Abstract
Background & Aims
The expression and role of methyltransferase SET and MYND domain-containing protein 5 (SMYD5) in inflammatory bowel disease (IBD) is completely unknown. Here, we investigated the role and underlying mechanism of epithelial SMYD5 in IBD pathogenesis and progression.
Methods
The expression levels of SMYD5 and the mitochondrial transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) were examined by Western blot, immunofluorescence staining, and immunohistochemistry in intestinal epithelial cells (IECs) and in colon tissues from human IBD patients and colitic mice. Mice with Smyd5 conditional knockout in IECs and littermate controls were subjected to dextran sulfate sodium–induced colitis and the disease severity was assessed. SMYD5-regulated mitochondrial biogenesis was examined by quantitative reverse-transcription polymerase chain reaction and transmission electron microscopy, and the mitochondrial oxygen consumption rate was measured in a Seahorse Analyzer system (Agilent, Santa Clara, CA). SMYD5 and PGC-1α interaction was determined by co-immunoprecipitation assay. PGC-1α degradation and turnover (half-life) were analyzed by cycloheximide chase assay. SMYD5-mediated PGC-1α methylation was assessed via in vitro methylation assay followed by mass spectrometry for identification of methylated lysine residues.
Results
Up-regulated SMYD5 and down-regulated PGC-1α were observed in intestinal epithelia from IBD patients and colitic mice. Smyd5 depletion in IECs protected mice from dextran sulfate sodium–induced colitis. SMYD5 was critically involved in regulating mitochondrial biology such as mitochondrial biogenesis, respiration, and apoptosis. Mechanistically, SMYD5 regulates mitochondrial functions in a PGC-1α–dependent manner. Furthermore, SMYD5 mediates lysine methylation of PGC-1α and subsequently facilitates its ubiquitination and degradation.
Conclusions
SMYD5 attenuates mitochondrial functions in IECs and promotes IBD progression by enhancing PGC-1α degradation in a methylation-dependent manner. Strategies to decrease SMYD5 expression and/or increase PGC-1α expression in IECs might be a promising therapeutic approach to treat IBD patients.
Keywords: IBD, Colitis, SMYD5, PGC-1α, Mitochondrion
Abbreviations used in this paper: CHX, cycloheximide; COX I/II, cytochrome c oxidase I/II; DSS, dextran sulfate sodium; FITC, fluorescein isothiocyanate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GST, Glutathione S-transferase; HA, hemagglutinin; H+L, heavy + light chain; IBD, inflammatory bowel disease; IEC, intestinal epithelial cell; IF, immunofluorescence; IFN, interferon; IHC, immunohistochemical; K223R, lysine 223 to arginine; KO, knockout; L3MBTL1/3, lethal (3) malignant brain tumor-like protein 1/3; mRNA, messenger RNA; mtDNA, mitochondrial DNA; OCR, oxygen consumption rate; OE, overexpression; PAS, periodic acid–Schiff; PBS, phosphate-buffered saline; pcDNA, plasmid complementary DNA; PCNA, proliferating cell nuclear antigen; PCR, polymerase chain reaction; PGC-1α, peroxisome proliferator-activated receptor γ coactivator 1α; PHF20L1, plant homeodomain finger protein 20-like 1; ROS, reactive oxygen species; RT-qPCR, quantitative reverse-transcription polymerase chain reaction; SDHA, succinate dehydrogenase complex subunit A; SMYD5, SET and MYND domain-containing protein 5; SOD, superoxide dismutase; Tfam/TFAM, mitochondrial transcription factor A; TNF, tumor necrosis factor; UCP, uncoupling protein; ZO-1, zonula occludens 1
Graphical abstract
Summary.
The methyltransferase SET and MYND domain-containing protein 5, whose expression is up-regulated in intestinal epithelia of inflammatory bowel disease, exaggerates colitis severity by suppressing epithelial mitochondrial functions via down-regulating the stability of peroxisome proliferator-activated receptor γ coactivator-1α, the master mitochondrial transcriptional coactivator.
Inflammatory bowel disease (IBD), mainly composed of Crohn’s disease and ulcerative colitis, is a chronic, relapsing inflammatory disorder of the gastrointestinal tract.1 The increasing prevalence of IBD in North America and around the world,2 coupled with the significant lifetime morbidity and financial burden,3 clearly highlights the urgent need for IBD research to identify novel therapeutic targets and develop effective treatments. The intestinal epithelial cells (IECs) constitute a critical line of defense that plays a pivotal role in regulating host–microbiota interaction and intestinal homeostasis.4 Although IBD is a multifactorial disease, accumulating studies have linked mitochondrial dysfunction and oxidative stress to IBD pathogenesis, suggesting a bioenergetics failure of the intestinal mitochondria, leading to disruption of epithelial integrity and intestinal inflammation.5,6
Mitochondrion is a highly complex organelle that undergoes constant turnover in response to stimuli in a process known as mitochondrial dynamics.7 Mitochondrial biogenesis, the process by which new mitochondria are generated and repaired, plays a significant role in maintaining cellular metabolic homeostasis.8 Peroxisome proliferator-activated receptor-γ coactivator 1-α (PGC-1α) is the master transcriptional coactivator of genes encoding proteins responsible for the regulation of mitochondrial biogenesis and function.9 The expression and transcriptional activity of PGC-1α are tightly regulated by multiple signaling pathways and post-translational modifications including phosphorylation, acetylation, methylation, or ubiquitination.10,11 Accumulating evidence has shown that specificity of PGC-1α to interact with its binding partners is modulated by post-translational modifications of the PGC-1α protein.10 For example, phosphorylation of PGC-1α by AMP-activated protein kinase promotes expression of PGC-1α target genes that are involved in mitochondrial function and cellular metabolism.12 In addition. methyltransferases such as protein arginine methyltransferase 1 and SET domain containing lysine methyltransferase 7/9 have been shown to methylate PGC-1α to regulate its functions, including mitochondrial biogenesis.13,14
The SET and MYND domain containing proteins (SMYDs), currently composed of 5 members (SMYD1–5), are a special class of protein methyltransferases that mediate lysine methylation of histones and nonhistone proteins and are involved in transcriptional regulation, signaling cascades, and cellular functions.15 SMYD5 has been reported to methylate histone H4 and regulate macrophage inflammation,16 stem cell self-renewal and differentiation,17 and zebrafish hematopoiesis.18 To date, the only substrate methylated by SMYD5 is histone H4 at lysine 20.16,17 It has been reported that PGC-1α is highly expressed in intestinal epithelia,19 but is reduced significantly in colitic mice and IBD patients.20,21 Furthermore, PGC-1α induction in IECs was shown to maintain mitochondrial integrity and barrier function and decrease intestinal inflammation.20 However, the molecular mechanism underlying the regulation of PGC-1α expression and activity in intestinal homeostasis remains elusive. A recent study showed that loss of SMYD1, a muscle-specific SMYD member, leads to a decrease in PGC-1α expression and down-regulation of mitochondrial energetics in cardiomyocytes.22 The study further showed that SMYD1 epigenetically regulates expression of PGC-1α at the transcriptional level via histone H3 lysine K4 trimethylation to up-regulate PGC-1α transcription. However, whether and how SMYD5 also may regulate PGC-1α expression and mitochondrial function in intestinal homeostasis and diseases (such as IBD) remains unknown. Herein, we sought to explore the potential role and underlying mechanism of epithelial SMYD5 in IBD pathology. Our study uncovered PGC-1α as a novel nonhistone target of SMYD5. Furthermore, Smyd5 ablation in IECs protects mice from experimental colitis by preserving PGC-1α protein and thus promoting mitochondrial biogenesis and functions in IECs. Therefore, targeting SMYD5 expression in IECs may be a potential therapeutic strategy for IBD treatment.
Results
Clinical Relevance of SMYD5 and PGC-1α in Colon Mucosa of IBD Patients
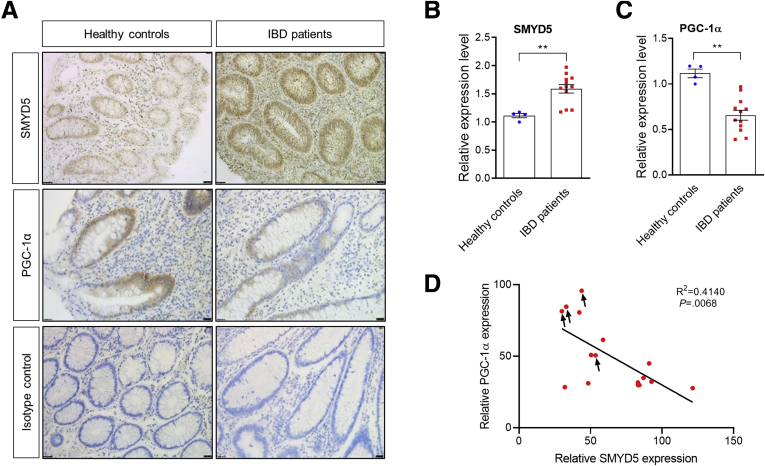
Recent studies have shown that SMYD5 epigenetically regulates expression of Toll-like receptor 4 target genes including IL1β, TNF, and CXCL10 during macrophage immune response,16 suggesting a critical role of SMYD5 in immunity and inflammatory diseases. To investigate if SMYD5 is involved in IBD, which is characterized by chronic inflammation of the gastrointestinal tract, we first examined its expression in human colonic tissues. Immunohistochemical (IHC) staining of colonic mucosa showed SMYD5 immunopositivity in the colon tissues and SMYD5 was expressed mainly in colonic epithelia (ie, IECs) and lamina propria/stroma (Figure 1A), with significantly up-regulated expression in IBD patients with active inflammation compared with healthy controls (Figure 1A and B), suggesting the possible involvement of SMYD5 in IBD pathogenesis.
Figure 1.
Expression of SMYD5 and PGC-1α in colonic mucosa of IBD patients and healthy controls. (A) IHC staining of SMYD5 and PGC-1α expression in colon sections from healthy controls and IBD patients with active inflammation. Isotope controls with no primary antibodies also are shown (bottom panel). Scale bars: 50 μm. Quantification of IHC staining for expression of (B) SMYD5 and (C) PGC-1α in colonic epithelia from healthy controls (n = 4) and IBD patients (8 ulcerative colitis patients and 4 Crohn’s disease patients; n = 12). ∗∗P < .01. (D) Scatterplots between the relative expression (IHC staining intensity in arbitrary units) of SMYD5 and PGC-1α in colonic epithelia from healthy controls (indicated by black arrows) and IBD patients. P = .0068; n = 16.
Because mitochondrial dysfunction is associated with IBD,23 we also examined the expression of PGC-1α, a key regulator of mitochondrial biogenesis and function,9 in colonic mucosa. It has been reported that PGC-1α is highly expressed in intestinal epithelia,19 whose expression was reduced significantly in colitic mice and IBD patients.20,21 Interestingly, we also observed a significant reduction of PGC-1α expression in colonic epithelia from IBD patients vs healthy subjects (Figure 1A and C), which is consistent with a previous study.20 Recently, it was reported that SMYD1 modulates mitochondrial respiration in cardiomyocytes via epigenetically up-regulating gene expression of master regulators of cardiac energetics including PGC-1α.22 To reveal the potential relationship between SMYD5 and PGC-1α expression levels in intestinal tissue, we plotted SMYD5 expression level against PGC-1α level in colonic epithelia from both healthy controls and IBD patients. Quantification of SMYD5 and PGC-1α expression levels in colonic epithelia showed a significant inverse correlation between these 2 proteins (Figure 1D), which is opposite to the relationship between SMYD1 and PGC-1α in cardiomyocytes as reported before.22 These results clearly indicate that SMYD5 is up-regulated in inflamed intestinal epithelia and may be involved in IBD pathogenesis and progression.
Generation and Basal Characterization of an Intestine-Specific Smyd5-Deficient Mouse Line
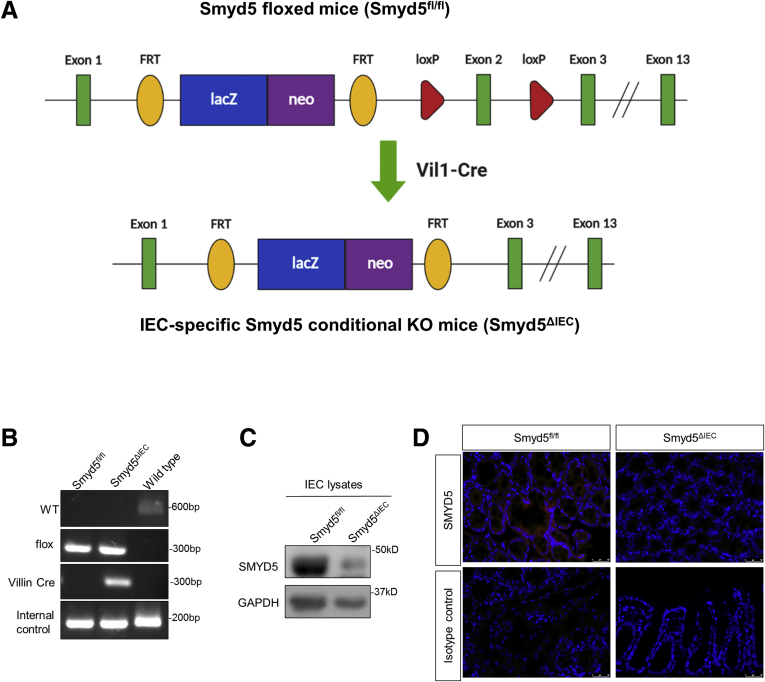
To determine the potential importance of epithelial SMYD5 in IBD pathogenesis, we generated IEC-specific Smyd5 knockout (KO) mice. Smyd5 floxed mice (Smyd5fl/fl) were bred with Villin-Cre transgenic mice to produce IEC-specific Smyd5 conditional KO mice (Smyd5ΔIEC) (Figure 2A). IEC-specific ablation of Smyd5 gene was confirmed by polymerase chain reaction (PCR) analysis (Figure 2B). Lack of SMYD5 protein specifically in IECs and intestinal epithelia also was validated by immunoblot analysis (Figure 2C) and immunofluorescence (IF) staining (Figure 2D), respectively.
Figure 2.
Generation of an intestine-specific Smyd5-deficient mouse line. (A) Smyd5 allele containing LacZ-reporter-promoter (blue box)-driven neo targeting cassette (purple box), FLP-FRT sites (yellow oval), Cre-loxP sites (red triangle), and Smyd5 exons (green rectangle). Breeding of Smyd5 floxed mice (Smyd5fl/fl) with Vil-Cre transgenic mice resulted in truncated Smyd5 copy (with exon 2 deleted), denoted IEC-specific Smyd5 depletion (Smyd5ΔIEC). (B) Genotyping of IEC-specific Smyd5 conditional KO mice (Smyd5ΔIEC) and Smyd5 floxed mice (Smyd5fl/fl) by PCR analysis. (C) Western blot analysis of SMYD5 expression in murine IECs isolated from Smyd5fl/fl and Smyd5ΔIEC mice. (D) IF staining of SMYD5 expression in intestinal epithelia from Smyd5fl/fl and Smyd5ΔIEC mice. Isotope controls with no primary antibody also are shown (bottom panel). Scale bars: 25 μm.
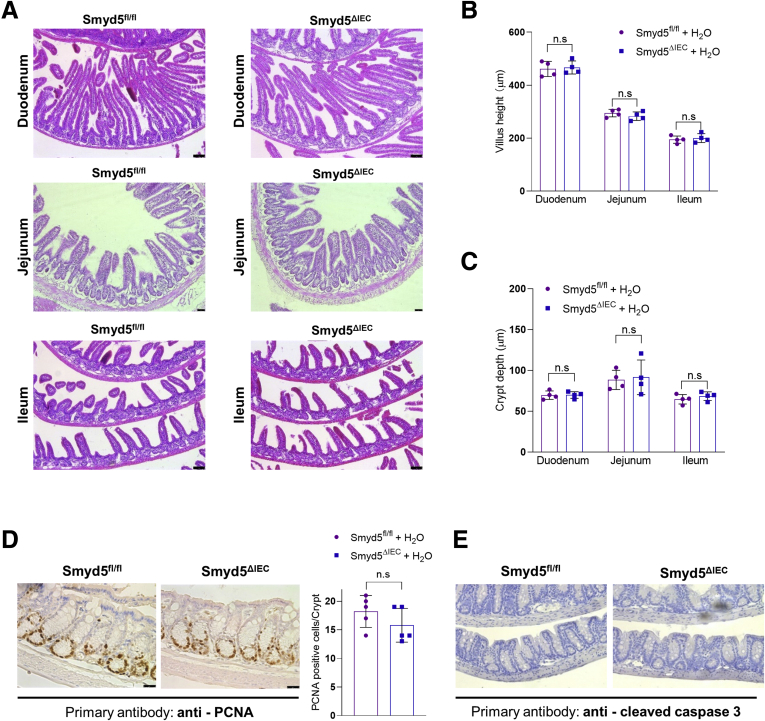
Of note, Smyd5ΔIEC mice were born at normal Mendelian frequencies and developed normally to adulthood. We thoroughly examined the Smyd5ΔIEC mice for any possible alterations of intestinal architecture, and, in particular, epithelial structure at baseline. Our examination showed that the Smyd5ΔIEC mice were phenotypically normal and showed no abnormality in morphology of different organs, including the gastrointestinal tract (Figure 3A), compared with Smyd5fl/fl control mice. Histologic analysis of intestinal architecture in both mouse strains did not show any irregularity of the size and shape of crypts or crypt branching in the basal state (Figure 3A). We also measured the crypt depth and villus height in duodenum, jejunum, and ileum from both mouse strains, and the quantification did not show any significant difference in villus height (Figure 3A and B) and crypt depth (Figure 3A and C). In addition, Smyd5 deficiency did not elicit the development of spontaneous intestinal inflammation in mice because no evidence of colitis (ie, immune cell infiltration) in the basal state in either genotype was shown by H&E staining of colonic/intestinal sections (Figure 3A). Furthermore, we evaluated cell proliferation and apoptosis in colon tissues of both Smyd5fl/fl and Smyd5ΔIEC mice. The IHC staining of proliferation marker proliferating cell nuclear antigen (PCNA) showed a comparable PCNA positivity between the 2 groups (Figure 3D). However, IHC staining of apoptosis marker cleaved caspase-3 was barely detectable (Figure 3E) at baseline.
