Abstract
Despite the wealth of knowledge gained on intrinsically disordered proteins (IDPs) since their discovery, there are several aspects that remain unexplored and hence, poorly understood. A living cell is a complex adaptive system that can be described as a wetware – a metaphor used to describe the cell as a computer comprising both hardware and software and attuned to logic gates – capable of ‘making’ decisions. In this focused review, we discuss how IDPs, as critical components of the wetware, influence cell-fate decisions by wiring protein interaction networks to keep them minimally frustrated. Since IDPs lie between order and chaos, we explore the possibility that they can be modeled as attractors. Further, we discuss how conformational dynamics of IDPs manifests itself as conformational noise, which can potentially amplify transcriptional noise to stochastically switch cellular phenotypes. Finally, we explore the potential role of IDPs in prebiotic evolution, in forming proteinaceous membrane-less organelles, in the origin of multicellularity, and protein conformation-based transgenerational inheritance of acquired characteristics. Together, these ideas provide a new conceptual framework to discern how IDPs may perform critical biological functions despite lack of structure.
Keywords: Intrinsically disordered proteins, dark matter, dark proteome, protein universe, protein interaction networks, conformational noise, prebiotic evolution, protein conformation-based inheritance, Lorentz attractor, weak Pinsker conjecture
Graphical Abstract
“As far as the laws of mathematics refer to reality, they are not certain; and as far as they are certain, they do not refer to reality.”
Albert Einstein (Address to the Prussian Academy of Sciences, 1921)
1. INTRODUCTION
1.1. Dark matter in biology
The plethora of proteins from all the extinct and extant organisms, is defined as the protein ‘universe’, a concept introduced by István Ladunga in the 1990s.1 While a significant fraction of the protein universe is well studied, and the structures of thousands of proteins have been determined, the nature of remainder of this universe remains poorly understood. The latter fraction of proteins with unknown structures is referred to as the ‘dark’ proteome, being a constituent of the biological dark matter.2 Indeed, significant fractions of the proteomes of archaea and bacteria, and almost half of the eukaryotic and viral proteomes, are estimated to be dark.3 Like the dark matter in the physical universe, the dark matter in biology is not easily detectable by traditional approaches of structural biology. However, the dark matter is essential to life, is involved in performing functions that are perceptible and complement the functions performed by their ordered (visible) counterparts.
Several computational studies on the molecular evolution of the protein universe3–6 suggest that the protein universe is expanding, and proteins with common ancestors they shared billions of years ago, are diverging in their molecular composition.7 Further, they also reveal that within the dark proteome, even the foldable domains have specific features. For example, they are generally shorter in length and have unique distributions of hydrophobic residues when compared to known domain families. More importantly, the studies indicate that the constituents of the dark proteome exhibit a higher propensity for intrinsic disorder.6 Indeed, intrinsically disordered proteins (IDPs) (and intrinsically disordered protein regions within ordered proteins, or IDPRs) that lack 3-Dimensional structure under physiological conditions,8–10 comprise a large fraction of the dark matter in the protein universe.4,8,11–16
In line with these observations, bioinformatics analyses reveal that the proteomes of organisms across kingdoms including viral proteomes, are considerably enriched in IDPs/IDPRs.8,11–16 Furthermore, the length and frequency of IDPRs appear to positively correlate with organismal complexity. For example, ~33% of eukaryotic proteins contain long IDPRs, and ~10% of proteins in eukaryotes are fully disordered.17 In other studies, noticeably broader penetrance of intrinsic disorder into the various proteomes was indicated, where fractions of disordered residues range from 12 to 21% and from ~18% to ~28% in archaea and bacteria increasing to 20–40% and 35–45% in viruses and eukaryotes, respectively.11 Whole genome analyses from several organisms from bacteria to mammals revealed that their genomes encode proteins which are predicted to be highly disordered.18 Furthermore, it was recently shown that intrinsic disorder is not only correlated with organismal complexity but also appears to contribute to clade-specific functions.19
1.2. IDPs and their interactions
IDPs lack rigid structure and exist as diverse conformational ensembles. These ensembles are malleable (dynamic), which facilitates the interactions of IDPs with multiple partners in response to the changing environment.20–21 All of these interactions form the interaction networks (PINs) that are scale-free,22–23 and channelize the flow of information within the cell. Consistent with this role, PIN organization and properties are evolutionarily conserved.24 IDPs occupy hub positions in the PINs25–30 and play important roles in transcription, translation, signal transduction,13,31–36, cell cycle regulation,37–38 circadian rhythmicity,39–42 and in regulating phenotypic plasticity.11,43–44 Furthermore, IDPs tend to engage in “promiscuous” interactions which can lead to pathological states.45–48 Therefore it is unsurprising that, overexpression of IDPs has emerged as a major cause for several chronic human diseases, e.g. diabetes, cancer and neurodegenerative illnesses.32,49–56
With regard to the molecular mechanisms underlying the functions of IDPs, it is generally held that at least some IDPs transition from disordered conformational ensembles to (at least partially) ordered structures upon interacting with their targets, a process referred to as “coupled folding and binding”.57 Possible mechanism that underlie this phenomenon58 include the “induced fit” hypothesis, which suggests that folding occurs after binding to the target, and the “conformational selection” hypothesis which envisages that potential conformations pre-exist in the unbound state and the target selects the most appropriate one. However, the binding mechanisms of many IDPs possibly involve some combination of the above two models. On the other hand, many IDPs retain significant intrinsic disorder in their bound states,59–65 with some of them being capable of forming ultrahigh-affinity but highly disordered complexes (see below). Finally, some IDPs have been observed to stochastically switch conformational states while remaining intrinsically disordered.66
Together, these observations suggest that some IDPs are on the verge of structural instability and can readily adopt functional conformations to become biologically active. This is conceptually analogous to the conformational (fold) switching events seen in some metastable folded proteins (referred to as ‘metamorphic’ proteins) subsequent to mutagenesis or environmental perturbations, giving rise to new functions (Fig. 1).67 On the other hand, some IDPs remain mostly disordered even when they interact with other proteins.60,68–70 In fact, recent computational studies seem to indicate that less compact protein structures are essential for biological activity due to their capacity of weak yet dynamic interactions with the target proteins.71–72
Fig. 1:
Proteins on the brink of stability can undergo a continuum of order/disorder transitions. (A) Examples of transitions from top left to bottom right: Transition between the extended and collapsed disordered states of prostate associated Gene 4 (PAGE4), modulated by phosphorylation; disorder-to-order transition of 4E-BP2 induced by phosphorylation; order-to-order fold switching between GA98 and GB98, triggered by single amino acid changes or ligand binding. In contrast, stable proteins such as subtilisin (shown in dark blue) do not undergo such changes. (B) Approximate energy well diagrams for each protein from PAGE4 (top) to subtilisin (bottom). Reproduced with permission from Kulkarni et al. Structural metamorphism and polymorphism in proteins on the brink of thermodynamic stability. Protein Sci. 2018 Sep;27(9):1557–1567. Published by Wiley © 2018 The Protein Society.253
Notwithstanding the multiplicity of mechanisms, both coupled folding and binding73 and the degree of disorder in the “fuzzy complexes”,62 constitute the majority of the atomistic mechanisms underlying the interactions promoted by IDPs. Indeed, IDPs can interact with very high affinity (Kd =picomolar range)74 while remaining disordered and maintaining their flexibility and dynamic natures.74–75 Interactions of the above kind were observed in IDPs with regions of considerable opposite net charges in their sequences. Protein-protein interaction between these IDPs are governed by the complementarily charged, unstructured regions (long-range electrostatic attraction) rather than specific inter-residue contacts or binding sites within folded domains.74 Since eukaryotic proteomes are enriched in proteins with high net charge,32,76 it is possible that such an interaction mechanism is quite abundant in nature.74 Consistent with this reasoning, it is now increasingly evident that IDPs are key functional constituents of proteinaceous membrane-less organelles (PMLOs). The opposite charge interactions are likely crucial in IDP mediated liquid-liquid phase separation forming these organellies.77–82 1.3. Scope of this Focused Review
Despite this wealth of knowledge regarding IDPs, since they were first discovered >20 years ago,83 as well as the mechanisms by which they accomplish their biological functions in the absence of apparent structure, there are several unique aspects that remain relatively unexplored and poorly understood. In this Focused Review, we examine IDPs from a dynamical systems perspective, since this unique aspect of the IDPs though acknowledged, has not been investigated in sufficient detail. First, we examine how IDPs, as complex systems that self-organize, may influence cell-fate decisions by wiring PINs to drive the system toward a stable attractor. Next, we discuss how conformational dynamics of IDPs could potentially manifest itself as conformational noise, which in turn can amplify transcriptional noise resulting in PIN rewiring, phenotypic plasticity, and adaptive evolution. Finally, we highlight the potential role of IDPs in the origin and evolution of life from a primordial, Chemoton-like entity,84–85 to the last universal common ancestor (LUCA), and eventually through the major evolutionary transitions,86 to multicellular forms and beyond. In concluding, we note that a deeper understanding of the IDPs that likely pre-dated life as we know it, could shed new light on how these constituents of the dark matter of biology evolved as critical components of the wetware and guide cellular decision making.
2. IDPs, SELF-ORGANIZATION AND SCALE-FREE NETWORKS
2.1. Self-organization
Living systems, and many non-living systems, fall in the category of complex systems that follow two primary characteristics 1) ability to self-organize, and 2) manifestation of nonlinear dynamics.87 Self-organization is defined as a process by which global order emerges from local interactions between the components of a system that is initially disordered or chaotic. Such systems are open systems that reside far from thermodynamic equilibrium.88 Furthermore, self-organized systems exhibit emergent properties that are different from those of their individual components and are collectively governed by the interactions among the constituent parts within a global topology. Emergent properties are not predictable based on the behavior of the individual components unless their mutual interactions are considered as well. Further, systems with emergent properties respond to external perturbations in non-linear ways, leading to complex coordinated dynamics.89
2.2. Self-organizing properties of scale-free networks
Previously, it was generally held that biological networks follow a random architecture, where the nodes interact with one another with a fixed probability that is independent of the other edges in the network.90 The modern concept of scale-free networks was introduced by Barabasi et al.22–23,91 who discovered that the degree distribution (probability P(k) of a random network node to be connected to k other nodes, degree being the number of connections) of biological networks often follow a power law. Specifically, P(k) ~ k−γ (exponent γ is a constant), where most of the network connectivity is concentrated among a few of the nodes (hubs), while the rest of the nodes have very low degree. Furthermore, these insightful studies also revealed that scale-free networks are resilient and remain functional even in response to the failure of random nodes. However, scale-free networks are vulnerable and can become suboptimal in response to failure of hubs.92
As self-organizing systems, scale-free networks are comprised of self-repeating patterns (fractals) across diverse length scales.93 Curiously, the same principles are also applicable to individual proteins since the spatiotemporal structural organization can be envisaged as an intra-protein network, wherein the individual amino acids participate in interactions that are transient or stable.94 In fact, as it was recently shown that a protein molecule may be visualized as a complex system existing as a dynamic, multilevel network of networks analogous to a nesting doll (Matryoshka). In this metaphor, one can think of the different regions of the polypeptide chain as forming secondary structure elements through local networks of inter-residue contacts, as the lowest level or organization. Interactions between the elements of secondary structure would give rise to the next level to form foldons, non-foldons, and unfoldons etc. These interactions give rise to higher levels of the network namely proteins domains and finally, a network including inter-domain and between domains interactions to result in second-tier networks. Therefore, from this Russian nesting doll perspective, a typical PIN represents a highly dynamic and multilevel network of networks.94
3. IDPs, CONFORMATIONAL NOISE, AND PHENOTYPIC SWITCHING
3.1. Conformational noise
Based on the characteristics discussed in section 2, a model was proposed95 that is known as the MRK hypothesis.96–97 The authors postulated that the conformational dynamics of IDPs may contribute to differential signaling noise in cells. The authors referred to this noise as ‘conformational noise’ to distinguish it from the well-recognized transcriptional noise. Thus, in contrast to transcriptional noise, the MRK hypothesis states that conformational noise may be defined by the randomness in the confirmations that an IDP ensemble populates. Although interconversion of IDP conformations exhibit fast exchange, covalent modifications that occur post-translationally or changes in their environment, can enable the ensemble to preferentially occupy particular conformations and/or conformational dynamics. Thus, variations in the conformational ensembles can have significantly physiological effects.98 Furthermore, conformational noise can promote promiscuous interactions of the IDPs.48 Given that, many transcription factors (TFs) are IDPs,99–102 the authors of the MRK hypothesis posited that conformational noise can be a fundamental aspect of transcriptional variation, and IDPs could potentially propagate or enhance the stochasticity of cellular response in reaction to intrinsic and/or extrinsic perturbations.
3.2. IDPs and phenotypic switching
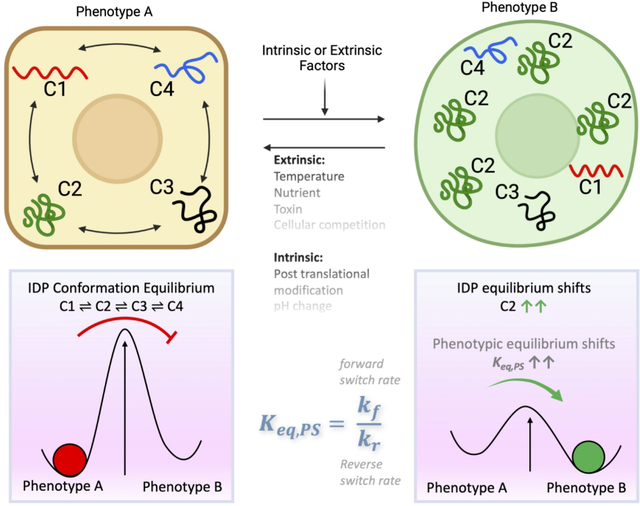
As per the MRK hypothesis, conformational noise due to stochastic IDP interactions in conjunction with intrinsic or extrinsic stimuli, would enable the system to explore the broader network configurational space. Therefore, this heuristic enables network rewiring and consequently, phenotypic switching, giving rise to phenotypic heterogeneity. Stated differently, the hypothesis implies that IDPs unmask latent network configurations to cause phenotypic switching to alternate states (Fig. 2).95 Indeed, stochastic phenotypic switching is observed during differentiation,103 somatic cell reprogramming (induced pluripotent (iPS) cell generation),104–109 and cancer cells with stem cell-like properties.110–111
Fig. 2:
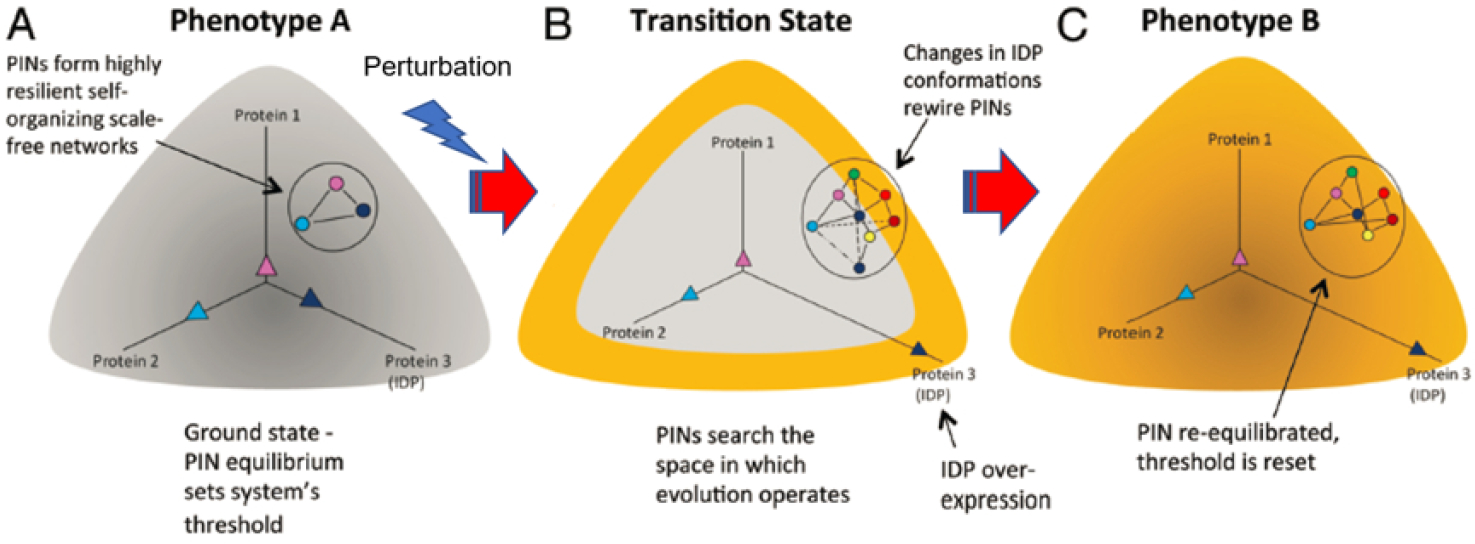
Rewiring of protein networks facilitates state-switching by activating latent pathways. (A) The state of a cell with phenotype A is depicted in grey and shows a simple protein network with three proteins (1–3), of which one is an IDP (indicated in dark blue) and expressed at different levels represented by the three vectors. This configuration represents the protein network’s ground state threshold. (B) Depicts a transition state. A perturbation causes increased IDP expression (protein 3). Overexpression of the IDP results in promiscuity and the protein network explores the network search space shown by the various dashed lines. This transition state is depicted state in yellow around the grey area. (C) The state of the cell after it has transitioned to phenotype B from phenotype A represented in yellow. A particular configuration of the protein network that increased its fitness is “selected,” which now represents the new ground state. Reproduced with permission from with permission from Mahmoudabadi et al. Intrinsically disordered proteins and conformational noise: Implications in cancer. Cell Cycle. 2013 12(1):26–3, Taylor & Francis Online.95
Of note, the MRK model implied that information encoding cellular phenotype, resides in the PIN configuration, and every cell has the potential to switch its phenotype in response to a specific input. Per this model, although the network is relatively flexible to physiological changes, it is robust in resisting non-physiological perturbations. More importantly, per the MRK model IDPs can rewire the network, unmasking latent network configurations (and consequently, alternative phenotypes) in response to stress. However, the model envisaged that the PIN can return to the normal (default) setting when the stress is relieved.
