Abstract
Background:
Penicillin and ciprofloxacin are important for invasive meningococcal disease (IMD) management and prevention. IMD cases caused by penicillin- and ciprofloxacin-resistant Neisseria meningitidis containing a ROB-1 β-lactamase gene (blaROB-1) and a mutated DNA gyrase gene (gyrA), have been recently reported in the United States.
Methods:
We examined 2097 meningococcal genomes collected through US population-based surveillance from January 2021 to February 2020 to identify IMD cases caused by strains with blaROB-1 or gyrA-mediated resistance. Antimicrobial resistance was confirmed phenotypically. The US isolate genomes were compared to non-US isolate genomes containing blaROB-1. Interspecies transfer of ciprofloxacin resistance was assessed by comparing gyrA among Neisseria species.
Results:
Eleven penicillin- and ciprofloxacin-resistant isolates were identified after December 2018; all were serogroup Y, sequence type 3587, clonal complex (CC) 23, and contained blaROB-1 and a T91I-containing gyrA allele. An additional 22 penicillin-resistant, blaROB-1-containing US isolates with wild-type gyrA were identified from 2013–2020. All 33 blaROB-1-containing isolates formed a single clade, along with 12 blaROB-1-containing isolates from six other countries. Two-thirds of blaROB-1-containing US isolates were from Hispanic individuals. Twelve additional ciprofloxacin-resistant isolates with gyrA T91 mutations were identified. Ciprofloxacin-resistant isolates belonged to six CCs and contained 10 unique gyrA alleles; seven were similar or identical to alleles from N. lactamica or N. gonorrhoeae.
Conclusions:
Recent IMD cases caused by a dual resistant serogroup Y suggest changing antimicrobial resistance patterns in the United States. The emerging dual-resistance is due to acquisition of ciprofloxacin resistance by β-lactamase-containing N. meningitidis. Routine antimicrobial resistance surveillance will effectively monitor resistance changes and spread.
Keywords: Neisseria meningitidis, ciprofloxacin, β-lactamase, antibiotic resistance, meningococcal disease
INTRODUCTION
Neisseria meningitidis causes life-threatening invasive meningococcal disease (IMD). Suspected meningococcal disease is treated empirically with cefotaxime or ceftriaxone, while penicillin and ampicillin have historically been treatment options only after meningococcal disease is confirmed [1]. Penicillin is also sometimes used for long-term meningococcal disease chemoprophylaxis among patients using complement inhibitors such as eculizumab or ravulizumab [2–5]. Ciprofloxacin is commonly used for antibiotic prophylaxis for close contacts of meningococcal disease patients; rifampin, or ceftriaxone can also be used for prophylaxis [1].
Antimicrobial resistance has historically been rare among N. meningitidis. While isolates with reduced susceptibility to penicillin have been reported in multiple countries [6–10], only intermediate susceptibility phenotypes are commonly detected; penicillin-resistant isolates remain rare [11, 12]. Resistance to ciprofloxacin is relatively uncommon globally, with the notable exception in China, where the majority of invasive and carriage isolates were ciprofloxacin-resistant [6, 7, 13–18]. A novel ciprofloxacin- and penicillin-resistant N. meningitidis strain was recently identified in several US cases [19, 20]; all isolates were serogroup Y (NmY) and belonged to the sequence type (ST)-23 clonal complex (CC23). Six meningococcal isolates, collected during 2017–2019, with similar characteristics (dual-resistance, belonging to CC23, and serogroup Y) were also identified in El Salvador [21].
Nearly all previously characterized ciprofloxacin-resistant N. meningitidis isolates have isoleucine (I) or phenylalanine (F) mutations at position 91 in the quinolone resistance determining region (QRDR) of gyrA [6, 13, 14, 16, 17, 21–26]. Additional mutations in gyrA (D95N and T193A) and also in parC (D86N, S87R, and E91G) have also been associated with higher ciprofloxacin minimum inhibitory concentrations (MICs) in N. meningitidis [17, 22, 25–27]. These mutations are frequently introduced by homologous recombination from other Neisseria species, as reported for three US ciprofloxacin-resistant N. meningitidis serogroup B (NmB) isolates [13] and several ciprofloxacin-resistant isolates from China [25].
Recently, penicillin resistance due to the β-lactamase gene blaROB-1 has been observed in N. meningitidis despite being rare historically. The β-lactamase-containing isolates were identified in Canada, France, the United States and El Salvador [11, 12, 19, 21, 28]. β-lactamase-independent mechanisms of penicillin resistance are poorly understood in N. meningitidis; intermediate susceptibility can be caused by mosaic penA alleles, in which inter-species recombination introduces four to five amino acid substitutions within penicillin-binding protein 2 [10, 29].
Here, we evaluated penicillin and/or ciprofloxacin resistance among IMD cases reported through meningococcal disease surveillance in the United States and explored the origins of the resistance mechanisms observed in the dual-resistant CC23 isolates.
METHODS
Invasive meningococcal disease surveillance and strain collection
Epidemiologic information on all US IMD cases was submitted to the Centers for Disease Control and Prevention (CDC) through the National Notifiable Diseases Surveillance System; state health departments provided supplemental epidemiologic data for cases with isolates containing blaROB-1. Meningococcal isolates from cases reported from January 1st 2011 to February 14th 2020 were submitted by jurisdiction health departments through three surveillance programs, including the Active Bacterial Core Surveillance during 2011–20, which covers ~14% of the US population [30], expanded surveillance sites in 2013–14 [31] and Enhanced Meningococcal Disease Surveillance 2015–20, with isolates submitted for over 75% of US IMD cases by 2018 [32]. If two or more isolates were submitted for a single meningococcal case, only one was included in the analysis.
Collection of meningococcal case information is still on-going; total meningococcal case counts during 2019–20 are not currently available.
Whole Genome Sequencing (WGS) and Identification of Antimicrobial Resistance Determinants
All submitted meningococcal isolates were sequenced as described previously [33]. Each isolate genome was characterized using standard molecular typing methods (multilocus sequence typing [MLST] and typing of PorA and FetA) [33]. Genes involved in antibiotic resistance were identified by a BLAST search of genome assemblies using reference sequences for blaROB-1 from the Haemophilus influenzae plasmid pB1000 (NC_019178) and the full-length gyrA (NEIS1320), parC (NEIS1525), penA (NEIS1753), and blaTEM-1 (NEIS2357) genes from the PubMLST Neisseria database [34]. Amino-acid mutations in gyrA, parC, and penA were identified by aligning the translated sequence to allele 1 from PubMLST using BioPython [35]. Alleles for gyrA were identified according to the PubMLST locus definition for a 525bp fragment that contains the QRDR [22, 34] (defined as gyrA-QRDR). Isolates with blaROB-1 or T91 mutations in gyrA are listed in Supplemental Table 1 and genome assemblies were submitted to PubMLST.
