Abstract
Verotoxin (VT)-producing Escherichia coli O157:H7 was culturable on agar media after being left in water for 21 months. However, there were a number of colonies which had lost O157 O antigenicity. These colonies produced VTs, which are pathogenic to humans. These observations suggest that the immunologic methods based on O157 O antigenicity are unable to detect and isolate VT-producing E. coli in foods and other environments if the organism has been under starvation conditions for a long period.
Transmission of Escherichia coli O157:H7 via water, such as drinking water and lake water, is important in E. coli O157:H7 infection (1, 4, 7, 9, 11, 14). It is suggested that water is contaminated with E. coli O157:H7 by humans and animals harboring the organism. There are several studies showing survival of E. coli O157:H7 in water for a long time under starvation conditions (12, 15, 16). For E. coli serotypes other than E. coli O157, expression of genes has been shown to be changed relative to the survival of the organism in water (3, 10). However, for E. coli O157, it is uncertain whether such changes occur with survival.
In the present study, we investigated the viability and change in the biochemical and antigenic properties characteristic of E. coli O157:H7 in distilled water (DW) and phosphate-buffered saline (PBS), which were chosen as a model for starvation conditions.
The bacterial strains (given by K. Tamura, Department of Bacteriology, National Institute of Infectious Diseases) used in this study are shown in Table 1. The 19 strains were cultured separately in 10 ml of tryptic soy broth (TSB; Difco, Detroit, Mich.) for 18 h at 37°C. The TSB cultures were centrifuged at 4,000 × g for 20 min. The supernatant was removed, and the pellet was washed three times with 10 ml of PBS (Nissui Co. Ltd., Tokyo, Japan) or DW (Nihon Millipore Ltd., Tokyo, Japan). Finally, the cells of all 19 strains, washed with PBS, were resuspended in 10 ml of PBS at 107 CFU/ml. For three strains (A, B, and C), cells washed with DW were resuspended in 10 ml of DW at 102, 103, and 105 CFU/ml. They were then incubated statically at 4 or 18°C.
TABLE 1.
Strains of E. coli O157:H7 used in this study
| Strain | Reference no. | Source | VT production |
|---|---|---|---|
| A | 2 | Patient | VT1, VT2 |
| B | 126 | Beef | VT2 |
| C | 23 | Patient | VT1, VT2 |
| D | 212 | Patient | VT1, VT2 |
| E | 298 | Patient | VT2 |
| F | 133 | Beef | VT1, VT2 |
| G | 1291 | Salad | VT1, VT2 |
| H | KE5 | Patient | VT1, VT2 |
| I | 758 | Patient | VT1, VT2 |
| J | 1102 | Beef | VT1, VT2 |
| K | 1646 | Salad | VT2 |
| L | 970056 | Radish sprouts | VT1, VT2 |
| M | 18 | Patient | VT1, VT2 |
| N | 42 | Patient | VT1, VT2 |
| O | 60 | Patient | VT1, VT2 |
| P | 132 | Beef | VT2 |
| Q | 174 | Patient | VT2 |
| R | 307 | Patient | VT1, VT2 |
| S | 434 | Patient | VT1, VT2 |
E. coli O157:H7 bacteria in PBS and in DW were incubated for 230 and 635 days, respectively. During incubation, 0.1 ml of the cell suspension was taken and serially diluted with PBS, and 0.5 ml of the suspension was plated onto five plates of tryptic soy agar (Difco). The colonies formed on the plates were counted after 18 h of incubation at 37°C to determine cell numbers.
Cell numbers decreased gradually with time in PBS at 4°C (Table 2). The cell numbers for P and the other 14 strains (B, C, D, F, G, J, K, L, M, N, O, Q, R, and S) decreased to below the detectable level (2 CFU/ml) on days 105 and 230, respectively. The remaining three strains (A, E, and H) survived till day 230. The numbers for some of these strains tended to decrease with time at 18°C, but the decrease was not as remarkable as that at 4°C.
TABLE 2.