Another salient feature of this model is that information generated by network rewiring may operate across different temporal scales. Therefore, while transient information resides within the PIN, information related to slower changes (e.g., evolutionary processes), is transferred to the genome accounting for transgenerational inheritance. Alternatively, the model postulated that a mechanism similar to genetic assimilation of the acquired character as suggested by Waddington may be involved.112 Furthermore, it was postulated that the macroscopic behavior of a system such as, phenotype switching and adaptive evolution, are driven by PIN rewiring initiated by the IDPs.95–97,113
From the foregoing it follows that cell fate specification need not be only deterministic but can also be stochastic and thus account for reversible phenotypic switching as seen in epithelial-mesenchymal transition (EMT) and mesenchymal-epithelial transition (MET), the switching of a drug-sensitive cell to one that is drug resistant or vice versa108,114–117 or the malignant transformation of a normal cell and its reversal to a non-malignant phenotype.118–119 Interestingly, many transcription factors involved in EMT and/or MET,43 and in drug resistance such as paxillin120 are IDPs. A theoretical study elucidating how transformation of a normal cell into a cancer cell, and its reversal, may be actuated by modulating the levels of the oncogene c-Myc, an illustrative IDP,121 further supports many aspects of the MRK hypothesis.
The significance of IDPs in PIN rewiring warrants development of statistical or dynamical models to further elucidate such mechanisms. The complexity in developing any such model is contributed by the multiple cellular processes (e.g., protein dynamics, PTMs, intracellular diffusion, and gene expression) manifested at various spatiotemporal levels. One has to therefore come up with a suitable multiscale approach which integrates the different time and length scales using appropriate models and parameterizations. Development of such approaches has been the topic of several reviews122–123.
4. CAN IDPs BE REPRESENTED AS COMPLEX DYNAMICAL SYSTEMS?
4.1. IDPs as complex or “edge of chaos” systems
In dynamical systems theory pioneered by Poincaré124 that describes complex systems, an “attractor” may be defined as a set of values of the variables, towards which the system tends to evolve from diverse initial states (Box 1). As discussed above, the configuration of the PIN (in conjunction with the environment) defines a cell’s phenotype.95 A PIN could be represented as a dynamical system in an appropriate configuration space. It could then be thought of as starting from a context-dependent initial configuration, evolving in time due to mutual interactions and eventually settling down into an “attractor”, defined here as a stable cell phenotype.43,125 The different steady states that a PIN can potentially occupy, can be predicted by modeling its dynamics. Therefore, at steady state, the probability that the system will occupy an attractor is proportional to the stability of the PIN configuration of the attractor. Thus, a set of attractors and their probabilities of being occupied by the system, represent a high-dimensional ‘landscape’ as envisioned by Waddington in the epigenetic landscape metaphor (Fig. 3).126
Box 1.
Attractor can be described as a stable state towards which a system would tend to get ‘attracted’ in long run. Strange attractor is strange because it has fractal structure. Often, but not always, they are chaotic in nature. A chaotic attractor is very sensitive to initial conditions. It means that two very close initial conditions inside a chaotic attractor can lead to two locally diverging trajectories thus showing local instability. But once the trajectory is inside its basin of attraction then it will not depart from the attractor, which shows global stability.
Fig. 3:
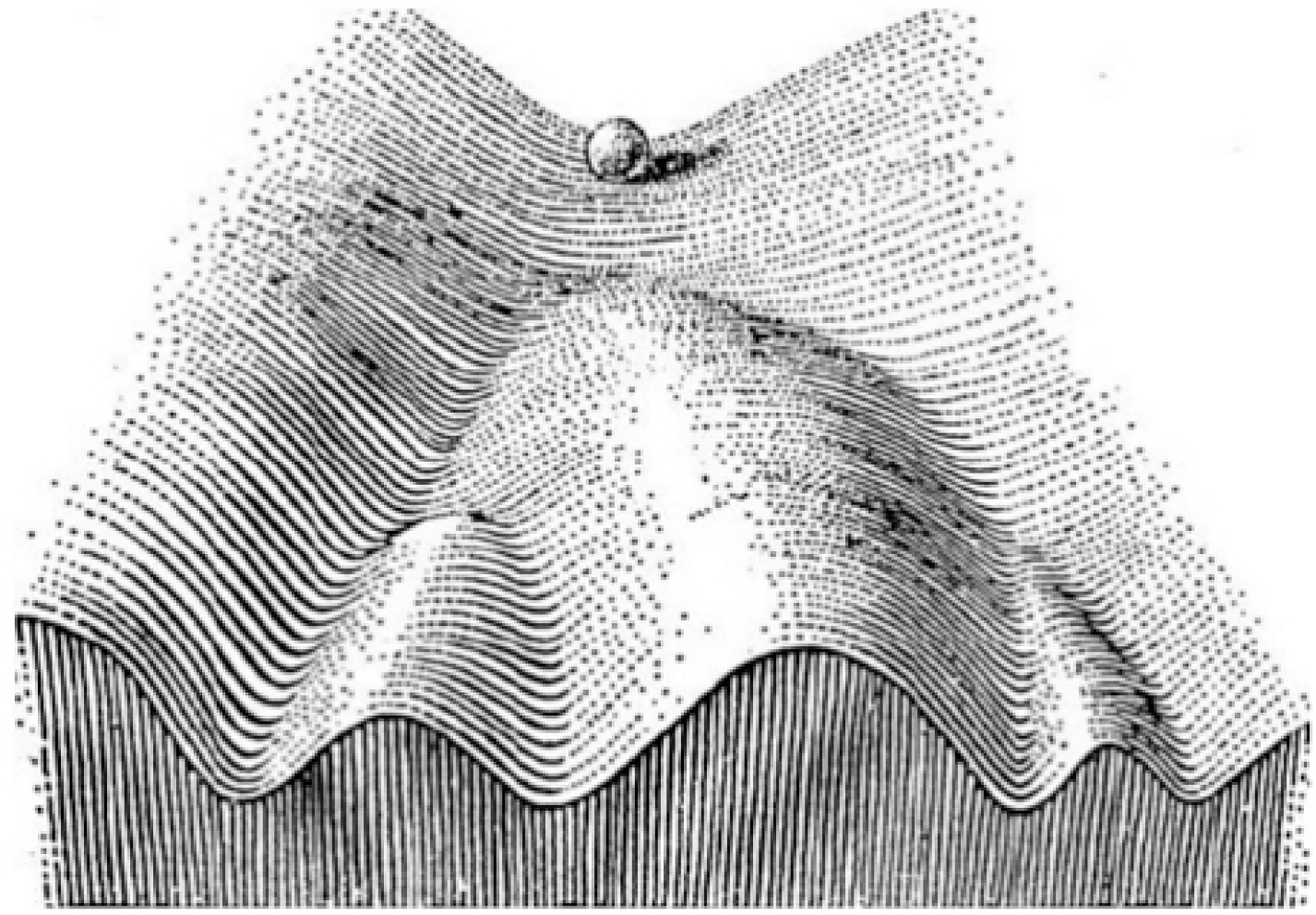
Schematic illustration of Waddington’s epigenetic landscape (adopted and from Schematic illustration of Waddington’s epigenetic landscape. Adopted from Waddington, 1957126). Reproduced with permission from Kulkarni P. Intrinsically Disordered Proteins: Insights from Poincaré, Waddington, and Lamarck. Biomolecules. 2020. 10(11):1490. MDPI Publishers.
The concept of an “attractor” has been adopted in cancer as well.127–129 Here, ‘cancer attractors’, defined as stable states of latent PIN configurations not occupied by normal cells, drive a normal cell to switch to a malignant phenotype. Access to cancer attractors can be determined by genetic or non-genetic mechanisms in which the IDPs play a crucial role, especially as occupants of key hubs in the PIN.
To elaborate further, complex systems are thought to have some of the following properties:130
They contain many nonlinearly interacting heterogeneous components Thus, the behavior of complex systems may not be defined by the sum of its parts
There is interdependency of constituents
Constitute a nested entity that encompasses multiple components (each of which can be complex systems themselves) spanning diverse scales
Is capable of emergent behavior
The system enables a dynamic exchange between order and chaos (disorder)
Can involve cooperation as well as positive (reinforcing) and negative (damping) feedbacks
Likely to have a memory (hysteresis). Previous states may influence future states.
Thus, it follows that IDPs could be considered as complex systems.131 For example, IDPs are heterogeneous (see above) with constituents that are autonomous or dependent on each other, and can interact nonlinearly. The functional misfolding may be thought of as a product of coupled competition and cooperation. The fact that the IDPs can undergo excursions between order and disorder underscores their spatiotemporal complexity. IDPs exhibit conformational changes in response to various stimuli to exhibit the ‘butterfly effect’ where minute thermodynamic perturbations might produce significant conformational and eventually functional changes (Fig. 4). Finally, IDPs display emergent behavior. Under certain conditions (e.g. post-translational modifications or contact with other proteins) they can self-organize by undergoing disorder-to-order transitions.131
Fig. 4. IDPs, attractors and stable states.
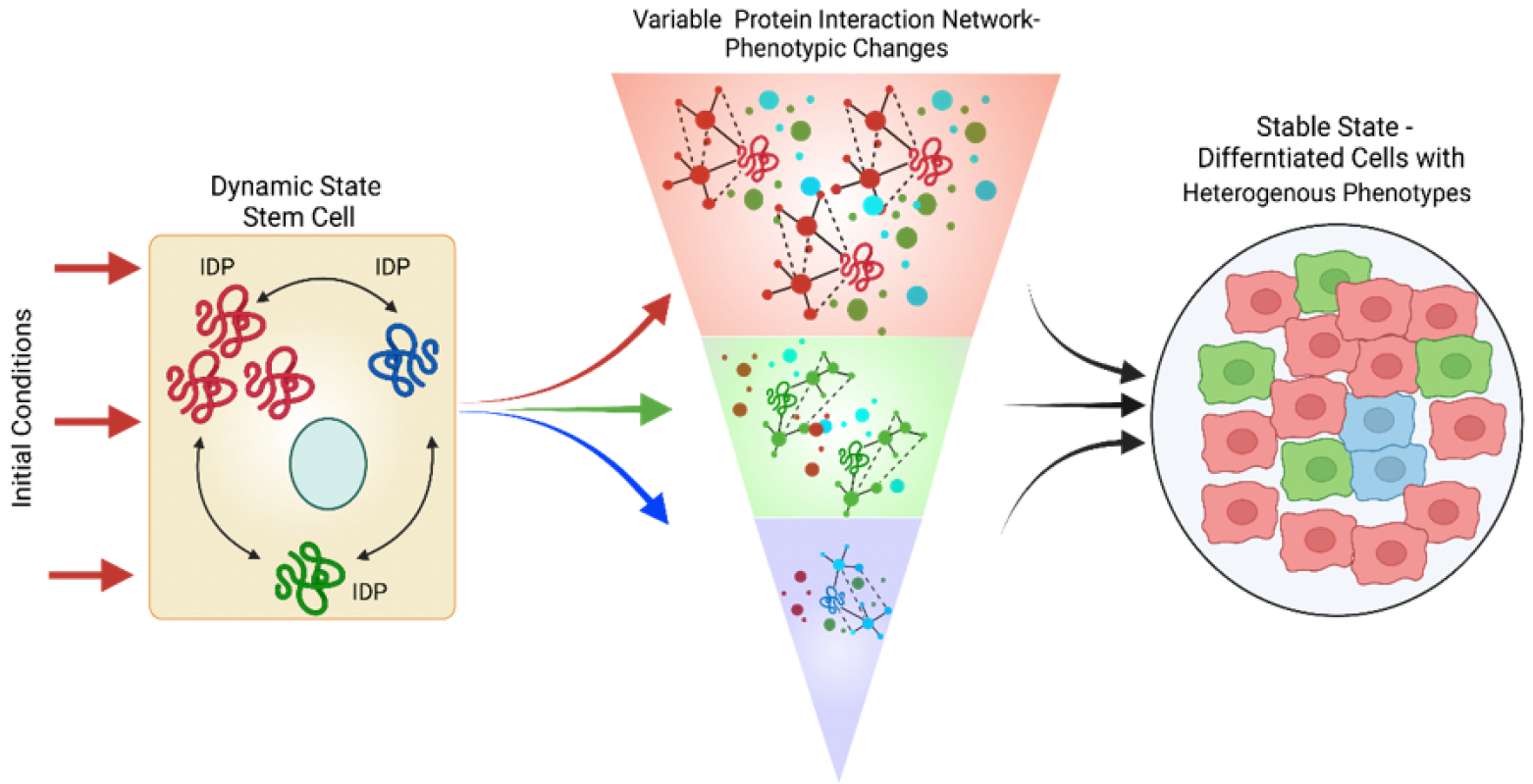
A key IDP is expressed in a stem cell that and exhibits a high degree of conformational dynamics. The various conformations are shown in red to blue to green. If the initial conditions favor the red conformation more than the green or blue, the red conformation induces specific protein interaction that leads to the differentiation of the stem cell (e.g., the red phenotype). In addition, the initial conditions favor green conformation partially followed by blue conformation. The green and blue conformations initiate distinct interaction and give rise to the green and blue phenotypic state. The net result is a heterogeneous population with a mixture of phenotypes.
4.2. IDPs: strange attractors?
From the foregoing, it is tempting to conjecture that, in an appropriate space, the dynamics of the constantly changing IDP network configuration can be described as a chaotic system,131–132 where the system neither converges to a steady state nor does it diverge to infinity; it will stay in a bounded region with chaotic motion in that space (Fig. 5) (Box 1). Under some conditions, it is likely that the system trajectory could be analogous a strange attractor; i.e. small changes in initial conditions can cause significant changes in the outcome (butterfly effect).133
Fig. 5:
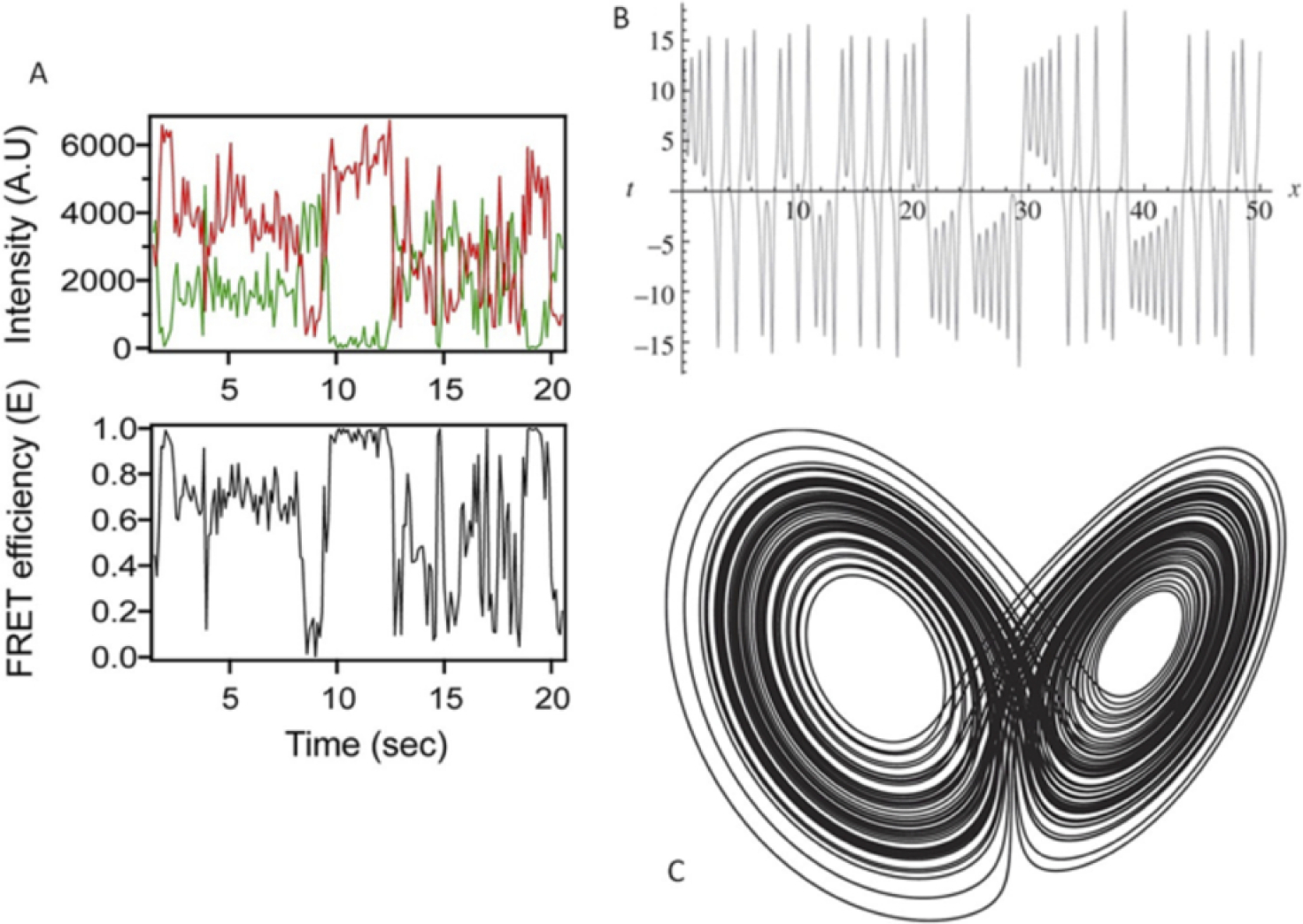
Similarity of the dynamic conformational behavior of an IDP with the behavior of a typical chaotic system (the Lorenz attractor). (A) Single molecule FRET trajectory of intrinsically disordered neuroligin cytoplasmic do-main as a representation of conformational dynamics. The top plot depicts the time evolution of donor (green) and acceptor (red) intensities. The bottom plot shows the FRET efficiency over time. A. U.: arbitrary unit. (B) The dynamics trajectory of a chaotic system; i.e., the Lorentz attractor, which shows qualitative resemblance with the conformational dynamics of an IDP. The Lorentz system is comprised of three independent variables and coupled differential equations to describe their dynamics. Here, the time evolution of one of the independent variable is depicted. (C) The phase-space representation of one of the variables of the Lorentz system. The variable is plot-ted against its rate of change. The overlapping loops represent the attractor basins of the strange attractor, where the system does not converge to a single state, neither diverges to an infinitely large space, but hovers within a defined domain of the attractor basin. Reproduced with permission from Uversky VN. Unusual biophysics of intrinsically disordered proteins. Biochim Biophys Acta. 2013. 1834(5):932–51. Elsevier Publisher.