The PubMLST database was queried on April 21, 2020 to identify genomes containing blaROB-1 or blaTEM-1 using the BLAST function and to identify isolates with gyrA T91 mutations using the API [22, 34].
Phenotypic antimicrobial susceptibility testing
Isolates containing blaROB-1 or a mutation at position T91 of gyrA were assessed for β-lactamase activity by nitrocefin testing (BD BBL DrySlide; Franklin Lakes, NJ). All isolates with a mutation at position T91 of gyrA and a subset of blaROB-1 -containing isolates were tested by reference broth microdilution (BMD) in accordance with the Clinical and Laboratory Standards (CLSI) guidelines; susceptibility interpretations were assigned according to M100, 30th Edition [36]. Frozen reference microdilution panels were prepared at CDC [37] and contained the following antimicrobial dilution series (μg/mL): ampicillin (0.03–16), azithromycin (0.06–4), cefotaxime (0.015–8), ceftriaxone (0.015–8), chloramphenicol (1–16), ciprofloxacin (0.015–4), levofloxacin (0.03–8), meropenem (0.06–4), minocycline (0.25–2), penicillin (0.015–8), rifampin (0.06–4), and trimethoprim-sulfamethoxazole (0.06/1.14 – 4/76).
Whole genome sequence comparisons
Whole-genome phylogenetic analysis was performed with blaROB-1-containing isolates and closely related isolates with matching STs (Supplemental Table 1). Assemblies were aligned to the complete genome sequence of a ST-23 isolate collected in 2003 from the United States (M10868) using Snippy v4.3.8 [38]. Recombinant regions were removed from the 1,706,760 bp core-genome alignment using Gubbins v1.4.1 [39]. Single nucleotide polymorphisms (SNPs) were counted as the number of nucleotide differences between each pair of genomes in the alignment, both before and after running Gubbins. Finally, a maximum likelihood phylogenetic tree rooted on M10868 was created using RAxML-NG v0.9 [40] with GTR+G substitution model, a minimum branch length of 10−8 substitutions/site, Stamatakis ascertainment correction, and autoMRE bootstopping, which determined that bootstrap support values converged after 1000 replicates.
gyrA-QRDR sequence comparisons
Neighbor-net analysis of gyrA-QRDR was performed with SplitsTree4 [41], including the seven Neisseria spp. in the PubMLST database [34] that had gyrA-QRDR alleles documented for at least 10 isolates. Alleles from the US invasive meningococcal disease isolate collection were included if they contained either the T91I or T91F mutations.
RESULTS
Characterization of US penicillin-and/or ciprofloxacin-resistant meningococcal isolates
A total of 2,097 isolates were available from US IMD cases occurring between January 2011 and February 2020, including 773 NmB (36.9%), 580 NmC (27.7%), 374 NmY (17.8%), 207 NmW (9.9%), 153 nongroupable (7.3%), 9 NmE and 1 NmZ.
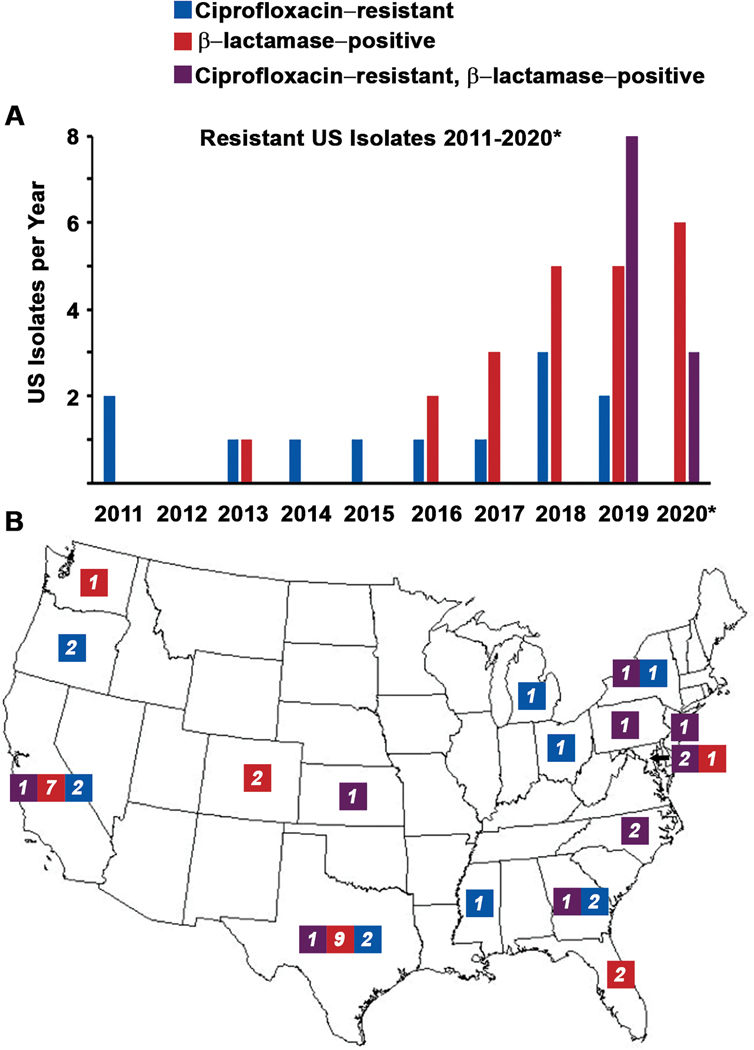
Whole genome analysis identified 45 isolates that have blaROB-1 encoding penicillin resistance and/or a gyrA mutation causing ciprofloxacin resistance (T91), with at least one of these isolates detected in every year except 2012 (Figure 1A). Eleven isolates collected from 2019–2020 contained both blaROB-1 and gyrA T91I (gyrA-QRDR allele 242). Twenty-two isolates collected from 2013–2020 contained blaROB-1 and a wild type gyrA (gyrA-QRDR allele 2). Twelve isolates from 2011–2019 without β-lactamase genes contained a gyrA T91 mutation (9 with T91I and 3 with T91F); nine different gyrA-QRDR alleles with T91 mutations were identified.
Figure 1.
Temporal (A) and geographic (B) distribution of Neisseria meningitidis isolates with β-lactamase activity (red: contain the blaROB-1 gene), ciprofloxacin resistance (blue: T91I or T91F mutations in gyrA), or both (purple) in the United States. States are labeled with the number of isolates identified with each resistance profile. *2020 only includes isolates collected through February 14th, 2020.
