Abstract
Strain DMS-S1 isolated from seawater was able to utilize dimethyl sulfide (DMS) as a sulfur source only in the presence of light in a sulfur-lacking medium. Phylogenetic analysis based on 16S ribosomal DNA genes indicated that the strain was closely related to Marinobacterium georgiense. The strain produced dimethyl sulfoxide (DMSO), which was a main metabolite, and small amounts of formate and formaldehyde when grown on DMS as the sole sulfur source. The cells of the strain grown with succinate as a carbon source were able to use methyl mercaptan or methanesulfonate besides DMS but not DMSO or dimethyl sulfone as a sole sulfur source. DMS was transformed to DMSO primarily at wavelengths between 380 and 480 nm by heat-stable photosensitizers released by the strain. DMS was also degraded to formaldehyde in the presence of light by unidentified heat-stable factors released by the strain, and it appeared that strain DMS-S1 used the degradation products, which should be sulfite, sulfate, or methanesulfonate, as sulfur sources.
Dimethyl sulfide (DMS) released from the sea is an important compound in global sulfur circulation (28) and global climate regulation (4). DMS is generated by the degradation of dimethylsulfoniopropionate, which is present in many species of marine algae and plants, including dinoflagellates and coccolithophores (20). However, more than 10 times the amount of DMS released to the atmosphere is degraded in the sea by microorganisms (23). DMS is degraded or transformed by terrestrial and marine microorganisms via methyl mercaptan (MM) or dimethyl sulfoxide (DMSO). Some strains of sulfur oxidizers (5, 19, 29, 32, 33) and methylotrophs (7, 9, 30, 38) degrade DMS via MM. Ammonia oxidizers (18), methanotrophs (11), algae (12), and some strains of phototrophs (15, 31, 36) transform DMS to DMSO. Some terrestrial heterotrophic bacterial strains also have been found to degrade or transform DMS via DMSO. Comamonas acidovorans DMR-11 (originally Pseudomonas acidovorans) isolated from peat biofilters transformed DMS to DMSO in the medium containing other organic carbon sources, such as sodium malate (37). A dibenzothiophene-desulfurizing bacterium, Rhodococcus sp. strain SY1, was able to degrade DMS in the oxidative pathway via DMSO, dimethyl sulfone, methanesulfonate, and sulfate, and genes encoding an enzyme that oxidized DMS to DMSO have been cloned from Acinetobacter sp. strain 20B, which was able to grow on DMS as the sole sulfur source (17, 26). Recently, several marine isolates in the α subclass of the class Proteobacteria have been found to be able to transform DMS to DMSO or MM (14). There have been no reports on marine heterotrophic isolates other than the α subclass of the class Proteobacteria that are able to degrade DMS aerobically. This paper reports the isolation and characterization of a marine heterotrophic bacterium that belongs to the γ subclass of the class Proteobacteria and is able to utilize DMS as the sole sulfur source only in the presence of light.
Isolation and characterization of strain DMS-S1.
The basal medium (NSYE) for isolation and cultivation of strain DMS-S1 contained 25 g of NaCl, 0.7 g of KCl, 50 mg of KH2PO4, 1 g of NH4NO3, 0.2 g of MgCl2 · 6H2O, 20 mg of CaCl2 · 2H2O, 5 mg of FeEDTA, 1 g of Tris, 5 mg of yeast extract, and 5 g of sodium succinate in 1 liter of distilled water. The final pH was adjusted to 7.7 to 8.0 with NaOH solution. The basal medium was autoclaved at 110°C for 10 min. ZoBell medium (27) with 1.5% agar was also used for isolation by streaking. Isolation and cultivation were done at 21°C with shaking at 100 rpm under illumination (45 to 57 μmol · m−2 · s−1) provided by 20-W fluorescent lamps. Strain DMS-S1 was isolated from a marine sample taken from Edauchi Bay (Hiroshima, Japan) in November 1992. The cells of strain DMS-S1 are gram-negative, non-spore-forming, aerobic, short rods with single polar flagella. They are oxidase positive, catalase positive, and O-F test negative, and they require a seawater base for growth. The activities of DNA hydrolysis and gelatin hydrolysis were not found. The quinone type of the cells was Q-8. The G+C content of the DNA was 56 mol%. To test growth on various carbon sources, carbon sources were added to SWNC medium (1 g of NH4NO3, 0.5 g of KH2PO4, 5 mg of FeEDTA, and 10 mg of yeast extract in 1 liter of filtered seawater, pH 7.7 to 8.0) or to NSYE medium without sodium succinate and containing 2 to 10 mM Na2SO4. The strain was cultured in 25- by 200-mm (71-ml capacity) test tubes containing 20 ml of media with Teflon-lined screw caps. Strain DMS-S1 was able to utilize succinate, acetate, ethanol, propanol, and butanol as carbon sources. The strain was not able to utilize glucose, glycerol, methanol, DMS, DMSO, dimethyl sulfone, methanesulfonate, diethyl sulfide, tetrahydrothiophene, diethyl sulfone, ethanesulfonate, methionine, or (2-carboxyethyl)dimethylsulfonium chloride as carbon sources.
