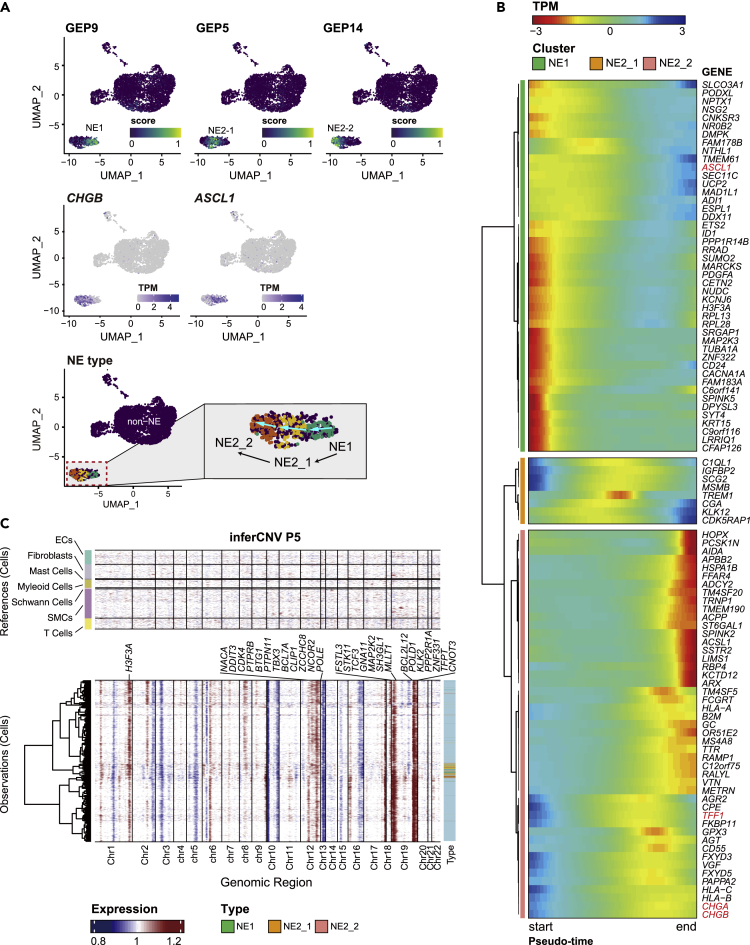
Figure 3.
NE trans-differentiation in P5
(A) The min-max normalized activity score of one NE1 GEP and two NE2 GEPs were colored. UMAP Plots were arranged in the assumed chronological order of differentiation. Expression of marker genes of note, ASCL1 and CHGB, for NE1 and NE2, respectively, were shown. In the right panel, the arrows denote the transition direction inferred from velocity analysis by Dong et al. (2020). TPM, transcript per million (normalized value).
(B) Dynamic expression along the trajectory identified 100 genes that vary significantly over trans-differentiation pseudo-time (likelihood ratio test of nested models adjusted by FDR <5%). Cells were arranged in column by pseudo-time series. Hierarchical clustering of these genes at row via Ward D2’s method recovered three nonredundant groups that covary over the trajectory. Cluster analysis indicated large shifts in gene expression occurred as NE progenitor progressed toward maturation. TPM, transcript per million (scaled value).
(C) inferCNV heatmap with hierarchical clustering of P5. The oncogenes or tumor suppressor genes in Cancer Gene Census were depicted in CNV regions. The top panel indicates a lack of CNV events in reference cells. The bottom panel is the malignant cells. Different types of NE cells were annotated on the sidebar (also see Figure S6).