Fate of E. coli O157:H7 strains in PBS
| Temp (°C) | Day | No. of cells (log CFU/ml) for each strain
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | ||
| 4 | 0 | 6.4 | 6.1 | 6.5 | 6.5 | 6.5 | 6.5 | 6.7 | 7.0 | 6.5 | 6.4 | 6.3 | 6.5 | 5.9 | 6.1 | 6.4 | 6.4 | 6.7 | 6.6 | 6.6 |
| 2 | 5.8 | 5.2 | 5.8 | 5.8 | 5.8 | 5.3 | 5.5 | 6.1 | 5.9 | 5.8 | 5.8 | 6.2 | 5.6 | 5.7 | 6.0 | 5.9 | 6.0 | 5.6 | 5.8 | |
| 21 | 5.1 | 4.9 | 5.2 | 5.2 | 5.0 | 5.2 | 5.3 | 6.0 | 4.4 | 5.1 | 5.0 | 5.4 | 5.0 | 4.1 | 5.4 | 4.8 | 5.4 | 5.4 | 4.9 | |
| 42 | 5.4 | 5.2 | 4.9 | 5.3 | 5.3 | 5.5 | 5.5 | 5.8 | 5.4 | 5.1 | 5.1 | 5.8 | 4.7 | 5.2 | 5.5 | 4.5 | 5.7 | 5.3 | 4.3 | |
| 105 | 3.8 | 2.9 | 2.2 | 4.5 | 3.7 | 3.9 | 2.2 | 5.1 | 1.3 | 1.5 | 1.2 | 4.0 | 2.0 | 2.0 | 1.0 | ND | 4.0 | 3.5 | 1.9 | |
| 230 | 1.0 | NDa | ND | ND | 1.7 | ND | ND | 4.7 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| 18 | 0 | 5.8 | 5.4 | 5.9 | 6.2 | 6.4 | 6.0 | 6.1 | 6.8 | 6.4 | 6.3 | 6.0 | 6.8 | 5.7 | 5.9 | 6.5 | 5.7 | 6.2 | 5.7 | 6.2 |
| 2 | 5.8 | 5.0 | 6.1 | 5.4 | 6.0 | 5.5 | 6.1 | 6.2 | 6.0 | 5.9 | 5.5 | 7.0 | 6.1 | 5.1 | 6.1 | 6.1 | 6.0 | 5.7 | 5.9 | |
| 21 | 5.0 | 3.6 | 5.4 | 5.3 | 5.2 | 5.8 | 5.9 | 6.1 | 6.4 | 5.3 | 4.0 | 5.3 | 4.3 | 4.3 | 5.7 | 3.6 | 5.1 | 5.3 | 5.4 | |
| 42 | 5.4 | 4.9 | 5.3 | 5.4 | 5.5 | 5.4 | 5.4 | 5.7 | 5.4 | 5.4 | 4.1 | 4.3 | 4.3 | 4.9 | 5.4 | 4.0 | 4.9 | 4.6 | 5.3 | |
| 105 | 5.0 | 4.9 | 4.8 | 4.6 | 4.9 | 4.9 | 4.8 | 4.4 | 5.0 | 5.3 | 4.3 | 4.6 | 4.7 | 4.6 | 4.6 | 4.7 | 4.9 | 4.6 | 4.9 | |
| 230 | 5.6 | 4.6 | 5.0 | 4.8 | 4.7 | 5.3 | 5.4 | 5.0 | 5.0 | 5.6 | 4.6 | 4.6 | 5.0 | 5.3 | 4.3 | 4.5 | 4.9 | 4.9 | 5.4 | |
ND, not detected (<0.3 log CFU/ml).
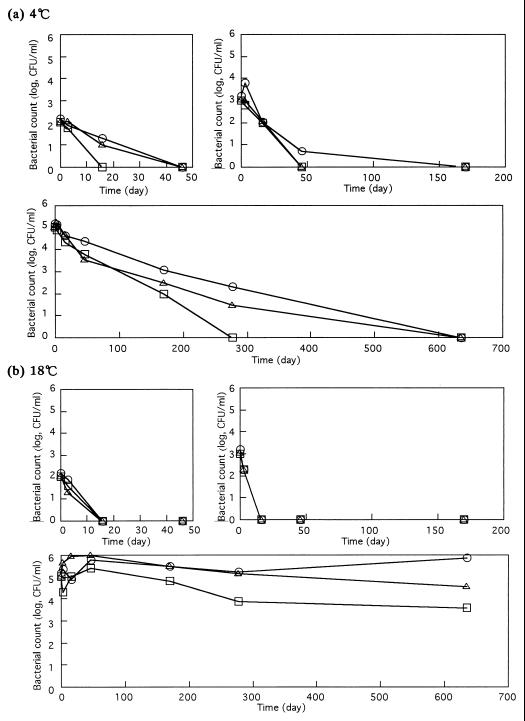
The numbers of bacteria decreased with time in DW also at 4°C (Fig. 1a). They tended to decrease more slowly when 105 CFU/ml was inoculated than when 102 and 103 CFU/ml were inoculated. When incubated at 18°C (Fig. 1b), E. coli O157:H7 that had been inoculated at the levels of 102 and 103 CFU/ml decreased to fewer than 2 CFU/ml within 48 days, but those bacteria inoculated at the level of 105 CFU/ml decreased by less than 1 order of magnitude during 48 days of incubation (Fig. 1b). The fate of the E. coli O157:H7 population depended on the initial number, indicating the importance of bacterial concentration in water. This phenomenon may relate to the interaction of bacterial cells. As found on PBS, E. coli O157:H7 at high cell concentrations survived better at 18°C than at 4°C (Fig. 1a and b).
