatty acid β-oxidation, and gene knockdown in a colon carcinoma cell line decreased cell proliferation and increased sensitivity to the monoclonal antibody cetuximab72. Regarding the aforementioned nucleoporins, even though there is a known link to cancer73, targeting membrane proteins remains a challenge74.
Radial glia (RG) cells are located in the ventricular zone during embryonic development and in the subventricular zone in the adult brain. In Wang et al., RG-like cells were found to play a role in the initiation or maintenance of adult GBM54. Specifically, two subpopulations of RG cells were identified: ventricular RG-like and outer RG-like, and upregulated RG genes are consistent with our results (Fig. 4C). Firstly, only one gene for ventricular RG-like was found in this study. ACSL6 (acyl-CoA Synthetase Long Chain Family Member 6) is predominately expressed in the brain, where it participates in omega-3 fatty acid metabolism (crucial for correct brain function)75. Six genes were highlighted as outer RG-like: ABHD6 (Abhydrolase Domain-Containing Protein 6), CYP4V2 (cytochrome P450 family 4 subfamily V member 2), FASN (fatty acid synthase), POLH (DNA polymerase eta), MGLL and ACSBG154. ABHD6 is an endocannabinoid hydrolase with oncogenic functions, knockdown of which, reduces metastatic seeding in murine models76,77. CYP4V2 metabolizes fatty acids; however, as mutants with an impaired activity are the cause of multiple diseases (e.g. Bietti’s crystalline dystrophy), this gene may not be a suitable drug target78. Regarding FASN, it is known to be highly expressed in GBM, where its inhibition in vitro and in vivo by means of a saffron-derived compound, induced cell death and impaired migration79. POLH is a DNA repair gene associated with correcting UV-induced damage, and its deficiency increases the risk of developing skin cancer80. Conversely, ovarian cancer stem cells survive cisplatin treatment due to an enhanced POLH expression, leading to tumor relapse81. Lastly, the remaining two genes identified as outer RG-like (MGLL and ACSBG1) are also marker genes for intra-GSC clusters, as discussed above.
Targeting transcription factors driving characteristic biological capabilities of cancer remains an attractive therapeutic approach21. 47/168 genes of interest had entries in RegNetwork (Fig. 4D), a database for regulatory information last updated in 201967. Interestingly, 21 of these genes were related to the MYC family oncogene, which regulates a plethora of cellular functions and has been subjected to targeting studies at different levels82. Specifically, MYC maintains GSC83 and downregulating MYC promotes GBM autophagy84. Similarly, 13 genes are modulated by PPARα67, a transcription factor whose depletion inhibits tumor cell proliferation and induces senescence85. Overall, genes of interest identified by the workflow agreed with published attempts to find potential drug targets for GBM. Therefore, the validation of its predictive capabilities should foster more research into the aforementioned genes whose relevance to GBM has been poorly studied (e.g., ACSBG1). Improving the workflow by adding more biological information (such as epigenomics, proteomics or survival data) would allow further refining of potential drug targets to enable in vitro and in vivo efficacy characterizations.
Selection of chemicals and in vitro testing
The goal of TISMAN is to facilitate drug repurposing projects by generating a prioritized list of potential drug targets to identify chemicals for testing in an efficient and timely manner. Multiple gene–chemical interactions databases were combined and used to identify relevant compounds, ranking them by the number of genes they interacted with (directly and/or indirectly, Fig. 2). Next, a literature search allowed selection based upon proven efficacy for other cancer types, but which have not yet been tested for GBM (Fig. 1).This approach reduced thousands of gene-chemical interactions to a ranked list of 30 compounds, allowing a detailed inspection of each one of them. Upon considering reported results, five chemicals were selected based on their potential effects on cell viability and invasion in GBM: afuresertib, pyrogallol, isorhamnetin, formononetin, and taxifolin (Table 1).
Table 1.