A 16S ribosomal DNA sequence containing 1,478 bp of strain DMS-S1 was analyzed as described previously (11). Strain DMS-S1 was closely related to Marinobacterium georgiense, which was isolated from a marine pulp mill effluent enrichment culture (13). The similarity of the 1,423 bp of strain DMS-S1 to the M. georgiense sequence was 98.2%. The characteristics of strain DMS-S1 observed here resembled those of M. georgiense reported by Gonzalez et al. (13) except for the utilization of glucose, glycerol, and methanol as carbon sources, though the media used for the tests were different. M. georgiense ATCC 700074 was not able to grow on DMS as a sulfur source under the same culture conditions as strain DMS-S1 within 5 days after inoculation. Based on these findings, strain DMS-S1 was identified as a Marinobacterium sp.
Growth of strain DMS-S1 on organic sulfur compounds as sole sulfur sources.
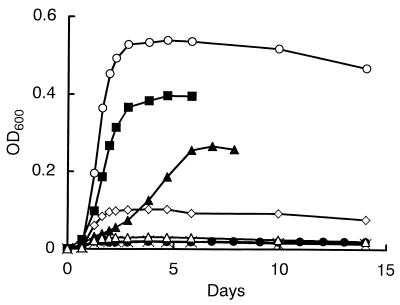
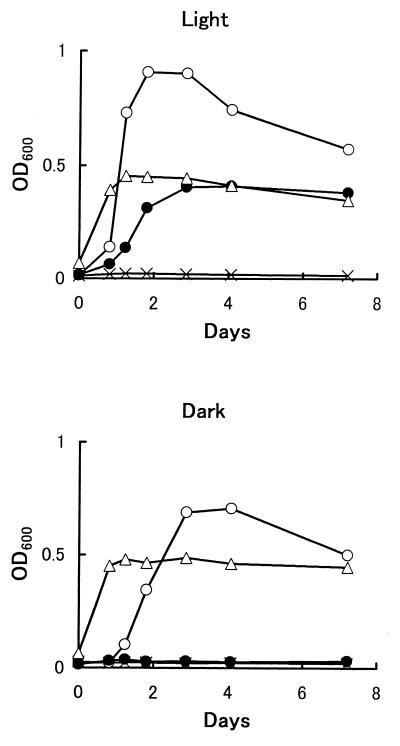
The growth of strain DMS-S1 on DMS as a sulfur source was compared with growth on Na2SO4. For the experiments assessing growth on organic sulfur compounds as sulfur sources, a sulfur source was added to the medium after filter sterilization with a 0.2-μm-pore-size membrane filter. Precultured strain DMS-S1 in sulfate-containing medium was inoculated after dilution with the basal medium. Growth of the strain was monitored at 600 nm with a Spectronic 20D spectrophotometer (Milton Roy Co., Rochester, N.Y.). Cultures were always run in pairs under the same conditions, and values for growth are shown here as the means for the two cultures. We have not analyzed the sulfur concentration of basal medium exactly. However, the results of the growth experiment (Fig. 1) showed that the sulfur compounds present in the basal medium and carried over from preculture were less effective than 0.2 μM Na2SO4. The maximum growth of the strain on 4 μmol of DMS, which is calculated to be 0.16 mM in the medium after equilibrium (6), was less than that on 400 nmol of Na2SO4 (20 μM in the medium) and more than that on 40 nmol of Na2SO4 (2 μM in the medium) (Fig. 1). This suggests that the strain requires over 10-fold more DMS than Na2SO4 to support growth. Effects of light on the growth of the strain were examined (Fig. 2). The strain needed light only when DMS was the sole sulfur source.
FIG. 1.
Growth of strain DMS-S1 utilizing Na2SO4 or DMS as a sulfur source. Four nanomoles of Na2SO4 (▵), 40 nmol of Na2SO4 (◊), 400 nmol of Na2SO4 (○), 400 nmol of DMS (●), 4 μmol of DMS (▴), or 40 μmol of DMS (■) was added to 71-ml test tubes containing 20 ml of NSYE medium. The concentrations in the medium after equilibrium were calculated to be as follows: 0.2 μM Na2SO4 (▵), 2 μM Na2SO4 (◊), 20 μM Na2SO4 (○), 16 μM DMS (●), 160 μM DMS (▴), and 1.6 mM DMS (■). ×, NSYE medium without added sulfur sources. OD600, optical density at 600 nm.
FIG. 2.
