Abstract
An outbreak caused by salted salmon roe contaminated with enterohemorrhagic Escherichia coli O157 occurred in Japan in 1998. Since about 0.75 to 1.5 viable cells were estimated to cause infection, we presumed that O157 might enter the viable but nonculturable (VNC) state in salted salmon roe and consequently that viable cell numbers might be underestimated. Although patient-originating O157 cells could not grow on agar plates after 72 h of incubation in 13% NaCl, they were resuscitated in yeast extract broth, and more than 90% of the cells were shown to be viable by fluorescent staining, suggesting that almost all of them could enter the VNC state in NaCl water. Roe-originating O157 was resistant to NaCl because it could grow on agar after 72 h of incubation in NaCl water, but about 20% of cells appeared to enter the VNC state. Therefore, germfree mice were infected with O157 to examine the resuscitation of cells in the VNC state and the retention of pathogenicity. O157 that originated in roe, but not patients, killed mice and was isolated from the intestine. However, these isolates had become sensitive to NaCl. O157 cells of roe origin incubated in normal media also killed mice and were isolated from the intestine, but they also became transiently NaCl sensitive. We therefore propose that bacterial cells might enter the VNC state under conditions of stress, such as those encountered in vivo or in high salt concentrations, and then revive when those conditions have eased. If so, the VNC state in food is potentially dangerous from a public health viewpoint and may have to be considered at the time of food inspection. Finally, the establishment of a simple recovery system for VNC cells should be established.
In Japan, salmon roe is soaked in a liquid seasoning, which consists of soy sauce (79.0%), water (14.0%), chemical seasoning (6.5%), synthetic sake (0.3%), and a fermented seasoning (0.2%); its salt content is equal to a 13% NaCl concentration. This salted salmon roe is a popular component of Japanese sushi. The preservation of foods by high salt concentrations has been viewed historically as an effective means of preventing food-borne infections (11, 13). However, an outbreak caused by salted salmon roe which was contaminated with enterohemorrhagic Escherichia coli (EHEC) O157 occurred in four independent places in Japan in 1998, with 62 cases reported (1). Since all the causative foods were manufactured by the same company, the salmon roe was probably contaminated with O157 during the production process. Although the definitive source of O157 could not be identified because the roe was stored frozen for 9 months, it appeared that O157 could survive freezing and a high concentration of NaCl (22) and retain its pathogenicity for humans. In addition, it was proved by the most probable number method that about 0.75 to 1.5 viable cells of O157 could cause infection (1). This number was considered to be very low for infection (23). Recently, it has been reported that Vibrio cholerae, E. coli, Campylobacter jejuni, and Salmonella spp. enter the viable but nonculturable (VNC) state in environments such as seawater, ground water, and rivers (3, 5, 6, 17, 18, 19, 26), resulting in an underestimation of viable cell numbers. Therefore, viable cell numbers of O157 in salted salmon roe might be considerably underestimated due to O157 entering the VNC state because of lengthy preservation under a high concentration of NaCl. In this study, to clarify whether O157 isolates entered the VNC state in salted salmon roe, their NaCl sensitivities and pathogenicity for mice were examined.
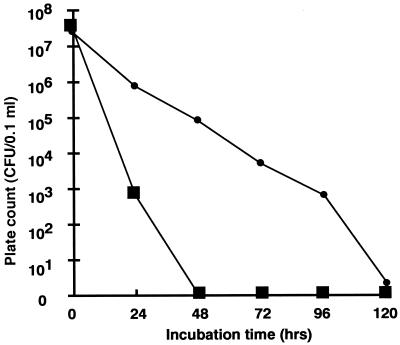
Seven EHEC O157 strains used in this study were isolated from the causative food, salted salmon roe, or from patients (Table 1). We first considered that O157 might be able to grow in salted salmon roe because of the nutrients contained therein, but those O157 isolates could not grow at 37°C even in mashed roe (data not shown). However, the possibility exists that O157 grows in the liquid seasoning and consequently causes infection if the water activity of the food increases. Therefore, the changes in numbers of CFU of roe origin (strain 2) and patient origin (strain 5) in liquid seasoning were examined (Fig. 1). About 5.0 × 107 cells of both strains, which were cultivated in L broth (Funakoshi Co., Ltd., Tokyo, Japan) at 37°C for 16 h with shaking, were added to the liquid seasoning, followed by measurement of CFU on L agar (LA; Funakoshi Co., Ltd.) plates every 24 h. The CFU of strain 2 decreased gradually, but colonies could be detected on LA after 120 h of incubation. However, in strain 5, no colonies could be detected on LA after 48 h of incubation.