FIG. 1.
Fate of three E. coli O157:H7 strains in DW at 4°C (a) and 18°C (b). Strain and inoculation level symbols: ○, strain A; □, strain B; ▵, strain C.
Colonies on tryptic soy agar at day 230 of incubation in PBS and those at days 277 and 635 of incubation in DW were tested for O antigenicity with an agglutination kit (UNI; Oxoid) and O157 antiserum (Denka Seiken Co. Ltd., Tokyo, Japan) without heat treatment of the colonies. The colonies were also tested for cellobiose and lactose fermentation and indole- and glucuronidase-positive reactions with cellobiose-lactose-indole-d-glucuronidase agar (Kyokuto, Tokyo, Japan) culture. Nonagglutinated colonies were found in strain A at 4°C and strains B, K, P, Q, R, and S at 18°C on day 230 in PBS and in strain A at 18°C on days 277 and 635 in DW. Shimizu et al. (13) showed that orf2 is an important gene for the synthesis of the O157 O antigen. To confirm genetically the loss of O157 antigenicity by detection of orf2, the colonies were inoculated in TSB and cultured at 37°C for 18 h. The TSB culture was then centrifuged at 40,000 × g for 20 min. The supernatants were assayed for the presence of verotoxins (VTs) using a latex agglutination assay kit (Denka Seiken) by following the manufacturer's instructions. Cell pellets were assayed for orf2 genes and VT genes by PCR according to the method of Shimizu et al. (13) and the manufacturer's instructions (O157 PCR screening set; Takara, Tokyo, Japan), respectively. The amplified gene was analyzed by electrophoresis in a 2% agarose gel. Table 3 shows the changes in properties characteristic of inoculated strains after incubation in PBS at 4 or 18°C for 230 days or in DW at 18°C for 277 and 635 days. A number of isolates of strains A, B, K, Q, and S lost O157 O antigenicity, although the orf2 gene encoding O157 O antigen existed in these isolates. However, some isolates of strains P and R lost the orf2 gene as well as O157 O antigenicity (data not shown). These findings suggest that E. coli O157 may occasionally lose O157 O antigenicity not only with the loss of the orf2 gene but also without it.
TABLE 3.