To make matters more concrete, consider the Lorenz system The Lorenz attractor is a non-linear dynamical system that was developed to describe atmospheric convection in response to perturbations in gravity and temperature.134–135 Lorenz and his coworkers modeled this system using coupled differential equations involving three independent variables x, y and z. This system exhibits a strange attractor behavior (Fig. 5). The time evolution of an independent variable in the Lorentz system (Fig. 5B) shows a striking resemblance with the conformational dynamics trajectory of an IDP, represented by the FRET characteristics plotted over time (Fig. 5A). The donor-acceptor distance stochastically fluctuates as the IDP adopts various conformations over time, leading to different FRET efficiencies.
How IDP dynamics can be modeled in a similar way using nonlinear differential equations to reproduce chaotic behavior (analogous to the Lorenz system) is currently unclear. However, we want to provide the readers with some possible avenues for the investigation in this direction. At first glance, the analogy between the two systems (weather and IDP) is not at all apparent. One is a model for fluid convection and atmospheric changes over time, while the other is about the changes in protein conformation over time. However, upon closer inspection, one can see that the two systems indeed have certain common aspects.
Like the weather model, proteins are multidimensional systems involving numerous variables (e.g., the coordinates of thousands of protein atoms and the surrounding solvent and ions). Due to the frequent inter-atomic interactions, the dynamics of many of these variables are correlated, thus making dimensionality reduction feasible.136 It is the coupled dynamics of correlated variables that makes it possible to represent complex systems like the weather using simpler mathematical models, such as the one devised by Lorenz and coworkers. Notably, the strange attractor was also recently discussed in the context of cellular networks (which too involve thousands of interacting variables) that drive diseases such as cancer.137 Protein dynamics features resulting from methods like molecular dynamics can be approximated by linear models in reduced dimensions using approaches, such as principal component analysis or time-lagged independent component analysis.138–139 This raises several important questions that warrant further investigations. For example, can the IDP dynamics involving millions of atoms be expressed as a mathematical model in reduced dimensions, analogous to the system of ordinary differential equations (ODEs) that exemplifies the strange attractor? Could certain thresholds in the parameters of this model represent the transition from the nearly deterministic behavior of a structured protein to the chaotic behavior of an IDP?
It is likely that a mathematical model involving IDPs may not be feasible using only three independent variables that were invoked to create the Lorenz attractor. Instead of two attractors of the Lorenz system, most IDPs will likely transition among multiple attractor basins. Perhaps, it may be possible to develop a mathematical system involving a greater number of dimensions. However, whether such a model is feasible for describing IDP dynamics is not presently known. We leave it to the enthusiastic researchers of this and the future generation to figure it out.
4.3. IDP dynamics: A combination of deterministic and random components?
It is well known that some dynamical systems can be completely deterministic. Therefore, if one knows the system’s position a priori, one can predict its position in the future. However, some dynamical systems can be completely random. In such cases, even if one knows the history of the system up to a certain point in time a priori, that information cannot precisely predict the behavior of the system in the future. Most dynamical systems, however, fall somewhere in between these two extremes, where some aspects of the dynamics can be described as deterministic, and the rest as stochastic. Recently, the weak Pinsker conjecture, which states that all ergodic stationary processes can be expressed as a combination of an almost deterministic component and a completely random component (Box 2) was proven by Austin.140–141 Such endeavors, and the promising development of artificial intelligence in reproducing chaotic behavior142–143 have rekindled the interest in understanding complex systems by deconvolving their dynamics into elementary components.
Box 2.
A key property of dynamical systems, irrespective of the complexity, is that they can be expressed as a combination of deterministic and random elements. Indeed, Pinsker140 surmised (the Pinsker conjecture) that most, if not all, dynamical systems are a fusion of a stochastic dynamical system and a deterministic one. This idea was challenged by Ornstein.254 However, Thouvenot suggested that Pinsker’s conjecture may be valid if the deterministic component in Pinsker’s original description had some residual randomness. This led to the Weak Pinsker conjecture. Recently, the Weak Pinsker conjecture was mathematically proven by Austin141 on the foundation of the “stationary stochastic process” (where a dynamical system is described as a sequence of events that are individually random, but with fixed probabilities). The uncertainty of each event in the process (i.e. it’s decoupling from the past events) can be quantified by entropy, where zero entropy makes the system entirely deterministic. Austin’s proof analyzes the self-clustering (deterministic) property of a dynamical system trajectory on a hamming cube by discretizing it in space and time, simultaneously resolving the inherent randomness within each cluster. This shows that the deterministic component of the system is always associated with some randomness that varies depending on system characteristics.
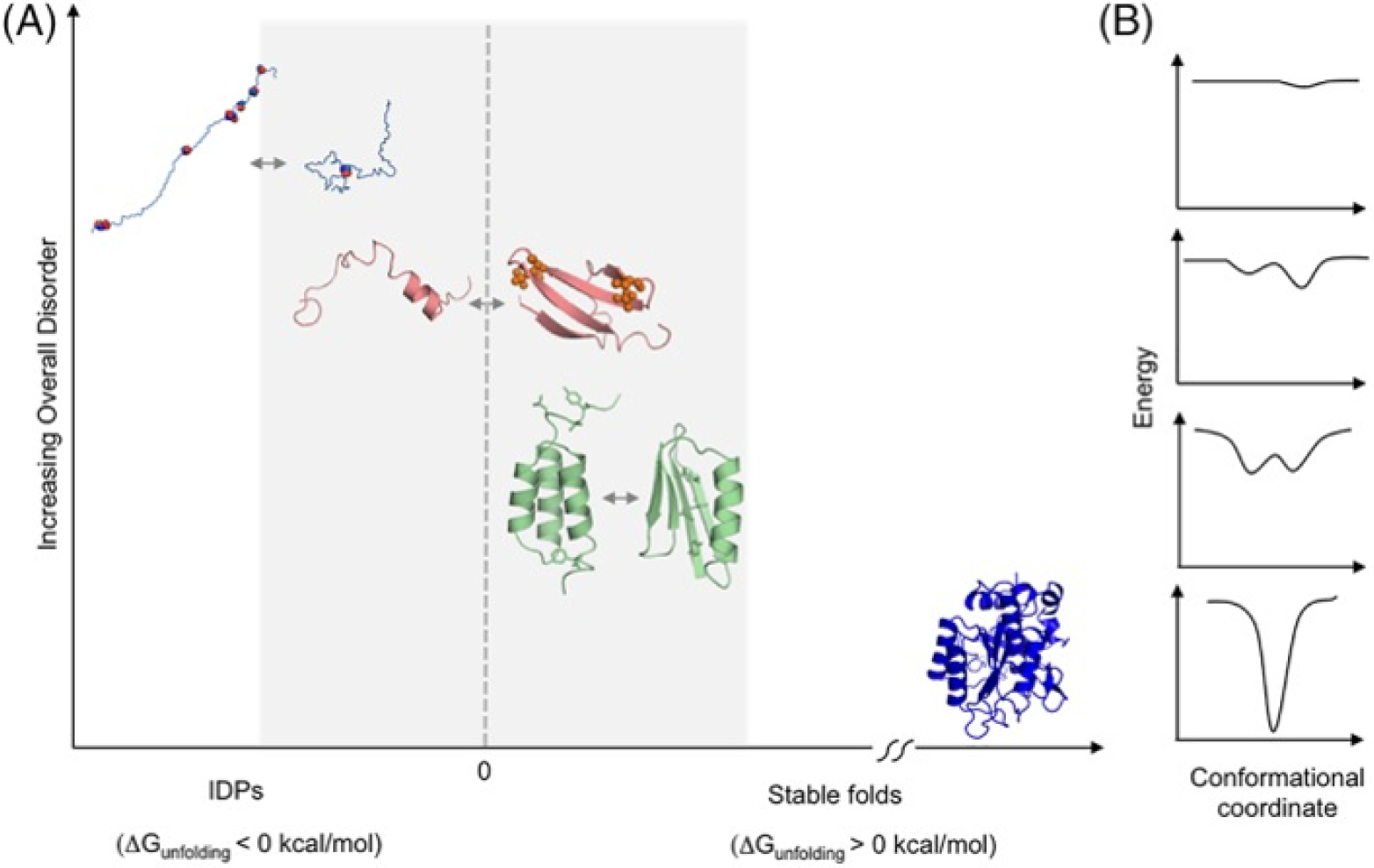
Since IDPs are complex dynamical systems97 that exist as flexible ensembles in their physiological state, a logical question is, whether it is possible to understand the complex motion of IDPs in more systematic ways by elucidating the dynamics using statistical theories of dynamical systems. Protein motion, in general, is a combination of dynamics at multiple lengths and timescales ranging from bond vibrations in the picoseconds to domain motions (of ordered proteins) in the micro to millisecond timescales.144 This complex multi-scale dynamics is a result of solvent viscosity that influences the slower motions of entire protein domains through friction, as well as thermal noise that affects the high frequency components of protein dynamics, such as side chain fluctuations. Consequently, all proteins exist as ensembles of conformations with frequent exchanges in real time. The difference between folded/ordered proteins and IDPs is that, for folded proteins/domains, this ensemble of conformations is tightly restricted to a small region of the conformational space, whereas in case of IDPs, the conformational ensemble is highly diverse.
The ensemble of protein conformations can be visualized via their corresponding free energy landscapes (Fig. 6),145 which is analogous to Waddington’s epigenetic landscape originally proposed for studying cellular phenotypes.126 The energy landscapes provide a mapping of possible states of the dynamical system, which, in the context of proteins, describe all possible conformations and their corresponding stabilities. While the free energy landscape of a structured protein typically shows a single deep attractor basin (also known as the folding funnel, Fig. 6C), the energy landscape of an IDP can have multiple attractors within a large overall basin (Fig. 6D). Therefore, the dynamics of an IDP can be envisaged as excursions to different attractors over time. Within each attractor well, the protein traverses a trajectory of closely related conformations, before transitioning to a neighboring well. However, the overall IDP dynamics is bounded within a well-defined region of the conformational space (the wider basin in Fig. 6D that includes the three attractors). In contrast, the free energy landscape of a completely unfolded (random coil-like) polypeptide can be thought of as relatively flat (Fig. 6B). Of note, because of the enormous spatiotemporal heterogeneity, some IDPs can also be characterized by nearly flat free energy landscapes.
Fig. 6.
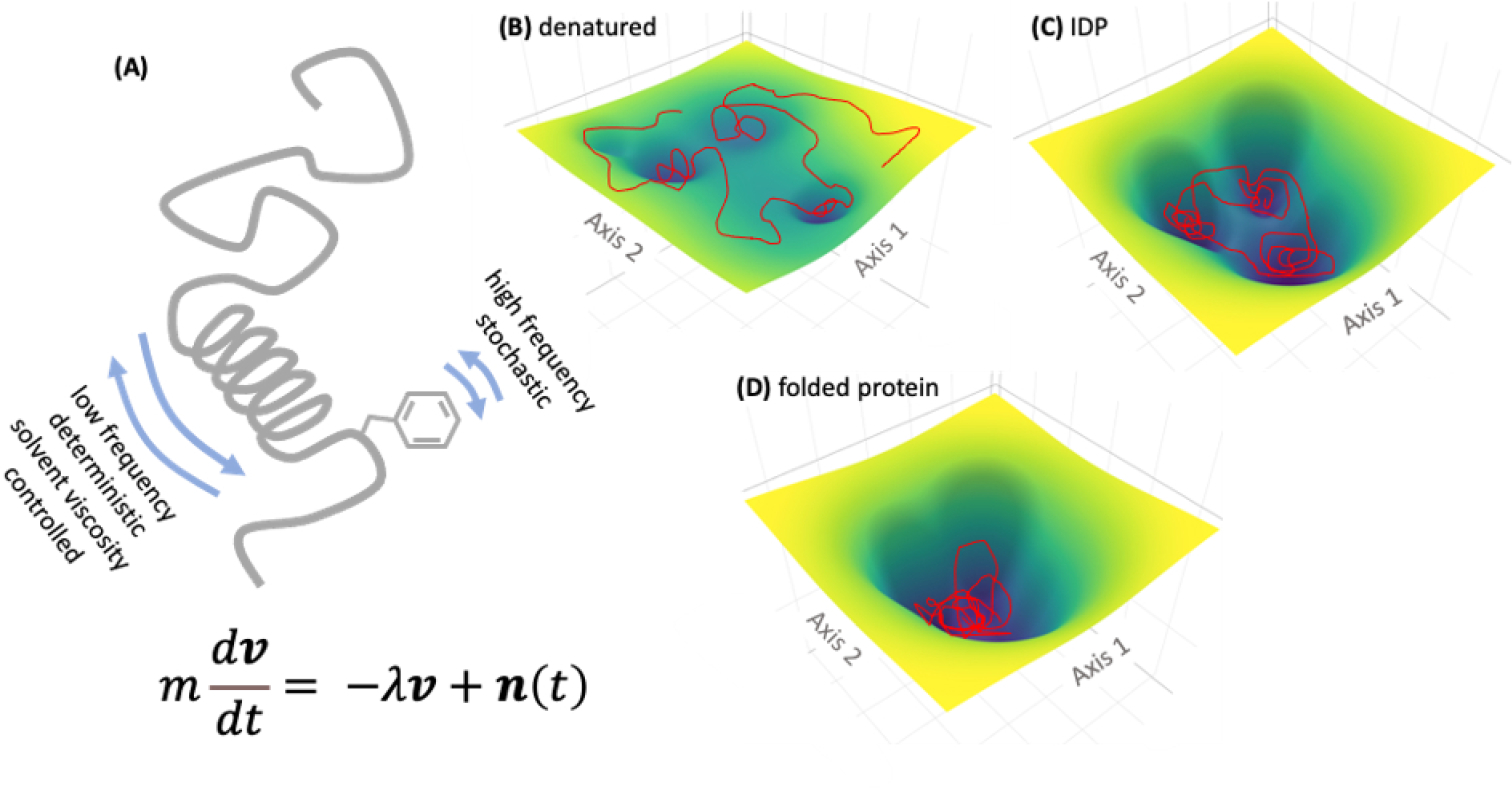
A) Schematic of IDP with random coil and transient secondary structures. The Langevin equation modeling the deterministic and stochastic components of protein dynamics is shown below. B-D) model attractor landscapes for denatured, disordered, and folded proteins. The protein conformational trajectories over time are shown as red scribbles.
How can such complex dynamics be broken into elementary components, for example, nearly deterministic and stochastic components? Although, we envision more research will be required in this direction, we will simply conjecture here that the IDP excursions between different attractor wells can be described in terms of stochastic dynamics. Under steady state, the probability of finding the IDP in a given attractor well is governed by the nature of the energy landscape. However, at a sufficiently long timescale, the transitions between these wells can be considered to be completely random. In contrast, the motion of an IDP within each attractor well can be described as clusters of closely related trajectories and therefore nearly deterministic. As a concrete but simplistic example, a hybrid system consisting of deterministic and random components can be modeled using the Langevin equation146:
| (1) |
where, m is the mass of a protein particle (usually a coarse-grain cluster of protein atoms), v is the velocity vector, −∇U(x) is the force-field-dependent term that describes intra and intermolecular forces (i.e. the gradient of potential energy U, which is a function of the protein atomic/coarse-grain particle coordinates, represented by x), the term λν represents the viscous force due to solvent, and n(t) is a noise term describing the stochastic component (Fig. 6A). Models, such as described by the Langevin equation, and similar but more complicated and realistic models, are suitable for modeling protein and polymer dynamics because they include both deterministic component (i.e., −∇U(x) and λν) and a random component (n(t)). By varying the strength of the noise term, it is possible to adjust the contribution of the random component in such hybrid models. Inclusion of a protein conformation dependent component (−∇U(x)) allows excursions of the different attractor wells.
Such approaches in modeling IDP dynamics could have multiple applications. Deconvoluting the IDP dynamics into underlying components that are easier to conceptualize, allows us to complement existing computationally intensive methods for predicting IDP dynamics (e.g., Molecular Dynamics) with novel approaches. Such approaches have practical applications in understanding how the IDPs alter their conformations in response to cellular environments and interact with other proteins as well as in designing therapeutics. One potential application is the deconvolution of the ensemble-average experimental properties of IDPs, such as NMR shifts or small angle X-ray scattering intensities to derive the underlying conformational ensemble. For example, the intermolecular contacts made by the IDPs can be encoded by a Hopfield network to model the free energy landscape, where each attractor is characterized by a different cluster of intermolecular contacts.147 The resulting network can be trained using existing data to predict conformational ensembles in new cases, even with incomplete or noisy data. Such an approach has been applied in modeling Waddington’s epigenetic landscape, where the inner architecture of gene regulatory networks (GRNs) could be derived by training against single cell transcriptomics data.148 Therefore, such approaches could also prove highly advantageous in modeling the IDP conformational landscapes. However, one should keep in mind that as a complex dynamical system, IDPs are not easily subjected to the reductionist approach. In fact, as it follows from the general understanding of such systems, their heterogeneous components are interconnected and interact nonlinearly, indicating that a perturbation does not necessarily cause a proportional effect, and that the system’s behavior is not equal to the sum of its parts. Clearly, further studies along these lines are needed to shed new light on the remarkable correlation between complexity and intrinsic disorder; they could also reveal why IDPs, as complex dynamical systems, occupy key nodes in PINs and hence, were selected during evolution.