TABLE 1.
Serotype O157:H7 strains used in this study
| Strain | Origin | Place of isolation |
|---|---|---|
| 2 | Salmon roe | Hokkaido, Japan |
| 3 | Salmon roe | Hokkaido, Japan |
| 4 | Salmon roe | Hokkaido, Japan |
| 5 | Patient | Kanagawa, Japan |
| 6 | Patient | Tokyo, Japan |
| 7 | Patient | Toyama, Japan |
| 8 | Patient | Chiba, Japan |
Toxin typing was performed by PCR and reverse passive latex agglutination (VTEC-RPLA “SEIKEN”; Denka Seiken Co., Ltd., Tokyo, Japan) as described previously (2). The toxin types of all strains were Shiga toxins 1 and 2.
FIG. 1.
Bacterial cell numbers of O157 in liquid seasoning. Colony counts of roe-originating O157 strain 2 (●) and patient-originating O157 strain 5 (■) were performed on LA plates.
Since the CFU of strains 2 and 5 in liquid seasoning were different, we examined the genetic relatedness of all isolates. First, disk diffusion susceptibility tests using Sensi-Disc (Becton Dickinson, Detroit, Mich.) were performed according to the supplier's instructions using 12 antibiotics (fosfomycin, tetracycline, minocycline, streptomycin, ampicillin, penicillin G, chloramphenicol, kanamycin, nalidixic acid, norfloxacin, cefdinir, and trimethoprim). Strain 6 was resistant to penicillin G, but the other six isolates were sensitive to all drugs used in this study. In addition, all strains had a common large plasmid, which is frequently carried by O157 (21), although strain 6 had an additional small plasmid, and by pulsed-field gel electrophoresis analysis (10) using XbaI, they seemed to be genetically identical to each other (data not shown). The results suggested that these strains might derive from a common source, although their NaCl sensitivities differed.
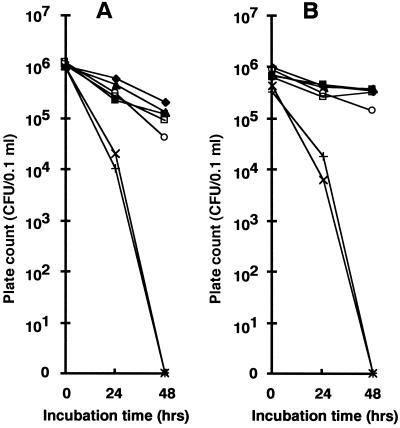
To measure bacterial numbers in NaCl water, 0.1-ml samples of O157 cultures grown in nutrient broth (N broth; Difco Laboratories, Detroit, Mich.), which contained no NaCl, were centrifuged at 37°C for 16 h and then washed once with saline, suspended in 10 ml of 13 or 7% NaCl water, and incubated at 37°C for 48 h. One-tenth milliliter of each culture was spread on LA plates every 24 h. Strains 5 and 6 of patient origin formed no colonies after 48 h of incubation in NaCl water (Fig. 2), showing that both strains were sensitive to NaCl. Three roe-originating and the two other patient-originating EHEC O157 strains were resistant to NaCl, though the patient-originating strains were slightly more sensitive than the roe-originating strains (Fig. 2). Although all the isolates were subcultured in N broth for 100 generations, their NaCl sensitivities remained stable (data not shown).
FIG. 2.
