Abstract
Assembly of the dimeric complex III (CIII2) in the mitochondrial inner membrane is an intricate process in which several accessory proteins are involved as assembly factors. Despite numerous studies, this process has yet to be fully understood. Here we report the identification of human OCIAD2 (ovarian carcinoma immunoreactive antigen–like protein 2) as an assembly factor for CIII2. OCIAD2 was found to be deregulated in several carcinomas and also in some neurogenerative disorders; however, its nonpathological role had not been elucidated. We have shown that OCIAD2 localizes to mitochondria and interacts with electron transport chain (ETC) proteins. Complete loss of OCIAD2 using gene editing in HEK293 cells resulted in abnormal mitochondrial morphology, a substantial decrease of both CIII2 and supercomplex III2+IV, and a reduction in CIII enzymatic activity. Identification of OCIAD2 as a protein required for assembly of functional CIII2 provides a new insight into the biogenesis and architecture of the ETC. Elucidating the mechanism of OCIAD2 action is important both for the understanding of cellular metabolism and for an understanding of its role in malignant transformation.
INTRODUCTION
Mitochondria are ubiquitous organelles, essential for ATP production by oxidative phosphorylation (OXPHOS). In mammals, OXPHOS relies on five multisubunit enzyme complexes and two mobile electron carriers embedded within the inner mitochondrial membrane. The first four enzyme complexes (CI–IV) form the electron transport chain (ETC) that transfers electrons from electron donors through a series of electron acceptors to molecular oxygen, which is coupled to the generation of a proton gradient across the inner membrane that is used by the ATP synthase (CV) to drive ATP synthesis. With the exception of CII, each OXPHOS complex has a dual genetic origin. Subunits encoded by the mitochondrial DNA (mtDNA) are cotranslationally inserted into the membrane by matrix-located ribosomes tethered to OXA1 insertase (Hell et al., 2001; Haque et al., 2010; Ott and Herrmann, 2010; Itoh et al., 2021). These mitochondrially encoded subunits act as a “seed” around which the rest of the nuclear-encoded subunits are assembled after being imported from the cytosol through the outer-membrane (TOM) and inner-membrane (TIM) translocase machineries (Neupert and Herrmann, 2007; Chacinska et al., 2009; Friedman and Nunnari, 2014; Wiedemann and Pfanner, 2017). In mammalian cells, individual OXPHOS CI, III, and IV physically interact to form a variety of supramolecular structures called supercomplexes (SCs I-III2, III2-IV, I-III2-IV1-4, or “respirasome”) (Schafer et al., 2006; Althoff et al., 2011; Cogliati et al., 2016; Gu et al., 2016; Letts and Sazanov, 2017). It was proposed that supercomplex formation stabilizes ETC complexes, facilitates electron transfer, and decreases formation of reactive oxygen species (ROS), but the exact function of supercomplexes is still under debate (Dudkina et al., 2010; Milenkovic et al., 2017; Lobo-Jarne and Ugalde, 2018). The assembly of ETC complexes is a convoluted process, due to the nature of the ETC complexes themselves. First, due to the dual genetic origin of ETC complexes, a tight coregulation of nuclear and mitochondrially encoded gene expression is required (Mick et al., 2012; Rampelt and Pfanner, 2016; Richter-Dennerlein et al., 2016; Priesnitz and Becker, 2018; Singh et al., 2020). Second, many subunits are embedded in the inner mitochondrial membrane and they are prone to aggregation before and during assembly (Schmitt et al., 1996; Vendruscolo et al., 2003; Calvo and Mootha, 2010; Bender et al., 2011; Voos et al., 2016; Wilkening et al., 2018). Furthermore, each protein complex contains multiple redox active cofactors that can promote the formation of harmful radicals if not properly assembled into the final complex (Perry et al., 2003). As a result, a number of proteins, so-called assembly factors, are needed to coordinate different steps of the ETC assembly process (Perez-Perez et al., 2016; Aich et al., 2018; Lobo-Jarne et al., 2020; Stephan and Ott, 2020; Timon-Gomez et al., 2020).
Assembly factors represent a functionally heterogeneous group of proteins, most of which are specific for the assembly of each ETC complex, but they are not part of the final complex. Many assembly factors are conserved between yeast and humans, but new factors unrelated to the yeast proteins are being discovered in human cells (Signes and Fernandez-Vizarra, 2018). Mutations in assembly factors leading to subsequent impaired respiratory complex assembly are associated with a number of pathologies that mainly affect tissues with high energy requirements (Diaz et al., 2011; Rodenburg, 2016; Ghezzi and Zeviani, 2018; Hock et al., 2020; Protasoni and Zeviani, 2021). Thus, full understanding of ETC assembly regulation is essential for understanding the molecular mechanisms underlying mitochondrial pathology. Despite extensive studies, the exact mechanism by which ETC complexes are assembled is not fully described and some putative assembly factor proteins remain unknown.
Complex III (or ubiquinol:cytochrome c oxidoreductase) plays a central role in the ETC, transferring electrons from coenzyme Q to cytochrome c. All available x-ray diffraction structures (Xia et al., 1997; Iwata et al., 1998; Zhang et al., 1998; Hunte et al., 2000) and the Cryo-EM structure of CIII (Gu et al., 2016) are identical in both shape and subunit composition. CIII is always a symmetrical dimer (CIII2), with each monomer being composed of 11 different subunits. Three of them contain the catalytic centers: cytochrome b (MT-CYB), cytochrome c1 (CYC1), and the Rieske protein (UQCRFS1), whereas the other eight subunits (UQCRC1, UQCRC2, UQCRH, UQCRB, UQCRQ, subunit 9, UQCR10, and UQCR11) do not possess prosthetic groups and do not directly participate in electron transport or proton pumping (Smith et al., 2012). The formation of CIII starts with the assembly of two factors, UQQC1 and UQQC2, to newly synthesized mtDNA-encoded cytochrome b (MT-CYTB) (Tucker et al., 2013). The binding of the first nuclear-encoded subunits releases these early assembly factors from cytochrome b and the rest of the structural subunits are then incorporated into the nascent complex, resulting in the formation of dimeric pre-CIII (Smith et al., 2012). Then, UQCRFS1 protein is bound with the assistance of three assembly factors: BCS1L, LYRM7, and TTC19 (Cruciat et al., 1999; Atkinson et al., 2011; Ghezzi et al., 2011; Sanchez et al., 2013; Bottani et al., 2017). Following import into mitochondria, UQCRFS1 is bound and stabilized in the mitochondrial matrix by the LYRM7 chaperone (Mzm1 in yeast) (Cui et al., 2012; Sanchez et al., 2013). Bcs1/BCS1L translocates UQCRFS1 from the matrix to help in the incorporation of UQCRFS1 into premature CIII2 (Wagener and Neupert, 2012; Tang et al., 2020). This step is crucial for CIII2 maturation because it activates the catalytic activity of the enzyme. The last subunit (UQCR11) then binds to the CIII2, and the assembly is completed. Finally, TTC19 interacts with fully assembled CIII2 to facilitate the removal of subunit 9 and the cleaved N-terminal fragments of UQCRFS1 that remain bound between UQCRC1 and UQCRC2 (Bottani et al., 2017).
Given the structural similarity between yeast and mammalian CIII, as well as the identification of several orthologues of yeast assembly factors in humans, the assembly of CIII in mammals has so far been considered to be similar to that in yeast. However, it has been shown in recent protemic data of a human cell line with CIII2 assembly defect that CYC1, UQCR10, and UQCRH form assembly intermediates that accumulate during the failure of incorporation into pre-CIII2, which is not described in yeast (Protasoni et al., 2020). Moreover, assembly factors specific only for mammalian CIII, such as TTC19 and Brawnin, have also been described (Ghezzi et al., 2011; Bottani et al., 2017; Zhang et al., 2020). Therefore, the challenge now is to unravel unknown mechanisms and factors specific to mammalian CIII biogenesis.
In the present study, we identified and characterized a novel CIII assembly factor specific to metazoan cells, ovarian carcinoma immunoreactive antigen–like protein 2 (OCIAD2). Recent studies explored the origin, evolution, and function of the OCIAD2, which has been shown to be expressed in endosomes and mitochondria (Sinha et al., 2018). In the present study, we explored the functional role of this protein in mitochondria. So far OCIAD2 expression has been implicated in several cancers and neurodegenerative diseases (Han et al., 2014; Wu et al., 2017; Sinha et al., 2018), but its precise function in nonpathological contexts remains unknown. In this report we propose OCIAD2 as a novel CIII assembly factor of the mitochondrial respiratory chain in human cells.
RESULTS
OCIAD2 is a mitochondrial membrane protein
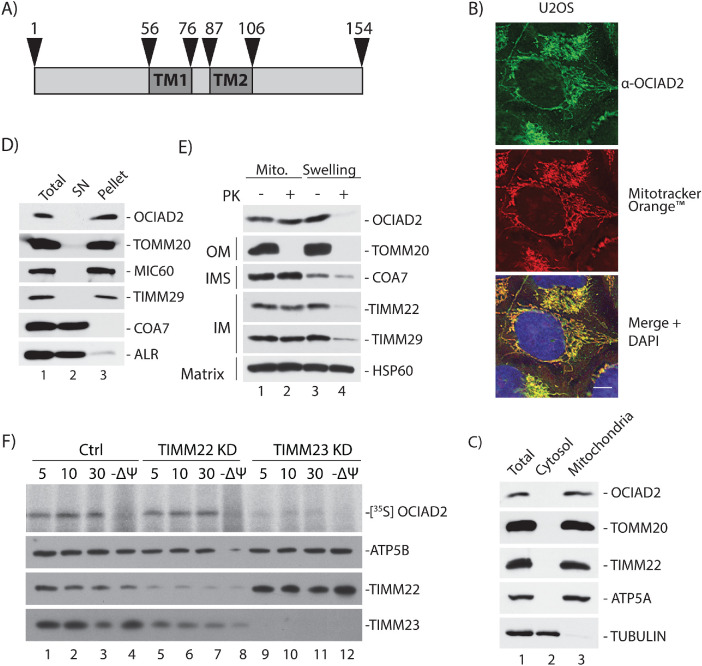
Sequence analysis revealed that OCIAD2 is conserved in metazoa (Supplemental Figure S1), and no yeast homologue could be identified. Moreover, the OCIAD2 amino-acid sequence is devoid of a canonical cleavable N-terminal mitochondrially targeted presequence and indicates the presence of two α-helical stretches of high hydrophobicity that are probably transmembrane (TM) domains (Figure 1A). To check whether OCIAD2 is mitochondrially localized despite the lack of a predicted cleavable presequence, we analyzed its cellular localization by immunofluorescence microscopy using an antibody specifically directed against native OCIAD2. The OCIAD2 signal largely colocalized with that of Mitotracker Orange (Figure 1B). We additionally proved the mitochondrial localization by performing cellular fractionation, where native OCIAD2 was predominantly localized to mitochondria (Figure 1C, lane 3). The association of OCIAD2 with the mitochondrial membranes was analyzed by treatment of mitochondria with sodium carbonate (pH 10.8), which causes the release of peripheral membrane-associated proteins. OCIAD2 was resistant to carbonate extraction (Figure 1D, lane 3), in line with the membrane integrated proteins TOMM20, MIC60, and TIMM29, indicating that it is very likely an integral membrane protein. Next, we examined the submitochondrial localization of OCIAD2 by performing a hypo-osmotic swelling (Figure 1E). In intact mitochondria, native OCIAD2 was protected from degradation by the outer membrane (OM), similar to other proteins localized inside mitochondria, such as COA7 (intermembrane space, IMS), TIMM22, TIMM29 (inner membrane, IM), and HSP60 (matrix) (Figure 1E, lane 1). As expected, the OM protein TOMM20 was efficiently degraded by proteinase K in intact mitochondria (Figure 1E, lane 2). Upon disruption of the OM by hypo-osmotic swelling, the IMS becomes accessible to protease treatment. Under these conditions, using an antibody against the N-terminal part of OCIAD2, we observed the digestion of OCIAD2 by proteinase k similarly to known IMS and IM proteins (Figure 1E, compare lanes 2 and 4). Consistently, we concluded that OCIAD2 is an inner membrane protein with its N-terminal part exposed to the IMS.