5. IDPs AND EVOLUTION
5.1. IDPs and prebiotic evolution
Starting from the prebiotic origins of life to the Last Universal Common Ancestor (LUCA), that gave rise to all living beings are thought to have evolved, several evolutionary innovations occurred that are thought to be guided by self-organization.149 Furthermore, it was postulated that short, unstructured peptides (possible precursors to IDPs) may have been the key components of a flexible cellular network with the capacity to readily evolve.150 The inclusion of primitive IDPs into discrete membranous structures, may have given rise to a self-sustaining unit that could replicate itself. Such a hypothetical entity is conceptually similar to the Chemoton (or chemical automaton, a basic entity satisfying the sufficient and necessary conditions for sustaining life).84–85 Thus, it is quite likely that IDPs had self-templating activity. In subsequent steps, interactions between IDPs and other biopolymers, through self-organization, may have formed an interaction network. Through many rounds of selection, such a structure, may have given rise to the predecessor of LUCA.151–152
Consistent with their involvement in prebiotic evolution, IDPs across lineages are enriched in polar and charged amino acids but show a paucity in hydrophobic residues.76,153–155 Furthermore, the amino acids are thought to be added to the genetic code in a temporal order: G/A, V/D, P, S, E/L, T, R, N, K, Q, I, C, H, F, M, Y, and W. Notably, the first few amino acids to emerge (until S in the above list) are disorder inducing, while the later amino acids are order promoting (e.g. Y, F, W, C), suggesting that protein disorder played a critical evolutionary role in early life.(Fig. 7).156–157 These observations were confirmed by Brooks et al., whom estimated the distributions of different amino acids in the last universal ancestral genomes.158
Fig. 7.
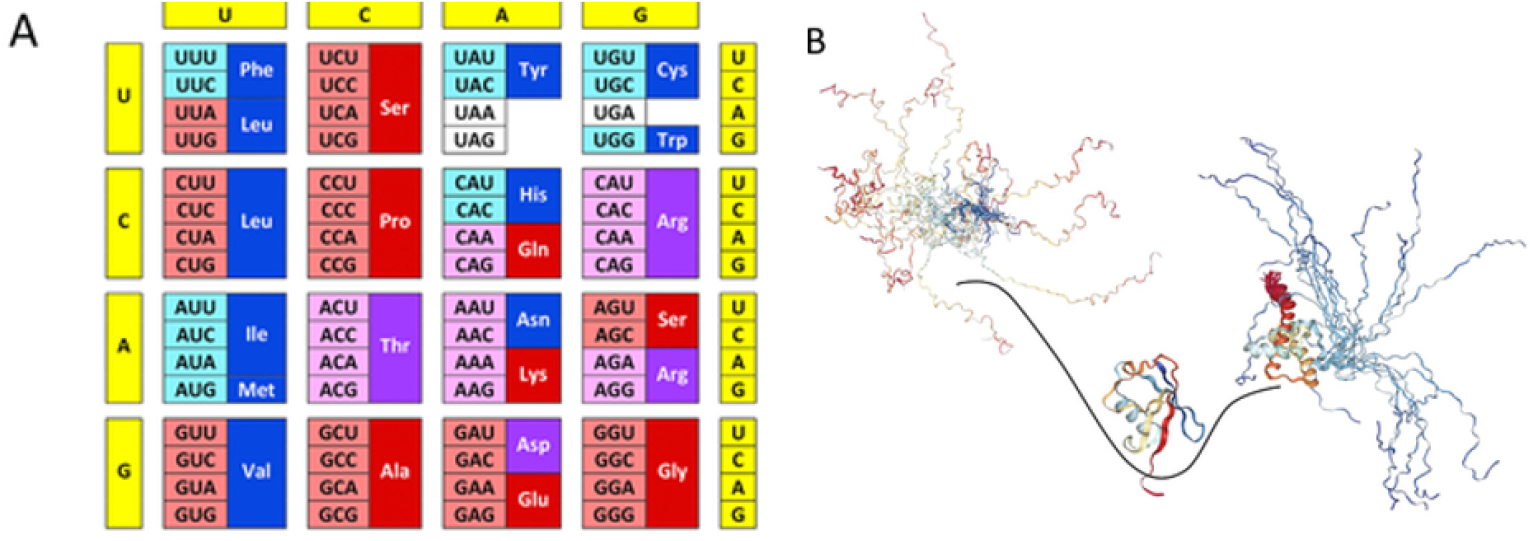
Intrinsic disorder and protein evolution. A) Modern genetic code with information on the early and late codons (shown by light pink and light cyan colors, respectively) and disorder- and order-promoting residues (shown by dark red and dark blue colors, respectively). Codons with intermediate ages are shown by light violet color. Disorder-neutral residues are shown by dark violet color. B) Evolution of intrinsic disorder in proteins has a characteristic wavy pattern. X-axis represents evolutionary time and Y-axis shows global disorder content in proteins at given evolutionary time point. Here, primordial proteins are expected to be mostly disordered (left-hand side of the plot), proteins in LUA likely are mostly structured (center of the plot), whereas many proteins in eukaryotes are either totally disordered or represent hybrids containing both ordered and disordered regions (right-hand side of the plot). Reproduced with permission from Kulkarni and Uversky. Intrinsically disordered proteins: The dark horse of the dark proteome. Proteomics 18(21–22):e1800061. ©2018 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.150
It is generally held that the primordial IDPs in prebiotic evolution may not have catalytic activity.159 However, it is equally unlikely that the transition from disordered primordial polypeptides to modern enzymes with highly ordered domains occurred in quick succession during evolution. Thus, it is possible that some primordial IDPs evolved limited catalytic functions. In fact, a challenge, the Janus challenge,160 seeks to address this particular facet of IDPs. Indeed, it has been reported that some modern enzymes or their mutated variants behave as molten globular structures (i.e., structures with a loose tertiary topology, but lacking intricate inter-residue contacts).161–164 It is also possible that primordial enzymes were more disordered than the molten globule state. In line with this argument, a ligase evolved in vitro lacked a hydrophobic core synonymous with structured enzymes; instead, it contained a flexible, catalytic loop which was supported by a polar core with zinc atoms.165 Indeed, emerging evidence suggests that so-called classical enzymes may lack a unique structure and yet be functional.163–164,166–172 Therefore, the idea that prototypical IDPs could have had catalytic activity seems plausible, and the chances that the Janus challenge will be met in the near future are high.
Yet another salient feature of the prototypic IDPs that may be of significant relevance to the origin of life, is the potential for liquid–liquid phase transitions (LLPTs) or liquid–liquid phase separation (LLPS) or coacervation, resulting in protein-enriched liquid phases. Proteins together with nucleic acids, or by themselves, can phase separate resulting in proteinaceous membrane-less organelles (PMLOs). PMLOs exist as liquid droplets and are found in chloroplasts.77,173–179 Recently, it was pointed out that LLPS serves as a fundamental mechanism of compartmentalizing intracellular space and bio-membranes, thus enhancing the cellular adaptation to environmental variations.180 Furthermore, it seems that LLPS can take place in 1D, 2D, and 3D, where the 3D-condensation is associated with the PMLO biogenesis, 2D-films may be formed near membrane surfaces whereas 1D phase separation occurs on DNA and/or the cyto- and nucleoskeleton.180 PMLOs serve many important biological functions such as protein degradation, phosphorylation, splicing and transcriptional regulation.79 Thus, it is conceivable that PMLO functionality is predetermined by the nature of its constituentIDPs.79 Alternatively, it is also possible that the properties of PMLOs could enable the functions of their constituents.79
PMLOs may also act as micro-reactors that brings the RNA and protein molecules in close proximity to accelerate cytoplasmic reactions.178,181–182 Thus, we speculate that, primordial IDPs equipped the ability to phase separate, gave rise to protein-enriched phases183 (Fig. 8). This conjecture is consistent with the model proposed by Oparin in 1922.184 Indeed, the demonstration of peptide-based synthons can form catalytically active coacervates, lends further credence to this hypothesis.185 For a recent article on the conformational plasticity of IDPs from a physical chemistry standpoint see a recent comprehensive review by Mukhopadhyay.186
Fig. 8:
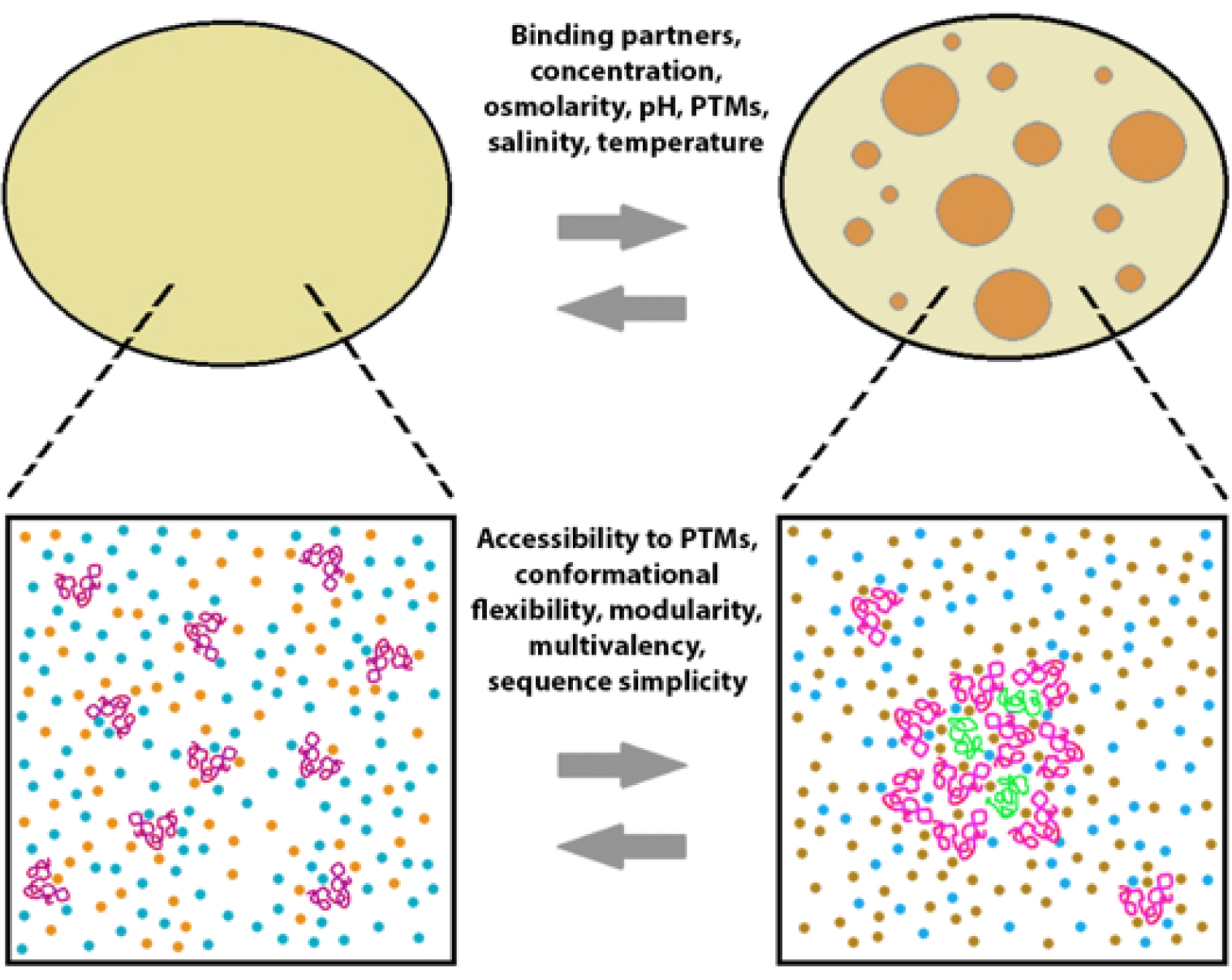
Thermodynamic factors (top) and disorder-related features controlling liquid-liquid phase transitions in protein solutions.
Additionally, it was recently hypothesized that spaces between mica sheets may have been potential sites for the origin of life as it may have provided shelters to the primitive PMLOs.187 Taken together, these examples underscore the critical role of IDPs in the life’s origin and evolution on the earth,188 which ultimately led to the evolution of multicellular forms and their expansion.
5.2. IDPs and multicellularity
One of the key milestones in the timeline of evolution is the emergence of multicellularity. It is estimated that multicellularity has evolved multiple times independently in evolutionary distinct lineages from bacterial biofilms to plants and metazoans with the most ancient eukaryotic instances dating back to ~1.6 billion years ago.189 It is thought to be driven by the life history trade-offs between survival and reproduction in response to stress and/or predation. Adaptive changes, such as increase in size, division of labor and increase in complexity, enable the multicellular organisms escape from predators and cope with environmental and nutritional stress. Comparative and functional genomics data suggest a recurring theme in the evolution of multicellularity is the co-option of ancestral pathways from unicellular organisms190–192. But the key question is how were the ancestral pathways and genes adopted for the new functions of the multicellular organisms? The facts that IDPs, by virtue of their conformation flexibility and propensity for post-translational modification (e.g., phosphorylation), can rewire the PINs and several of stress response proteins are IDPs, suggest that they can be a source of adaptive plasticity required for bringing about this evolutionary transition.
Cell specification and specialization is thought to be facilitated by modification of the GRNs,193 involving differential gene expression and cell signaling. TFs and signaling molecules are important components of the GRNs, and many of them are IDPs.36,194–196 IDPs and hybrid proteins with ordered domains and IDPRs, can regulate cell-specific transcription by forging promiscuous interactions with different partners via changes in conformations in the IDPs/IDPRs. In addition to the stochastic changes in conformations, PTMs of IDPs can cause changes in their conformational ensembles resulting in interactions with different partners. IDPs can also change protein interaction networks via the addition, deletion, or modification of binding sites by means of alternate splicing of pre-mRNA coding IDPRs.9,197–198 Transcriptional noise originating from the inherent stochasticity of gene expression can result in heterogeneity in isogenic cellular populations. Therefore, IDP overexpression in response to a specific extrinsic perturbation (e.g., stress), results in amplification of transcriptional noise and rewiring of PINs that result in modification of the GRNs to actuate phenotypic switching and facilitate adaptive evolution.97,98
Several proteins that are key to the development of innovations required for multicellularity, such as extra cellular matrix expansion, cell adhesion, cell communication, cell cycle modifications, asymmetric cell division, and cell differentiation, are IDPs. For example, cadherins, which play important roles in cell segregation and boundary formation during development, are IDPs199 and were co-opted from the unicellular progenitor of animals to function as adhesion receptors in epithelia. Similarly, retinoblastoma (RB) and retinoblastoma-related proteins (RBRs) that regulate the cell cycle, are IDPs.200 RBs and RBRs are transcriptional repressors that modulate cell cycle regulated gene expression through binding to E2F-DP transcription factors. The disordered linker domain of the RB-like protein, that is phosphorylated by cyclin-CDK,201 is different between Chlamydomonas, a unicellular organism and Gonium and Volvox which are multicellular,192 suggesting that different protein-protein interactions of RBs and their interacting partners regulate cell cycle in these unicellular and multicellular organisms. HSP70, a chaperon protein, that regulates asymmetric cell division in Volvox is also an IDP202 and so is hydroxyproline-rich glycoprotein (HRGP), a constituent of the extra-cellular matrix (ECM), which is also highly disordered203. Disorder in the ECM interactome could provide the structural flexibility required for the ECM to interact with membrane proteins and soluble proteins in more complex organisms.
Interestingly, some HRGPs of both multicellular and unicellular algal forms have evolved into pheromones deployed for sexual signaling 204–207 The HRGPs are also present in much greater numbers in Volvox than in unicellular algae. Therefore, it is likely that during evolution, intrinsically disordered ECM/cell wall proteins diversified to adapt to developmental processes. Therefore, these proteins may represent a source of adaptive plasticity that is specifically observed in the volvocine algae.190 The increase in the disordered residues in proteomes from bacteria to single-celled and multicellular eukaryotes, lends further credence to the involvement of IDPs in the origin of multicellularity.19,208–210
In order for unicellular organisms to exist as stable multicellular clusters, the individual cells need to adapt to changing environments and cooperate with neighboring cells. We envision that IDPs, due to their unique structural and dynamic properties, facilitated the necessary phenotypic plasticity and adaptation that were critical to the origin of multicellularity. It is well known that IDPs can couple with multiple binding partners, and these promiscuous interactions could have been advantageous in PIN rewiring and switching of phenotypes. Moreover, IDPs alternate between multiple structures in a stochastic manner, as evidenced from the time-resolved Förster Resonance Energy Transfer (FRET) measurements.211 This conformational noise coupled with their interaction with multiple signaling proteins could produce the phenotypic diversity that enabled the multicellular clusters to exist.
