Abstract
Homeostatic regulation of plasma membrane (PM) phosphatidylinositol 4,5-bisphosphate (PIP2) in receptor-stimulated cells is mediated by the lipid transfer protein Nir2. Nir2 is dynamically recruited to endoplasmic reticulum–plasma membrane (ER–PM) junctions to facilitate replenishment of PM PIP2 hydrolyzed during receptor-mediated signaling. However, our knowledge regarding the activation and sustainment of Nir2-mediated replenishment of PM PIP2 is limited. Here, we describe the functions of Nir1 as a positive regulator of Nir2 and PIP2 homeostasis. In contrast to the family proteins Nir2 and Nir3, Nir1 constitutively localizes at ER–PM junctions. Nir1 potentiates Nir2 targeting to ER–PM junctions during receptor-mediated signaling and is required for efficient PM PIP2 replenishment. Live-cell imaging and biochemical analysis reveal that Nir1 interacts with Nir2 via a region between the FFAT motif and the DDHD domain. Combined, results from this study identify Nir1 as an ER–PM junction localized protein that promotes Nir2 recruitment for PIP2 homeostasis.
INTRODUCTION
Phosphatidylinositol 4,5 bisphosphate (PIP2) is a critical lipid molecule involved in the regulation of a diverse array of cell processes such as endocytosis and exocytosis, ion channel function, and store-operated Ca2+ entry (Balla, 2013). PIP2 is enriched in the inner leaflet of the plasma membrane (PM) and functions as a localization and signaling hub by recruiting and/or activating specific proteins. Additionally, PIP2 is hydrolyzed following receptor-induced activation of phospholipase C (PLC) to generate the secondary signaling molecules inositol trisphosphate, diacylglycerol (DAG), and downstream phosphatidic acid (PA), to activate distinct signaling pathways (Berridge, 1984; Wang et al., 2006; Tanguy et al., 2019). PIP2 can also be converted into PI 3,4,5-trisphosphate via PI 3-kinases (PI3Ks) to mediate growth factor signaling (Fruman et al., 2017). Therefore, replenishment of PIP2 at the PM following receptor-induced hydrolysis is essential for cells to sustain such diverse signaling outputs.
The homeostasis of PIP2 during receptor stimulation is maintained by a cyclical metabolic pathway, referred to as the phosphatidylinositol (PI) cycle (Hokin and Hokin, 1964). Lipid intermediates are sequentially catalyzed by enzymes assigned to each metabolic step in the PI cycle. Interestingly, the enzymatic machinery required to sustain the PI cycle are spatially segregated between two organelles. On the one hand, PI synthase and cytidine diphosphate DAG synthase reside on the endoplasmic reticulum (ER) and are required for the conversion of PA to PI (Agranoff et al., 1958; Benjamins and Agranoff, 1969). On the other hand, PI 4-kinases (PI4K), PI 4-phosphate 5-kinases (PIP5K), PLC, and DAG kinases that collectively convert PI to PA via production of PIP2 and DAG reside on the PM (Balla, 2013). Consequently, the spatial segregation of PI cycle enzymes requires a mechanism to transfer the lipid precursors PA and PI between the ER and the PM.
While vesicular transport between membranes has been known to play a role in lipid trafficking (Voelker, 1990), many studies have revealed the importance of lipid transfer proteins (LTPs) in this process (Lev, 2012). A class of LTPs called PI transfer proteins (PITPs) are implicated in the transfer of PI through a lipid-binding PITP domain that facilitates lipid transport (Cockcroft, 2012). The type IIA PITPs Nir2 (PITPNM1/RdgBαI) and Nir3 (PITPNM2/RdgBαII) were shown to promote efficient replenishment of PM PIP2 following receptor-mediated depletion (Chang et al., 2013; Kim et al., 2013; Chang and Liou, 2015; Kim et al., 2015). Following receptor-induced PLC activation, Nir2 and Nir3 are dynamically recruited from the cytoplasm to endoplasmic reticulum–plasma membrane (ER–PM) junctions, subcellular loci that enable close apposition and short-range exchange of lipids between the ER and the PM (Toulmay and Prinz, 2011; Chang et al., 2017). Therefore, the PITP domain of Nir2 and Nir3 has been proposed to facilitate direct PI/PA exchange at ER–PM junctions (Chang and Liou, 2015; Kim et al., 2015; Yadav et al., 2015). In support of this model, the PITP domain of the Drosophila homologue of Nir2, RDGBα, was shown to bind and transfer PI and PA in vitro (Yadav et al., 2015).
A third mammalian Nir homologue, Nir1 (PITPNM3/RdgBαIII), has been linked with several diseases. In humans, Nir1 mutations have been identified in patients with autosomal dominant cone dystrophy (CORD5; Kohn et al., 2007, 2010; Bakhoum et al., 2018). Nir1 has also been demonstrated to promote metastasis and epithelial to mesenchymal transition in breast cancer (Chen et al., 2011; Lin et al., 2016). Lastly, a zebrafish homologue of Nir1 was capable of slowing retinal degeneration in an rdgB mutant of Drosophila (Elagin et al., 2000). Nir1 does not contain a PITP domain, yet it shares high sequence identity with Nir2 and Nir3. Nonetheless, the functional role of Nir1 or its impact on disease is unclear. Here, we report that Nir1 localizes at ER–PM junctions, interacts with the LTP Nir2, and positively regulates PM PIP2 homeostasis.