To confirm the genomic susceptibility predictions, phenotypic testing was completed. Of the 19 blaROB-1-containing isolates tested by reference BMD (Table 1), all were resistant to penicillin and ampicillin. Eleven of the 19 isolates had gyrA-QRDR allele 242 and where resistant to ciprofloxacin and levofloxacin; the remaining 8 isolates had a wildtype gyrA-QRDR allele and were fluoroquinolone susceptible. All 19 isolates were resistant to trimethoprim-sulfamethoxazole but susceptible to all other antibiotics tested, including cefotaxime and ceftriaxone. β-lactamase activity was confirmed by nitrocefin testing for these 19 isolates plus an additional 14 blaROB-1-containing isolates that were not assessed by BMD. Twelve isolates with a gyrA-QRDR T91 mutation lacked β-lactamase activity and were resistant to ciprofloxacin and levofloxacin. One of these isolates contained four previously characterized mutations (gyrA T91I, D95N, T173A; parC S87I) and had an MIC of 4 μg/mL for both fluoroquinolones. All other isolates with gyrA T91I or T9IF mutations had MICs ranging from 0.12–0.5 μg/mL (Table 2).
Table 1:
Antimicrobial susceptibility testing results for the isolates containing the blaROB-1 gene
| blaROB-1-containing and T91I gyrA mutation (n=11) | blaROB-1-containing and wild type gyrA-QRDR (n=8ǂ) | T91 gyrA mutation without blaROB-1 (n=12) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
CLSI
Breakpoints |
No. of
Isolates |
No. of Isolates | No. of Isolates | ||||||||||||
| Antibiotic | S | I | R | MIC Range | S | I | R | MIC Range | S | I | R | MIC Range | S | I | R |
| Penicillin* | ≤0.06 | 0.12–0.25 | ≥0.5 | 8 - >8 | 0 | 0 | 11 | 8 - >8 | 0 | 0 | 8 | ≤0.016 – 0.5 | 6 | 5 | 1 |
| Ampicillin* | ≤0.12 | 0.25–1 | ≥2 | 8 - >16 | 0 | 0 | 11 | 16 - >16 | 0 | 0 | 8 | ≤0.03 – 1 | 7 | 5 | 0 |
| Cefotaxime* | ≤0.12 | -- | -- | ≤0.015 | 11 | -- | 0 | ≤0.015 | 8 | -- | 0 | ≤0.015 – 0.03 | 12 | -- | 0 |
| Ceftriaxone*δ | ≤0.12 | -- | -- | ≤0.015 | 11 | -- | 0 | ≤0.015 | 8 | -- | 0 | ≤0.015 | 12 | -- | 0 |
| Meropenem* | ≤0.25 | -- | -- | ≤0.06 | 11 | -- | 0 | ≤0.06 | 8 | -- | 0 | ≤0.06 | 12 | -- | 0 |
| Ciprofloxacin δ | ≤0.03 | 0.06 | ≥0.12 | 0.12–0.25 | 0 | 0 | 11 | ≤0.015 | 8 | 0 | 0 | 0.12 – 4 | 0 | 0 | 12 |
| Levofloxacin | ≤0.03 | 0.06 | ≥0.12 | 0.25 | 0 | 0 | 11 | ≤0.03 | 8 | 0 | 0 | 0.12 – 4 | 0 | 0 | 12 |
| Chloramphenicol | ≤2 | 4 | ≥8 | ≤1 | 11 | 0 | 0 | ≤1 – 2 | 8 | 0 | 0 | ≤1 – 2 | 12 | 0 | 0 |
| Azithromycin | ≤2 | -- | -- | 0.25–0.5 | 11 | -- | 0 | 0.5–1.0 | 8 | -- | 0 | ≤0.06 – 1 | 12 | -- | 0 |
| Minocycline | ≤2 | -- | -- | ≤0.25 | 11 | -- | 0 | ≤0.25 | 8 | -- | 0 | ≤0.25 – 0.5 | 12 | -- | 0 |
| Rifampinδ | ≤0.5 | 1 | ≥2 | ≤0.06 – 0.5 | 11 | 0 | 0 | ≤0.06 | 8 | 0 | 0 | ≤0.06 – 0.25 | 12 | 0 | 0 |
| Trimethoprim-sulfamethoxazole | ≤0.12/2.4 | 0.25/4.75 | ≥0.5/9.5 | 2/38 - >4/76 | 0 | 0 | 11 | 4/76 - >4/76 | 0 | 0 | 8 | 1/19 - >4/76 | 0 | 0 | 12 |
The antibiotics recommended for patient treatment
and for chemoprophylaxis
are denoted in the table [1]. MICs were determined using reference broth microdilution according to CLSI guidelines and interpretive criteria were applied according to the M100. S = susceptible, I = Intermediate (a dash indicates that an intermediate breakpoint is not defined for that antibiotic), and R = resistant.
Eight blaROB-1-containing isolates, collected in seven different years from six states, were selected for reference broth microdilution.
Table 2.
Invasive Ciprofloxacin-Resistant, β-lactamase-negative Neisseria meningitidis Isolates with gyrA Mutations at Position T91 in the United States, 2011–19
| MIC Range (μg/mL) | ||||||
|---|---|---|---|---|---|---|
| ST, CC | No. of Isolates | Collection Years |
gyrA Mutations (Allele No.) |
parC Mutations | Ciprofloxacin | Levofloxacin |
| NmB | ||||||
| ST-6595, CC4821 | 2 | 2014, 2019 | T91F, D95G (363) | -- | 0.12–0.25 | 0.25 |
| ST-5662, unassigned | 1 | 2017 | T91I (146) | -- | 0.25 | 0.5 |
| ST-13904, CC41/44 | 1 | 2018 | T91I, T173A (98) | -- | 0.12 | 0.12 |
| NmC | ||||||
| ST-273, CC231 | 1 | 2018 | T91I, T173A (368) | -- | 0.25 | 0.25 |
| NmW | ||||||
| ST-11, CC11 | 1 | 2016 | T91I, D95N, T173A (211) | S87I | 4 | 4 |
| NmY | ||||||
| ST-767, CC167 | 1 | 2011 | T91I (8) | -- | 0.25 | 0.25 |
| ST-2533, CC23 | 4 | 2011, 2013, 2015, 2018 | T91I (348) | -- | 0.12–0.25 | 0.12–0.25 |
| NmNG | ||||||
| ST-11, CC11 | 1 | 2019 | T91F, D95G (140) | -- | 0.5 | 0.25 |
MICs were determined using reference broth microdilution according to CLSI guidelines and interpretive criteria were applied according to the M100.