While a sophisticated model connecting IDP conformational noise with PIN rewiring is beyond the scope of this review, we present a simple conceptual framework elucidating the role of IDPs, where conformational noise acts as intrinsic noise in the cellular network and phenotypic transitions are controlled by the level of disorder in the cell (Fig. 9A). Consider that within the cell, the IDP A is expressed at low levels (at any time, the expression of A is denoted by A as well) and is produced at a rate R1. For simplicity, we will assume R1 to be constant. We also propose that A stochastically switches between two alternative conformations Aa and  (this is a simplified description, since IDPs typically will adopt multiple conformational states, rather than just two). While the conformation Aa couples with other proteins to carry out cell signaling and phenotypic transition, the conformation  gets phosphorylated and degraded. This model bears similarity to the PAGE4 circuit described previously, where the IDP PAGE4 is degraded upon hyperphosphorylation.212 Due to the structural flexibility of IDPs, the level of  in the cell will fluctuate with time and these fluctuations will be more prominent at low protein concentrations, which is typical for many IDPs in the cell.213 The time averaged  concentration will depend on the depth of the attractor well representing  in the IDP free energy landscape. Under the above assumptions, the temporal rate of change of the IDP concentration can be expressed as
| (2) |
| (3) |
Fig. 9:
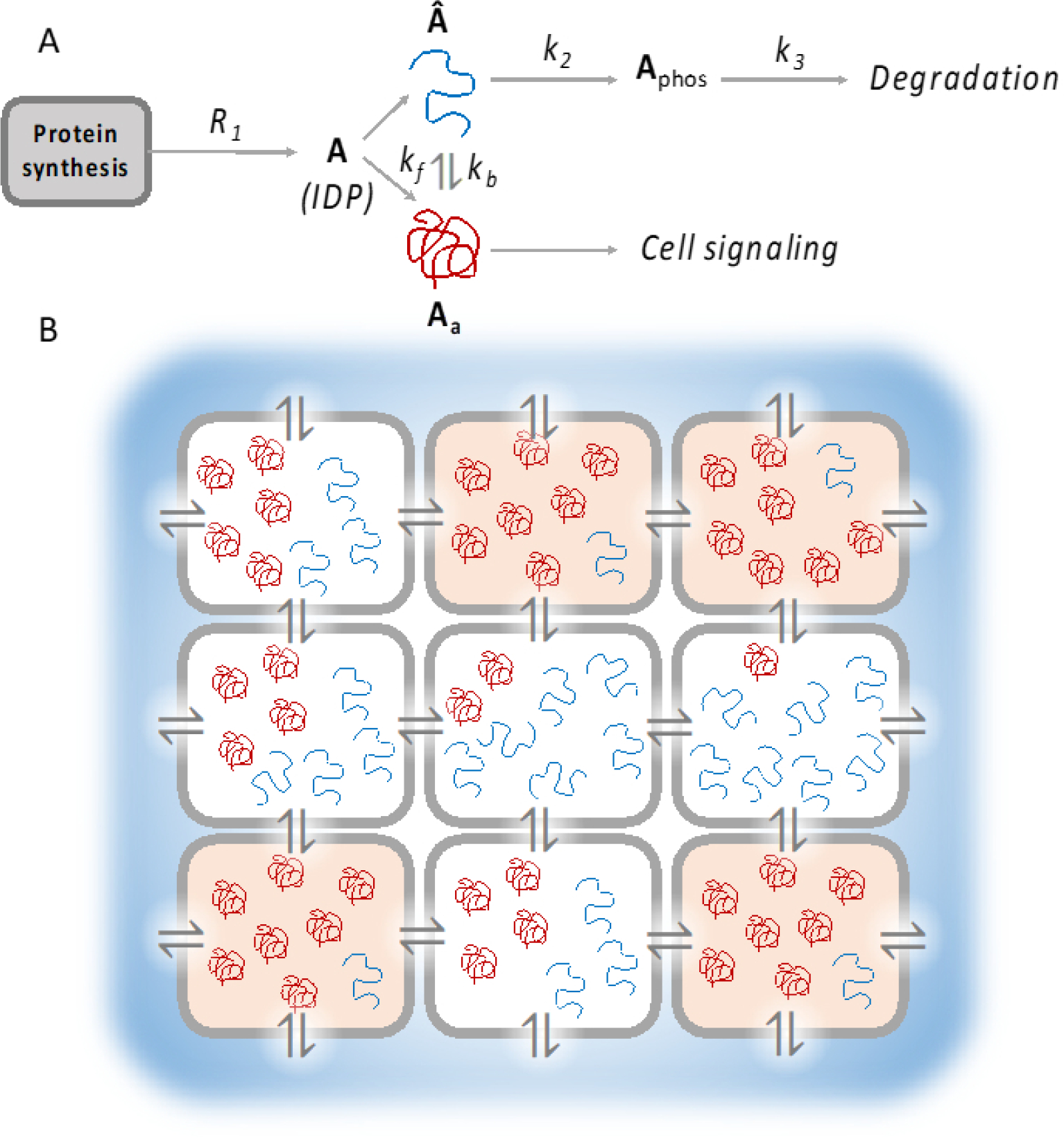
A model for IDP conformational noise contributing to multicellularity. A) A conceptual reaction network where conformational noise leads to fluctuating levels of IDP conformation. In this model, the IDP A (where A also denotes its concentration in the cell) can assume two alternative conformations Aa (denoted by the red string, which participates in cellular signaling) and  (blue string, which is phosphorylated and subsequently degraded). A stochastically switches between Aa and Â, leading to fluctuating levels of the two conformations over time. B) A schematic representing an array of cells with varying levels of the IDP A. High level of the functional conformation Aa allows the cells to switch to a different phenotype, represented in orange. The level of Aa is further modulated by exchange of diffusible factors with neighboring cells and the environment (indicated by the ⇌ symbol). Such a system can give rise to spatiotemporal patterns and cooperative development of cellular clusters, as a precursor to multicellularity.
Here, k2 is the rate constant for converting  to Aphos, the phosphorylated form of A, by a kinase. In general, such enzymatic reactions will follow Michaelis-Menten kinetics,214 but at low substrate concentrations, the reaction rate can be approximated to be linearly proportional to the substrate concentration. Also, we model the degradation rate as linearly proportional to the level of phosphorylated A, with a rate constant k3. Since  fluctuates due to intrinsic, conformational noise, the above equations are stochastic in nature.
Coupled differential equations have been widely used in the literature to model transcriptional regulation and catalytic events such as phosphorylation, involving multiple partners, potentially in a feedback loop. Equation 3 demonstrates how such models can be extended to incorporate stochastic effect of IDP conformational fluctuations that may lead to distinct cellular states. Fig. 9 demonstrate how IDP conformational fluctuations, in conjunction with environmental perturbations may stabilize distinct phenotypes in a multicellular environment. We suggest that single cell models exemplified by equation 3 can be coupled together to numerically simulate such multicellular systems and analyze the emergence of stable phenotypes. Quantitative demonstration of such models is an exciting idea, but unfortunately is beyond the scope of this manuscript. One of the overarching goals of this article is to stimulate exploration in this direction by aspiring scientists.
It would be an interesting exercise to extend this model further by imagining a cluster of cells, each exhibiting a similar set of reactions. We can also introduce additional terms in the rate equations to represent the influence of soluble factors exchanged with neighboring cells or the effect of the environment. Such a model is schematically depicted in Fig. 9B. It remains to be seen whether the solution to the above system of differential equations can yield multiple stable states, where each stable state is represented by a different proportion of Aa to Â. Cells that achieve a stable state with a Aa level beyond a certain threshold may undergo transition to a different phenotype (Fig. 9B). Additionally, conformational noise can make these phenotypic transitions more feasible. The above model can be easily extended to incorporate multiple functional conformations instead of just two, which may lead to increased phenotypic heterogeneity. We may therefore envision that increased disorder may lead to more stable states in the phenotypic landscape. Thus, in a multicellular system such as the one described above, IDP conformational noise, in conjunction with a variable environment can enable the cells to adopt different phenotypes. Such heterogeneity superposed with intercellular signaling may lead to interesting spatiotemporal patterns or cooperative phenotype development in an organized community of cells.
5.3. IDPs and inheritance of acquired characteristics
Yet another discovery that is of immense fundamental importance but appears to challenge the central dogma of molecular biology,215 is the facilitation of transgenerational information transfer by IDPs. Emerging evidence 216 that prions (proteins that are capable of self-replication by inducing prion-like conformations in other proteins), apart from nucleic acids, can drive organismal phenotypes by self-templating conformations, and thereby serve as vehicles of inheritance. Consistent with this observation, it was recently reported that in the yeast, several IDPs resemble prions from functional perspective.68 Indeed, the transient overexpression of several of these proteins yielded heritable traits even after their expressions returned to normal levels. Most intriguingly, these proteins were not identified as prions previously, and they did not form amyloids. However, they contained hallmarks of nucleic acid-binding proteins with long IDPRs and were evolutionarily conserved.68 Taken together, the data established a general form of protein-based inheritance wherein IDPs give rise to new traits and adaptive opportunities.
At least one of these prion-like proteins was reported to drive self-assembly giving rise to gel-like condensates.217 Nonetheless, such protein-rich particles did not form amyloid fibrils but were infectious apparently using a protein-based epigenetic element. Cells expressing these proteins were found to repress gene expression patterns in order to facilitate improved growth when nutrient supply was limited. Thus, these non-amyloid proteins appeared to modulate a form of non-chromosomal epigenetics to modulate the expression of genes that are heritable over significantly long biological timescales. Since IDPs undergo LLPTs or coacervation giving rise to PMLOs,79,174 it may be expected that the prion-like particles self-assemble into gel-like condensates. In line with the observations in yeast, the IDP from C. elegans called PGL-1, was observed to form aggregate-like structures in the germ cells. Amazingly enough, such aggregates were found to be inherited for multiple generations even after the original mutation triggering their formation is not observed any more. Therefore, these observations suggest that even in animals, IDPs generate self-propagating aggregates that may serve as vehicles for transgenerational inheritance (Kennedy S.G. Available: https://grantome.com/grant/NIH/R21-AG061850-01A1. [Ref list] [(accessed on 1 September 2020)].
6. CONCLUSIONS AND FUTURE DIRECTIONS
Despite several excellent reviews on IDPs that appeared in just this past year alone that cover many interesting aspects of these multifaceted proteins with pleotropic functions,218–232 very little is known about the IDPs from a dynamical systems perspective.43,97,125,131,233–234 However, from the foregoing, it follows that a rigorous theoretical foundation addressing how interactions between an organism and its environment can shape its phenotype,125,235 and how IDPs may modulate the attractor landscape in driving the phenotypic states,131 (guide cellular decisions), warrant further investigations. Although, the idea that a cell (or a protist) is capable of making decisions is often met with skepticism, and perhaps may even be bantered, several tantalizing reports suggest that cells can ‘anticipate’, and even ‘learn’ from, environmental fluctuations and survival hurdles.236–243 Together with the demonstration that noise plays an important role in cellular decision making,244 these observations should inspire new research in this next frontier. The wetware (i.e., the combined hardware and software in biology) metaphor should help invigorate scientists from across disciplines to study these fascinating molecules, IDPs, which are critical, yet poorly understood.
From an experimental perspective, elucidating the structure and dynamics of an IDP ensemble, and how the preferences are shaped by the environment, may help to better understand the mechanism(s), by which IDPs interact with their partners to carry out their respective functions. With the recognition of the IDPs and the critical roles they perform, a new frontier in the biology, biochemistry, and biophysics of proteins has dawned. Furthermore, recent developments in targeting IDPs that were previously marginalized as ‘undruggable’, with small molecule inhibitors/activators,71,245–252 has ushered a new era in biomedical research.
Acknowledgements
MKJ was supported by Ramanujan Fellowship (SB/S2/RJN-049/2018) awarded by SERB (Science and Engineering Research Board), DST (Department of Science & Technology), Government of India
Abbreviations
- ECM
Extra-cellular matrix
- EMT
Epithelial-Mesenchymal Transition
- FRET
Förster Resonance Energy Transfer
- GRNs
Gene Regulatory Networks
- HRGP
Hydroxyproline Rich Glycoprotein
- IDPs
Intrinsically Disordered Proteins
- IDPRs
Intrinsically Disordered Protein Regions
- iPSCs
Induced Pluripotent Stem cells
- LLPTs
Liquid–liquid phase transitions
- LLPS
Liquid–liquid phase separation
- LUCA
Last Universal Common Ancestor
- MET
Mesenchymal-Epithelial Transition
- MRK
Mahmoudabadi, Rangarajan, Kulkarni hypothesis
- ODEs
Ordinary differential equations
- PAGE4
Prostate associated Gene 4
- PINs
Protein Interaction Networks
- PMLOs
Proteinaceous Membrane-less Organelles
- PTMs
Post-translational modifications
- RB
Retinoblastoma
- RBRs
Retinoblastoma-related proteins
- TFs
Transcription factors
Biographies
Biographies
Prakash Kulkarni, PhD, FRSB is Professor and Director of Translational Research at the City of Hope National Medical Center, California. He obtained his PhD in biochemistry from India and completed postdoctoral training at New York University School of Medicine. He then held Staff Scientist positions at the California Institute of Technology and Yale University. He started his independent academic career as an Assistant Professor at Johns Hopkins University. His research interests are interdisciplinary and across spatiotemporal scales and are focused on understanding how conformational dynamics of intrinsically disordered proteins contributes to phenotypic switching, especially in evolution of multicellularity, disease pathology and in non-genetic heterogeneity in cancer.
Supriyo Bhattacharya, PhD is a Research Assistant Professor at City of Hope. After obtaining his BTech degree from the Indian Institute of Technology, Kharagpur, he did his PhD in Chemical Engineering from North Carolina State University and completed his postdoctoral training in computational biophysics at City of Hope. He has more than 15 years of experience in computational science, statistics, algorithms development, data visualization, statistical analysis of biological data using linear regression, multivariate analysis, innovative clustering and stochastic methods. His research interests are diverse, ranging from protein dynamics to evolutionary theories. One of his major contributions was in the development of algorithms to analyze allosteric communication pipelines in proteins from molecular dynamics simulations.
Srisairam Achuthan, PhD, is Director, Biomedical Data Science at the City of Hope National medical Center, California. He obtained in undergraduate degree in mathematics from the Indian Institute of Technology, Kharagpur, and his master’s degree also in mathematics from the Indian Institute of Technology, Madras. He then did a master’s in applied mathematics from University of Southern California before he completed his PhD in applied mathematics from Florida State University. He is particularly interested in modeling biological processes, in utilizing scientific data for predictive analysis, in developing and supporting data driven scientific applications.
Amita Behal, PhD, is currently in the Department of Medical Oncology at the City of Hope National Medical Center, California. She obtained her PhD in biochemistry from the Indian Institute of Science and completed her postdoctoral training in cell biology at New York University and developmental biology at the California Institute of Technology. Subsequently, she worked in cancer biology at Yale University, at the Genome Database, Johns Hopkins University, and at the National Institutes of Health, Bethesda. She is interested in understanding the role of intrinsically disordered proteins in evolution and multicellularity.
Mohit Kumar Jolly, PhD, is an Assistant Professor at the Centre for BioSystems Science and Engineering, Indian Institute of Science. He has made seminal contributions to decoding the emergent dynamics of epithelial-mesenchymal plasticity (EMP) in cancer metastasis, through mathematical modeling of regulatory networks implicated in EMP. Currently, his lab focuses on decoding mechanisms and implications of non-genetic heterogeneity in cancer metastasis and therapy resistance, with specific focus on mechanism-based and data-based mathematical modeling in close collaboration with experimental cancer biologists and clinicians. He won the coveted 2016 iBiology Young Scientist Seminar Series award for communicating his research to diverse audiences. He currently co-chairs the Mathematical Oncology subgroup at Society for Mathematical Biology (SMB).
Sourabh Kotnala, MS, obtained his master’s degree in Aeronautical Engineering from the Indian Institute of Science. He is completing his PhD in fluid dynamics from Rijksuniversiteit Groningen, the Netherlands, working on modelling of stochastic differential equations. His current research interests are in mathematical biology, especially in modeling conformational noise and phenotypic heterogeneity.
Atish Mohanty, PhD, is a Research Assistant Professor at City of Hope. He obtained his PhD in molecular cell biology from the University of Tokyo and completed his postdoctoral training in oncology at City of Hope. His main research interests are to understand molecular mechanism of drug resistance and identify key targets which can suppress the evolution of resistant clones. He identified integrin β4 and paxillin as not only being required for adhesion associated signaling but that they are also essential for platinum resistance. His studies revealed that that the disordered region in the cytoplasmic tail of integrin β4 drives the interaction with paxillin whose amino terminus is intrinsically disordered.
Govindan Rangarajan, PhD, is Professor of Applied Mathematics, JC Bose National Fellow, and Director, Indian Institute of Science, Bangalore. He obtained an Integrated MSc (Hons) degree from the Birla Institute of Technology and Science, Pilani, and a PhD in physics from the University of Maryland. He completed his postdoctoral training at the Lawrence Berkeley Lab, University of California, Berkeley. Prof. Rangarajan’s research interests include nonlinear dynamics and chaos and time series analysis, and mathematical biology. He is currently the Director of the Indo-French Centre for Applied Mathematics (IFCAM). He is also a Fellow of the Indian Academy of Sciences and the National Academy of Sciences, India. He was awarded the Chevalier dans l’Ordre des Palmes Academiques by the Government of France and is a recipient of the Homi Bhabha Fellowship, Government of India.
Ravi Salgia, MD, PhD, is the Arthur & Rosalie Kaplan Chair in Medical Oncology and Professor and Chair, Department of Medical Oncology & Therapeutics Research at City of Hope National Medical Center. Prior to City of Hope, Dr. Salgia was Professor and Vice Chair at University of Chicago School of Medicine. He began his career as a faculty at Harvard Medical School following his postgraduate training in internal medicine at Johns Hopkins University School of Medicine. His research interests focus on novel therapeutics against lung cancer and other solid tumors. His other interest lies in exploring fractals and chaos theory and how they can be used in cancer. Dr. Salgia is also utilizing various strategies to understand tumor heterogeneity, including the role of intrinsically disordered proteins, and mathematical modeling.
Vladimir N. Uversky, PhD, DSc, is a Professor in the Department of Molecular Medicine at the University of South Florida (USF). He obtained his Ph.D. from Moscow Institute of Physics and Technology and D.Sc. from Institute of Experimental and Theoretical Biophysics, Russia. He is the discoverer of intrinsically disordered proteins (IDPs). His research is focused on protein physics including protein structure, stability, dynamics, function, folding, misfolding, and non-folding, and are trying to establish relationships between the protein sequence and function in the light of pharmaceutical biophysics. His laboratory uses a combination of experimental and bioinformatics approaches to describe structural properties, functions, dynamics, and conformational stability of different proteins; characterize their partially folded states and analyze the molecular mechanisms of their folding and misfolding; quantify structural consequences of protein interaction with various binding partners; uncover the relationship between protein structure, stability and pathogenesis of various protein conformation-based diseases.
Footnotes
Conflicts of interests: Authors declare they have no conflict of interests.
