Abstract
The antifungal activity spectrum of Lactobacillus coryniformis subsp. coryniformis strain Si3 was investigated. The strain had strong inhibitory activity in dual-culture agar plate assays against the molds Aspergillus fumigatus, A. nidulans, Penicillium roqueforti, Mucor hiemalis, Talaromyces flavus, Fusarium poae, F. graminearum, F. culmorum, and F. sporotrichoides. A weaker activity was observed against the yeasts Debaryomyces hansenii, Kluyveromyces marxianus, and Saccharomyces cerevisiae. The yeasts Rhodotorula glutinis, Sporobolomyces roseus, and Pichia anomala were not inhibited. In liquid culture the antifungal activity paralleled growth, with maximum mold inhibition early in the stationary growth phase, but with a rapid decline in antifungal activity after 48 h. The addition of ethanol to the growth medium prevented the decline and gave an increased antifungal activity. The activity was stable during heat treatment and was retained even after autoclaving at 121°C for 15 min. Maximum activity was observed at pH values of between 3.0 and 4.5, but it decreased rapidly when pH was adjusted to a level between 4.5 and 6.0 and was lost at higher pH values. The antifungal activity was fully regained after readjustment of the pH to the initial value (pH 3.6). The activity was irreversibly lost after treatment with proteolytic enzymes (proteinase K, trypsin, and pepsin). The antifungal activity was partially purified using ion-exchange chromatography and (NH4)2SO4 precipitation, followed by gel filtration chromatography. The active compound(s) was estimated to have a molecular mass of approximately 3 kDa. This is the first report of the production of a proteinaceous antifungal compound(s) from L. coryniformis subsp. coryniformis.
Molds and yeasts are important spoilage organisms in different food and feed systems. During the last few years there has been a growing interest in biopreservation, i.e., the use of microorganisms and/or their metabolites to prevent spoilage and to extend the shelf-life of foods (20). Lactic acid bacteria (LAB) are of particular interest as biopreservation organisms. Their preserving effect mainly relates to the formation of lactic acid, acetic acid, and hydrogen peroxide; competition for nutrients; and the production of bacteriocins (13, 20). The bacteriocins from LAB are bioactive peptides, derived from ribosomally synthesized precursors and with a bacteriocidal effect on a number of different gram-positive bacteria (11, 17). While many studies have assessed their antibacterial effects (6), there are very few reports on specific antifungal compounds from LAB. Early research suggested antifungal activities from a Lactobacillus casei strain that inhibited both the growth and the aflatoxin production of Aspergillus parasiticus (7). Production of fungal inhibitory compounds from L. casei subsp. rhamnosus, all with molecular masses of <1,000 Da, was described elsewhere (22). The antifungal activity of a Leuconostoc mesenteroides strain from cheese has been reported, but no antifungal substance could be isolated (21). A mixture of Lactobacillus spp. from a commercial silage inoculum was found to reduce both mold growth and spore germination, as well as aflatoxin production by Aspergillus flavus subsp. parasiticus (9). An antifungal Lactobacillus sanfrancisco CBI, isolated from sour dough inhibited bread spoilage molds from the genera Fusarium, Penicillium, Aspergillus, and Monilia. The antifungal activity was caused by formation of several short-chained fatty acids, among which caproic acid was the most important molecule (4). Niku-Paavola et al. (15) reported the production of antimicrobial low-molecular-weight compounds other than organic acids from Lactobacillus plantarum. The active fraction containing, for example, benzoic acid, methylhydantoin, mevalonolactone, and cyclo-(glycyl-l-leucyl) and acting synergistically with lactic acid, was active against both Fusarium avenacum and the gram-negative bacterium Pantoea agglomerans. Lavermicocca et al. (12) found that phenyl-lactic acid and 4-hydroxy-phenyl-lactic acid from a sourdough isolate of L. plantarum had broad spectrum fungicidal activity. Recently, Okkers et al. (18) characterized the peptide pentocin TV35b from Lactobacillus pentosus, with a fungistatic effect on Candida albicans and with inhibitory effect against a number of gram-positive bacteria.
We have identified strain Si3, an antifungal LAB strain, as Lactobacillus coryniformis subsp. coryniformis. The antifungal isolate Si3, previously isolated from grass silage in our laboratory, has been found to inhibit yeast growth in grass silage (I. Thylin and S. Lindgren, submitted for publication). There are no other literature reports on antifungal effects, nor of any bacteriocin-like activity, for this species.
The aims of the present study were to describe the antifungal spectrum, the basal biochemical characteristics, and the production conditions for the fungal inhibitory compound(s) from L. coryniformis subsp. coryniformis Si3.