RESULTS AND DISCUSSION
Nir1 constitutively localizes at ER–PM junctions
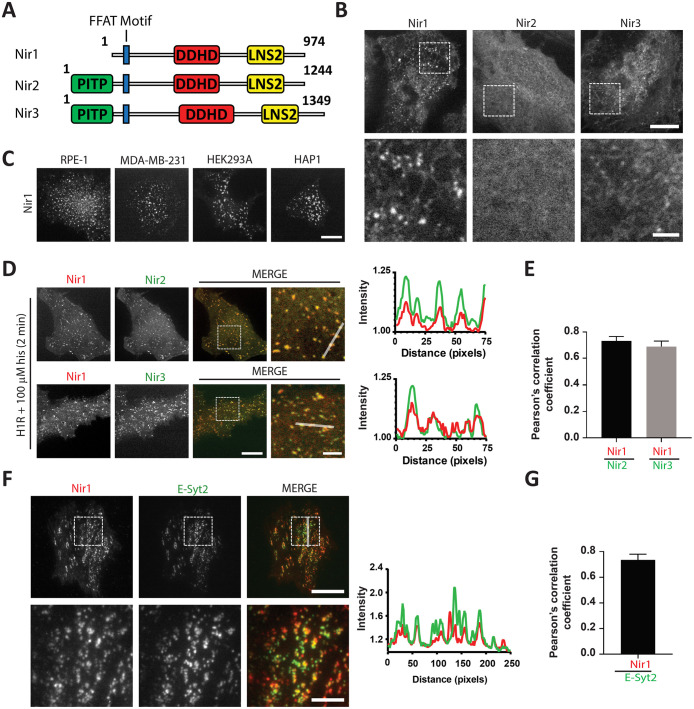
Nir1 is a homologue of the mammalian LTPs Nir2 and Nir3 that were shown to facilitate PM PIP2 homeostasis and phosphoinositide signaling. While Nir1 lacks an N-terminal PITP domain (Figure 1A), the remainder of Nir2 and Nir3 sequences share 53.3% and 60.1% identity to Nir1. To determine Nir1’s cellular localization, we expressed mCherry-tagged Nir1 (Nir1-mCh) in HeLa cells and performed live-cell confocal microscopy. In contrast to Nir2 and Nir3, which have cytosolic and ER localizations in nonstimulated HeLa cells (Chang and Liou, 2015), Nir1 localized to discrete puncta (Figure 1B). Nir1 puncta were observed in RPE-1, MDA-MB-231, HEK293A, and HAP1 cells, ruling out the possibility of a HeLa cell–specific localization (Figure 1C). These puncta highly resembled Nir2 and Nir3 localizations at ER–PM junctions following PIP2 hydrolysis at the PM (Chang et al., 2013; Chang and Liou, 2015; Kim et al., 2015). To explore this possibility, we coexpressed Nir1-mCh with Nir2-YFP or Nir3-YFP and the histamine H1 receptor (H1R) in HeLa cells. H1R is a G-protein (Gq) coupled receptor that activates PLC to hydrolyze PIP2 following histamine stimulation (Panula et al., 2015). We have previously shown that histamine stimulation can trigger Nir2 and Nir3 translocation to ER–PM junctions via PLC and diacylglycerol kinases in HeLa cells overexpressing H1R (Chang and Liou, 2015). We observed robust colocalization of Nir1 with Nir2 or Nir3 puncta following treatment with histamine exemplified by line scan analyses of puncta (Figure 1D). In support of this observation, quantification using Pearson’s correlation coefficient revealed a high degree of colocalization between Nir1-mCh with Nir2-YFP or Nir3-YFP (Figure 1E). We observed similar colocalization of Nir1 with Nir2 or Nir3 in HAP1 and RPE-1 cells as well (Supplemental Figure 1A).
FIGURE 1:
Nir1 constitutively localizes at ER–PM junctions. (A) A diagram of Nir protein domains. (B) Confocal images of HeLa cells expressing Nir1-mCh, Nir2-mCh, or Nir3-mCh. Nir1 was observed in puncta in all cells (n = 60; three experiments). Scale bars: 10 µm and 3 µm (magnified images, bottom row). (C) Confocal images of RPE-1, MDA-MB-231, HEK293A, and HAP1 cells expressing Nir1-mCh or Nir1-YFP. Nir1 was observed in puncta in all cells (n = 30; two experiments). Scale bar: 10 μm. (D) Confocal images of HeLa cells coexpressing Nir1-mCh and Nir2-YFP (top row) or Nir3-YFP (bottom row) and H1R taken 2 min after 100 μM histamine (his) treatment and corresponding line scan histogram. Scale bars: 10 µm and 3 µm (magnified images, right panels). (E) Colocalization of Nir1-mCh with either Nir2-YFP (mean ± SEM; n = 30) or Nir3-YFP (mean ± SEM; n = 15) was quantified using Pearson’s correlation coefficient (PCC). (F) TIRF images of a HeLa cell coexpressing Nir1-mCh and GFP-E-Syt2 and corresponding line scan histogram. Scale bars: 20 µm and 5 µm (magnified images, bottom row). (G) Colocalization between Nir1-mCh and GFP-E-Syt2 was quantified using PCC (mean ± SEM; n = 6).
Next, we coexpressed Nir1-mCh with an ER–PM junction localized protein, extended synaptotagmin 2 (E-Syt2; Giordano et al., 2013). Indeed, Nir1 puncta colocalized with GFP-E-Syt2 as detected using total internal reflection fluorescent (TIRF) microscopy that selectively excites and detects fluorophores within ∼100 nm of the PM to facilitate detection of ER–PM junctions (Figure 1, F and G). In addition to E-Syt2, we observed Nir1 puncta colocalization with a mutant of stromal interaction molecule 1, STIM1-D76A, that constitutively localizes at ER–PM junctions (Liou et al., 2005), and with the genetically encoded ER–PM junction marker MAPPER (Supplemental Figure 1, B and C; Chang et al., 2013). Together, these results reveal that Nir proteins have distinct cellular localizations. While Nir2 and Nir3 are inducibly targeted to ER–PM junctions during receptor stimulation, Nir1 is constitutively localized at these sites.
Nir1 localization at ER–PM junctions requires the FFAT motif and the LNS2 domain
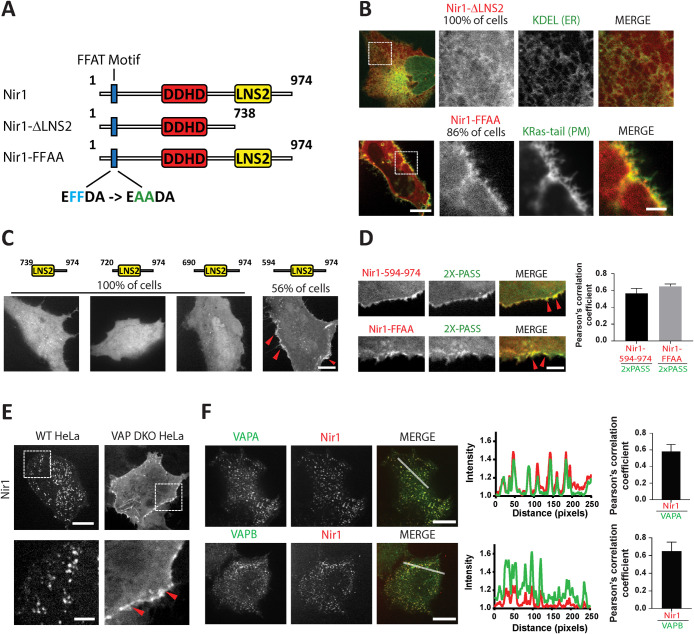
Given the high degree of sequence identity between Nir proteins and their ability to localize to ER–PM junctions, it is reasonable to predict that membrane targeting mechanisms are conserved among this protein family. Nir1 contains a C-terminal LNS2 (Lipin/Ned1/Smp2) domain and a putative FFAT (two phenylalanines in an acid tract) motif (Figure 2A). In Nir2 and Nir3, the LNS2 domain is involved in PA binding and PM association triggered by receptor stimulation of PIP2 hydrolysis (Chang et al., 2013; Kim et al., 2013; Chang and Liou, 2015; Kim et al., 2015). To explore the relevance of the conserved LNS2 domain in Nir1, we generated a Nir1 deletion construct lacking amino acids (aa) 739–974, named Nir1-∆LNS2 (Figure 2A). As expected, Nir1-∆LNS2 lacked ER–PM junction localization and instead localized to the cytosol and the ER, as indicated by its colocalization with the ER marker YFP-KDEL in 100% of cells we examined (Figure 2B). To further define the region of Nir1 that mediates PM localization, we generated a series of LNS2 domain-containing constructs. Nir1-739–974, which begins at the NCBI Gene Database annotated N-terminal LNS2 boundary (aa 739) and spans the remaining C-terminal region, displayed cytosolic localization in 100% of cells (Figure 2C). The same was observed in slightly longer versions, Nir1-720–974 and Nir1-690–974. Notably, the longest construct, Nir1-594–974, exhibited localization at the cell periphery in 56% of cells. These results revealed that the currently defined LNS2 domain is not sufficient for PM targeting. This observation is in agreement with several studies of Nir2 and Nir3. C-terminal fragments of Nir2 and Nir3 containing the LNS2 domain and an extended upstream region have been shown to bind PA in vitro (Kim et al., 2013; Chang and Liou, 2015). In addition, a region upstream of the LNS2 domain was shown to be required for targeting the C-terminal half of Nir2 to the PM (Kim et al., 2015). Together, these observations suggest that Nir proteins interact with the PM by binding to PA via their LNS2-containing C-terminal region.