Figure 3.
Basal characterization of the intestine-specific Smyd5-deficient mice. (A) Representative H&E staining of mouse duodenum (top panel), jejunum (middle panel), and ileum (bottom panel) from Smyd5fl/fl and Smyd5ΔIEC mice. Quantitative analysis of (B) villus height and (C) crypt depth in the duodenum, jejunum, and ileum from Smyd5fl/fl and Smyd5ΔIEC mice. n = 4 per group. (D) Representative IHC staining (left) and quantitative analysis (right) for PCNA in colon tissues from Smyd5fl/fl and Smyd5ΔIEC mice. n = 5 per group. (E) Representative IHC staining of cleaved caspase-3 in colon tissues from Smyd5fl/fl and Smyd5ΔIEC mice.
We also examined the expression of stem cell markers or cell numbers in the colon tissues of Smyd5fl/fl and Smyd5ΔIEC mice. Immunofluorescence staining of an intestinal stem cell marker B cell-specific Moloney murine leukemia virus insertion site 1 (BMI1)24 in the colon tissues did not show any significant change of BMI1-positive cell per crypt (Figure 4A). Western blot analysis of leucine-rich repeat-containing G-protein coupled receptor 5 (LGR5) expression in isolated primary IECs did not reveal any significant difference between the 2 groups (Figure 4B). Kruppel like factor 5 (KLF5) has been reported as a critical regulator for crypt cell differentiation and self-renewal25; however, IHC staining of KLF5 expression in colonic tissues did not show any significant difference between the 2 mouse groups (Figure 4C). In addition, we performed periodic acid–Schiff (PAS) staining and analyzed the number of crypt goblet cells in the colon tissues. However, PAS staining did not reveal any significant difference in counts of goblet cells per crypt between Smyd5fl/fl and Smyd5ΔIEC mice in the basal state (Figure 4D). These results suggest that epithelial Smyd5 ablation does not cause basal changes in the numbers of stem cells or goblet cells.
Figure 4.
Epithelial Smyd5 ablation does not cause basal changes in the numbers of stem or goblet cells. (A) Representative images of IF staining of BMI1 (green; left) and quantitative analysis (right) of BMI1-positive cells per crypt in colon tissues from Smyd5fl/fl and Smyd5ΔIEC mice. Nucleus was stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). n = 5 per group. (B) Western blot analysis of LGR5 expression in primary IECs isolated from Smyd5fl/fl (n = 4) and Smyd5ΔIEC mice (n = 5). (C) Representative IHC staining (left) and quantitative analysis (right) for KLF5 in colon tissues from Smyd5fl/fl and Smyd5ΔIEC mice. n = 9 per group. (D) Representative images of PAS staining (left) and quantitative analysis (right) of PAS-positive cells per crypt in colon tissues from Smyd5fl/fl and Smyd5ΔIEC mice. n = 5 per group.
In conclusion, conditional deletion of Smyd5 in IECs does not alter intestinal homeostasis in the basal state.
SMYD5 Depletion in IECs Protects Mice From Dextran Sulfate Sodium–Induced Experimental Colitis
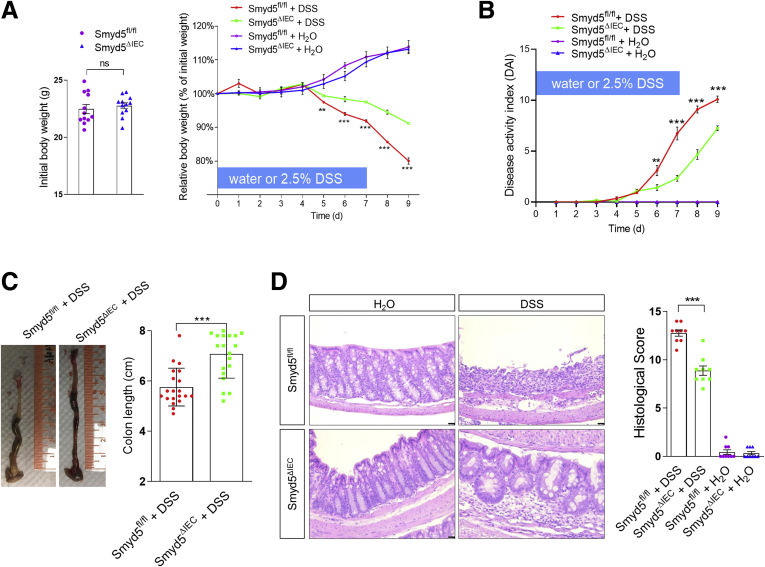
To investigate whether Smyd5 deficiency affects the development of IBD, both floxed and KO mice were subjected to colitis induced by dextran sulfate sodium (DSS), a colitogenic chemical widely used to induce colitis in experimental animals.26 Smyd5fl/fl and Smyd5ΔIEC mice administered only water had gained weight to a similar extent (Figure 5A). Although both mouse lines lost weight after DSS administration, Smyd5ΔIEC mice had significantly less weight loss during days 5 to 9, with an approximately 11% difference in weight change at day 9 between the 2 mouse lines (Figure 5A). Furthermore, Smyd5ΔIEC mice had better-shaped stools and less rectal bleeding than Smyd5fl/fl mice during days 6 to 9, resulting in a lower disease activity index (Figure 5B). In addition, Smyd5ΔIEC mice showed less colon shortening upon DSS exposure (Figure 5C). Histologic analysis on day 9 after DSS administration revealed less extensive ulceration and erosion, less severe inflammatory cell infiltration, and less thickening of mucosa with edema in colon sections of Smyd5ΔIEC mice compared with that of Smyd5fl/fl mice, leading to lower histologic scores (Figure 5D). Together, these data show that IEC-specific Smyd5 deficiency reduces the severity of DSS-induced colitis in mice.
Figure 5.
Smyd5 depletion in IECs protects mice from DSS-induced experimental colitis. (A) Initial body weight (left) and percentage body weight (relative to initial body weight) after DSS or water treatment (right) of Smyd5fl/fl and Smyd5ΔIEC mice. ∗∗P < .01, ∗∗∗P < .001, DSS-treated Smyd5fl/fl vs DSS-treated Smyd5ΔIEC mice, n = 12. (B) Changes in disease activity index (DAI). ∗∗P < .01 on day 6, and ∗∗∗P < .001 on days 7–9, Smyd5ΔIEC vs Smyd5fl/fl mice upon DSS treatment, n = 12. (C) Gross morphology of representative colons (left) and colon length measurements (right) in Smyd5fl/fl and Smyd5ΔIEC mice after DSS administration. n = 19–20. (D) Representative H&E staining (left) and histology scores (right) of colon sections from Smyd5fl/fl and Smyd5ΔIEC mice after DSS or water treatment. Scale bars: 50 μm. n = 9.
SMYD5 Negatively Regulates PGC-1α Expression at Post-Transcriptional Level
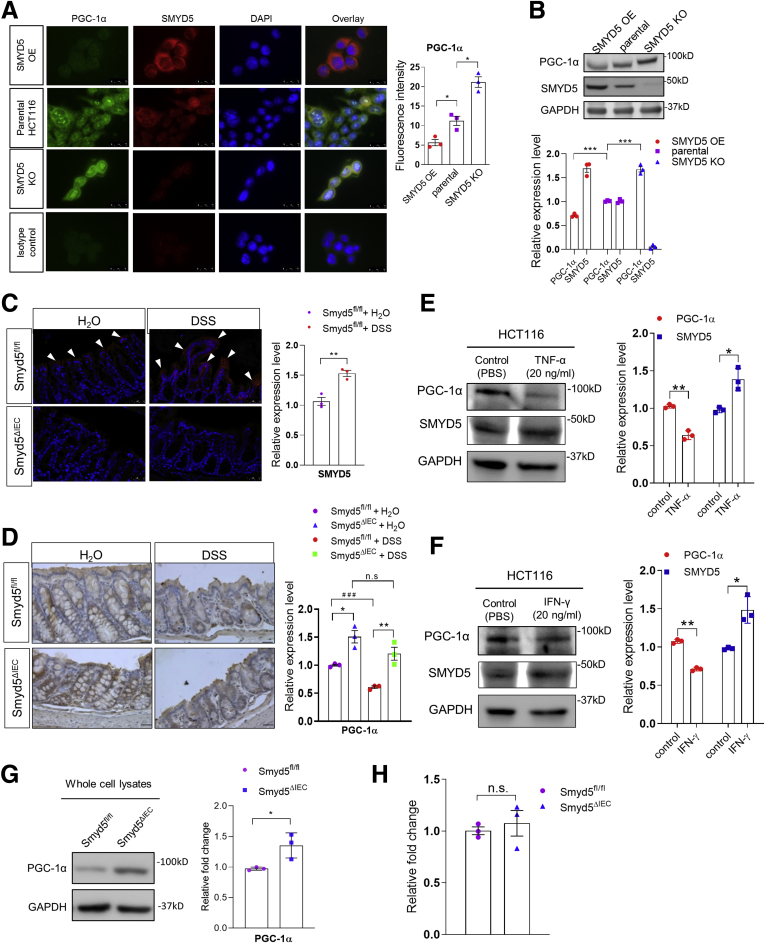
Accumulating studies have shown that members of the SMYD family not only mediate the methylation of histone substrates but also nonhistone targets.15 Moreover, the SMYD family of methyltransferases have been reported to modulate protein expression at both transcriptional and post-transcriptional levels.15,27,28 Based on the earlier-described findings as observed in human colonic tissues (Figure 1), we next examined whether SMYD5 is involved in regulation of PGC-1α expression in vitro using the human colonic epithelial cell line HCT116. The results showed that overexpressing SMYD5 in HCT116 cells substantially suppressed PGC-1α expression; conversely, SMYD5 knockout resulted in a significant increase in PGC-1α level (Figure 6A and B), indicating that SMYD5 negatively regulates PGC-1α expression in IECs. Interestingly, similar changes of SMYD5 and PGC-1α expression also were observed in vivo in colitic mice as shown by immunofluorescence staining (Figure 6C) and immunohistochemistry (Figure 6D). Immunofluorescence staining showed that DSS administration resulted in significantly up-regulated SMYD5 in epithelia of inflamed mucosa of Smyd5fl/fl control mice compared with water administration (Figure 6C, top panel), as reflected by increased fluorescence intensity (red color as indicated by white arrowheads). However, DSS treatment induced a decrease in expression of PGC-1α in epithelial mucosa in both Smyd5fl/fl and Smyd5ΔIEC mice as compared with water treatment (Figure 6D), which is consistent with a previous study.20 Of note, PGC-1α expression level in intestinal epithelia of Smyd5ΔIEC mice was significantly higher than that from Smyd5fl/fl mice at both baseline (water treatment) and upon DSS-colitis induction (Figure 6D). This is consistent with the findings from human colon sections (Figure 1A–C). Furthermore, treatment of HCT116 cells with tumor necrosis factor (TNF)-α or interferon (IFN)-γ, the proinflammatory cytokines up-regulated in intestinal mucosa in human IBD and murine colitis,29 resulted in up-regulated SMYD5 and down-regulated PGC-1α (Figure 6E and F).
Figure 6.
SMYD5 negatively regulates PGC-1α at the post-transcriptional level. (A) Representative images (left) and quantitative analysis (right) of IF staining of SMYD5 and PGC-1α expression in HCT116 cells with either SMYD5 OE or KO. Isotope controls with no primary antibodies also are shown (left, bottom panel). Scale bars: 25 μm. ∗P < .05, n = 3. (B) Representative Western blots (top) and quantitative analysis (bottom) of SMYD5 and PGC-1α expression (normalized to GAPDH) in SMYD5 OE or KO HCT116 cells. The expression levels of SMYD5 and PGC-1α in parental HCT116 cells were set as 1. ∗∗∗P < .001, comparison of PGC-1α levels among groups; n = 3. (C) Representative IF staining (left) and quantitative analysis (right) of SMYD5 (red staining indicated by white arrowheads) in intestinal mucosa epithelia of Smyd5fl/fl mice after water or DSS administration. SMYD5 was not detected in Smyd5ΔIEC mice. Scale bars: 50 μm. ∗∗P < .01, n = 3. (D) Representative IHC staining (left) and quantitative analysis (right) of PGC-1α (brown color) in intestinal epithelia from Smyd5fl/fl and Smyd5ΔIEC mice after water or DSS administration. Scale bars: 50 μm. DSS vs water-treated, ###P < .001; Smyd5ΔIEC vs Smyd5fl/fl, ∗P < .05, ∗∗P < .01; n = 3. (E) HCT116 cells were treated with TNF-α (20 ng/mL) or vehicle control (PBS) for 24 hours, and the expression levels of SMYD5, PGC-1α, and GAPDH were determined by Western blot analysis. The band intensity of SMYD5 and PGC-1α expression in TNF-α–treated HCT116 cells was quantified, normalized to GAPDH, and presented as fold changes relative to the vehicle control-treated cells. ∗P < .05, ∗∗P < .01; n = 3. (F) HCT116 cells were treated with IFN-γ (20 ng/mL) or vehicle control (PBS) for 24 hours, and the expression levels of SMYD5, PGC-1α, and GAPDH were determined by Western blot analysis. The band intensity of SMYD5 and PGC-1α expression in IFN-γ–treated HCT116 cells was quantified, normalized to GAPDH, and presented as fold changes relative to the vehicle control-treated cells. ∗P < .05, ∗∗P < .01; n = 3. (G) Representative Western blots (left) of PGC-1α and GAPDH expression in whole-cell lysates of IECs isolated from Smyd5fl/fl and Smyd5ΔIEC mice. PGC-1α expression (right) was quantified (normalized to GAPDH) and presented as the fold change relative to the level in Smyd5fl/fl IECs. ∗P < .05, n = 3. (H) Pgc-1α mRNA was assessed by RT-qPCR (presented as relative fold change) in IECs isolated from Smyd5fl/fl and Smyd5ΔIEC mice. P = .4346, n = 3.
Western blot analysis of cell lysates of IECs isolated from Smyd5ΔIEC and Smyd5fl/fl mice revealed that the protein level of PGC-1α in IECs from Smyd5ΔIEC mice was significantly higher than that from Smyd5fl/fl mice (Figure 6G). However, quantitative reverse-transcription PCR (RT-qPCR) analysis showed that there was no significant difference in PGC-1α messenger RNA (mRNA) in IECs from Smyd5fl/fl vs Smyd5ΔIEC mice (Figure 6H), suggesting that SMYD5 regulates PGC-1α expression at the post-transcriptional level.
SMYD5 Is Not Involved in Mitochondrial Fission, Fusion, or Autophagy
In addition to PGC-1α, we also examined the expression levels of other proteins that are critical for mitochondrial functions (ie, biogenesis, electron transport chain, antioxidant), such as the mitochondrial DNA packaging and transcription factor, mitochondrial transcription factor A (TFAM); the critical players of electron transport chain, succinate dehydrogenase complex subunit A (SDHA) and cytochrome c oxidase subunit IV (COX IV); the mitochondrial inner membrane transporters, uncoupling protein 2 and 3 (UCP2 and UCP3); and the mitochondrial antioxidant enzymes, superoxide dismutase 1 (SOD1) and 2 (SOD2), by RT-PCR and Western blot in IECs isolated from Smyd5fl/fl and Smyd5ΔIEC mice. Our results did not reveal any significant difference in the expression of UCP2/3, SDHA, COX IV, and SOD1/2 between the 2 groups (Figure 7A–C).
Figure 7.