All 33 blaROB-1-containing isolates had an intact NmY capsule locus, but one was nongroupable (NmNG) phenotypically. One molecular profile was shared by 30 of the 33 blaROB-1-containing isolates (PorA P1.5–2,10–2: FetA F4–1: ST-3587: CC23), including all 11 dual-resistant isolates. The remaining three isolates were ST-15379 or ST-13034, each differing from ST-3587 at a single typing locus. The ST-13034 isolate had the same PorA and FetA types as the ST-3587 isolates, while the two ST-15379 isolates contained P1.5–2,10–25. The 12 β-lactamase-negative, ciprofloxacin-resistant isolates were genetically diverse (Table 2); five serogroups and six CCs were detected. Four NmY CC23 isolates were detected across four separate years; all contained a novel gyrA-QRDR allele (348) and the molecular profile (P1.5–2,10–1: F4–1: ST-2533: CC23), which differed from the blaROB-1-containing isolates at three MLST loci and the PorA locus.
Invasive Meningococcal Disease caused by penicillin- and/or ciprofloxacin-resistant isolates
The 33 IMD cases caused by blaROB-1-containing isolates were reported from 11 US states (Figure 1B) across several age groups. The highest proportions of cases were in persons aged ≥45 years (45%) and aged <1 year (18%); the age distribution was similar for cases caused by blaROB-1-containing isolates that were ciprofloxacin resistant and ciprofloxacin-susceptible. Of the 33 cases, 22 (67%) occurred in Hispanic individuals, including 8 of the 11 cases with ciprofloxacin-resistant isolates. There were no known epidemiologic links among cases. One patient, with IMD associated with a ciprofloxacin-susceptible isolate, died (case-fatality ratio: 3.0%).
Twenty-one of 33 IMD patients with blaROB-1-containing isolates had additional epidemiologic data available. Six (29%) had resided in or traveled to Mexico shortly before meningococcal disease onset (all had isolates containing blaROB-1 and a wild type gyrA-QRDR). Among 18/33 cases with treatment information, none were treated with penicillin or ampicillin.
The antibiotic used for prophylaxis of close contacts was reported for five of the 11 cases with dual-resistant isolates. Ciprofloxacin was initially used as the primary prophylaxis antibiotic for contacts of four cases; however, prophylaxis was repeated for the contacts of two cases with an alternate antibiotic once susceptibility results were available. No prophylaxis failures were reported.
The 12 IMD cases caused by β-lactamase negative, ciprofloxacin-resistant isolates were reported from eight states (Figure 1B) and occurred across several age groups, with the highest proportion in adults aged ≥45 years (33%) and 22–44 years (33%). Only one case (8%) occurred in a Hispanic individual. One case was fatal.
Comparison between US and non-US meningococcal isolates
The PubMLST database included 12 additional blaROB-1-containing N. meningitidis genomes (11 NmY and 1 NmNG) [11, 12]. All twelve isolates were ST-3587 (CC23) with a wild type gyrA-QRDR allele 2 and were collected during 2016–19 from six countries: 7 from Mexico, 4 from four European countries, and 1 from Canada (Supplemental Table 2). A screen for a second β-lactamase encoding gene identified six blaTEM-1-containing meningococcal genomes in PubMLST, collected during 2013–18 from five countries. The blaTEM-1 isolates were diverse, belonging to six CCs (Supplemental Table 3); blaTEM-1 was not identified in the US collection.
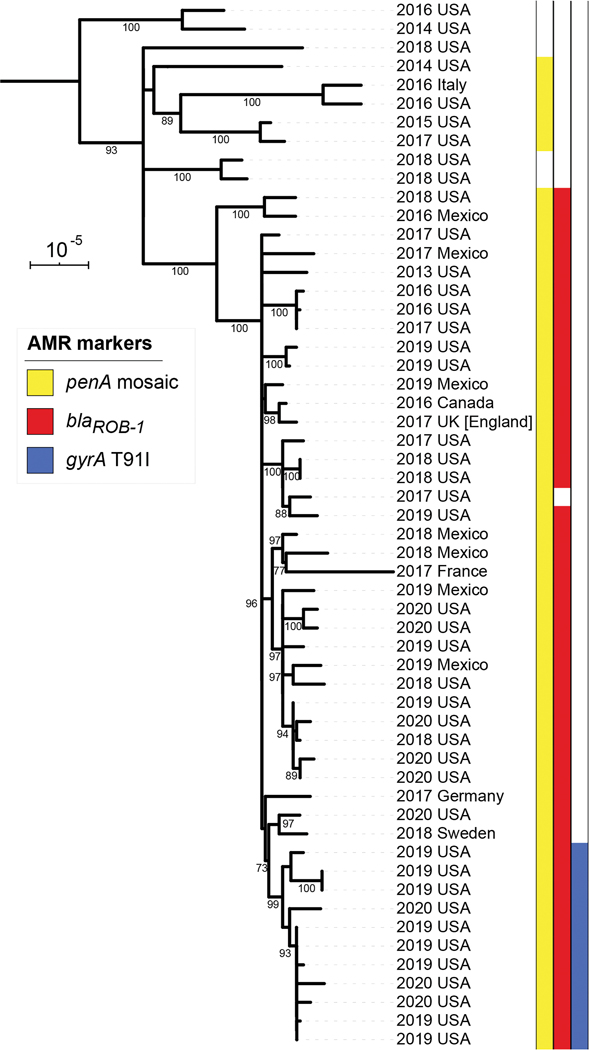
The genomes of the blaROB-1-containing isolates (33 US, 12 non-US) were compared to invasive ST-3587 isolates lacking blaROB-1 (10 US, 1 non-US). Phylogenetic analysis based on 23,286 polymorphic positions (Figure 2, Supplemental Tables 4 & 5) showed that the 11 US isolates with the blaROB-1 gene and the gyrA T91I mutation (allele 242) formed a single subclade (bootstrap = 99%) within a larger clade of blaROB-1-containing isolates (bootstrap = 100%). Outside of the dual-resistant subclade, the US isolates were intermixed with isolates from Mexico, Canada, and Europe (Figure 2). One isolate in the blaROB-1-containing clade did not contain the blaROB-1 gene; this US isolate was β-lactamase-negative and collected in 2017. All 45 blaROB-1 genes were identical to the H. influenzae reference sequence and located between the NEIS0803 and NEIS0807 loci, which are both present in the β-lactamase negative ST-3587 genomes, indicating chromosomal integration. Additionally, all isolates within this blaROB-1-containing clade contained a mosaic penA gene with 5 amino acid mutations (A510V, N512Y, I515V, H541N, I566V), which are associated with intermediate penicillin susceptibility [10], as did five other ST-3587 isolates outside of the clade.