Bacterial cell numbers of O157 in NaCl water. Colony counts of roe-originating O157 strains 2 (⧫), 3 (▴), and 4 (■) and patient-originating O157 strains 5 (×), 6 (+), 7 (□), and 8 (○), which were incubated in 13% (A) and 7% (B) NaCl water, were determined on TSA plates.
Moreover, strain 2 of roe origin and strain 5 of patient origin were incubated in 13% NaCl water for 48 h and their viabilities were then determined on different solid media (Table 2). The numbers of CFU depended on the medium. With strain 2, although the largest numbers of bacterial colonies were detected on Trypticase soy agar (TSA) plates, only 0.085% of the inoculum size was detected on DHL agar plates (Eiken Co., Ltd., Tokyo, Japan) after 48 h of incubation (Table 2), but strain 2 formed colonies on DHL agar plates after 144 h of incubation, as did other O157 isolates listed in Table 1 (data not shown). With strain 5, only 0.0008% of the inoculum size was detected on TSA plates, while no colonies were detected on DHL agar, MacConkey agar, and LA plates (Table 2) or TSA plates after 72 h of incubation (data not shown). Similar results were observed in 3.5 and 7% NaCl water (data not shown). Next, both suspensions were fluorescently stained by using a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes Inc., Eugene, Oreg.) (9). To count the numbers of live (green fluorescence) and dead (red fluorescence) bacterial cells, 1,500 cells were selected randomly. With strain 2, percentages of green- and red-fluorescing cells were 99.3 and 0.7%, respectively. With strain 5, percentages of green- and red-fluorescing cells were 96.5 and 3.5%, respectively. In addition, after 72 h of incubation in 13% NaCl water, 0.1 ml of the strain 5 suspension, which formed no colonies on TSA plates, was inoculated into yeast extract (YE) (1 mg/ml) broth and then incubated at 37°C for 48 h, with the result that live O157 cells were recovered from the broth. The bacterial cells had levels of toxin production and pulsed-field gel electrophoresis patterns identical to those of their parent (data not shown). Cell elongation in YE broth supplemented with nalidixic acid was also observed in at least 5% of the cells counted (data not shown). From these results, it was concluded that patient-originating strain 5 enters the VNC state in 13% NaCl water in spite of this concentration being sublethally injurious (12). If the fluorescent kit reliably discriminated bacteria of the VNC state by fluorescent staining, 20 to ∼30% of NaCl-resistant strain 2 cells and almost all of the NaCl-sensitive strain 5 cells incubated in NaCl water appeared to enter the VNC state (Table 2). Although O157 might be distributed heterogeneously in the foods because 0.75 to 1.5 O157 cells can cause disease (1), we presumed that NaCl-resistant and VNC O157 cells coexisted in the food and consequently that viable cell numbers might be underestimated.
TABLE 2.
Viable cell numbers of O157 on various media after 48 h of incubation in 13% NaCl
| Strain | Inoculum size (107) | Bacterial number (CFU/0.1 ml) ona:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| LA | DHL | MC | TSA | HI | BHI | NA | MH | ||
| 2 | 1.0 | 1.1 × 106 | 8.5 × 103 | 5.0 × 103 | 7.8 × 106 | 1.1 × 106 | 8.7 × 105 | 1.1 × 106 | 1.0 × 106 |
| 5 | 1.4 | 0 | 0 | 0 | 112 | 1 | 24 | 98 | 10 |
DHL, DHL agar (Eiken Co., Ltd.); MC, MacConkey agar (Eiken Co., Ltd.); HI, heart infusion agar (Eiken Co., Ltd.); BHI, brain heart infusion agar (Difco Laboratories); NA, agar (Difco Laboratories); MH, Mueller-Hinton agar (Eiken Co., Ltd.).