SMYD5 is not involved in mitochondrial fission and fusion or autophagy. (A) Western blots of proteins critical for mitochondrial functions (TFAM, SDHA, COX IV, UCP2, UCP3) in primary IECs isolated from SMYD5fl/fl and SMYD5ΔIEC mice. GAPDH was used as loading control. Left: each blot represents 1 mouse of specific genotype (n = 3). (B) RT-qPCR analysis of Sod1 and Sod2 mRNA levels in primary IECs isolated from SMYD5fl/fl and SMYD5ΔIEC mice. n = 3. (C) Western blots of protein expression of SOD2 in primary IECs isolated from SMYD5fl/fl and SMYD5ΔIEC mice. GAPDH was used as loading control. Each blot represents 1 mouse of specific genotype (n = 4 for SMYD5fl/fl mice; n = 5 for SMYD5ΔIEC mice). (D) Western blots of proteins critical for mitochondrial dynamics (mitofusin [MFN]1, MFN2, dynamin-related protein 1 [DRP1], LC3) in SMYD5 KO or parental HCT116 cells. SMYD5 protein abundance was examined to validate KO efficacy. GAPDH was used as loading control. (E) Western blots of FUN14 domain containing 1 (FUNDC1) expression in primary IECs isolated from SMYD5fl/fl and SMYD5ΔIEC mice. GAPDH was used as loading control. Each blot represents 1 mouse of specific genotype (n = 3 for each genotype).
In addition, we assessed the expression levels of proteins that are important for mitochondrial dynamics such as mitophagy, fusion, and fission, specifically, mitofusin 1 and 2, dynamin-related protein 1, and microtubule-associated protein 1A/1B-light chain 3 (LC3), in HCT116 parental cells and HCT116 cells with SMYD5 KO (Figure 7D). We also compared the expression of the FUN14 domain containing 1, the recently identified mitophagy regulator,30 in IECs isolated from Smyd5fl/fl and Smyd5ΔIEC mice (Figure 7E). The results showed that the expression of these proteins was comparable between the 2 groups (Figure 7D and E).
In addition to PGC-1α (Figure 6G), we also found that the expression of TFAM was increased in IECs with SMYD5 deficiency (Figure 7A). It has been well documented that PGC-1α can activate the transcription of TFAM.31,32 The increased expression of TFAM could have resulted from PGC-1α up-regulation resulting from SMYD5 depletion.
SMYD5 Is Critically Involved in Regulating Mitochondrial Biogenesis and Respiration
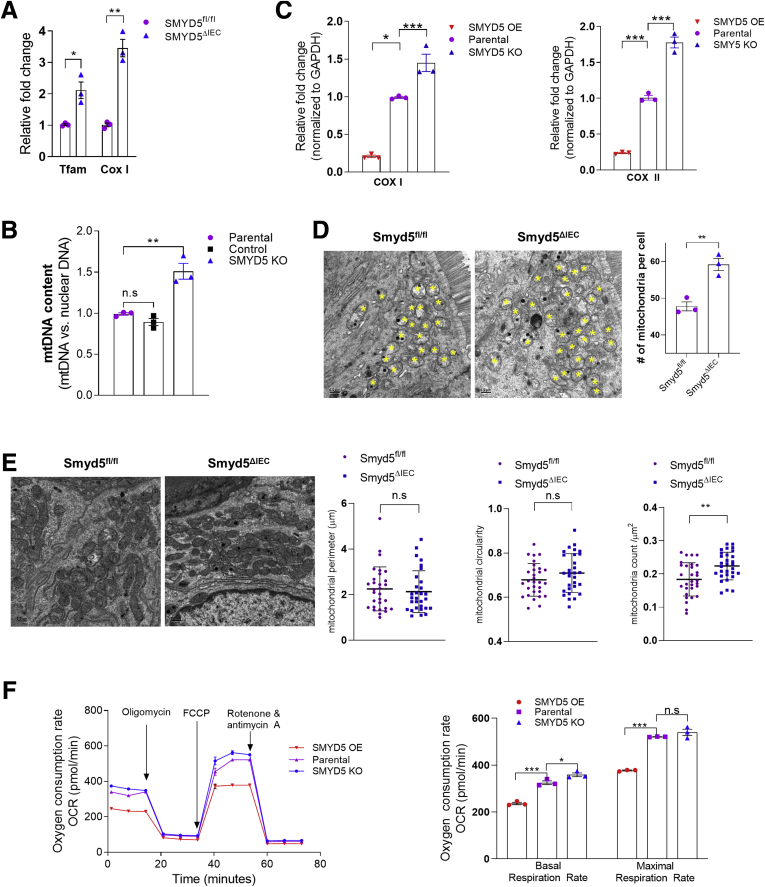
PGC-1α is a multifunctional transcriptional co-activator critically involved in mitochondrial biogenesis and oxidative phosphorylation to modulate diverse cellular functions and processes.33,34 Accumulating evidence from recent studies has implicated mitochondrial dysfunction within the intestinal epithelium in the onset and progression of IBD.5,6,20 We speculated that up-regulation of SMYD5 in IECs during IBD may result in a decrease of PGC-1α level, leading to attenuated mitochondrial biogenesis and function. To test this hypothesis, we examined the expression of markers of mitochondrial biogenesis in IECs from Smyd5fl/fl and Smyd5ΔIEC mice. The results showed that Smyd5 deficiency led to up-regulated mitochondrial biogenesis as revealed by significantly increase expression of Tfam, which controls mitochondrial DNA (mtDNA) replication and transcription and thus promotes mitochondrial biogenesis,35 and COX I, which is a mtDNA-encoded protein36 (Figure 8A). Similar findings also were observed for mtDNA content (Figure 8B) and expression of mtDNA-encoded genes (COX I and COX II) (Figure 8C) in HCT116 cells with overexpressing or knockout SMYD5.
Figure 8.
SMYD5 critically regulates mitochondrial biogenesis and function. (A) RT-qPCR analysis of mRNA (presented as relative fold change) of mitochondrial biogenesis markers Tfam (n = 3) and Cox I (n = 3) in IECs isolated from Smyd5fl/fl and Smyd5ΔIEC mice. (B) RT-qPCR analysis of the abundance of mtDNA content in HCT116 cells with KO SMYD5 or control vector. The relative mtDNA content was determined using the Comparative Ct method quantification (2-ΔCt method) method. The primers for human mtDNA-encoded Cox II gene and for human nuclear DNA-encoded GAPDH were used for the analysis. The results are presented as the ratio of mtDNA relative to nuclear DNA. Results are presented as fold changes relative to parental HCT116 cells. n = 3. (C) RT-qPCR analysis of the expression of the mtDNA-encoded genes, the mitochondrial biogenesis markers COX I (left) and COX II (right) in HCT116 cells with OE or KO SMYD5. Results are presented as fold changes relative to parental HCT116 cells. n = 3. (D) Representative TEM images (left) of IECs in sections of colon tissues from Smyd5fl/fl and Smyd5ΔIEC mice and quantitative analyses of the number of mitochondria per cell (right). The “n” refers to the number of mice per genotype. n = 3 mice per group. Yellow asterisks indicate mitochondria. Scale bars: 500 nm. (E) Representative TEM images (left) of IECs from Smyd5fl/fl and Smyd5ΔIEC mice (n = 3 mice for each group). Right: Quantitative analyses of mitochondrial perimeter, circularity, and the number of mitochondria per unit area (μm2). For mitochondrial perimeter and circularity, n = 30 mitochondria per genotype group. For mitochondria counts per μm2, n = 30 random visual fields per genotype group. (F) OCR measured by a Seahorse XF96 analyzer (left) in HCT116 cells with SMYD5 OE, KO, or unchanged (parental). Glucose (25 mmol/L) and pyruvate (1 mmol/L) were supplied as substrates. Oxygen consumption was measured under basal conditions, after the sequential addition of oligomycin, the pharmacologic uncoupler carbonyl cyanide phenylhydrazone (FCCP), the Complex III and I inhibitors antimycin A, and rotenone. Right: Quantitation of basal and maximal respiration capacity. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001; n = 3.
Furthermore, we performed transmission electron microscopy (TEM) of IECs in sections of colon tissues from Smyd5fl/fl and Smyd5ΔIEC mice, and analyzed the TEM images for measurements of mitochondrial parameters, including mitochondrial perimeter, circularity, and the number of mitochondria per cell or per unit area (μm2). Mitochondrial counts examined by TEM showed that Smyd5 depletion resulted in a slight but significant increase in the number (count) of mitochondria in IECs (per cell in Figure 8D, and per μm2 in Figure 8E, right). However, mitochondrial perimeter and circularity in IECs did not show any significant differences between Smyd5fl/fl and Smyd5ΔIEC mice (Figure 8E).
It has been reported that augmented oxidative phosphorylation and increased intestinal adenosine triphosphate protected mice from DSS and trinitrobenzene sulfonate–induced experimental colitis.37 We next examined the effect of SMYD5 expression on mitochondrial respiratory function of IECs. HCT116 cells with SMYD5 KO or overexpression (OE) were subjected to Seahorse assays (Agilent, Santa Clara, CA) to measure oxygen consumption rate (OCR)38 (Figure 8F, left). SMYD5 OE significantly reduced basal OCR and maximal respiratory capacity after carbonyl cyanide phenylhydrazone treatment, while SMYD5 KO led to the opposite effect (Figure 8F, right).
SMYD5 Modulates Intestinal Barrier Integrity
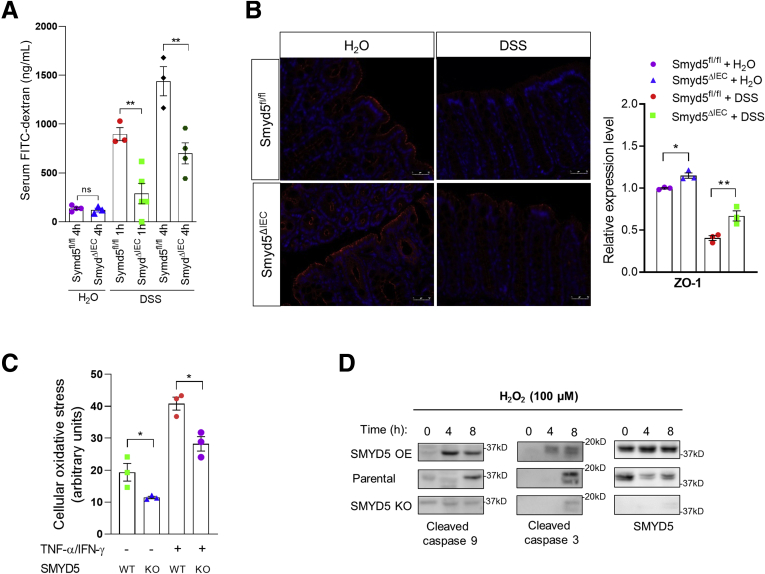
One of the characteristic features of IBD is loss of intestinal barrier integrity.39 Epithelial mitochondrial dysfunction has been implicated as a predisposing factor for increased gut epithelial permeability and disrupted barrier function, resulting in intestinal inflammation.23 Thus, it is possible that SMYD5 may be involved in modulating epithelial monolayer integrity by regulating mitochondrial function of IECs. To examine the effect of SMYD5 on epithelial monolayer integrity, an in vivo intestinal permeability assay was performed by oral gavage of fluorescein isothiocyanate (FITC)-dextran in Smyd5fl/fl and Smyd5ΔIEC mice, followed by measurement of FITC-dextran in the serum after 1 or 4 hours postgavage. The results showed that at basal condition without DSS injury, Smyd5fl/fl and Smyd5ΔIEC mice maintained a comparable barrier integrity reflected by low levels of plasma FITC-dextran in both groups (Figure 9A). DSS administration disrupted intestinal barrier integrity in both mouse strains as revealed by the drastic increase of serum FITC-dextran in a time-dependent manner. However, serum FITC-dextran in Smyd5ΔIEC mice was significantly lower than that in Smyd5fl/fl mice, suggesting a protective role of Smyd5 ablation in epithelial barrier integrity (Figure 9A). To investigate the possible other mechanism by which SMYD5 deficiency might alter epithelial permeability in colitic mice, the effect of SMYD5 deficiency on tight junction proteins involved in intestinal epithelial barrier also was examined. Immunofluorescence staining of zonula occludens 1 (ZO-1) protein in intestinal epithelia showed a slight increase at baseline in Smyd5ΔIEC mice vs Smyd5fl/fl mice (Figure 9B). Upon DSS challenge, ZO-1 staining was decreased drastically in intestinal epithelia of both Smyd5fl/fl and Smyd5ΔIEC mice, but was less decreased in Smyd5ΔIEC compared with Smyd5fl/fl mice (Figure 9B).
Figure 9.
SMYD5 is critically involved in regulating intestinal barrier integrity. (A) Smyd5fl/fl and Smyd5ΔIEC mice were subjected to DSS-induced acute colitis for 7 days. On the last day of DSS feeding, mice were fasted for 4 hours and gavaged with FITC-dextran (4 kilodaltons) and blood was collected for measurement of FITC-dextran at 1 or 4 hours postgavage. Intestinal permeability was measured by the concentration of FITC-dextran in the blood serum. n = 3–5 per group. (B) Representative IF staining of ZO-1 expression (left) in colon tissues of Smyd5ΔIEC or Smyd5fl/fl mice administered water or DSS. Red fluorescence, ZO-1; blue fluorescence, 4′,6-diamidino-2-phenylindole. Scale bars: 50 μm. Quantification of ZO-1 staining (right) in colon tissues of water or DSS-administered Smyd5fl/fl or Smyd5ΔIEC mice. n = 3. (C) The cellular oxidative stress was measured using a fluorescence-based assay as described in the Methods section without or with exposure to proinflammatory cytokines (20 ng/mL TNF-α and 20 ng/mL IFN-γ, 24 h) in SMYD5 KO HCT116 cells and parental HCT116 cells (SMYD5 WT). n = 3. (D) Parental HCT116, SMYD5 OE HCT116, or SMYD5 KO HCT116 cells were treated with H2O2 (100 μmol/L) for 4 or 8 hours, and the expression of cleaved caspase 9 and 3 as well as SMYD5 was examined by Western blot analysis using respective antibodies. ∗P < .05, ∗∗P < .01.
Proinflammatory cytokine-induced oxidative stress test using a fluorescence-based assay40 in SMYD5 KO HCT116 cells and parental cells showed that SMYD5 deficiency reduced cellular oxidative stress at the basal level, and exposure to proinflammatory cytokines (TNF-α and IFN-γ) increased oxidative stress in both groups as revealed by increased fluorescence intensity (Figure 9C). However, SMYD5 depletion attenuated the increase of oxidative stress induced by proinflammatory cytokines (Figure 9C). Moreover, SMYD5 overexpression in HCT116 cells exaggerated apoptosis (indicated by a time-dependent increase in cleaved caspase 3 and 9 levels) compared with HCT116 parental cells, while SMYD5 deletion reduced apoptotic response in HCT116 cells (Figure 9D). Collectively, these results indicate that SMYD5 modulates intestinal barrier integrity, at least partially, by regulating mitochondrial functions, probably through modulating the epithelial PGC-1α protein level.
PGC-1α Is a Substrate of SMYD5 Methyltransferase
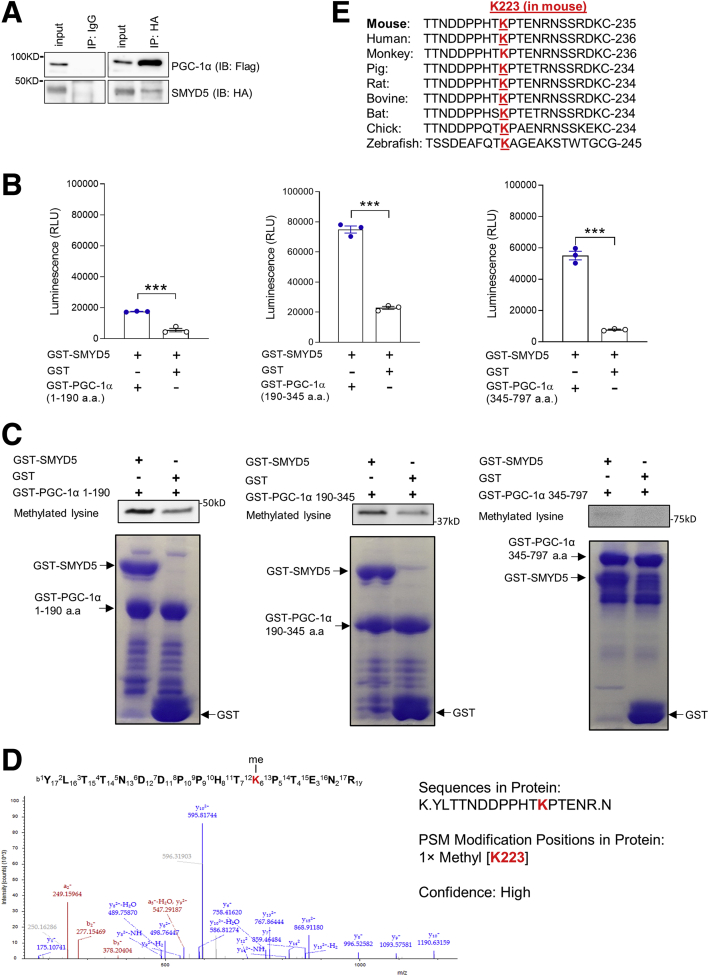
Accumulating studies have suggested that protein lysine methylation has been linked to their proteasomal degradation.41, 42, 43 For example, methylation of lysine (K) 185 of E2 promoter binding factor 1 (E2F1) induced its proteasomal degradation, and thus inhibited E2F1 apoptotic activity.41 To determine whether SMYD5 mediates methylation of PGC-1α to regulate its proteasomal degradation, we first evaluated the potential physical interaction between SMYD5 and PGC-1α. Co-immunoprecipitation assays using human embryonic kidney 293T (HEK293T) cells co-transfected with hemagglutinin (HA)-SMYD5 and Flag-PGC-1α showed that PGC-1α was co-precipitated with SMYD5 (Figure 10A). To investigate if SMYD5 can methylate PGC-1α in vitro, Glutathione S-transferase (GST)-SMYD5 was mixed with murine PGC-1α fragments (GST–PGC-1α aa 1–190, aa 190–345, and aa 345–797) or GST protein in a luminescence-based in vitro methyltransferase assay.44,45 Compared with GST alone, all GST–PGC-1α fragments showed significantly increased methylation in the presence of SMYD5 (Figure 10B). Because SMYD5 is known as a lysine methyltransferase, we next investigated if SMYD5 mediates methylation of lysine residues in PGC-1α protein. An in vitro methylation assay46 using purified GST–PGC-1α fragments mixed with GST-SMYD5 or GST alone showed that, compared with GST alone, GST-SMYD5 led to enhanced lysine methylation in PGC-1α aa 1–190 (Figure 10C, left) and PGC-1α aa 190–345 (Figure 10C, middle). Of note, PGC-1α fragments also showed basal lysine methylation signal even in the absence of GST-SMYD5 (Figure 10C).
