Abstract
J-domain protein cochaperones drive much of the functional diversity of Hsp70-based chaperone systems. Sis1 is the only essential J-domain protein of the cytosol/nucleus of Saccharomyces cerevisiae. Why it is required for cell growth is not understood, nor how critical its role is in regulation of heat shock transcription factor 1 (Hsf1). We report that single-residue substitutions in Tti1, a component of the heterotrimeric TTT complex, a specialized chaperone system for phosphatidylinositol 3-kinase-related kinase (PIKK) proteins, allow growth of cells lacking Sis1. Upon depletion of Sis1, cells become hypersensitive to rapamycin, a specific inhibitor of TORC1 kinase. In addition, levels of the three essential PIKKs (Mec1, Tra1, and Tor2), as well as Tor1, decrease upon Sis1 depletion. Overexpression of Tti1 allows growth without an increase in the other subunits of the TTT complex, Tel2 and Tti2, suggesting that it can function independent of the complex. Cells lacking Sis1, with viability supported by Tti1 suppressor, substantially up-regulate some, but not all, heat shock elements activated by Hsf1. Together, our results suggest that Sis1 is required as a cochaperone of Hsp70 for the folding/maintenance of PIKKs, making Sis1 an essential gene, and its requirement for Hsf1 regulation is more nuanced than generally appreciated.
INTRODUCTION
Hsp70-based molecular chaperone machineries function in a wide range of cellular processes. They play critical roles in protein homeostasis, including facilitating folding of nascent polypeptide chains and promoting maintenance of protein structure upon stress (Balchin et al., 2020). J-domain protein cochaperones are responsible for much of this functional versatility (Kampinga and Craig, 2010; Rosenzweig et al., 2019). Their J-domains bind partner Hsp70s, driving hydrolysis of bound ATP, and thereby triggering conformational changes that lead to stabilization of Hsp70-substrate interactions.
Most cellular compartments have multiple J-domain proteins that partner with the same Hsp70. Some show no sequence similarity outside the J-domain, while others have substantial structural similarity, particularly in domains that bind substrate (Craig and Marszalek, 2017). In the budding yeast Saccharomyces cerevisiae, 12 different J-domain proteins function with the Ssa type of Hsp70 in the cytosol/nucleus. Of these, four have a double ß-barrel substrate–binding domain that follows an N-terminal J-domain and the adjacent glycine-rich region, which is often called the G/F region. Sis1 is unique; it is essential even under optimal growth conditions (Luke et al., 1991). For its essential role(s), Sis1 acts as an Hsp70 cochaperone, as substitutions that disrupt J-domain function render Sis1 nonfunctional (Yan and Craig, 1999). None of the other J-domain proteins of the cytosol or nucleus can substitute for Sis1 to rescue viability, even the more abundant double β-barrel Ydj1, which partners with the same Hsp70 (Sahi and Craig, 2007). However, Sis1 homologues from other species, including human DnaJB1, can rescue the viability of sis1∆ cells (Lopez et al., 2003), indicating functional conservation in the evolution of this class of double β-barrel J-domain proteins.
Neither the specific critical biological processes in which involvement of Sis1 is required, nor the substrate proteins that need its action for cell viability, have been identified. However, many cellular functions of Sis1 are known. Some, such as facilitating transport of nascent polypeptides for translocation across the endoplasmic reticulum and mitochondrial membranes, overlap with those of Ydj1 (Jores et al., 2018; Cho et al., 2021). Some, such as targeting certain proteins for degradation (Shiber et al., 2013; Summers et al., 2013; Prasad et al., 2018) and maintaining prions (Sondheimer et al., 2001), are unique to Sis1, but are not essential. Sis1 has also been identified as a regulator of heat shock transcription factor Hsf1 (Klaips et al., 2020; Feder et al., 2021), which drives expression from promoters having a heat shock element (HSE). It is well established that Hsf1 activity is down-regulated by Hsp70 binding (Masser et al., 2020; Pincus, 2020), but how critical and unique the role of Sis1 is as the J-domain protein cochaperone in this regulation by driving the Hsp70-Hsf1 interaction remains unresolved.
To better understand Sis1 functionality, we undertook a genetic approach, isolating mutations that allowed cells to grow in the absence of Sis1. We previously reported that substitutions in Ydj1 or Hsp70 Ssa1 can overcome the requirement for Sis1 (Schilke et al., 2017). While analysis of these Ydj1 and Ssa1 suppressor variants is informative regarding how tuning of the initial step of the Hsp70–substrate interaction cycle can diversify Hsp70 system function, they do not provide insight into the Hsp70 substrates that normally specifically require Sis1 cochaperone function for cell viability.
RESULTS AND DISCUSSION
Tti1 variants allow growth of cells having a deletion of SIS1
With the goal of gaining insight into the cellular processes that require Sis1 function for viability, we continued our selection for genomic mutations that permit cell growth in the absence of Sis1. Cells having a chromosomal deletion of the SIS1 gene (sis1Δ) and expressing Sis1 from a plasmid containing the URA3 gene were plated on media containing 5-fluoroorotic acid (5-FOA). Because 5-FOA is toxic to cells expressing Ura3, those that have lost the plasmid but obtained chromosomal mutations that allow growth in the absence of Sis1 can form colonies. To eliminate from consideration suppressors in the YDJ1 gene, the site of the initially identified suppressors (Schilke et al., 2017), we focused on two isolates whose suppressor mutations segregated independently from SIS1 (Figure 1A; Supplemental Figure 1A), as SIS1 and YDJ1 are genetically linked. These two new suppressor alleles were genetically linked to each other. Sequencing revealed that both had point mutations encoding single-residue changes in the TTI1 gene, albeit at different positions—T598R in suppressor #1, G858V in suppressor #2—called tti1sup#1 and tti2sup#2 throughout. Much of Tti1, which contains 1038 residues, is composed of α-helical repeats of the common Armadillo superfamily (Figure 1B). Both suppressor substitutions lie within this segment, which extends from residue 76 to 1016.