FIGURE 2:
Nir1 localization at ER–PM junctions requires the FFAT motif and the LNS2 domain. (A) A diagram of wild-type (WT) and mutated Nir1 constructs. (B) Confocal images of HeLa cells coexpressing Nir1-∆LNS2-mCh and YFP-KDEL (top row), or Nir1-FFAA-mCh and YFP-KRas-tail (bottom row). Nir1-∆LNS2-mCh displayed cytosolic and ER localization in 100% of cells (n = 58; three experiments). Nir1-FFAA-mCh displayed PM localization in 86% of cells (49 out of 57; three experiments). Scale bars: 10 μm and 3 μm (magnified images). (C) Confocal images of HeLa cells expressing Nir1-739-974-mCh, Nir1-720-974-mCh, Nir1-690-974-mCh, or Nir1-594-974-mCh. Nir1-594-974-mCh enrichment at the PM was observed in 56% of cells (18 out of 32; two experiments), while the other constructs displayed cytosolic localization in 100% of cells (n = 30; two experiments). Scale bar: 10 μm. Red arrows indicate regions on the cell periphery with Nir1-594-974-mCh signal. (D) Confocal images of HeLa cells coexpressing GFP-2X-PASS and Nir1-594-974-mCh or Nir1-FFAA-mCh. Scale bar: 3 µm. Red arrows indicate regions on the cell periphery with overlapping GFP-2X-PASS and Nir1-594-974-mCh or Nir1-FFAA-mCh signal. Colocalization of GFP-2X-PASS with either Nir1-594-974-mCh (mean ± SEM; n = 23) or Nir1-FFAA-mCh (mean ± SEM; n = 27) was quantified using PCC. (E) Confocal images of either WT or VAP DKO HeLa cells expressing Nir1-mCh. Localization patterns depicted were observed in 100% of cells (n = 40; two experiments). Scale bars: 10 μm and 3 μm (bottom row). Red arrows indicate regions on the cell periphery with Nir1-mCh signal. (F) Confocal images of VAP DKO HeLa cells expressing Nir1-mCh and either VAPA-YFP (top row) or VAPB-YFP (bottom row) and corresponding line scan histograms (right). Scale bars: 10 μm. Colocalization was observed in 100% of cells (n = 30; two experiments) and quantified using PCC (mean ± SEM).
Nir proteins also contain an N-terminal FFAT motif. FFAT motifs target proteins to the ER by serving as interaction sites for the ER transmembrane proteins, vesicle-associated membrane protein-associated protein isoforms A and B (VAPA and VAPB) (Loewen et al., 2003). All three Nir proteins have been previously shown to interact with VAPB (Amarilio et al., 2005). To explore the role of the two phenylalanines in an acidic tract (FFAT) in Nir1 localization in cells, we generated a Nir1-FFAA construct by making point mutations within the FFAT motif of Nir1 (Figure 2A). Nir1-FFAA failed to localize at ER–PM junctions and instead displayed PM localization in 86% of cells as indicated by colocalization with the PM marker YFP-KRas-tail (Figure 2B). Consistent with previous findings that Nir proteins target to the PM by binding to PA, both Nir1-FFAA and Nir1-594–974 colocalized with GFP-2X-PASS (phosphatidic acid biosensor with superior sensitivity) that selectively detects PA constituting 1–2% of total PM phospholipids in resting cells (Figure 2D; Ferrell and Huestis, 1984; Mitchell et al., 1986; Bohdanowicz et al., 2013; Zhang et al., 2014).
To further validate the importance of the FFAT motif and VAP proteins in Nir1 localization at ER–PM junctions, we employed a previously generated VAPA and VAPB double knockout (VAP DKO) HeLa cell line (Dong et al., 2016). In the VAP DKO line, Nir1 lacked ER–PM junction localization in 100% of cells and instead localized to the PM (Figure 2E), resembling Nir1-FFAA and Nir1-594–974 localization. Expression of either VAPA or VAPB fully rescued Nir1’s localization at ER–PM junctions in VAP DKO cells and Nir1 puncta colocalized with both VAPA and VAPB in 100% of cells (Figure 2F). These results indicate that Nir proteins have conserved ER and PM targeting mechanisms. Namely, the FFAT motif associates with VAP proteins in the ER, while the C-terminal region including the LNS2 domain associates with PA at the PM.
The PITP domain limits ER–PM junction targeting of Nir proteins
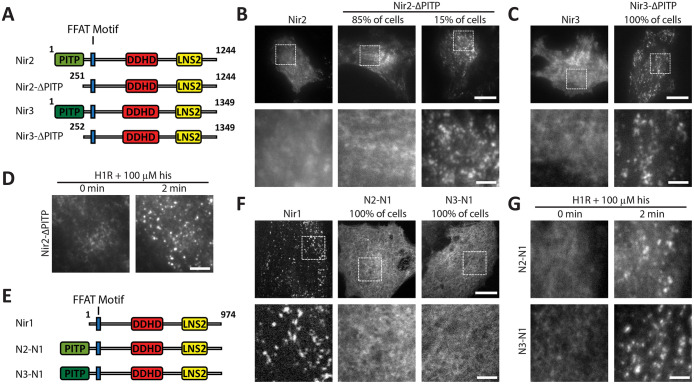
Our findings up to this point revealed that Nir proteins have conserved mechanisms for targeting to the ER and the PM. However, in contrast to Nir2 and Nir3, Nir1 is constitutively localized at ER–PM junctions. We speculated whether the PITP domain plays a role in regulating protein localization. To test this hypothesis, we generated Nir2 and Nir3 constructs lacking the N-terminal PITP domain, named Nir2-ΔPITP and Nir3-ΔPITP, respectively (Figure 3A). Interestingly, Nir2-ΔPITP-mCh localized to puncta indicative of ER–PM junctions in 15% of cells (Figure 3B), while Nir3-ΔPITP-mCh localized to puncta indicative of ER–PM junctions in 100% of cells (Figure 3C). We previously demonstrated that Nir2 and Nir3 have differential strengths in PA binding in their C-terminal regions and that Nir3 has a higher PA-binding ability than Nir2 (Chang and Liou, 2015). This difference likely underlies the discrepancy in ER–PM junction localization of the Nir2-ΔPITP and Nir3-ΔPITP constructs. In line with previous findings (Kim et al., 2015), we observed robust Nir2-ΔPITP-mCh translocation to ER–PM junctions in response to receptor stimulation that hydrolyzes PM PIP2 (Figure 3D).