Figure 10.
Murine PGC-1α is mono-methylated at lysine 223 by SMYD5. (A) Co-immunoprecipitation and immunoblot analysis of SMYD5-PGC-1α interaction in HEK293T cells co-transfected with HA-SMYD5 and Flag–PGC-1α. Whole-cell lysates were immunoprecipitated with anti-HA or control IgG, and the immunocomplexes were immunoblotted using anti-Flag (for PGC-1α) and anti-HA (for SMYD5) antibodies. n = 3. (B) Luminescence-based in vitro methyltransferase assay of PGC-1α methylation mediated by SMYD5. Bacterially purified GST-SMYD5 was mixed with GST alone or GST–PGC-1α fragments (GST–PGC-1α aa 1–190, GST–PGC-1α aa 190–345, and GST–PGC-1α aa 345–797) in an assay reaction that detects S-adenosyl-L-homocysteine, the universal reaction products of all methyltransferases, as described in the Methods section. ∗∗∗P < .001; n = 3. (C) In vitro methyltransferase assay of lysine methylation of PGC-1α and immunoblot analysis using a methyl lysine–specific antibody to detect methylated lysine(s) of PGC-1α. Bacterially purified GST-SMYD5 or GST alone was mixed with GST–PGC-1α fragments (GST–PGC-1α aa 1–190, aa 190–345, and aa 345–797) in a methylation reaction as described in the Methods section. Bottom: Coomassie blue staining showing equal amounts of GST–PGC-1α fragments mixed with either GST-SMYD5 protein or GST alone used in the in vitro methylation assay. (D) Liquid chromatography with tandem mass spectrometry (LC-MS/MS) spectrum of the mono-methylated lysine 223 (K223) of murine PGC-1α fragment/peptide by SMYD5. The in vitro methylation assay was conducted as described in panel C, followed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. LC-MS/MS analysis was conducted after digestion of samples by trypsin. LC-MS/MS analysis was repeated once with the same results (n = 2). (E) Alignment of peptide sequences spanning K223 residue (highlighted in red) of murine PGC-1α among the indicated species. IB, immunoblotting; IP, immunoprecipitation; RLU, relative light units.
To identify the specific lysine residues that have been methylated by SMYD5, the in vitro methylation assay was conducted as described earlier, and mass spectrometric analysis was performed to identify the methylated lysine residue in all of the PGC-1α fragments. The results showed SMYD5 mediated mono-methylation of lysine 223 (K223) of murine PGC-1α (Figure 10D), an evolutionally conserved lysine residue among different species (from zebrafish to human) (Figure 10E). We could not detect any lysine methylation within fragments of PGC-1α aa 1–190 or PGC-1α aa 345–797. Taken together, these results suggest that SMYD5 mediates lysine methylation of PGC-1α at K223 and may modulate its degradation.
SMYD5 Mediates Proteasomal Degradation of PGC-1α
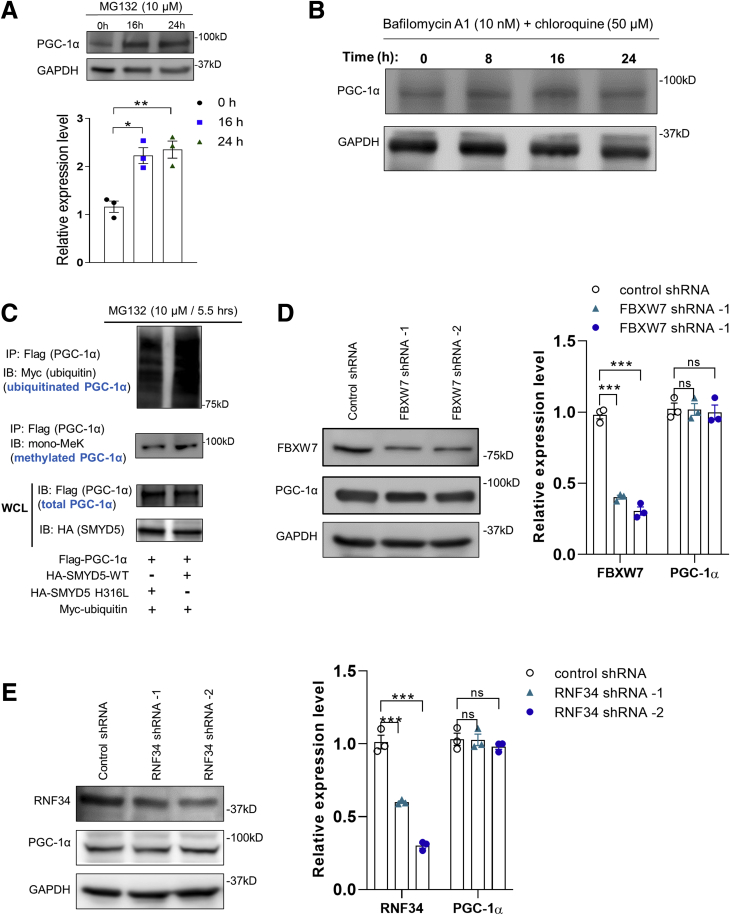
There are 2 major fundamentally distinct mechanisms by which proteins are degraded in eukaryotic cells: the ubiquitin-proteasome pathway and the autophagy-lysosomal pathway. Previous studies have shown that PGC-1α is a short-lived, unstable protein primarily targeted for ubiquitin-proteasome–dependent degradation at the basal level.47, 48, 49 We next sought to explore how PGC-1α protein level is regulated by SMYD5. In alignment with previous studies,48 treatment with proteasomal inhibitor MG132 resulted in significant up-regulation of PGC-1α protein level in HEK293T cells (Figure 11A). However, blockade of the autophagy-lysosomal degradation pathway with bafilomycin A1 (an inhibitor of vacuolar H+–adenosine triphosphatase) and chloroquine (which inhibits autophagic flux by decreasing autophagosome-lysosome fusion),50 did not affect PGC-1α protein level in HCT116 cells (Figure 11B), suggesting that SMYD5-induced PGC-1α degradation may be mediated by the ubiquitin-proteasome pathway. Because proteasomal degradation is mediated by protein ubiquitination,51 we next investigated whether SMYD5-mediated methylation affects PGC-1α ubiquitination and degradation. Western blot analysis of ubiquitinated, methylated, or total PGC-1α protein in HEK293T cells co-expressed with myc-ubiquitin, Flag-PGC-1α, and wild-type SMYD5 (HA-SMYD5-WT) or enzymatically inactive SMYD5 (HA-SMYD5-H316L)16 showed that wild-type SMYD5 significantly increased PGC-1α lysine methylation and ubiquitination compared with inactive SMYD5 (Figure 11C). Of note, the total PGC-1α amount was reduced substantially in cells co-expressed with HA-SMYD5-WT vs inactive mutant HA-SMYD5-H316L, even in the presence of MG132 (Figure 11C, lower band). To date, there have been a few E3 ligases, such as SCFCdc4 (FBXW7)47 and RNF3449 that have been shown to mediate PGC-1α ubiquitination and degradation. However, knocking down FBXW7 or RNF34 expression in IECs did not cause any significant change in PGC-1α protein levels (Figure 11D and E), suggesting that FBXW7 and RNF34 are not involved in the regulation of PGC-1α stability in IECs.
Figure 11.
SMYD5 regulates PGC-1α proteasomal degradation in an FBXW7- or RNF34-independent manner. (A) HEK293T cells were treated with the proteasome inhibitor MG132 (10 μmol/L) for the indicated times and total cell extracts were analyzed by Western blot for PGC-1α abundance. n = 3. (B) HCT116 cells were treated with a combination of inhibitors of autophagy-lysosomal pathway: bafilomycin A1 (10 nmol/L) and chloroquine (50 μmol/L), for 0, 8, 16, and 24 hours. Whole-cell lysates were immunoblotted for the expression of PGC-1α and GAPDH using respective antibodies. (C) HEK293T cells were co-transfected with myc-tagged ubiquitin (myc-ubiquitin), Flag–PGC-1α, and HA-SMYD5-WT or enzymatically inactive SMYD5 (HA-SMYD5-H316L), followed by treatment with MG132 (10 μmol/L) for 5.5 hours. Then, whole-cell lysates were subjected to immunoprecipitation with anti-Flag antibody (for Flag–PGC-1α) and the immunoprecipitated PGC-1α was analyzed by Western blot for ubiquitination using anti-myc antibody (for myc-ubiquitin) and for lysine methylation using anti–mono methyl lysine (mono-MeK) antibody. A small fraction of whole-cell lysates (WCL) before immunoprecipitation also was immunoblotted for PGC-1α (anti-Flag) and SMYD5 (anti-HA). (D) HCT116 cells were transduced with short hairpin RNA (shRNA) viruses targeting the E3 ligase FBXW7 (FBXW7 shRNA-1 or -2) or control shRNA viruses, and whole-cell lysates were immunoblotted for FBXW7, PGC-1α, and GAPDH. n = 3. (E) HCT116 cells were transduced with shRNA viruses targeting the E3 ligase RNF34 (RNF34 shRNA-1 or -2) or control shRNA viruses, and whole-cell lysates were immunoblotted for RNF34, PGC-1α, and GAPDH. n = 3. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
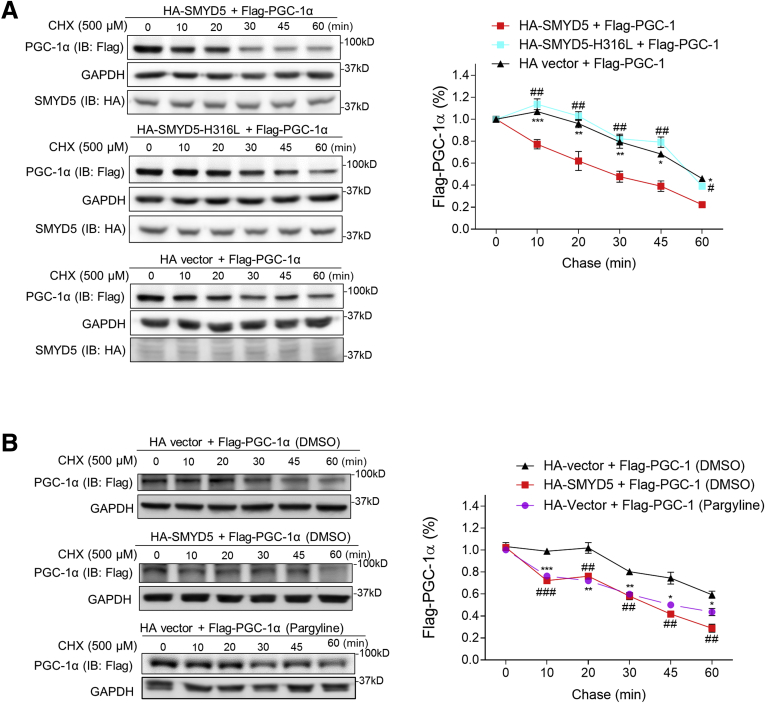
Next, we monitored the half-life of PGC-1α protein by pulse-chase analysis. HEK293T cells were transfected with Flag–PGC-1α, together with HA-SMYD5-WT, or HA-SMYD5-H316L, or HA alone vector, and the cells were treated with translation elongation inhibitor cycloheximide (CHX) to block protein synthesis, followed by chasing the remaining PGC-1α. The results showed that co-expression of wild-type SMYD5 with PGC-1α significantly reduced PGC-1α half-life (Figure 12A). However, co-expressing the inactive SMYD5 mutant with PGC-1α showed a similar effect on PGC-1α half-life as HA alone vector (Figure 12A), indicating that the reduction of PGC-1α half-life by SMYD5 overexpression is SMYD5 enzymatic activity–dependent. Many proteins are being regulated dynamically by methylation and demethylation, and the interplay of the methylation–demethylation machinery controls various processes such as gene expression, protein function, modification, and degradation.52 Therefore, it is possible that PGC-1α degradation also could be dynamically regulated by the lysine methylation–demethylation cycle. We postulated that inhibiting lysine demethylation might facilitate PGC-1α degradation, an effect similar to SMYD5 overexpression-mediated methylation. Treatment of HEK293T cells with a lysine-specific demethylase 1 inhibitor, pargyline,53 led to a comparable reduction of PGC-1α half-life as did SMYD5 overexpression (Figure 12B), suggesting dynamic control of PGC-1α stability by lysine methylation–demethylation balance. In all, these results clearly indicate that methylation of PGC-1α by SMYD5 leads to increased ubiquitination and consequently promotes proteasomal degradation of PGC-1α.
Figure 12.
SMYD5 regulates PGC-1α proteasomal degradation in an enzymatic activity–dependent manner. (A) HEK293T cells were transfected with Flag–PGC-1α along with HA-SMYD5-WT, HA-SMYD5-H316L, or HA alone vector. Then, cells were treated with CHX (500 μmol/L) for the indicated time periods before whole-cell lysates were immunoblotted for PGC-1α (with anti-Flag antibody), SMYD5 (with anti-HA antibody), and GAPDH. The relative levels of Flag–PGC-1α protein at each time point (normalized to GAPDH) is plotted as a percentage of the amount at 0 minutes. HA-SMYD5 vs HA vector, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001; HA-SMYD5 vs HA-SMYD5-H316L, #P < .05, ##P < .01; n = 3. (B) HEK293T cells were transfected with Flag–PGC-1α along with HA alone vector or HA-SMYD5-WT. Cells then were treated with either the lysine-specific demethylase 1 inhibitor, pargyline (2.5 mmol/L), or vehicle control (DMSO) as indicated in the figures for 24 hours. Thereafter, cells were treated with CHX (500 μmol/L) for the indicated time periods before whole-cell lysates were immunoblotted for PGC-1α (with anti-Flag antibody) and GAPDH. The relative levels of Flag–PGC-1α protein at each time point (normalized to GAPDH) is plotted as a percentage of the amount at 0 minutes. Pargyline treatment vs DMSO treatment, ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001; HA-SMYD5 vs HA vector, ##P < .01, ###P < .001; n = 3. IB, immunoblotting.