FIGURE 1:
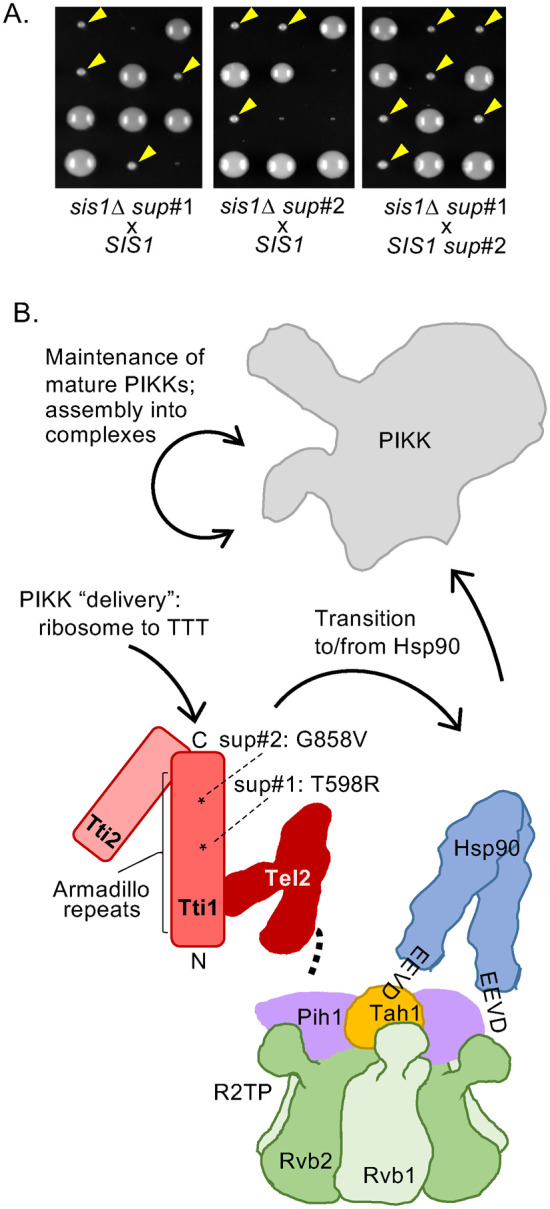
Isolation of spontaneous TTI1 suppressors of cells lacking Sis1. (A) sis1∆ (sis1::LEU2) cells carrying putative suppressor mutations were crossed with a WT SIS1 strain and sporulated and resulting asci dissected onto rich medium. Plates were incubated at 30°C for 4 d (left, suppressor #1 [sup#1]; center, suppressor #2 [sup#2]) and then replica plated to leucine omission media. Colonies that grew (suppressors) are indicated by yellow arrowheads. Right, an sis1∆ sup#1 and a SIS1 sup#2 strain of opposite mating type were crossed and treated as above. All resulting sis1::LEU2 deletion haploids were suppressed for lethality, indicating close linkage of sup#1 and sup#2. (B) Cartoon of TTT complex and possible points of action of Sis1 in PIKK proteostasis (solid arrows). TTT subunits in shades of red, indicating interaction of Tti1 with Tel2 and Tti2. Expanse of α-helical Armadillo Tti1 repeats indicated by bracket; asterisks indicate position of suppressors—sup#1 T598R; sup#2 G858V. Both TTT and Hsp90 have been implicated in folding of PIKKs in coordination with Rvb1/2 in complex with Pih1 and Tah1 as the R2TP complex (or with accessory protein Asa1, not shown). Tel2 interacts physically with the Pih1 subunit (heavy dotted line) and Hsp90 with the Tah1 subunit (via C-terminal EEVD).
Tti1 is the largest subunit of the heterotrimeric TTT chaperone complex (Hurov et al., 2010), a specialized chaperone system dedicated to facilitating folding and/or maintenance of the phosphatidylinositol 3-kinase-related kinase (PIKK) proteins (Takai et al., 2007, 2010; Sugimoto, 2018; Elias-Villalobos et al., 2019). PIKKs play important roles in diverse cellular processes in eukaryotes, from response to nutritional stress to regulation of cell growth and transcriptional regulation (Villa et al., 2016; Gonzalez and Hall, 2017; Cheung and Diaz-Santin, 2019; Lustig, 2019). We decided to investigate the relationship between Sis1 and the TTT complex, as we considered PIKKs as plausible essential substrates of the Sis1–Ssa1 Hsp70 chaperone system. Even though Sis1 is present at ∼30,000 molecules/cell (Ho et al., 2018), only a small fraction of that is needed to maintain robust cell growth (Aron et al., 2007), consistent with a critical chaperone role for relatively low abundance substrates such as PIKKs. In addition, PIKKs are among the largest proteins in the cell. The 5 PIKKs of S. cerevisiae—Tel1, Tor1, Tor2, Mec1, and Tra1—range in size from 233 to 431 kDa, each containing a characteristic kinase, FAT, and HEAT domain architecture common to all PIKKs (Imseng et al., 2018). Tor2, Mec1, and Tra1 are essential proteins, as are all three TTT subunits—Tti1, Tti2, and Tel2.
That PIKK proteostasis is an elaborate process is underscored by the fact that the TTT complex coordinates with a more elaborate chaperone system, the R2TP complex, which interacts not only with several other specialized chaperone systems, but also with Hsp90 (Figure 1B; Houry et al., 2018). It has been suggested to be important at initial stages of PIKK folding, as well as in the maintenance of active, functional PIKKs and the assembly of multimeric complexes in which they function.
Phosphatidylinositol 3-kinase-related kinase protein levels decrease as Sis1 is depleted
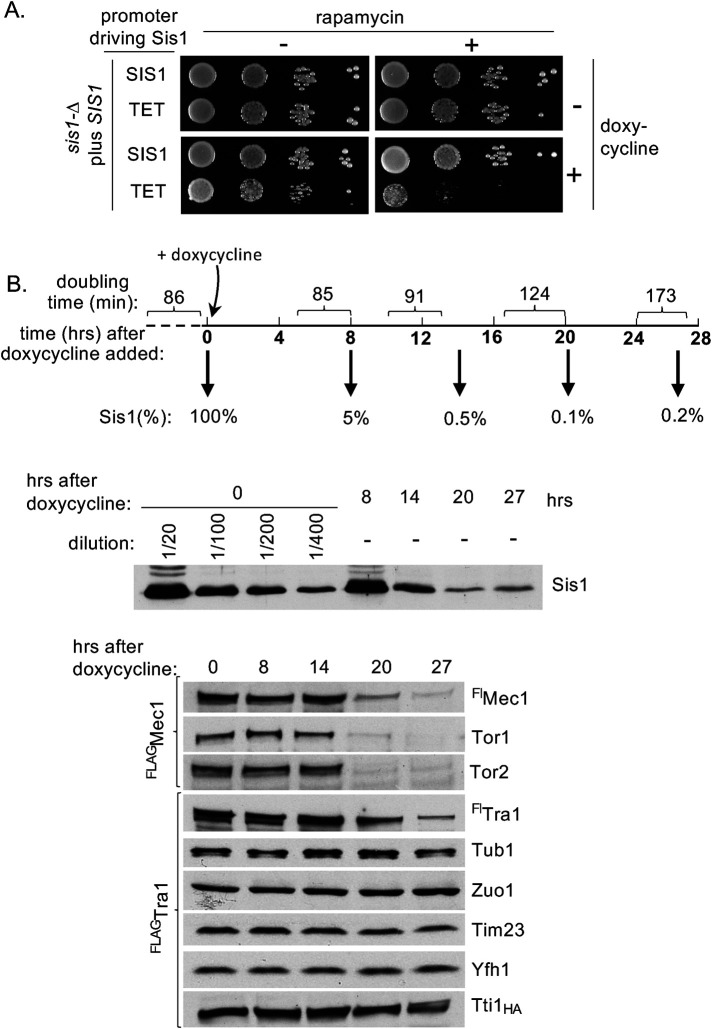
To test for additional connections between Sis1 and PIKKs, we took advantage of the fact that rapamycin is a specific inhibitor of the TORC1 kinase complex, which includes either PIKK Tor1 or Tor2 as the catalytic subunit (Martin and Hall, 2005). We tested the effect of Sis1 depletion on cell sensitivity to rapamycin using a plate assay. We chose a borderline concentration of rapamycin—one that had no obvious effect on the growth of cells expressing normal amounts of Sis1—whether driven from the native SIS1 or from the doxycycline-repressible TET promoter (Figure 2A), which result in similar expression of Sis1 (Supplemental Figure 1B). As expected, because of the small amount of Sis1 required for normal growth (Aron et al., 2007), cells expressing Sis1 from the TET promoter grew only slightly more slowly than those expressing normal levels of Sis1 when plates contained doxycycline. However, growth was very poor on plates containing both doxycycline and rapamycin. This more severe effect of rapamycin on growth of cells having reduced expression of Sis1 is consistent with the idea that cells expressing low levels of Sis1 have compromised TORC1 function.