FIGURE 3:
The PITP domain of Nir proteins limits targeting to ER–PM junctions. (A) A diagram of WT and deletion constructs of Nir2 and Nir3. (B) TIRF images of HeLa cells expressing Nir2-mCh or Nir2-∆PITP-mCh. Cells (85%; 34 out of 40) expressing Nir2-∆PITP-mCh displayed diffuse localization resembling WT Nir2, while 15% of cells (6 out of 40; two experiments) displayed enrichment in puncta. Scale bars: 20 μm and 5 μm (bottom row). (C) TIRF images of HeLa cells expressing Nir3-mCh or Nir3-∆PITP-mCh. Nir3-∆PITP-mCh puncta was observed in 100% of cells (n = 30; two experiments). Scale bars: 20 μm and 5 μm (bottom row). (D) TIRF images of HeLa cells expressing Nir2-∆PITP-mCh treated with 100 μM his. Scale bar: 5 μm. (E) A diagram of WT Nir1 and fusion constructs N2-N1 and N3-N1. (F) TIRF images of HeLa cells expressing Nir1-mCh, N2-N1-mCh, or N3-N1-mCh. Cells (100%) expressing N2-N1-mCh or N3-N1-mCh lost puncta localization (n = 35; two experiments). Scale bars: 20 μm and 5 μm (bottom row). (G) TIRF images of HeLa cells expressing N2-N1 or N3-N1 treated with 100 μM his (n = 20; two experiments). Scale bar: 5 μm.
Next, we tested whether the addition of a PITP domain to Nir1 would limit its targeting to ER–PM junctions. Indeed, fusion of either the Nir2 or Nir3 PITP domain to the N-terminus of Nir1, named N2-N1 and N3-N1, respectively (Figure 3E), resulted in a complete loss of targeting to ER–PM junctions in 100% of cells (Figure 3F). However, targeting of N2-N1 and N3-N1 to ER–PM junctions could be induced in the context of H1R overexpression and stimulation with histamine (Figure 3G). These results suggest that the PITP domain may provide an autoinhibitory mechanism to limit Nir2 and Nir3 targeting to ER–PM junctions. Alternatively, the PITP domain may limit targeting of Nir proteins to ER–PM junctions by clearance of PA from the PM.
Nir1 promotes Nir2 recruitment to ER–PM junctions and regulates PM PIP 2 replenishment
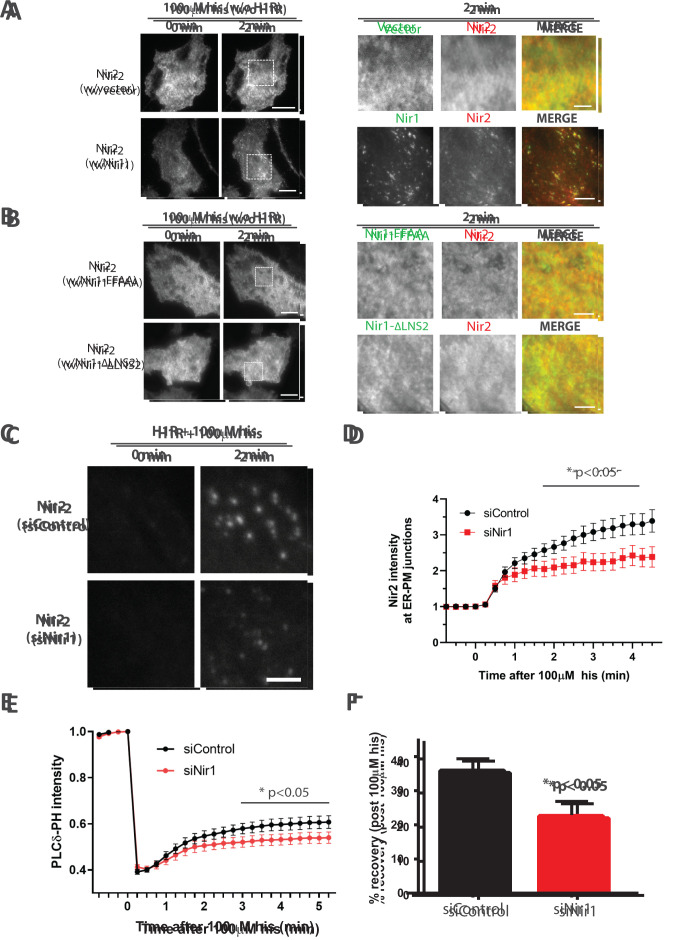
We previously demonstrated that the PITP domain of Nir2 is more effective at mediating PM PIP2 replenishment than the PITP domain of Nir3 (Chang and Liou, 2015). This finding, coupled with Nir2’s weaker association to PA, creates a potential problem. Nir2 is critical for efficient replenishment of PM PIP2, yet the threshold of activation during receptor-mediated signaling is relatively high. Therefore, we speculated whether additional factors may support or bolster Nir2’s ability to target to ER–PM junctions. Given Nir1’s constitutive localization at ER–PM junctions, we examined whether Nir1 overexpression would promote Nir2’s targeting to ER–PM junctions. In our HeLa cell system, Nir2 translocation to ER–PM junctions requires robust PIP2 hydrolysis achieved by overexpression of H1R (Chang et al., 2013). To assess the impact of Nir1 overexpression, we measured Nir2 targeting to ER–PM junctions; however, we omitted H1R overexpression. As expected, histamine stimulation was not sufficient to trigger Nir2 translocation in cells cotransfected with the YFP vector without H1R (Figure 4A, top row). Strikingly, cells cotransfected with Nir1 displayed robust translocation of Nir2 to ER–PM junctions following histamine stimulation (Figure 4A, bottom row), suggesting that Nir1 coexpression could subvert the requirement of overexpressing H1R to generate high levels of PA for Nir2 targeting to ER–PM junctions upon receptor activation. Further, this potentiating effect was not observed in cells coexpressing either Nir1-FFAA or Nir1-∆LNS2 (Figure 4B), suggesting that Nir1 localization at ER–PM junctions is required for its support of Nir2 targeting to ER–PM junctions. Additionally, we observed a significant reduction in overexpressed Nir2 translocation to ER–PM junctions in H1R-overexpressing cells treated with small interfering RNA (siRNA) targeting Nir1 (siNir1) compared with cells treated with a control siRNA (siControl; Figure 4, C and D). Knockdown efficiency and specificity of siRNA targeting Nir1 were validated by performing quantitative RT-PCR on Nir1, Nir2, Nir3, and VAPA (Supplemental Figure 2). These results indicate that endogenous Nir1 promotes Nir2 translocation to ER–PM junctions.