MATERIALS AND METHODS
Cultures and media.
The strain Si3, originally isolated from grass silage (Thylin and Lindgren, submitted), was grown on MRS agar (Oxoid Ltd., Basingstoke, England) at 30°C in anaerobic jars under a CO2 + N2 atmosphere (GasPak System; BBL, Cockeysville, Md.). Working cultures were kept on MRS agar plates at 5°C, while long-term storage was done either at −70°C in 15% glycerol or as lyophilized cultures in skimmed milk powder. MRS broth (Oxoid) was used as liquid growth medium, unless otherwise stated.
Identification of strain Si3.
The strain Si3, isolated from grass silage, was identified from fermentation patterns, confirmed by sequence analysis of 16S ribosomal DNA (rDNA). The fermentation pattern was determined using the API 50CH system (BioMérieux), with additional confirmation tests for fermentation of raffinose and rhamnose. Results from the API test were compared with the API database, and the fermentation pattern was further evaluated according to the method of Kandler and Weiss (10). Bacterial DNA was isolated according to the method of Axelsson and Lindgren (1). The almost complete 16S rRNA gene was amplified by PCR using slightly modified domain Bacteria-specific primers (23). The primer sequences used were 5′-AGAGTTTGATYMTGGC-3′ (E. coli numbering 8 to 23) and 5′-AGAAAGGAGGTGATCC-3′ (E. coli numbering 1544 to 1529). PCR reactions were performed under the following conditions: 94°C for 30 s, 54°C for 30 s, and 72°C for 80 s, for 35 cycles. The resulting PCR product was purified from an agarose gel. Both strands of the purified fragment were partially sequenced using the Thermo Sequenase dye terminator cycle sequencing pre-mix (Amersham) and the automated sequence analyzer ABI PRISM 377XL (Perkin-Elmer). The same primers that were used for the amplification were used for sequencing of the PCR product, together with additional customised internal primers.
Preparation of concentrated culture filtrate.
L. coryniformis subsp. coryniformis Si3 was inoculated to a concentration of 105 cells/ml of 800 ml of MRS broth in 1,000-ml Erlenmeyer flasks, plugged with cotton to allow air access, and incubated as a still culture at 30°C for 48 h. The culture was then centrifuged (15,000 × g, 10 min), followed by filter sterilization (0.45-μm pore size; Millipore). The sterile cell-free supernatant was freeze-dried and resuspended (to a 15-fold concentration) in either 10 mM acetic acid (HAc) or 20 mM citrate-phosphate buffer (pH 3.4). Culture filtrate from the type strain of L. coryniformis subsp. coryniformis (ATCC 25602) was used as a control and prepared in the same manner as that from strain Si3.
Fungal inocula.
The molds Aspergillus fumigatus J9, Aspergillus nidulans J10, Penicillium commune J238, Penicillium roqueforti J229, Mucor hiemalis J42, Talaromyces flavus J37, Fusarium poae J24, Fusarium graminearum J114, Fusarium culmorum J300, and Fusarium sporotrichoides J319 and the yeasts Debaryomyces hansenii J136 and J187, Kluyveromyces marxianus J186, Pichia anomala J121, Rhodotorula glutinis J195, Saccharomyces cerevisiae J122, Sporobolomyces roseus J104, and Zygosaccharomyces rouxii J107 came from our own culture collection. They were grown on malt extract agar (MEA) slants (Oxoid) at 25°C for 7 days and then stored at 5°C. Inocula containing spores or conidia were prepared by growing the molds on MEA slants for 7 to 10 days (or until sporulation) and then collecting spores or conidia after vigorously shaking the slants with sterile peptone water (0.2% [wt/vol]). Yeast cell inocula were prepared from washed cultures grown in malt extract broth (Oxoid) as still cultures at 30°C for 24 h. Mold (spores or conidia) and yeast concentrations were determined using a Buerkner hemocytometer, and adjusted to 105 per ml of sterile peptone water (0.2%).
Antifungal activity assays.
Three different assays, the overlay method, the agar-well diffusion method, and the microtiter plate well assay, were used to detect antifungal activity. All experiments assaying inhibitory activity were, unless stated otherwise, performed in duplicate. The overlay method was performed using MRS agar plates on which LAB were inoculated as two 2-cm-long lines and incubated at 30°C for 48 h in anaerobic jars. The plates were then overlaid with 10 ml of malt extract soft agar (2% malt extract, 0.7% agar; Oxoid) containing 104 yeast cells or fungal spores (conidia) per ml. The plates were then incubated aerobically at 30°C for 48 h. The plates were examined for clear zones of inhibition around the bacterial streaks, and the area of the zones was scored as follows: −, no suppression; +, no fungal growth on 0.1 to 3% of the plate area per bacterial streak; ++, no fungal growth on 3 to 8% of plate area per bacterial streak; or +++, no fungal growth on >8% of plate area per bacterial streak.