Growth of strain DMS-S1 with or without light. Twenty milliliters of medium was used. ×, basal medium without added sulfur sources; ○, basal medium with 2 mM Na2SO4; ●, basal medium with 16 mM DMS (total, 400 μmol in a tube); ▵, ZoBell medium.
Organic sulfur compounds utilized as sulfur sources by the strain are shown in Table 1. When MM gas was added to the test tubes, screw caps with a valve for a syringe were used. This strain was able to utilize diethyl sulfide but not di-n-propyl sulfide or di-n-butyl sulfide. It was not able to grow on DMSO or dimethyl sulfone but was able to grow on MM as a sulfur source, unlike Rhodococcus sp. strain SY1 and Acinetobacter sp. strain 20B, which were able to grow on DMSO and dimethyl sulfone but not on MM as a sulfur source (17, 26). Strain DMS-S1 also utilized alkanesulfonates as a sulfur source. MM and methanesulfonate were utilized even in the absence of light.
TABLE 1.
Growth of strain DMS-S1 on various sulfur compounds as sulfur sources
| Sulfur compounda (purity and sourceb) | Growth in:
|
|
|---|---|---|
| Light | Dark | |
| MM (>98.5%, SS) | + | + |
| DMS (>99%, TK) | + | − |
| DMSO (>99%, WA) | − | |
| Dimethyl sulfone (>99%, WA) | − | |
| Methanesulfonic acid sodium salt (>98%, AL) | + | + |
| Dimethyl disulfide (>98%, TK) | + | |
| Ethyl mercaptan (>98%, WA) | + | |
| Diethyl sulfide (>97%, TK) | + | |
| Diethyl sulfone (>97%, AL) | + | |
| Ethanesulfonic acid sodium salt (>98%, TK) | + | + |
| Diethyl disulfide (>99%, TK) | − | |
| n-Propanethiol (>95%, WA) | − | |
| Di-n-propyl sulfide (>98%, TK) | − | |
| Di-n-propyl sulfone (>99%, TK) | − | |
| 1-Propanesulfonic acid sodium salt (>98%, TK) | + | |
| n-Butyl mercaptan (>95%, WA) | − | |
| Di-n-butyl sulfide (>95%, TK) | − | |
| Di-n-butyl sulfoxide (>96%, TK) | − | |
| Di-n-butyl sulfone (>98%, TK) | ± | |
| 1-Butanesulfonic acid sodium salt (>98%, TK) | + | |
| Methylphenyl sulfide (>99%, TK) | − | |
| Diphenyl sulfide (>98%, TK) | − | |
| Tetrahydrothiophene (>99%, TK) | + | |
| Tetramethylene sulfoxide (>95%, TK) | − | |
| Sulfolane (>99%, TK) | − | |
| l-Methionine (>99%, WA) | + | |
| (2-Carboxyethyl)dimethylsulfonium chloride (>98%, TK) | − | |
For each compound except MM, 40 μmol was added to 20 ml of basal medium; 4 μmol of gaseous MM was added to each medium.
Guaranteed purity and the company from which the chemical was purchased: AL, Aldrich; SS, Sumitomo Seika; TK, Tokyo Kasei Kogyo; WA, Wako Pure Chemical.
Metabolites from DMS produced by growth of strain DMS-S1.
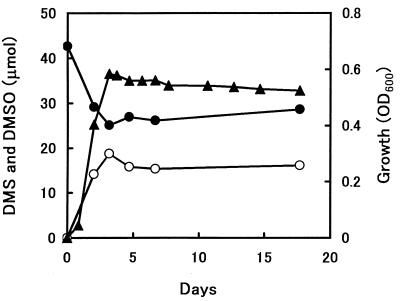
The culture supernatant of strain DMS-S1 grown on DMS was analyzed by gas chromatography-mass spectrometry (GC-MS). For GC-MS, a JMS Automass 150 (JEOL, Tokyo, Japan) or a QP-5000 (Shimadzu, Kyoto, Japan) with a 30-m capillary column (DB-5; J&W Scientific, Folsom, Calif.) was used. The obtained fragmentation of a metabolite from DMS occurring at m/z (%) 78 (58) and 63 (100) had a pattern similar to that from authentic DMSO, whose fragments were at m/z (%) 78 (63) and 63 (100). The metabolite was identified as DMSO. Residual DMS and accumulated DMSO produced by the growth of strain DMS-S1 on DMS were quantified by GC with a flame photometric detector as described previously (12). DMSO was the main product when strain DMS-S1 was grown on DMS (Fig. 3). Accumulation of DMSO in the medium without inoculation of the strain was negligible. There were no metabolites corresponding to dimethyl sulfone on the gas chromatogram.
FIG. 3.