Selected chemicals from the priority list for in vitro testing. Interactions for afuresertib were derived from Comparative Toxicogenomics Database (CTD) Oct 202046, and interactions for all other compounds derived from The Drug-Gene Interaction database (DGIdb) Nov 2020, v4.2.045. Gene relevance refers to the summation of the occurrence (extended choke point) and the topological importance (centrality score by PageRank). The stoichiometry of a reaction related to ACSF2 can be found in Supplementary Table S2.
| Position in priority list (relevance) | Drug/chemical | Gene name | Gene symbol | Gene relevance | Interaction(s) (predicted “–”) |
|---|---|---|---|---|---|
| 4 (385) | Afuresertib | Acyl-CoA synthetase family member 2 | ACSF2 | 119 | Increases exp |
| Acyl-CoA synthetase long chain family member 5 | ACSL5 | 86 | Increases exp | ||
| Phenylalanyl-TRNA synthetase subunit beta | FARSB | 9 | Decreases exp | ||
| Lipin 1 | LPIN1 | 9 | Increases exp | ||
| Metabolism of cobalamin associated B | MMAB | 26 | Increases exp | ||
| Patatin like phospholipase domain containing 2 | PNPLA2 | 3 | Increases exp | ||
| DNA polymerase alpha 2, accessory subunit | POLA2 | 4 | Decreases exp | ||
| DNA polymerase delta 4, accessory subunit | POLD4 | 4 | Increases exp | ||
| Solute carrier family 27 member 1. Long-chain fatty acid transport protein 1 | SLC27A1 | 119 | Increases exp | ||
| Solute carrier family 29 member 1. Equilibrative nucleoside transporter 1 | SLC29A1 | 6 | Decreases exp | ||
| 8 (123) | Pyrogallol | Hydroxysteroid 17-beta dehydrogenase 10 | HSD17B10 | 119 | – |
| DNA polymerase kappa | POLK | 4 | – | ||
| 9 (119) | Formononetin | Hydroxysteroid 17-beta dehydrogenase 10 | HSD17B10 | 119 | – |
| 13 (16) | 3-O-methylquercetin (Isorhamnetin) | DNA polymerase beta | POLB | 4 | – |
| DNA polymerase eta | POLH | 4 | – | ||
| DNA polymerase iota | POLI | 4 | – | ||
| DNA polymerase kappa | POLK | 4 | – | ||
| 13 (16) | Taxifolin | DNA polymerase beta | POLB | 4 | – |
| DNA polymerase eta | POLH | 4 | – | ||
| DNA Polymerase iota | POLI | 4 | – | ||
| DNA Polymerase kappa | POLK | 4 | – |
Afuresertib is an orally bioavailable inhibitor of the serine/threonine protein kinase Akt pathway86 that can resensitize ovarian cancer to platinum-based chemotherapy87 and enhanced cisplatin treatment of mesothelioma cells88. The PI3K/AKT/mTOR pathway is related to metabolism, proliferation and migration, and is generally active in GBM89. Pyrogallol is a natural polyphenol with antitumor effects in hepatocellular carcinoma via the upregulation of miR-13490. This microRNA is deregulated in GBM (U87 cells), and overexpressing it inhibited cell proliferation and invasion91. Formononetin is an isoflavone affecting multiple signaling pathways with proven antitumorigenic properties in a rat glioma cell line92,93. 3-O-methylquercetin (isorhamnetin) is a flavonoid that inhibits cell proliferation and invasion, and induces apoptosis in triple-negative breast cancer cells94. Finally, taxifolin (also a flavonoid) down-regulates β-catenin, thus promoting mesenchymal to epithelial transition—the reverse process is the epithelial to mesenchymal transition, related to metastasis. Specifically, this compound inhibited cell proliferation and invasion in breast cancer cell lines95. Knockdown of PELP1, a β-catenin coactivator, proved both elements to be crucial for GBM progression in a murine model96. Even though interesting candidates were identified in the databases, some instances referred to predicted interactions with DNA polymerases rather than stating the aforementioned actual effects (Table 1). Well-curated and maintained databases would greatly benefit drug repurposing efforts, and researchers should consider updating the chemical-gene interaction entries based upon new data.
