Abstract
Transfer RNAs (tRNAs) are ubiquitous adapter molecules that link specific codons in messenger RNA (mRNA) with their corresponding amino acids during protein synthesis. The tRNA genes of Drosophila have been investigated for over half a century but have lacked systematic identification and nomenclature. Here, we review and integrate data within FlyBase and the Genomic tRNA Database (GtRNAdb) to identify the full complement of tRNA genes in the D. melanogaster nuclear and mitochondrial genomes. We apply a logical and informative nomenclature to all tRNA genes, and provide an overview of their characteristics and genomic features.
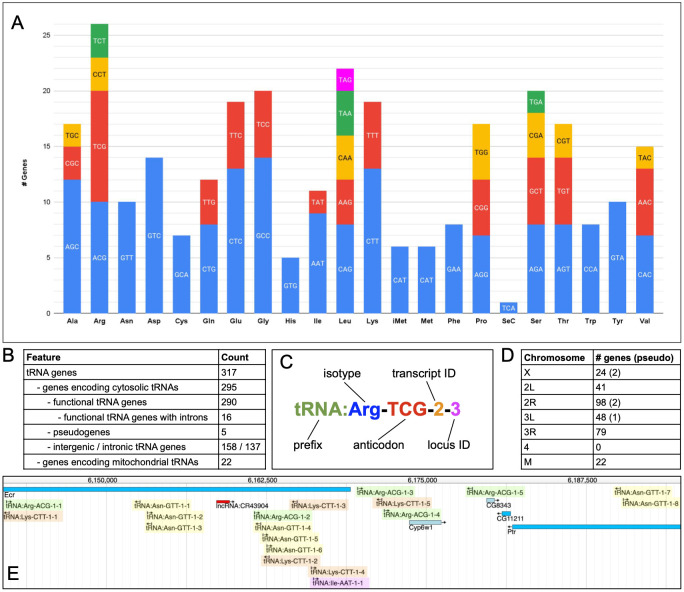
Figure 1. Drosophila tRNA genes.
(A) Isoacceptor counts of functional cytosolic tRNA genes in Drosophila - pseudogenes and mitochondrial tRNA genes are excluded. Colors distinguish distinct anticodons within each isoacceptor family. Counts for initiator (iMet) and elongator tRNA:Met are shown separately. (B) Summary statistics for all Drosophila tRNA genes. (C) Example of tRNA gene nomenclature syntax. (D) Distribution of tRNA genes across major chromosomal scaffolds; pseudogene numbers are in parentheses. ‘M’ = mitochondrial genome. (E) Example of a tRNA gene cluster from the 2R:6,144,000..6,195,000 genomic region, corresponding to cytological region 42A shown in Figure 3 of Kubli 1982. (Notably, the only significant change to tRNA annotations in this region is the addition of tRNA:Arg-ACG-1-4 in the current version.) Green highlight: tRNA:Arg-ACG-1 isodecoders; orange highlight: tRNA:Lys-CTT-1; yellow highlight: tRNA:Asn-GTT-1; magenta highlight: tRNA:Ile-AAT-1; blue rectangles: protein-coding genes; red rectangle is a lncRNA gene.
Description
tRNAs are universal to all cellular life and provide the essential molecular link between mRNA codons and their corresponding amino acids during translation (reviewed by Suzuki 2021). The main functional regions in a tRNA are the anticodon triplet, which base pairs with mRNA codons, and the 3′ end to which the cognate amino acid is attached. Codon degeneracy for the 21 amino acids (20 standard amino acids plus selenocysteine) means that up to six tRNAs with distinct anticodons (‘isoacceptors’) are required depending on the amino acid. tRNA diversity is further increased through the existence of tRNAs that share the same anticodon but differ in the sequence of their body structure (Goodenbour and Pan 2006). Such ‘isodecoders’ may differ from each other by just one or several nucleotides. Moreover, each specific isodecoder sequence can be present in multiple copies within a genome. This combination of diversity and redundancy results in eukaryotic nuclear genomes having hundreds of genes encoding tRNAs functioning in cytosolic translation (cytosolic tRNAs). An additional set of tRNAs functioning in mitochondria are encoded by the mitochondrial genome of eukaryotes: in vertebrates and many other metazoa, there are often 22 mitochondrial tRNA genes with tRNA:Leu and tRNA:Ser represented by two different isoacceptors.