FIGURE 1:
OCIAD2 is an inner mitochondrial membrane protein. (A) Primary sequence of OCIAD2. Dark gray boxes area depicts the predicted transmembrane segments. (B) U2OS cells were immunolabeled using antibody against OCIAD2 and stained with Mitotracker Orange to visualize the mitochondria. Scale bar, 8 μm. (C) Subcellular fractionation of HEK293 cells. (D) Carbonate extraction of HEK293 cells at pH 10.8. After differential centrifugation, samples were analyzed by SDS–PAGE and Western blotting. (E) Isolated mitochondria from HEK293 cells were subjected to submitochondrial fractionation. Mitochondria (lanes 1 and 2); hypotonic swelled mitochondria (lanes 3 and 4) were treated with (+) or without (−) proteinase K (PK). (F) Radiolabeled OCIAD2 precursors were imported into mitochondria isolated from control and TIMM22KD or TIMM23KD cells for the indicated times in the presence or absence of a membrane potential (Ψ) across the IM and treated with proteinase K (PK; 50 mg/ml). Samples were analyzed by SDS–PAGE followed by autoradiography or immunoblotting using ATP5B-, TIMM22-, or TIMM23-specific antibodies.
The majority of inner membrane protein precursors are synthesized in the cytosol and imported into the mitochondria by TIM22 or TIM23 complexes. TIM22 is responsible for the import of precursor proteins carrying internal targeting signals, whereas TIM23 facilitates the import of presequence-containing proteins (Neupert and Herrmann, 2007; Chacinska et al., 2009). It should also be noted that many proteins from the inner membrane do not resemble typical substrates for the TIM22 or TIM23 translocases (Reinhold et al., 2012; Turakhiya et al., 2016; Rampelt et al., 2020). To distinguish the import pathway that OCIAD2 uses for integration into the inner membrane, we applied an in vitro import assay. In brief, radioactive [35S]-OCIAD2 was synthesized in rabbit reticulocyte lysate and imported into mitochondria isolated from control (Ctrl) HEK293 cells and HEK293 cells depleted of TIM22 or TIM23 translocases by small interfering RNA (siRNA) (Figure 1F). In the case of the Ctrl cells (Figure 1F, lanes 1–4) and TIMM22 knockdown cells (Figure 1F, lanes 5–8), OCIAD2 was imported into a protease-protected location in a time-dependent manner. In contrast, OCIAD2 import into mitochondria lacking TIMM23 was inhibited (Figure 1F, lanes 9–12). This indicates that despite the absence of a presequence, OCIAD2 requires TIM23 for its import into mitochondria.
OCIAD2 interacts with respiratory chain complexes
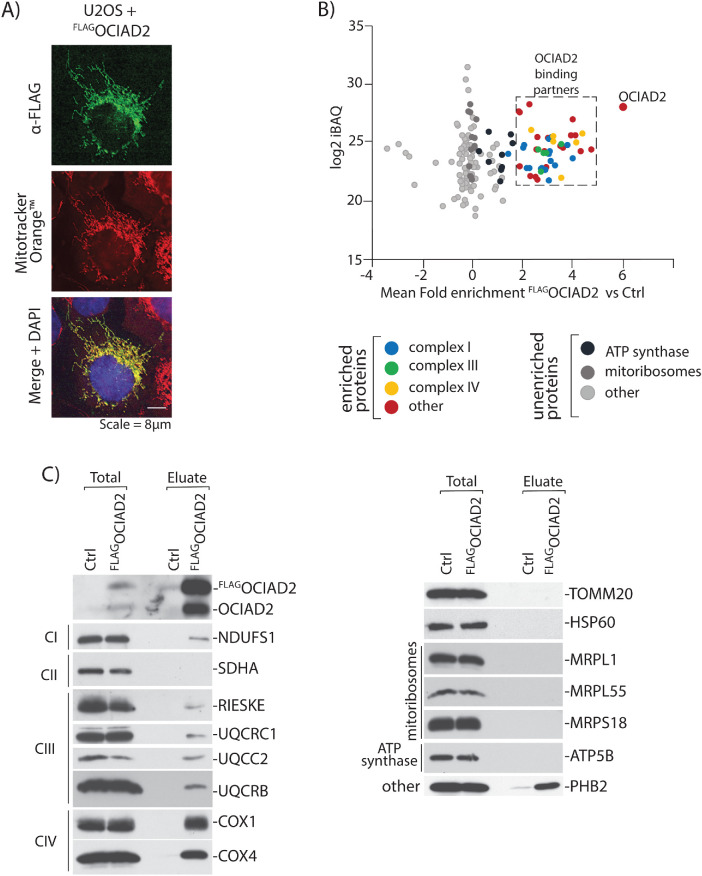
To functionally characterize OCIAD2 in human mitochondria, we catalogued OCIAD2 binding partners by performing affinity purification of FLAGOCIAD2 under nondenaturing conditions, followed by quantitative mass spectrometry. We first generated a stable tetracycline-inducible Flp-In T-REx 293 cell line expressing OCIAD2 with an N-terminal FLAG-tag. Confocal microscopy showed colocalization of FLAGOCIAD2 with Mitotracker Orange, thus confirming mitochondrial localization of the fusion protein (Figure 2A). The FLAGOCIAD2-expressing cell line and the control cells were then differentially labeled via stable isotope labeling by amino acids (SILAC) and mixed early in the protocol to minimize potential experimental errors resulting from parallel mitochondrial isolation and subsequent processing steps. Following FLAG-based enrichment, the proteins were digested with trypsin and analyzed by shotgun proteomics. Next, we filtered the list of identified proteins based on their annotation in the Integrated Mitochondrial Protein Index (IMPI) reference set of genes deposited in the MitoMiner database v. 4.0 (Smith and Robinson, 2019). Among 180 mitochondrial proteins identified in our analysis, we assigned 48 as putative binding partners of OCIAD2, based on a mean fold enrichment value >2 in the comparison of intensities of proteins derived from FLAGOCIAD2 versus control cells. The list of identified mitochondrial proteins (Supplemental Table S1) was then subjected to gene ontology (GO) annotation and to a term enrichment analysis (Supplemental Figure S2). One of the most enriched GO terms was related to the respiratory ETC. Indeed, a large pool (29) of proteins comprising ETC CI, III, and IV copurified with FLAGOCIAD2 (Figure 2B). In contrast, some large groups of proteins sharing common terms, for example, ATP synthase (9) and mitoribosomal protein (37) were not bound specifically to the FLAGOCIAD2 as judged by the apparent lack of enrichment (fold enrichment <2) (Figure 2B). Next, we validated the proteomic results by Western blot analysis employing antibodies specific toward selected proteins identified in the mass spectrometric analysis (Figure 2C). Indeed, under these experimental conditions, we confirmed that proteins of ETC complexes I, III, and IV were efficiently copurified with the bait, in contrast to mitoribosomal proteins or ATP synthase (Figure 2C).
FIGURE 2:
OCIAD2 interacts with proteins from ETC. (A) U2OS cells expressing FLAGOCIAD2 were immunolabeled using antibodies against FLAG. Mitotracker Orange was used to visualize the mitochondria. Scale bar, 8 µm. (B) Mitochondria isolated from control HEK293 Flp-in WT cells (SILAC light) and from the cells expressing FLAGOCIAD2 protein (SILAC heavy) were mixed and solubilized in digitonin-containing buffer following a FLAG-targeting immunoprecipitation. Fold enrichment of proteins coimmunoprecipitated with FLAGOCIAD2 vs. Ctrl (x-axis) was correlated with intensity-based absolute quantification (iBAQ) values, a measure of protein abundance (y-axis) categories. For the entire mitochondrial protein list, see Supplemental Table S1. (C) Control mitochondria and mitochondria expressing FLAGOCIAD2 were solubilized in digitonin-containing buffer and subjected to immunoprecipitation with FLAG affinity resin. Samples were analyzed using SDS–PAGE and immunoblotting with the indicated antibodies. Total 1%; Eluate 100%.