It has been reported that VNC forms of V. cholerae retain their pathogenicity in rabbits by resuscitating in the rabbit intestine (6). In order to examine this possibility for O157, germfree BALB/c female mice aged 3 to ∼4 months, which were bred in a vinyl isolator, were used for the infection experiments. One-tenth milliliter of strain 2 or 5 culture after 72 h of incubation in 13% NaCl water at 37°C was administered orally to mice using a metal catheter. When 0.1 ml of suspension was spread on TSA plates, colony formation was observed for strain 2 but not for strain 5. After 4 days, all mice injected with strain 2 died and O157 cells were isolated from the cecum contents but all mice injected with strain 5 survived for the 2-week experimental period and no bacterial colonies were isolated from the intestinal contents, showing that strain 5 incubated in NaCl water did not resuscitate in the mouse intestine. Next, the NaCl sensitivities of 50 O157 colonies, which were originally derived from strain 2 and recovered from the intestine, were examined. Although 28 isolates were resistant to NaCl, like their parental strain, strain 2, another 22 isolates became more sensitive to NaCl than strain 2 was; in particular, 9 of them formed no colonies after 72 h of incubation (Table 3). However, the NaCl sensitivities of those 22 isolates were unstable, because after passage for 100 generations in N broth, all isolates became resistant to NaCl (data not shown). In addition, when mice were infected with O157 strains without prior exposure to NaCl, all mice died and O157 cells were recovered from the cecum contents. In this case, the NaCl sensitivities of 50 strain 2 colonies recovered from the intestine were unstable and became sensitive to NaCl (data not shown). Since the NaCl sensitivity of strain 2 changed after animal passage, the conversions of the NaCl sensitivities of strains 2 and 5 in 13% NaCl water were determined. Strain 2 cells after 72 h of incubation in 13% NaCl water were stably resistant to NaCl. When 20 tubes of 13% NaCl water containing strain 5 (patient origin) were inoculated at 37°C for 72 h and 0.2 ml of each was then spread on TSA plates, only two colonies were isolated. Both colonies became more resistant to NaCl than their parental strain 5 was, but these changes were unstable after passage in N broth for 100 generations (data not shown).
TABLE 3.
NaCl sensitivities of O157 isolates from mouse intestine
| Strain(s) | No. of culturable strain 2 cells (CFU/0.1 ml)a after incubation in 13% NaCl water for:
|
||
|---|---|---|---|
| 24 h | 48 h | 72 h | |
| Controls | |||
| 2 | UC | UC | UC |
| 5 | 103 | 0 | 0 |
| Intestine-originating isolates (of a total of 50 isolates) | |||
| 28 isolates | UC | UC | UC |
| 3 isolates | UC | 106–278 | 54–110 |
| 10 isolates | UC | 8–83 | 1–23 |
| 9 isolates | 86–111 | 0–10 | 0 |
UC, uncountable because the numbers of colonies on TSA were too many to count.
Furthermore, of four O157 strains of patient origin, two were sensitive to NaCl (Fig. 2), suggesting that O157 cells with different NaCl sensitivities coexisted in the causative foods. Although NaCl-sensitive O157 in the foods formed no colonies on normal media, they were resuscitated in YE broth in vitro, and the fact that they were human isolates suggested that resuscitation might occur in humans in vivo. Simultaneously, cells might also recover their pathogenicity in the body. In the germfree-mouse model system for O157 infections, strain 2 of roe origin incubated in NaCl water killed mice but strain 5 of patient origin incubated in NaCl water did not. These results suggested that VNC O157 cells might lose virulence for humans, but we could not strictly determine whether VNC O157 retained pathogenicity for humans because O157 cells do not normally colonize the mouse intestine in the same way as they do the human intestine (K. Itoh, personal communication).
In this study, although it was not possible to clarify how the NaCl sensitivity of O157 changed, NaCl-sensitive O157 converted to NaCl-resistant O157 after lengthy preservation under a high concentration of NaCl and O157 became sensitive to NaCl after animal passage. However, this change was readily reversed by passage in N broth for 100 generations, suggesting that the original phenotype of O157 might be NaCl resistant. Under various stress conditions such as NaCl, heating, freezing (4, 11, 22, 24), and animal passage, reported here, bacterial cells might enter the VNC state as a protective mechanism. Resuscitation might then occur under certain conditions (25), meaning that the NaCl sensitivity of O157 might be one protective mechanism against foreign stress. In the future, since genetic changes induced by NaCl have been reported for several species of bacteria (5, 7, 8, 14, 15, 16), the mechanisms of NaCl sensitivity in EHEC O157 may well be elucidated by means of the genetic analysis of O157 reported in this study.