Growth and DMSO transformed from DMS by strain DMS-S1. The concentration of DMS in the medium was calculated to be 1.7 mM at the start. Growth of strain DMS-S1 is expressed as the optical density at 600 nm (OD600) (▴). DMSO (○) and residual DMS (●) are expressed in micromoles. Values are means for two samples.
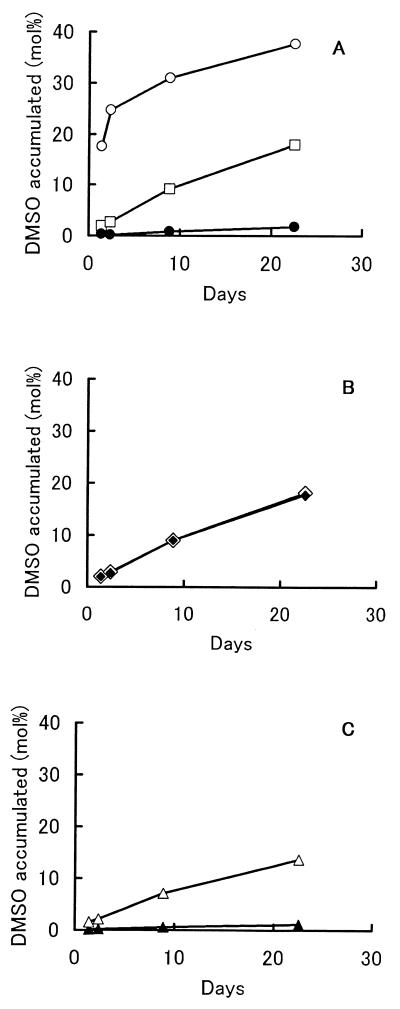
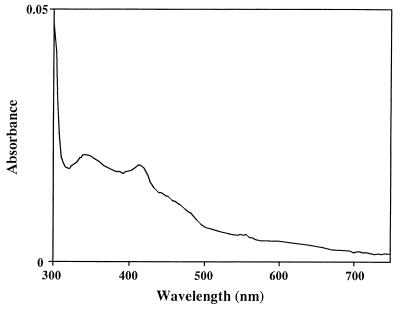
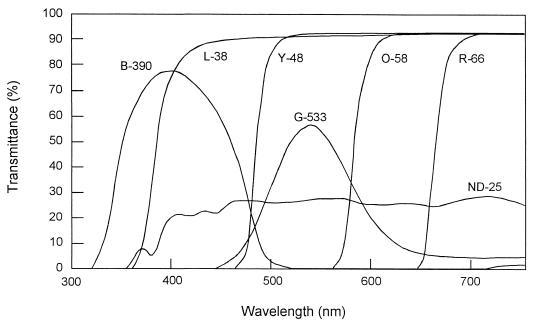
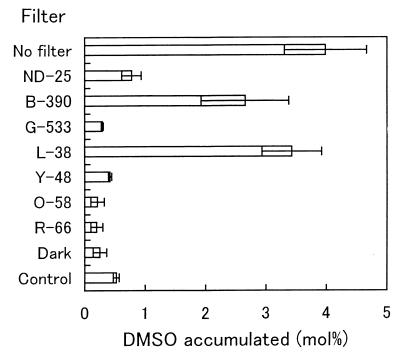
The factor for transforming DMS to DMSO in the culture was investigated. Strain DMS-S1 was cultured in NSYE medium containing 20 μM Na2SO4 for several days. The broth was separated into supernatant and cells by centrifugation (6,300 to 9,800 × g , 10 min) and filtration with 0.2-μm-pore-size Nucleopore filters. The supernatant was subjected to ultrafiltration using USY-1 (Advantec, Tokyo, Japan), which separated molecules larger than 10,000. The retained concentrate of the ultrafiltration was made up with NS buffer (25 g of NaCl, 0.7 g of KCl, 0.2 g of MgCl2 · 6H2O, 20 mg of CaCl2 · 2H2O, and 0.5 g of Tris in 1 liter of distilled water, pH 7.7) to the volume of the supernatant subjected to ultrafiltration. Cells separated by centrifugation were washed with NS buffer and suspended in the buffer at the same volume as the broth. The factor was found in the culture supernatant but not in the cells and was stable to heat treatment for 5 min at 105°C. The molecular weight was lower than 10,000 (Fig. 4). An absorption spectrum of the culture supernatant which was prepared from the culture grown on 40 μM Na2SO4 was measured in a Hitachi 150-20 spectrophotometer. The culture supernatant of the strain was almost colorless, though its absorption spectrum had two local maxima at about 340 and 412 nm (Fig. 5). Wavelengths of fluorescent lamps needed for the oxidation of DMS to DMSO in the heated culture supernatants were investigated by using seven optical filters purchased from Kenko. Absorption characteristics of the filters are shown in Fig. 6. The oxidation of DMS occurred predominantly between 380 and 480 nm (Fig. 7). This wavelength is almost coincident with the result for seawater (21). These facts suggested that the factor for transforming DMS to DMSO was a photosensitizer found in algae and seawater (2, 12).