Loss of OCIAD2 causes mitochondrial morphology and cell growth defect and affects mitochondrial bioenergetics
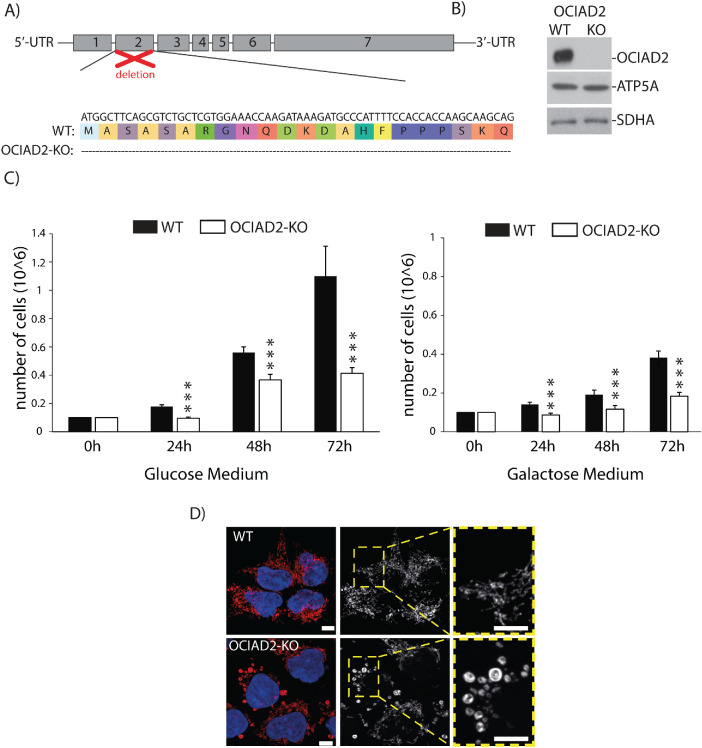
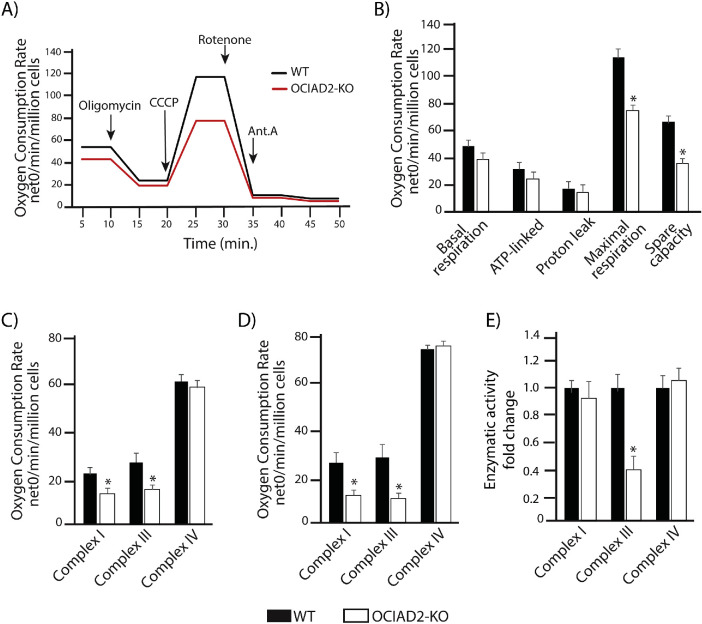
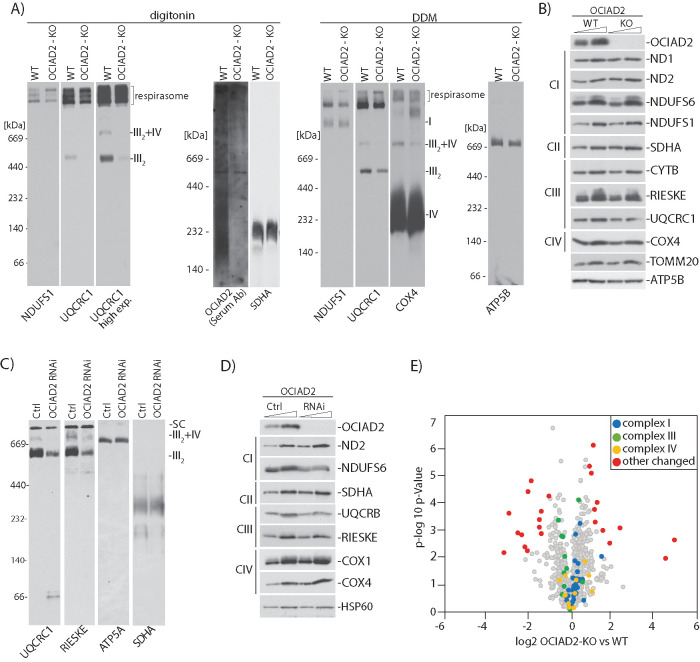
Our observation that OCIAD2 binds to many proteins from the ETC complexes led us to investigate whether OCIAD2 has an influence on the activity of these complexes. To this end, we generated a knockout of OCIAD2 (OCIAD2-KO) using CRISPR/Cas9-mediated disruption of both the alleles in HEK293 cells. The second exon of the gene was targeted, and the selected knockout clone was a compound homozygote for a 66-nucleotide deletion (1_66del) encoding the N-terminal part of OCIAD2 (Figure 3A). The OCIAD2-KO cell line was verified by sequencing of the genomic locus (Figure 3A) and Western blot analysis (Figure 3B). In this model, reduction of OCIAD2 levels resulted in a growth phenotype (Figure 3C) and abnormal mitochondrial morphology compared with wild-type (WT) cells (Figure 3D), which suggests alterations in mitochondrial function. We also confirmed this phenotype in a knockdown of OCIAD2 using siRNA (Supplemental Figure S5, A and B). Next, we aimed to analyze in detail the performance of mitochondria in the OCIAD2-KO cells. Therefore, we analyzed mitochondrial respiration by measuring the oxygen consumption rates (OCRs) in WT and OCIAD2-KO cells. First, we used intact cells to maintain the cellular environment and to minimize potential artifacts due to mitochondrial preparation (Figure 4, A and B). Under these conditions, deletion of OCIAD2 had no significant impact on basal respiration compared with WT cells (82% residual respiration). However, the maximal respiration induced by the protonophore CCCP was significantly lower in OCIAD2-KO cells compared with WT (66% residual respiration, p < 0.05) (Figure 4B and Supplemental Figure S4C). To confirm this reduction in maximal respiration of OCIAD2-KO cells, we assessed CI, CIII, and IV activities coupled to oxygen consumption by two different protocols as described in Hofhaus et al. (1996) and Doerrier et al. (2018). Mitochondria depleted of OCIAD2 exhibited decreased oxygen consumption driven by pyruvate, malate, and glutamate (CI substrate electron donors) and by succinate and glycerol-3-phosphate in the presence of rotenone (CIII substrate electron donor). No alteration was observed in CIV activity (Figure 4C). Using the second protocol, mitochondria depleted of OCIAD2 exhibited a similar decreased oxygen consumption related to CI and CIII activity, with no alteration in CIV activity (Figure 4D). As the oxygen consumption related to CI activity depends on the electron transfer through CIII, we wondered whether CIII was the only ETC component altered in this situation. Therefore, the enzymatic activities of uncoupled CI, CIII, and CIV were examined. Mitochondria lacking OCIAD2 exhibited decreased enzymatic activity for CIII, but no alterations in CI and CIV activity (Figure 4E and Supplemental Figure S4C). Altogether, OCIAD2 does not affect the basal activity of mitochondria or oxygen consumption coupled to ATP production, indicating no influence on mitochondrial activity under a metabolic nonstress situation. Nevertheless, a reduction in maximal respiration and spare capacity, coupled with a reduction in CIII activity, indicates that OCIAD2-depleted cells have less capacity to cope with metabolic stress. These data suggest that OCIAD2 is essential for cellular bioenergetics, potentially through regulation of the ETC.
FIGURE 3:
OCIAD2-KO cells display abnormal mitochondria morphology and proliferation. (A) Knockout cell line of OCIAD2 (OCIAD2-KO) was created in HEK293 cells by using CRISPR/Cas9 technology. Alignment of respective mutated region compared with WT shows deletion of 66 amino acids from the N-terminal part of protein. (B) Western blot results confirming OCIAD2 deletion. (C) WT and OCIAD2-KO cells were seeded 24 h before experiment at concentration of 0.1 × 106/well in the growth medium. After 24, 48, and 72 h of culture at 37°C with 5% CO2, cells were harvested by trypsinization and counted. Data are shown as the mean ± SD (n = 5); ***p < 0.01 (two-tailed Student’s t test). (D) WT and OCIAD2-KO cells were immunolabeled using antibody against cyclophilin D to visualize the mitochondria. Scale bar, 10 μm.
FIGURE 4:
OCIAD2 Influences bioenergetis in mitochondria. (A) Oxygen consumption rates (OCR) in intact OCIAD2-KO cells cultured in glucose. Oligomycin (ATPsynthase blocker), CCCP (uncoupler), rotenone (CI inhibitor), and antimycin A (CIII inhibitor) were added at the indicated time points. OCR was normalized to number of cells. Data are presented as the mean ± SD (n = 3); *p < 0.05 (two-tailed Student’s t test). (B) Basal, ATP-linked, proton leak, maximal OCR, and spare respiratory capacity were quantified from original data presented in A. Data are shown as mean ± SD (n = 3); *p < 0.05 (two-tailed Student’s t test). (C) Respiration of digitonin-permeabilized cells to assess isolated complex I, complex III, and complex IV activity. (D) Substrate-uncoupler-inhibitor titration protocol to assess complex I, complex III, and complex IV activity in digitonin-permeabilized cells. (E) Complex I, complex III, and complex IV activity assessed by enzymatic assay mean ± SD (n = 3); *p < 0.05 (two-tailed Student’s t test).
OCIAD2 is required for the assembly of respiratory chain complexes
Because in our experimental model OCIAD2 showed an impact on the activity of the ETC complexes, we further analyzed their assembly by Blue Native-PAGE (BN–PAGE). Mitochondria from OCIAD2-KO cells were solubilized in digitonin or in n-dodecyl-β-d-maltoside (DDM)-containing buffer, mild detergents commonly used in the isolation of membrane protein complexes before analysis by BN–PAGE and immunodetection (Figure 5A). We used both buffers because digitonin better preserves native supercomplexes, whereas the harsher DDM induces supercomplex separation into subcomplexes (Reisinger and Eichacker, 2008). These experiments revealed that fully assembled CIII2, as well as III2+IV–containing supercomplexes, were down-regulated in the OCIAD2-KO cell line, as shown in digitonin- and DDM-treated mitochondria (Figure 5A and Supplemental Figure S3A). The fully assembled CIV and CV were not affected by OCIAD2 deletion, as shown in DDM- and digitonin-treated mitochondria (Figure 5A and Supplemental Figure S3A). To assess whether changes in the assembly of ETC complexes result from a decrease in steady-state levels of the proteins constituting these complexes, we performed Western blot analysis using specific antibodies against selected ETC proteins (Figure 5B). However, these analyses showed no major impact of OCIAD2 deletion on steady-state levels of individual ETC proteins. These results suggest that OCIAD2 is rather required for the structural rearrangements of ETC complexes instead of participating in ETC protein homeostasis. We attempted to rescue this phenotype by overexpressing FLAGOCIAD2 in OCIAD2-KO cells, but we did not observe the expected rescue of CIII2, likely due to the adaptation of the cells to the OCIAD2-KO environment (Supplemental Figure S4, A and B). To validate the CIII2 down-regulation phenotype of OCIAD2-KO cells, we also performed a transient knockdown of the OCIAD2 gene using siRNA. Importantly, the alterations in ETC complex assembly and their steady-state levels in OCIAD2 siRNA-mediated knockdown cells were identical to the ones observed in the CRISPR/Cas9 OCIAD2-KO cell line (Figure 5, C and D), thus validating the causative gene for this phenotype. To further study the influence of OCIAD2 on CIII and to find the intermediate subassemblies blocked by deletion of OCIAD2, we performed two-dimensional (2D) gel analysis. However, no accumulation of intermediates was found. Nevertheless, CIII subunits in CIII2 and CIII2+CIV were significantly reduced (Supplemental Figure S3B). With the aim to better understand the molecular changes induced in mitochondria upon deletion of OCIAD2, we performed a comparative label-free quantitative proteomic analysis of protein levels in mitochondria isolated from OCIAD2-KO and WT HEK293 cells (Figure 5E). Our analysis revealed that among 998 mitochondrial proteins quantified (Supplemental Table S2), no significant changes in the steady-state levels of proteins constituting CI, CIII, and CIV were discovered, which is in the line with our Western blot results (Figure 5B).
FIGURE 5:
OCIAD2 is needed for the proper assembly of CIII2 and SC CIII2+IV. (A) Isolated WT or OCIAD2-KO mitochondria were solubilized in 1% digitonin or 0.5% DDM and resolved using BN–PAGE, followed by Western blotting and detection using the indicated antibodies. (B) WT and OCIAD2-KO mitochondria were analyzed by Western blotting. (C) Isolated WT or OCIAD2 siRNA mitochondria were solubilized in 1% digitonin and resolved by BN–PAGE, followed by Western blotting and immunodetection. (D) Isolated WT and OCIAD2 siRNA mitochondria were analyzed by Western blotting. (E) Volcano plot representation of proteins changed upon OCIAD2 deletion. Mitochondria isolated from WT and OCIAD2-KO were lysed in 0.1% trifluoroacetic acid (TFA), desalted, and analyzed by LC-MS/MS (n = 3). Blue, green, and color indicate proteins belonging to complex I, III, and IV of ETC, respectively.