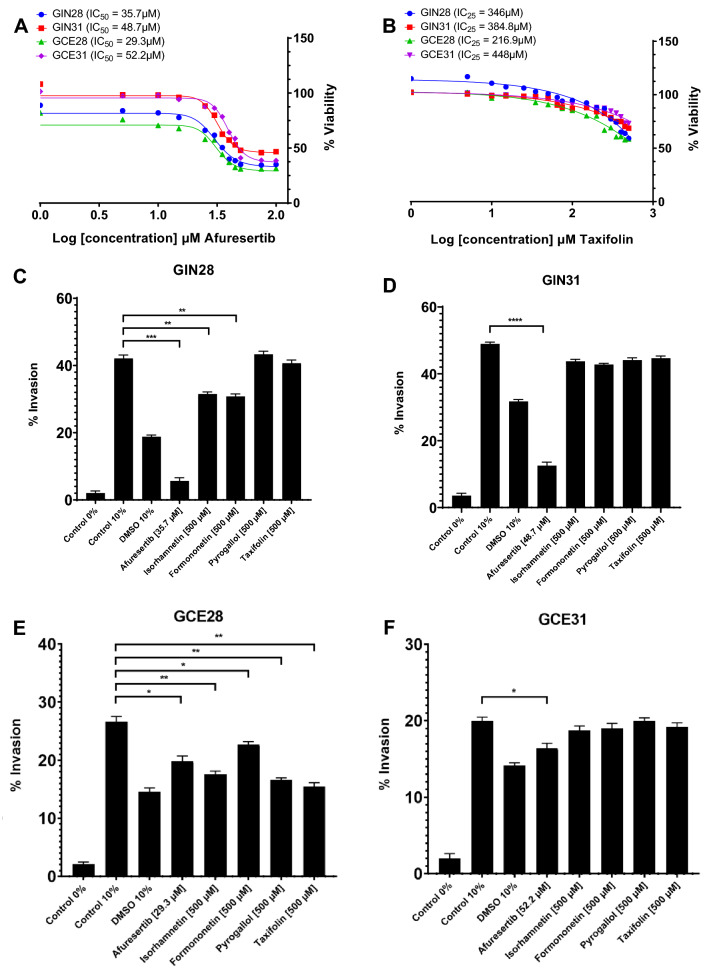
Two in vitro tests were performed to assess the effects of the selected chemicals on metabolic viability and invasiveness of four patient-derived GBM cell lines, as a means to validate this innovative in silico pipeline. Afuresertib was identified as the most potent drug inhibiting proliferation (Fig. 5A), while taxifolin showed a modest but significant effect on cell sensitization (Fig. 5B). No significant effect on proliferation was observed after treatment with isorhamnetin, formononetin and pyrogallol at 72 h. Regarding the invasion assays, afuresertib was the only compound to significantly inhibit invasion at IC50, impairing invasion by 86.7% (p < 0.001) and 74.5% (p < 0.0001) in GIN28 and GIN31 GBM invasive margin cells, respectively, and by 25.6% (p = 0.017) and 18% (p = 0.028) in corresponding GCE28 and GCE31 counterpart cell lines derived from the central tumor core. At 500 μM, isorhamnetin significantly reduced invasion by 25.2% (p = 0.0036) in GIN28 and 34.2% (p = 0.0011) in GCE28. A significant reduction in invading cells was also observed in GIN28 and GCE28 cell lines upon treatment with formononetin at 500 μM, impairing invasion by 26.8% (p = 0.0042) and 14.7% (p = 0.0193), respectively. While taxifolin demonstrated an antiproliferative effect across all cell lines, a significant effect on invasion was only observed in GCE28 (p = 0.0034) when used at 500 μM, reducing invasion to 15.5% (Fig. 5C–F). Therefore, afuresertib should be considered for in vitro studies to understand the underlying mechanisms to different responses, as well to assess the effect on GSCs, potentially leading to further in vivo studies to characterize its pre-clinical suitability. Collectively, in vitro assessment identifies 2/5 tested compounds as impairing metabolic viability, and 3/5 tested compounds as impairing invasion. Considering both intra- and inter-tumor heterogeneity which manifests in GBM, and thereby the likely biological variation in TCGA transcriptomics data compared to the patient-derived GBM lines used in this study, validation of 40% and 60% of repurposed compounds for anti-proliferative and anti-invasion effects, represents an effective hit rate and validates TISMAN as an in silico workflow to expedite compound prioritization at a drug discovery phase. Furthermore, identified genes lacking targeting molecules should be considered in structural studies to enable and exploit molecular docking efforts.
Figure 5.