FIG. 4.
Transformation of DMS by culture components of strain DMS-S1. The strain was grown in NSYE medium with 20 μM Na2SO4, and the culture medium was separated into components as described in the text. Forty-five micromoles of DMS was added to 4.5 ml of culture component solution in 22-ml vials (7.8 mM DMS in the solution). (A) Whole culture medium containing cells of strain DMS-S1 (○), culture supernatant (□), and cells in buffer (●). (B) Whole culture medium containing cells heated for 5 min at 105°C (◊) and culture supernatant heated for 5 min at 105°C (⧫). (C) Components of culture supernatant whose molecular weight was higher than 10,000 in buffer (▴) and culture supernatant after exclusion of the components with a molecular weight higher than 10,000 (▵).
FIG. 5.
Absorption spectrum of the culture supernatant of strain DMS-S1 grown on 40 μM Na2SO4.
FIG. 6.
Absorption characteristics of optical filters.
FIG. 7.
DMSO accumulation by heated culture supernatants under light passed through optical filters. Twenty-nine micromoles of DMS was added to 3.6 ml of heated (105°C, 5 min) culture supernatants in 22-ml vials. The vials were kept under light passed through optical filters, and the DMSO that accumulated in the supernatants was quantified after 4 days. NSYE medium containing 40 μM Na2SO4 and 29 μmol of DMS was used as a control and kept under light without a filter. Error bars indicate standard deviations for three samples.
Production of formate by the strain was investigated via high-pressure liquid chromatography after derivatization of products with 2-nitrophenylhydrazine (1). When formate in the cultured medium was to be quantified, 1 to 2 ml of ethanol, butanol, or propanol was substituted for 5 g of sodium succinate in 1 liter of NSYE medium because large amounts of succinate interfered with the derivatization of formate and made the detection of formate difficult. Accumulation of formate was detected only in the cultures grown on DMS as a sulfur source, and it was negligible in the cultures grown on sulfate no matter what alcohol was used as a carbon source. This fact suggested that formate was produced during DMS utilization by this strain or that DMS might have inhibited the metabolism of formate derived from other sources. Formate accumulated from DMS was about 3 to 5% (mol/mol) of the added 40 μmol of DMS after cultivation for 13 to 18 days. Accumulation of formate from DMS by the culture grown on sulfate as a sulfur source was suppressed in the absence of light (data not shown).
In addition, we examined the production of formaldehyde by the culture components of the strain. A product from DMS was identified via GC-MS and quantified via high-pressure liquid chromatography after derivatization to formaldehyde-2,4-dinitrophenylhydrazone (22). The obtained fragmentation of the derivatized metabolite from DMS occurring at m/z (%) 210 (50), 180 (9), 152 (11), 122 (22), 91 (18), 79 (77), 63 (100), and 51 (91) had a pattern similar to that obtained from the authentic formaldehyde-2,4-dinitrophenylhydrazone, with fragments at m/z (%) 210 (47), 180 (11), 152 (8), 122 (20), 91 (22), 79 (93), 63 (100), and 51 (94). Thus, the metabolite was identified as formaldehyde. A larger amount of formaldehyde was accumulated by the culture supernatant than by the cells in buffer, and it was also accumulated by the culture supernatant heated for 5 min at 105°C, though formate was not accumulated by the culture supernatant (Table 2). These facts confirmed that formaldehyde and formate were produced during DMS utilization by this strain. Accumulation of MM was not observed with the production of formaldehyde by the heat-treated culture components.
TABLE 2.
Accumulation of formaldehyde and formate by culture components of strain DMS-S1a
| Culture component(s) | Heat treatmentb | Added DMS | Amt (μmol) of accumulated:
|
|
|---|---|---|---|---|
| Formaldehyde | Formate | |||
| Cells and supernatant | − | − | 0.01 | 0.29 |
| + | 0.21 | 1.51 | ||
| + | − | 0.14 | 0.35 | |
| + | 0.37 | 0.47 | ||
| Cells in NS buffer | − | − | 0.03 | 0.10 |
| + | 0.12 | 0.24 | ||
| + | − | 0.05 | 0.02 | |
| + | 0.09 | 0.08 | ||
| Supernatant | − | − | 0.10 | 0.28 |
| + | 0.25 | 0.28 | ||
| + | − | 0.15 | 0.37 | |
| + | 0.44 | 0.53 | ||
Strain DMS-S1 was grown on 0.2% (34 mM) ethanol with 10 μM Na2SO4 for 6 days. Then the culture was separated into culture components, and 40 μmol of DMS was added to 20 ml of culture component solution in 71-ml test tubes. After cultivation for 11 to 17 days, formate and formaldehyde in a pair of culture component solutions were quantified. Mean values are shown.