FIGURE 2:
Reduced Sis1 levels result in rapamycin hypersensitivity and reduction in PIKK levels. (A) Tenfold serial dilutions of sis1∆ cells expressing Sis1 from either the native promoter (SIS1) or doxycycline repressible Tet promoter (TET) were spotted on rich media with no addition (–), 1.5 nM rapamycin, and/or 10 µg/ml doxycycline and incubated for 2 d at 30°C. (B) Doxycycline was added at time zero to log phase cultures of BY4741 sis1∆ having Sis1 expressed from the TET promoter. Cells were maintained in log phase over the 27-h time course by dilution into prewarmed medium. (Top) Doubling times were determined by measuring OD600 at 3–4 h intervals, indicated by brackets on timeline. Samples were removed for lysate preparation at times indicated in bold after doxycycline addition. (Bottom) Lysates were subjected to electrophoresis and immunoblot analysis using antibodies specific for the proteins indicated on right. Cells had in the chromosome either Flag-tagged Mec1 (FlagMec1) or Tra1 (FlagTra1), as indicated on left, as well as HA-tagged Tti1 (Tti1HA). Growth rate of FlagTra1 strains is shown (see Supplemental Figure 1C for complete growth data). In the case of Sis1, indicated dilutions of extract were made to estimate relative amounts remaining after repression.
This increased rapamycin sensitivity prompted us to assess amounts of four PIKKs directly (Tor1, Tor2, Mec1, and Tra1) in cells during depletion of Sis1. We used two strains, both having Sis1 driven by the TET promoter, but one having FLAG-tagged TRA1 and the other having FLAG-tagged MEC1 in the chromosome. Samples were removed from liquid cultures over time to measure growth rate. Not surprisingly, cells expressing Sis1 from the TET promoter continued to grow at the same rate for many hours after addition of doxycycline before slowing. It was not until the 17–20 and 24–27 h intervals that doubling times increased—about a 50% and 100% increase, respectively (Supplemental Figure 1C). Samples for protein analysis were taken at 0, 8, 14, 20, and 27 h after doxycycline addition. The Sis1 levels dropped to less than 10% at 8 h and less than 1% by 14 h (Figure 2B). The amounts of control proteins (e.g., tubulin, the ribosomal-associated chaperone Zuo1, and the mitochondrial proteins Tim23 and Yfh1) remained static. However, the levels of PIKK proteins, though similar in the first samples, were substantially reduced by 20 h (Figure 2B; Supplemental Figure 1D).
These results showing reduction in Tor1, Tor2, Mec1, and Tra1 levels upon depletion of Sis1 are consistent with Sis1 playing a role in PIKK proteostasis. A reduction in PIKKs has been reported when the levels of individual TTT subunits were reduced in both fungal and mammalian cells (Takai et al., 2007; Genereaux et al., 2012; Hoffman et al., 2016; Goto et al., 2017). However, due to intrinsic difficulties in studying PIKKs, it is not known at what stage(s) of protein biogenesis/homeostasis the TTT complex functions. Initial steps of protein folding and maintenance of “mature” PIKKs, as well as roles in assembly of multimeric complexes of which they are a part, have been suggested (von Morgen et al., 2015; Sugimoto, 2018). Sis1 could plausibly function in any of these roles as well. However, the ability of both Mec1 and Tra1 to be immunoprecipitated by Sis1-specific antibodies (Supplementary Figure 1E) suggests that Sis1 participates directly in PIKK homeostasis.
Overexpression of Tti1 permits growth of sis1∆
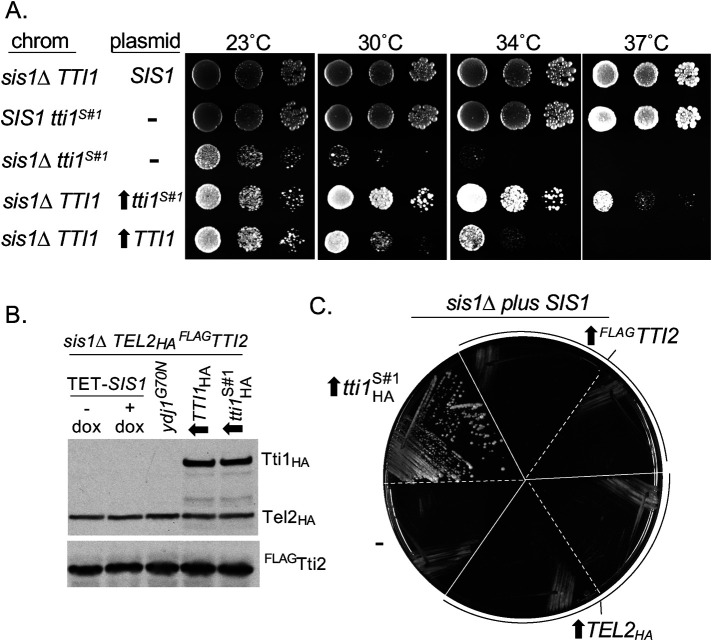
Extending our analysis of the involvement of TTT complex subunits in Sis1 function, we first asked if overexpression of Tti1 affects the degree of suppression. Both WT TTI1 and tti1sup#1 were placed under the control of the strong ADH1 promoter. ADH1-driven tti1sup#1 allowed better growth of sis1∆ cells than the rather poor growth enabled by expression from the native promoter—colony formation occurred even at 37°C. WT TTI1 driven by ADH1 also allowed growth, though not as robustly as tti1sup#1 (Figure 3A; Supplemental Figure 2, A and B). Analysis, after addition of an HA tag to allow detection, revealed that placement under the ADH1 promoter resulted in about a sixfold increase in both Tti1 and tti1sup#1 (Supplemental Figure 2C). To test whether suppression by tti1sup#1 was recessive or dominant, the growth of homozygous and heterozygous diploids was evaluated. TTI1/tti1sup#1 cells grew nearly as well as tti1sup#1/tti1sup#1 cells, particularly at lower temperatures, indicating that tti1sup#1 is substantially dominant (Supplemental Figure 2D).
FIGURE 3:
Overexpression of Tti1, but not Tel2 or Tti2, allows growth of sis1∆ cells. (A) Tenfold serial dilutions of cells having indicated TTI1 and SIS1 on the chromosome (chrom) or expressed from a plasmid were plated on rich media. Overexpression from the ADH1 promoter indicated by an upward arrow. Plates were incubated for 3 d at indicated temperatures, except 23°C plates for 5 d. (B) Lysates of sis1∆ cells having HA-tagged TEL2 and Flag-tagged TTI2 in the chromosome and indicated proteins expressed from centromeric plasmids were subjected to electrophoresis and immunoblot analysis using antibodies against HA and Flag tags. Sis1 under control of TET promoter (TET-SIS1) with doxycycline added 21 h before harvest (+) or, as a control, no addition (–); suppressor Ydj1G70N from its native promoter; HA-tagged TTI1 or tti1sup#1 (tti1S#1) from the strong ADH1 promoter (upward arrow). (C) Transformants of sis1∆ strain carrying SIS1 on a URA3-based plasmid expressing the indicated protein or vector control (–) were streaked onto plates containing 5-FOA and incubated for 4 d at 30°C.