Figure 2.
Phylogeny of Neisseria meningitidis isolates containing blaROB-1 and others with the same sequence types (ST-3587, 15379 or 13034) available in the PubMLST database as of April 21, 2020, based on a core genome alignment with recombinant regions removed. Isolates are labeled with the year and country of collection. Colored bars to the right of the phylogeny indicate the presence of antimicrobial resistance markers: penA mosaic allele (yellow), blaROB-1 (red), and gyrA T91I (blue). The scale bar is 10−5 substitutions per site. Branches with bootstrap percentages over 70% are labeled. The tree is rooted on a ST-23 isolate collected in the USA in 2003.
The phylogenetic analysis of blaROB-1-containing isolates identified a 2,115 bp recombination event in the ancestor of the dual-resistant subclade that included the entire gyrA gene and introduced the T91I and T193A mutations associated with reduced susceptibility to ciprofloxacin. The recombination introduced 120 single-nucleotide polymorphisms, replacing gyrA-QRDR allele 2 with allele 242. In the PubMLST database, allele 242 was only present in three N. lactamica genomes.
PubMLST contained 63 N. meningitidis genomes with a gyrA T91 mutation and MLST results; these were collected in 2011–2020 from 14 countries (Supplemental Table 6). Only one genome had a T91F mutation [26]. Similar to the US isolates, the 62 T91I genomes in PubMLST were diverse, with 11 different CCs and all six common invasive serogroups plus NmNG detected (Supplemental Table 6). Nearly half of the T91I genomes belonged to CC4281 with diverse serogroups (NmB, NmC, NmW, NmY) and were collected predominantly from China. The one CC23 isolate was NmY with a different molecular profile than the β-lactamase positive strains (P1.5–1,2–2: F5–8: ST-23: CC23).
Tracing the genetic origin of gyrA alleles
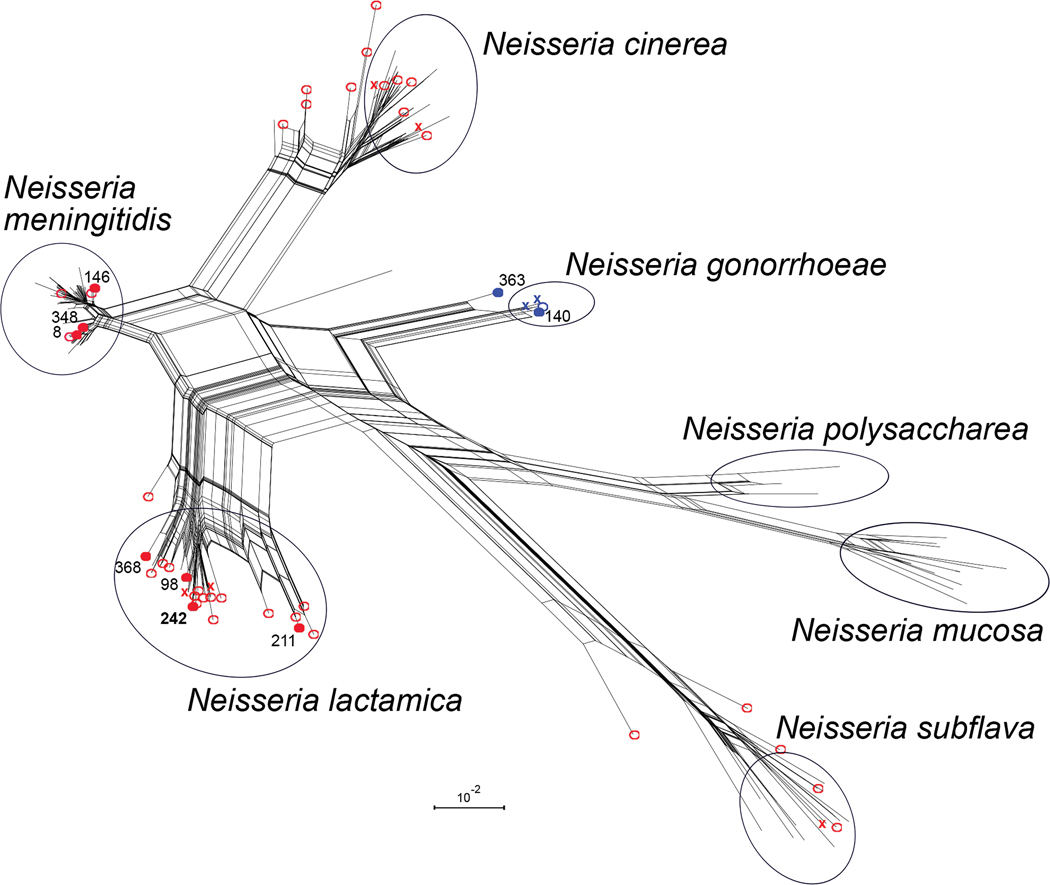
The T91-mutant gyrA-QRDR allele sequences from invasive US isolates, including allele 242 from the dual-resistant isolates, were compared to all gyrA-QRDR sequences from seven Neisseria species in the PubMLST database (Figure 3). For each Neisseria species examined, the most common gyrA-QRDR alleles clustered together based on sequence similarity; N. meningitidis alleles containing T91I or T91F clustered with four other species: N. lactamica, N. gonorrhoeae, N. cinerea, and N. subflava (Figure 3; colored open circles). The 2011–20 US isolates included 7 unique T91I gyrA-QRDR alleles; three clustered with the most common N. meningitidis alleles and four clustered with the most common N. lactamica alleles (Figure 3; filled red circles). The two T91F/D95G alleles from US isolates belonged to the N. gonorrhoeae cluster: allele 140 was found in 1,552 of the 4,354 N. gonorrhoeae isolates in PubMLST and allele 363 was similar but not identical to the N. gonorrhoeae alleles.
Figure 3.
Neighbor-net of 177 gyrA gene fragments containing the QRDR. Alleles with ciprofloxacin resistance-associated mutations are marked in blue (T91F) or red (T91I). Filled circles indicate T91F and T91I alleles that were present among the 2,097 invasive meningococcal isolates collected in the United States between January 1st, 2011 and February 14th, 2020. Open circles indicate additional T91F and T91I alleles from 18,249 N. meningitidis isolates in the PubMLST database. Colored “X”s indicate T91F and T91I alleles that were only identified in isolates from other species in the PubMLST database: 4,354 N. gonorrhoeae, 363 N. lactamica, 39 N. cinerea, 33 N. subflava, 22 N. mucosa, and 21 N. polysaccharea. Allele numbers detected in US isolates are written adjacent to filled circles, with allele 242 from blaROB-1-containing isolates in bold. Black circles indicate clusters in the network that contain all moderate frequency alleles (>1%) for the named species. Scale bar represents 1% sequence divergence and only sequences present in the PubMLST database, as of April 21, 2020, are shown.