For the agar well diffusion assay, MRS agar plates containing 104 A. fumigatus conidia per ml agar were prepared. Wells, with a diameter of either 3 or 5 mm, were then cut in the agar using a sterile cork-borer. A droplet of agar was added to each well in order to seal it to avoid leakage. Then, either 40- or 70-μl samples were added to the wells and allowed to diffuse into the agar during a 5-h preincubation period at room temperature, followed by aerobic incubation at 30°C for 48 h. The antifungal effects recorded were graded as follows: −, no suppression; +, weak suppression around the wells; ++, strong suppression, with detectable clear zones around the wells; or +++, very strong suppression, with large, clear zones around the wells.
For the microtiter plate well assay, a 30-μl sample and 50 μl of MRS broth containing 104 A. fumigatus spores per ml were added to each well. The plate was incubated in a humid chamber at 30°C for 48 h. The degree of inhibition was either measured as the optical density at 550 nm in a Microplate Autoreader EL 309 (Biotek Instruments), measured by using an inverted microscope for estimating the growth of the indicator fungi, or measured by using the naked eye. The antifungal effects were given numerical values as follows: no mold growth = an inhibition factor of 1.0; one or a few mold colonies/well = an inhibition factor of 0.6; mycelium monolayer in the wells = an inhibition factor of 0.3; or complete mycelium coverage of the wells = an inhibition factor of 0. Scaled antifungal units were calculated as follows: antifungal unit = (the inhibition value × the reciprocal of the highest dilution at which inhibitory activity could be detected).
Spectrum of antifungal activity.
The overlay method described above was used to determine the ability of L. coryniformis subsp. coryniformis Si3 to inhibit growth of various species of molds and yeasts at temperatures between 25 and 30°C.
Effects of temperature, pH, and proteolytic enzymes on antifungal activity.
The antifungal activity remaining after exposure to high temperatures, different pH values, or proteolytic enzymes was determined using either the agar well diffusion assay or the microtiter plate assay. Aliquots (10 ml) of 15-fold-concentrated culture filtrate, prepared as described above, were heated to either 50, 70, 96, or 121°C for 10 min. The samples were allowed to cool and then tested for antifungal activity. The pH effect was investigated with 15-fold-concentrated culture filtrate, in 10-ml aliquots, adjusted to pH values of 2.5, 3.0, 4.0, 4.5, 5.0, 6.0, 7.0, and 9.0 with 1 M HCl and 2 M NaOH before evaluating the antifungal activity. MRS broth, concentrated 15-fold and adjusted to the same pH values, served as a control. The effect of proteolytic enzymes on antifungal activity was investigated with 10-ml aliquots of 15-fold-concentrated culture filtrate treated with one of the following proteolytic enzymes: proteinase K (Sigma), trypsin (Sigma), or pepsin (Sigma). Samples were adjusted with 1 M HCl and 2 M NaOH to the optimum pH value for each enzyme, i.e., 7.6, 7.6, and 2.0 for proteinase K, trypsin, and pepsin, respectively. After adjustment of the pH, the supernatants were treated with 100 μg of the respective enzyme per ml and incubated at 37°C for 1 h. Before evaluating the antifungal activity the pH of the supernatants was readjusted to the initial pH value 3.6. Both 15-fold-concentrated MRS broth treated with enzymes and pH-adjusted 15-fold-concentrated samples served as controls.
Influence of temperature and aeration on production of antifungal activity.
Growth, antifungal activity, and pH were monitored over time with 200-ml cultures of MRS broth inoculated with 105 bacteria per ml. The flasks were incubated at 25 or 30°C, either as still cultures in 250-ml anaerobic flasks sealed with butyl rubber membranes or in 250-ml Erlenmeyer flasks plugged with cotton (to allow air access) on a rotary shaker (100 rpm). Every second hour, a sample was collected for the determination of pH, the numbers of cells (Buerkner hemocytometer), and the antifungal activity (microtiter plate assay).