Because Tti1 functions in the heterotrimeric TTT complex, we decided to assess levels of the two other subunits, Tel2 and Tti2. We constructed sis1∆ strains expressing HA-tagged Tel2 and Flag-tagged Tti2 from the chromosome—kept viable by the presence of a plasmid carrying either TET-SIS1, a previously isolated ydj1 suppressor mutant (Schilke et al., 2017), or either TTI1 or tti1sup#1 driven by the ADH1 promoter. Levels of Tel2 and Tti2 were indistinguishable in these four strains, as well as after repression of Sis1 synthesis (Figure 3B). Because overexpression of WT Tti1 allows growth of sis1∆ cells, we tested whether overexpression of Tel2 or Tti2 can also rescue growth. TEL2 and TTI2 were placed under the control of the ADH1 promoter. While, as expected, sis1∆ cells lacking the URA3-based plasmid carrying the SIS1 gene were recovered after plating on 5-FOA when tti1sup#1was expressed, this was not the case for Tel2 or Tti2, even though they both were overexpressed more than Tti1 (Figure 3C, Supplemental Figure 2E).
The results above suggest that Tti1 can act independently from the other components of the TTT complex to enable growth of cells in the absence of Sis1. This is somewhat surprising, as there is little evidence supporting the idea that an individual TTT subunit, separate from the complex, is functional. Reduction of one of the subunits in human cells has been reported to result in reduction of the others (Hurov et al., 2010). The ability of increased expression of Tti1 alone to suppress sis1∆ was also unexpected because it has not been directly implicated in PIKK binding. Rather, the Tel2 subunit had been shown to interact with a PIKK in vitro (Takai et al., 2010). However, recently, in vitro interaction between the Tti1–Tti2 dimer and a PIKK has been reported (Pal et al., 2021), leaving open the possibility that Tti1 can interact directly with PIKKs.
Hsf1-dependent expression is variable in sis1∆ cells
Sis1 has been implicated as a key regulator of heat shock transcription factor Hsf1 (Klaips et al., 2020; Alford et al., 2021; Feder et al., 2021). We took advantage of our ability to manipulate the amount of Sis1 in cells to probe the extent of its role. Using a set of antibodies specific for heat shock proteins (Hsps), we tested extracts from cells after shutoff of Sis1 expression, cells that lacked Sis1 but expressed Tti1sup#1, and, as a control, ssa1∆ ssa2∆ cells. ssa1∆ ssa2∆ lacks the two constitutively expressed Ssa Hsp70 genes, causing Hsf1 to be constitutively activated (Boorstein and Craig, 1990), but retains heat-inducible SSA3 and SSA4. Except for Hsp42, increased amounts of Hsps were observed upon Sis1 depletion. These amounts were further enhanced in the absence of Sis1, with Hsp26 having the most dramatic increase (Figure 4A). However, in no case did levels approach those found in ssa1∆ ssa2∆ cells (Figure 4B).
FIGURE 4:
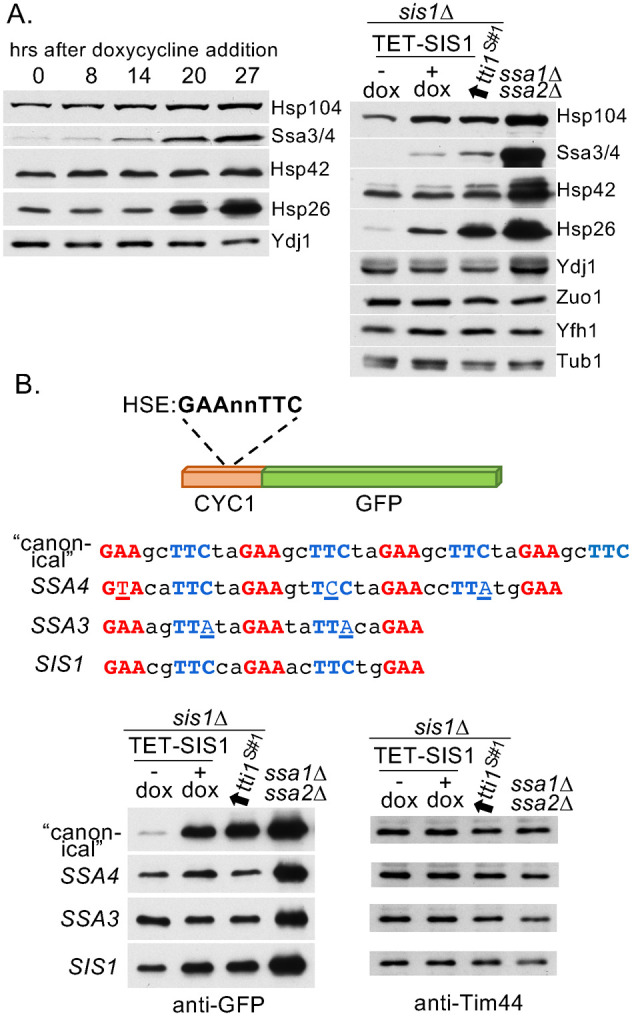
Absence of Sis1 results in increased Hsf1 activity from some, but not all, HSEs. (A) Immunoblot analysis using antibodies specific for indicated Hsps indicated on right. (Left) Extracts analyzed in Figure 2B taken at indicated times after addition of doxycycline to shut off Sis1 expression. (Right) Extracts from sis1∆ cells carrying a plasmid with SIS1 under the TET promoter in the absence (–) or presence (+) of doxycycline for 21 h or having tti1sup#1 (tti1S#1) under control of ADH1 promoter. ssa1∆ ssa2∆ extracts included as a control for constitutive high activation of Hsf1. (B) Extracts from cells carrying a plasmid having the CYC1 promoter–Green fluorescent protein (GFP) fusion with indicated HSEs, and ssa1∆ ssa2∆ cells as a control were subjected to immunoblot analysis with antibodies to GFP and, as a loading control, Tim44.
Hsp gene promoters often have multiple HSEs with a varying number of inverted repeats of nGAAn, some with gaps or imperfect repeats (Yamamoto et al., 2005), as well as other stress-responsive elements (Ruis and Schuller, 1995). To help clarify the picture of Hsf1 activation linked to Sis1, we tested the response of isolated HSEs to the reduction or absence of Sis1. We used a fusion between green fluorescent protein (GFP) and a CYC1 promoter whose activation sequence was replaced by one of 4 HSEs that had been studied previously—Ssa3 (Liu et al., 1997), Ssa4 (Young and Craig, 1993), Sis1 (Zhong et al., 1996), or a “canonical” HSE with four “perfect” nGAAnnTTCn repeats (Brandman et al., 2012). The canonical HSE showed the most dramatic difference in GFP levels between normal and low/no Sis1 levels, with the amount in the latter approaching that found in ssa1∆ ssa2∆ cells (Figure 4B). Ssa3 and Ssa4 HSE activity changed little upon varying Sis1 levels.