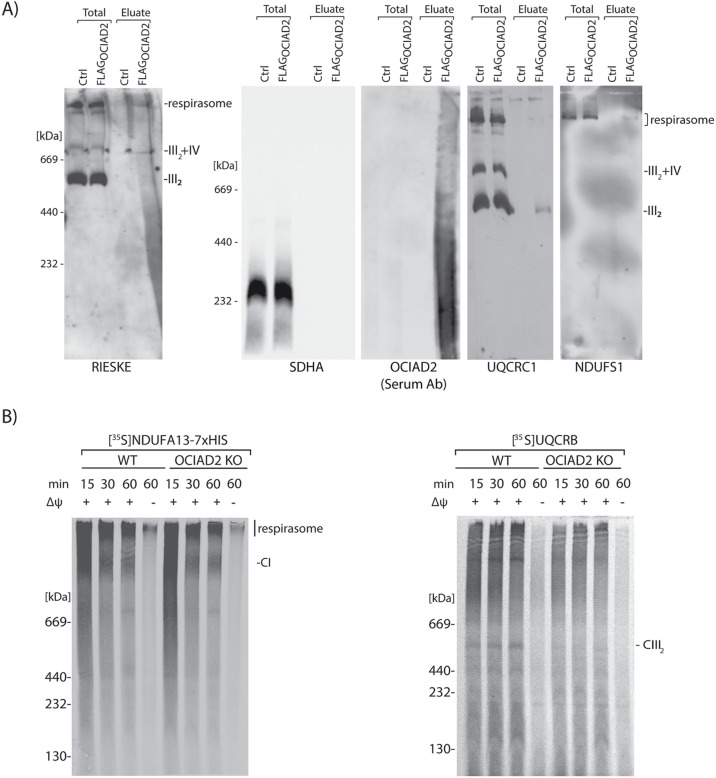
The identification of a large pool of CI, CIII, and CIV proteins as binding partners of OCIAD2 led us to check whether OCIAD2 forms a common complex with ETC subunits. We performed immunopurification of FLAGOCIAD2 and resolved the eluate fraction by BN–PAGE. Using antibodies against rieske and UQCRC1, both components of CIII, we detected OCIAD2 interaction with CIII2 and SC III2+IV and with the respirasome (I+III2+IV). We also detected an interaction of OCIAD2 with the respirasome using an antibody against NDUFS1, a component of CI. (Figure 6A and Supplemental Figure S3C). We then concluded that OCIAD2 interacts with both the dimeric complex III and the respirasome. This prompted us to distinguish whether OCIAD2 is a factor needed for the assembly of a particular complex (I or III) or whether it is dispensable for supercomplex formation. We focused on CI and CIII because we observed alterations in their composition and activities, which was not the case for CIV (Figures 5A and 4, C–E). Thus, we assessed import and assembly of the radiolabeled NDUFA13 (a component of CI) and UQCRB (a component of CIII) into isolated WT and OCIAD2-KO mitochondria by BN–PAGE analyses (Figure 6B). As a negative control we used mitochondria with dissipated membrane potential to ensure that protein import and assembly depends on the membrane potential across the inner mitochondrial membrane. We noticed that in OCIAD2-KO cells, the import of UQCRB into CIII2 was diminished, whereas NDUFA13 import and assembly into CI was unaltered. Thus, we concluded that OCIAD2 is specifically needed for the proper assembly of CIII2 in human mitochondria.
FIGURE 6:
OCIAD2 is involved in the import and assembly of CIII2. (A) FLAGOCIAD2 was immunopurified, and the elute was resolved by BN–PAGE. (B) NDUFA13-7XHIS and UQCRB were imported into isolated mitochondria from WT HEK293 and OCIAD2-KO cells, for the indicated times, in the presence or absence of membrane potential (Δψ) at 37°C. Samples were analyzed by BN–PAGE and digital radiography.
DISCUSSION
CIII, or the cytochrome bc1 complex, is structurally similar in yeast and mammals, and current knowledge about CIII assembly factors was obtained mostly by extrapolating the function of the well-studied yeast orthologues to mammalian systems. However, some factors specific to higher eukaryotes have been discovered. Two mammalian assembly factors known to play a role in CIII2 biogenesis are TTC19 and BRAWNIN. The former was discovered in patient samples associated with a CIII-defective, progressive mitochondrial encephalopathy (Ghezzi et al., 2011), and the latter was found during screening of the small open reading frame (sORF)-encoded peptides (SEPs) in the human peptidome (Zhang et al., 2020). In this report we propose a new molecular role of the metazoan-specific protein, OCIAD2, in the stabilization and assembly of CIII in human cells.
OCIAD2 was originally discovered as an immunoreactive antigen in patients with ovarian cancer (Luo et al., 2001). Later, OCIAD2 was reported to be dysregulated in various types of cancer; however, its regulatory roles in cancer progression are still unclear and appear to be tumor type–dependent (Nagata et al., 2012; Wu et al., 2017; Sakashita et al., 2018). In addition, OCIAD2 was implicated in Alzheimer’s disease (Han et al., 2014). OCIAD2 has been previously shown to localize predominantly to the mitochondria (Sinha et al., 2018), but its physiological role in the nonpathological context of this organelle remained obscure.
In this work we used a combination of OCIAD2 immunodetection, mass spectrometry–based proteomics, and mitochondrial in vitro import assays to demonstrate that OCIAD2 associates with different CIII-containing structures, including CIII2, SC III2+IV, and SC I+III2+IV (respirasome). In our experimental model, the absence of OCIAD2 causes a decrease in the level of assembled CIII2 and the SC III2+IV, with no changes in respirasome formation. This is in accordance with previous observations suggesting that the biogenesis of the SC III2+IV is not necessary for respirasome formation (Perez-Perez et al., 2016). The fact that respirasomes (but not SC III2+IV) are present in OCIAD2-KO mitochondria provides support for a role of OCIAD2 in the stabilization of SC III2+IV. These findings also suggest the existence of independent regulatory mechanisms for the biogenesis and turnover of SC III2+IV and the respirasomes. Possibly, there is a specific, yet-to-be-understood, physiological importance of SC III2+IV. We also observed a reduction in the oxygen consumption activities of CI and CIII, which is in line with the well-established correlation between severe CIII deficiency and CI impairment (Lamantea et al., 2002; Acin-Perez et al., 2004; Barel et al., 2008; Tucker et al., 2013; Wanschers et al., 2014; Feichtinger et al., 2017; Protasoni et al., 2020). Nevertheless, the enzymatic activity of ETC complexes in OCIAD2-KO cells confirmed reduced activity only in CIII, indicating no compromise of CI. The respirasomes are important mitochondrial architecture serving various purposes such as cell survival and averting aggregation (Milenkovic et al., 2017). In OCIAD2-KO cells, the total amount of CIII2 is low and seems to preferentially associate with the respirasomes (Lapuente-Brun et al., 2013; Tropeano et al., 2020). Furthermore, OCIAD2-KO cells exhibit proper formation and assembly of individual CI and CIV, which may result from the fact that the respirasome is also not affected in OCIAD2-KO cells. This is in agreement with a study by Protasoni et al. (2020) in which association of respirasome formation was shown to precede the maturation of stable individual CI and CIV. We also show that the deletion of OCIAD2 does not affect steady-state levels of protein components of CIII but rather the amount of fully assembled CIII2. In fact, OCIAD2 deletion does not affect steady-state levels of all detected OXHPOS proteins. Because CIII2 is the necessary component of the respirasome, and the formation of the respirasome was not affected by OCIAD2 deletion, it is conceivable that this protein may act as a factor for stabilizing matured CIII2, rather than as a factor needed for its maturation. This would make the function of OCIAD2 similar to that of COX7A2L or UQCC3A, both interacting with CIII2 and stabilizing SC III2+IV (Desmurs et al., 2015; Perez-Perez et al., 2016). The similarities between these proteins may also suggest their cooperation; however, further studies are needed to prove such functional interactions. The mechanism by which OCIAD2 promotes CIII assembly and/or stability requires further investigation.
We also report here abnormal mitochondrial morphology in cells depleted of OCIAD2, which is not surprising because mitochondrial morphology has been reported to be tightly connected with mitochondrial function. While we cannot entirely exclude that OCIAD2 has some direct role in maintaining the morphology of mitochondria, our results suggest a simpler explanation in that the altered respiration of OCIAD2-KO mitochondria is the cause of morphological changes. These results are in line with the results of previous studies, which showed a change in mitochondrial morphology in patients with CIII deficiency (Slipetz et al., 1991).
The biogenesis of the respirasome involves a CI assembly intermediate, which acts as a scaffold for the incorporation of CIII and CIV subunits (Moreno-Lastres et al., 2012). However, CIII plays a central role in the formation of supercomplexes and misassembly of the CIII subunits affects biogenesis of CIV (Protasoni et al., 2020). It is also known that the mitochondrial inner membrane is especially protein rich, leading to tight protein packaging. Accordingly, proteins contact each other by weak interactions to avoid any deleterious effect of a strong interaction such as aggregation or protein nucleation (Milenkovic et al., 2017). These weak interactions are highly coordinated by various assembly factors, and we hypothesize that OCIAD2 could act as one of them, potentially exclusively for CIII.
Until now, there are no data on patients with mitochondrial dysfunction resulting from mutations in OCIAD2. Nevertheless, identification of OCIAD2 as an assembly/stabilizing factor for CIII2 and SC III2+IV opens up the possibility of screening for the presence of pathogenic mutations in the OCIAD2 gene in as-yet-unresolved cases of mitochondrial disease associated with CIII2 deficiency. Additionally, because OCIAD2 has been implicated in various cancer types with no common mechanism reported, and mitochondrial dysfunction is a hallmark of cancer, it cannot be excluded that dysfunctional mitochondria due to OCIAD2 loss leads to the cellular changes resulting in cancer. However, the causal relationship between OCIAD2 function in mitochondrial CIII2 and cancer requires further in-depth investigation.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell lines and growth conditions
For these studies we used human embryonic kidney cell lines (HEK293T-Flp-InTM T-RExTM) or HEK293 or U2OS (used only where specifically mentioned) cells purchased from the American Type Culture Collection (ATCC; www.atcc.org). Cells were cultured at 37°C in a 5% CO2 atmosphere. The cell lines were sustained in DMEM with high glucose content (4500 mg/l), supplemented with 10% (vol/vol) fetal bovine serum (FBS), 2 mM l-glutamine and 1% (vol/vol) penicillin–streptomycin. For SILAC analysis, cells were grown for five passages on DMEM lacking arginine and lysine, supplemented with 10% (vol/vol) dialyzed FBS, 600 mg/l proline, 42 mg/l arginine hydrochloride (13C6, 15N4-arginine in “heavy” media), and 146 mg/l lysine hydrochloride (13C6, 15N2-lysine in “heavy” media) (Cambridge Isotope Laboratories, Tewksbury, MA). The stable tetracycline-inducible cell line (FLAGOCIAD2) was generated using the Flp-IN T-REx System (Thermo Fisher Scientific). Briefly, Flp-IN T-REx HEK293T cells were cotransfected with the pcDNA5/FRT/TO vector containing the sequence of FLAGOCIAD2 and the pOG44 vector using GeneJuice (Novagen) as transfection reagent. After 24 hours of transfection, cells were selected with 200 µg/ml hygromycin B (Thermo Fisher) to select stable transformants. Individual colonies were further expanded and screened for FLAGOCIAD2 expression under the regulation of tetracycline (1 µg/ml). The OCIAD2-KO cell line was generated using the CRISPR/Cas9 as described (Ran et al., 2013). The guide RNAs targeting the OCIAD2 gene were designed using software provided by the laboratory of Feng Zhang (Ran et al., 2013). The selected guide was cloned into the pX330 vector using the oligos 5′ CACCGATGGCTTCAGCGTCTGCTCG 3′, 5′ AAACCGAGCAGACGCTGAAGCCATC 3′. HEK293 cells were transfected with pX330 vector together with an EGFP-containing plasmid (pEGFP-N1; Clontech Laboratories). Single cells were sorted using a FACS Vantage SE machine and the knockouts confirmed by sequencing, SDS–PAGE, and Western blotting. For overexpression of FLAGOCIAD2, the plasmid pcDNA 3.1(+) containing the FLAGOCIAD2 sequence was transfected using jetPRIME polyplus transfection reagent. After 5 days incubation, cells were harvested for further experiments.
siRNA-mediated knockdown
Cells were reverse-transfected with 25 nM OCIAD2 siRNA (sense strand: 5′ AGG-UUA-UUU-GGC-AGC-UAA-U 3′; Eurogentech), 16 nM TIMM23 (5′ CCCUCUGUCUCCUUAUUUA 3′; Eurogentech), or 16 nM TIM22 (5′ GUGAGGAGCAGAAGAUGAU 3′; Eurogentech) using the Invitrogen Lipofectamine RNAiMAX Reagent (Invitrogen, Carlsbad, CA) diluted with serum-free Opti-MEM I (Life Technologies, Thermo Fisher, Waltham, MA) as per the manufacturer’s recommended protocol. On the third day posttransfection, cells were collected for subsequent assays. As a control, the cells were transfected with Mission siRNA universal negative control (SIC001; Sigma).