In vitro assessment of identified compounds for drug repurposing as validation of in silico workflow. (A,B) Metabolic viability was assessed for afuresertib (A) and taxifolin (B) in a 96-well 16-point dose response assay across four patient-derived GBM lines. PrestoBlue was used to assess metabolic viability after 72 h and data is presented as the mean percent viability compared to DMSO-treated controls (n = 6). Afuresertib was identified as the most potent drug inhibiting proliferation at IC50 values of 35.7 μM, 48.7 μM, 29.3 μM and 52.2 μM for the GIN28, GIN31, GCE28 and GCE31 cell lines, respectively (A). Taxifolin showed a modest but significant effect on cell sensitisation generating IC25 values of 346 μM, 384.8 μM, 216.9 μM, 448 μM on GIN28, GIN31, GCE28 and GCE31 cell lines, respectively (B). (C–F) Effects on invasion in GBM invasive margin (GIN28, GIN31) and GBM Contrast-Enhanced core (GCE28, GCE31) cell lines following treatment with four compounds at 500 μM and afuresertib at IC50 concentration. Invasion towards 0% FBS (Control 0%) on untreated cells and DMSO treated (DMSO 10%) controls were also included. Data is presented as mean percent invasion ± SEM compared to number of seeded cells after 24 h incubation. Significance was tested by one-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001, **** p < 0.0001) comparing each data set to control 10% (invasion towards 10% FBS; n = 3).
Supplementary Information
Acknowledgements
This work was supported by the Biotechnology and Biological Sciences Research Council [Grant number BB/L013940/1], the Engineering and Physical Sciences Research Council [Grant number BB/L013940/1] and The Stoneygate Trust [Grant number CARO/SS/2016/CC1], through its Career Catalyst to RR. We thank the University of Nottingham for supporting the PhD studentship of CTA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
C.T.A. and D.H.K. designed research; C.T.A. designed and performed the modelling; A.P. and R.R. designed the experimental setup; A.P. performed the experiments; C.T.A. and A.P. analyzed data; C.T.A., A.P., K.W., J.K., R.R., and D.H.K. wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-14721-w.
References
- 1.World Health Organization. (Geneva, 2020).
- 2.Hanahan D, Weinberg R. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Hausser J, Alon U. Tumour heterogeneity and the evolutionary trade-offs of cancer. Nat. Rev. Cancer. 2020;20:247–257. doi: 10.1038/s41568-020-0241-6. [DOI] [PubMed] [Google Scholar]
- 4.Chen P-H, et al. Metabolic diversity in human non-small cell lung cancer cells. Mol. Cell. 2019;76:838–851. doi: 10.1016/j.molcel.2019.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J. Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 6.Petrecca K, Guiot M, Panet-Raymond V, Souhami L. Failure pattern following complete resection plus radiotherapy and temozolomide is at the resection margin in patients with glioblastoma. J. Neurooncol. 2013;111:19–23. doi: 10.1007/s11060-012-0983-4. [DOI] [PubMed] [Google Scholar]
- 7.Neftel C, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–849.e821. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards LM, et al. Gradient of developmental and injury response transcriptional states defines functional vulnerabilities underpinning glioblastoma heterogeneity. Nat. Cancer. 2021;2:157–173. doi: 10.1038/s43018-020-00154-9. [DOI] [PubMed] [Google Scholar]
- 10.Couturier CP, et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat. Commun. 2020;11:3406. doi: 10.1038/s41467-020-17186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castellan M, et al. Single-cell analyses reveal YAP/TAZ as regulators of stemness and cell plasticity in glioblastoma. Nat. Cancer. 2021;2:174–188. doi: 10.1038/s43018-020-00150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polson ES, et al. KHS101 disrupts energy metabolism in human glioblastoma cells and reduces tumor growth in mice. Sci. Transl. Med. 2018;10:eaar2718. doi: 10.1126/scitranslmed.aar2718. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, et al. Targeting pyrimidine synthesis accentuates molecular therapy response in glioblastoma stem cells. Sci. Transl. Med. 2019;11:eaau4972. doi: 10.1126/scitranslmed.aau4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCrorie P, et al. Biomedical engineering approaches to enhance therapeutic delivery for malignant glioma. J. Control. Release. 2020;328:917–931. doi: 10.1016/j.jconrel.2020.11.022. [DOI] [PubMed] [Google Scholar]