References
- (1).Ladunga I Phylogenetic Continuum Indicates “Galaxies” in the Protein Universe: Preliminary Results on the Natural Group Structures of Proteins. J. Mol. Evol. 1992, 34, 358–375. [DOI] [PubMed] [Google Scholar]
- (2).Ross JL The Dark Matter of Biology. Biophys. J. 2016, 111, 909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Perdigao N; Heinrich J; Stolte C; Sabir KS; Buckley MJ; Tabor B; Signal B; Gloss BS; Hammang CJ; Rost B; Schafferhans A; O’Donoghue SI Unexpected Features of the Dark Proteome. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 15898–15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Illuminating the Dark Proteome. Cell 2016, 166, 1074–1077. [DOI] [PubMed] [Google Scholar]
- (5).Baboo S; Cook PR “Dark Matter” Worlds of Unstable Rna and Protein. Nucleus 2014, 5, 281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Bitard-Feildel T; Callebaut I Exploring the Dark Foldable Proteome by Considering Hydrophobic Amino Acids Topology. Sci. Rep. 2017, 7, 41425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Povolotskaya IS; Kondrashov FA Sequence Space and the Ongoing Expansion of the Protein Universe. Nature 2010, 465, 922–926. [DOI] [PubMed] [Google Scholar]
- (8).Peng Z; Yan J; Fan X; Mizianty MJ; Xue B; Wang K; Hu G; Uversky VN; Kurgan L Exceptionally Abundant Exceptions: Comprehensive Characterization of Intrinsic Disorder in All Domains of Life. Cell. Mol. Life Sci. 2015, 72, 137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Oldfield CJ; Dunker AK Intrinsically Disordered Proteins and Intrinsically Disordered Protein Regions. Annu. Rev. Biochem. 2014, 83, 553–584. [DOI] [PubMed] [Google Scholar]
- (10).Dyson HJ; Wright PE Intrinsically Unstructured Proteins and Their Functions. Nat. Rev. Mol. Cell. Biol. 2005, 6, 197–208. [DOI] [PubMed] [Google Scholar]
- (11).Xue B; Dunker AK; Uversky VN Orderly Order in Protein Intrinsic Disorder Distribution: Disorder in 3500 Proteomes from Viruses and the Three Domains of Life. J. Biomol. Struct. Dyn. 2012, 30, 137–149. [DOI] [PubMed] [Google Scholar]
- (12).Ward JJ; Sodhi JS; McGuffin LJ; Buxton BF; Jones DT Prediction and Functional Analysis of Native Disorder in Proteins from the Three Kingdoms of Life. J. Mol. Biol. 2004, 337, 635–645. [DOI] [PubMed] [Google Scholar]
- (13).Uversky VN; Dunker AK Understanding Protein Non-Folding. Biochim. Biophys. Acta 2010, 1804, 1231–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Uversky VN The Mysterious Unfoldome: Structureless, Underappreciated, yet Vital Part of Any Given Proteome. J. Biomed. Biotechnol. 2010, 2010, 568068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Dunker AK; Obradovic Z; Romero P; Garner EC; Brown CJ Intrinsic Protein Disorder in Complete Genomes. Genome Inform. Ser. Workshop Genome Inform. 2000, 11, 161–171. [PubMed] [Google Scholar]
- (16).Dunker AK; Lawson JD; Brown CJ; Williams RM; Romero P; Oh JS; Oldfield CJ; Campen AM; Ratliff CM; Hipps KW; Ausio J; Nissen MS; Reeves R; Kang C; Kissinger CR; Bailey RW; Griswold MD; Chiu W; Garner EC; Obradovic Z Intrinsically Disordered Protein. J. Mol. Graph. Model. 2001, 19, 26–59. [DOI] [PubMed] [Google Scholar]
- (17).Bogatyreva NS; Finkelstein AV; Galzitskaya OV Trend of Amino Acid Composition of Proteins of Different Taxa. J. Bioinform. Comput. Biol. 2006, 4, 597–608. [DOI] [PubMed] [Google Scholar]
- (18).Oldfield CJ; Cheng Y; Cortese MS; Brown CJ; Uversky VN; Dunker AK Comparing and Combining Predictors of Mostly Disordered Proteins. Biochemistry 2005, 44, 1989–2000. [DOI] [PubMed] [Google Scholar]
- (19).Gao C; Ma C; Wang H; Zhong H; Zang J; Zhong R; He F; Yang D Intrinsic Disorder in Protein Domains Contributes to Both Organism Complexity and Clade-Specific Functions. Sci. Rep. 2021, 11, 2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Uversky VN The Multifaceted Roles of Intrinsic Disorder in Protein Complexes. FEBS Lett. 2015, 589, 2498–2506. [DOI] [PubMed] [Google Scholar]
- (21).Uversky VN Intrinsic Disorder, Protein-Protein Interactions, and Disease. Adv. Protein Chem. Struct. Biol. 2018, 110, 85–121. [DOI] [PubMed] [Google Scholar]
- (22).Barabasi AL; Albert R Emergence of Scaling in Random Networks. Science 1999, 286, 509–512. [DOI] [PubMed] [Google Scholar]
- (23).Barabasi AL Scale-Free Networks: A Decade and Beyond. Science 2009, 325, 412–413. [DOI] [PubMed] [Google Scholar]
- (24).Rangarajan N; Kulkarni P; Hannenhalli S Evolutionarily Conserved Network Properties of Intrinsically Disordered Proteins. PLoS One 2015, 10, e0126729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Patil A; Kinoshita K; Nakamura H Hub Promiscuity in Protein-Protein Interaction Networks. Int. J. Mol. Sci. 2010, 11, 1930–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Oldfield CJ; Meng J; Yang JY; Yang MQ; Uversky VN; Dunker AK Flexible Nets: Disorder and Induced Fit in the Associations of P53 and 14–3-3 with Their Partners. BMC Genomics 2008, 9 Suppl 1, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Hu G; Wu Z; Uversky VN; Kurgan L Functional Analysis of Human Hub Proteins and Their Interactors Involved in the Intrinsic Disorder-Enriched Interactions. Int. J. Mol. Sci. 2017, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Haynes C; Oldfield CJ; Ji F; Klitgord N; Cusick ME; Radivojac P; Uversky VN; Vidal M; Iakoucheva LM Intrinsic Disorder Is a Common Feature of Hub Proteins from Four Eukaryotic Interactomes. PLoS Comput. Biol. 2006, 2, e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Gsponer J; Babu MM The Rules of Disorder or Why Disorder Rules. Prog. Biophys. Mol. Biol. 2009, 99, 94–103. [DOI] [PubMed] [Google Scholar]
- (30).Dunker AK; Cortese MS; Romero P; Iakoucheva LM; Uversky VN Flexible Nets. The Roles of Intrinsic Disorder in Protein Interaction Networks. FEBS J. 2005, 272, 5129–5148. [DOI] [PubMed] [Google Scholar]
- (31).Wright PE; Dyson HJ Intrinsically Disordered Proteins in Cellular Signalling and Regulation. Nat. Rev. Mol. Cell. Biol. 2015, 16, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Uversky VN; Oldfield CJ; Dunker AK Intrinsically Disordered Proteins in Human Diseases: Introducing the D2 Concept. Annu. Rev. Biophys. 2008, 37, 215–246. [DOI] [PubMed] [Google Scholar]
- (33).Uversky VN Functional Roles of Transiently and Intrinsically Disordered Regions within Proteins. FEBS J. 2015, 282, 1182–1189. [DOI] [PubMed] [Google Scholar]
- (34).Urakami K; Takahashi K; Adachi Y; Awaki E; Mura T; Ikawa S Apolipoprotein Abnormalities in Dementia. Jpn. J. Psychiatry Neurol. 1989, 43, 63–65. [DOI] [PubMed] [Google Scholar]
- (35).Shammas SL Mechanistic Roles of Protein Disorder within Transcription. Curr. Opin. Struct. Biol. 2017, 42, 155–161. [DOI] [PubMed] [Google Scholar]
- (36).Bondos SE; Dunker AK; Uversky VN On the Roles of Intrinsically Disordered Proteins and Regions in Cell Communication and Signaling. Cell. Commun. Signal. 2021, 19, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Yoon MK; Mitrea DM; Ou L; Kriwacki RW Cell Cycle Regulation by the Intrinsically Disordered Proteins P21 and P27. Biochem. Soc. Trans. 2012, 40, 981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Galea CA; Wang Y; Sivakolundu SG; Kriwacki RW Regulation of Cell Division by Intrinsically Unstructured Proteins: Intrinsic Flexibility, Modularity, and Signaling Conduits. Biochemistry 2008, 47, 7598–7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Michael AK; Fribourgh JL; Van Gelder RN; Partch CL Animal Cryptochromes: Divergent Roles in Light Perception, Circadian Timekeeping and Beyond. Photochem. Photobiol. 2017, 93, 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Hurley JM; Loros JJ; Dunlap JC Circadian Oscillators: Around the Transcription-Translation Feedback Loop and on to Output. Trends Biochem. Sci. 2016, 41, 834–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Hurley JM; Larrondo LF; Loros JJ; Dunlap JC Conserved Rna Helicase Frh Acts Nonenzymatically to Support the Intrinsically Disordered Neurospora Clock Protein Frq. Mol. Cell 2013, 52, 832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Dong P; Fan Y; Sun J; Lv M; Yi M; Tan X; Liu S A Dynamic Interaction Process between Kaia and Kaic Is Critical to the Cyanobacterial Circadian Oscillator. Sci. Rep. 2016, 6, 25129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Mooney SM; Jolly MK; Levine H; Kulkarni P Phenotypic Plasticity in Prostate Cancer: Role of Intrinsically Disordered Proteins. Asian J. Androl. 2016, 18, 704–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Xue B; Oldfield CJ; Van YY; Dunker AK; Uversky VN Protein Intrinsic Disorder and Induced Pluripotent Stem Cells. Mol. Biosyst. 2012, 8, 134–150. [DOI] [PubMed] [Google Scholar]
- (45).Babu MM The Contribution of Intrinsically Disordered Regions to Protein Function, Cellular Complexity, and Human Disease. Biochem. Soc. Trans. 2016, 44, 1185–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Marcotte EM; Tsechansky M Disorder, Promiscuity, and Toxic Partnerships. Cell 2009, 138, 16–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Vavouri T; Semple JI; Garcia-Verdugo R; Lehner B Intrinsic Protein Disorder and Interaction Promiscuity Are Widely Associated with Dosage Sensitivity. Cell 2009, 138, 198–208. [DOI] [PubMed] [Google Scholar]
- (48).Cumberworth A; Lamour G; Babu MM; Gsponer J Promiscuity as a Functional Trait: Intrinsically Disordered Regions as Central Players of Interactomes. Biochem. J. 2013, 454, 361–369. [DOI] [PubMed] [Google Scholar]
- (49).Cheng Y; LeGall T; Oldfield CJ; Dunker AK; Uversky VN Abundance of Intrinsic Disorder in Protein Associated with Cardiovascular Disease. Biochemistry 2006, 45, 10448–10460. [DOI] [PubMed] [Google Scholar]
- (50).Uversky VN Amyloidogenesis of Natively Unfolded Proteins. Curr. Alzheimer Res. 2008, 5, 260–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Du Z; Uversky VN A Comprehensive Survey of the Roles of Highly Disordered Proteins in Type 2 Diabetes. Int. J. Mol. Sci. 2017, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Iakoucheva LM; Brown CJ; Lawson JD; Obradovic Z; Dunker AK Intrinsic Disorder in Cell-Signaling and Cancer-Associated Proteins. J. Mol. Biol. 2002, 323, 573–584. [DOI] [PubMed] [Google Scholar]
- (53).Uversky VN Intrinsic Disorder in Proteins Associated with Neurodegenerative Diseases. Front. Biosci. (Landmark Ed.) 2009, 14, 5188–5238. [DOI] [PubMed] [Google Scholar]
- (54).Kulkarni P; Uversky VN Intrinsically Disordered Proteins in Chronic Diseases. Biomolecules 2019, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Midic U; Oldfield CJ; Dunker AK; Obradovic Z; Uversky VN Protein Disorder in the Human Diseasome: Unfoldomics of Human Genetic Diseases. BMC Genomics 2009, 10 Suppl 1, S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Uversky VN The Triple Power of D(3): Protein Intrinsic Disorder in Degenerative Diseases. Front. Biosci. (Landmark Ed.) 2014, 19, 181–258. [DOI] [PubMed] [Google Scholar]
- (57).Dyson HJ; Wright PE Coupling of Folding and Binding for Unstructured Proteins. Curr. Opin. Struct. Biol. 2002, 12, 54–60. [DOI] [PubMed] [Google Scholar]
- (58).Boehr DD; Nussinov R; Wright PE The Role of Dynamic Conformational Ensembles in Biomolecular Recognition. Nat. Chem. Biol. 2009, 5, 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Permyakov SE; Millett IS; Doniach S; Permyakov EA; Uversky VN Natively Unfolded C-Terminal Domain of Caldesmon Remains Substantially Unstructured after the Effective Binding to Calmodulin. Proteins 2003, 53, 855–862. [DOI] [PubMed] [Google Scholar]
- (60).Borg M; Mittag T; Pawson T; Tyers M; Forman-Kay JD; Chan HS Polyelectrostatic Interactions of Disordered Ligands Suggest a Physical Basis for Ultrasensitivity. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 9650–9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Sigalov AB; Zhuravleva AV; Orekhov VY Binding of Intrinsically Disordered Proteins Is Not Necessarily Accompanied by a Structural Transition to a Folded Form. Biochimie 2007, 89, 419–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Tompa P; Fuxreiter M Fuzzy Complexes: Polymorphism and Structural Disorder in Protein-Protein Interactions. Trends Biochem. Sci. 2008, 33, 2–8. [DOI] [PubMed] [Google Scholar]
- (63).Sigalov AB Protein Intrinsic Disorder and Oligomericity in Cell Signaling. Mol. Biosyst. 2010, 6, 451–461. [DOI] [PubMed] [Google Scholar]
- (64).Sigalov AB Uncoupled Binding and Folding of Immune Signaling-Related Intrinsically Disordered Proteins. Prog. Biophys. Mol. Biol. 2011, 106, 525–536. [DOI] [PubMed] [Google Scholar]
- (65).Fuxreiter M Fuzzy Protein Theory for Disordered Proteins. Biochem. Soc. Trans. 2020, 48, 2557–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Choi UB; McCann JJ; Weninger KR; Bowen ME Beyond the Random Coil: Stochastic Conformational Switching in Intrinsically Disordered Proteins. Structure 2011, 19, 566–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Bryan PN; Orban J Proteins That Switch Folds. Curr. Opin. Struct. Biol. 2010, 20, 482–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Chakrabortee S; Meersman F; Kaminski Schierle GS; Bertoncini CW; McGee B; Kaminski CF; Tunnacliffe A Catalytic and Chaperone-Like Functions in an Intrinsically Disordered Protein Associated with Desiccation Tolerance. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 16084–16089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).He Y; Chen Y; Mooney SM; Rajagopalan K; Bhargava A; Sacho E; Weninger K; Bryan PN; Kulkarni P; Orban J Phosphorylation-Induced Conformational Ensemble Switching in an Intrinsically Disordered Cancer/Testis Antigen. J. Biol. Chem. 2015, 290, 25090–25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Andresen C; Helander S; Lemak A; Fares C; Csizmok V; Carlsson J; Penn LZ; Forman-Kay JD; Arrowsmith CH; Lundstrom P; Sunnerhagen M Transient Structure and Dynamics in the Disordered C-Myc Transactivation Domain Affect Bin1 Binding. Nucleic Acids Res. 2012, 40, 6353–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Gianni S; Freiberger MI; Jemth P; Ferreiro DU; Wolynes PG; Fuxreiter M Fuzziness and Frustration in the Energy Landscape of Protein Folding, Function, and Assembly. Acc. Chem. Res. 2021, 54, 1251–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Freiberger MI; Wolynes PG; Ferreiro DU; Fuxreiter M Frustration in Fuzzy Protein Complexes Leads to Interaction Versatility. J. Phys. Chem. B 2021, 125, 2513–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Wright PE; Dyson HJ Linking Folding and Binding. Curr. Opin. Struct. Biol. 2009, 19, 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Borgia A; Borgia MB; Bugge K; Kissling VM; Heidarsson PO; Fernandes CB; Sottini A; Soranno A; Buholzer KJ; Nettels D; Kragelund BB; Best RB; Schuler B Extreme Disorder in an Ultrahigh-Affinity Protein Complex. Nature 2018, 555, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Mittag T; Kay LE; Forman-Kay JD Protein Dynamics and Conformational Disorder in Molecular Recognition. J. Mol. Recognit. 2010, 23, 105–116. [DOI] [PubMed] [Google Scholar]
- (76).Uversky VN; Gillespie JR; Fink AL Why Are “Natively Unfolded” Proteins Unstructured under Physiologic Conditions? Proteins 2000, 41, 415–427. [DOI] [PubMed] [Google Scholar]
- (77).Uversky VN Intrinsically Disordered Proteins in Overcrowded Milieu: Membrane-Less Organelles, Phase Separation, and Intrinsic Disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [DOI] [PubMed] [Google Scholar]
- (78).Darling AL; Liu Y; Oldfield CJ; Uversky VN Intrinsically Disordered Proteome of Human Membrane-Less Organelles. Proteomics 2018, 18, e1700193. [DOI] [PubMed] [Google Scholar]
- (79).Uversky VN Protein Intrinsic Disorder-Based Liquid-Liquid Phase Transitions in Biological Systems: Complex Coacervates and Membrane-Less Organelles. Adv. Colloid. Interface Sci. 2017, 239, 97–114. [DOI] [PubMed] [Google Scholar]
- (80).Uversky VN The Roles of Intrinsic Disorder-Based Liquid-Liquid Phase Transitions in the “Dr. Jekyll-Mr. Hyde” Behavior of Proteins Involved in Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration. Autophagy 2017, 13, 2115–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Chu WT; Wang J Thermodynamic and Sequential Characteristics of Phase Separation and Droplet Formation for an Intrinsically Disordered Region/Protein Ensemble. PLoS Comput. Biol. 2021, 17, e1008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Mammen Regy R; Zheng W; Mittal J Using a Sequence-Specific Coarse-Grained Model for Studying Protein Liquid-Liquid Phase Separation. Methods Enzymol. 2021, 646, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Uversky VN Per Aspera Ad Chaos: A Personal Journey to the Wonderland of Intrinsic Disorder. Biochem. J. 2021, 478, 3015–3024. [DOI] [PubMed] [Google Scholar]
- (84).Gánti’ T Biogenesis Itself. J. Theor. Biol. 1997, 187, 583–593. [DOI] [PubMed] [Google Scholar]
- (85).Gánti’ T The Principles of Life; Oxford University Press: Oxford, 2003, p. 224. [Google Scholar]