Proliferation assay
Twenty-four hours before the experiment, WT and OCIAD2-KO cells were seeded into six-well format at a concentration of 0.1 × 106/well (for OCIAD2 silencing experiments, 0.2 × 106/well) in the growth medium. At day 0 and then after 24 , 48, and 72 h of culture at 37°C with 5% CO2, cells were harvested by trypsinization and counted in a cell counter (Countess II; Life Technologies). Cell growth was expressed in absolute cell count numbers.
Electrophoresis and immunoblot analysis
For BN–PAGE, mitochondrial pellets were solubilized in digitonin-containing buffer (1% digitonin, 20 mM Tris-HCl, pH 7.4, 0.1 mM EDTA, 50 mM NaCl, 10% [wt/vol] glycerol, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) to a final concentration of 1 µg/µl for 30 min at 4°C or DDM-containing buffer (10% DDM, 1.5 M aminocaproic acid, 50 mM Bis-Tris–HCl pH 7.0). Lysates were cleared by centrifugation at 14,000 × g for 10 min at 4°C followed by the addition of loading dye (5% Coomassie brilliant blue G-250, 500 mM 6-aminohexanoic acid, and 100 mM Bis-Tris, pH 7.0). Samples were loaded onto self-made 4–10%, 4–13%, or 4–16% polyacrylamide gradient gels separated and transferred onto PVDF membranes (Merck Millipore) followed by detection by specific antibodies. The High Molecular Weight Calibration Kit for native electrophoresis (Amersham) was used as a molecular weight standard. For 2D gel analysis, BN–PAGE gels incubated in a complex disruption buffer (containing 1% SDS [wt/vol] and 2-mercaptoethanol [vol/vol]) for 30 min and sealed with 12% SDS–PAGE resolving gel with the help of stacking gel. The proteins were resolved using SDS–PAGE and Western blotted, followed by detection using the indicated antibodies (Calvaruso et al., 2008).
Mitochondrial isolation and mitochondrial procedures
Cells were harvested in phosphate-buffered saline (PBS) and suspended in THE buffer (300 mM trehalose; 10 mM KCl; 10 mM HEPES, pH 7.4; 1 mM EDTA; 0.5 mM PMSF) with 0.1 mg/ml bovine serum albumin (BSA). Cells were homogenized twice using a Potter S dounce homogenizer (Sartorius). After each homogenization step, cells were pelleted at 400 × g, 10 min at 4°C. The supernatant was collected and the remaining debris removed by centrifugation (10 min, 4°C, 800 × g). The supernatant was collected, and mitochondria were recovered by centrifugation at 11,000 × g for 10 min, then washed with BSA-free THE buffer, and finally resuspended in THE BSA-free buffer. Protein determination was determined by Bradford assay. The mitochondrial pellet was suspended in BSA-free THE buffer or stored at −80°C until further use. Membrane integration of proteins was determined by 30 min incubation of mitochondria with freshly prepared Na2CO3, pH = 10.8, followed by centrifugation for 30 min at 100,000 × g, 4°C. Submitochondrial localization was analyzed by a protease protection assay. Mitochondrial pellets were resuspended in either sucrose buffer (250 mM sucrose and 20 mM HEPES/KOH [pH 7.4]) or swelling buffer (25 mM sucrose and 20 mM HEPES/KOH [pH 7.4]). Samples were split and either left untreated or treated with proteinase K (50 µg/ml) for 5 min on ice. The reaction was stopped by the addition of 2 mM PMSF.
Antibodies
The antibodies against the following proteins were used in the study: ALR (Santa Cruz Biotechnology; Sc-134869; 1:500), ATP5A (Abcam; ab14748; 1:500), ATP5B (rabbit serum; Rehling laboratory, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany; 1:500), COA7 (Sigma; HPA029926; 1:500), COX1 (rabbit serum; Rehling laboratory; 1:2000), COX4 (Cell Signaling Technology; 4850; 1:2000), cytochrome B (rabbit serum; Rehling laboratory; 1:1000), HSP60 (Sigma; H4149; 1:500), MIC60 (Novus Biologicals; NB100-1919; 1:1000), MRPL1 (rabbit serum; Rehling laboratory; 1:1000), MRPL55 (Proteintech; 17679-1-AP; 1:500); MRPS18 (Proteintech; 16139-1-AP; 1:500), ND1 (rabbit serum; Rehling laboratory; 1:1000), ND2 (rabbit serum; Rehling laboratory; 1:1000), NDUFS1 (Santa Cruz Biotechnology; Sc-50132; 1:1000), prohibitin 2 (Sigma Aldrich; HPA039874; 1:1000), OCIAD2 (rabbit serum; Chacin´ska laboratory, IMol Polish Academy of Sciences, Warsaw, Poland; 1:1000), Rieske (rabbit serum; Rehling laboratory; 1:1000), TIMM22 (Proteintech; 14927-1-AP; 1:500), TIMM23 (BD Biosciences; 611222; 1:1000), TIMM29 (Proteintech; 25652-1-AP; 1:500), TOMM20 (Santa Cruz Biotechnology; Sc-11415; 1:500), tubulin (Santa Cruz Biotechnology; sc-134239; 1:2000), SDHA (Santa Cruz Biotechnology; Sc-166947; 1:1000), UQCC2 (Novus Biological; NBP2-14240; 1:500), UQCRB (Sigma Aldrich; HPA002815; 1:500), UQCR1 (Sigma; HPA002815; 1:500), Total OXPHOS Blue Native WB Antibody Cocktail (abcam; ab110412; 1:1000), OCIAD2 (Invitrogen; PA5-59375; 1:100).
Immunopurification of proteins
Isolated mitochondria were solubilized in digitonin-containing buffer (150 mM NaCl, 10% glycerol [vol/vol], 20 mM MgCl2, 1 mM PMSF, 50 mM Tris-HCl, pH 7.4, with 1% digitonin [vol/wt]) at a protein concentration of 1 μg/μl on a rotary wheel for 30–60 min at 4°C. Mitochondrial lysates were cleared by centrifugation at 20,000 × g at 4°C and transferred to preequilibrated anti-FLAG M2 agarose beads (Sigma-Aldrich) for FLAG immunoprecipitation or onto protein A-Sepharose (PAS) containing cross-linked OCIAD2 antibody in a Mobicol spin column (MoBiTec) and incubated at 4°C for 2 h under mild agitation. The unbound proteins were removed by spinning out the supernatant for 1 min, 150 × g at 4°C. The beads were washed eight times with washing buffer (150 mM NaCl, 10% glycerol [vol/vol], 20 mM MgCl2, 1 mM PMSF, 50 mM Tris-HCl, pH 7.4, with 0.1% digitonin [vol/vol]). The column-bound proteins were eluted either with 3× FLAG peptide for mass spectrometry or with 0.2 M glycine, pH 2.5, for Western blotting.
Immunofluorescence
For immunofluorescence, U2OS/HEK293 cells were fixed with prewarmed (37°C) 4% paraformaldehyde (Sigma-Aldrich) for 10 min and permeabilized by incubation with PBS containing 0.5% Triton X-100 and blocked with 5% BSA in PBS for 20 min. Subsequently, the cells were incubated with affinity-purified anti-OCIAD2 serum or monoclonal mouse anti-FLAG or cyclophilin D. After being washed, cells were incubated with Alexa Fluor 488– or 568–conjugated secondary antibodies. For mitochondria staining, cells were incubated with Mitotracker Orange (500 nM) for 15 min at 37°C prior to fixation and following incubation with secondary antibodies. Confocal images were recorded with a Leica SP8 confocal microscope (Leica Microsystems, Wetzlar, Germany). Z-image series were taken, and maximum projections of the stacks are displayed.
Radioactive precursor synthesis and in organello import
Radiolabeled OCIAD2, TIMM22, TIMM23, UQCRB, and NDUFA13 precursors were synthesized using rabbit reticulocyte lysate (Promega) in the presence of [35S]methionine. Isolated mitochondria were diluted in import buffer (250 mM sucrose, 80 mM postassium acetate, 5 mM magnesium acetate, 5 mM methionine, 10 mM sodium succinate, 20 mM HEPES/KOH [pH 7.4], supplemented with 2 mM ATP). Import reactions were initiated by the addition of either 2% lysate (for SDS–PAGE analysis) or 8% lysate (assembly on BN–PAGE), and samples were incubated for the indicated times. To stop the reaction, membrane potential was dissipated on ice using 8 mM antimycin A, 1 mM valinomycin, and 10 mM oligomycin. Nonimported proteins were removed by proteinase K (20 μg/ml) treatment for 10 min on ice. PMSF (2 mM) was added to inactivate proteinase K for 10 min on ice. Mitochondria were collected, washed with SEM buffer (250 mM sucrose, 1 mM EDTA, 20 mM MOPS [pH 7.2]), and used for SDS–PAGE or BN–PAGE analyses. Results were visualized using digital autoradiography.
High-resolution respirometry and OXPHOS complex activities
Oxygen consumption was measured in the intact or permeabilized HEK293 WT or OCIAD2-KO cell line using an Oxygraph-2 k high-resolution respirometer (Oroboros Instruments, Innsbruck, Austria). Data were digitally recorded using DatLab v. 5.1.0.20 (Oroboros Instruments, Innsbruck, Austria) expressed as pmol of O2/min per 106 cells and then normalized to protein content. Before each assay, air calibration and background correction were done according to the manufacturer’s protocol. For endogenous cell respiration, intact cells were trypsinized, suspended at 0.5 × 106 cells/ml in 2 ml in MiR05 medium (ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid [EGTA] 0.5 mM, MgCl2 3 mM, lactobionic acid 60 mM, taurine 20 mM, KH2PO4 10 mM, HEPES 20 mM, d-sucrose 110 mM, 1 mg/ml BSA–fatty acid free added freshly, pH 7.1). The cell suspension was immediately placed into the Oxygraph chamber to measure endogenous respiration. Respiration connected to proton leak was determined by adding ATP synthase inhibitor oligomycin (final concentration 1.25 μM). Subsequently, the mitochondrial inner membrane uncoupler CCCP (carbonyl cyanide m-chlorophenyl hydrazone) was titrated (0.20 μM/step) to reach maximum respiration (electron transport system [ETS] capacity). The complex I inhibitor rotenone (0.5 μM) and the complex III inhibitor antimycin A (5 μM) were subsequently added to detect oxygen consumption related to each complex and to completely block mitochondrial respiration, resulting residual oxygen consumption (ROX). Stable plateaus of oxygen flux in each experimental step were corrected for ROX afterward. Substrate-driven respiration was assessed in digitonin-permeabilized cells by two different protocols. The first protocol was based on Hofhaus et al. (1996) and Sladowska et al. (2021) and consists of CI stimulation by malate (0.5 mM), pyruvate (5 mM), and glutamate (10 mM) in the presence of ADP+Mg+2 (2.5 mM). The CIII was stimulated by succinate (10 mM) and glycerol-3-phosphate (5 mM) after CI inhibition by rotenone (0.05 μM). The outer membrane integrity of mitochondria was assessed by cytochrome c (10 μM) before CIII inhibition by antimycin A (2.5 μM). The CIV was stimulated by ascorbate (2 mM) and N,N,N′,N′-tetramethyl-p-phenylenediamine dihydrochloride (TMPD; 0.5 mM). After CIV inhibition by sodium azide (50 mM), the measurements were concluded.