The results above present a complex picture of the relationship between Sis1 and cellular stress responses. It is difficult to parse what effects of reduced/absent Sis1 levels are due to the reduced function of PIKKs, some of which are involved directly or indirectly in broad cellular stress responses, and what are due to Sis1’s direct involvement in Hsf1 activation. On one hand, the increase in expression driven by a canonical HSE having four contiguous nGGAnnTTCn repeats is consistent with the proposed major role of Sis1 in repressing Hsf1 activity by targeting Hsp70 to it. On the other, the differences in expression driven by the naturally occurring HSEs are much less dramatic and indicate the adaptability and diversity of the Hsf1 regulatory network, pointing to the importance of Sis1 in PIKK protein homeostasis. In addition, these results also lend a cautionary note—exclusive dependence on the canonical HSE reporter may fail to give a complete picture of the complexities found in the cell. In this regard, it is well established that Hsf1 responds in diverse ways to different HSEs (Verghese et al., 2012) and few natural HSEs have four contiguous repeats. However, the “code” to the complex regulation is not well understood. In the future, such understanding will help to clarify the underlying complexity of cellular responses to stress.
SUMMARY
The results presented here establish that Tti1 can partially substitute for the essential function(s) of Sis1. That levels of PIKK proteins, the known substrates of the TTT complex, decrease upon Sis1 depletion, such as occurs upon inactivation of components of the TTT complex, points to a role in PIKK protein homeostasis. However, when in the lifetime of PIKKs Sis1 is important remains unclear. Indeed, the role of the TTT complex itself, which is part of a much larger, intricate machinery—as one of the handful of “adaptors” for the R2TP AAA+ complex that cooperates with Hsp90 in generation of a number of multimeric complexes (Figure 1B)—is unclear as well. It is also possible that Sis1 has a role in the folding of Tti1 itself, even though PIKKs decrease much more upon Sis1 depletion than Tti1. Considerably more work will be needed to understand the role of Sis1, Tti1, and the entire machinery in PIKK homeostasis, as well as the uniqueness of Sis1’s ability to regulate Hsf1.
MATERIALS AND METHODS
Yeast strains, plasmids, and culture manipulations
Yeast strains and plasmids used are listed in Supplemental Tables 1 and 2, respectively. Cells were grown in a rich medium, YPD (1% yeast extract, 2% peptone (Difco Laboratories, Detroit, MI), 2% dextrose), or in selective minimal media from which particular amino acids could be omitted (0.67% yeast nitrogen base without amino acids [US Biological, Marblehead, MA], 2% dextrose), supplemented with required amino acids (Sherman et al., 1986). W303 strains were sporulated on potassium acetate plates; BY4743 strains were sporulated in potassium acetate liquid media (doi:10.1101/pdb.rec090613, Cold Spring Harb Protoc 2016, 2016 Cold Spring Harbor Laboratory Press) after growth in presporulation liquid medium (1% YPA; Elrod et al., 2009). Strains used were of the W303 genetic background unless stated otherwise in the figure legend.
Genomic DNA was isolated using the MasterPure yeast DNA purification kit from Lucigen (Madison, WI). Yeast was transformed using a previously developed protocol (Chen et al., 1992). To analyze cell growth, 10-fold serial dilutions of cells were spotted onto YPD or selective minimal media and grown for the numbers of days and temperatures indicated in figure legends. Representative examples are shown for both serial dilution and plating on 5-FOA (Toronto Research Chemicals, Canada), with all experiments repeated a minimum of three times. Doxycycline (Sigma-Aldrich, St. Louis, MO) was added to media at 10 µg/ml. Independent transformants were used in the liquid-media Sis1 “shut-off” experiments and for the testing of HSE activity, with representative immunoblots shown.
Selection of suppressors
Spontaneous suppressors allowing growth of strains lacking Sis1 were obtained by streaking transformants of W303 sis1∆ cells carrying SIS1 on the URA3 plasmid YCp50 (WY26) with the TRP1 vector pRS314 from a Trp omission plate to a plate containing 5-FOA (Sikorski and Boeke, 1991), as previously described for the initial isolation of the suppressor having a mutation in YDJ1 (Schilke et al., 2017). After incubation for 3 d at 30°C, followed by 3 d at room temperature, colonies that developed were tested for the presence of Sis1 by immunoblot analysis of lysates using anti-Sis1 antibodies. Two that did not express detectable levels of Sis1 (i.e., suppressors) were backcrossed to a WT haploid and sporulated. Resulting asci were dissected to determine the tetrad segregation pattern of the suppressor mutation(s) with the sis1Δ::LEU2 allele. Both suppressors segregated independently from sis1∆::LEU2. Linkage of these two suppressors to each other was assessed by crossing haploids from this first cross—sis1∆ sup#1 to SIS1 sup#2. The resulting diploid was sporulated and resulting asci dissected.
Identification of the mutation allowing suppression of sis1∆ was performed using pooled linkage analysis and whole-genome sequencing (Birkeland et al., 2010) with modifications as described in MacDiarmid et al. (2013). After the original mutant strain (containing sup#1) was backcrossed twice to the isogenic WT strain, 24 haploids possessing the WT genotype and 24 haploids possessing the mutant genotype were grown up to saturation, and equivalent amounts of cells of each genotype were placed together into two pools before the purification of genomic DNA. The two pools of DNA were submitted to the UW Biotechnology Center for whole-genome sequencing on an Illumina HiSeq2500 using 2 × 250-bp reads. The genomic DNA from the mutant pool contained a single missense mutation in TTI1. After confirmation that sup#2 was linked to sup#1, the TTI11 gene from sup#2 was isolated, as described below, and sequenced. Sup#1 encoded a change in codon 598, T to R (ACA to AGA); sup#2 encoded a change in codon 858, G to V (GGG to GTG). Cloning and transformation of the isolated tti1 genes confirmed the ability of the variants to allow colony formation of sis1∆ cells.
Strain and plasmid construction
For testing the effect of depletion of Sis1, a sis1∆ strain having SIS1 under the control of the doxycycline repressible promoter was isolated by transforming WY26, which carries YCp50-SIS1, with the TETrSIS1 plasmid (called TET-SIS1). Colonies having lost YCp50-SIS1 were selected for on complete minimal media plates containing 5-FOA. Because of the difficulty of isolating W303 cell extracts having full-length PIKKs, a sis1∆ strain in the BY4743 background was constructed, as this background had previously been used for such experiments (Hoffman et al., 2016). To obtain an appropriate BY4743 strain, a LEU2 marked deletion of SIS1 was amplified by PCR using WY26 genomic DNA as a template with primers annealing 200 bp upstream of the ATG and 470 bp downstream of the stop codon and used to transform BY4743 trp1-∆ (Open Biosystems, Huntsville, AL; Winzeler et al., 1999). Correct replacement of SIS1 with LEU2 was confirmed by colony PCR using a primer internal to LEU2 and a primer 255 bp upstream of the ATG start codon of Sis1. A confirmed isolate (BY4743sis1∆) was transformed with TET-SIS1, sporulated, and dissected to obtain a haploid strain deleted for SIS1 and carrying TET-SIS1 (BYsis1∆). BYsis1∆ strains containing either tagged MEC1 or TRA1 in the chromosome were constructed—BYsis1∆Flag5-MEC1 and BYsis1∆FLAG5-TRA1, respectively. CY6808, a Trp+ strain isogenic to BY4741 that contains URA3-Flag5-TRA1 (tag at N-terminus; Berg et al., 2018), was first crossed to a trp1∆ haploid that was obtained by dissecting asci from BY4743trp∆. The resulting diploid was sporulated and asci dissected to isolate a haploid strain containing URA3-Flag5TRA and trp1∆. This haploid was crossed to BYsis1∆ TET-SIS1. The resulting diploid was sporulated and asci dissected to isolate a haploid containing the sis1∆ deletion, URA3-Flag5TRA1, and the TET-SIS1 plasmid (BYsis1∆ Flag5-TRA1 TET-SIS1). MEC1 was Flag tagged at the N-terminus in BY4743sis1∆ by transforming it with a restriction fragment from pCB2363 containing URA3-Flag5MEC1 (DaSilva et al., 2013). A confirmed isolate was transformed with TET-SIS1, sporulated, and dissected to obtain a haploid strain deleted for SIS1, possessing URA3-Flag5-MEC1 and carrying TET-SIS1 (BYsis1∆Flag5-MEC1 TET-SIS1). Diploids homozygous for sis1∆ and homozygous or heterozygous for TTI1 or tti1sup#1 were created by mating WY26 or sis1-∆tti1sup#1 carrying pRS316-SIS1 with haploids obtained by dissecting the diploid isolated from crossing sis1∆tti1sup#1 with PJ51-3A carrying pRS316-SIS1.