Change in properties characteristic of E. coli O157:H7 after incubation in watera
| Strain | Results before incubation
|
Incubating conditions | Results after incubation
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O157 O antigenicity
|
Orf2 | PCR-VT1 | PCR-VT2 | Latex-VT1 | Latex-VT2 | O157 O antigenicity
|
Orf2 | PCR-VT1 | PCR-VT2 | Latex-VT1 | Latex-VT2 | ||||
| Antiserum | Latex | Antiserum | Latex | ||||||||||||
| A | + (10/10)b | + (10/10) | + | + | + | + | + | In PBS at 4°C (230 days) | + (4/8) − (4/8) | + (4/8) − (4/8) | + (4/4) − (0/4) | + (4/4) − (0/4) | + (4/4) − (0/4) | + (4/4) − (0/4) | + (0/4) − (4/4) |
| In DW at 18°C (277 days) | + (0/3) − (3/3) | + (0/3) − (3/3) | + (3/3) − (0/3) | + (3/3) − (0/3) | + (3/3) − (0/3) | + (3/3) − (0/3) | + (0/3) − (3/3) | ||||||||
| In DW at 18°C (635 days) | + (0/3) − (3/3) | + (0/3) − (3/3) | + (3/3) − (0/3) | + (3/3) − (0/3) | + (3/3) − (0/3) | + (3/3) − (0/3) | + (0/3) − (3/3) | ||||||||
| B | + (10/10) | + (10/10) | + | − | + | − | + | In PBS at 18°C (230 days) | + (13/34) − (21/34) | + (13/34) − (21/34) | + (21/21) − (0/21) | + (0/21) − (21/21) | + (21/21) − (0/21) | + (0/21) − (21/21) | + (21/21) − (0/21) |
| K | + (10/10) | + (10/10) | + | − | + | − | + | In PBS at 18°C (230 days) | + (2/30) − (28/30) | + (2/30) − (28/30) | + (28/28) − (0/28) | + (0/28) − (28/28) | + (28/28) − (0/28) | + (0/28) − (28/28) | + (28/28) − (0/28) |
| Q | + (10/10) | + (10/10) | + | − | + | − | + | In PBS at 18°C (230 days) | + (9/30) − (21/30) | + (9/30) − (21/30) | + (21/21) − (0/21) | + (0/21) − (21/21) | + (0/21) − (0/21) | + (0/21) − (21/21) | + (21/21) − (0/21) |
| S | + (10/10) | + (10/10) | + | + | + | + | + | In PBS at 18°C (230 days) | + (20/30) − (10/30) | + (20/30) − (10/30) | + (10/10) − (0/10) | + (10/10) − (0/10) | + (10/10) − (0/10) | + (10/10) − (0/10) | + (10/10) − (0/10) |
O157 O antigenicity, agglutination test for anti-O157 serum and a latex agglutination kit; Orf2, detection of the orf2 O157 lipopolysaccharide gene by PCR; PCR-VT1, detection of the VT1 gene by PCR; Latex-VT1: detection of VT1 using the reversed passive latex agglutination test; +, positive reaction; −, negative reaction.
Number of colonies with reactions/number of colonies tested.
The lost O157 O antigenicity was not recovered by culture in TSB. These strains were confirmed to possess such properties of E. coli O157 as cellobiose nonfermentation, lactose fermentation, and indole- and β-glucuronidase-positive reactions. The VT gene was maintained in these strains, which possessed the gene initially. The strains maintained VT production except for the VT2 production of strain A. It was suggested that the expression of the VT2 gene was inhibited in strain A.
Many O antigen serotypes of E. coli are known to produce VTs or to be pathogenic. The E. coli O157:H7 strain which lost O157 O antigenicity was tested for agglutination to the other O antigen antisera (Denka Seiken). However, the O antigen of the E. coli O157:H7 strain which lost O157 O antigenicity did not change to the other types, such as O1, O6, O8, O15, O18, O20, O25, O26, O27, O28, O29, O44, O55, O63, O78, O86, O111, O112, O114, O115, O119, O124, O125, O126, O127, O128, O136, O142, O143, O144, O146, O148, O151, O152, O153, O157, O158, O159, O164, O166, O167, O168, and O169 of pathogenic E. coli.
Wang and Doyle (15) showed changes in outer membrane proteins of E. coli O157:H7 cells surviving in water. In the present study, we found a loss of immunologic response against O157 O antigen. O antigenicity is a very important characteristic for detection of O157, a most frequently isolated type of enterohemorrhagic E. coli (EHEC) (6). Many kinds of detection kits and systems utilizing O157 O antigenicity have been developed (2, 5, 8) and widely used to detect a small number of E. coli O157 bacteria in food and environmental samples contaminated with a large number of competitive bacteria. However, a method which does not depend on immunologic techniques should be used to test for EHEC in foods and other environments, when the organism is suspected to have been under starvation conditions for a long period.