DISCUSSION
Widespread resistance to either penicillin or ciprofloxacin could have important implications for treatment and prevention of meningococcal disease. Ciprofloxacin or penicillin resistance in N. meningitidis was uncommon in the United States prior to 2012 [6]. However, reports of ciprofloxacin resistance and intermediate susceptibility to penicillin around the world demonstrate that reduced susceptibility can develop in N. meningitidis populations. Here, we report 45 US IMD cases during 2011–20 that were caused by penicillin-resistant and/or ciprofloxacin-resistant N. meningitidis, including 11 cases due to penicillin- and ciprofloxacin-resistant N. meningitidis isolates; all dual-resistant meningococcal isolates were serogroup Y, a serogroup that is vaccine preventable and accounted for 14.6% of U.S. IMD cases in 2018 [32]. While single resistant IMD isolates were detected in almost every year assessed, the 11 dual-resistant isolates were only detected in a 14-month period beginning in January 2019, from a diverse array of states with no known epidemiological links. Altogether, the data indicate that the recent increase in cases caused by ciprofloxacin- and penicillin-resistant N. meningitidis is due to the emergence of a blaROB-1-containing meningococcal strain within the United States.
Other than the 11 dual-resistant isolates, ciprofloxacin resistance in meningococcal cases remains uncommon in the United States. Consistent with previous US trends, only a few ciprofloxacin-resistant isolates that lacked a β-lactamase were identified each year of this study, indicating that acquisition of ciprofloxacin resistance remains an uncommon event in N. meningitidis [6, 13]. In contrast to the closely related dual-resistant isolates, the ciprofloxacin-resistant isolates were from diverse genetic lineages, suggesting that the increased ciprofloxacin resistance observed in 2019–20 was specifically caused by the expansion of the β-lactamase positive ST-3587 (CC23) strain that acquired ciprofloxacin resistance from N. lactamica. Notably, dual-resistant ST-3587 isolates were reported from El Salvador, in Central America, while this manuscript was under review [21]; a phylogenetic comparison demonstrated that they belong to a single dual-resistant subclade with the US isolates and are likely part of this emerging strain (data not shown). In addition to ST-3587, expansion of ciprofloxacin resistance within a specific CC had only been previously observed in serogroup B and C strains of CC4821 [17, 42]. Overall, the ciprofloxacin-resistant isolates analyzed in this study developed resistance from either mutation events or interspecies recombination from other Neisseria species, which is consistent with previous reports [13, 25, 26].
Even though β-lactamase acquisition by N. meningitidis has been rare historically, this analysis identified 45 phylogenetically-related isolates with the blaROB-1 gene that were collected from a total of 7 countries, including the United States. However, this analysis is limited to the data released through PubMLST, which may not receive a representative dataset from submitter countries. All blaROB-1-containing isolates also contained an identical blaROB-1 sequence, indicating expansion of a single strain that possibly acquired the gene from H. influenzae. However, novel acquisition of β-lactamases remains rare in N. meningitidis; no blaROB-1 sequences were detected outside ST-3587 and the blaTEM-1 gene was rare among N. meningitidis isolates.
By screening the genomes of invasive US isolates for the blaROB-1 gene or a T91 mutation in gyrA, this analysis accurately predicted penicillin- and ciprofloxacin-resistant isolates. The expanding genomic surveillance programs in many countries provide an opportunity for rapid identification of isolates with known mechanisms of antimicrobial resistance. However, genomic screens will be unable to detect novel resistance mechanisms and may have reduced accuracy when predicting susceptibility phenotypes from complex resistance mechanisms such as the mosaic penA alleles. Thus, a combination of phenotypic susceptibility studies in addition to genomic screens will be necessary to effectively monitor future trends.
Acquisition of blaROB-1 and ciprofloxacin resistance within ST-3587 in US IMD cases has emphasized the importance of antimicrobial resistance surveillance for N meningitidis, particularly with cases detected in both Central and North America. Susceptibility to penicillins should be confirmed prior to use of penicillin or ampicillin for meningococcal disease treatment [20]. In addition, clinicians and public health staff in areas that have experienced IMD cases caused by ciprofloxacin-resistant strains within the past 1–2 years should consider performing AST on meningococcal isolates to inform prophylaxis decisions; however, AST should not delay initiation of prophylaxis [20]. Accurate and timely antimicrobial resistance surveillance, using AST and/or WGS, is essential to understanding the prevalence and spread of resistance and informing future meningococcal disease treatment and prophylaxis recommendations.
Supplementary Material
Supplemental Table 1. Accession numbers and descriptions for genome sequences described in Figure 1, Figure 2, and Table 1. The 2003 isolate from the United States (M10868) was included as a reference for genome alignment and phylogenetic rooting.
Supplemental Table 2: List of ST-3587 isolates identified on PubMLST, on the query date April 21, 2020, including all Neisseria meningitidis isolates with the blaROB-1 gene.
Supplemental Table 3: List of Neisseria meningitidis isolates on PubMLST with the blaTEM-1 gene on the query date April 21, 2020.
Supplemental Table 4: Single Nucleotide Polymorphism (SNP) distances between genomes based on core genome alignment with 23,286 polymorphic positions. Dual-resistant isolates are highlighted in green. Supp Table 5 shows SNP distances after filtering of recombinant regions.
Supplemental Table 5: Single Nucleotide Polymorphism (SNP) distances between genomes based on recombination-masked core genome alignment with 929 polymorphic positions. Dual-resistant isolates are highlighted in green. Supp Table 4 shows SNP distances in core genome alignment before filtering of recombinant regions.
Supplemental Table 6: Counts of Neisseria meningitidis isolates on PubMLST, on the query date April 21, 2020, with the gyrA T91I mutation collected 2011–2020, tabulated according to country and clonal complex.
Key Points.
Ciprofloxacin- and penicillin-resistant N. meningitidis caused 11 cases of invasive meningococcal disease in the US during a 14-month period. The dual-resistant N. meningitidis were not detected before 2019 and belong to a single genetic lineage, suggesting changing antimicrobial resistance patterns.