Samples were heated for 5 min at 105°C.
Proposed pathway of DMS degradation by strain DMS-S1.
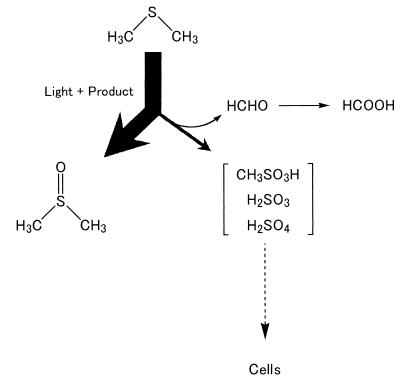
DMS is readily oxidized to DMSO by photochemical reactions in the presence of photosensitizers such as humic acid, methylene blue, and rose bengal (2), though DMS does not undergo appreciable photo-oxygenation in the absence of photosensitizers. Our results for DMSO production from DMS suggested that strain DMS-S1 excreted substances that served as photosensitizers. However, formaldehyde, which was an unexpected product in the photosensitizing reaction, was also produced from DMS by the supernatant of this strain in the presence of light. DMS is also photooxidized to formaldehyde, sulfur dioxide, sulfate, and methanesulfonate under light in the presence of NOX in the air (16, 35). Hatakeyame et al. (16) reported that methanesulfonate was the main product and the yield was more than 50%. On the other hand, according to Yin et al. (35), sulfur dioxide was the main product and the yield was 62 to 71%. We have not been able to detect sulfite, sulfate, or methanesulfonate in this reaction because high concentrations of salts prevented detection of these compounds by the methods available to us. On the other hand, strain DMS-S1 could utilize sulfate and methanesulfonate as a sulfur source but not DMSO, and production of MM was not observed in this photooxidation of DMS to formaldehyde. Therefore, sulfite, sulfate, and methanesulfonate are the most likely intermediates that serve as sources of sulfur during the utilization of DMS by this strain. Based on the results and the speculations described above, we propose the degradation pathway of DMS by Marinobacterium sp. strain DMS-S1 shown in Fig. 8. This pathway corresponds with the results of growth experiments. DMS is transformed mainly to unusable DMSO and only a small percentage of DMS is used for growth, which was why more than 10 times more DMS than sulfate was required for growth. The compounds that aid in photolysis of DMS need to be purified to clarify the photooxidation of DMS by this strain.
FIG. 8.
Proposed pathway of DMS degradation by Marinobacterium sp. strain DMS-S1.
Sulfate is assimilated after reduction to sulfite (25). Methanesulfonate is decomposed and sulfite is released from it by monooxygenases in Methylosulfonomons methylovora (8) and E. coli (10). Some marine bacteria belonging to the α subclass of the class Proteobacteria are able to release MM from dimethylsulfoniopropionate and DMS and incorporate it directly into methionine (14, 24). They use MM in preference to sulfate, which is present at 106- to 107-fold-higher concentrations. The merit of usage of MM is to save the reducing power needed for conversion of sulfate to sulfide. As for strain DMS-S1, the assimilation of sulfite or methanesulfonate instead of sulfate could also provide an energetic advantage, though they are less effective than MM.
The oxidation of DMS to other compounds in the sea plays an important role in sulfur circulation because the oxidation reduces the release of DMS into the air. The reaction decomposing DMS to formaldehyde is irreversible, while DMSO oxidized from DMS can be reduced back to DMS again (34). Kieber et al. pointed out that only 14% of DMS photolyzed was converted to DMSO and that the relatively low conversion was not due to losses of DMSO (21). The reaction decomposing DMS to formaldehyde may play some role in the loss of DMS. Photolysis of DMS accounted for 7 to 40% of the total turnover of DMS in the photic zone of the equatorial Pacific Ocean (21). On the other hand, 88% of the DMS was photolyzed in the top 10 m of the water column of the northern Adriatic Sea (3). Not only the quantity but also the quality of dissolved organic carbon affects the photolysis of DMS, as mentioned by Brugger et al. (3). This study showed that marine bacteria excrete substances which could transform DMS to DMSO and decompose DMS, releasing formaldehyde photochemically. Marine algae were also shown to produce photosensitizers (12). These facts suggest that photolysis of DMS is also affected by biological activities, although we are not able to estimate the effects yet.
Acknowledgments
We thank Terumi Tanimoto in Chugoku National Industrial Research Institute for his help in collecting marine samples. We also thank Misaki Ohta at Towa Kagaku Co., Ltd., for her help in determining the 16S ribosomal DNA sequences.