- 15.Moffat JG, Vincent F, Lee JA, Eder J, Prunotto M. Opportunities and challenges in phenotypic drug discovery: An industry perspective. Nat. Rev. Drug Discov. 2017;16:531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- 16.Faubert B, et al. Lactate metabolism in human lung tumors. Cell. 2017;171:358–371. doi: 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nilsson A, Haanstra J, Teusink B, Nielsen J. Metabolite depletion affects flux profiling of cell lines. Trends Biochem. Sci. 2018;43:395–397. doi: 10.1016/j.tibs.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Rancati G, Moffat J, Typas A, Pavelka N. Emerging and evolving concepts in gene essentiality. Nat. Rev. Genet. 2018;19:34–49. doi: 10.1038/nrg.2017.74. [DOI] [PubMed] [Google Scholar]
- 19.Lin A, Sheltzer JM. Discovering and validating cancer genetic dependencies: Approaches and pitfalls. Nat. Rev. Genet. 2020;21:671–682. doi: 10.1038/s41576-020-0247-7. [DOI] [PubMed] [Google Scholar]
- 20.Adasme MF, Parisi D, Sveshnikova A, Schroeder M. Structure-based drug repositioning: Potential and limits. Semin. Cancer Biol. 2020 doi: 10.1016/j.semcancer.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Bushweller JH. Targeting transcription factors in cancer—From undruggable to reality. Nat. Rev. Cancer. 2019;19:611–624. doi: 10.1038/s41568-019-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schellenberger J, et al. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox v2.0. Nat. Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orth JD, Thiele I, Palsson BO. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahn E, Kumar P, Mukha D, Tzur A, Shlomi T. Temporal fluxomics reveals oscillations in TCA cycle flux throughout the mammalian cell cycle. Mol. Syst. Biol. 2017;13:953. doi: 10.15252/msb.20177763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richelle A, Joshi C, Lewis NE. Assessing key decisions for transcriptomic data integration in biochemical networks. PLoS Comput. Biol. 2019 doi: 10.1371/journal.pcbi.1007185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Opdam S, et al. A systematic evaluation of methods for tailoring genome-scale metabolic models. Cell Syst. 2017;4:318–329. doi: 10.1016/j.cels.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pires Pacheco M, et al. Identifying and targeting cancer-specific metabolism with network-based drug target prediction. EBioMedicine. 2019;43:98–106. doi: 10.1016/j.ebiom.2019.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Csermely P, Korcsmáros T, Kiss H, London G, Nussinov R. Structure and dynamics of molecular networks: A novel paradigm of drug discovery. A comprehensive review. Pharmacol. Ther. 2013;138:333–408. doi: 10.1016/j.pharmthera.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colaprico A, et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubert, E. & Rousseeuw, P. J. In Similarity Search and Applications. (eds. Giuseppe, A., Claudio, G., Vincent, O., & Miloš, R.) 171–187 (Springer International Publishing).
- 31.Yoshimaru T, Nakamura Y, Katagiri T. Functional genomics for breast cancer drug target discovery. J. Hum. Genet. 2021;66:927–935. doi: 10.1038/s10038-021-00962-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maié, T. & Ticconi, F. Analysis of Cancer Genome Atlas in R, Retrieved from https://costalab.ukaachen.de/open_data/Bioinformatics_Analysis_in_R_2019/BIAR_D3/handout.html (2019). Accessed 20 Nov 2020.