The influence of ethanol on the recovery of antifungal activity was evaluated using batches of 200 ml of MRS broth, inoculated with 5 × 105 bacteria per ml in 250-ml Erlenmeyer flasks, plugged with cotton, and incubated as still cultures at 30°C. Ethanol was added to reach a maximum (theoretical) value of 2 mg/ml at 7 h, 3 mg/ml at 12 h, and 5 mg/ml at 15 h to a final concentration of 7 mg/ml (early stationary phase). The actual ethanol concentration was not measured, but evaporation was assumed to be minor during the experimental conditions. Samples were collected every second hour for determination of the numbers of cells, the pH, and the antifungal activity.
The growth and antifungal activity of L. coryniformis subsp. coryniformis Si3 was evaluated at different pH values under controlled fermentor conditions. Here, 105 bacteria per ml was inoculated in MRS broth at pH 4.5, 5.5, and 6.5 and was grown as 800-ml cultures at 30°C without aeration in a 1.0-liter fermentor (Bioreactor BR0.4; Belach Bioteknik). The pH was controlled and adjusted with 2 M KOH. After 48 h of growth the bacterial cells were removed by centrifugation (15,000 × g, 10 min), followed by filter sterilization. The cell-free culture filtrate was freeze-dried and dissolved in 53 ml of 10 mM HAc, resulting in a 15-fold concentration. The antifungal activity was tested with the microtiter plate assay at pH 4.0. The 15-fold-concentrated culture filtrate was also evaluated for stability after storage in 1-ml aliquots in Eppendorf tubes at 25, 4, and −28°C. The antifungal activity against A. fumigatus remaining after 1 to 15 days storage was determined using the microtiter plate assay. The stability of a precipitate resulting from 100% (NH4)2SO4 saturation was also evaluated.
Primary purification.
To obtain a larger amount of material, 800 ml of MRS broth in a cotton-plugged 1,000-ml Erlenmeyer flask was inoculated with 105 cells of L. coryniformis subsp. coryniformis Si3 per ml and grown as a still culture at 30°C for 48 h. After incubation the broth was centrifuged (15,000 × g, 10 min), sterile filtered (0.45-μm pore size; Millipore), and the filtrate was freeze-dried and adjusted to 15 times the original concentration in 20 mM citrate-phosphate buffer (pH 5.0).
The antifungal substance(s) from the 15-fold-concentrated culture filtrate of L. coryniformis subsp. coryniformis Si3 was partially purified. The first purification step was ion-exchange chromatography using Q-Sepharose (Pharmacia, Uppsala, Sweden) with 20 mM citrate-phosphate buffer (pH 5.0). Samples were eluted in three steps with 0.2, 0.5, or 1.0 M NaCl in 20 mM citrate-phosphate buffer (pH 5.0). The fractions were evaluated with the microtiter plate assay. The second step was precipitation at 60, 80, and 100% (NH4)2SO4 saturations. The precipitates were pelleted (15,000 × g, 10 min) and dissolved in 5 ml of 10 mM HAc each. Then, 1 ml of each fraction was dialyzed in a Spectra/Pore 1000 Da Membrane (Spectrum Medical Industries, Inc.) against 20 mM citrate-phosphate buffer (pH 5.0) and was evaluated with the microtiter plate assay. The dissolved pellets from the 80 and 100% (NH4)2SO4 saturations were pooled and run on a gel filtration column (Superdex Peptide PC 3.2/30) using the SMART chromatography system (Pharmacia, Uppsala, Sweden) with 10 mM HAc as buffer. Fractions from the gel filtration were evaluated with the microtiter plate assay.
RESULTS
Identification of strain Si3.
The fermentation pattern identified Si3 as L. coryniformis subsp. coryniformis (results not shown); the positive fermentation of rhamnose differentiated our isolate from L. coryniformis subsp. torquens. This identification was confirmed by the 16S ribosomal DNA (rDNA) sequence data, where 588 nucleotides corresponding to positions 86 to 674 of the L. coryniformis subsp. coryniformis ATCC 25602 (GenBank accession no. M58813) sequence were determined for strain Si3 (GenBank accession no. AF228698). The 16S rDNA sequences of strains Si3 and ATCC 25602 were found to be identical at all positions, except for 11 ambiguous nucleotides in the published sequence M58813 from strain ATCC 25602.
Spectrum of antifungal activity.
L. coryniformis subsp. coryniformis Si3 had a broad antifungal inhibitory spectrum, with activity against several taxonomic groups of mold and yeast (Table 1). Generally, molds seemed to be more sensitive than yeasts. However, the fast-growing zygomycete M. hiemalis was only marginally affected. The antifungal activity differed only slightly between dual cultures incubated at 25 or 30°C. After 2 days of bacterial growth the pH values in the inhibitory zone between and outside the bacterial streaks were 4.5 and 4.7, respectively. Outside the inhibition zone the pH in the A. fumigatus-containing MRS plates was 5.6.