The second protocol was based on Doerrier et al. (2018) and consisted of a substrate-uncoupler-inhibition titration strategy. The sequence of compounds to reach full CoenzymeQ pool activation was applied to the Oxygraph-2K chamber in the following order: malate (0.5 mM), pyruvate (5 mM), cytochrome c (10 μM), ADP+Mg+2 (2.5 mM), stepwise addition of carbonyl cyanide 3-chlorophenylhydrazone (0.1 mM), glutamate (10 mM), succinate (10 mM), octanoylcarnitine (0.2 mM). At this point CI was inhibited by rotenone (0.05 μM) and CIII was further stimulated by glycerol-3-phosphate (5 mM). After CIII inhibition by antimycin A (2.5 μM), the CIV was evaluated. The CIV was stimulated by ascorbate (2 mM) and TMPD (0.5 mM). After CIV inhibition by sodium azide (50 mM), measurements were concluded.
Digitonin-solubilized samples were used to evaluate CI, CIII, and CIV activities (Tiranti et al., 1995). CI (NADH:decylubiquinone, rotenone sensitive) and CIII (decylubiquinol:cytochrome c oxidoreductase) enzymatic activities were measured as previously described (Kirby et al., 2007). As previously described, CIV enzymatic activity was measured (Mohanraj et al., 2019). Briefly, the CI activity rate was expressed as NADH oxidation per minute per milligram of protein. The NADH oxidation was detected for 2 min in the absorbance of 340 nm. The CIII activity rate was expressed in nanomoles of cytochrome c per minute per milligram of protein. The cytochrome c reduction was detected for 2 min in the absorbance of 550 nm. The CIV activity was expressed in nanomoles of cytochrome c per minute per milligram of protein. The cytochrome c oxidation was detected for 3 min in the absorbance of 550 nm. Linear changes in absorbance were observed for each reaction. The protein concentrations of digitonized cell suspension to evaluate CI, CIII, and CIV activities were measured by Bradford assay. The reactions were performed in 96-well plates in a final volume of 200 μl (Protasoni et al., 2020). The data were analyzed by Student’s t test and are presented as means ± SD of three experiments; *p = 0.05, **p < 0.01, ***p < 0.001.
LC-MS/MS analysis
Chromatographic separation was performed on an Easy-Spray Acclaim PepMap column 25 or 50 cm length × 75 µm inner diameter (Thermo Fisher Scientific) at 35°C by applying 60 min (mitochondrial protein levels) or 90 min (FLAGOCIAD2 coimmunoprecipitation [co-IP]) acetonitrile gradients in 0.1% aqueous formic acid at a flow rate of 450 or 300 nl/min, respectively. An UltiMate 3000 nano-LC system was coupled to a Q Exactive HF-X mass spectrometer via an Easy-Spray source (all Thermo Fisher Scientific). For the FLAGOCIAD2 co-IP experiments, the Q Exactive HF-X was operated in data-dependent mode with survey scans acquired at a resolution of 60,000 at m/z 200. Up to 12 of the most abundant isotope patterns with charge 2 or higher from the survey scan were selected with an isolation window of 1.3 m/z and fragmented by higher-energy collision dissociation (HCD) with normalized collision energies of 27, while the dynamic exclusion was set to 15 s. The maximum ion injection times for the survey scan and the MS/MS scans (acquired with a resolution of 15,000 at m/z 200) were 45 and 22 ms, respectively. The ion target value for MS was set to 3 × 10e6 and for MS/MS to 10e5, and the intensity threshold for MS/MS was set to 8.3 × 10e2. For the determination of the mitochondrial protein levels, the Q Exactive HF-X was operated in a “BoxCar” mode through MaxQuant Live (ref) with survey scans acquired at a resolution of 120,000 at m/z 200 within m/z range 350–1650, followed by three BoxCarScans (12 BoxCarBoxes) within m/z range 400–1200. Up to five of the most abundant isotope patterns with charge states 2–5 from the survey scan were selected with an isolation window of 1.4 m/z and fragmented by HCD with normalized collision energies of 27, while the dynamic exclusion was set to 40 s. The maximum ion injection times for the survey scan and the MS/MS scans (acquired with a resolution of 15,000 at m/z 200) were 250 and 28 ms, respectively. The ion target value for MS was set to 10e6 and for MS/MS to 10e5.
Proteomics data processing
The data were processed with MaxQuant v. 1.6.6.0 (Cox and Mann, 2008), and the peptides were identified from the MS/MS spectra searched against Human Reference Proteome UP000005640 using the built-in Andromeda search engine. Cysteine carbamidomethylation was set as a fixed modification, and methionine oxidation as well as protein N-terminal acetylation were set as variable modifications. For in silico digests of the reference proteome, cleavages of arginine or lysine followed by any amino acid were allowed (trypsin/P), and up to two missed cleavages were allowed. The false discovery rate (FDR) was set to 0.01 for peptides, proteins, and sites. For SILAC samples (FLAGOCIAD2 Co-IP samples), a multiplicity of 2 was selected and light labels set as Arg0 & Lys0 and heavy labels set as Arg10 & Lys8, the maximal number of labeled amino acids per peptide set to 3, and the requantify option was set to on. Match between runs was enabled. Other parameters were used as preset in the software. Unique and razor peptides were used for quantification enabling protein grouping (razor peptides are the peptides uniquely assigned to protein groups and not to individual proteins). Data were further analyzed using Perseus v. 1.6.6.0 and Microsoft Office Excel 2016.
LFQ-based differential analysis of mitochondrial protein levels
Label-free quantitation (LFQ) values for protein groups were loaded into Perseus v. 1.6.6.0. Standard filtering steps were applied to clean up the data set: reverse (matched to decoy database), identified only by site, and potential contaminant (from a list of commonly occurring contaminants included in MaxQuant) protein groups were removed. Protein accessions were matched to accessions generated from gene names constituting the IMPI data set (Smith and Robinson, 2019) and successfully matched protein groups retained for further analysis. LFQ intensities were log2 transformed, protein groups with LFQ values in fewer than three samples were removed, and all remaining missing values were imputed from normal distribution (width = 0.3, downshift = 1.8 × SD). Gaussian distribution of log2-transformed LFQ intensities were confirmed by histogram analysis, preventing the unbiased introduction of small values. Student’s t test (permutation-based FDR with 250 randomizations = 0.01, S0 = 0.25) was performed to return proteins of which levels were statistically significantly changed in response to OCIAD2-KO (Supplemental Table S2).
SILAC-based FLAGOCIAD2 co-IP enrichment analysis
Ratio H/L normalized values for protein groups were loaded into Perseus v. 1.6.6.0. Standard filtering steps were applied to clean up the data set: reverse (matched to decoy database), identified only by site, and potential contaminant (from a list of commonly occurring contaminants included in MaxQuant) protein groups were removed. Protein accessions were matched to accessions generated from gene names constituting the IMPI data set (Smith and Robinson, 2019) and successfully matched protein groups retained for further analysis. Ratio H/L normalized values were log2 transformed, and mean fold enrichment values for FLAGOCIAD2 versus Ctrl were calculated for all protein groups; the mean fold enrichment threshold for was set to 2 (Supplemental Table S1).
Proteomic data deposition
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the data set identifier PXD023025.
Supplementary Material
Acknowledgments
We thank Sven Dennerlein, Vanessa Linke, Urszula Nowicka and Michał Wasilewski for scientific discussion and Albert Roethel for experimental assistance. Aleksandra Gosk is currently affiliated with the Institute of Polish Culture, Faculty of Polish Studies, University of Warsaw, Poland, and we thank her for generating OCIAD2-KO cell line. The work was funded by “Regenerative Mechanisms for Health” project MAB/2017/2 carried out within the International Research Agendas program of the Foundation for Polish Science cofinanced by the European Union under the European Regional Development Fund, by National Science Centre grants 2015/19/B/NZ3/03272, 2015/18/A/NZ1/00025, and 2016/20/S/NZ1/00423, by Polish Ministry of Science and Higher Education funds for science Diamond Grant 2/0050/DIA/2017/46 and Ideas Plus 0002/ID1/2014/63, by Deutsche Forschungsgemeinschaft (DFG) SFB1190 (P13, P. R.), FOR2848 (P04, P. R.), and the Max Planck Society (P. R.), supported by the DFG, by the German Research Foundation under Germany’s Excellence Strategy EXC 2067/1-390729940, and the Copernicus Award by the DFG and Foundation for Polish Science.