REFERENCES
- 1.Ackman D, Marks S, Mack P, Caldwell M, Root T, Brikhead D. Swimming-associated haemorrhagic colitis due to in Escherichia coli O157:H7 infection: evidence of prolonged contamination of a fresh water lake. Epidemiol Infect. 1997;119:1–8. doi: 10.1017/s095026889700770x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett A R, MacPhee S, Betts R P. The isolation and detection of Escherichia coli O157:H7 by use of immunomagnetic separation and immunoassay procedures. Lett Appl Microbiol. 1996;22:237–243. doi: 10.1111/j.1472-765x.1996.tb01151.x. [DOI] [PubMed] [Google Scholar]
- 3.Chai T-J. Characteristics of Escherichia coli grown in bay water as compared with rich medium. Appl Environ Microbiol. 1983;45:1316–1323. doi: 10.1128/aem.45.4.1316-1323.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson S G, Goodbrand R B, Johnson R P, Odorico V G, Alves D, Rahn K, Wilson J B, Welch M K, Khakhria R. Escherichia coli O157:H7 diarrhoea associated with well water and infected cattle on an Ontario farm. Epidemiol Infect. 1998;120:17–20. doi: 10.1017/s0950268897008479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson J L, Rose B E, Sharar A K, Ranson G M, Lattuada C P, McNamara A M. Methods used for detection and recovery of Escherichia coli O157:H7 associated with a food-borne disease outbreak. J Food Prot. 1994;58:597–603. doi: 10.4315/0362-028X-58.6.597. [DOI] [PubMed] [Google Scholar]
- 6.Karmail M A. Infection by verotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2:15–38. doi: 10.1128/cmr.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keene W E, McAnulty J M, Hoesly F C, Williams L P, Jr, Hedberg K, Oxman G L, Barrett T J, Pfaller M A, Fleming D W. A swimming-associated outbreak of hemorrhagic colitis caused by Escherichia coli O157:H7 and Shigella sonnei. N Engl J Med. 1994;331:579–584. doi: 10.1056/NEJM199409013310904. [DOI] [PubMed] [Google Scholar]
- 8.Meer R R, Park D L. Immunochemical detection methods for Salmonella spp., Escherichia coli O157:H7, and Listeria monocytogenes in foods. Rev Environ Contam Toxicol. 1995;142:1–12. doi: 10.1007/978-1-4612-4252-9_1. [DOI] [PubMed] [Google Scholar]
- 9.Michel P, Wilson J B, Martin S W, McEwen S A, Gyles C L. Temporal and geographical distributions of reported cases of Escherichia coli O157:H7 infection in Ontario. Epidemiol Infect. 1999;122:193–200. doi: 10.1017/s0950268899002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozkanca R, Flint K P. Relationship between respiratory enzymes and survival of Escherichia coli under starvation stress in lake water. J Appl Microbiol. 1997;82:301–309. doi: 10.1046/j.1365-2672.1997.00360.x. [DOI] [PubMed] [Google Scholar]
- 11.Paunio M, Pebody R, Keskimaki M, Kokki M, Ruutu P, Oinonen S, Vuotari V, Siitonen A, Lahti E, Leinikki P. Swimming-associated outbreak of Escherichia coli O157:H7. Epidemiol Infect. 1999;122:1–5. doi: 10.1017/s0950268898001927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigsbee W, Simpson L M, Oliver J D. Detection of the viable but nonculturable state in Escherichia coli O157:H7. J Food Safety. 1997;16:255–262. [Google Scholar]
- 13.Shimizu T, Yamasaki S, Tsukamoto T, Takeda Y. Analysis of the genes responsible for the O-antigen synthesis in enterohaemorrhagic Escherichia coli O157. Microb Pathog. 1999;26:235–247. doi: 10.1006/mpat.1998.0253. [DOI] [PubMed] [Google Scholar]
- 14.Swerdlow D L, Woodruff B A, Brady R C, Griffin P M, Tippen S, Donnell H D, Geldreich E, Payne B J, Meyer A, Wells J G, Greene K D, Bright M, Bean N H, Blake P A. A waterborne outbreak in Missouri of Escherichia coli O157:H7 associated with bloody diarrhea and death. Ann Intern Med. 1992;117:812–819. doi: 10.7326/0003-4819-117-10-812. [DOI] [PubMed] [Google Scholar]
- 15.Wang G, Doyle M P. Survival of enterohemorrhagic Escherichia coli O157:H7 in water. J Food Prot. 1998;61:662–667. doi: 10.4315/0362-028x-61.6.662. [DOI] [PubMed] [Google Scholar]
- 16.Warburton D W, Austin J W, Harrison B H, Sanders G. Survival and recovery of Escherichia coli O157:H7 in inoculated bottled water. J Food Prot. 1998;61:948–952. doi: 10.4315/0362-028x-61.8.948. [DOI] [PubMed] [Google Scholar]