ACKNOWLEDGEMENTS
We would like the acknowledge Veronica Pinell-McNamara, Keegan Rudmann, and members of the Bacterial Meningitis Laboratory, CDC; Amanda Metz, Emily Spence-Davizon, Karen Xavier, Colorado Department of Public Health and Environment; Scott Pritchard, Florida Department of Health; Chelsea Raybern, Kansas Department of Health and Environment; Mink Antwi, Paula Del Rosso, Marie Dorisinville, Don Weiss, Jessica Sell, Jennifer Rakeman-Cagno, Ulrike Siemetzki-Kapoor, New York City Department of Health and Mental Hygiene; Lisa Dettinger, Venkata Vapchedu, Pennsylvania Department of Health Bureau of Laboratories; John Faherty Philadelphia Department of Health; Greg Leos, Texas Department of State Health Services; vaccine-preventable diseases surveillance staff of the Connecticut, Hawaii, Illinois, Indiana, Louisiana, Massachusetts, Michigan, Minnesota, Mississippi, Montana, Nevada, Oklahoma, South Carolina, Tennessee, Utah, West Virginia, and Wisconsin state health departments. This publication made use of the PubMLST website (http://pubmlst.org/).
FUNDING
This work was supported by the Centers for Disease Control and Prevention
Footnotes
DISCLAIMER
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Authors in the Antimicrobial-resistant Neisseria meningitidis team
Nirmala Dhungana, California Department of Public Health; Ryan Gabrio-Brannon, California Department of Public Health; Jennifer Kyle, California Department of Public Health; Brittany Martin, California Department of Public Health; Joseph Campos, Children’s National Hospital Washington, DC; Benjamin Hanisch, Children’s National Hospital Washington, DC; Gillian Taormina, Children’s National Hospital Washington, DC; Meghan Barnes, Colorado Department of Public Health and Environment; Ashley Moore, Georgia Department of Public Health; Catherine E. Dominguez, Maryland Department of Health; Kristy Lunquest, Maryland Department of Health; Ami A. Patel, Maryland Department of Health; David Torpey, Maryland Department of Health; Susan Hannagan, New Jersey Department of Health; Page Keating, New York City Department of Health and Mental Hygiene; Sandy Li, New York City Department of Health and Mental Hygiene; Justin Albertson, North Carolina Department of Health and Human Services; Wayne Fleming, Bureau of Epidemiology Pennsylvania Department of Health; Christina Russell, Bureau of Laboratories Pennsylvania Department of Health; Kara Reid, Philadelphia Department of Health; Kelsey Sanders, Texas Department of State Health Services; Chas DeBolt, Washington State Department of Health; Nicholas Graff, Washington State Department of Health; Esther Lam, Washington State Department of Health.
CONFLICT OF INTEREST
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.AAP Committee on Infectious Diseases. Red Book (2018), 2018. [Google Scholar]
- 2.Parikh SR, Lucidarme J, Bingham C, et al. Meningococcal B Vaccine Failure With a Penicillin-Resistant Strain in a Young Adult on Long-Term Eculizumab. Pediatrics 2017; 140(3). [DOI] [PubMed] [Google Scholar]
- 3.Bozio CH, Isenhour C, McNamara LA. Characteristics of and meningococcal disease prevention strategies for commercially insured persons receiving eculizumab in the United States. PLoS One 2020; 15(11): e0241989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crew PE, McNamara L, Waldron PE, McCulley L, Christopher Jones S, Bersoff-Matcha SJ. Antibiotic prophylaxis in vaccinated eculizumab recipients who developed meningococcal disease. J Infect 2020; 80(3): 350–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNamara LA, Topaz N, Wang X, Hariri S, Fox L, MacNeil JR. High Risk for Invasive Meningococcal Disease Among Patients Receiving Eculizumab (Soliris) Despite Receipt of Meningococcal Vaccine. MMWR Morbidity and mortality weekly report 2017; 66(27): 734–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harcourt BH, Anderson RD, Wu HM, et al. Population-Based Surveillance of Neisseria meningitidis Antimicrobial Resistance in the United States. Open Forum Infect Dis 2015; 2(3): ofv117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vacca P, Fazio C, Neri A, Ambrosio L, Palmieri A, Stefanelli P. Neisseria meningitidis Antimicrobial Resistance in Italy, 2006 to 2016. Antimicrob Agents Chemother 2018; 62(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bijlsma MW, Bekker V, Brouwer MC, Spanjaard L, van de Beek D, van der Ende A. Epidemiology of invasive meningococcal disease in the Netherlands, 1960–2012: an analysis of national surveillance data. Lancet Infect Dis 2014; 14(9): 805–12. [DOI] [PubMed] [Google Scholar]
- 9.Bertrand S, Carion F, Wintjens R, Mathys V, Vanhoof R. Evolutionary changes in antimicrobial resistance of invasive Neisseria meningitidis isolates in Belgium from 2000 to 2010: increasing prevalence of penicillin nonsusceptibility. Antimicrob Agents Chemother 2012; 56(5): 2268–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taha MK, Vazquez JA, Hong E, et al. Target gene sequencing to characterize the penicillin G susceptibility of Neisseria meningitidis. Antimicrob Agents Chemother 2007; 51(8): 2784–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong E, Deghmane AE, Taha MK. Acquisition of Beta-Lactamase by Neisseria meningitidis through Possible Horizontal Gene Transfer. Antimicrob Agents Chemother 2018; 62(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsang RSW, Ahmad T, Jamieson FB, Tyrrell GJ. WGS analysis of a penicillin-resistant Neisseria meningitidis strain containing a chromosomal ROB-1 β-lactamase gene. Journal of Antimicrobial Chemotherapy 2018: dky391-dky. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu HM, Harcourt BH, Hatcher CP, et al. Emergence of ciprofloxacin-resistant Neisseria meningitidis in North America. N Engl J Med 2009; 360(9): 886–92. [DOI] [PubMed] [Google Scholar]
- 14.Tsang RS, Law DK, Deng S, Hoang L. Ciprofloxacin-resistant Neisseria meningitidis in Canada: likely imported strains. Can J Microbiol 2017; 63(3): 265–8. [DOI] [PubMed] [Google Scholar]
- 15.Gorla MC, Pinhata JMW, Dias UJ, de Moraes C, Lemos AP. Surveillance of antimicrobial resistance in Neisseria meningitidis strains isolated from invasive cases in Brazil from 2009 to 2016. J Med Microbiol 2018; 67(6): 750–6. [DOI] [PubMed] [Google Scholar]
- 16.Bukovski S, Vacca P, Anselmo A, et al. Molecular characterization of a collection of Neisseria meningitidis isolates from Croatia, June 2009 to January 2014. J Med Microbiol 2016; 65(9): 1013–9. [DOI] [PubMed] [Google Scholar]