Bacteria, which enter the VNC state following stress, generally exhibit altered physiological function and cell division (6). We showed here that O157 entered the VNC state in a high concentration of NaCl. If this conversion occurred in food, it would be difficult to isolate VNC bacterial cells using routine microbiological detection systems. Moreover, the retention of pathogenicity by VNC pathogens and subsequent resuscitation in vivo are significant for human infection. Accordingly, attempts to isolate the causative agent from outbreaks of food poisoning should take into account the possibility of VNC cells. In addition, although selective media supplemented with bile salts are usually used for the detection of Enterobacteriaceae, such media used in this study were not appropriate for the detection of either VNC O157 cells or NaCl-resistant O157 cells in salted foods. An accurate and simple system of recovering VNC cells needs to be established for the isolation of pathogenic bacteria from foods.
Acknowledgments
We are grateful to B. Adler for critical suggestions on the manuscript.
This work was supported, in part, by a grant from Grants-in-Aid for Scientific Research (10357003), Japan Society for the Promotion of Science.
REFERENCES
- 1.Asai Y, Murase T, Osawa R, Okitsu T, Suzuki R, Sata S, Yamai S, Terajima J, Izumiya H, Tamura K, Watanabe H. Isolation of Shiga toxin-producing Escherichia coli O157:H7 from processed salmon roe associated with the outbreaks in Japan, 1998, and a molecular typing of the isolates by pulsed-field gel electrophoresis. Kansenshogaku Zasshi. 1999;73:20–24. doi: 10.11150/kansenshogakuzasshi1970.73.20. [DOI] [PubMed] [Google Scholar]
- 2.Asakura H, Makino S, Shirahata T, Tsukamoto T, Kurazono H, Ikeda T, Takeshi K. Detection and long-term existence of Shiga toxin (Stx)-producing Escherichia coli in sheep. Microbiol Immunol. 1998;42:683–687. doi: 10.1111/j.1348-0421.1998.tb02340.x. [DOI] [PubMed] [Google Scholar]
- 3.Barcina I, Lebaron P, Vives-Rego J. Survival of allochthonous bacteria in aquatic systems: a biological approach. FEMS Microbiol Ecol. 1997;23:1–9. [Google Scholar]
- 4.Blackburn C W, Curtis L M, Humpheson L, Billon C, McClure P J. Development of thermal inactivation models for Salmonella enteritidis and Escherichia coli O157:H7 with temperature, pH and NaCl as controlling factors. Int J Food Microbiol. 1997;38:31–44. doi: 10.1016/s0168-1605(97)00085-8. [DOI] [PubMed] [Google Scholar]
- 5.Choa J C, Kim S J. Viable, but non-culturable, state of a green fluorescence protein-tagged environmental isolate of Salmonella typhi in groundwater and pond water. FEMS Microbiol Lett. 1999;170:257–264. doi: 10.1111/j.1574-6968.1999.tb13382.x. [DOI] [PubMed] [Google Scholar]
- 6.Colwell R R, Brayton P R, Grimes D J, Roszak D B, Huq S A, Palmer L M. Viable but non-culturable Vibrio cholerae and related pathogens in the environment: implications for release of genetically engineered microorganisms. Bio/Technology. 1985;3:817–820. [Google Scholar]
- 7.Csonka L N. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dallmier A W, Martin S E. Catalase, superoxide dismutase, and hemolysin activities and heat susceptibility of Listeria monocytogenes after growth in media containing sodium chloride. Appl Environ Microbiol. 1990;56:2807–2810. doi: 10.1128/aem.56.9.2807-2810.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Defives C, Guyard S, Oulare M M, Mary P, Hornez J P. Total counts, culturable and viable, and non-culturable microflora of a French mineral water: a case study. J Appl Microbiol. 1999;86:1033–1038. doi: 10.1046/j.1365-2672.1999.00794.x. [DOI] [PubMed] [Google Scholar]