- 33.Robinson J, et al. An atlas of human metabolism. Sci. Signal. 2020 doi: 10.1126/scisignal.aaz1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agren R, et al. The RAVEN toolbox and its use for generating a genome-scale metabolic model for Penicillium chrysogenum. PLoS Comput. Biol. 2013 doi: 10.1371/journal.pcbi.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vlassis N, Pires Pacheco M, Sauter T. Fast reconstruction of compact context-specific metabolic network models. PLoS Comput. Biol. 2014 doi: 10.1371/journal.pcbi.1003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zielinski D, et al. Systems biology analysis of drivers underlying hallmarks of cancer cell metabolism. Sci. Rep. 2017 doi: 10.1038/srep41241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner B, Venkataraman S, Buettner G. The rate of oxygen utilization by cells. Free Radic. Biol. Med. 2011;51:700–712. doi: 10.1016/j.freeradbiomed.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furse S, et al. Lipid membrane curvature induced by distearoyl phosphatidylinositol 4-phosphate. Soft Matter. 2012;8:3090–3093. doi: 10.1039/C2SM07358G. [DOI] [Google Scholar]
- 39.Chuang Y, et al. Role of synaptojanin 2 in glioma cell migration and invasion. Cancer Res. 2004;64:8271–8275. doi: 10.1158/0008-5472.CAN-04-2097. [DOI] [PubMed] [Google Scholar]
- 40.Ben-Chetrit N, et al. Synaptojanin 2 is a druggable mediator of metastasis and the gene is overexpressed and amplified in breast cancer. Sci. Signal. 2015 doi: 10.1126/scisignal.2005537. [DOI] [PubMed] [Google Scholar]
- 41.Budinich M, Bourdon J, Larhlimi A, Eveillard D. A multi-objective constraint-based approach for modeling genome-scale microbial ecosystems. PLoS One. 2017;12:e0171744. doi: 10.1371/journal.pone.0171744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marler RT, Arora JS. Survey of multi-objective optimization methods for engineering. Struct. Multidiscip. Optim. 2004;26:369–395. doi: 10.1007/s00158-003-0368-6. [DOI] [Google Scholar]
- 43.Page, L. B. & Motwani, R. W. (ed Stanford InfoLab) (1998).
- 44.Tomi-Andrino C, et al. Physicochemical and metabolic constraints for thermodynamics-based stoichiometric modelling under mesophilic growth conditions. PLoS Comput. Biol. 2021;17:e1007694. doi: 10.1371/journal.pcbi.1007694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freshour S, et al. Integration of the Drug–Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acid Res. 2020 doi: 10.1093/nar/gkaa1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis A, et al. Comparative toxicogenomics database (CTD): Update 2021. Nucleic Acid Res. 2020 doi: 10.1093/nar/gkaa891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilson M, et al. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acid Res. 2016;44:D1045–D1053. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piñeiro-Yáñez E, et al. PanDrugs: A novel method to prioritize anticancer drug treatments according to individual genomic data. Genome Med. 2018 doi: 10.1186/s13073-018-0546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheils T, et al. TCRD and Pharos 2021: Mining the human proteome for disease biology. Nucleic Acid Res. 2020 doi: 10.1093/nar/gkaa993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitsopoulos C, et al. canSAR: Update to the cancer translational research and drug discovery knowledgebase. Nucleic Acids Res. 2020;49:D1074–D1082. doi: 10.1093/nar/gkaa1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greco C, et al. Development of Pyrazolo[3,4-d]pyrimidine kinase inhibitors as potential clinical candidates for glioblastoma multiforme. ACS Med. Chem. Lett. 2020;11:657–663. doi: 10.1021/acsmedchemlett.9b00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vasey CE, et al. Polymer pro-drug nanoparticles for sustained release of cytotoxic drugs evaluated in patient-derived glioblastoma cell lines and in situ gelling formulations. Pharmaceutics. 2021 doi: 10.3390/pharmaceutics13020208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rogawski D, Vitanza N, Gauthier A, Ramaswamy V, Koschmann C. Integrating RNA sequencing into neuro-oncology practice. Transl. Res. 2017;189:93–104. doi: 10.1016/j.trsl.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang R, et al. Adult human glioblastomas harbor radial glia-like cells. Stem Cell Rep. 2020;14:338–350. doi: 10.1016/j.stemcr.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Folger O, et al. Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol. 2011 doi: 10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larsson I, Uhlén M, Zhang C, Mardinoglu A. Genome-scale metabolic modeling of glioblastoma reveals promising targets for drug development. Front. Genet. 2020 doi: 10.3389/fgene.2020.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szklarczyk D, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jung SH, et al. Acyl-CoA thioesterase 7 is involved in cell cycle progression via regulation of PKCζ–p53–p21 signaling pathway. Cell Death Dis. 2017;8:e2793–e2793. doi: 10.1038/cddis.2017.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.You BJ, et al. Orlistat displays antitumor activity and enhances the efficacy of paclitaxel in human hepatoma Hep3B cells. Chem. Res. Toxicol. 2019;32:255–264. doi: 10.1021/acs.chemrestox.8b00269. [DOI] [PubMed] [Google Scholar]
- 61.Grube S, et al. Overexpression of fatty acid synthase in human gliomas correlates with the WHO tumor grade and inhibition with Orlistat reduces cell viability and triggers apoptosis. J. Neurooncol. 2014;118:277–287. doi: 10.1007/s11060-014-1452-z. [DOI] [PubMed] [Google Scholar]
- 62.Ganipineni LP, et al. Paclitaxel-loaded multifunctional nanoparticles for the targeted treatment of glioblastoma. J. Drug Target. 2019;27:614–623. doi: 10.1080/1061186x.2019.1567738. [DOI] [PubMed] [Google Scholar]
- 63.Huang Y-Q, Wang Y, Hu K, Lin S, Lin X-H. Hippocampal glycerol-3-phosphate acyltransferases 4 and BDNF in the progress of obesity-induced depression. Front. Endocrinol. 2021 doi: 10.3389/fendo.2021.667773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pei Z, et al. Acyl-CoA synthetase VL3 knockdown inhibits human glioma cell proliferation and tumorigenicity. Cancer Res. 2009;69:9175–9182. doi: 10.1158/0008-5472.CAN-08-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin J, et al. ARS2/MAGL signaling in glioblastoma stem cells promotes self-renewal and M2-like polarization of tumor-associated macrophages. Nat. Commun. 2020;11:2978. doi: 10.1038/s41467-020-16789-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verma H, Cholia RP, Kaur S, Dhiman M, Mantha AK. A short review on cross-link between pyruvate kinase (PKM2) and Glioblastoma Multiforme. Metab. Brain Dis. 2021;36:751–765. doi: 10.1007/s11011-021-00690-y. [DOI] [PubMed] [Google Scholar]
- 67.Liu Z-P, Wu C, Miao H, Wu H. RegNetwork: An integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database. 2015 doi: 10.1093/database/bav095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oliveros, J. C. Venny. An interactive tool for comparing lists with Venn's diagrams. https://bioinfogp.cnb.csic.es/tools/venny/index.html (2007–2015). Accessed 1 Aug 2021.
- 69.Xie S, et al. Metabolic control by heat stress determining cell fate to ferroptosis for effective cancer therapy. ACS Nano. 2021;15:7179–7194. doi: 10.1021/acsnano.1c00380. [DOI] [PubMed] [Google Scholar]
- 70.Yee PP, et al. Neutrophil-induced ferroptosis promotes tumor necrosis in glioblastoma progression. Nat. Commun. 2020;11:5424. doi: 10.1038/s41467-020-19193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eun SY, et al. Identification of cytochrome c oxidase subunit 6A1 as a suppressor of Bax-induced cell death by yeast-based functional screening. Biochem. Biophys. Res. Commun. 2008;373:58–63. doi: 10.1016/j.bbrc.2008.05.178. [DOI] [PubMed] [Google Scholar]
- 72.Hu T-T, 2020. Detection of genes responsible for cetuximab sensitization in colorectal cancer cells using CRISPR-Cas9. Biosci. Rep. [DOI] [PMC free article] [PubMed]
- 73.Köhler A, Hurt E. Gene regulation by nucleoporins and links to cancer. Mol. Cell. 2010;38:6–15. doi: 10.1016/j.molcel.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 74.The International Transporter Consortium Membrane transporters in drug development. Nat. Rev. Drug Discov. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fernandez RF, et al. Acyl-CoA synthetase 6 enriches the neuroprotective omega-3 fatty acid DHA in the brain. Proc. Natl. Acad. Sci. 2018;115:12525. doi: 10.1073/pnas.1807958115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miralpeix C, et al. Carnitine palmitoyltransferase 1C negatively regulates the endocannabinoid hydrolase ABHD6 in mice, depending on nutritional status. Br. J. Pharmacol. 2021;178:1507–1523. doi: 10.1111/bph.15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tang Z, et al. Enhanced monoacylglycerol lipolysis by ABHD6 promotes NSCLC pathogenesis. EBioMedicine. 