The tRNAs and tRNA genes of Drosophila melanogaster (hereafter ‘Drosophila’) have been investigated for over half a century. Early work identified the sequences and locations of many individual cytosolic tRNA genes, demonstrating that several isoacceptor families appear as clusters in cytological (polytene chromosome) views of the genome (reviewed in Kubli 1982 and Sharp et al. 1983). Subsets of mitochondrial tRNA genes were also reported (de Bruijn 1983; Garesse 1988). Subsequently, the analysis of the sequenced Drosophila genome predicted 292 cytosolic tRNA genes (Adams et al. 2000). This number was later refined in release 4 of the genome to 297 cytosolic tRNA genes, including four pseudogenes (Drosophila 12 Genomes Consortium 2007; Bergman and Ardell 2014). However, none of these studies fully classified or specifically named tRNA genes by their molecular features, and the genomic data were not fully rationalized with the earlier information on tRNAs present within FlyBase, the primary database for Drosophila research (Larkin et al. 2021).
We revisited cytosolic tRNA gene annotations in the current version of the Drosophila genome (release 6), integrating gene predictions from the Genomic tRNA database (GtRNAdb; Chan and Lowe 2016; Chan et al. 2021) with existing data within FlyBase. We find there are a total of 295 cytosolic tRNA genes, of which 289 encode tRNAs decoding the 20 standard amino acids, one encodes a selenocysteine tRNA, and five are classified as tRNA-like genes/pseudogenes (Figure 1A, 1B; Extended Data Table 1). Importantly, this rationalization exercise resulted in improvements within both databases. For example, ~20 records pertaining to unlocalized tRNA genes were merged/deleted within FlyBase, two tRNAs with undetermined isotypes within the GtRNAdb were corrected, and pseudogene classifications within both databases were made consistent.
Prior to our analysis, greater than 50% of Drosophila cytosolic tRNA genes were unnamed in FlyBase, while the named genes used an ambiguous and esoteric nomenclature incorporating the single letter amino acid code and cytogenetic map information but lacking anticodon information. We therefore implemented the logical and systematic nomenclature used by the GtRNAdb within FlyBase (Figure 1C; Extended Data Table 1). This syntax comprises the 3-letter code of the cognate amino acid (isotype), the anticodon triplet, a number identifying each unique transcript (isodecoder) sequence, followed by a second number to specify each copy (locus) of that sequence within the genome. This is preceded by the standard ‘ tRNA: ’ prefix used for tRNA genes in FlyBase. For example, tRNA:Arg-TCG-2-3 is the systematic name given to the gene encoding the third copy of the second unique transcript of tRNA:Arg-TCG. tRNA pseudogenes are named using the standard FlyBase syntax for pseudogenes, with a greek Psi character appended to the gene name, e.g. tRNA:His-GTG-2-1Ψ .
Mitochondrial tRNA genes are not currently included within the GtRNAdb. We therefore compared existing FlyBase annotations against Drosophila entries in the mitotRNAdb (Jühling et al . 2009) and verified these using the tRNAscan-SE program (Chan et al . 2021). This resulted in minor edits to five mitochondrial tRNA sequences in FlyBase. We also applied the standardized nomenclature to the 22 mitochondrial tRNA genes, including the standard ‘ mt: ’ prefix used for genes of the mitochondrial genome in FlyBase (Figure 1B; Extended Data Table 2).
Extended Data Tables 1 and 2 provide detailed information on all the cytosolic and mitochondrial tRNA genes, respectively. Among the functional, cytosolic tRNAs decoding the 20 standard amino acids, each isoacceptor is encoded by between five (tRNA:His) and 26 (tRNA:Arg) genes, with the number of distinct anticodons in each isoacceptor family ranging from one (e.g. tRNA:Asn-GTT) to five (tRNA:Leu-CAG, tRNA:Leu-AAG, tRNA:Leu-CAA, tRNA:Leu-TAA, tRNA:Leu-TAG). Up to four distinct transcript sequences exist for a given anticodon (as is the case for tRNA:Arg-TCG, tRNA:Cys-GCA, tRNA:Gln-CTG and tRNA:Leu-TAA), and a given transcript sequence may be present in up to 13 exact gene copies (tRNA:Gly-GCC-1 and tRNA:Lys-CTT-1). Overall, there are 44 different isoacceptors and 84 distinct tRNA transcripts encoded by the Drosophila nuclear genome. Cytosolic tRNA genes are present on all major chromosome arms (Figure 1D) and are frequently found within clusters, harboring members of either the same or different isoacceptor families (Figure 1E; Kubli 1982; Phillips and Ardell 2021). 46% of cytosolic tRNA genes are located within introns of protein-coding genes, with the remainder being intergenic. A minority (5%) of cytosolic tRNA genes contain an intron, namely two tRNA:Ile-TAT, four tRNA:Leu-CAA and ten tRNA:Tyr-GTA genes (Figure 1B; Bergman and Ardell 2014). These characteristics are largely comparable with the cytosolic tRNA gene complement of other metazoa ( http://gtrnadb.ucsc.edu/ ). However, the Drosophila genome is distinguished by its relative paucity of non-functional tRNA-like genes: <2% of Drosophila tRNA genes are classed as pseudogenes or repetitive element derivatives, compared to 20% in C. elegans , 30% in humans or >99% in rodents.