Abbreviations used:
- Ab
antibody
- ALR
augmenter of liver regeneration
- ATP
adenosine triphosphate
- ATP5A
ATP synthase lipid-binding protein
- ATP5B
ATP synthase subunit beta
- Bcs1/BCS1L
ubiquinol-cytochrome c reductase complex chaperone
- BN–PAGE
blue native-PAGE
- BRAWNIN
protein BRAWNIN
- BSA
bovine serum albumin
- CCCP
carbonyl cyanide m-chlorophenyl hydrazone
- 6C
carbon-6 isotope
- CI
NADH:ubiquinone oxidoreductase (Complex I)
- CII
succinate-coenzyme Q reductase (Complex II)
- CIII
cytochrome bc1, coenzyme Q: cytochrome C oxidoreductase (Complex III)
- CIII 2
complex III dimer
- CIV
cytochrome c oxidase (Complex IV)
- co-IP
coimmunoprecipitation
- COA7
cytochrome c oxidase assembly factor 7
- COX1
cytochrome c oxidase subunit 1
- COX4
cytochrome c oxidase subunit 4
- COX7A2L
cytochrome c oxidase subunit 7A2 like
- Cryo-EM
cryo-electron microscopy
- Ctrl
control
- CV
ATP synthase (Complex V)
- CYC1
cytochrome c1
- DAPI
4′,6-diamidino-2-phenylindole
- 2D
two-dimensional
- DDM
n-dodecyl-β- d-maltoside
- DMEM
Dulbecco’s Modified Eagle Medium
- EDTA
ethylenediaminetetraacetic acid
- EGTA
ethylene glycol-bis(β-aminoethyl ether)- N, N, N′, N′-tetraacetic acid
- ETC
electron transport chain
- ETS
electron transport system
- FACS
fluorescence-activated cell sorting
- FBS
fetal bovine serum
- FDR
false discovery rate
- FLAG
FLAG-tag protein
- g
gravity
- GO
gene ontology
- H/L
heavy/light
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- His
histidine
- HSP60
heat shock protein 60
- iBAQ
intensity-based absolute quantification
- IM
inner membrane
- IMPI
integrated mitochondrial protein index
- IMS
intermembrane space
- KCl
potassium chloride
- KH 2PO 4
potassium dihydrogen phosphate
- KO
knockout
- KOH
potassium hydroxide
- LC-MS/MS
liquid chromatography mass spectrometry
- LFQ
label-free quantitation
- LYRM7
LYR motif containing 7
- Ψ
membrane potential
- MgCl 2
magnesium chloride
- MIC60
MICOS complex subunit MIC60
- min
minute
- mM
micromolar
- MoBiTec
mobicol spin column
- MOPS
4-morpholinepropanesulfonic acid, 3-propanesulfonic acid
- MRPL1
mitochondrial Ribosomal Protein L1
- MRPL55
mitochondrial ribosomal protein L55
- MRPS18
mitochondrial ribosomal protein S18C
- MT-CYB
cytochrome b
- mtDNA
mitochondrial DNA
- Mzm1
mitochondrial zinc maintenance protein 1
- 14N
nitrogen-14 isotope
- NaCl
sodium chloride
- NADH
nicotinamide adenine dinucleotide
- ND1
NADH dehydrogenase subunit 1
- ND2
NADH dehydrogenase subunit 2
- NDUFA13
NADH:Ubiquinone Oxidoreductase Subunit A13
- NDUFS1
NADH:ubiquinone oxidoreductase core subunit S1
- nM
nanomolar
- O 2
oxygen
- OCIAD2
ovarian carcinoma immunoreactive antigen–like protein 2
- OCRs
oxygen consumption rates
- OM
outer membrane
- OXA1
oxidase assembly protein1
- OXPHOS
oxidative phosphorylation
- PAS
protein A-sepharose
- PBS
phosphate-buffered saline
- Pmol
picomoles
- PMSF
phenylmethylsulfonyl fluoride
- PVDF
polyvinylidene difluoride
- RNAi
RNA interference
- ROS
reactive oxygen species
- ROX
residual oxygen consumption
- 35S
Sulfur-35 isotope
- S.C./SC
supercomplex
- SDHA
succinate dehydrogenase complex flavoprotein subunit A
- SDS
sodium dodecyl sulfate
- SEPs
small open reading frame (sORF)-encoded peptides
- SILAC
stable isotope labeling by amino acids in cell culture
- siRNA
small interfering RNA
- sORF
small open reading frame
- TFA
trifluoroacetic acid
- TIM
inner-membrane translocase
- TIMM22
translocase of inner mitochondrial membrane 22
- TIMM23
translocase of inner mitochondrial membrane 23
- TIMM29
translocase of inner mitochondrial membrane 29
- TM
transmembrane
- TMPD
N, N, N′, N′-tetramethyl-p-phenylenediamine dihydrochloride
- TOM
outer-membrane translocase
- TOMM20
translocase of outer mitochondrial membrane 20
- TTC19
tetratricopeptide repeat domain 19
- UQCC3
ubiquinol-cytochrome c reductase complex assembly factor 3
- UQCR10
ubiquinol-cytochrome c reductase, Complex III Subunit X
- UQCR11
ubiquinol-cytochrome c reductase, Complex III Subunit XI
- UQCRB
ubiquinol-cytochrome c reductase binding protein
- UQCRC1/ UQCR1
ubiquinol-cytochrome c reductase core protein 1
- UQCRC2
ubiquinol-cytochrome c reductase core protein 2
- UQCRFS1
ubiquinol-cytochrome c reductase, rieske iron-sulfur polypeptide 1
- UQCRH
ubiquinol-cytochrome c reductase hinge protein
- UQCRQ
ubiquinol-cytochrome c reductase Complex III Subunit VII
- UTR
untranslated region
- Vol
volume
- WB
western blotting
- WT
Wild Type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-03-0143) on January 26, 2022.
REFERENCES
- Acin-Perez R, Bayona-Bafaluy MP, Fernandez-Silva P, Moreno-Loshuertos R, Perez-Martos A, Bruno C, Moraes CT, Enriquez JA (2004). Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol Cell 13, 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aich A, Wang C, Chowdhury A, Ronsor C, Pacheu-Grau D, Richter-Dennerlein R, Dennerlein S, Rehling P (2018). COX16 promotes COX2 metallation and assembly during respiratory complex IV biogenesis. eLife 7, e32572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althoff T, Mills DJ, Popot JL, Kuhlbrandt W (2011). Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J 30, 4652–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson A, Smith P, Fox JL, Cui TZ, Khalimonchuk O, Winge DR (2011). The LYR protein Mzm1 functions in the insertion of the Rieske Fe/S protein in yeast mitochondria. Mol Cell Biol 31, 3988–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barel O, Shorer Z, Flusser H, Ofir R, Narkis G, Finer G, Shalev H, Nasasra A, Saada A, Birk OS (2008). Mitochondrial complex III deficiency associated with a homozygous mutation in UQCRQ. Am J Hum Genet 82, 1211–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender T, Lewrenz I, Franken S, Baitzel C, Voos W (2011). Mitochondrial enzymes are protected from stress-induced aggregation by mitochondrial chaperones and the Pim1/LON protease. Mol Biol Cell 22, 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottani E, Cerutti R, Harbour ME, Ravaglia S, Dogan SA, Giordano C, Fearnley IM, D’Amati G, Viscomi C, Fernandez-Vizarra E, Zeviani M (2017). TTC19 plays a husbandry role on UQCRFS1 turnover in the biogenesis of mitochondrial respiratory complex III. Mol Cell 67, 96–105.e104. [DOI] [PubMed] [Google Scholar]
- Calvaruso MA, Smeitink J, Nijtmans L (2008). Electrophoresis techniques to investigate defects in oxidative phosphorylation. Methods 46, 281–287. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Mootha VK (2010). The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet 11, 25–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N (2009). Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati S, Calvo E, Loureiro M, Guaras AM, Nieto-Arellano R, Garcia-Poyatos C, Ezkurdia I, Mercader N, Vazquez J, Enriquez JA (2016). Mechanism of super-assembly of respiratory complexes III and IV. Nature 539, 579–582. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cruciat CM, Hell K, Folsch H, Neupert W, Stuart RA (1999). Bcs1p, an AAA-family member, is a chaperone for the assembly of the cytochrome bc(1) complex. EMBO J 18, 5226–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui TZ, Smith PM, Fox JL, Khalimonchuk O, Winge DR (2012). Late-stage maturation of the Rieske Fe/S protein: Mzm1 stabilizes Rip1 but does not facilitate its translocation by the AAA ATPase Bcs1. Mol Cell Biol 32, 4400–4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmurs M, Foti M, Raemy E, Vaz FM, Martinou JC, Bairoch A, Lane L (2015). C11orf83, a mitochondrial cardiolipin-binding protein involved in bc1 complex assembly and supercomplex stabilization. Mol Cell Biol 35, 1139–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F, Kotarsky H, Fellman V, Moraes CT (2011). Mitochondrial disorders caused by mutations in respiratory chain assembly factors. Semin Fetal Neonatal Med 16, 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerrier C, Garcia-Souza LF, Krumschnabel G, Wohlfarter Y, Mészáros AT, Gnaiger E (2018). High-resolution fluorespirometry and OXPHOS protocols for human cells, permeabilized fibers from small biopsies of muscle, and isolated mitochondria. Methods Mol Biol 1782, 31–70. [DOI] [PubMed] [Google Scholar]
- Dudkina NV, Kouril R, Peters K, Braun HP, Boekema EJ (2010). Structure and function of mitochondrial supercomplexes. Biochim Biophys Acta 1797, 664–670. [DOI] [PubMed] [Google Scholar]
- Feichtinger RG, Brunner-Krainz M, Alhaddad B, Wortmann SB, Kovacs-Nagy R, Stojakovic T, Erwa W, Resch B, Windischhofer W, Verheyen S, et al. (2017). Combined respiratory chain deficiency and UQCC2 mutations in neonatal encephalomyopathy: defective supercomplex assembly in complex III deficiencies. Oxid Med Cell Longev 2017, 7202589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Nunnari J (2014). Mitochondrial form and function. Nature 505, 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi D, Arzuffi P, Zordan M, Da Re C, Lamperti C, Benna C, D’Adamo P, Diodato D, Costa R, Mariotti C, et al. (2011). Mutations in TTC19 cause mitochondrial complex III deficiency and neurological impairment in humans and flies. Nat Genet 43, 259–263. [DOI] [PubMed] [Google Scholar]
- Ghezzi D, Zeviani M (2018). Human diseases associated with defects in assembly of OXPHOS complexes. Essays Biochem 62, 271–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Wu M, Guo R, Yan K, Lei J, Gao N, Yang M (2016). The architecture of the mammalian respirasome. Nature 537, 639–643. [DOI] [PubMed] [Google Scholar]
- Han J, Jung S, Jang J, Kam TI, Choi H, Kim BJ, Nah J, Jo DG, Nakagawa T, Nishimura M, Jung YK (2014). OCIAD2 activates gamma-secretase to enhance amyloid beta production by interacting with nicastrin. Cell Mol Life Sci 71, 2561–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque ME, Spremulli LL, Fecko CJ (2010). Identification of protein-protein and protein-ribosome interacting regions of the C-terminal tail of human mitochondrial inner membrane protein Oxa1L. J Biol Chem 285, 34991–34998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K, Neupert W, Stuart RA (2001). Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J 20, 1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock DH, Robinson DRL, Stroud DA (2020). Blackout in the powerhouse: clinical phenotypes associated with defects in the assembly of OXPHOS complexes and the mitoribosome. Biochem J 477, 4085–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofhaus G, Shakeley RM, Attardi G (1996). Use of polarography to detect respiration defects in cell cultures. Methods Enzymol 264, 476–483. [DOI] [PubMed] [Google Scholar]
- Hunte C, Koepke J, Lange C, Rossmanith T, Michel H (2000). Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure 8, 669–684. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Andrell J, Choi A, Richter U, Maiti P, Best RB, Barrientos A, Battersby BJ, Amunts A (2021). Mechanism of membrane-tethered mitochondrial protein synthesis. Science 371, 846–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, Link TA, Ramaswamy S, Jap BK (1998). Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281, 64–71. [DOI] [PubMed] [Google Scholar]
- Kirby DM, Thorburn DR, Turnbull DM, Taylor RW (2007). Biochemical assays of respiratory chain complex activity. Methods Cell Biol 80, 93–119. [DOI] [PubMed] [Google Scholar]
- Lamantea E, Carrara F, Mariotti C, Morandi L, Tiranti V, Zeviani M (2002). A novel nonsense mutation (Q352X) in the mitochondrial cytochrome b gene associated with a combined deficiency of complexes I and III. Neuromuscul Disord 12, 49–52. [DOI] [PubMed] [Google Scholar]
- Lapuente-Brun E, Moreno-Loshuertos R, Acín-Pérez R, Latorre-Pellicer A, Colás C, Balsa E, Perales-Clemente E, Quirós PM, Calvo E, Rodríguez-Hernández MA, et al. (2013). Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340, 1567–1570. [DOI] [PubMed] [Google Scholar]