Tti1, Tti1sup#1 and Tel2 were HA tagged on their C-termini, while Tti2 was tagged at the N-terminus with Flag in both the chromosome and on plasmids driven by the ADH1 promoter. TTI1 open reading frames were cloned from genomic DNA by PCR amplification followed by restriction digestion with primers that introduced a BamHI site 5′ of the ATG start codon and an XhoI site 3′ to the stop codon into similarly digested p414ADH (Mumberg et al., 1995). The open reading frames for TEL2 and TTI2 were similarly cloned into p414ADH from PCR amplified genomic DNA except the 5′ primer for TEL2 introduced a BglII site which can ligate with a BamHI site. In the case of Tti1, a 3x-HA tag, contained on a NotI fragment, was placed at the C-terminus of TTI1 by cloning into a NotI restriction site that was introduced just before the stop codon using the QuikChange protocol (Stratagene). p414ADH-TEL2-HA was constructed by amplifying genomic DNA from a yeast strain containing a 3x-HA tag at the C-terminus of TEL2 with a forward primer (+602–+630) and a primer downstream of the HA-tag and stop codon. The resulting DNA fragment was used to replace the WT fragment from the target plasmid after both were digested with PstI and XhoI and then ligated together. p414ADH-Flag3-TTI2 was constructed by subcloning TTI2 from p414ADH-TTI2 into p414ADH-Flag3 using BamHI and XhoI.
TTI1 and TEL2 were epitope-tagged at their C-termini on the chromosome in W303 using pFa6 3xHA:HIS3MX6 as a template and 60-mer primers to target recombination just upstream of their stop codons. To verify that correct fusions were obtained, a DNA fragment was amplified for each and sequenced. To move TTI1-HA-HIS3 (WT and sup#1) into BYsis1∆ TET-SIS1, inserts were amplified from chromosomal DNA using primers 1480 bp upstream and 265 bp downstream of the stop codon for TTI1 and transformed into BYsis1∆. His+ candidates were tested by immunoblot analysis. Tti2 was Flag tagged at its N-terminus on the chromosome using Crispr. A target-specific sgRNA was created by using a 60-mer bridging primer containing a 20-nucleotide target sequence of Tti2 (+38 to +57) which was cloned into a NotI digested pXIPHOS vector (accession MG897154; GenBank; Higgins et al., 2018; Kuang et al., 2018) using NEBuilder HiFi DNA assembly master mix from New England BioLabs. A 461-bp rescue DNA was designed with 187 bp upstream of the ATG start codon followed by the 3x-Flag tag (Ueda et al., 2011) and ending with 199 bp of Tti1 sequence (+1 to +199). The pXIPHOS-TTI2 sgRNA plasmid, which carries the natamycin resistance marker, was cotransformed into PJ51-3A with a 20× molar excess of rescue DNA. Natamycin-resistant transformants were selected for on YPD with 100 µg/ml nourseothricin (Werner BioAgents GmbH, Jena, Germany). Transformants were tested by colony PCR using primers 187 bp upstream of the start codon and 199 bp downstream of the start codon for the insertion of the Flag tag.
HSE testing
HSE-Cyc-GFP plasmids were constructed by first subcloning the 4XHSE + crippled CYC1 promoter + Emerald GFP fragment from a Ura marked plasmid (Brandman et al., 2012) into pRS313 using SacI-XhoI. The 4XHSE (CTAGAAGCTTCTAGAAGCTTCTAGAAGCTTCTAGAAGCTTCTAGG) was replaced with Ssa4HSE (CAATGAAGTACATTCTAGAAGTTCCTAGAACCTTATGGAAGCAC), Ssa3HSE (CGCTGTGGAAAGTTATAGAATATTACAGAAGCAGCCA), or Sis1HSE (TTATATGAACGTTCCAGAAACTTCTGGAAAAAGAATG) by using 500-bp G blocks manufactured by Integrated DNA Technologies (Coralville, IA) that replace the XhoI-NcoI of pRS313-4XHSE-CYC1-GFP with the aid of NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, Ipswich, MA).
Immunoblot and coimmunoprecipitation analysis
Cell lysates were made from 8–10 OD600 units of log phase cells (OD600 of 0.8–1.0) that were pelleted and washed with water using bead beating as follows. Cell pellets were disrupted by bead beating (5 × 1 min vortexing at 4°C with 1 min on ice in between) by resuspending in 200 µl of lysis buffer (25 mM HEPES-KOH, pH 7.5, 150 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM MgCl2, 10% glycerol, and 0.1% NP40) and 100 µl of 0.5 mm glass beads (BioSpec products). Lysates were cleared by centrifugation at 16 rcf for 15 min. Protein concentration of the lysates were determined using the Bio-Rad protein assay reagent (Bio-Rad, Hercules, CA). Equivalent amounts of cell lysates (μg of protein) were separated on SDS–PAGE and transferred to nitrocellulose. For analysis of PIKK proteins, lysates were separated using Tris-acetate polyacrylamide 4–10% gradient gels as described in https://doi.org/10.1007/978-1-4939-8793-1_22 but adapted to the Bio-Rad mini Protean system using 1.5 mm spacers. All immunoblot analyses were carried out using the Enhanced Chemiluminescence system (Pittsburgh, PA) according to the manufacturer’s suggestion.
Lysates, prepared from 40 OD600 of cells as described above, were used in coimmunoprecipitation experiments as previously described with slight modifications (Anderson et al., 2008). Purified polyclonal Sis1 polyclonal antibodies (4.5 µg) were added to 1 mg of lysate in a volume of 200 µl and rotated at 4°C for two hours. Equilibrated Dynabeads Protein G (Invitrogen by Thermo Fisher Scientific; 35 µl) were added, and rotation continued for 1 h. The beads were collected and washed three times with lysis buffer with the aid of a MagRack 6 (GE life Sciences) followed by boiling for 5 min in 20 µl of 2x-LDS buffer (diluted from 4x-LDS NuPAGE buffer from Invitrogen by Thermo Fisher Scientific). Samples were loaded on Tris-Acetate gels and processed for immunoblot analysis as described above. Five percent of starting lysates were loaded as input controls.