FIGURE 4:
Nir1 promotes Nir2 recruitment to ER–PM junctions and regulates PM PIP2 replenishment. (A) TIRF images of HeLa cells coexpressing Nir2-mCh and YFP vector (top row) or Nir1-YFP (bottom row) treated with 100 μM his (n = 40; four experiments). Scale bars: 20 μm and 5 μm (right). (B) TIRF images of HeLa cells coexpressing Nir2-YFP and Nir1-FFAA-mCh (top row) or Nir1-∆LNS2-mCh (bottom row) treated with 100 μM his (n = 30; three experiments). Scale bars: 20 μm and 5 μm (right). (C) TIRF images of HeLa cells expressing Nir2-mCh treated with siControl (top) or siNir1 (bottom) treated with 100 μM his. Scale bar: 3 μm. (D) Relative change in Nir2-mCh fluorescence intensity in puncta following 100 μM his stimulation monitored by TIRF microscopy in HeLa cells as described in C (mean ± SEM; p < 0.05, two-way ANOVA; n values: siNir1, 18; siControl, 23; four experiments). (E) Relative change in GFP-PLCδ-PH fluorescence intensity following 100 μM his stimulation monitored by TIRF microscopy in HeLa cells overexpressing H1R and treated with siNir1 or siControl (mean ± SEM; p < 0.05, two-way ANOVA; n values: siNir1, 22; siControl, 18; three experiments). (F) Percent recovery of GFP-PLCδ-PH intensity following 100 μM his stimulation in HeLa cells as described in E (mean ± SEM; p < 0.05, t test).
Given Nir1’s potentiation effect on Nir2, we next examined whether Nir1 may be involved in the regulation of the PI cycle. To investigate this possibility, we employed an assay we previously developed to monitor PM PIP2 levels in live cells using TIRF microscopy (Chang et al., 2013; Chang and Liou, 2019). HeLa cells coexpressing a genetically encoded PIP2 biosensor, GFP-PLCδ-PH (Stauffer et al., 1998), and H1R to enhance PIP2 hydrolysis were treated with siNir1 or a control siRNA. Treatment with 100 µM histamine induced a rapid depletion of PIP2 followed by a gradual recovery to a new steady state. Compared to control, siNir1-treated cells displayed reduced PM PIP2 replenishment following histamine stimulation (Figure 4, E and F), indicating that endogenous Nir1 regulates PM PIP2 replenishment in receptor-stimulated cells despite the lack of a PITP domain. Similar results were observed using GFP-Tubby (Supplemental Figure 3), another biosensor that binds PIP2 selectively (Quinn et al., 2008). Together, these results suggest that Nir1 lowers the threshold of activation for Nir2 targeting to ER–PM junctions. While Nir1 lacks the machinery to engage in lipid transfer on its own, it can modulate the activity of Nir2, which is a main driver in PM PIP2 replenishment.
Nir1 interacts with Nir2 via a region between the FFAT motif and the DDHD domain
Nir1’s ability to modulate Nir2’s targeting to ER–PM junctions raised the possibility that Nir1 and Nir2 may be interacting. As a first test, we designed a live-cell imaging assay to assess whether Nir2 coexpression and targeting to ER–PM junctions could recruit Nir1-∆LNS2, a Nir1 mutant lacking PM binding. HeLa cells were cotransfected with either Nir2-YFP or YFP vector, H1R, and Nir1-∆LNS2-mCh, treated with 100 µM histamine and imaged using TIRF microscopy. As expected, Nir1-∆LNS2-mCh displayed no ER–PM junction targeting pre- or poststimulation in YFP vector coexpressing cells (Figure 5A, left panels). However, in cells cotransfected with Nir2-YFP, Nir1-∆LNS2-mCh rapidly translocated to ER–PM junctions following histamine stimulation (Figure 5A, right panels). These results suggest that Nir2 can recruit this PM-binding deficient Nir1 mutant and supports the notion that Nir1 and Nir2 interact.
FIGURE 5:
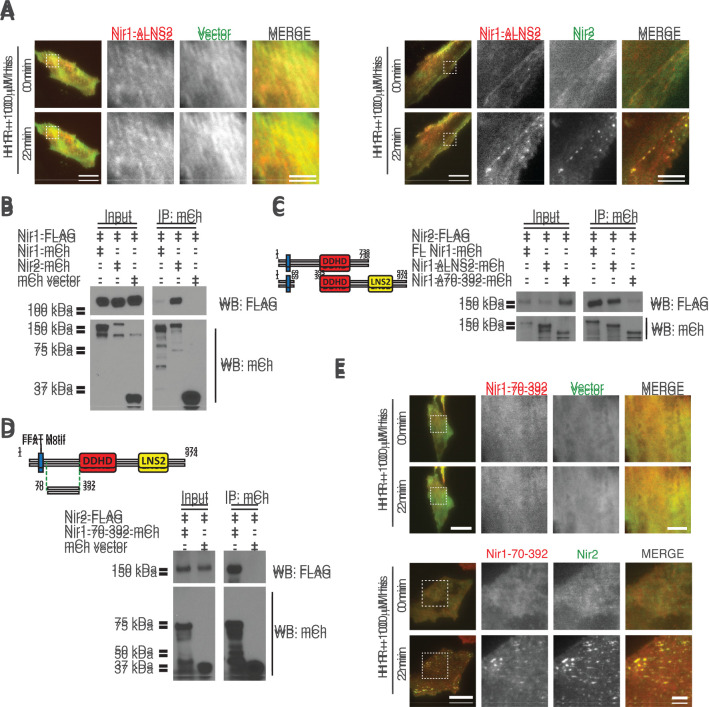
Nir1 interacts with Nir2 via a region between the FFAT motif and the DDHD domain. (A) Translocation of Nir1-∆LNS2-mCh to ER–PM junctions following 100 μM his monitored by TIRF microscopy in HeLa cells cotransfected with YFP vector (left) or Nir2-YFP (right; n = 40; four experiments). Scale bars: 20 μm and 5 μm (magnified images). (B) Coimmunoprecipitation of Nir1-FLAG with Nir1-mCh, Nir2-mCh, or mCherry vector in HeLa cells. (C) Coimmunoprecipitation of Nir2-FLAG with Nir1-mCh, Nir1-∆LNS2-mCh, or Nir1-∆70-392-mCh in HeLa cells. (D) Coimmunoprecipitation of Nir2-FLAG with Nir1-70-392-mCh or mCherry vector in HeLa cells. (E) Translocation of Nir1-70-392-mCh to ER–PM junctions following 100 μM his treatment monitored by TIRF microscopy in HeLa cells transfected with YFP vector (top) or Nir2-YFP (bottom; n = 35; four experiments). Scale bars: 20 μm and 5 μm (magnified images).