Methyl-Binding Protein Plant Homeodomain Finger Protein 20-Like 1 Is Involved in SMYD5-Mediated PGC-1α Degradation
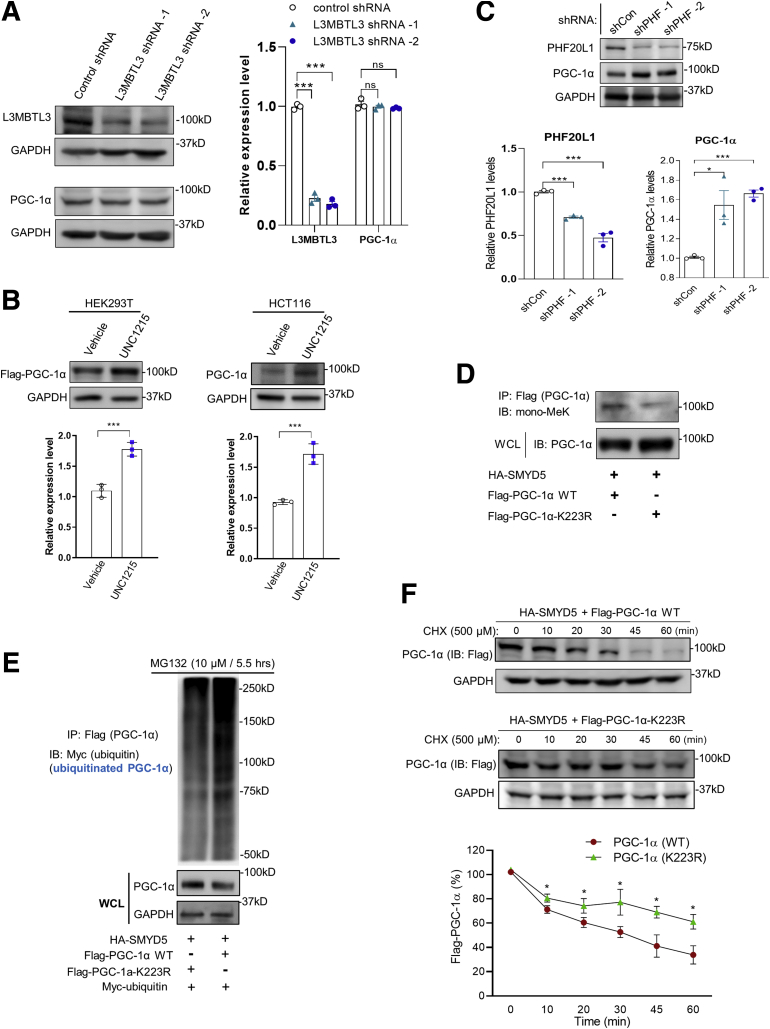
We have shown so far that SMYD5-mediated PGC-1α methylation triggers ubiquitin-dependent PGC-1α proteasomal degradation. However, it remains unclear how SMYD5-catalyzed PGC-1α methylation leads to its accelerated degradation. It has been reported that methylated lysine residues in substrate proteins interact with certain methyl-binding domain containing readers, which subsequently recruit, directly or indirectly, specific E3 ubiquitin ligases to regulate protein stability and turnover.43 We speculated that inhibiting the methyl lysine readers may block the binding/recognition of methylated lysine residues of PGC-1α by methyl lysine readers, thus preventing E3 ubiquitin ligase-mediated PGC-1α degradation. A previous study reported that the mono-methylated histone 4 lysine 20 catalyzed by SMYD2 was recognized by the methyl-lysine reader lethal (3) malignant brain tumor-like protein 1/3 (L3MBTL1).54 UNC1215, a potent methyl-lysine binding protein inhibitor, has been shown to inhibit the cognate reader L3MBTL3 for binding to mono- or dimethyl lysine-containing peptides.55 However, knockdown of L3MBTL3 did not cause any significant change of PGC-1α protein level (Figure 13A). It has been shown that UNC1215, at high concentrations, inhibited other methyl-binding proteins, such as plant homeodomain finger protein 20-like 1 (PHF20L1).56 Treatment of HEK293T and HCT116 cells with UNC1215 led to a significant increase of PGC-1α protein (Figure 13B), indicating that PGC-1α degradation is mediated, at least partially, by PHF20L1. To determine whether PHF20L1 might be involved in mediating PGC-1α degradation, clones of HCT116 cells stably expressing short hairpin RNA targeting PHF20L1 were generated and immunoblot analysis showed that PHF20L1 silencing substantially increased PGC-1α expression (Figure 13C), indicating that PHF20L1 may mediate PGC-1α degradation.
Figure 13.
Methyl-binding protein PHF20L1 is involved in SMYD5-mediated PGC-1α degradation. (A) HCT116 cells were transduced with short hairpin RNA (shRNA) viruses targeting the methyl-binding protein L3MBTL3 (L3MBTL3 shRNA-1 or -2) or control shRNA viruses, and whole-cell lysates were immunoblotted for L3MBTL3, PGC-1α, and GAPDH. (n = 3). (B) HEK293T cells overexpressing Flag–PGC-1α and HCT116 cells were treated with vehicle control or the methyl-binding protein inhibitor, UNC1215 (80 μmol/L), for 24 hours, and the whole-cell lysates were immunoblotted for the expression of PGC-1α (anti-Flag for HEK293T) and GAPDH. n = 3. (C) HCT116 cells were transduced with shRNA viruses targeting the methyl-binding protein PHF20L1 (shPHF-1 and shPHF-2) or control shRNA viruses (shCon), and whole-cell lysates were immunoblotted for PHF20L1, PGC-1α, and GAPDH. n = 3. (D) HEK293T cells were transfected with HA-SMYD5 along with Flag–PGC-1α–WT or Flag–PGC-1α–K223R. Then, whole-cell lysates were subjected to immunoprecipitation with anti-Flag antibody (for Flag–PGC-1α WT or K223R) and the immunoprecipitated Flag–PGC-1α (WT or K223R) was analyzed by Western blot for lysine methylation using anti-mono methyl lysine (mono-MeK) antibody. Whole-cell lysates (WCL) also were immunoblotted for PGC-1α (anti-Flag). (E) HEK293T cells were co-transfected with myc-ubiquitin, HA-SMYD5, and Flag–PGC-1α WT or Flag–PGC-1α–K223R followed by treatment with MG132 (10 μmol/L) for 5.5 hours. Then, whole-cell lysates were subjected to immunoprecipitation with anti-Flag antibody (for Flag–PGC-1α WT or K223R) and the immunoprecipitated Flag–PGC-1α (WT or K223R) was analyzed by Western blot for ubiquitination using anti-myc antibody (for myc-ubiquitin). WCL also were immunoblotted for PGC-1α (anti-Flag) and GAPDH. (F) HEK293T cells were transfected with HA-SMYD5 along with Flag–PGC-1α–WT or Flag–PGC-1α–K223R. Then, cells were treated with CHX (500 μmol/L) for the indicated time periods before whole-cell lysates were immunoblotted for PGC-1α (anti-Flag) and GAPDH. The relative protein levels of Flag–PGC-1α WT or K223R at each time point (normalized to GAPDH) is plotted as a percentage of the amount at 0 minutes. Flag–PGC-1α–WT vs Flag–PGC-1α–K223R. n = 3. ∗P < .05, ∗∗∗P < .001.
We next explored the role that lysine residue K223 plays in PHF20L1-mediated PGC-1α ubiquitination and degradation. First, we constructed lysine 223 to arginine (K223R) point mutation in PGC-1α (Flag–PGC-1α–K223R) and co-expressed SMYD5 and wild-type PGC-1α (Flag–PGC-1α) or Flag–PGC-1α–K223R in HEK293T cells. Immunoblot analysis showed that K223R point mutation of PGC-1α significantly reduced SMYD5-mediated PGC-1α methylation and increased PGC-1α protein level (Figure 13D), indicating a critical role of K223 mono-methylation in PGC-1α stability. To test whether K223R mutation would render PGC-1α to be less ubiquitinated owing to failure of methylation of K223R mutant, we co-expressed myc-ubiquitin, HA-SMYD5, and Flag–PGC-1α WT or Flag–PGC-1α–K223R in HEK293T cells and performed a co-immunoprecipitation assay. The result showed that K223R methylation mutation indeed led to reduced PGC-1α ubiquitination (Figure 13E, top) and increased PGC-1α protein level (Figure 13E, lower band), suggesting that K223 residue plays an important role in mediating PGC-1α ubiquitination. Furthermore, CHX chase assay showed a significantly increased half-life of PGC-1α K223R mutant compared with PGC-1α WT (Figure 13F), suggesting that K223R mutation, at least partially, protects PGC-1α from ubiquitin-dependent proteasomal degradation.
Altogether, these results clearly showed that SMYD5-catalyzed PGC-1α methylation at K223 residue, probably via methyl-binding protein PHF20L1, promotes PGC-1α ubiquitination and proteasomal degradation.
Discussion
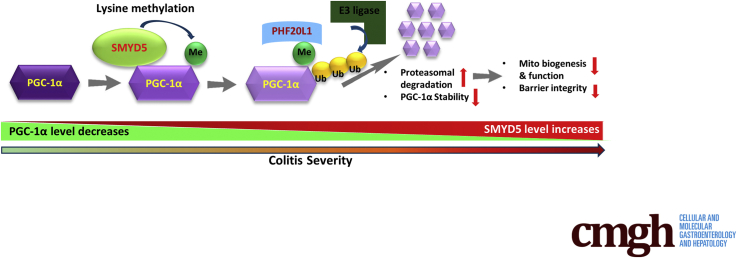
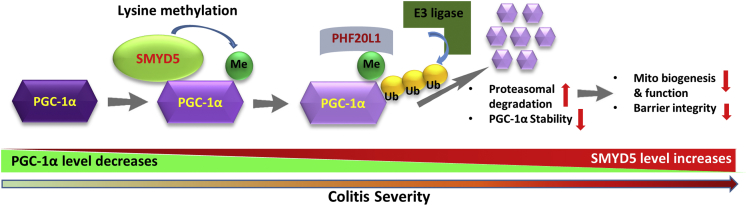
Mitochondrial dysfunction is implicated in the pathogenesis and progression of IBD.23 It has been shown that decreased PGC-1α in IECs resulted in mitochondrial dysfunction in murine colitis.20 However, the underlying molecular mechanism remains elusive. In this study, we have provided evidence that SMYD5, which initially was identified as an epigenetic modifier that trimethylates H4 at K20 to regulate TLR4 target genes in macrophages,16 functions as a critical negative regulator of epithelial integrity and intestinal homeostasis by suppressing mitochondrial biogenesis and function through methylation-mediated degradation of PGC-1α (Figure 14). We observed that epithelial SMYD5 was up-regulated in intestinal mucosa from IBD patients and colitic mice. SMYD5 knockdown in human IECs resulted in increased mitochondrial biogenesis and function and reduced reactive oxygen species (ROS) production. Furthermore, IEC-specific SMYD5 deficiency protected mice against DSS-induced colitis and intestinal inflammation. Mechanistically, we showed that SMYD5 interacts directly with and methylates PGC-1α, and methylation of PGC-1α in IECs leads to its ubiquitination and proteasome-mediated degradation. The expression of SMYD5 in intestinal epithelia, along with its ability to interact with and methylate PGC-1α, suggests that SMYD5 and PGC-1α may work together to regulate mitochondrial function and intestinal homeostasis.
Figure 14.
A schematic model depicting SMYD5-catalyzed methylation, ubiquitination, and degradation of PGC-1α protein in regulating intestinal health and colitis. By controlling the methylation status of PGC-1α, SMYD5 modulates its stability and turnover, which further regulates mitochondria functions and epithelial barrier integrity. Me, methylation; Ub, ubiquitinylation.
Our finding that SMYD5 is up-regulated in colon mucosa from IBD patients is consistent with a previous study that, by using genome-wide association study, showed SMYD5 up-regulation in human IBD colonic biopsy specimens.57 Recent proteomic studies have shown that compared with other members of the SMYD family, SMYD5 is broadly expressed, but with a medium level in colon and rectum in healthy tissues.58 Meanwhile, it has been reported that in cancer patients, especially in patients with gastrointestinal cancers, SMYD5 transcription is up-regulated significantly compared with healthy individuals.59 It is well documented that IBD is the primary risk factor for the development of gastrointestinal cancer.60 Together, this suggests that SMYD5 may play a critical role in intestinal epithelia during the pathogenesis of various gastrointestinal diseases, especially IBD.
A more recent study reported that mitochondria play a key role in determining cell fate and mitochondrial down-regulation (via fission) promotes stem cell differentiation into secretory lineages (goblet cells and Paneth cells).61 To check for the possible effect of Smyd5 ablation on cell fate decisions, we compared the number of stem cells and goblet cells in the colons of Smyd5fl/fl and Smyd5ΔIEC mice. We did not observe any difference in BMI1-positive cells per crypt or LGR5 staining between Smyd5ΔIEC mice and Smyd5fl/fl mice. In addition, PAS staining did not reveal any significant difference in crypt goblet cell counts between Smyd5fl/fl and Smyd5ΔIEC mice in the basal state. We also measured the crypt depth and villus height of small intestines from both mouse strains, and the quantification did not reveal any significant difference in villus height and crypt depth between them. These results suggest that conditional ablation of Smyd5 in IECs does not alter intestinal homeostasis (such as epithelial cell type or stem cell numbers) in the basal state (before insult with DSS). We speculate that SMYD5 may be redundant to normal intestinal development and function, either owing to the compensatory effects of other factors and/or alternative pathways, or the minor, fine-tuning effects of the target proteins of SMYD5-mediated methylation. In pathologic situations (such as DSS insult), however, when specific biological processes are hyperactivated or repressed, the modulatory effects of post-translational modifications may be manifested in a more dominant manner. This is likely the case with SMYD5 function because under cellular damage conditions triggering intestinal injury and inflammation, SMYD5-deficient mice showed a significantly alleviated clinical phenotype.
Post-translational modifications of the PGC-1α protein have been shown to alter its stability and transcriptional specificity.10 Recent studies have shown that methyltransferases such as SET7/9 and PRMT1 can methylate PGC-1α to regulate its functions including mitochondrial biogenesis.13,14 Our present study uncovered PGC-1α as a novel nonhistone target of SMYD5. Mechanistically, we have identified a lysine residue, K223, of murine PGC-1α, that was mono-methylated by SMYD5. The mutation of this lysine residue to arginine (K223R) significantly reduced SMYD5-mediated PGC-1α methylation and ubiquitination. More importantly, K223R mutation has increased the PGC-1α half-life. These findings have supported the notion that SMYD5-mediated PGC-1α methylation is involved in regulating PGC-1α turnover in IECs. Interestingly, a recent study also showed the mono-methylation of human PGC-1α at K224 (equivalent to the K223 of mouse PGC-1α) induced by hypoxia in human glioblastoma cells and PGC-1α K224 showed limited mono-methylation under normoxic conditions.62 However, they did not detect any changes of PGC-1α mRNA or protein expression. Instead, they found that hypoxia decreases PGC-1α activity via inhibition of the lysine demethylase 3A–mediated demethylation of human PGC-1α at K224, which consequently results in reduced mitochondrial biogenesis.62 They also observed that treatment of human glioblastoma cells with 5-carboxy-8-hydroxyquinoline, a potent broad-spectrum inhibitor of the Jumonji C domain-containing family of 2-oxoglutarate-dependent demethylases, or deferoxamine, an iron chelator that blocks iron-dependent demethylation, drastically increased PGC-1α K224 monomethylation, suggesting that PGC-1α K224 monomethylation induced by hypoxia is owing to inhibition of a demethylase. However, in our present study, treatment of IECs with either 5-carboxy-8-hydroxyquinoline or deferoxamine failed to induce any change in PGC-1α protein level, suggesting that the mechanism and effect of PGC-1α methylation on mitochondrial biogenesis and function may be cell type– or context-dependent.
In our study, immunoblot analysis showed methylated lysine signals at N-terminal, middle, and C-terminal (weak signal) regions of PGC-1α. However, our mass spectrometry analysis only detected a lysine methylation at K223 within the middle fragment of PGC-1α (aa 190–345). We speculate this may be owing to technique challenge. Interestingly, the N-terminal domain of PGC-1α has been reported to have no effect on protein stability and subcellular distribution.48 Recently, a lysine residue within the C-terminal (K779) of PGC-1α was reported to be mono-methylated by SET7/9 in Hepa 1–6 cells; however, this methylation had no effect on PGC-1α protein stability.14 Nonetheless, methylation on other lysine residues of PGC-1α also may contribute to its ubiquitination and degradation. Of note, K223R mutation failed to completely reverse SMYD5-mediated PGC-1α methylation and degradation. Therefore, it is possible that SMYD5 catalyzes methylation of multiple lysine residues within PGC-1α, which collectively contribute to PGC-1α degradation, although K223 methylation may play a major role. It will be interesting to identify all the lysine residues in PGC-1α methylated by SMYD5 and define their distinct functional significance.
It is possible that SMYD5 may exert its role in the intestinal epithelium through other mechanisms as well. It has been reported that SMYD5 depletion leads to increased cell growth owing to decreased genome-wide histone 4 lysine 20 trimethylation.17 In the present study, the IHC staining of proliferation marker PCNA revealed a comparable PCNA positivity between the 2 groups in the basal state; however, upon DSS administration, IECs from Smyd5ΔIEC mice showed increased cell proliferation (PCNA staining) compared with those from Smyd5fl/fl mice (data not shown). This suggests that SMYD5 also may play a PGC-1α–independent role during the pathogenesis of IBD. It has been reported that the MYND domain within the SMYD proteins are involved in interactions with a preference for binding to a proline-rich motif (PXLXP).63 For instance, SMYD1 has been reported to interact with the PPLIP motif of skeletal and heart muscle–specific variant of the α subunit of nascent polypeptide–associated complex. Moreover, the PXL motif in heat shock protein 90 and 23 kDa cochaperone protein (p23) has been reported to mediate their binding with SMYD2.64 Interestingly, PGC-1α also contains a proline-rich region.65 Therefore, it is possible that the proline-rich motif may mediate the interaction between PGC-1α and SMYD5, which warrants further investigation.