Notably, this project enabled several additional improvements to the representation of tRNAs within FlyBase. All functional (Gene Ontology) annotations were reviewed and revised as necessary. Reciprocal links between tRNA gene/transcript reports in FlyBase and corresponding pages at the GtRNAdb and RNAcentral (RNAcentral Consortium 2021) have been established, and 2D structural images for Drosophila tRNAs (Sweeney et al. 2021) have been added. Finally, a ‘Gene Group’ report (Attrill et al. 2016) for the Drosophila tRNA genes has been generated ( https://flybase.org/reports/FBgg0000459.html ), which provides easy access to all classes of tRNA gene and their associated data within FlyBase.
In conclusion, we have generated definitive sets of cytosolic and mitochondrial tRNA genes present in the Drosophila genome and implemented a systematic and informative nomenclature for them. The improved datasets are available from several databases, including FlyBase, GtRNAdb, RNAcentral, and the Alliance of Genome Resources (Alliance of Genome Resources Consortium 2022). Our work will facilitate further exploration of tRNA biology within Drosophila as well as new comparative studies with other species.
Methods
Data on cytosolic tRNA genes were accessed and downloaded from FlyBase ( http://flybase.org ) and the GtRNAdb ( http://gtrnadb.ucsc.edu/ ). Data were initially compared and rationalized between FlyBase release FB2015_04 and GtRNAdb release 16. Necessary revisions were made in subsequent database releases, and the data presented herein are from FlyBase release FB2022_02 and GtRNAdb release 19 (which uses tRNAscan-SE 2.0). tRNAscan-SE uses multiple score thresholds to discriminate functional tRNA genes from likely non-functional tRNA-like genes/pseudogenes based on their primary sequence, secondary structure and isotype-specific models (Chan et al. 2021).
Data on mitochondrial tRNA genes were accessed and downloaded from FlyBase ( http://flybase.org ) and mitotRNAdb http://mttrna.bioinf.uni-leipzig.de/mtDataOutput/ . Additionally, the Drosophila mitochondrial genome (RefSeq NC_024511) was used as a query sequence in tRNAscan-SE 2.0 ( http://trna.ucsc.edu/tRNAscan-SE/ ) with parameters ‘other mitochondrial’ as the sequence source, ‘Invertebrate Mito’ as the genetic code and a score cutoff of zero. Necessary revisions were made in FlyBase and data presented herein are from FlyBase release FB2022_02.
Extended Data
Description: Cytosolic tRNA genes. Resource Type: Dataset. DOI: 10.22002/D1.20161
Description: Mitochondrial tRNA genes. Resource Type: Dataset. DOI: 10.22002/D1.20162
Acknowledgments
Funding
SJM is funded by a grant from the National Human Genome Research Institute, National Institutes of Health (U41HG000739) to Norbert Perrimon (PI), Nicholas Brown (co-PI). PPC and TML are funded by a grant from the National Human Genome Research Institute, National Institutes of Health (R01HG006753) to TML.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Sidén-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000 Mar 24;287(5461):2185–2195}. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Alliance of Genome Resources Consortium . Harmonizing model organism data in the Alliance of Genome Resources. Genetics. 2022 Apr 4;220(4) doi: 10.1093/genetics/iyac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attrill H, Falls K, Goodman JL, Millburn GH, Antonazzo G, Rey AJ, Marygold SJ, FlyBase Consortium. FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 2015 Oct 13;44(D1):D786–D792}. doi: 10.1093/nar/gkv1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman C, Ardell D. 2014. Methods for nuclear tRNA gene predictions for 12 species in the genus Drosophila. figshare. Journal contribution.