- 10.Gautom R K. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J Clin Microbiol. 1997;35:2977–2980. doi: 10.1128/jcm.35.11.2977-2980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guraya R, Frank J F, Hassan A N. Effectiveness of salt, pH, and diacetyl as inhibitors for Escherichia coli O157:H7 in dairy foods stored at refrigeration temperatures. J Food Prot. 1998;61:1098–1102. doi: 10.4315/0362-028x-61.9.1098. [DOI] [PubMed] [Google Scholar]
- 12.Kogure K, Simidu U, Taga N. A tentative direct microscopic method for counting living marine bacteria. Can J Microbiol. 1979;25:415–420. doi: 10.1139/m79-063. [DOI] [PubMed] [Google Scholar]
- 13.Masuda S, Hara-Kudo Y, Kumagai S. Reduction of Escherichia coli O157:H7 populations in soy sauce, a fermented seasoning. J Food Prot. 1998;61:657–661. doi: 10.4315/0362-028x-61.6.657. [DOI] [PubMed] [Google Scholar]
- 14.McIver K S, Heath A S, Scott J R. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect Immun. 1995;63:4540–4542. doi: 10.1128/iai.63.11.4540-4542.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myers E R, Dallmier A W, Martin S E. Sodium chloride, potassium chloride and virulence in Listeria monocytogenes. Appl Environ Microbiol. 1993;59:2082–2086. doi: 10.1128/aem.59.7.2082-2086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patchett R A, Kelly A F, Kroll R G. Effect of sodium chloride on the intracellular solute pools of Listeria monocytogenes. Appl Environ Microbiol. 1992;58:3959–3963. doi: 10.1128/aem.58.12.3959-3963.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rollins D M, Colwell R R. Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl Environ Microbiol. 1986;52:531–538. doi: 10.1128/aem.52.3.531-538.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roszak D B, Colwell R R. Metabolic activity of bacterial cells enumerated by direct viable count. Appl Environ Microbiol. 1987;53:2889–2893. doi: 10.1128/aem.53.12.2889-2893.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roszak D B, Colwell R R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasakawa C, Kamata K, Sakai T, Murayama S, Makino S, Yoshikawa M. Molecular alteration of the 140-mega-dalton plasmid associated with loss of virulence and Congo red binding activity in Shigella flexneri. Infect Immun. 1986;51:467–482. doi: 10.1128/iai.51.2.470-475.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semanchek J J, Golden D A. Influence of growth temperature on inactivation and injury of Escherichia coli O157:H7 by heat, acid, and freezing. J Food Prot. 1998;61:395–401. doi: 10.4315/0362-028x-61.4.395. [DOI] [PubMed] [Google Scholar]
- 23.Tilden J, Jr, Younz W, McNamara A M, Custer C, Boesel B, Lambert-Fair M A, Majkowski J, Vugia D, Werner S B, Hollingsworth J, Morris J G., Jr A new route of transmission for Escherichia coli: infection from dry fermented salami. Am J Public Health. 1996;86:1142–1145. doi: 10.2105/ajph.86.8_pt_1.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Velazquez M, Feirtag J M. Helicobacter pylori: characteristics, pathogenicity, detection methods and mode of transmission implicating foods and water. Int J Food Microbiol. 1999;53:95–104. doi: 10.1016/s0168-1605(99)00160-9. [DOI] [PubMed] [Google Scholar]
- 25.Wai S N, Moriya T, Kondo K, Misumi H, Amono K. Resuscitation of Vibrio cholerae O1 strain TSI-4 from a viable but nonculturable state by heat shock. FEMS Microbiol Lett. 1996;136:187–191. doi: 10.1111/j.1574-6968.1996.tb08047.x. [DOI] [PubMed] [Google Scholar]
- 26.Xu H-S, Roberts N, Singleton F L, Attwell R W, Grimes D J, Colwell R R. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb Ecol. 1982;8:313–323. doi: 10.1007/BF02010671. [DOI] [PubMed] [Google Scholar]