To further address the possibility of interaction, we performed coimmunoprecipitation experiments. HeLa cells were transfected with FLAG-tagged Nir1 (Nir1-FLAG), and Nir2-mCh, Nir1-mCh, or mCherry vector, lysed, and immunoprecipitated with RFP-Trap magnetic agarose affinity beads. Nir1-FLAG coimmunoprecipitated with Nir2-mCh, yet only weakly coimmunoprecipitated with Nir1-mCh (Figure 5B). Our results from Figure 5A suggested that the LNS2 domain is not required for interaction, and results from Figure 2 indicated that the FFAT motif serves to associate with VAP proteins. Therefore, we centered our analysis on an uncharacterized region between the FFAT motif and the DDHD domain. In line with results from Figure 5A, Nir2-FLAG coimmunoprecipitated with Nir1-mCh or Nir1-∆LNS2-mCh (Figure 5C, middle lane). However, a Nir1 construct lacking amino acids 70–392 (Nir1-∆70-392-mCh) did not coimmunoprecipitate Nir2-FLAG, suggesting this region is necessary for interaction with Nir2 (Figure 5C, right lane).
Next, to test whether this region is sufficient for interaction with Nir2, we employed a Nir1 construct composed of only amino acids 70–392, named Nir1-70–392-mCh. Indeed, we observed robust coimmunoprecipitation of Nir2-FLAG with Nir1-70–392-mCh (Figure 5D). To examine whether Nir2 could recruit the minimal Nir1-70–392 fragment to ER–PM junctions in live cells, we cotransfected with either Nir2-YFP or YFP vector, H1R, and Nir1-70–392-mCh, and treated HeLa cells with 100 µM histamine. Nir1-70–392-mCh displayed no ER–PM junction localization pre- or poststimulation in cells cotransfected with YFP vector (Figure 5E, top panels), yet rapidly translocated to ER–PM junctions following stimulation in cells cotransfected with Nir2-YFP (Figure 5E, bottom panels). Collectively, these findings identify Nir1 as an interactor of the LTP Nir2. Importantly, our live-cell and biochemical analysis map the interaction interface to a previously uncharacterized region in Nir1.
Overall, our study uncovers Nir1 as a novel ER–PM junction resident protein and regulator of the LTP Nir2. We demonstrated that Nir proteins have conserved targeting mechanisms for their localization at ER–PM junctions. In addition, we showed that the PITP domain provides an autoinhibitory mechanism to limit Nir2 and Nir3 targeting to ER–PM junctions. Moreover, we provide evidence suggesting that Nir1 indirectly impacts PM PIP2 homeostasis by interacting with and modulating the activation threshold of Nir2 targeting to ER–PM junctions. By potentiating Nir2 translocation to ER–PM junctions, Nir1 exerts a positive regulation on the PI cycle. It would be interesting to study Nir1 modulation of Nir2 in cell or tissue types with varying expression ratios. Also, it is enticing to hypothesize that certain physiological contexts that require greater PM PIP2 turnover would benefit from higher levels of Nir1 expression and potentiation of Nir2 activity. In summary, this study expands our understanding of the PI cycle and may help define the pathological mechanisms underlying cancer metastasis and retinal dystrophy associated with Nir1 defects in humans.
MATERIALS AND METHODS
Reagents
All chemicals for extracellular buffer (ECB; 125 mM NaCl, 5 mM KCl, 1.5 mM MgCl2, 20 mM HEPES, 10 mM glucose, and 1.5 mM CaCl2 [pH 7.4]) and histamine were purchased from Sigma. siRNA targeting control and Nir1 were generated as described previously (Liou et al., 2005). Primers used for siRNA generation are listed in Supplemental Table S1.
Cell culture and transfection
HeLa cells purchased from American Type Culture Collection were cultured in minimal essential medium supplemented with 10% fetal bovine serum (FBS; Hyclone) and 1% penicillin/streptomycin solutions at 37°C and 5% CO2. DNA plasmids (25–50 ng/well) and 40 ng siRNAs/well were transfected with Trans-IT-LT1 reagent for 16–20 h and TransIT-TKO reagent for 48–72 h, respectively (Muris Bio). RPE-1, MDA-MB-231, HEK293A, HAP1, and VAP DKO HeLa cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS (Hyclone) and 1% penicillin/streptomycin solutions at 37°C and 5% CO2. VAP DKO HeLa cells were obtained from Pietro De Camilli’s group (Yale University, New Haven, CT).
DNA constructs
Nir1-mCherry was cloned by replacing the Nir2 part of Nir2-mCherry cloned previously (Chang et al., 2013) with PCR fragments retrieved from a human cDNA library containing full-length Nir1. Nir1-YFP was cloned by replacing the mCherry part of Nir1-mCherry with YFP. Nir1-FFAA-mCherry, Nir1-∆LNS2-mCherry, Nir1-∆70-392-mCherry, Nir1-70–392-mCherry, Nir1-594–974-mCherry, Nir1-690–974-mCherry, Nir1-720–974-mCh, and Nir1-739–974-mCherry were generated using site-directed mutagenesis. N2-N1-mCherry and N3-N1-mCherry were generated using Gibson Assembly (New England Bio-Rad) adding the amino acids 1–315 of Nir2 or amino acids 1–310 of Nir3 to the N-terminus of Nir1-mCherry, respectively. Nir2-∆PITP-mCherry was generated previously (Chang et al., 2013). Nir3-∆PITP-mCherry was cloned by replacing the Nir2 part of Nir2-mCherry with PCR fragments retrieved from a human cDNA library containing full-length Nir3 (isoform, AB385472). Nir1-FLAG and Nir2-FLAG were cloned using site-directed mutagenesis to replace the mCherry part with the FLAG tag from Nir1-mCherry and Nir2-mCherry, respectively. YFP-KDEL and YFP-STIM1-D76A were generated previously (Liou et al., 2005). VAPA-YFP, VAPB-YFP, and MAPPER were generated previously (Chang et al., 2013). GFP-PLCδ-PH was obtained from Tobias Meyer’s group (Weill Cornell Medicine, New York, NY). The H1R plasmid was obtained from Elliott Ross’ group (UT Southwestern Medical Center, Dallas, TX). GFP-E-Syt2 was obtained from Pietro De Camilli’s group (Yale University, New Haven, CT). GFP-2X-PASS was obtained from Guangwei Du’s group (UT Health Science Center, Houston, TX). All constructs listed here were verified by Sanger sequencing (Sanger Sequencing Core, McDermott Center, UT Southwestern Medical Center, Dallas, TX). Primers used for cloning are listed in Supplemental Table S1.