2020;53:102696. doi: 10.1016/j.ebiom.2020.102696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jarrar YB, Lee S-J. Molecular functionality of cytochrome P450 4 (CYP4) genetic polymorphisms and their clinical implications. Int. J. Mol. Sci. 2019 doi: 10.3390/ijms20174274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Colapietro A, et al. Crocetin extracted from saffron shows antitumor effects in models of human glioblastoma. Int. J. Mol. Sci. 2020 doi: 10.3390/ijms21020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Di Lucca J, et al. Variants of the xeroderma pigmentosum variant gene (POLH) are associated with melanoma risk. Eur. J. Cancer. 2009;45:3228–3236. doi: 10.1016/j.ejca.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 81.Srivastava AK, et al. Enhanced expression of DNA polymerase eta contributes to cisplatin resistance of ovarian cancer stem cells. Proc. Natl. Acad. Sci. U. S. A. 2015;112:4411–4416. doi: 10.1073/pnas.1421365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target. Ther. 2018;3:5. doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang J, et al. c-Myc is required for maintenance of glioma cancer stem cells. PLoS One. 2008;3:e3769. doi: 10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ke XX, Zhang R, Zhong X, Zhang L, Cui H. Deficiency of G9a inhibits cell proliferation and activates autophagy via transcriptionally regulating c-Myc expression in glioblastoma. Front. Cell Dev. Biol. 2020;8:593964. doi: 10.3389/fcell.2020.593964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Y, et al. PPARα regulates tumor cell proliferation and senescence via a novel target gene carnitine palmitoyltransferase 1C. Carcinogenesis. 2017;38:474–483. doi: 10.1093/carcin/bgx023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Spencer A, et al. The novel AKT inhibitor afuresertib shows favorable safety, pharmacokinetics, and clinical activity in multiple myeloma. Blood. 2014;124:2190–2195. doi: 10.1182/blood-2014-03-559963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blagden SP, et al. Phase IB dose escalation and expansion study of AKT inhibitor afuresertib with carboplatin and paclitaxel in recurrent platinum-resistant ovarian cancer. Clin. Cancer Res. 2019;25:1472–1478. doi: 10.1158/1078-0432.Ccr-18-2277. [DOI] [PubMed] [Google Scholar]
- 88.Yamaji M, et al. Novel ATP-competitive Akt inhibitor afuresertib suppresses the proliferation of malignant pleural mesothelioma cells. Cancer Med. 2017;6:2646–2659. doi: 10.1002/cam4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Le Rhun E, et al. Molecular targeted therapy of glioblastoma. Cancer Treat. Rev. 2019 doi: 10.1016/j.ctrv.2019.101896. [DOI] [PubMed] [Google Scholar]
- 90.Ahn H, et al. Antitumor effect of pyrogallol via miR-134 mediated S phase arrest and inhibition of PI3K/AKT/Skp2/cMyc signaling in hepatocellular carcinoma. Int. J. Mol. Sci. 2019 doi: 10.3390/ijms20163985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Niu CS, Yang Y, Cheng CD. MiR-134 regulates the proliferation and invasion of glioblastoma cells by reducing Nanog expression. Int. J. Oncol. 2013;42:1533–1540. doi: 10.3892/ijo.2013.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tay KC, et al. Formononetin: A review of its anticancer potentials and mechanisms. Front. Pharmacol. 2019;10:820. doi: 10.3389/fphar.2019.00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ong SKL, et al. Focus on formononetin: Anticancer potential and molecular targets. Cancers (Basel). 2019 doi: 10.3390/cancers11050611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu J, et al. ROS-mediated activation and mitochondrial translocation of CaMKII contributes to Drp1-dependent mitochondrial fission and apoptosis in triple-negative breast cancer cells by isorhamnetin and chloroquine. J. Exp. Clin. Cancer Res. 2019;38:225. doi: 10.1186/s13046-019-1201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li J, et al. Taxifolin inhibits breast cancer cells proliferation, migration and invasion by promoting mesenchymal to epithelial transition via β-catenin signaling. Life Sci. 2019;232:116617. doi: 10.1016/j.lfs.2019.116617. [DOI] [PubMed] [Google Scholar]
- 96.Sareddy GR, et al. PELP1 promotes glioblastoma progression by enhancing Wnt/β-catenin signaling. Neurooncol. Adv. 2019;1:vdz042. doi: 10.1093/noajnl/vdz042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Hu T-T, 2020. Detection of genes responsible for cetuximab sensitization in colorectal cancer cells using CRISPR-Cas9. Biosci. Rep. [DOI] [PMC free article] [PubMed]