TABLE 1.
Inhibition of molds and yeasts by L. coryniformis subsp. coryniformis Si3 in a dual-culture overlay system
| Mold or yeast strain | Activitya at:
|
|
|---|---|---|
| 25°C | 30°C | |
| Molds | ||
| Aspergillus fumigatus J9 | +++ | +++ |
| Aspergillus nidulans J10 | +++ | ++ |
| Penicillium commune J238 | ++ | ++ |
| Penicillium roqueforti J229 | + | + |
| Mucor hiemalis J42 | − | ++ |
| Talaromyces flavus J37 | +++ | ++ |
| Fusarium poae J24 | +++ | +++ |
| Fusarium graminearum J114 | +++ | +++ |
| Fusarium culmorum J300 | +++ | +++ |
| Fusarium sporotrichoides J319 | +++ | +++ |
| Yeasts | ||
| Rhodotorula glutinis J195 | − | ND |
| Sporobolomyces roseus J104 | − | ND |
| Pichia anomala J121 | − | − |
| Debaryomyces hansenii var. hansenii J136 | ++ | + |
| Debaryomyces hansenii var. hansenii J187 | + | + |
| Zygosaccharomyces rouxii J107 | − | − |
| Saccharomyces cerevisiae J122 | − | + |
| Kluyveromyces marxianus var. marxianus J186 | + | + |
Activity was scored as follows: −, no suppression; +, weak suppression around the streaks; ++, strong suppression, with detectable clear zones around the streaks; +++, very strong suppression, with large, clear zones around the streaks. ND, not determined.
Effects of temperature, pH, and proteolytic enzymes on antifungal activity.
The antifungal activity was found to be heat stable. Freeze-dried supernatant autoclaved for 15 min at 121°C retained full inhibitory activity against yeast and mold growth. The activity was stable at pH values that were between 3.0 and 4.5 but rapidly decreased between pH 4.5 and 6.0 (data not shown). No inhibitory activity was detected at a pH above 6.0. The activity was fully regained after readjustment of the pH to the starting value. The inhibitory activity of the freeze-dried supernatant was totally lost after treatment with proteinase K (Fig. 1) and trypsin and was radically decreased after treatment with pepsin.
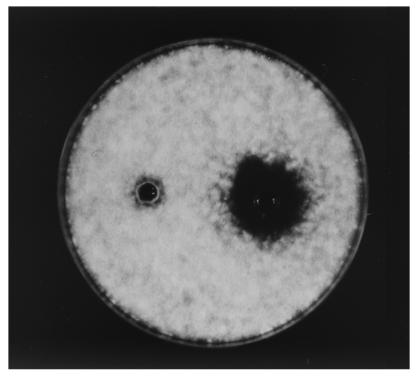
FIG. 1.
Effect of proteinase K on antifungal activity against A. fumigatus of a 15-fold-concentrated (freeze-dried) culture filtrate of L. coryniformis subsp. coryniformis Si3. The control sample (left well) was pH adjusted in the same manner as the proteinase K-treated sample (right well).
Influence of temperature and aeration on production of antifungal activity.
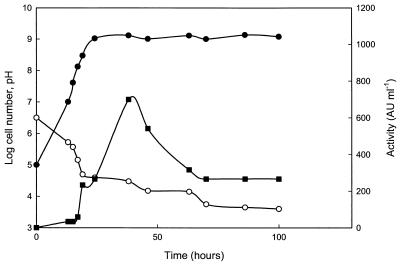
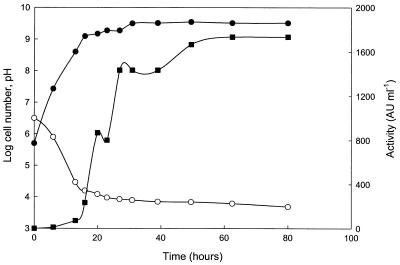
The maximum antifungal activity was observed as a distinct peak after about 40 h growth at 30°C, i.e., early in the stationary phase (Fig. 2). Only minor differences in antifungal activity were observed between cultures incubated as still cultures in capped flasks (data not shown) and those incubated with air access on a rotary shaker (Fig. 2). The addition of ethanol during growth doubled the recovered antifungal activity, and no decline was observed during the stationary phase (Fig. 3).
FIG. 2.