In summary, our study showed a protective role of SMYD5 ablation in IECs against IBD and identified the first nonhistone substrate of SMYD5, PGC-1α, the master regulator of mitochondrial biogenesis and functions. By controlling the methylation status of PGC-1α, SMYD5 modulates its stability and turnover, which further regulates mitochondria functions and epithelial barrier integrity (Figure 14). Limited research has been performed on SMYD5, with focus solely on histone biology and epigenetics. Our findings have broadened understanding of SMYD family members, and provided evidence that targeting the SMYD5/PGC-1α axis in IECs could be a potential therapeutic target for IBD treatment, as well as other mitochondria-implicated disease conditions such as neurodegenerative diseases, cancers, obesity, and diabetes.
Materials and Methods
Human Colonic Samples
To examine SMYD5 and PGC-1α expression in human intestinal epithelium in health and disease, paraffin-embedded specimens of human colonic mucosa samples of control individuals (n = 4) and IBD patients with active inflammation (n = 12) were obtained from US Biomax, Inc (Rockville, MD). The tissue samples included normal colon tissue (n = 4), inflamed colon tissues of chronic ulcerative colitis (n = 8), and Crohn's disease of the ileocecal junction (n = 4).
Generation and Genotyping of IEC-Specific Smyd5 Conditional KO Mice
To generate IEC-specific Smyd5 KO mice (denoted Smyd5ΔIEC), embryonic stem cell clones with conditional potential targeting exon were obtained from the European Mouse Mutant Archive (EM:06942). Male embryonic stem cells were injected into C57BL/6 blastocysts at the Georgia State University Transgenic and Gene Targeting Core facility. The targeted Smyd5 allele contains a LacZ reporter, the flippase - flippase recombinase target (FLP-FRT) sites, the neomycin-resistant marker, and the Cre- locus of X-over P1 (loxP) sites flanking exon 2 of the murine Smyd5 gene (Figure 2A). The pups with transmission of heterozygous Smyd5fl/+ was confirmed by PCR analysis. The mice carrying the Smyd5fl/+ allele were mated until the Smyd5fl/fl colony was obtained. Smyd5fl/fl mice then were bred with mice expressing intestinal-specific Cre-recombinase under the control of the villin promoter (catalog number 4586; The Jackson Laboratory, Bar Harbor, ME) to generate both littermate control (Smyd5fl/fl) mice and IEC-specific Smyd5 KO (Smyd5ΔIEC) mice. Both Smyd5fl/fl and Smyd5ΔIEC mice were genotyped by PCR using primers listed in Table 1. In this study, all the mice used were on a C57BL/6 background, and the mutant lines were backcrossed for at least 6 generations and littermates were used as controls in all experiments. The animal studies were approved by the Institutional Animal Care and Use Committee of Georgia State University.
Table 1.
Primer Sequences for Genotyping
| Primer | Sequence 5’ → 3’ | Primer type | Note |
|---|---|---|---|
| Primers for Smyd5 genotyping | |||
| Smyd5_F | GGTCTCATGGGGAACTGAGG | WT and mutant forward | Designed by EMMA (European Mouse Mutant Archive) |
| Smyd5_R | GCTTTCAGCCAAGCCAAGTC | WT reverse | |
| CAS_R1_Term | TCGTGGTATCGTTATGCGCC | Mutant reverse | |
| Primers for Vil1-Cre genotyping | |||
| 18960 | TTCTCCTCTAGGCTCGTCCA | Transgene reverse | Designed by The Jackson Laboratory |
| 14506 | CATGTCCATCAGGTTCTTGC | Transgene forward | |
| oIMR7338 | CTAGGCCACAGAATTGAAAGATCT | Internal positive control forward | |
| oIMR7339 | GTAGGTGGAAATTCTAGCATCATCC | Internal positive control reverse | |
DSS-Induced Colitis in Mice and Disease Severity Assessment
Mice at 8–10 weeks of age were treated with 2.5% (wt/vol) DSS (colitis grade, molecular weight of 36–50 kilodaltons; MP Biochemicals, Irvine, CA) in drinking water ad libitum for 7 days and DSS-free water for another 2 days. In water control groups, mice were administered DSS-free drinking water for 9 days. The severity of colitis was recorded daily based on body weight loss, stool consistency, and the presence of blood in the stools, and the daily disease activity index was scored based on a combination of body weight loss, stool consistency, and the presence or absence of blood in stool as we previously reported.66 On day 9, the colonic tissues were collected and fixed in 10% formaldehyde for 24 hours, embedded in paraffin, and sectioned for H&E or IHC staining as described later.
Histopathology Scoring of Colonic Inflammation
For histologic analysis of colonic inflammation, the Swiss roll method was used to prepare the sections of the mouse colons for histology (exclusive of cecum), as we reported previously.66 Then, the tissues were fixed overnight in paraformaldehyde, embedded in paraffin, and cut into 5-μm–thick sections for H&E staining. The colonic inflammation was evaluated by 4 simply detectable pathologic measures such as cellular infiltration (0–5), deterioration of crypt architecture (crypt damage, 0–4), extent of mucosal ulceration (0–3), and the absence or presence of submucosal edema (0 and 1), as described previously67 and as we recently reported.66 The evaluation was performed by an experienced IBD pathologist who was blinded to the experimental design. The data were generated by averaging 5 sections from each animal, and these sections were from similar locations among groups.
Intestinal Permeability Assay
An intestinal permeability assay was performed by evaluation of FITC-dextran (4 kilodaltons, Sigma Aldrich, St. Louis, MO) in the blood as previously described.68 Briefly, Smyd5fl/fl and Smyd5ΔIEC mice were subjected to DSS-induced acute colitis as described earlier. On the last day of DSS treatment, mice were fasted for 4 hours and administered FITC-dextran tracer (0.4 mg/g body weight) dissolved in phosphate-buffered saline (PBS) by oral gavage. Hemolysis-free serum then was collected after 1 and 4 hours post-gavage. A standard curve for FITC-dextran concentration was established by serial dilution of known amount of FITC-dextran, and serum from mice administered PBS only was used to determine the background signal. The samples were diluted in PBS and the absorbance at 520 nm was measured.
Isolation of Mouse Primary IECs
IECs were isolated as reported previously,69 with minor modifications. Briefly, colonic tissues were removed from the mice and opened longitudinally and cut into pieces followed by washing with PBS. IECs then were isolated after incubation of tissue pieces in PBS containing 30 mmol/L EDTA and 2 mmol/L dithiothreitol for 20 minutes at 37°C with gentle shaking at 200 rpm. Cells then were subjected to vigorous shaking for 30 seconds. After removal of the tissue debris, the isolated cells were pelleted by centrifugation at 1000 × g for 5 minutes.
IF and IHC
For IF and IHC analysis, antigen retrieval was performed by a heat-induced epitope retrieval method using sodium citrate buffer (pH 6.0) in a water bath. Endogenous peroxidase was inhibited by blocking solution (SP-6000; Vector Laboratories, Newark, CA) according to the manufacturer’s instructions. The sections were blocked for 1 hour with Tris-buffered saline, 0.1% Tween 20 (TBST) containing 5% normal goat serum (G9023; Sigma). The sections were incubated with the following primary antibodies overnight at 4°C: anti-SMYD5 (NBP1-31222; Novus, Centennial, CO), 1:100 for IHC analysis; anti-SMYD5 (ab81419; Abcam, Cambridge, UK), 1:100 for IF imaging; anti–PGC-1α (NBP1-04676; Novus), 1:150 for IF imaging; and anti–ZO-1 (MABT11; Sigma), 1:100 for IF imaging. Thereafter, the sections were washed with TBST 3 times and incubated for 1 hour at room temperature with Alexa Fluor 568 goat anti-rabbit IgG (heavy + light chain H+L) (A11036; Invitrogen, Carlsbad, CA) or Alexa Fluor 488 goat anti-rabbit IgG (H+L) (A11034; Invitrogen). The sections then were washed, stained with 4′,6-diamidino-2-phenylindole for 5 minutes, and covered with antifade mountant (P36970; Invitrogen).
For IHC staining, goat anti-rabbit IgG (H+L)-peroxidase conjugated secondary antibody (A0545; Invitrogen) was used. Then, the sections were washed and incubated with peroxidase substrate (SK-4105; Vector Laboratories) for 10 minutes. Sections then were washed with water to remove extra substrate, processed for dehydration, and covered with Permount Mounting Medium (SP-15; Fisher Scientific). The images were evaluated with ImageJ (National Institutes of Health, Bethesda, MD) by an experienced researcher who was blinded to the study design. Quantification of the IHC staining of SMYD5 and PGC-1α expression was performed using ImageJ software according to a published protocol.70 The protocol gives detailed steps on how to deconvolute IHC images stained with hematoxylin and 3,3-diaminobenzidine tetrahydrochloride (DAB) and how to quantify their expression using ImageJ. The graphs then were plotted using GraphPad Prism software (GraphPad, San Diego, CA).
TEM and Examination of Mitochondrial Ultrastructure
Sample embedding, sectioning, and TEM were performed by the Robert P. Apkarian Integrated Electron Microscopy Core at Emory University (Atlanta, GA) as described previously.71 Colon tissues obtained from Smyd5fl/fl mice and Smyd5ΔIEC mice (n = 3 mice/group) were fixed with 2.5% glutaraldehyde in 0.1 mol/L cacodylate buffer (pH 7.4), followed by postfixation with 1% osmium and 1.5% potassium ferrocyanide in the same buffer. Tissues then were dehydrated in ethanol and embedded in Eponate 12 resin (Ted Pella, Inc., Redding, CA). Ultrathin (70-nm) sections were cut with an ultramicrotome and stained with 5% uranyl acetate and 2% lead citrate. Images were acquired using a Hitachi H-7500 TEM (Chiyoda-ku, Tokyo) equipped with a SIA L12C (Duluth, GA) 16 megapixel charged-coupled device (CCD) camera. All measurement analyses were performed by laboratory personnel blinded to sample identities using ImageJ software. The Robert P. Apkarian Integrated Electron Microscopy Core at Emory University is subsidized by the School of Medicine and Emory College of Arts and Sciences. Additional support was provided by the Georgia Clinical & Translational Science Alliance. Quantitative TEM was used to image enterocytes/IECs in sections of colon tissues. Specifically, a total of 35 images were taken for each mouse. Thirty images were randomly selected per mouse for analysis according to the protocol described previously.72 Of note, more than 30 cells were examined for quantification of the number of mitochondria per cell.
Cell Culture, Transfection, and Treatments
Human colorectal epithelial cells (HCT116) and HEK293T cells were cultured under standard culture conditions as described previously.66,73 In brief, cells were cultured in McCoy’s 5A (SH30200.1; Hyclone, Logan, UT) or Dulbecco’s modified Eagle medium (11965092; Gibco, Waltham, MA) medium supplemented with 10% fetal bovine serum and antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin) and maintained in a 37°C incubator with 5% CO2. Cells were used for transfection when they reached 80% confluence. For transient transfection, plasmids encoding wild-type SMYD5 (plasmid pcDNA3.1-HA-SMYD5) (Genscript Biotech, Piscataway, NJ) or enzymatically inactive SMYD5 (pcDNA3.1-HA-SMYD5-H316L), wild-type PGC-1α (pcDNA–Flag–PGC-1α) (1026; Addgene, Watertown, MA) or methylation-resistant PGC-1α (pcDNA–Flag–PGC-1α K223R), ubiquitin (pcDNA3.1-Myc-ubiquitin), or empty vector plasmid were transfected into cells using Lipofectamine LTX (Waltham, MA) according to the manufacturer’s instructions. To generate methylation-resistant PGC-1α, pcDNA–Flag–PGC-1α, which encodes the Flag-tagged wild-type PGC-1α, was used as template. The Q5 Site-Directed Mutagenesis Kit (E0554S; New England Biolabs, Ipswich, MA) was used to generate the K223R mutation (lysine to arginine) using primers synthesized by Sigma-Aldrich according to the manufacturer’s instructions.
For evaluating cellular oxidative stress induced by inflammatory response and changes of expression of PGC-1α and SMYD5 upon exposure to proinflammatory cytokines, cells were treated with TNF-α (Prospec, East Brunswick, NJ) (20 ng/mL) and/or IFN-γ (Prospec) (20 ng/mL) or vehicle control (PBS) for 24 hours. To define the role of SMYD5 in oxidative stress–induced cell apoptosis, cells were treated with hydrogen peroxide (100 μmol/L) for 0, 4, or 8 hours. Then, cells were subjected to Western blot analysis of SMYD5 expression and the cleavage of caspases (caspases 3 and 9).
To investigate the pathways for PGC-1α degradation, cells were treated with proteasomal inhibitor (MG132, 10 μmol/L, for 0, 5.5, 16, and 24 h) or autophagy inhibitors (10 nmol/L Bafilomycin A1 and 50 μmol/L chloroquine for 0, 8, 16, and 24 h), respectively, followed by Western blot. To study the role of SMYD5 in PGC-1α ubiquitination, cells were treated with MG132 (10 μmol/L) for 5.5 hours and then subjected to immunoblotting analysis with respective antibodies. To study the effect of methylation on the proteasomal degradation of PGC-1α, cells were treated with lysine demethylase inhibitor (2.5 mmol/L pargyline) or vehicle control (DMSO) for 24 hours, and then treated with protein synthesis inhibitor (500 μmol/L cycloheximide) for 0, 10, 20, 30, 45, and 60 minutes. Thereafter, the cells were used for Western blot analysis. To determine the role of PHF20L1 in SMYD5-mediated PGC-1α degradation, cells were incubated with a small-molecule inhibitor of methyl-lysine readers (80 μmol/L UNC1215) or vehicle control (DMSO) for 24 hours followed by Western blot to detect the expression of PGC-1α.
RT-qPCR Analysis
Total RNA was extracted from cells with TRIzol reagent (AM9738; Invitrogen) and complementary DNA was synthesized using a rAMP Complementary DNA Synthesis kit (CC1151; Denville) according to the manufacturer’s instructions. RT-qPCR analysis was performed to determine the expression levels of mitochondrial biogenesis markers (Tfam, Cox I, and Cox II) in cultured human IEC lines and mouse primary IECs. RT-PCR was performed in triplicate using power-up SYBR Green master mix (A25741; Applied Biosystems, Waltham, MA). The expression levels of Tfam, Cox I, and Cox II were normalized to the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The respective primers used are listed in Table 2.
Table 2.
Primer Sequences for RT-qPCR
| Gene | Forward 5’ → 3’ | Reverse 5’ → 3’ |
|---|---|---|
| Tfam (human) | AAGATTCCAAGAAGCTAAGGGTGA | CAGAGTCAGACAGATTTTTTCCAGTTT |
| Cox I (human) | CCCACCGGCGTCAAAGTATT | TACAATGCCAGTCAGGCCAC |
| Gapdh (human) | ACCCACTCCTCCACCTTTGA | CTGTTGCTGTAGCCAAATTCGT |
| NADH dehydrogenase (human) | CGATTCCGCTACGACCAACT | GTTTGAGGGGGAATGCTGGA |
| GAPDH (human) | TTTCTTTGCAGCAATGCCTCC | CCATTCCCCAGCTCTCATACC |
| Cox I (mouse) | GGTCAACCAGGTGCACTTTT | TGGGGCTCCGATTATTAGTG |
| Cox II (mouse) | CCACTTCAAGGGAGTCTGGA | AGTCATCTGCTACGGGAGGA |
| Gapdh (mouse) | CATCGTGGAAGGGCTCATGAC | CTTGGCAGCACCAGTGGATG |
| Pgc1a (mouse) | TGAATGCAGCGGTCTTAGCA | TGCTCCATGAATTCTCGGTCTTA |
Detection of mtDNA Copy Number
Total DNA was extracted from HCT116 cells (parental HCT116, SMYD2 KO HCT116, and control HC116) using the Blood & Cell Culture DNA Mini Kit (13323; Invitrogen) according to the manufacturer’s instructions. mtDNA copy number was determined by RT-qPCR as previously reported.74 In general, the mitochondrial gene (Cox II or reduced nicotinamide adenine dinucleotide dehydrogenase) and nuclear gene (GAPDH) were determined and the respective primers used are listed in Table 2. The results are presented as the ratio of mitochondrial DNA relative to nuclear DNA.
Fluorescence-Based Analysis of Cellular Oxidative Stress
The cellular oxidative stress in SMYD5 KO HCT116 and parental HCT116 cells (SMYD5 WT) was measured using CellROX Orange Reagent (C10443; Fisher Scientific) as reported previously.40 CellROX Orange Reagent is a novel fluorogenic probe for measuring oxidative stress in live cells. This cell-permeant dye is nonfluorescent while in a reduced state and shows bright orange fluorescence upon oxidation by ROS. HCT116 cells (WT and SMYD5 KO) were treated with proinflammatory cytokines TNF-α (20 ng/mL; Prospec) and IFN-γ (20 ng/mL; Prospec), or vehicle control (PBS) for 24 hours. Then cells were stained with CellROX Orange reagent to detect the cellular ROS following the manufacturer’s protocol.