- (86).Szathmary E; Smith JM The Major Evolutionary Transitions. Nature 1995, 374, 227–232. [DOI] [PubMed] [Google Scholar]
- (87).Kaneko K Life: An Introduction to Complex Systems Biology; Springer: Berlin, Heidelberg, 2006, p. 374. [Google Scholar]
- (88).Prigogine I The End of Certainty : Time, Chaos, and the New Laws of Nature; The Free Press: New York, 1997, p. 240. [Google Scholar]
- (89).Maithreye R; Suguna C; Sinha S Collective Dynamics of Multicellular Systems. Pramana 2011, 77, 843–853. [Google Scholar]
- (90).Erdös P; Rényi A On Random Graphs I. Publ. Math. Debr. 1959, 6, 290. [Google Scholar]
- (91).Barabasi AL; Bonabeau E Scale-Free Networks. Sci. Am. 2003, 288, 60–69. [DOI] [PubMed] [Google Scholar]
- (92).Albert R; Jeong H; Barabasi AL Error and Attack Tolerance of Complex Networks. Nature 2000, 406, 378–382. [DOI] [PubMed] [Google Scholar]
- (93).Strogatz SH Complex Systems: Romanesque Networks. Nature 2005, 433, 365–366. [DOI] [PubMed] [Google Scholar]
- (94).Uversky VN; Giuliani A Networks of Networks: An Essay on Multi-Level Biological Organization. Front. Genet. 2021, 12, 706260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Mahmoudabadi G; Rajagopalan K; Getzenberg RH; Hannenhalli S; Rangarajan G; Kulkarni P Intrinsically Disordered Proteins and Conformational Noise: Implications in Cancer. Cell Cycle 2013, 12, 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Kulkarni V; Kulkarni P Intrinsically Disordered Proteins and Phenotypic Switching: Implications in Cancer. Prog. Mol. Biol. Transl. Sci. 2019, 166, 63–84. [DOI] [PubMed] [Google Scholar]
- (97).Kulkarni P Intrinsically Disordered Proteins: Insights from Poincare, Waddington, and Lamarck. Biomolecules 2020, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Biswas K; Jolly MK; Ghosh A Stability and Mean Residence Times for Hybrid Epithelial/Mesenchymal Phenotype. Phys. Biol. 2019, 16, 025003. [DOI] [PubMed] [Google Scholar]
- (99).Clark S; Myers JB; King A; Fiala R; Novacek J; Pearce G; Heierhorst J; Reichow SL; Barbar EJ Multivalency Regulates Activity in an Intrinsically Disordered Transcription Factor. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Niklas KJ; Bondos SE; Dunker AK; Newman SA Rethinking Gene Regulatory Networks in Light of Alternative Splicing, Intrinsically Disordered Protein Domains, and Post-Translational Modifications. Front. Cell. Dev. Biol. 2015, 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Staby L; O’Shea C; Willemoes M; Theisen F; Kragelund BB; Skriver K Eukaryotic Transcription Factors: Paradigms of Protein Intrinsic Disorder. Biochem. J. 2017, 474, 2509–2532. [DOI] [PubMed] [Google Scholar]
- (102).Strzyz P Concentrating on Intrinsic Disorder. Nat. Rev. Mol. Cell. Biol. 2018, 19, 544. [DOI] [PubMed] [Google Scholar]
- (103).Eldar A; Elowitz MB Functional Roles for Noise in Genetic Circuits. Nature 2010, 467, 167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Yamanaka S Elite and Stochastic Models for Induced Pluripotent Stem Cell Generation. Nature 2009, 460, 49–52. [DOI] [PubMed] [Google Scholar]
- (105).Wakao S; Kitada M; Dezawa M The Elite and Stochastic Model for Ips Cell Generation: Multilineage-Differentiating Stress Enduring (Muse) Cells Are Readily Reprogrammable into Ips Cells. Cytometry A 2013, 83, 18–26. [DOI] [PubMed] [Google Scholar]
- (106).MacArthur BD; Please CP; Oreffo RO Stochasticity and the Molecular Mechanisms of Induced Pluripotency. PLoS One 2008, 3, e3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Lin YT; Hufton PG; Lee EJ; Potoyan DA A Stochastic and Dynamical View of Pluripotency in Mouse Embryonic Stem Cells. PLoS Comput. Biol. 2018, 14, e1006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Emran AA; Marzese DM; Menon DR; Stark MS; Torrano J; Hammerlindl H; Zhang G; Brafford P; Salomon MP; Nelson N; Hammerlindl S; Gupta D; Mills GB; Lu Y; Sturm RA; Flaherty K; Hoon DSB; Gabrielli B; Herlyn M; Schaider H Distinct Histone Modifications Denote Early Stress-Induced Drug Tolerance in Cancer. Oncotarget 2018, 9, 8206–8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Chung KM; Kolling F. W. t.; Gajdosik MD; Burger S; Russell AC; Nelson CE Single Cell Analysis Reveals the Stochastic Phase of Reprogramming to Pluripotency Is an Ordered Probabilistic Process. PLoS One 2014, 9, e95304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Sehl ME; Shimada M; Landeros A; Lange K; Wicha MS Modeling of Cancer Stem Cell State Transitions Predicts Therapeutic Response. PLoS One 2015, 10, e0135797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Gupta PB; Fillmore CM; Jiang G; Shapira SD; Tao K; Kuperwasser C; Lander ES Stochastic State Transitions Give Rise to Phenotypic Equilibrium in Populations of Cancer Cells. Cell 2011, 146, 633–644. [DOI] [PubMed] [Google Scholar]
- (112).Waddington CH Canalization of Development and the Inheritance of Acquired Characters. Nature 1942, 150, 563–565. [Google Scholar]
- (113).Sonnenschein C; Soto AM; Rangarajan A; Kulkarni P Competing Views on Cancer. J. Biosci. 2014, 39, 281–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Su Y; Wei W; Robert L; Xue M; Tsoi J; Garcia-Diaz A; Homet Moreno B; Kim J; Ng RH; Lee JW; Koya RC; Comin-Anduix B; Graeber TG; Ribas A; Heath JR Single-Cell Analysis Resolves the Cell State Transition and Signaling Dynamics Associated with Melanoma Drug-Induced Resistance. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 13679–13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Sharma SV; Lee DY; Li B; Quinlan MP; Takahashi F; Maheswaran S; McDermott U; Azizian N; Zou L; Fischbach MA; Wong KK; Brandstetter K; Wittner B; Ramaswamy S; Classon M; Settleman J A Chromatin-Mediated Reversible Drug-Tolerant State in Cancer Cell Subpopulations. Cell 2010, 141, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Sahoo S; Mishra A; Kaur H; Hari K; Muralidharan S; Mandal S; Jolly MK A Mechanistic Model Captures the Emergence and Implications of Non-Genetic Heterogeneity and Reversible Drug Resistance in Er+ Breast Cancer Cells. NAR Cancer 2021, 3, zcab027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Hammerlindl H; Schaider H Tumor Cell-Intrinsic Phenotypic Plasticity Facilitates Adaptive Cellular Reprogramming Driving Acquired Drug Resistance. J. Cell. Commun. Signal. 2018, 12, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Shachaf CM; Kopelman AM; Arvanitis C; Karlsson A; Beer S; Mandl S; Bachmann MH; Borowsky AD; Ruebner B; Cardiff RD; Yang Q; Bishop JM; Contag CH; Felsher DW Myc Inactivation Uncovers Pluripotent Differentiation and Tumour Dormancy in Hepatocellular Cancer. Nature 2004, 431, 1112–1117. [DOI] [PubMed] [Google Scholar]
- (119).Shachaf CM; Felsher DW Tumor Dormancy and Myc Inactivation: Pushing Cancer to the Brink of Normalcy. Cancer Res. 2005, 65, 4471–4474. [DOI] [PubMed] [Google Scholar]
- (120).Neerathilingam M; Bairy SG; Mysore S Deciphering Mode of Action of Functionally Important Regions in the Intrinsically Disordered Paxillin (Residues 1–313) Using Its Interaction with Fat (Focal Adhesion Targeting Domain of Focal Adhesion Kinase). PLoS One 2016, 11, e0150153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Rangarajan N; Fox Z; Singh A; Kulkarni P; Rangarajan G Disorder, Oscillatory Dynamics and State Switching: The Role of C-Myc. J. Theor. Biol. 2015, 386, 105–114. [DOI] [PubMed] [Google Scholar]
- (122).Deisboeck TS; Wang Z; Macklin P; Cristini V Multiscale Cancer Modeling. Annu. Rev. Biomed. Eng. 2011, 13, 127–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Meier-Schellersheim M; Fraser IDC; Klauschen F Multiscale Modeling for Biologists. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009, 1, 4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Poincar’e H Sur Le ProblÈme Des Trois Corps Et Les ‘Equations De La Dynamique. Acta Math. 1890, 13, 1–270. [Google Scholar]
- (125).Jia D; Jolly MK; Kulkarni P; Levine H Phenotypic Plasticity and Cell Fate Decisions in Cancer: Insights from Dynamical Systems Theory. Cancers (Basel) 2017, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Waddington CH; Kacser H The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology; Allen & Unwin: London, 1957. [Google Scholar]
- (127).Li Q; Wennborg A; Aurell E; Dekel E; Zou JZ; Xu Y; Huang S; Ernberg I Dynamics inside the Cancer Cell Attractor Reveal Cell Heterogeneity, Limits of Stability, and Escape. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 2672–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Huang S; Kauffman S How to Escape the Cancer Attractor: Rationale and Limitations of Multi-Target Drugs. Semin. Cancer Biol. 2013, 23, 270–278. [DOI] [PubMed] [Google Scholar]
- (129).Huang S; Ernberg I; Kauffman S Cancer Attractors: A Systems View of Tumors from a Gene Network Dynamics and Developmental Perspective. Semin. Cell. Dev. Biol. 2009, 20, 869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Baranger M; Institute NECS Chaos, Complexity, and Entropy: A Physics Talk for Non-Physicists; New England Complex Systems Institute, 2001. [Google Scholar]
- (131).Uversky VN Unusual Biophysics of Intrinsically Disordered Proteins. Biochim. Biophys. Acta 2013, 1834, 932–951. [DOI] [PubMed] [Google Scholar]
- (132).Ruelle D; Takens F On the Nature of Turbulence. Commun. Math. Phys. 1971, 20, 167–192. [Google Scholar]
- (133).Gribbin J Deep Simplicity: Chaos, Complexity and the Emergence of Life; Penguin Adult, 2005. [Google Scholar]
- (134).Lorenz EN Deterministic Nonperiodic Flow. J. Atmos. Sci. 1963, 20, 130–141. [Google Scholar]
- (135).Lorenz EN Section of Planetary Sciences: The Predictability of Hydrodynamic Flow*,†. Trans. N. Y. Acad. Sci. 1963, 25, 409–432. [Google Scholar]
- (136).Bu Z; Cook J; Callaway DJE Dynamic Regimes and Correlated Structural Dynamics in Native and Denatured Alpha-Lactalbumin11edited by M. F. Moody. J Mol. Biol. 2001, 312, 865–873. [DOI] [PubMed] [Google Scholar]
- (137).Uthamacumaran A A Review of Dynamical Systems Approaches for the Detection of Chaotic Attractors in Cancer Networks. Patterns (N Y) 2021, 2, 100226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (138).Jolliffe IT; Cadima J Principal Component Analysis: A Review and Recent Developments. Philos. Trans. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Molgedey L; Schuster HG Separation of a Mixture of Independent Signals Using Time Delayed Correlations. Phys. Rev. Lett. 1994, 72, 3634–3637. [DOI] [PubMed] [Google Scholar]
- (140).Pinsker MS Dynamical Systems with Completely Positive or Zero Entropy. Soviet Math. Dokl. 1960, 133, 1025–1026. [Google Scholar]
- (141).Austin T Measure Concentration and the Weak Pinsker Property. Publ. Math. IHÉS 2018, 128, 1–119. [Google Scholar]
- (142).Pathak J; Hunt B; Girvan M; Lu Z; Ott E Model-Free Prediction of Large Spatiotemporally Chaotic Systems from Data: A Reservoir Computing Approach. Phys. Rev. Lett. 2018, 120, 024102. [DOI] [PubMed] [Google Scholar]
- (143).Pathak J; Lu Z; Hunt BR; Girvan M; Ott E Using Machine Learning to Replicate Chaotic Attractors and Calculate Lyapunov Exponents from Data. Chaos 2017, 27, 121102. [DOI] [PubMed] [Google Scholar]
- (144).Haran G; Mazal H How Fast Are the Motions of Tertiary-Structure Elements in Proteins? J. Chem. Phys. 2020, 153, 130902. [DOI] [PubMed] [Google Scholar]
- (145).Burkoff NS; Varnai C; Wells SA; Wild DL Exploring the Energy Landscapes of Protein Folding Simulations with Bayesian Computation. Biophys. J. 2012, 102, 878–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).Lemons DS; Gythiel A Paul Langevin’s 1908 Paper “on the Theory of Brownian Motion” [“Sur La Théorie Du Mouvement Brownien,” C. R. Acad. Sci. (Paris) 146, 530–533 (1908)]. Am. J. Phys. 1997, 65, 1079–1081. [Google Scholar]
- (147).Pritchard L; Dufton MJ Do Proteins Learn to Evolve? The Hopfield Network as a Basis for the Understanding of Protein Evolution. J. Theor. Biol. 2000, 202, 77–86. [DOI] [PubMed] [Google Scholar]
- (148).Fard AT; Srihari S; Mar JC; Ragan MA Not Just a Colourful Metaphor: Modelling the Landscape of Cellular Development Using Hopfield Networks. NPJ Syst. Biol. Appl. 2016, 2, 16001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Kauffman SA The Origins of Order: Self-Organization and Selection in Evolution; Oxford University Press: Oxford, New York, Toronto, 1993, p. 734 [Google Scholar]
- (150).Kulkarni P; Uversky VN Intrinsically Disordered Proteins: The Dark Horse of the Dark Proteome. Proteomics 2018, 18, e1800061. [DOI] [PubMed] [Google Scholar]
- (151).Kunnev D; Gospodinov A Possible Emergence of Sequence Specific Rna Aminoacylation Via Peptide Intermediary to Initiate Darwinian Evolution and Code through Origin of Life. Life (Basel) 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (152).Chatterjee S; Yadav S The Origin of Prebiotic Information System in the Peptide/Rna World: A Simulation Model of the Evolution of Translation and the Genetic Code. Life (Basel) 2019, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Vacic V; Uversky VN; Dunker AK; Lonardi S Composition Profiler: A Tool for Discovery and Visualization of Amino Acid Composition Differences. BMC Bioinformatics 2007, 8, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Romero P; Obradovic Z; Li X; Garner EC; Brown CJ; Dunker AK Sequence Complexity of Disordered Protein. Proteins 2001, 42, 38–48. [DOI] [PubMed] [Google Scholar]
- (155).Campen A; Williams RM; Brown CJ; Meng J; Uversky VN; Dunker AK Top-Idp-Scale: A New Amino Acid Scale Measuring Propensity for Intrinsic Disorder. Protein Pept. Lett. 2008, 15, 956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (156).Seckbach J Precursors of Life, Chemical Models and Early Biological Evolution; Springer, Dordrecht, 2012. [Google Scholar]
- (157).Uversky VN A Decade and a Half of Protein Intrinsic Disorder: Biology Still Waits for Physics. Protein Sci. 2013, 22, 693–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (158).Brooks DJ; Fresco JR; Lesk AM; Singh M Evolution of Amino Acid Frequencies in Proteins over Deep Time: Inferred Order of Introduction of Amino Acids into the Genetic Code. Mol. Biol. Evol. 2002, 19, 1645–1655. [DOI] [PubMed] [Google Scholar]
- (159).Poole AM; Jeffares DC; Penny D The Path from the Rna World. J. Mol. Evol. 1998, 46, 1–17. [DOI] [PubMed] [Google Scholar]
- (160).Kulkarni P; Uversky VN Intrinsically Disordered Proteins and the Janus Challenge. Biomolecules 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Zambelli B; Cremades N; Neyroz P; Turano P; Uversky VN; Ciurli S Insights in the (Un)Structural Organization of Bacillus Pasteurii Ureg, an Intrinsically Disordered Gtpase Enzyme. Mol. Biosyst. 2012, 8, 220–228. [DOI] [PubMed] [Google Scholar]
- (162).Vamvaca K; Vogeli B; Kast P; Pervushin K; Hilvert D An Enzymatic Molten Globule: Efficient Coupling of Folding and Catalysis. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 12860–12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (163).Uversky VN; Kutyshenko VP; Protasova N; Rogov VV; Vassilenko KS; Gudkov AT Circularly Permuted Dihydrofolate Reductase Possesses All the Properties of the Molten Globule State, but Can Resume Functional Tertiary Structure by Interaction with Its Ligands. Protein Sci. 1996, 5, 1844–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (164).Pervushin K; Vamvaca K; Vogeli B; Hilvert D Structure and Dynamics of a Molten Globular Enzyme. Nat. Struct. Mol. Biol. 2007, 14, 1202–1206. [DOI] [PubMed] [Google Scholar]
- (165).Pohorille A; Wilson MA; Shannon G Flexible Proteins at the Origin of Life. Life (Basel) 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Woycechowsky KJ; Choutko A; Vamvaca K; Hilvert D Relative Tolerance of an Enzymatic Molten Globule and Its Thermostable Counterpart to Point Mutation. Biochemistry 2008, 47, 13489–13496. [DOI] [PubMed] [Google Scholar]
- (167).Vamvaca K; Jelesarov I; Hilvert D Kinetics and Thermodynamics of Ligand Binding to a Molten Globular Enzyme and Its Native Counterpart. J Mol Biol 2008, 382, 971–977. [DOI] [PubMed] [Google Scholar]
- (168).Stojanovski BM; Breydo L; Hunter GA; Uversky VN; Ferreira GC Catalytically Active Alkaline Molten Globular Enzyme: Effect of Ph and Temperature on the Structural Integrity of 5-Aminolevulinate Synthase. Biochim. Biophys. Acta 2014, 1844, 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Protasova N; Kireeva ML; Murzina NV; Murzin AG; Uversky VN; Gryaznova OI; Gudkov AT Circularly Permuted Dihydrofolate Reductase of E. Coli Has Functional Activity and a Destabilized Tertiary Structure. Protein Eng. 1994, 7, 1373–1377. [DOI] [PubMed] [Google Scholar]
- (170).Palombo M; Bonucci A; Etienne E; Ciurli S; Uversky VN; Guigliarelli B; Belle V; Mileo E; Zambelli B The Relationship between Folding and Activity in Ureg, an Intrinsically Disordered Enzyme. Sci. Rep. 2017, 7, 5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (171).Larion M; Miller B; Bruschweiler R Conformational Heterogeneity and Intrinsic Disorder in Enzyme Regulation: Glucokinase as a Case Study. Intrinsically Disord. Proteins 2015, 3, e1011008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (172).DeForte S; Uversky VN Not an Exception to the Rule: The Functional Significance of Intrinsically Disordered Protein Regions in Enzymes. Mol. Biosyst. 2017, 13, 463–469. [DOI] [PubMed] [Google Scholar]