Live-cell confocal and TIRF microscopy
HeLa cells were seeded on an eight-well Lab-Tek chambered cover glass (Nunc) at a low density 24 h before transfection. At 16 h posttransfection, cells were washed once with ECB and imaged in ECB. Live-cell confocal and TIRF imaging experiments were performed at room temperature with a CFI Apo 100× objective (1.49 NA) and a custom spinning-disk confocal-TIRF imaging system built around an Eclipse Ti microscope (Nikon) with an EMCCD camera (Hamamatsu Photonics). The microscope was controlled by µManager software (Edelstein et al., 2010). Excitation of mCherry, and YFP or GFP fusion proteins, was achieved using a Sapphire 561 (561 nm; Coherent, Santa Clara, CA) and an LS200 Argon (514 nm/488 nm; Dynamic Laser, Salt Lake City, UT) laser, respectively.
Translocation to ER–PM junctions assay
TIRF microscopy (100×) was used to image single HeLa cells before and after 100 µM histamine stimulation. Intensities of 10–20 stable Nir2 puncta per cell were measured over time, background subtracted, normalized to time zero, and averaged.
PIP 2 replenishment assay
TIRF microscopy (100×) was used to image single HeLa cells. The relative intensity of GFP-PLCδ-PH was monitored and used to assess dynamic changes in PM PIP2 following receptor stimulation in cells overexpressing histamine H1 receptor (H1R). To prevent photobleaching, an ND8 filter was introduced into the light path. GFP-PLCδ-PH intensity was measured over time, background subtracted, and normalized to time zero.
Colocalization analysis
Pearson’s correlation coefficients between two fluorescence images were calculated using the colocalization plugin JACoP (just another colocalization plugin) of the ImageJ software (Bolte and Cordelières, 2006).
Coimmunoprecipitation
HeLa cells seeded in six-well cell culture plates (CellStar, Cat. No. 657 160) were cotransfected with FLAG and mCherry-tagged proteins. Posttransfection (16 h), cells were washed twice with ice-cold phosphate-buffered saline, and then lysed with Pierce IP Lysis Buffer (Prod. #87787) containing 1X protease inhibitor cocktail (Sigma-Aldrich; P8340). Lysates were centrifuged for 15 min 12,000 × g for 15 min at 4°C. Supernatants were incubated overnight at 4°C in RFP-Trap Magnetic Agarose affinity beads (Chromotek product code rtma-20) washed three times with dilution/wash buffer (10 mM Tris HCl, 150 mM NaCl, pH 7.5). Input and immunoprecipitates were boiled for 10 min at 100°C in 10% β-mercaptoethanol laemmli’s sample buffer, run on 7.5% or 10% Mini-PROTEAN TGX stain-free gels (Cat. #4568026 and #4568036) and transferred to polyvinylidene difluoride membranes. Membranes were blocked in 4% milk Tris-buffered saline with 0.1% TWEEN (TBST) for 1 hour at room temperature, then incubated overnight in 1:2000 mouse anti-FLAG (Sigma-Aldrich; F1804) or 1:2000 mouse anti-mCherry (abcam; ab125096) 4% milk TBST at 4°C. Membranes were washed three times in TBST pre- and postincubation in 1:2000 horse anti-mouse HRP-linked antibody (Cell Signaling Technology; #7076) in 4% milk TBST for 1 h at room temperature, then developed on film using Clarity Western ECL Substrate (Bio-Rad; cat. #170-5060).
Quantitative reverse transcription PCR analysis
HeLa cells were treated with siRNA targeting Nir1 or control for 48 h. Total RNA was then extracted using the RNAeasy Protect MiniKit (QIAGEN; cat. #74124). To quantify mRNA expression levels, equal amounts of cDNA were synthesized using the High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific; cat. #4368814). Reactions (10 µl) included cDNA, iTaq Universal SYBR Green Supermix (Bio-Rad; cat. #1725120), and 5 µM of both forward and reverse primers, and were performed at 95°C for 3 min, and then 44 cycles of 95°C for 10 s and 60°C for 30 s. Reactions were set up in triplicate on a C1000 Touch Thermal Cycler (Bio-Rad) with a CFX96 Real-Time PCR Detection System (Bio-Rad) operated by Bio-Rad Maestro software (Bio-Rad). GAPDH was amplified as an internal control and the relative quantitation of Nir1 was performed using the comparative Ct method. Primer sets are listed in Supplemental Table 1.
Statistical analysis
Data were statistically analyzed by t test or two-way analysis of variance with Fisher’s LSD test with a 95% confidence interval using GraphPad Prism version 8.0.0 for Windows (GraphPad Software, San Diego, CA), www.graphpad.com.
Supplementary Material
Acknowledgments
We thank the Liou Laboratory members for valuable discussions. We are grateful to Tobias Meyer for the GFP-PLCδ-PH construct, to Elliott Ross for the H1R plasmid, to Guangwei Du for the GFP-2X-PASS construct, and to Pietro De Camilli for the VAP DKO cell line and the GFP-E-Syt2 construct. This work was supported by National Institutes of Health R01 Grant no. GM-113079, Welch Foundation Grant no. I-1789, National Institutes of Health Cell and Molecular Biology Training Program T32 GM008203, and National Institutes of Health Cancer Biology Training Program T32 CA124334. J.L. is a Sowell Family Scholar in Medical Research.
Abbreviations used:
- CORD5
cone-rod dystrophy-5
- DAG
diacylglycerol
- ER
endoplasmic reticulum
- FFAT
two phenylalanines in an acidic tract
- LNS2
Lipin/Ned1/Smp2
- H1R
histamine 1 receptor
- LTP
lipid transfer protein
- PA
phosphatidic acid
- PI
phosphatidylinositol
- PIP 2
phosphatidylinositol 4,5-bisphosphate
- PITP
phosphatidylinositol transfer protein
- PLC
phospholipase C
- PM
plasma membrane
- TIRF
total internal reflection fluorescence
- VAPA/B
vesicle-associated membrane protein-associated protein isoforms A and B.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-07-0356) on January 12, 2022.