OCR Analysis
OCR was measured in an XFe96 extracellular flux analyzer (Seahorse Bioscience, Santa Clara, CA) as previously described.75 Briefly, parental HCT116 cells and HCT116 cells with SMYD5 OE or KO were seeded in a 96-well plate (in triplicate) (101085-004; Seahorse Bioscience) the day before measurement. On the day of measurement, the cells were incubated at 37°C and the medium was replaced with 180 μL XF assay medium (102365-100; Seahorse Bioscience). Oligomycin, trifluoromethoxy carbonyl cyanide phenylhydrazone, and rotenone/antimycin A provided with the XF Cell Mito Stress Test Kit (103015-100; Seahorse Bioscience) were prepared in XF assay medium (100840-000; Seahorse Bioscience). Measurement was performed at 37°C to detect basal respiration, maximal respiration, proton leak, and coupled respiration. Data were analyzed using Wave software provided by Seahorse Bioscience.
Western Blot Analysis
Western blot analysis was performed according to standard procedures using lysates from tissues or cells as described previously.66,76 In brief, tissues or cells were solubilized in M-PER mammalian protein extraction reagent (78501; Thermo Scientific, Waltham, MA) supplemented with protease inhibitor cocktail (P8340; Sigma). After the protein concentration was measured using the Bradford method, the same amounts of total proteins for each sample or treatment were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and transferred to polyvinylidene difluoride membranes (#1620177; Bio-Rad, Hercules, CA). The membranes were blocked with 5% goat serum at room temperature for 1 hour before the addition of primary antibodies for overnight incubation at 4°C. The following day, the membranes were incubated with secondary antibodies for 1 hour at room temperature. The enhanced chemiluminescent kit was used for development, and the images were captured using the Amersham Imager 600 RGB system (GE Health Care, Chicago, IL).
The primary antibodies used were as follows: anti-SMYD5 (ab81419; Abcam), anti-ubiquitin (13-1600; Invitrogen), anti-PHF20L1 (NBP1-79401), anti–PGC-1α (NBP1-04676; Novus), anti–COX I (sc-19998; Santa Cruz, Santa Cruz, CA), anti–COX II (12282; Cell Signaling Technology, Danvers, MA), anti–COX IV (4850; Cell Signaling Technology), anti-HSP60 (12165; Cell Signaling Technology), anti–pyruvate dehydrogenase (3205; Cell Signaling Technology), anti-SDHA (11998; Cell Signaling Technology), anti-VDAC (4661; Cell Signaling Technology), anti-UCP2 (89326; Cell Signaling Technology), anti-UCP3 (14670; Cell Signaling Technology), anti–cleaved caspase 3 (9664; Cell Signaling Technology), anti–cleaved caspase 9 (7237; Cell Signaling Technology), anti-TFAM (ABE483; Millipore, St. Louis, MO), anti-myc (05-724, Millipore), anti-Flag (F9291; Sigma), anti-HA (H9658; Sigma), and anti-GAPDH (AM4300; Invitrogen).
Immunoprecipitation
Immunoprecipitation assay was used for detecting the methylation status of lysine residues in PGC-1α. Briefly, HEK293T cells were co-transfected with HA-tagged SMYD5 and wild-type Flag-tagged PGC-1α or co-transfected with HA-tagged SMYD5 and mutated Flag-tagged PGC-1α (K223R). Forty-eight hours after transfection, cells were lysed with M-PER mammalian protein extraction reagent supplemented with protease inhibitor cocktail, and the cleared lysates were incubated with anti-Flag magnetic beads (M8823; Sigma) at 4°C overnight. After washing, the bound proteins were eluted by boiling in Laemmli sample buffer. Immunoblot analysis was conducted to detect the mono-methylated lysine (16479; Cell Signaling Technology), dimethylated lysine (14117; Cell Signaling Technology), and trimethylated lysine (14680; Cell Signaling Technology), as described earlier.
Co-immunoprecipitation
Co-immunoprecipitation assay was used for detecting the interaction between SMYD5 and PGC-1α, and detecting the ubiquitination of PGC-1α. To detect SMYD5–PGC-1α interaction, HEK293T cells were co-transfected with HA-SMYD5 and Flag–PGC-1α. Forty-eight hours after transfection, cells were lysed as described earlier and whole-cell lysates were immunoprecipitated with anti-HA (A2095; Sigma) or control IgG, and the immunocomplexes were immunoblotted using anti-Flag (for PGC-1α) and anti-HA (for SMYD5) antibodies.
For an in vivo ubiquitination assay, HEK293T cells were co-transfected with myc-tagged ubiquitin (myc-ubiquitin), Flag-tagged PGC-1α (Flag–PGC-1α), and HA-tagged wild-type SMYD5 (HA-SMYD5-WT) or enzymatically inactive SMYD5 (HA-SMYD5-H316L) as indicated in the respective Figure legends. Forty-eight hours after transfection, cells were treated with proteasome inhibitor MG132 (10 μmol/L) for 5.5 hours. In other experiments, HEK293T cells were co-transfected with myc-ubiquitin, HA-SMYD5, and Flag–PGC-1α WT or Flag–PGC-1α–K223R followed by treatment with MG132 as described earlier. Cells were lysed as described earlier and whole-cell lysates were subjected to immunoprecipitation with anti-Flag antibody (M8823; Sigma) for Flag–PGC-1α (WT or K223R) and the immunoprecipitated Flag–PGC-1α (WT or K223R) was analyzed by Western blot for ubiquitination using anti-myc antibody (for myc-ubiquitin), or for lysine methylation in PGC-1α using anti–mono-methyl lysine antibody. A small fraction of whole-cell lysates before immunoprecipitation also was immunoblotted for PGC-1α (anti-Flag), SMYD5 (anti-HA), or GAPDH.
PGC-1α Protein Stability (Half-Life) Analysis
HEK293T cells were transfected with Flag–PGC-1α along with HA alone vector, or HA-SMYD5-WT, or HA-SMYD5-H316L using Lipofectamine 2000. In parallel, HEK293T cells were transfected with HA-SMYD5 along with Flag–PGC-1α–WT or Flag–PGC-1α–K223R. In some groups, cells transfected with Flag–PGC-1α as described earlier were treated with lysine-specific demethylase 1 inhibitor, pargyline (2.5 mmol/L), or vehicle control (DMSO) for 24 hours. Then, cells were treated with CHX (500 μmol/L) to inhibit protein translation initiation for the indicated time periods as described in the Figure legends before whole-cell lysates were immunoblotted for PGC-1α (with anti-Flag antibody), SMYD5 (with anti-HA antibody), and GAPDH. The intensity of Flag–PGC-1α protein bands (WT or K223R) at each time point were quantified, normalized to GAPDH, and plotted as a percentage relative to levels observed at time 0.
Methyltransferase-Glo Assay
Methyltransferase-Glo assays were performed to examine the methylation signal in PGC-1α by multistep format in a 96-well plate using the Methyltransferase (MTase)-Glo methyltransferase assay kit (V7601; Promega, Madison, WI ) according to the manufacturer’s instructions.44 Briefly, GST-tagged PGC-1α and GST-tagged SMYD5 were added into 20 μL reaction buffer (20 mmol/L Tris, pH 8.0, 50 mmol/L NaCl, 1 mmol/L EDTA, 3 mmol/L MgCl2, and 0.1 mg/mL bovine serum albumin) containing S-Adenosyl Methionine (10 μmol/L final concentration), and incubated for 30 minutes at room temperature. After the incubation, 5 μL MTase-Glo Reagent provided in the kit was added into the mixture and incubated for 30 minutes at room temperature. Once the incubation was completed, 25 μL MTase-Glo Detection solution was added and incubated for another 30 minutes at room temperature. Afterward, the luminescence was measured with a plate-reading luminometer. In the control group, identical reaction conditions were applied except purified GST protein was used instead of GST-tagged SMYD5. For all experiments, all reactions were performed in triplicate.
In Vitro Methyltransferase Assay of PGC-1α Lysine Methylation
Bacterially purified GST-SMYD5 or GST alone was mixed with GST–PGC-1α fragments (GST–PGC-1α aa 1–190, GST–PGC-1α aa 190–345, and GST–PGC-1α aa 345–797) in a methylation reaction buffer (20 mmol/L Tris, pH 8.0, 50 mmol/L NaCl, 1 mmol/L EDTA, 3 mmol/L MgCl2, and 0.1 mg/mL bovine serum albumin) containing SAM (10 μmol/L final concentration) and incubated for 30 minutes at room temperature. Then, the reaction mixtures were subjected to Western blot using a methyl lysine-specific antibody (ab23366; Abcam; and cat. 14117 and 14679; Cell Signaling Technology) to detect methylation signal in PGC-1α fragments, or subjected to mass spectrometry analysis as described later to identify specific methylated lysine residues.
Mass Spectrometry
Liquid chromatography with tandem mass spectrometry was used to identify the SMYD5-methylated lysine residues in PGC-1α. Briefly, in vitro methylated PGC-1α as described earlier was subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and the gels were stained with 0.5% Coomassie Blue R-250. The gels were stored in 5% acetic acid and subsequently processed and analyzed by the System Mass Spectrometry Core Facility within the Georgia Institute of Technology (Atlanta, GA)
Statistical Analysis
Data were expressed as means ± SEM. Graphs representing means ± SEM were obtained from at least 3 independent experiments. Statistical analyses were performed using GraphPad Prism 8.0. Pairwise comparisons were made using the unpaired Student t test, while 1- or 2-way analysis of variance with the Bonferroni post hoc test was used for multiple comparisons, and asterisk and pound symbols were used to denote statistical significance. The Pearson correlation coefficient was used to assess the relationship between the expression levels of SMYD5 and PGC-1α in colon samples from IBD patients and healthy controls.
Access to Data
All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
The authors thank Drs Didier Merlin and Ming-Hui Zou for their intellectual input of the study, and Qian (Zoe) Liu for technical support for mouse breeding.
This manuscript was posted to bioRxiv (doi: https://doi.org/10.1101/2020.11.16.385765) on November 17, 2020.
CRediT Authorship Contributions
Yuning Hou, MD, PhD (Conceptualization: Equal; Formal analysis: Lead; Investigation: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Xiaonan Sun, MD (Investigation: Supporting)
Pooneh Tavakoley Gheinani, MS (Investigation: Supporting)
Xiaoqing Guan, MD, PhD (Investigation: Supporting)
Shaligram Sharma, MS (Formal analysis: Supporting)
Yu Zhou, MD, PhD (Formal analysis: Supporting)
Chengliu Jin, PhD (Investigation: Supporting; Resources: Supporting)
Zhe Yang, PhD (Conceptualization: Supporting; Writing – review & editing: Supporting)
Anjaparavanda P Naren, PhD (Writing – review & editing: Supporting)
Jun Yin, PhD (Conceptualization: Supporting; Writing – review & editing: Supporting)
Timothy L Denning, PhD (Conceptualization: Supporting; Writing – review & editing: Supporting)
Andrew T Gewirtz, PhD (Conceptualization: Supporting; Writing – review & editing: Supporting)
Yuan Liu, PhD (Conceptualization: Supporting)
Zhonglin Xie, MD, PhD (Conceptualization: Supporting; Funding acquisition: Supporting; Methodology: Supporting; Resources: Supporting; Writing – review & editing: Supporting)
Chunying Li, PhD (Conceptualization: Lead; Formal analysis: Equal; Funding acquisition: Lead; Investigation: Equal; Methodology: Equal; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – review & editing: Lead)
Footnotes
Current addresses of Y.H.: Cancer Animal Models Shared Resource, Winship Cancer Institute, Emory University, Atlanta, Georgia; of A.P.N.: Division of Pulmonary Medicine and Gastroenterology, Department of Medicine, Cedars-Sinai Medical Center, Los Angeles, California
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by National Institutes of Health grants R01HL128647 (C.L.) and R01HL128014 (Z.X.); Transformational Project Award 20TPA35410012 from the American Heart Association (C.L.); and a predoctoral Fellowship (917128) from the American Heart Association and the Ahmed T. Abdelal Fellowship in Molecular Genetics and Biotechnology from Georgia State University (S.S.). Additional support was provided by the Georgia Clinical & Translational Science Alliance of the National Institutes of Health (UL1TR000454).
References
- 1.Abraham C., Cho J.H. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kappelman M.D., Rifas-Shiman S.L., Kleinman K., Ollendorf D., Bousvaros A., Grand R.J., Finkelstein J.A. The prevalence and geographic distribution of Crohn's disease and ulcerative colitis in the United States. Clin Gastroenterol Hepatol. 2007;5:1424–1429. doi: 10.1016/j.cgh.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Kappelman M.D., Rifas-Shiman S.L., Porter C.Q., Ollendorf D.A., Sandler R.S., Galanko J.A., Finkelstein J.A. Direct health care costs of Crohn's disease and ulcerative colitis in US children and adults. Gastroenterology. 2008;135:1907–1913. doi: 10.1053/j.gastro.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson L.W., Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 5.Ho G.T., Aird R.E., Liu B., Boyapati R.K., Kennedy N.A., Dorward D.A., Noble C.L., Shimizu T., Carter R.N., Chew E.T.S., Morton N.M., Rossi A.G., Sartor R.B., Iredale J.P., Satsangi J. MDR1 deficiency impairs mitochondrial homeostasis and promotes intestinal inflammation. Mucosal Immunol. 2018;11:120–130. doi: 10.1038/mi.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson D.N., Panopoulos M., Neumann W.L., Turner K., Cantarel B.L., Thompson-Snipes L., Dassopoulos T., Feagins L.A., Souza R.F., Mills J.C., Blumberg R.S., Venuprasad K., Thompson W.E., Theiss A.L. Mitochondrial dysfunction during loss of prohibitin 1 triggers Paneth cell defects and ileitis. Gut. 2020;69:1928–1938. doi: 10.1136/gutjnl-2019-319523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liesa M., Palacin M., Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiol Rev. 2009;89:799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- 8.Cherry A.D., Piantadosi C.A. Regulation of mitochondrial biogenesis and its intersection with inflammatory responses. Antioxid Redox Signal. 2015;22:965–976. doi: 10.1089/ars.2014.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Spiegelman B.M. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Marcos P.J., Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr. 2011;93:884s–890. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodgers J.T., Lerin C., Gerhart-Hines Z., Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jager S., Handschin C., St-Pierre J., Spiegelman B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teyssier C., Ma H., Emter R., Kralli A., Stallcup M.R. Activation of nuclear receptor coactivator PGC-1alpha by arginine methylation. Genes Dev. 2005;19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguilo F., Li S., Balasubramaniyan N., Sancho A., Benko S., Zhang F., Vashisht A., Rengasamy M., Andino B., Chen C.H., Zhou F., Qian C., Zhou M.M., Wohlschlegel J.A., Zhang W., Suchy F.J., Walsh M.J. Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1alpha. Cell Rep. 2016;14:479–492. doi: 10.1016/j.celrep.2015.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spellmon N., Holcomb J., Trescott L., Sirinupong N., Yang Z. Structure and function of SET and MYND domain-containing proteins. Int J Mol Sci. 2015;16:1406–1428. doi: 10.3390/ijms16011406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stender J.D., Pascual G., Liu W., Kaikkonen M.U., Do K., Spann N.J., Boutros M., Perrimon N., Rosenfeld M.G., Glass C.K. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell. 2012;48:28–38. doi: 10.1016/j.molcel.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kidder B.L., Hu G., Cui K., Zhao K. SMYD5 regulates H4K20me3-marked heterochromatin to safeguard ES cell self-renewal and prevent spurious differentiation. Epigenetics Chromatin. 2017;10:8. doi: 10.1186/s13072-017-0115-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujii T., Tsunesumi S., Sagara H., Munakata M., Hisaki Y., Sekiya T., Furukawa Y., Sakamoto K., Watanabe S. Smyd5 plays pivotal roles in both primitive and definitive hematopoiesis during zebrafish embryogenesis. Sci Rep. 2016;6 doi: 10.1038/srep29157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Errico I., Salvatore L., Murzilli S., Lo Sasso G., Latorre D., Martelli N., Egorova A.V., Polishuck R., Madeyski-Bengtson K., Lelliott C., Vidal-Puig A.J., Seibel P., Villani G., Moschetta A. Peroxisome proliferator-activated receptor-gamma coactivator 1-alpha (PGC1alpha) is a metabolic regulator of intestinal epithelial cell fate. Proc Natl Acad Sci U S A. 2011;108:6603–6608. doi: 10.1073/pnas.1016354108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunningham K.E., Vincent G., Sodhi C.P., Novak E.A., Ranganathan S., Egan C.E., Stolz D.B., Rogers M.B., Firek B., Morowitz M.J., Gittes G.K., Zuckerbraun B.S., Hackam D.J., Mollen K.P. Peroxisome proliferator-activated receptor-gamma coactivator 1-alpha (PGC1alpha) protects against experimental murine colitis. J Biol Chem. 2016;291:10184–10200. doi: 10.1074/jbc.M115.688812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ussakli C.H., Ebaee A., Binkley J., Brentnall T.A., Emond M.J., Rabinovitch P.S., Risques R.A. Mitochondria and tumor progression in ulcerative colitis. J Natl Cancer Inst. 2013;105:1239–1248. doi: 10.1093/jnci/djt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren J.S., Tracy C.M., Miller M.R., Makaju A., Szulik M.W., Oka S.I., Yuzyuk T.N., Cox J.E., Kumar A., Lozier B.K., Wang L., Llana J.G., Sabry A.D., Cawley K.M., Barton D.W., Han Y.H., Boudina S., Fiehn O., Tucker H.O., Zaitsev A.V., Franklin S. Histone methyltransferase Smyd1 regulates mitochondrial energetics in the heart. Proc Natl Acad Sci U S A. 2018;115:E7871–E7880. doi: 10.1073/pnas.1800680115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novak E.A., Mollen K.P. Mitochondrial dysfunction in inflammatory bowel disease. Front Cell Dev Biol. 2015;3:62. doi: 10.3389/fcell.2015.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan K.S., Chia L.A., Li X., Ootani A., Su J., Lee J.Y., Su N., Luo Y., Heilshorn S.C., Amieva M.R., Sangiorgi E., Capecchi M.R., Kuo C.J. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim C.K., Saxena M., Maharjan K., Song J.J., Shroyer K.R., Bialkowska A.B., Shivdasani R.A., Yang V.W. Kruppel-like factor 5 regulates stemness, lineage specification, and regeneration of intestinal epithelial stem cells. Cell Mol Gastroenterol Hepatol. 2020;9:587–609. doi: 10.1016/j.jcmgh.2019.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okayasu I., Hatakeyama S., Yamada M., Ohkusa T., Inagaki Y., Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- 27.Du S.J., Tan X., Zhang J. SMYD Proteins: key regulators in skeletal and cardiac muscle development and function. Anat Rec (Hoboken) 2014;297:1650–1662. doi: 10.1002/ar.22972. [DOI] [PubMed] [Google Scholar]