Changes in cell numbers (●), antifungal activity (■), and pH (○) over time. Erlenmeyer flasks (250 ml), plugged with cotton (to allow air access), with 200 ml of MRS broth were inoculated with 105 L. coryniformis subsp. coryniformis Si3 per ml and incubated at 30°C on a rotary shaker.
FIG. 3.
Effect of gradual addition of ethanol on antifungal activity of L. coryniformis subsp. coryniformis Si3. Erlenmeyer flasks (250 ml), plugged with cotton (to allow air access), with 200 ml of MRS broth were inoculated with 5 × 105 cells per ml and incubated as still cultures at 30°C. Ethanol was added to reach a theoretical concentration of 2 mg/ml at 7 h, 3 mg/ml at 12 h, and 5 mg/ml at 15 h and a final concentration of 7 mg/ml at the early stationary phase. Cell numbers (●), antifungal activity (■), and pH (○) results are shown.
The influence of substrate pH on activity under controlled fermentor conditions was evaluated using the microtiter plate assay at pH 4.0. Cells grown for 48 h at pH 6.5 gave a substantially higher antifungal activity, i.e., 3,000 antifungal units (AU) ml−1, than cells cultivated at pH 4.5 or 5.5, which gave 267 and 433 AU ml−1, respectively.
Stability.
The antifungal activity of freeze-dried culture filtrate was lost during prolonged storage. The activity was stable during storage for 7 days at either 4 or 25°C, but it rapidly decreased after 7 days at both temperatures. No activity could be recovered after storage for 2 days at −28°C. However, the activity remained during 14 days of storage in 100% saturated (NH4)2SO4 at 4°C.
Primary purification.
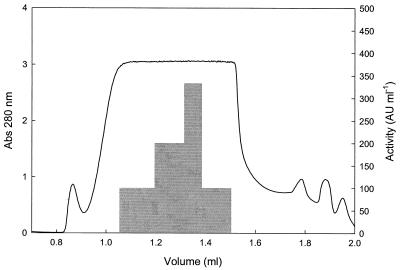
It was possible to follow the activity during several purification steps. After ion-exchange chromatography, the antifungal activity was detected in the fraction containing 20 mM citrate-phosphate buffer (pH 5.0) and 0.5 M NaCl. Ammonium sulfate precipitation of this fraction gave antifungal activities with dialyzed precipitates of both 80 and 100% (NH4)2SO4 saturations. When dissolved pellets were applied to a gel filtration column, the peak in antifungal activity was consistently found to be between elution volumes of 1.3 and 1.4 ml, indicating a molecular mass of about 3 kDa (Fig. 4).
FIG. 4.
Antifungal activity against A. fumigatus of fractions from gel filtration on Superdex Peptide PC 3.2/30, after ion-exchange chromatography and (NH4)2SO4 precipitation. The activity was evaluated with the microtiter plate assay (shaded area), and the protein concentration was determined as the absorbance at 280 nm (line).
DISCUSSION
Both molds and yeasts are important spoilage organisms in different food and feed systems. The molds evaluated in this study, such as P. roqueforti and P. commune, commonly spoil hard cheese, while different Fusarium species can produce mycotoxins in cereal grains (8). The yeasts Candida parapsilosis and D. hansenii are common spoilage organisms of yogurt and other fermented dairy products (19). There is thus a need for efficient and safe procedures to prevent fungal growth in various raw materials and food products. LAB are known to produce antimicrobial substances, but these mainly in the form of organic acids and bacteriocins. Very few reports have been published about the production of specific antifungal substances from LAB. The present study and a recent publication by Okkers et al. (18) clearly document the production of proteinaceous antifungal substances by LAB. However, Okkers et al. (18) only reported the fungistatic effect against the yeast C. albicans and not against filamentous fungi. Our study shows that L. coryniformis subsp. coryniformis Si3 is inhibitory against a broad range of filamentous fungi (molds) and, to a lesser extent, against spoilage yeasts. We have not found any previous literature reports on the antimicrobial activity of L. coryniformis subsp. coryniformis.