Alpha-tubulin (12G10) antibodies were obtained from the monoclonal antibody facility at the University of Iowa (University Heights, IA). Anti-GFP monoclonal antibody (GF28R) was produced by Invitrogen and purchased from ThermoFisher. Anti-HA polyclonal antibody came from Proteintech Group (Rosemont, IL); anti-FLAG monoclonal M2 antibody from Sigma-Aldrich (St. Louis, MO). Rabbit polyclonal antibodies generated for use in the Craig laboratory: anti-Sis1 (1–121) (Yu et al., 2015), anti-Ydj1 (Yan and Craig, 1999), anti-Tim44 (Liu et al., 2001), anti-Zuo1 (Hundley et al., 2002), anti-Yfh1 (Aloria et al., 2004), anti-Ssa3/4 (Baxter and Craig, 1998) and anti-Tim23 (D’Silva et al., 2008). Polyclonal antibodies to Tor1 (Alarcon et al., 1996) and Tor2 (Lorenz and Heitman, 1995) were gifts from Joseph Heitman. Polyclonal antibodies to Hsp104 (Ab 8-2) were a gift from Susan Lindquist (Parsell et al., 1991). Polyclonal antibodies to Hsp26 and Hsp42 (Haslbeck et al., 2004) were gifts from Johannes Buchner.
Supplementary Material
Acknowledgments
We thank Chris Brandl, Joseph Heitman, Johannes Buchner, Chris Hittinger, and Onn Brandman for gifts of strains, plasmids, and antibodies, and Jaroslaw Marszalek and Tom Ziegelhoffer for helpful comments. This work was supported by National Institutes of Health Grant R35 GM127009 (EAC).
Abbreviations used:
- 5-FOA
5-fluoroorotic acid
- HSE
heat shock element
- PIKK
phosphatidylinositol 3-kinase-related kinase.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-10-0493) on December 22, 2021.
REFERENCES
- Alarcon CM, Cardenas ME, Heitman J (1996). Mammalian RAFT1 kinase domain provides rapamycin-sensitive TOR function in yeast. Genes Dev 10, 279–288. [DOI] [PubMed] [Google Scholar]
- Alford BD, Tassoni-Tsuchida E, Khan D, Work JJ, Valiant G, Brandman O (2021). ReporterSeq reveals genome-wide dynamic modulators of the heat shock response across diverse stressors. Elife 10, e57376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloria K, Schilke B, Andrew A, Craig EA (2004). Iron-induced oligomerization of yeast frataxin homologue Yfh1 is dispensable in vivo. EMBO Rep 5, 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CM, Korkin D, Smith DL, Makovets S, Seidel JJ, Sali A, Blackburn EH (2008). Tel2 mediates activation and localization of ATM/Tel1 kinase to a double-strand break. Genes Dev 22, 854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron R, Higurashi T, Sahi C, Craig EA (2007). J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J 26, 3794–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balchin D, Hayer-Hartl M, Hartl FU (2020). Recent advances in understanding catalysis of protein folding by molecular chaperones. FEBS Lett 594, 2770–2781. [DOI] [PubMed] [Google Scholar]
- Baxter BK, Craig EA (1998). Suppression of an Hsp70 mutant phenotype in Saccharomyces cerevisiae through loss of function of the chromatin component Sin1p/Spt2p. J Bacteriol 180, 6484–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MD, Genereaux J, Karagiannis J, Brandl CJ (2018). The pseudokinase domain of Saccharomyces cerevisiae Tra1 is required for nuclear localization and incorporation into the SAGA and NuA4 complexes. G3 (Bethesda) 8, 1943–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland SR, Jin N, Ozdemir AC, Lyons RH Jr, Weisman LS, Wilson TE (2010). Discovery of mutations in Saccharomyces cerevisiae by pooled linkage analysis and whole-genome sequencing. Genetics 186, 1127–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein WR, Craig EA (1990). Transcriptional regulation of SSA3, an HSP70 gene from Saccharomyces cerevisiae. Mol Cell Biol 10, 3262–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. (2012). A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DC, Yang BC, Kuo TT (1992). One-step transformation of yeast in stationary phase. Curr Genet 21, 83–84. [DOI] [PubMed] [Google Scholar]
- Cheung ACM, Diaz-Santin LM (2019). Share and share alike: The role of Tra1 from the SAGA and NuA4 coactivator complexes. Transcription 10, 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Shim WJ, Liu Y, Shan SO (2021). J-domain proteins promote client relay from Hsp70 during tail-anchored membrane protein targeting. J Biol Chem 296, 100546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Marszalek J (2017). How do J-proteins get Hsp70 to do so many different things? Trends Biochem Sci 42, 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaSilva LF, Pillon S, Genereaux J, Davey MJ, Gloor GB, Karagiannis J, Brandl CJ (2013). The C-terminal residues of Saccharomyces cerevisiae Mec1 are required for its localization, stability, and function. G3 (Bethesda) 3, 1661–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Silva PR, Schilke B, Hayashi M, Craig EA (2008). Interaction of the J-protein heterodimer Pam18/Pam16 of the mitochondrial import motor with the translocon of the inner membrane. Mol Biol Cell 19, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias-Villalobos A, Fort P, Helmlinger D (2019). New insights into the evolutionary conservation of the sole PIKK pseudokinase Tra1/TRRAP. Biochem Soc Trans. [DOI] [PubMed] [Google Scholar]
- Elrod SL, Chen SM, Schwartz K, Shuster EO (2009). Optimizing sporulation conditions for different Saccharomyces cerevisiae strain backgrounds. In: Meiosis. Methods in Molecular Biology (Methods and Protocols), vol. 557, ed. S. Keeney: Humana Press. [DOI] [PubMed] [Google Scholar]
- Feder ZA, Ali A, Singh A, Krakowiak J, Zheng X, Bindokas VP, Wolfgeher D, Kron SJ, Pincus D (2021). Subcellular localization of the J-protein Sis1 regulates the heat shock response. J Cell Biol 220, e202005165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genereaux J, Kvas S, Dobransky D, Karagiannis J, Gloor GB, Brandl CJ (2012). Genetic evidence links the ASTRA protein chaperone component Tti2 to the SAGA transcription factor Tra1. Genetics 191, 765–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Hall MN (2017). Nutrient sensing and TOR signaling in yeast and mammals. EMBO J 36, 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto GH, Ogi H, Biswas H, Ghosh A, Tanaka S, Sugimoto K (2017). Two separate pathways regulate protein stability of ATM/ATR-related protein kinases Mec1 and Tel1 in budding yeast. PLoS Genet 13, e1006873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Braun N, Stromer T, Richter B, Model N, Weinkauf S, Buchner J (2004). Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J 23, 638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DA, Young MKM, Tremaine M, Sardi M, Fletcher JM, Agnew M, Liu L, Dickinson Q, Peris D, Wrobel RL, et al. (2018). Natural variation in the multidrug efflux pump SGE1 underlies ionic liquid tolerance in yeast. Genetics 210, 219–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B, Baryshnikova A, Brown GW (2018). Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst 6, 192–205.e193. [DOI] [PubMed] [Google Scholar]
- Hoffman KS, Duennwald ML, Karagiannis J, Genereaux J, McCarton AS, Brandl CJ (2016). Saccharomyces cerevisiae Tti2 Regulates PIKK Proteins and Stress Response. G3 (Bethesda) 6, 1649–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houry WA, Bertrand E, Coulombe B (2018). The PAQosome, an R2TP-based chaperone for quaternary structure formation. Trends Biochem Sci 43, 4–9. [DOI] [PubMed] [Google Scholar]