- Chan PP, Lin BY, Mak AJ, Lowe TM. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021 Sep 20;49(16):9077–9096}. doi: 10.1093/nar/gkab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2015 Dec 15;44(D1):D184–D189}. doi: 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn MH. Drosophila melanogaster mitochondrial DNA, a novel organization and genetic code. Nature. 1983 Jul 21;304(5923):234–241}. doi: 10.1038/304234a0. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, Pollard DA, Sackton TB, Larracuente AM, Singh ND, Abad JP, Abt DN, Adryan B, Aguade M, Akashi H, Anderson WW, Aquadro CF, Ardell DH, Arguello R, Artieri CG, Barbash DA, Barker D, Barsanti P, Batterham P, Batzoglou S, Begun D, Bhutkar A, Blanco E, Bosak SA, Bradley RK, Brand AD, Brent MR, Brooks AN, Brown RH, Butlin RK, Caggese C, Calvi BR, Bernardo de Carvalho A, Caspi A, Castrezana S, Celniker SE, Chang JL, Chapple C, Chatterji S, Chinwalla A, Civetta A, Clifton SW, Comeron JM, Costello JC, Coyne JA, Daub J, David RG, Delcher AL, Delehaunty K, Do CB, Ebling H, Edwards K, Eickbush T, Evans JD, Filipski A, Findeiss S, Freyhult E, Fulton L, Fulton R, Garcia AC, Gardiner A, Garfield DA, Garvin BE, Gibson G, Gilbert D, Gnerre S, Godfrey J, Good R, Gotea V, Gravely B, Greenberg AJ, Griffiths-Jones S, Gross S, Guigo R, Gustafson EA, Haerty W, Hahn MW, Halligan DL, Halpern AL, Halter GM, Han MV, Heger A, Hillier L, Hinrichs AS, Holmes I, Hoskins RA, Hubisz MJ, Hultmark D, Huntley MA, Jaffe DB, Jagadeeshan S, Jeck WR, Johnson J, Jones CD, Jordan WC, Karpen GH, Kataoka E, Keightley PD, Kheradpour P, Kirkness EF, Koerich LB, Kristiansen K, Kudrna D, Kulathinal RJ, Kumar S, Kwok R, Lander E, Langley CH, Lapoint R, Lazzaro BP, Lee SJ, Levesque L, Li R, Lin CF, Lin MF, Lindblad-Toh K, Llopart A, Long M, Low L, Lozovsky E, Lu J, Luo M, Machado CA, Makalowski W, Marzo M, Matsuda M, Matzkin L, McAllister B, McBride CS, McKernan B, McKernan K, Mendez-Lago M, Minx P, Mollenhauer MU, Montooth K, Mount SM, Mu X, Myers E, Negre B, Newfeld S, Nielsen R, Noor MA, O'Grady P, Pachter L, Papaceit M, Parisi MJ, Parisi M, Parts L, Pedersen JS, Pesole G, Phillippy AM, Ponting CP, Pop M, Porcelli D, Powell JR, Prohaska S, Pruitt K, Puig M, Quesneville H, Ram KR, Rand D, Rasmussen MD, Reed LK, Reenan R, Reily A, Remington KA, Rieger TT, Ritchie MG, Robin C, Rogers YH, Rohde C, Rozas J, Rubenfield MJ, Ruiz A, Russo S, Salzberg SL, Sanchez-Gracia A, Saranga DJ, Sato H, Schaeffer SW, Schatz MC, Schlenke T, Schwartz R, Segarra C, Singh RS, Sirot L, Sirota M, Sisneros NB, Smith CD, Smith TF, Spieth J, Stage DE, Stark A, Stephan W, Strausberg RL, Strempel S, Sturgill D, Sutton G, Sutton GG, Tao W, Teichmann S, Tobari YN, Tomimura Y, Tsolas JM, Valente VL, Venter E, Venter JC, Vicario S, Vieira FG, Vilella AJ, Villasante A, Walenz B, Wang J, Wasserman M, Watts T, Wilson D, Wilson RK, Wing RA, Wolfner MF, Wong A, Wong GK, Wu CI, Wu G, Yamamoto D, Yang HP, Yang SP, Yorke JA, Yoshida K, Zdobnov E, Zhang P, Zhang Y, Zimin AV, Baldwin J, Abdouelleil A, Abdulkadir J, Abebe A, Abera B, Abreu J, Acer SC, Aftuck L, Alexander A, An P, Anderson E, Anderson S, Arachi H, Azer M, Bachantsang P, Barry A, Bayul T, Berlin A, Bessette D, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Bourzgui I, Brown A, Cahill P, Channer S, Cheshatsang Y, Chuda L, Citroen M, Collymore A, Cooke P, Costello M, D'Aco K, Daza R, De Haan G, DeGray S, DeMaso C, Dhargay N, Dooley K, Dooley E, Doricent M, Dorje P, Dorjee K, Dupes A, Elong R, Falk J, Farina A, Faro S, Ferguson D, Fisher S, Foley CD, Franke