The production of the antifungal substance from L. coryniformis subsp. coryniformis Si3 starts during the exponential growth phase and reaches a maximum early in the stationary phase, after which the activity rapidly decreases. This kinetic is similar to that found for the bacteriocins amylovorin L471 from Lactobacillus amylovorous (5) and Lactocin S from Lactobacillus sake (14). The observed decrease in activity could be caused by proteolytic degradation. Alternatively, the antifungal substance might be a highly hydrophobic molecule that rapidly adsorbs to the producer cells or forms spontaneous aggregates, as has been suggested for the bacteriocins amylovorin L471 and Lactocin S. The addition of ethanol to the growing culture increased the recovery of antifungal activity and prevented the decline during the stationary phase. Similar results have been found with bacteriocins from L. amylovorus and L. sake (5, 14), while Nilsen et al. (16) found that the presence of ethanol was inhibitory to bacteriocin production from Enterococcus faecium. We also observed that the recovery of antifungal activity was increased by addition of formic or acetic acid to the fermentation medium after cessation of growth (data not shown). Similarly, De Vuyst et al. (5) found that bacteriocin inactivation, ascribed to protein aggregation and adsorption, could be overcome by switching the pH to 2.0 after it had reached the activity peak during a fermentation run.
Initially, we used a dual-culture agar system to evaluate the antifungal effects. The pH value in the inhibition zone was ca. 4.6 to 4.7, suggesting a limited contribution of undissociated lactic acid to the inhibitory effect. However, the observed reduction in antifungal activity of the culture filtrates at pH values exceeding 4.5 indicates synergistic effects between lactic acid and other antifungal compounds. On the other hand, the production of antifungals in liquid culture was 10 times higher at pH 6.5 than at pH 4.5. The possibility of increased desorption of antifungal compounds from bacterial cells at very low pH values suggested above further indicates a very complex interaction between the antifungal effects of L. coryniformis subsp. coryniformis Si3 and the pH.
Gel filtration chromatography indicates that the inhibitory substance(s) has a molecular mass of about 3 kDa. The active antifungal substance(s) was thus found to be small, heat stable, sensitive to proteolytic enzymes, and active within a narrow pH range. The same characteristic can be found among bacteriocins of subclass II (11). A substantial proportion of the antifungal activity was lost during each individual purification step. The activity was consistently detectable after two purification steps, regardless of the combination used. However, after a third purification step the activity was often below the detection level. We also observed a splitting of the activity into at least two different active fractions during several of the purification procedures.
The poor stability of the antifungal activity at reduced temperatures further complicates the purification process. The loss of activity after storage at −28°C for only 2 days might be due to an irreversible precipitation-denaturation process. We have also observed a loss of antifungal activity during unintentional thawing of culture filtrate during the freeze-drying procedure.
This is the first report of the production of proteinaceous antifungal compound(s), or indeed of any antimicrobial activity, from a L. coryniformis subsp. coryniformis strain. The type strain L. coryniformis subsp. coryniformis ATCC 25602 had virtually no inhibitory activity against A. fumigatus compared with our strain Si3. We are presently investigating a number of L. coryniformis subsp. coryniformis strains to establish the occurrence of antifungal properties within this species. The possibility of using LAB with GRAS (generally regarded as safe) status as a biotechnological solution to fungal spoilage and mycotoxin formation is a promising option for both the food industry and the agricultural sector.
ACKNOWLEDGMENTS
This study has been financed by MISTRA (The Swedish foundation for Strategic Environmental Research).
We thank Bo Ek for advice on protein purification and Lars Axelsson and Hans Jonsson for helpful comments on the manuscript. Stefan Roos assisted in the species identification and gave valuable suggestions for manuscript improvements.
REFERENCES
- 1.Axelsson L, Lindgren S. Characterization and DNA homology of Lactobacillus strains isolated from pig intestine. J Appl Bacteriol. 1987;62:433–438. doi: 10.1111/j.1365-2672.1987.tb02673.x. [DOI] [PubMed] [Google Scholar]
- 2.Batish V K, Roy U, Lal R, Grover S. Antifungal attributes of lactic acid bacteria. Crit Rev Biotechnol. 1997;17:209–225. doi: 10.3109/07388559709146614. [DOI] [PubMed] [Google Scholar]