- Hundley H, Eisenman H, Walter W, Evans T, Hotokezaka Y, Wiedmann M, Craig E (2002). The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc Natl Acad Sci USA 99, 4203–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurov KE, Cotta-Ramusino C, Elledge SJ (2010). A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev 24, 1939–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imseng S, Aylett CH, Maier T (2018). Architecture and activation of phosphatidylinositol 3-kinase related kinases. Curr Opin Struct Biol 49, 177–189. [DOI] [PubMed] [Google Scholar]
- Jores T, Lawatscheck J, Beke V, Franz-Wachtel M, Yunoki K, Fitzgerald JC, Macek B, Endo T, Kalbacher H, Buchner J, et al. (2018). Cytosolic Hsp70 and Hsp40 chaperones enable the biogenesis of mitochondrial beta-barrel proteins. J Cell Biol 217, 3091–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11, 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaips CL, Gropp MHM, Hipp MS, Hartl FU (2020). Sis1 potentiates the stress response to protein aggregation and elevated temperature. Nat Commun 11, 6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang MC, Kominek J, Alexander WG, Cheng JF, Wrobel RL, Hittinger CT (2018). Repeated cis-regulatory tuning of a metabolic bottleneck gene during evolution. Mol Biol Evol 35, 1968–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Krzewska J, Liberek K, Craig EA (2001). Mitochondrial Hsp70 Ssc1: Role in protein folding. J Biol Chem 276, 6112–6118. [DOI] [PubMed] [Google Scholar]
- Liu XD, Liu PC, Santoro N, Thiele DJ (1997). Conservation of a stress response: Human heat shock transcription factors functionally substitute for yeast HSF. EMBO J 16, 6466–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez N, Aron R, Craig EA (2003). Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+]. Mol Biol Cell 14, 1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Heitman J (1995). TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J Biol Chem 270, 27531–27537. [DOI] [PubMed] [Google Scholar]
- Luke MM, Sutton A, Arndt KT (1991). Characterization of SIS1, a Saccharomyces cerevisiae homologue of bacterial dnaJ proteins. J Cell Biol 114, 623–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig AJ (2019). Towards the mechanism of yeast telomere dynamics. Trends Cell Biol 29, 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDiarmid CW, Taggart J, Kerdsomboon K, Kubisiak M, Panascharoen S, Schelble K, Eide DJ (2013). Peroxiredoxin chaperone activity is critical for protein homeostasis in zinc-deficient yeast. J Biol Chem 288, 31313–31327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DE, Hall MN (2005). The expanding TOR signaling network. Curr Opin Cell Biol 17, 158–166. [DOI] [PubMed] [Google Scholar]
- Masser AE, Ciccarelli M, Andreasson C (2020). Hsf1 on a leash—controlling the heat shock response by chaperone titration. Exp Cell Res 396, 112246. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M (1995). Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122. [DOI] [PubMed] [Google Scholar]
- Pal M, Munoz-Hernandez H, Bjorklund D, Zhou L, Degliesposti G, Skehel JM, Hesketh EL, Thompson RF, Pearl LH, Llorca O, et al. (2021). Structure of the TELO2–TTI1–TTI2 complex and its function in TOR recruitment to the R2TP chaperone. Cell Rep 36, 109317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell DA, Sanchez Y, Stitzel JD, Lindquist S (1991). Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature 353, 270–273. [DOI] [PubMed] [Google Scholar]
- Pincus D (2020). Regulation of Hsf1 and the heat shock response. Adv Exp Med Biol 1243, 41–50. [DOI] [PubMed] [Google Scholar]
- Prasad R, Xu C, Ng DTW (2018). Hsp40/70/110 chaperones adapt nuclear protein quality control to serve cytosolic clients. J Cell Biol 217, 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig R, Nillegoda NB, Mayer MP, Bukau B (2019). The Hsp70 chaperone network. Nat Rev Mol Cell Biol 20, 665–680. [DOI] [PubMed] [Google Scholar]
- Ruis H, Schuller C (1995). Stress signaling in yeast. Bioessays 17, 959–965. [DOI] [PubMed] [Google Scholar]
- Sahi C, Craig EA (2007). Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci USA 104, 7163–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilke BA, Ciesielski SJ, Ziegelhoffer T, Kamiya E, Tonelli M, Lee W, Cornilescu G, Hines JK, Markley JL, Craig EA (2017). Broadening the functionality of a J-protein/Hsp70 molecular chaperone system. PLoS Genet 13, e1007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks J (1986). Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press. [Google Scholar]
- Shiber A, Breuer W, Brandeis M, Ravid T (2013). Ubiquitin conjugation triggers misfolded protein sequestration into quality control foci when Hsp70 chaperone levels are limiting. Mol Biol Cell 24, 2076–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Boeke JD (1991). In vitro mutagenesis and plasmid shuffling: From cloned gene to mutant yeast. Methods Enzymol 194, 302–318. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lopez N, Craig EA, Lindquist S (2001). The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J 20, 2435–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K (2018). Branching the Tel2 pathway for exact fit on phosphatidylinositol 3-kinase-related kinases. Curr Genet 64, 965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers DW, Wolfe KJ, Ren HY, Cyr DM (2013). The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS One 8, e52099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Wang RC, Takai KK, Yang H, de Lange T (2007). Tel2 regulates the stability of PI3K-related protein kinases. Cell 131, 1248–1259. [DOI] [PubMed] [Google Scholar]
- Takai H, Xie Y, de Lange T, Pavletich NP (2010). Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev 24, 2019–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Manabe Y, Mukai M (2011). The high performance of 3XFLAG for target purification of a bioactive metabolite: A tag combined with a highly effective linker structure. Bioorg Med Chem Lett 21, 1359–1362. [DOI] [PubMed] [Google Scholar]
- Verghese J, Abrams J, Wang Y, Morano KA (2012). Biology of the heat shock response and protein chaperones: Budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol Mol Biol Rev 76, 115–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa M, Cassani C, Gobbini E, Bonetti D, Longhese MP (2016). Coupling end resection with the checkpoint response at DNA double-strand breaks. Cell Mol Life Sci 73, 3655–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Morgen P, Horejsi Z, Macurek L (2015). Substrate recognition and function of the R2TP complex in response to cellular stress. Front Genet 6, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Mizukami Y, Sakurai H (2005). Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. J Biol Chem 280, 11911–11919. [DOI] [PubMed] [Google Scholar]
- Yan W, Craig EA (1999). The glycine–phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol Cell Biol 19, 7751–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MR, Craig EA (1993). Saccharomyces cerevisiae HSP70 heat shock elements are functionally distinct. Mol Cell Biol 13, 5637–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HY, Ziegelhoffer T, Osipiuk J, Ciesielski SJ, Baranowski M, Zhou M, Joachimiak A, Craig EA (2015). Roles of intramolecular and intermolecular interactions in functional regulation of the Hsp70 J-protein co-chaperone Sis1. J Mol Biol 427, 1632–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong T, Luke MM, Arndt KT (1996). Transcriptional regulation of the yeast DnaJ homologue SIS1. J Biol Chem 271, 1349–1356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.