REFERENCES
- 1.Albert D B, Martens C S. Determination of low-molecular-weight organic acid concentrations in seawater and pore-water samples via HPLC. Mar Chem. 1997;56:27–37. [Google Scholar]
- 2.Brimblecombe P, Shooter D. Photo-oxidation of dimethylsulphide in aqueous solution. Mar Chem. 1986;19:343–353. [Google Scholar]
- 3.Brugger A, Slezak D, Obernosterer I, Herndl G J. Photolysis of dimethylsulfide in the northern Adriatic Sea: dependence on substrate concentration, irradiance and DOC concentration. Mar Chem. 1998;59:321–331. [Google Scholar]
- 4.Charlson R J, Lovelock J E, Andreae M O, Warren S G. Ocean phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature. 1987;326:655–661. [Google Scholar]
- 5.Cho K S, Hirai M, Shoda M. Degradation characteristics of hydrogen sulfide, methanethiol, dimethyl sulfide and dimethyl disulfide by Thiobacillus thioparus DW44 isolated from peat biofilter. J Ferment Bioeng. 1991;71:384–389. [Google Scholar]
- 6.Dacey J H W, Wakeham S G, Howes B L. Henry's law constants for dimethylsulfide in freshwater and seawater. Geophys Res Lett. 1984;11:202–208. [Google Scholar]
- 7.De Bont J A M, van Dijken J P, Harder W. Dimethyl sulphoxide and dimethyl sulphide as a carbon, sulphur and energy source for growth of Hyphomicrobium S. J Gen Microbiol. 1981;127:315–323. [Google Scholar]
- 8.De Marco P, Moradas-Ferreira P, Higgins T P, McDonald I, Kenna E M, Murrell J C. Molecular analysis of a novel methanesulfonic acid monooxygenase from the methylotroph Methylosulfonomonas methylovora. J Bacteriol. 1999;181:2244–2251. doi: 10.1128/jb.181.7.2244-2251.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Zwart J M M, Nelisse P N, Kuenen J G. Isolation and characterization of Methylophaga sulfidovorans sp. nov.: an obligately methylotrophic, aerobic, dimethylsulfide oxidizing bacterium from a microbial mat. FEMS Microbiol Lett. 1996;20:261–270. [Google Scholar]
- 10.Eichhorn, E., J. R. van der Ploeg, and T. Leisinger. 1999. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. 274:26639–26646. [DOI] [PubMed]
- 11.Fuse H, Ohta M, Takimura O, Murakami K, Inoue H, Yamaoka Y, Oclarit J M, Omori T. Oxidation of trichloroethylene and dimethyl sulfide by a marine Methylomicrobium strain containing soluble methane monooxygenase. Biosci Biotechnol Biochem. 1998;62:1925–1931. doi: 10.1271/bbb.62.1925. [DOI] [PubMed] [Google Scholar]
- 12.Fuse H, Takimura O, Kamimura K, Murakami K, Yamaoka Y, Murooka Y. Transformation of dimethyl sulfide and related compounds by cultures and cell extracts of marine phytoplankton. Biosci Biotechnol Biochem. 1995;59:1773–1775. [Google Scholar]
- 13.Gonzalez J M, Mayer F, Moran M A, Hodson R E, Whitman W B. Microbulbifer hydrolyticus gen. nov., sp. nov., and Marinobacterium georgiense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int J Syst Bacteriol. 1997;47:369–376. doi: 10.1099/00207713-47-2-369. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez J M, Kiene R P, Moran M A. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl Environ Microbiol. 1999;65:3810–3819. doi: 10.1128/aem.65.9.3810-3819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanlon S P, Holt R A, Moore G R, McEwan A G. Isolation and characterization of a strain of Rhodobacter sulfidophilus: a bacterium which grows autotrophically with dimethylsulphide as electron donor. Microbiology. 1994;140:1953–1958. [Google Scholar]
- 16.Hatakeyama S, Okuda M, Akimoto H. Formation of sulfur dioxide and methanesulfonic acid in the photooxidation of dimethyl sulfide in the air. Geophys Res Lett. 1982;9:583–586. [Google Scholar]
- 17.Horinouchi M, Kasuga K, Nojiri H, Yamane H, Omori T. Cloning and characterization of genes encoding an enzyme which oxidizes dimethyl sulfide in Acinetobacter sp. Strain 20B. FEMS Microbiol Lett. 1997;155:99–105. doi: 10.1111/j.1574-6968.1997.tb12692.x. [DOI] [PubMed] [Google Scholar]
- 18.Juliette L Y, Hyman M R, Arp D J. Inhibition of ammonia oxidation in Nitrosomonas europaea by sulfur compounds: thioethers are oxidized to sulfoxides by ammonia monooxygenase. Appl Environ Microbiol. 1993;59:3718–3727. doi: 10.1128/aem.59.11.3718-3727.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanagawa T, Kelly D P. Breakdown of dimethyl sulphide by mixed cultures and by Thiobacillus thioparus. FEMS Microbiol Lett. 1986;34:13–19. [Google Scholar]