- 28.Doughan M., Spellmon N., Li C., Yang Z. SMYD proteins in immunity: dawning of a new era. AIMS Biophys. 2016;3:450–455. doi: 10.3934/biophy.2016.4.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez-Munoz F., Dominguez-Lopez A., Yamamoto-Furusho J.K. Role of cytokines in inflammatory bowel disease. World J Gastroenterol. 2008;14:4280–4288. doi: 10.3748/wjg.14.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L., Feng D., Chen G., Chen M., Zheng Q., Song P., Ma Q., Zhu C., Wang R., Qi W., Huang L., Xue P., Li B., Wang X., Jin H., Wang J., Yang F., Liu P., Zhu Y., Sui S., Chen Q. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 31.Gureev A.P., Shaforostova E.A., Popov V.N. Regulation of mitochondrial biogenesis as a way for active longevity: interaction between the Nrf2 and PGC-1alpha signaling pathways. Front Genet. 2019;10:435. doi: 10.3389/fgene.2019.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ventura-Clapier R., Garnier A., Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1alpha. Cardiovasc Res. 2008;79:208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- 33.LeBleu V.S., O'Connell J.T., Gonzalez Herrera K.N., Wikman H., Pantel K., Haigis M.C., de Carvalho F.M., Damascena A., Domingos Chinen L.T., Rocha R.M., Asara J.M., Kalluri R. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Safdar A., Little J.P., Stokl A.J., Hettinga B.P., Akhtar M., Tarnopolsky M.A. Exercise increases mitochondrial PGC-1alpha content and promotes nuclear-mitochondrial cross-talk to coordinate mitochondrial biogenesis. J Biol Chem. 2011;286:10605–10617. doi: 10.1074/jbc.M110.211466. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Larsson N.G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G.S., Clayton D.A. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 36.Chicherin I.V., Dashinimaev E., Baleva M., Krasheninnikov I., Levitskii S., Kamenski P. Cytochrome c oxidase on the crossroads of transcriptional regulation and bioenergetics. Front Physiol. 2019;10:644. doi: 10.3389/fphys.2019.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bar F., Bochmann W., Widok A., von Medem K., Pagel R., Hirose M., Yu X., Kalies K., Konig P., Bohm R., Herdegen T., Reinicke A.T., Buning J., Lehnert H., Fellermann K., Ibrahim S., Sina C. Mitochondrial gene polymorphisms that protect mice from colitis. Gastroenterology. 2013;145:1055–1063.e3. doi: 10.1053/j.gastro.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 38.Wu S., Lu Q., Ding Y., Wu Y., Qiu Y., Wang P., Mao X., Huang K., Xie Z., Zou M.H. Hyperglycemia-driven inhibition of AMP-activated protein kinase alpha2 induces diabetic cardiomyopathy by promoting mitochondria-associated endoplasmic reticulum membranes in vivo. Circulation. 2019;139:1913–1936. doi: 10.1161/CIRCULATIONAHA.118.033552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antoni L., Nuding S., Wehkamp J., Stange E.F. Intestinal barrier in inflammatory bowel disease. World J Gastroenterol. 2014;20:1165–1179. doi: 10.3748/wjg.v20.i5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang T., Lu W., Xu W., Anderson L., Bacanamwo M., Thompson W., Chen Y.E., Liu D. MicroRNA-27 (miR-27) targets prohibitin and impairs adipocyte differentiation and mitochondrial function in human adipose-derived stem cells. J Biol Chem. 2013;288:34394–34402. doi: 10.1074/jbc.M113.514372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kontaki H., Talianidis I. Lysine methylation regulates E2F1-induced cell death. Mol Cell. 2010;39:152–160. doi: 10.1016/j.molcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Elkouris M., Kontaki H., Stavropoulos A., Antonoglou A., Nikolaou K.C., Samiotaki M., Szantai E., Saviolaki D., Brown P.J., Sideras P., Panayotou G., Talianidis I. SET9-mediated regulation of TGF-beta signaling links protein methylation to pulmonary fibrosis. Cell Rep. 2016;15:2733–2744. doi: 10.1016/j.celrep.2016.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leng F., Yu J., Zhang C., Alejo S., Hoang N., Sun H., Lu F., Zhang H. Methylated DNMT1 and E2F1 are targeted for proteolysis by L3MBTL3 and CRL4(DCAF5) ubiquitin ligase. Nat Commun. 2018;9:1641. doi: 10.1038/s41467-018-04019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsiao K., Zegzouti H., Goueli S.A. Methyltransferase-Glo: a universal, bioluminescent and homogenous assay for monitoring all classes of methyltransferases. Epigenomics. 2016;8:321–339. doi: 10.2217/epi.15.113. [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson A.W., Diep J., Dai S., Liu S., Ooi Y.S., Song D., Li T.M., Horton J.R., Zhang X., Liu C., Trivedi D.V., Ruppel K.M., Vilches-Moure J.G., Casey K.M., Mak J., Cowan T., Elias J.E., Nagamine C.M., Spudich J.A., Cheng X., Carette J.E., Gozani O. SETD3 is an actin histidine methyltransferase that prevents primary dystocia. Nature. 2019;565:372–376. doi: 10.1038/s41586-018-0821-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X., Tanaka K., Yan J., Li J., Peng D., Jiang Y., Yang Z., Barton M.C., Wen H., Shi X. Regulation of estrogen receptor alpha by histone methyltransferase SMYD2-mediated protein methylation. Proc Natl Acad Sci U S A. 2013;110:17284–17289. doi: 10.1073/pnas.1307959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olson B.L., Hock M.B., Ekholm-Reed S., Wohlschlegel J.A., Dev K.K., Kralli A., Reed S.I. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 2008;22:252–264. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trausch-Azar J., Leone T.C., Kelly D.P., Schwartz A.L. Ubiquitin proteasome-dependent degradation of the transcriptional coactivator PGC-1{alpha} via the N-terminal pathway. J Biol Chem. 2010;285:40192–40200. doi: 10.1074/jbc.M110.131615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei P., Pan D., Mao C., Wang Y.X. RNF34 is a cold-regulated E3 ubiquitin ligase for PGC-1alpha and modulates brown fat cell metabolism. Mol Cell Biol. 2012;32:266–275. doi: 10.1128/MCB.05674-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsin I.L., Sheu G.T., Jan M.S., Sun H.L., Wu T.C., Chiu L.Y., Lue K.H., Ko J.L. Inhibition of lysosome degradation on autophagosome formation and responses to GMI, an immunomodulatory protein from Ganoderma microsporum. Br J Pharmacol. 2012;167:1287–1300. doi: 10.1111/j.1476-5381.2012.02073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lecker S.H., Goldberg A.L., Mitch W.E. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 52.Chakraborty A., Viswanathan P. Methylation-demethylation dynamics: implications of changes in acute kidney injury. Anal Cell Pathol (Amst) 2018;2018 doi: 10.1155/2018/8764384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 54.Boehm D., Jeng M., Camus G., Gramatica A., Schwarzer R., Johnson J.R., Hull P.A., Montano M., Sakane N., Pagans S., Godin R., Deeks S.G., Krogan N.J., Greene W.C., Ott M. SMYD2-mediated histone methylation contributes to HIV-1 latency. Cell Host Microbe. 2017;21:569–579 e6. doi: 10.1016/j.chom.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.James L.I., Barsyte-Lovejoy D., Zhong N., Krichevsky L., Korboukh V.K., Herold J.M., MacNevin C.J., Norris J.L., Sagum C.A., Tempel W., Marcon E., Guo H., Gao C., Huang X.P., Duan S., Emili A., Greenblatt J.F., Kireev D.B., Jin J., Janzen W.P., Brown P.J., Bedford M.T., Arrowsmith C.H., Frye S.V. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nat Chem Biol. 2013;9:184–191. doi: 10.1038/nchembio.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Esteve P.O., Terragni J., Deepti K., Chin H.G., Dai N., Espejo A., Correa I.R., Jr., Bedford M.T., Pradhan S. Methyllysine reader plant homeodomain (PHD) finger protein 20-like 1 (PHF20L1) antagonizes DNA (cytosine-5) methyltransferase 1 (DNMT1) proteasomal degradation. J Biol Chem. 2014;289:8277–8287. doi: 10.1074/jbc.M113.525279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu F., Dassopoulos T., Cope L., Maitra A., Brant S.R., Harris M.L., Bayless T.M., Parmigiani G., Chakravarti S. Genome-wide gene expression differences in Crohn's disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis. 2007;13:807–821. doi: 10.1002/ibd.20110. [DOI] [PubMed] [Google Scholar]
- 58.Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S., Thomas J.K., Muthusamy B., Leal-Rojas P., Kumar P., Sahasrabuddhe N.A., Balakrishnan L., Advani J., George B., Renuse S., Selvan L.D., Patil A.H., Nanjappa V., Radhakrishnan A., Prasad S., Subbannayya T., Raju R., Kumar M., Sreenivasamurthy S.K., Marimuthu A., Sathe G.J., Chavan S., Datta K.K., Subbannayya Y., Sahu A., Yelamanchi S.D., Jayaram S., Rajagopalan P., Sharma J., Murthy K.R., Syed N., Goel R., Khan A.A., Ahmad S., Dey G., Mudgal K., Chatterjee A., Huang T.C., Zhong J., Wu X., Shaw P.G., Freed D., Zahari M.S., Mukherjee K.K., Shankar S., Mahadevan A., Lam H., Mitchell C.J., Shankar S.K., Satishchandra P., Schroeder J.T., Sirdeshmukh R., Maitra A., Leach S.D., Drake C.G., Halushka M.K., Prasad T.S., Hruban R.H., Kerr C.L., Bader G.D., Iacobuzio-Donahue C.A., Gowda H., Pandey A. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song J., Liu Y., Chen Q., Yang J., Jiang Z., Zhang H., Liu Z., Jin B. Expression patterns and the prognostic value of the SMYD family members in human breast carcinoma using integrative bioinformatics analysis. Oncol Lett. 2019;17:3851–3861. doi: 10.3892/ol.2019.10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim E.R., Chang D.K. Colorectal cancer in inflammatory bowel disease: the risk, pathogenesis, prevention and diagnosis. World J Gastroenterol. 2014;20:9872–9881. doi: 10.3748/wjg.v20.i29.9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ludikhuize M.C., Meerlo M., Gallego M.P., Xanthakis D., Burgaya Julia M., Nguyen N.T.B., Brombacher E.C., Liv N., Maurice M.M., Paik J.H., Burgering B.M.T., Rodriguez Colman M.J. Mitochondria define intestinal stem cell differentiation downstream of a FOXO/Notch axis. Cell Metab. 2020;32:889–900 e7. doi: 10.1016/j.cmet.2020.10.005. [DOI] [PubMed] [Google Scholar]
- 62.Qian X., Li X., Shi Z., Bai X., Xia Y., Zheng Y., Xu D., Chen F., You Y., Fang J., Hu Z., Zhou Q., Lu Z. KDM3A senses oxygen availability to regulate PGC-1alpha-mediated mitochondrial biogenesis. Mol Cell. 2019;76:885–895 e7. doi: 10.1016/j.molcel.2019.09.019. [DOI] [PubMed] [Google Scholar]
- 63.Sims R.J., 3rd, Weihe E.K., Zhu L., O'Malley S., Harriss J.V., Gottlieb P.D. m-Bop, a repressor protein essential for cardiogenesis, interacts with skNAC, a heart- and muscle-specific transcription factor. J Biol Chem. 2002;277:26524–26529. doi: 10.1074/jbc.M204121200. [DOI] [PubMed] [Google Scholar]
- 64.Obermann W.M.J. A motif in HSP90 and P23 that links molecular chaperones to efficient estrogen receptor alpha methylation by the lysine methyltransferase SMYD2. J Biol Chem. 2018;293:16479–16487. doi: 10.1074/jbc.RA118.003578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vercauteren K., Gleyzer N., Scarpulla R.C. PGC-1-related coactivator complexes with HCF-1 and NRF-2beta in mediating NRF-2(GABP)-dependent respiratory gene expression. J Biol Chem. 2008;283:12102–12111. doi: 10.1074/jbc.M710150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Farooq S.M., Hou Y., Li H., O'Meara M., Wang Y., Li C., Wang J.M. Disruption of GPR35 exacerbates dextran sulfate sodium-induced colitis in mice. Dig Dis Sci. 2018;63:2910–2922. doi: 10.1007/s10620-018-5216-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Erben U., Loddenkemper C., Doerfel K., Spieckermann S., Haller D., Heimesaat M.M., Zeitz M., Siegmund B., Kuhl A.A. A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int J Clin Exp Pathol. 2014;7:4557–4576. [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen H.T., Dalmasso G., Torkvist L., Halfvarson J., Yan Y., Laroui H., Shmerling D., Tallone T., D'Amato M., Sitaraman S.V., Merlin D. CD98 expression modulates intestinal homeostasis, inflammation, and colitis-associated cancer in mice. J Clin Invest. 2011;121:1733–1747. doi: 10.1172/JCI44631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nowarski R., Jackson R., Gagliani N., de Zoete M.R., Palm N.W., Bailis W., Low J.S., Harman C.C., Graham M., Elinav E., Flavell R.A. Epithelial IL-18 equilibrium controls barrier function in colitis. Cell. 2015;163:1444–1456. doi: 10.1016/j.cell.2015.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crowe A.R., Yue W. Semi-quantitative determination of protein expression using immunohistochemistry staining and analysis: an integrated protocol. Bio Protoc. 2019;9:e3465. doi: 10.21769/BioProtoc.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee C.A., Chin L.S., Li L. Hypertonia-linked protein Trak1 functions with mitofusins to promote mitochondrial tethering and fusion. Protein Cell. 2018;9:693–716. doi: 10.1007/s13238-017-0469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lam J., Katti P., Biete M., Mungai M., AshShareef S., Neikirk K., Garza Lopez E., Vue Z., Christensen T.A., Beasley H.K., Rodman T.A., Murray S.A., Salisbury J.L., Glancy B., Shao J., Pereira R.O., Abel E.D., Hinton A., Jr. A universal approach to analyzing transmission electron microscopy with ImageJ. Cells. 2021;10:2177. doi: 10.3390/cells10092177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li C., Roy K., Dandridge K., Naren A.P. Molecular assembly of cystic fibrosis transmembrane conductance regulator in plasma membrane. J Biol Chem. 2004;279:24673–24684. doi: 10.1074/jbc.M400688200. [DOI] [PubMed] [Google Scholar]
- 74.Zheng P., Xie Z., Yuan Y., Sui W., Wang C., Gao X., Zhao Y., Zhang F., Gu Y., Hu P., Ye J., Feng X., Zhang L. Plin5 alleviates myocardial ischaemia/reperfusion injury by reducing oxidative stress through inhibiting the lipolysis of lipid droplets. Sci Rep. 2017;7 doi: 10.1038/srep42574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu S., Lu Q., Wang Q., Ding Y., Ma Z., Mao X., Huang K., Xie Z., Zou M.H. Binding of FUN14 domain containing 1 with inositol 1,4,5-trisphosphate receptor in mitochondria-associated endoplasmic reticulum membranes maintains mitochondrial dynamics and function in hearts in vivo. Circulation. 2017;136:2248–2266. doi: 10.1161/CIRCULATIONAHA.117.030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li C., Krishnamurthy P.C., Penmatsa H., Marrs K.L., Wang X.Q., Zaccolo M., Jalink K., Li M., Nelson D.J., Schuetz J.D., Naren A.P. Spatiotemporal coupling of cAMP transporter to CFTR chloride channel function in the gut epithelia. Cell. 2007;131:940–951. doi: 10.1016/j.cell.2007.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]