- (173).Zhu L; Brangwynne CP Nuclear Bodies: The Emerging Biophysics of Nucleoplasmic Phases. Curr. Opin. Cell. Biol. 2015, 34, 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (174).Uversky VN; Kuznetsova IM; Turoverov KK; Zaslavsky B Intrinsically Disordered Proteins as Crucial Constituents of Cellular Aqueous Two Phase Systems and Coacervates. FEBS Lett. 2015, 589, 15–22. [DOI] [PubMed] [Google Scholar]
- (175).Mitrea DM; Kriwacki RW Phase Separation in Biology; Functional Organization of a Higher Order. Cell Commun. Signal. 2016, 14, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (176).Feric M; Vaidya N; Harmon TS; Mitrea DM; Zhu L; Richardson TM; Kriwacki RW; Pappu RV; Brangwynne CP Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (177).Dundr M; Misteli T Biogenesis of Nuclear Bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (178).Brangwynne CP Phase Transitions and Size Scaling of Membrane-Less Organelles. J. Cell. Biol. 2013, 203, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (179).Brangwynne Clifford P.; Tompa P; Pappu Rohit V. Polymer Physics of Intracellular Phase Transitions. Nat. Phys. 2015, 11, 899–904. [Google Scholar]
- (180).Nesterov SV; Ilyinsky NS; Uversky VN Liquid-Liquid Phase Separation as a Common Organizing Principle of Intracellular Space and Biomembranes Providing Dynamic Adaptive Responses. Biochim. Biophys. Acta Mol. Cell. Res. 2021, 1868, 119102. [DOI] [PubMed] [Google Scholar]
- (181).Strulson CA; Molden RC; Keating CD; Bevilacqua PC Rna Catalysis through Compartmentalization. Nat. Chem. 2012, 4, 941–946. [DOI] [PubMed] [Google Scholar]
- (182).Sokolova E; Spruijt E; Hansen MM; Dubuc E; Groen J; Chokkalingam V; Piruska A; Heus HA; Huck WT Enhanced Transcription Rates in Membrane-Free Protocells Formed by Coacervation of Cell Lysate. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 11692–11697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (183).Asherie N Protein Crystallization and Phase Diagrams. Methods 2004, 34, 266–272. [DOI] [PubMed] [Google Scholar]
- (184).Oparin AI The Origin of Life; Dover Publications: Mineola, NY, 1938, p. 270. [Google Scholar]
- (185).Abbas M; Lipiński WP; Nakashima KK; Huck WTS; Spruijt E A Short Peptide Synthon for Liquid–Liquid Phase Separation. Nat. Chem. 2021, 13, 1046–1054. [DOI] [PubMed] [Google Scholar]
- (186).Mukhopadhyay S The Dynamism of Intrinsically Disordered Proteins: Binding-Induced Folding, Amyloid Formation, and Phase Separation. J. Phys. Chem. B 2020, 124, 11541–11560. [DOI] [PubMed] [Google Scholar]
- (187).Hansma HG Better Than Membranes at the Origin of Life? Life (Basel) 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (188).Jaeken L The Neglected Functions of Intrinsically Disordered Proteins and the Origin of Life. Prog. Biophys. Mol. Biol. 2017, 126, 31–46. [DOI] [PubMed] [Google Scholar]
- (189).Bengtson S; Rasmussen B; Ivarsson M; Muhling J; Broman C; Marone F; Stampanoni M; Bekker A Fungus-Like Mycelial Fossils in 2.4-Billion-Year-Old Vesicular Basalt. Nat. Ecol. Evol. 2017, 1, 0141. [DOI] [PubMed] [Google Scholar]
- (190).Prochnik SE; Umen J; Nedelcu AM; Hallmann A; Miller SM; Nishii I; Ferris P; Kuo A; Mitros T; Fritz-Laylin LK; Hellsten U; Chapman J; Simakov O; Rensing SA; Terry A; Pangilinan J; Kapitonov V; Jurka J; Salamov A; Shapiro H; Schmutz J; Grimwood J; Lindquist E; Lucas S; Grigoriev IV; Schmitt R; Kirk D; Rokhsar DS Genomic Analysis of Organismal Complexity in the Multicellular Green Alga Volvox Carteri. Science 2010, 329, 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (191).Herron MD Origins of Multicellular Complexity: Volvox and the Volvocine Algae. Mol. Ecol. 2016, 25, 1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (192).Hanschen ER; Marriage TN; Ferris PJ; Hamaji T; Toyoda A; Fujiyama A; Neme R; Noguchi H; Minakuchi Y; Suzuki M; Kawai-Toyooka H; Smith DR; Sparks H; Anderson J; Bakaric R; Luria V; Karger A; Kirschner MW; Durand PM; Michod RE; Nozaki H; Olson BJ The Gonium Pectorale Genome Demonstrates Co-Option of Cell Cycle Regulation During the Evolution of Multicellularity. Nat. Commun. 2016, 7, 11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (193).Britten RJ; Davidson EH Gene Regulation for Higher Cells: A Theory. Science 1969, 165, 349–357. [DOI] [PubMed] [Google Scholar]
- (194).Sun X; Jones WT; Rikkerink EH Gras Proteins: The Versatile Roles of Intrinsically Disordered Proteins in Plant Signalling. Biochem. J. 2012, 442, 1–12. [DOI] [PubMed] [Google Scholar]
- (195).Cornish J; Chamberlain SG; Owen D; Mott HR Intrinsically Disordered Proteins and Membranes: A Marriage of Convenience for Cell Signalling? Biochem. Soc. Trans. 2020, 48, 2669–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (196).Chau W; Lee KH Kyphosis Complicating Pregnancy. J. Obstet. Gynaecol. Br. Commonw. 1970, 77, 1098–1102. [DOI] [PubMed] [Google Scholar]
- (197).Dunker AK; Silman I; Uversky VN; Sussman JL Function and Structure of Inherently Disordered Proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [DOI] [PubMed] [Google Scholar]
- (198).Buljan M; Chalancon G; Dunker AK; Bateman A; Balaji S; Fuxreiter M; Babu MM Alternative Splicing of Intrinsically Disordered Regions and Rewiring of Protein Interactions. Curr. Opin. Struct. Biol. 2013, 23, 443–450. [DOI] [PubMed] [Google Scholar]
- (199).Dunker AK; Bondos SE; Huang F; Oldfield CJ Intrinsically Disordered Proteins and Multicellular Organisms. Semin. Cell. Dev. Biol. 2015, 37, 44–55. [DOI] [PubMed] [Google Scholar]
- (200).Desvoyes B; Gutierrez C Roles of Plant Retinoblastoma Protein: Cell Cycle and Beyond. EMBO J. 2020, 39, e105802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (201).Olson BJ; Oberholzer M; Li Y; Zones JM; Kohli HS; Bisova K; Fang SC; Meisenhelder J; Hunter T; Umen JG Regulation of the Chlamydomonas Cell Cycle by a Stable, Chromatin-Associated Retinoblastoma Tumor Suppressor Complex. Plant Cell 2010, 22, 3331–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (202).Cheng Q; Pappas V; Hallmann A; Miller SM Hsp70a and Glsa Interact as Partner Chaperones to Regulate Asymmetric Division in Volvox. Dev. Biol. 2005, 286, 537–548. [DOI] [PubMed] [Google Scholar]
- (203).Johnson KL; Cassin AM; Lonsdale A; Bacic A; Doblin MS; Schultz CJ Pipeline to Identify Hydroxyproline-Rich Glycoproteins. Plant Physiol. 2017, 174, 886–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (204).Lee JH; Waffenschmidt S; Small L; Goodenough U Between-Species Analysis of Short-Repeat Modules in Cell Wall and Sex-Related Hydroxyproline-Rich Glycoproteins of Chlamydomonas. Plant Physiol. 2007, 144, 1813–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (205).Ferris PJ; Waffenschmidt S; Umen JG; Lin H; Lee JH; Ishida K; Kubo T; Lau J; Goodenough UW Plus and Minus Sexual Agglutinins from Chlamydomonas Reinhardtii. Plant Cell 2005, 17, 597–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (206).Ender F; Hallmann A; Amon P; Sumper M Response to the Sexual Pheromone and Wounding in the Green Alga Volvox: Induction of an Extracellular Glycoprotein Consisting Almost Exclusively of Hydroxyproline. J. Biol. Chem. 1999, 274, 35023–35028. [DOI] [PubMed] [Google Scholar]
- (207).Domozych DS; Domozych CE Multicellularity in Green Algae: Upsizing in a Walled Complex. Front. Plant Sci. 2014, 5, 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (208).Niklas KJ; Dunker AK; Yruela I The Evolutionary Origins of Cell Type Diversification and the Role of Intrinsically Disordered Proteins. J. Exp. Bot. 2018, 69, 1437–1446. [DOI] [PubMed] [Google Scholar]
- (209).Knuttgen D; Bremerich D; Rings J; Curth A; Doehn M [Failure of Relaxometry in Diabetic Polyneuropathy]. Anaesthesist 1992, 41, 559–563. [PubMed] [Google Scholar]
- (210).Kastano K; Erdos G; Mier P; Alanis-Lobato G; Promponas VJ; Dosztanyi Z; Andrade-Navarro MA Evolutionary Study of Disorder in Protein Sequences. Biomolecules 2020, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (211).Uversky VN Dancing Protein Clouds: The Strange Biology and Chaotic Physics of Intrinsically Disordered Proteins. J. Biol. Chem. 2016, 291, 6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (212).Kulkarni P; Jolly MK; Jia D; Mooney SM; Bhargava A; Kagohara LT; Chen Y; Hao P; He Y; Veltri RW; Grishaev A; Weninger K; Levine H; Orban J Phosphorylation-Induced Conformational Dynamics in an Intrinsically Disordered Protein and Potential Role in Phenotypic Heterogeneity. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (213).Edwards YJK; Lobley AE; Pentony MM; Jones DT Insights into the Regulation of Intrinsically Disordered Proteins in the Human Proteome by Analyzing Sequence and Gene Expression Data. Genome Biol. 2009, 10, R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (214).Johnson KA; Goody RS The Original Michaelis Constant: Translation of the 1913 Michaelis–Menten Paper. Biochemistry 2011, 50, 8264–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (215).Crick FH On Protein Synthesis. Symp. Soc. Exp. Biol. 1958, 12, 138–163. [PubMed] [Google Scholar]
- (216).Prusiner SB Prions. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (217).Chakravarty AK; Smejkal T; Itakura AK; Garcia DM; Jarosz DF A Non-Amyloid Prion Particle That Activates a Heritable Gene Expression Program. Mol. Cell 2020, 77, 251–265 e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (218).Zhao B; Katuwawala A; Uversky VN; Kurgan L Idpology of the Living Cell: Intrinsic Disorder in the Subcellular Compartments of the Human Cell. Cell. Mol. Life Sci. 2021, 78, 2371–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (219).Yoshizawa T; Nozawa RS; Jia TZ; Saio T; Mori E Biological Phase Separation: Cell Biology Meets Biophysics. Biophys. Rev. 2020, 12, 519–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (220).Wang W Recent Advances in Atomic Molecular Dynamics Simulation of Intrinsically Disordered Proteins. Phys. Chem. Chem. Phys. 2021, 23, 777–784. [DOI] [PubMed] [Google Scholar]
- (221).Vantrappen G; Rutgeerts L; Schurmans P; Coenegrachts JL Omeprazole (40 Mg) Is Superior to Ranitidine in Short-Term Treatment of Ulcerative Reflux Esophagitis. Dig. Dis. Sci. 1988, 33, 523–529. [DOI] [PubMed] [Google Scholar]
- (222).Uversky VN Recent Developments in the Field of Intrinsically Disordered Proteins: Intrinsic Disorder-Based Emergence in Cellular Biology in Light of the Physiological and Pathological Liquid-Liquid Phase Transitions. Annu. Rev. Biophys. 2021, 50, 135–156. [DOI] [PubMed] [Google Scholar]
- (223).Shea JE; Best RB; Mittal J Physics-Based Computational and Theoretical Approaches to Intrinsically Disordered Proteins. Curr. Opin. Struct. Biol. 2021, 67, 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (224).Ramanathan A; Ma H; Parvatikar A; Chennubhotla SC Artificial Intelligence Techniques for Integrative Structural Biology of Intrinsically Disordered Proteins. Curr. Opin. Struct. Biol. 2021, 66, 216–224. [DOI] [PubMed] [Google Scholar]
- (225).Mu J; Liu H; Zhang J; Luo R; Chen HF Recent Force Field Strategies for Intrinsically Disordered Proteins. J. Chem. Inf. Model. 2021, 61, 1037–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Metskas LA; Rhoades E Single-Molecule Fret of Intrinsically Disordered Proteins. Annu. Rev. Phys. Chem. 2020, 71, 391–414. [DOI] [PubMed] [Google Scholar]
- (227).Liu H; Jeffery CJ Moonlighting Proteins in the Fuzzy Logic of Cellular Metabolism. Molecules 2020, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (228).Lermyte F Roles, Characteristics, and Analysis of Intrinsically Disordered Proteins: A Minireview. Life (Basel) 2020, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (229).Hong S; Choi S; Kim R; Koh J Mechanisms of Macromolecular Interactions Mediated by Protein Intrinsic Disorder. Mol. Cells 2020, 43, 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (230).Fuxreiter M Classifying the Binding Modes of Disordered Proteins. Int. J. Mol. Sci. 2020, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (231).Cohan MC; Pappu RV Making the Case for Disordered Proteins and Biomolecular Condensates in Bacteria. Trends Biochem. Sci. 2020, 45, 668–680. [DOI] [PubMed] [Google Scholar]
- (232).Bugge K; Brakti I; Fernandes CB; Dreier JE; Lundsgaard JE; Olsen JG; Skriver K; Kragelund BB Interactions by Disorder - a Matter of Context. Front. Mol. Biosci. 2020, 7, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (233).Xia K; Wei GW Molecular Nonlinear Dynamics and Protein Thermal Uncertainty Quantification. Chaos 2014, 24, 013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (234).Uversky VN Dancing Protein Clouds: The Strange Biology and Chaotic Physics of Intrinsically Disordered Proteins. J Biol Chem 2016, 291, 6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (235).Ferrell JE Jr. Bistability, Bifurcations, and Waddington’s Epigenetic Landscape. Curr. Biol. 2012, 22, R458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (236).Zhu L; Kim SJ; Hara M; Aono M Remarkable Problem-Solving Ability of Unicellular Amoeboid Organism and Its Mechanism. R. Soc. Open Sci. 2018, 5, 180396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (237).Tero A; Takagi S; Saigusa T; Ito K; Bebber DP; Fricker MD; Yumiki K; Kobayashi R; Nakagaki T Rules for Biologically Inspired Adaptive Network Design. Science 2010, 327, 439–442. [DOI] [PubMed] [Google Scholar]
- (238).Saigusa T; Tero A; Nakagaki T; Kuramoto Y Amoebae Anticipate Periodic Events. Phys. Rev. Lett. 2008, 100, 018101. [DOI] [PubMed] [Google Scholar]
- (239).Reid CR; MacDonald H; Mann RP; Marshall JA; Latty T; Garnier S Decision-Making without a Brain: How an Amoeboid Organism Solves the Two-Armed Bandit. J. R. Soc. Interface 2016, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (240).Nakagaki T; Kobayashi R; Nishiura Y; Ueda T Obtaining Multiple Separate Food Sources: Behavioural Intelligence in the Physarum Plasmodium. Proc. Biol. Sci. 2004, 271, 2305–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (241).Feldmann H; Kimiai K; Fondermann C; Haas M; Herwig R [Local Treatment of the Locomotion Apparatus Diseases with a Flufenamic Acid Ointment. Results of a Double-Blind Test]. Med. Monatsschr. 1975, 29, 406–407. [PubMed] [Google Scholar]
- (242).Boussard A; Fessel A; Oettmeier C; Briard L; Dobereiner HG; Dussutour A Adaptive Behaviour and Learning in Slime Moulds: The Role of Oscillations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20190757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (243).Beekman M; Latty T Brainless but Multi-Headed: Decision Making by the Acellular Slime Mould Physarum Polycephalum. J. Mol. Biol. 2015, 427, 3734–3743. [DOI] [PubMed] [Google Scholar]
- (244).Meyer B; Ansorge C; Nakagaki T The Role of Noise in Self-Organized Decision Making by the True Slime Mold Physarum Polycephalum. PLoS One 2017, 12, e0172933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (245).Tsafou K; Tiwari PB; Forman-Kay JD; Metallo SJ; Toretsky JA Targeting Intrinsically Disordered Transcription Factors: Changing the Paradigm. J. Mol. Biol. 2018, 430, 2321–2341. [DOI] [PubMed] [Google Scholar]
- (246).Metallo SJ Intrinsically Disordered Proteins Are Potential Drug Targets. Curr. Opin. Chem. Biol. 2010, 14, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (247).Kumar D; Sharma N; Giri R Therapeutic Interventions of Cancers Using Intrinsically Disordered Proteins as Drug Targets: C-Myc as Model System. Cancer Inform. 2017, 16, 1176935117699408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (248).Joshi P; Vendruscolo M Druggability of Intrinsically Disordered Proteins. Adv. Exp. Med. Biol. 2015, 870, 383–400. [DOI] [PubMed] [Google Scholar]
- (249).Hu G; Wu Z; Wang K; Uversky VN; Kurgan L Untapped Potential of Disordered Proteins in Current Druggable Human Proteome. Curr. Drug Targets 2016, 17, 1198–1205. [DOI] [PubMed] [Google Scholar]
- (250).Hosoya Y; Ohkanda J Intrinsically Disordered Proteins as Regulators of Transient Biological Processes and as Untapped Drug Targets. Molecules 2021, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (251).Chong B; Li M; Li T; Yu M; Zhang Y; Liu Z Conservation of Potentially Druggable Cavities in Intrinsically Disordered Proteins. ACS Omega 2018, 3, 15643–15652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (252).Biesaga M; Frigole-Vivas M; Salvatella X Intrinsically Disordered Proteins and Biomolecular Condensates as Drug Targets. Curr. Opin. Chem. Biol. 2021, 62, 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (253).Kulkarni P; Solomon TL; He Y; Chen Y; Bryan PN; Orban J Structural Metamorphism and Polymorphism in Proteins on the Brink of Thermodynamic Stability. Protein Sci. 2018, 27, 1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (254).Ornstein D Bernoulli Shifts with the Same Entropy Are Isomorphic. Adv. Math. 1970, 4, 337–352. [Google Scholar]