- 20.Keller M D, Bellows W K, Guillard R R L. Dimethyl sulfide production in marine phytoplankton. In: Saltzman E S, Cooper W J, editors. Biogenic sulfur in the environment. Washington, D.C.: American Chemical Society; 1989. pp. 167–182. [Google Scholar]
- 21.Kieber D J, Jiao J, Kiene R P, Bates T S. Impact of dimethylsulfide photochemistry on methyl sulfur cycling in equatorial Pacific Ocean. J Geophys Res. 1996;101:3715–3722. [Google Scholar]
- 22.Kieber R J, Mopper K. Determination of picomolar concentration of carbonyl compounds in natural water, including seawater, by liquid chromatography. Environ Sci Technol. 1990;24:1477–1481. [Google Scholar]
- 23.Kiene R P, Bates T S. Biological removal of dimethyl sulfide from sea water. Nature. 1990;345:702–705. [Google Scholar]
- 24.Kiene R P, Linn L J, González J, Moran M A, Bruton J A. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl Environ Microbiol. 1999;65:4549–4558. doi: 10.1128/aem.65.10.4549-4558.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montoya G, Svensson C, Savage H, Schwenn J D, Sinning I. Crystallization and preliminary X-ray diffraction studies of phospho-adenylylsulfate (PAPS) reductase from E. coli. Acta Crystallogr D. 1998;54:281–283. doi: 10.1107/s0907444997010019. [DOI] [PubMed] [Google Scholar]
- 26.Omori T, Saiki Y, Kasuga K, Kodama T. Desulfurization of alkyl and aromatic sulfides and sulfonates by dibenzothiophene-desulfurizing Phodococcus sp. strain SY1. Biosci Biotechnol Biochem. 1995;59:1195–1198. [Google Scholar]
- 27.Oppenheimer C H, ZoBell C E. The growth and viability of sixty-three species of marine bacteria as influenced by hydrostatic pressure. J Mar Res. 1952;11:10–18. [Google Scholar]
- 28.Schwartz S E. Are global cloud albedo and climate controlled by marine phytoplankton? Nature. 1988;336:441–445. [Google Scholar]
- 29.Smith N A, Kelly D P. Isolation and physiological characterization of autotrophic sulphur bacteria oxidizing dimethyl disulphide as sole source of energy. J Gen Microbiol. 1988;134:1407–1417. [Google Scholar]
- 30.Suylen G M H, Kuenen J G. Chemostat enrichment and isolation of Hyphomicrobium EG. Antonie Leeuwenhoek. 1986;52:281–293. doi: 10.1007/BF00428640. [DOI] [PubMed] [Google Scholar]
- 31.Visscher P T, van Gemerden H. Photo-autotrophic growth of Thiocapsa roseopersicina on dimethyl sulfide. FEMS Microbiol Lett. 1991;81:247–250. [Google Scholar]
- 32.Visscher P T, Taylor B F. A new mechanism for the aerobic catabolism of dimethyl sulfide. Appl Environ Microbiol. 1993;59:3784–3789. doi: 10.1128/aem.59.11.3784-3789.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visscher P T, Taylor B F. Aerobic and anaerobic degradation of a range of alkyl sulfides by a denitrifying marine bacterium. Appl Environ Microbiol. 1993;59:4083–4089. doi: 10.1128/aem.59.12.4083-4089.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiner J H, MacIsaac D P, Bishop R E, Bilous P T. Purification and properties of Escherichia coli dimethyl sulfoxide reductase, an iron-sulfur molybdoenzyme with broad substrate specificity. J Bacteriol. 1988;170:1505–1510. doi: 10.1128/jb.170.4.1505-1510.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin F, Grosjean D, Flagan R C, Seinfeld J H. Photooxidation of dimethyl sulfide and dimethyl disulfide. II. Mechanism evaluation. J Atmos Chem. 1990;11:309–364. [Google Scholar]
- 36.Zeyer J, Eicher P, Wakeham S G, Schwarzenbach R P. Oxidation of dimethyl sulfide to dimethyl sulfoxide by phototrophic purple bacteria. Appl Environ Microbiol. 1987;53:2026–2032. doi: 10.1128/aem.53.9.2026-2032.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Kuniyoshi I, Hirai M, Shoda M. Oxidation of dimethyl sulfide by Pseudomonas acidovorans DMR-11 isolated from peat biofilter. Biotechnol Lett. 1991;13:223–228. [Google Scholar]
- 38.Zhang L, Hirai M, Shoda M. Removal characteristics of dimethyl sulfide, methanethiol and hydrogen sulfide by Hyphomicrobium sp. I55 isolated from peat biofilter. J Ferment Bioeng. 1991;72:392–396. [Google Scholar]