- 17.Chen M, Guo Q, Wang Y, et al. Shifts in the Antibiotic Susceptibility, Serogroups, and Clonal Complexes of Neisseria meningitidis in Shanghai, China: A Time Trend Analysis of the Pre-Quinolone and Quinolone Eras. PLoS Med 2015; 12(6): e1001838; discussion e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu B, Fan Y, Xu Z, et al. Genetic diversity and clonal characteristics of ciprofloxacin-resistant meningococcal strains in China. J Med Microbiol 2014; 63(Pt 11): 1411–8. [DOI] [PubMed] [Google Scholar]
- 19.Taormina G, Campos J, Sweitzer J, Retchless AC, Lunquest K, McNamara LA, Reese N, Karlsson M, Hanisch B. β-lactamase-producing, ciprofloxacin-resistant Neisseria meningitidis isolated from a 5-month-old male in the United States. Journal of Pediatric Infectious Diseases Society In press. [DOI] [PubMed] [Google Scholar]
- 20.McNamara LA, Potts C, Blain AE, et al. Detection of Ciprofloxacin-Resistant, β-Lactamase-Producing Neisseria meningitidis Serogroup Y Isolates - United States, 2019–2020. MMWR Morbidity and mortality weekly report 2020; 69(24): 735–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marin JEO, Villatoro E, Luna MJ, et al. Emergence of MDR invasive Neisseria meningitidis in El Salvador, 2017–19. J Antimicrob Chemother 2021. [DOI] [PubMed] [Google Scholar]
- 22.Hong E, Thulin Hedberg S, Abad R, et al. Target gene sequencing to define the susceptibility of Neisseria meningitidis to ciprofloxacin. Antimicrob Agents Chemother 2013; 57(4): 1961–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorla MC, Cassiolato AP, Pinhata JMW, et al. Emergence of resistance to ciprofloxacin in Neisseria meningitidis in Brazil. J Med Microbiol 2018; 67(3): 286–8. [DOI] [PubMed] [Google Scholar]
- 24.Sorhouet-Pereira C, Efron A, Gagetti P, et al. Phenotypic and genotypic characteristics of Neisseria meningitidis disease-causing strains in Argentina, 2010. PLoS One 2013; 8(3): e58065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen M, Zhang C, Zhang X, Chen M. Meningococcal Quinolone Resistance Originated from Several Commensal Neisseria Species. Antimicrob Agents Chemother 2020; 64(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brooks A, Lucidarme J, Campbell H, et al. Detection of the United States Neisseria meningitidis urethritis clade in the United Kingdom, August and December 2019 - emergence of multiple antibiotic resistance calls for vigilance. Euro Surveill 2020; 25(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castanheira M, Deshpande LM, Jones RN, Farrell DJ. Evaluation of quinolone resistance-determining region mutations and efflux pump expression in Neisseria meningitidis resistant to fluoroquinolones. Diagn Microbiol Infect Dis 2012; 72(3): 263–6. [DOI] [PubMed] [Google Scholar]
- 28.McNamara LA, Potts C, Blain AE, et al. Detection of Ciprofloxacin-Resistant, beta-Lactamase-Producing Neisseria meningitidis Serogroup Y Isolates - United States, 2019–2020. MMWR Morbidity and mortality weekly report 2020; 69(24): 735–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deghmane AE, Hong E, Taha MK. Emergence of meningococci with reduced susceptibility to third-generation cephalosporins. J Antimicrob Chemother 2017; 72(1): 95–8. [DOI] [PubMed] [Google Scholar]
- 30.Langley G, Schaffner W, Farley MM, et al. Twenty Years of Active Bacterial Core Surveillance. Emerging infectious diseases 2015; 21(9): 1520–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang HY, Vuong J, Hu F, et al. Distribution of Neisseria meningitidis serogroup b (NmB) vaccine antigens in meningococcal disease causing isolates in the United States during 2009–2014, prior to NmB vaccine licensure. J Infect 2019; 79(5): 426–34. [DOI] [PubMed] [Google Scholar]
- 32.Enhanced Meningococcal Disease Surveillance Report. Available at: https://www.cdc.gov/meningococcal/downloads/NCIRD-EMS-Report-2018.pdf.
- 33.Potts CC, Joseph SJ, Chang HY, et al. Population structure of invasive Neisseria meningitidis in the United States, 2011–15. J Infect 2018; 77(5): 427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome open research 2018; 3: 124-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cock PJ, Antao T, Chang JT, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics (Oxford, England) 2009; 25(11): 1422–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI supplement M100. Wayne, PA. Clinical and Laboratory Standards Insitute; 2020. [Google Scholar]
- 37.CLSI. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 11th ed. M07. Clinical and Laboratory Standards Institute; 2018. [Google Scholar]
- 38.Seemann T. Snippy: Rapid haploid variant calling and core SNP phylogeny. [Google Scholar]
- 39.Croucher NJ, Page AJ, Connor TR, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic acids research 2015; 43(3): e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics (Oxford, England) 2019; 35(21): 4453–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bryant D, Moulton V. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Molecular biology and evolution 2004; 21(2): 255–65. [DOI] [PubMed] [Google Scholar]
- 42.Lucidarme J, Zhu B, Xu L, et al. Genomic analysis of the meningococcal ST-4821 complex-Western clade, potential sexual transmission and predicted antibiotic susceptibility and vaccine coverage. PLoS One 20; 12): e0243426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Accession numbers and descriptions for genome sequences described in Figure 1, Figure 2, and Table 1. The 2003 isolate from the United States (M10868) was included as a reference for genome alignment and phylogenetic rooting.
Supplemental Table 2: List of ST-3587 isolates identified on PubMLST, on the query date April 21, 2020, including all Neisseria meningitidis isolates with the blaROB-1 gene.
Supplemental Table 3: List of Neisseria meningitidis isolates on PubMLST with the blaTEM-1 gene on the query date April 21, 2020.
Supplemental Table 4: Single Nucleotide Polymorphism (SNP) distances between genomes based on core genome alignment with 23,286 polymorphic positions. Dual-resistant isolates are highlighted in green. Supp Table 5 shows SNP distances after filtering of recombinant regions.
Supplemental Table 5: Single Nucleotide Polymorphism (SNP) distances between genomes based on recombination-masked core genome alignment with 929 polymorphic positions. Dual-resistant isolates are highlighted in green. Supp Table 4 shows SNP distances in core genome alignment before filtering of recombinant regions.
Supplemental Table 6: Counts of Neisseria meningitidis isolates on PubMLST, on the query date April 21, 2020, with the gyrA T91I mutation collected 2011–2020, tabulated according to country and clonal complex.