- Letts JA, Sazanov LA (2017). Clarifying the supercomplex: the higher-order organization of the mitochondrial electron transport chain. Nat Struct Mol Biol 24, 800–808. [DOI] [PubMed] [Google Scholar]
- Lobo-Jarne T, Perez-Perez R, Fontanesi F, Timon-Gomez A, Wittig I, Penas A, Serrano-Lorenzo P, Garcia-Consuegra I, Arenas J, Martin MA, et al. (2020). Multiple pathways coordinate assembly of human mitochondrial complex IV and stabilization of respiratory supercomplexes. EMBO J 39, e103912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo-Jarne T, Ugalde C (2018). Respiratory chain supercomplexes: structures, function and biogenesis. Semin Cell Dev Biol 76, 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo LY, Soosaipillai A, Diamandis EP (2001). Molecular cloning of a novel human gene on chromosome 4p11 by immunoscreening of an ovarian carcinoma cDNA library. Biochem Biophys Res Commun 280, 401–406. [DOI] [PubMed] [Google Scholar]
- Mick DU, Dennerlein S, Wiese H, Reinhold R, Pacheu-Grau D, Lorenzi I, Sasarman F, Weraarpachai W, Shoubridge EA, Warscheid B, Rehling P (2012). MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation. Cell 151, 1528–1541. [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Blaza JN, Larsson NG, Hirst J (2017). The enigma of the respiratory chain supercomplex. Cell Metab 25, 765–776. [DOI] [PubMed] [Google Scholar]
- Mohanraj K, Wasilewski M, Beninca C, Cysewski D, Poznanski J, Sakowska P, Bugajska Z, Deckers M, Dennerlein S, Fernandez-Vizarra E, et al. (2019). Inhibition of proteasome rescues a pathogenic variant of respiratory chain assembly factor COA7. EMBO Mol Med 11, e9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Lastres D, Fontanesi F, García-Consuegra I, Martín MA, Arenas J, Barrientos A, Ugalde C (2012). Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab 15, 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata C, Kobayashi H, Sakata A, Satomi K, Minami Y, Morishita Y, Ohara R, Yoshikawa H, Arai Y, Nishida M, Noguchi M (2012). Increased expression of OCIA domain containing 2 during stepwise progression of ovarian mucinous tumor. Pathol Int 62, 471–476. [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM (2007). Translocation of proteins into mitochondria. Annu Rev Biochem 76, 723–749. [DOI] [PubMed] [Google Scholar]
- Ott M, Herrmann JM (2010). Co-translational membrane insertion of mitochondrially encoded proteins. Biochim Biophys Acta 1803, 767–775. [DOI] [PubMed] [Google Scholar]
- Perez-Perez R, Lobo-Jarne T, Milenkovic D, Mourier A, Bratic A, Garcia-Bartolome A, Fernandez-Vizarra E, Cadenas S, Delmiro A, Garcia-Consuegra I, et al. (2016). COX7A2L is a mitochondrial complex III binding protein that stabilizes the III2+IV supercomplex without affecting respirasome formation. Cell Rep 16, 2387–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G, Taddeo MA, Petersen RB, Castellani RJ, Harris PLR, Siedlak SL, Cash AD, Liu Q, Nunomura A, Atwood CS, Smith MA (2003). Adventiously-bound redox active iron and copper are at the center of oxidative damage in Alzheimer disease. Biometals 16, 77–81. [DOI] [PubMed] [Google Scholar]
- Priesnitz C, Becker T (2018). Pathways to balance mitochondrial translation and protein import. Genes Dev 32, 1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protasoni M, Pérez-Pérez R, Lobo-Jarne T, Harbour ME, Ding S, Peñas A, Diaz F, Moraes CT, Fearnley IM, Zeviani M, et al. (2020). Respiratory supercomplexes act as a platform for complex III-mediated maturation of human mitochondrial complexes I and IV. EMBO J 39, e102817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protasoni M, Zeviani M (2021). Mitochondrial structure and bioenergetics in normal and disease conditions. Int J Mol Sci 22, 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelt H, Pfanner N (2016). Coordination of two genomes by mitochondrial translational plasticity. Cell 167, 308–310. [DOI] [PubMed] [Google Scholar]
- Rampelt H, Sucec I, Bersch B, Horten P, Perschil I, Martinou JC, van der Laan M, Wiedemann N, Schanda P, Pfanner N (2020). The mitochondrial carrier pathway transports non-canonical substrates with an odd number of transmembrane segments. BMC Biol 18, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold R, Kruger V, Meinecke M, Schulz C, Schmidt B, Grunau SD, Guiard B, Wiedemann N, van der Laan M, Wagner R, et al. (2012). The channel-forming Sym1 protein is transported by the TIM23 complex in a presequence-independent manner. Mol Cell Biol 32, 5009–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisinger V, Eichacker LA (2008). Solubilization of membrane protein complexes for blue native PAGE. J Proteomics 71, 277–283. [DOI] [PubMed] [Google Scholar]
- Richter-Dennerlein R, Oeljeklaus S, Lorenzi I, Ronsor C, Bareth B, Schendzielorz AB, Wang C, Warscheid B, Rehling P, Dennerlein S (2016). Mitochondrial protein synthesis adapts to influx of nuclear-encoded protein. Cell 167, 471–483.e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenburg RJ (2016). Mitochondrial complex I-linked disease. Biochim Biophys Acta 1857, 938–945. [DOI] [PubMed] [Google Scholar]
- Sakashita M, Sakashita S, Murata Y, Shiba-Ishii A, Kim Y, Matsuoka R, Nakano N, Sato Y, Noguchi M (2018). High expression of ovarian cancer immunoreactive antigen domain containing 2 (OCIAD2) is associated with poor prognosis in lung adenocarcinoma. Pathol Int 68, 596–604. [DOI] [PubMed] [Google Scholar]
- Sanchez E, Lobo T, Fox JL, Zeviani M, Winge DR, Fernandez-Vizarra E (2013). LYRM7/MZM1L is a UQCRFS1 chaperone involved in the last steps of mitochondrial complex III assembly in human cells. Biochim Biophys Acta 1827, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer E, Seelert H, Reifschneider NH, Krause F, Dencher NA, Vonck J (2006). Architecture of active mammalian respiratory chain supercomplexes. J Biol Chem 281, 15370–15375. [DOI] [PubMed] [Google Scholar]
- Schmitt M, Neupert W, Langer T (1996). The molecular chaperone Hsp78 confers compartment-specific thermotolerance to mitochondria. J Cell Biol 134, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signes A, Fernandez-Vizarra E (2018). Assembly of mammalian oxidative phosphorylation complexes I-V and supercomplexes. Essays Biochem 62, 255–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Salvatori R, Aftab W, Aufschnaiter A, Carlstrom A, Forne I, Imhof A, Ott M (2020). Molecular connectivity of mitochondrial gene expression and OXPHOS biogenesis. Mol Cell 79, 1051–1065.e1010. [DOI] [PubMed] [Google Scholar]
- Sinha S, Bheemsetty VA, Inamdar MS (2018). A double helical motif in OCIAD2 is essential for its localization, interactions and STAT3 activation. Sci Rep 8, 7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladowska M, Turek M, Kim MJ, Drabikowski K, Mussulini BHM, Mohanraj K, Serwa RA, Topf U, Chacinska A (2021). Proteasome activity contributes to pro-survival response upon mild mitochondrial stress in Caenorhabditis elegans. PLoS Biol 19, e3001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slipetz DM, Aprille JR, Goodyer PR, Rozen R (1991). Deficiency of complex III of the mitochondrial respiratory chain in a patient with facioscapulohumeral disease. Am J Hum Genet 48, 502–510. [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Robinson AJ (2019). MitoMiner v4.0: an updated database of mitochondrial localization evidence, phenotypes and diseases. Nucleic Acids Res 47, D1225–D1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Fox JL, Winge DR (2012). Biogenesis of the cytochrome bc(1) complex and role of assembly factors. Biochim Biophys Acta 1817, 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan K, Ott M (2020). Timing of dimerization of the bc1 complex during mitochondrial respiratory chain assembly. Biochim Biophys Acta 1861, 148177. [DOI] [PubMed] [Google Scholar]
- Tang WK, Borgnia MJ, Hsu AL, Esser L, Fox T, de Val N, Xia D (2020). Structures of AAA protein translocase Bcs1 suggest translocation mechanism of a folded protein. Nat Struct Mol Biol 27, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timon-Gomez A, Garlich J, Stuart RA, Ugalde C, Barrientos A (2020). Distinct roles of mitochondrial HIGD1A and HIGD2A in respiratory complex and supercomplex biogenesis. Cell Rep 31, 107607. [DOI] [PubMed] [Google Scholar]
- Tiranti V, Munaro M, Sandonà D, Lamantea E, Rimoldi M, DiDonato S, Bisson R, Zeviani M (1995). Nuclear DNA origin of cytochrome c oxidase deficiency in Leigh’s syndrome: genetic evidence based on patient’s-derived rho degrees transformants. Hum Mol Genet 4, 2017–2023. [DOI] [PubMed] [Google Scholar]
- Tropeano CV, Aleo SJ, Zanna C, Roberti M, Scandiffio L, Loguercio Polosa P, Fiori J, Porru E, Roda A, Carelli V, et al. (2020). Fine-tuning of the respiratory complexes stability and supercomplexes assembly in cells defective of complex III. Biochim Biophys Acta 1861, 148133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker EJ, Wanschers BF, Szklarczyk R, Mountford HS, Wijeyeratne XW, van den Brand MA, Leenders AM, Rodenburg RJ, Reljic B, Compton AG, et al. (2013). Mutations in the UQCC1-interacting protein, UQCC2, cause human complex III deficiency associated with perturbed cytochrome b protein expression. PLoS Genet 9, e1004034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turakhiya U, von der Malsburg K, Gold VAM, Guiard B, Chacinska A, van der Laan M, Ieva R (2016). Protein import by the mitochondrial presequence translocase in the absence of a membrane potential. J Mol Biol 428, 1041–1052. [DOI] [PubMed] [Google Scholar]
- Vendruscolo M, Zurdo J, MacPhee CE, Dobson CM (2003). Protein folding and misfolding: a paradigm of self-assembly and regulation in complex biological systems. Philos Trans A Math Phys Eng Sci 361, 1205–1222. [DOI] [PubMed] [Google Scholar]
- Voos W, Jaworek W, Wilkening A, Bruderek M (2016). Protein quality control at the mitochondrion. Essays Biochem 60, 213–225. [DOI] [PubMed] [Google Scholar]
- Wagener N, Neupert W (2012). Bcs1, a AAA protein of the mitochondria with a role in the biogenesis of the respiratory chain. J Struct Biol 179, 121–125. [DOI] [PubMed] [Google Scholar]
- Wanschers BF, Szklarczyk R, van den Brand MA, Jonckheere A, Suijskens J, Smeets R, Rodenburg RJ, Stephan K, Helland IB, Elkamil A, et al. (2014). A mutation in the human CBP4 ortholog UQCC3 impairs complex III assembly, activity and cytochrome b stability. Hum Mol Genet 23, 6356–6365. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N (2017). Mitochondrial machineries for protein import and assembly. Annu Rev Biochem 86, 685–714. [DOI] [PubMed] [Google Scholar]
- Wilkening A, Rub C, Sylvester M, Voos W (2018). Analysis of heat-induced protein aggregation in human mitochondria. J Biol Chem 293, 11537–11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Yang X, Peng H, Guo D, Zhao W, Zhao C, Zhou X (2017). OCIAD2 suppressed tumor growth and invasion via AKT pathway in hepatocellular carcinoma. Carcinogenesis 38, 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Yu CA, Kim H, Xia JZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J (1997). Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science 277, 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Reljić B, Liang C, Kerouanton B, Francisco JC, Peh JH, Mary C, Jagannathan NS, Olexiouk V, Tang C, et al. (2020). Mitochondrial peptide BRAWNIN is essential for vertebrate respiratory complex III assembly. Nat Commun 11, 1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, Crofts AR, Berry EA, Kim SH (1998). Electron transfer by domain movement in cytochrome bc1. Nature 392, 677–684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.