REFERENCES
- Agranoff BW, Bradley RM, Brady RO (1958). The enzymatic synthesis of inositol phosphatide. J Biol Chem 233, 1077–1083. [PubMed] [Google Scholar]
- Amarilio R, Ramachandran S, Sabanay H, Lev S (2005). Differential regulation of endoplasmic reticulum structure through VAP-Nir protein interaction. J Biol Chem 280, 5934–5944. [DOI] [PubMed] [Google Scholar]
- Bakhoum MF, Sengillo JD, Cui X, Tsang SH (2018). Autoimmune ret-inopathy in a patient with a missense mutation in PITPNM3. Retin Cases Brief Rep 12 (Suppl 1), S72–S75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T (2013). Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev 93, 1019–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins JA, Agranoff BW (1969). Distribution and properties of CDP-diglyceride:inositol transferase from brain. J Neurochem 16, 513–527. [DOI] [PubMed] [Google Scholar]
- Berridge MJ (1984). Inositol trisphosphate and diacylglycerol as second messengers. Biochem J 220, 345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohdanowicz M, Schlam D, Hermansson M, Rizzuti D, Fairn GD, Ueyama T, Somerharju P, Du G, Grinstein S (2013). Phosphatidic acid is required for the constitutive ruffling and macropinocytosis of phagocytes. Mol Biol Cell 24, 1700–1712, S1701–S1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP (2006). A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224, 213–232. [DOI] [PubMed] [Google Scholar]
- Chang CL, Chen YJ, Liou J (2017). ER-plasma membrane junctions: why and how do we study them? Biochim Biophys Acta Mol Cell Res 1864, 1494–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CL, Hsieh TS, Yang TT, Rothberg KG, Azizoglu DB, Volk E, Liao JC, Liou J (2013). Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep 5, 813–825. [DOI] [PubMed] [Google Scholar]
- Chang CL, Liou J (2015). Phosphatidylinositol 4,5-bisphosphate homeostasis regulated by Nir2 and Nir3 at endoplasmic reticulum-plasma membrane junctions. J Biol Chem 290, 14289–14301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CL, Liou J (2019). Analysis of phosphatidylinositol transfer at ER-PM junctions in receptor-stimulated live cells. Methods Mol Biol 1949, 1–11. [DOI] [PubMed] [Google Scholar]
- Chen J, Yao Y, Gong C, Yu F, Su S, Chen J, Liu B, Deng H, Wang F, Lin L, et al. (2011). CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell 19, 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft S (2012). The diverse functions of phosphatidylinositol transfer proteins. Curr Top Microbiol Immunol 362, 185–208. [DOI] [PubMed] [Google Scholar]
- Dong R, Saheki Y, Swarup S, Lucast L, Harper JW, De Camilli P (2016). Endosome-ER contacts control actin nucleation and retromer function through VAP-dependent regulation of PI4P. Cell 166, 408–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N (2010). Computer control of microscopes using µManager. Curr Protoc Mol Biol Chapter 14, Unit14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elagin VA, Elagina RB, Doro CJ, Vihtelic TS, Hyde DR (2000). Cloning and tissue localization of a novel zebrafish RdgB homolog that lacks a phospholipid transfer domain. Vis Neurosci 17, 303–311. [DOI] [PubMed] [Google Scholar]
- Ferrell J, Jr, Huestis WH (1984). Phosphoinositide metabolism and the morphology of human erythrocytes. J Cell Biol 98, 1992–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT (2017). The PI3K pathway in human disease. Cell 170, 605–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P (2013). PI(4,5)P(2)-dependent and Ca2+-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 153, 1494–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokin MR, Hokin LE (1964). Interconversions of phosphatidylinositol and phosphatidic acid involved in the responses to acetylcholine in the salt gland. In: Metabolism and Physiological Significance of Lipids, New York: Wiley, 423–434. [Google Scholar]
- Kim S, Kedan A, Marom M, Gavert N, Keinan O, Selitrennik M, Laufman O, Lev S (2013). The phosphatidylinositol-transfer protein Nir2 binds phosphatidic acid and positively regulates phosphoinositide signalling. EMBO Rep 14, 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Guzman-Hernandez ML, Wisniewski E, Balla T (2015). Phosphatidylinositol-phosphatidic acid exchange by Nir2 at ER-PM contact sites maintains phosphoinositide signaling competence. Dev Cell 33, 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn L, Kadzhaev K, Burstedt MS, Haraldsson S, Hallberg B, Sandgren O, Golovleva I (2007). Mutation in the PYK2-binding domain of PITPNM3 causes autosomal dominant cone dystrophy (CORD5) in two Swedish families. Eur J Hum Genet 15, 664–671. [DOI] [PubMed] [Google Scholar]
- Kohn L, Kohl S, Bowne SJ, Sullivan LS, Kellner U, Daiger SP, Sandgren O, Golovleva I (2010). PITPNM3 is an uncommon cause of cone and cone-rod dystrophies. Ophthalmic Genet 31, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S (2012). Nonvesicular lipid transfer from the endoplasmic reticulum. Cold Spring Harb Perspect Biol 4, a013300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Li W, Zhang H, Wu W, Peng Y, Zeng Y, Wan Y, Wang J, Ouyang N (2016). CCL18/PITPNM3 enhances migration, invasion, and EMT through the NF-kappaB signaling pathway in hepatocellular carcinoma. Tumour Biol 37, 3461–3468. [DOI] [PubMed] [Google Scholar]
- Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE Jr, Meyer T (2005). STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol 15, 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen CJ, Roy A, Levine TP (2003). A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J 22, 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KT, Ferrell JE, Jr, Huestis WH (1986). Separation of phosphoinositides and other phospholipids by two-dimensional thin-layer chromatography. Anal Biochem 158, 447–453. [DOI] [PubMed] [Google Scholar]
- Panula P, Chazot PL, Cowart M, Gutzmer R, Leurs R, Liu WL, Stark H, Thurmond RL, Haas HL (2015). International union of basic and clinical pharmacology. XCVIII. Histamine receptors. Pharmacol Rev 67, 601–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn KV, Behe P, Tinker A (2008). Monitoring changes in membrane phosphatidylinositol 4,5-bisphosphate in living cells using a domain from the transcription factor tubby. J Physiol 586, 2855–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer TP, Ahn S, Meyer T (1998). Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol 8, 343–346. [DOI] [PubMed] [Google Scholar]
- Tanguy E, Wang Q, Moine H, Vitale N (2019). Phosphatidic acid: from pleiotropic functions to neuronal pathology. Front Cell Neurosci 13, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A, Prinz WA (2011). Lipid transfer and signaling at organelle contact sites: the tip of the iceberg. Curr Opin Cell Biol 23, 458–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker DR (1990). Lipid transport pathways in mammalian cells. Experientia 46, 569–579. [DOI] [PubMed] [Google Scholar]
- Wang X, Devaiah SP, Zhang W, Welti R (2006). Signaling functions of phosphatidic acid. Prog Lipid Res 45, 250–278. [DOI] [PubMed] [Google Scholar]
- Yadav S, Garner K, Georgiev P, Li M, Gomez-Espinosa E, Panda A, Mathre S, Okkenhaug H, Cockcroft S, Raghu P (2015). RDGBα, a PtdIns-PtdOH transfer protein, regulates G-protein-coupled PtdIns(4,5)P2 signalling during Drosophila phototransduction. J Cell Sci 128, 3330–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang Z, Lu M, Yonekubo Y, Liang X, Zhang Y, Wu P, Zhou Y, Grinstein S, Hancock JF, Du G (2014). Temporal production of the signaling lipid phosphatidic acid by phospholipase D2 determines the output of extracellular signal-regulated kinase signaling in cancer cells. Mol Cell Biol 34, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.