- 3.Callewaert R, Holo H, Devreese B, Van Beeumen J, Nes I F, De Vuyst L. Characterization and production of amylovorin L471, a bacteriocin purified from Lactobacillus amylovorus DCE 471 by a novel three-step method. Microbiology. 1999;145:2559–2568. doi: 10.1099/00221287-145-9-2559. [DOI] [PubMed] [Google Scholar]
- 4.Corsetti A, Gobbetti M, Rossi J, Damiani P. Antimould activity of sourdough lactic acid bacteria: identification of a mixture of organic acids produced by Lactobacillus sanfrancisco CB1. Appl Microb Biotechnol. 1998;50:253–256. doi: 10.1007/s002530051285. [DOI] [PubMed] [Google Scholar]
- 5.De Vuyst L, Callewaert R, Crabbé K. Primary metabolite kinetics of bacteriocin biosynthesis by Lactobacillus amylovorus and evidence for stimulation of bacteriocin production under unfavourable conditions. Microbiology. 1996;142:817–827. doi: 10.1099/00221287-142-4-817. [DOI] [PubMed] [Google Scholar]
- 6.Dodd H M, Gasson M J. Bacteriocins of lactic acid bacteria. In: Gasson M J, De Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. London, England: Blackie Academic & Professional; 1994. pp. 211–251. [Google Scholar]
- 7.El-Gendy S M, Marth E H. Growth and aflatoxin production by Aspergillus parasiticus in the presence of Lactobacillus casei. J Food Prot. 1981;44:211–212. doi: 10.4315/0362-028X-44.3.211. [DOI] [PubMed] [Google Scholar]
- 8.Filtenborg O, Frisvad J C, Thrane U. Moulds in food spoilage. Int J Food Microbiol. 1996;33:85–102. doi: 10.1016/0168-1605(96)01153-1. [DOI] [PubMed] [Google Scholar]
- 9.Gourama H, Bullerman L B. Inhibition of growth and aflatoxin production of Aspergillus flavus by Lactobacillus species. J Food Prot. 1995;58:1249–1256. doi: 10.4315/0362-028X-58.11.1249. [DOI] [PubMed] [Google Scholar]
- 10.Kandler O, Weiss N. Genus Lactobacillus Beijerinck 1901, 212AL. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey's manual of systematic bacteriology. 10th ed. Baltimore, Md: Williams & Wilkins; 1986. pp. 1209–1234. [Google Scholar]
- 11.Klaenhammer T R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–85. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 12.Lavermicocca P, Valerio F, Evidente A, Lazzaroni S, Corsetti A, Gobbetti M. Purification and characterization of novel antifungal compounds from the sourdough Lactobacillus plantarum strain 21B. Appl Environ Microbiol. 2000;66:4048–4090. doi: 10.1128/aem.66.9.4084-4090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindgren S E, Dobrogosz W J. Antagonistic activities of lactic acid bacteria in food and feed fermentations. FEMS Microbiol Rev. 1990;87:149–164. doi: 10.1111/j.1574-6968.1990.tb04885.x. [DOI] [PubMed] [Google Scholar]
- 14.Mørtvedt-Abildgaard C I, Nissen-Meyer J, Jelle B, Grenov B, Skaugen M, Nes I F. Production and pH-dependent bactericidal activity of lactocin S, a lantibiotic from Lactobacillus sake L45. Appl Environ Microbiol. 1995;61:175–179. doi: 10.1128/aem.61.1.175-179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niku-Paavola M-L, Laitila A, Mattila-Sandholm T, Haikara A. New types of antimicrobial compounds produced by Lactobacillus plantarum. J Appl Microbiol. 1999;86:29–35. doi: 10.1046/j.1365-2672.1999.00632.x. [DOI] [PubMed] [Google Scholar]
- 16.Nilsen T, Nes I F, Holo H. An exported inducer peptide regulates bacteriocin production in Enterococcus faecium CTC492. J Bacteriol. 1998;180:1848–1854. doi: 10.1128/jb.180.7.1848-1854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nissen-Meyer J, Nes I F. Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch Microbiol. 1997;167:67–77. [PubMed] [Google Scholar]
- 18.Okkers D J, Dicks L M T, Silvester M, Joubert J J, Odendaal H J. Characterization of pentocin TV35b, a bacteriocin-like peptide isolate from Lactobacillus pentosus with fungistatic effect on Candida albicans. J Appl Microbiol. 1999;87:726–734. doi: 10.1046/j.1365-2672.1999.00918.x. [DOI] [PubMed] [Google Scholar]
- 19.Pitt J I, Hocking A D. Fungi and food spoilage. New York, N.Y: Chapman & Hall; 1997. [Google Scholar]
- 20.Stiles E M. Biopreservation by lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:331–345. doi: 10.1007/BF00395940. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki I, Nomura M, Morachi T. Isolation of lactic acid bacteria which suppress mold growth and show antifungal action. Milchwissenschaften. 1991;46:635–639. [Google Scholar]
- 22.Vandenbergh, P. A. April 1993. Process for producing novel yeast and mould inhibiting products. European patent 0 302 300 B1.
- 23.Weizenegger M, Neumann M, Stackebrandt E, Weiss N, Ludwig W. Eubacterium alactolyticum phylogenetically groups with Eubacterium limosum, Acetobacterium woodii and Clostridium barkeri. Syst Appl Microbiol. 1992;15:32–36. [Google Scholar]