A, Friedrich D, Gadbois L, Gearin G, Gearin CR, Giannoukos G, Goode T, Graham J, Grandbois E, Grewal S, Gyaltsen K, Hafez N, Hagos B, Hall J, Henson C, Hollinger A, Honan T, Huard MD, Hughes L, Hurhula B, Husby ME, Kamat A, Kanga B, Kashin S, Khazanovich D, Kisner P, Lance K, Lara M, Lee W, Lennon N, Letendre F, LeVine R, Lipovsky A, Liu X, Liu J, Liu S, Lokyitsang T, Lokyitsang Y, Lubonja R, Lui A, MacDonald P, Magnisalis V, Maru K, Matthews C, McCusker W, McDonough S, Mehta T, Meldrim J, Meneus L, Mihai O, Mihalev A, Mihova T, Mittelman R, Mlenga V, Montmayeur A, Mulrain L, Navidi A, Naylor J, Negash T, Nguyen T, Nguyen N, Nicol R, Norbu C, Norbu N, Novod N, O'Neill B, Osman S, Markiewicz E, Oyono OL, Patti C, Phunkhang P, Pierre F, Priest M, Raghuraman S, Rege F, Reyes R, Rise C, Rogov P, Ross K, Ryan E, Settipalli S, Shea T, Sherpa N, Shi L, Shih D, Sparrow T, Spaulding J, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Strader C, Tesfaye S, Thomson T, Thoulutsang Y, Thoulutsang D, Topham K, Topping I, Tsamla T, Vassiliev H, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Young G, Yu Q, Zembek L, Zhong D, Zimmer A, Zwirko Z, Jaffe DB, Alvarez P, Brockman W, Butler J, Chin C, Gnerre S, Grabherr M, Kleber M, Mauceli E, MacCallum I. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007 Nov 8;450(7167):203–218}. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Garesse R. Drosophila melanogaster mitochondrial DNA: gene organization and evolutionary considerations. Genetics. 1988 Apr 1;118(4):649–663}. doi: 10.1093/genetics/118.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenbour JM, Pan T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006 Nov 6;34(21):6137–6146}. doi: 10.1093/nar/gkl725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2008 Oct 28;37(Database issue):D159–D162}. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubli E. The genetics of transfer RNA in Drosophila. Adv Genet. 1982;21:123–172}. doi: 10.1016/s0065-2660(08)60298-9. [DOI] [PubMed] [Google Scholar]
- Larkin A, Marygold SJ, Antonazzo G, Attrill H, Dos Santos G, Garapati PV, Goodman JL, Gramates LS, Millburn G, Strelets VB, Tabone CJ, Thurmond J, FlyBase Consortium. FlyBase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2021 Jan 8;49(D1):D899–D907}. doi: 10.1093/nar/gkaa1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JB, Ardell DH. Structural and Genetic Determinants of Convergence in the Drosophila tRNA Structure-Function Map. J Mol Evol. 2021 Feb 2;89(1-2):103–116}. doi: 10.1007/s00239-021-09995-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RNAcentral Consortium. RNAcentral 2021: secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res. 2021 Jan 8;49(D1):D212–D220}. doi: 10.1093/nar/gkaa921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S, Cooley L, DeFranco D, Dingermann T, Söll D. Organization and expression of tRNA genes in Drosophila melanogaster. Recent Results Cancer Res. 1983;84:1–14}. doi: 10.1007/978-3-642-81947-6_1. [DOI] [PubMed] [Google Scholar]
- Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat Rev Mol Cell Biol. 2021 Mar 3;22(6):375–392}. doi: 10.1038/s41580-021-00342-0. [DOI] [PubMed] [Google Scholar]
- Sweeney BA, Hoksza D, Nawrocki EP, Ribas CE, Madeira F, Cannone JJ, Gutell R, Maddala A, Meade CD, Williams LD, Petrov AS, Chan PP, Lowe TM, Finn RD, Petrov AI. R2DT is a framework for predicting and visualising RNA secondary structure using templates. Nat Commun. 2021 Jun 9;12(1):3494–3494. doi: 10.1038/s41467-021-23555-5. [DOI] [PMC free article] [PubMed] [Google Scholar]