Abstract
Objectives
To describe Delta/Omicron SARS-CoV-2 variants co-infection detection and confirmation during the fifth wave of COVID-19 pandemics in France in 7 immunocompetent and epidemiologically unrelated patients.
Methods
Since December 2021, the surveillance of Delta/Omicron SARS-CoV-2 variants of concern (VOC) circulation was performed through prospective screening of positive-samples using single nucleotide polymorphism (SNP) PCR assays targeting SARS-CoV-2 S-gene mutations K417N (Omicron specific) and L452R (Delta specific). Samples showing unexpected mutational profiles were further submitted to whole genome sequencing (WGS) using three different primer sets.
Results
Between weeks 49-2021 and 02-2022, SARS-CoV-2 genome was detected in 3831 respiratory samples, of which 3237 (84.5%) were screened for VOC specific SNPs. Unexpected mutation profiles suggesting a dual Delta/Omicron population were observed in 7 nasopharyngeal samples (0.2%). These co-infections were confirmed by WGS. For 2 patients, the sequence analyses of longitudinal samples collected 7 to 11 days apart showed that Delta or Omicron can outcompete the other variant during dual infection. Additionally, for one of these samples, a recombination event between Delta and Omicron was detected.
Conclusions
This work demonstrates that SARS-CoV-2 Delta/Omicron co-infections are not rare in high virus co-circulation periods. Moreover, co-infections can further lead to genetic recombination which may generate new chimeric variants with unpredictable epidemic or pathogenic properties that could represent a serious health threat.
Keywords: Delta and Omicron VOC, Recombination, SARS-CoV-2 co-infection, Single nucleotide polymorphism screening, Whole genome sequencing
Graphical abstract
Screening was performed with the TaqMan™ SARS-CoV-2 mutation panel molecular assay (ThermoFisher Scientific, Massachusetts, USA). For each mutation of interest, probes labelled with VIC or FAM detect the wild type (WT) and the mutated sequence, respectively. WT samples are represented as red dots along the x-axis and mutated samples as blue dots along the y-axis. A dual population is suspected by intermediate mutation profile with detection of WT and mutated sequences in the same sample and is represented by green dots. The figure at the bottom represents the variant allele frequency clade-defining mutations for 21J Delta and 21K Omicron along the SARS-CoV-2 genome for one patient (P6). Mutations are represented with the variant allele frequency on the first Y axis and the depth of each genomic position on the secondary Y axis. Recombination points are indicated by blue arrows and were confirmed by Single Molecule Real Time sequencing.Abbreviations: SNP, Single Nucleotide Polymorphism
Introduction
In Europe, as elsewhere, emergence of the Omicron Variant of Concern (VOC) of SARS-CoV-2, characterised by a significant growth advantage and potential immune escape, occurred while the Delta VOC was the most common variant, which it rapidly displaced after a 2 months-period of intense viral co-circulation [1]. Co-infection with different SARS-COV-2 lineages was only rarely reported during the precedent epidemic waves [[2], [3], [4], [5]]. To detect and investigate co-infection is important as it is the prerequisite for genetic recombination event, a widespread evolutionary mechanism among RNA viruses including coronaviruses [[6], [7], [8]], which can generate new variant with unpredictable properties. Here, we report Omicron/Delta co-infection in 7 immunocompetent and epidemiologically unrelated patients. We also aimed to provide insights to ease the detection and the confirmation of SARS-CoV-2 dual infections.
Methods
Detection of positive samples and VOC screening by RT-PCR
After RNA extraction (MGI-SP960 platform, BGI, Shenzhen, China), SARS-CoV-2 was detected in respiratory samples by RT-PCR with the TaqPath™ COVID-19 RT-PCR-Kit (ThermoFisher-Scientific, Massachusetts, USA), according to the manufacturer's instructions. SARS-CoV-2 positive samples were further screened for VOC-specific amino acid substitution by targeting the single nucleotide polymorphisms (SNPs) S:L452R and S:K417N with the TaqMan™ SARS-CoV-2 mutation panel molecular assay (ThermoFisher-Scientific) to rapidly detect 21I/J-Delta and 21K/L-Omicron, respectively. Amplification curves and genotyping results were analysed with the QuantStudio Design and Analysis 2.4 software which discriminates between wild type and mutated samples.
Whole genome sequencing (WGS)
All samples underwent WGS using the COVIDSeq-Test™ using the Artic v3 and v4 primer sets (Illumina, San Diego, USA) as detailed in supplementary data. The proportion estimations of each population were assessed by the VAF at the genomic position 23604 (S:P681) which differs between Omicron (23604A; S:P681H) and Delta (23604G, S:P681R), and the average of the VAF clade-defining mutations. The relevance of low frequency clade-defining mutations was verified on the Integrative-Genomics-Viewer (IGV) for all samples. Raw reads were deposited in the Sequence Read Archive database (reference PRJNA809680).
Ethical statement
This study was based on local surveillance data. Informed consent was obtained from all patients. The study was approved by the review committee of the University Hospital of Clermont-Ferrand, France 2022/CE06.
Results
Co-infection delta/omicron detection
The 21K-Omicron, first detected at the university hospital of Clermont-Ferrand on week 49-2021, co-circulated with Delta, and their respective proportions have varied inversely since (Supplementary Figure S1). Between weeks 49-2021 and 02-2022, SARS-CoV-2 was detected in 3831 respiratory samples, of which 3237 (84.5%) were screened for VOC specific SNPs. Unexpected mutation profiles suggesting a dual Delta/Omicron population (detection of both S:K417N and S:L452R mutations) were observed in 7 nasopharyngeal samples (0.2%) (Fig. 1 ).
Fig. 1.
Screening profiles for VOC-specific amino-acid substitution S:L452R and S:K417N suggestive of a dual population Screening was performed with the TaqMan™ SARS-CoV-2 mutation panel molecular assay (ThermoFisher Scientific, Massachusetts, USA). For each mutation of interest, probes labelled with VIC or FAM detect the wild type (WT) and the mutated sequence, respectively. WT samples are represented as red dots along the x-axis and mutated samples as blue dots along the y-axis. A dual population is suspected by intermediate mutation profile with detection of WT and mutated sequences in the same sample and is represented by green dots. Abbreviations: SNP, Single Nucleotide Polymorphismcreening profiles for VOC-specific amino-acid substitution S:L452R and S:K417N suggestive of a dual population. Screening was performed with the TaqManÔ SARS-CoV-2 mutation panel molecular assay (ThermoFisher Scientific, Massachusetts, USA). For each mutation of interest, probes labelled with VIC or FAM detect the wild type (WT) and the mutated sequence, respectively. WT samples are represented as red dots along the x-axis and mutated samples as blue dots along the y-axis. A dual population is suspected by intermediate mutation profile with detection of WT and mutated sequences in the same sample and is represented by green dots. Abbreviations: SNP, Single Nucleotide Polymorphism
Clinical characteristics of patients with suspected co-infection delta/omicron
The seven patients were immunocompetent adults. Six were outpatients under 35 years old presenting with mild (n = 5) or asymptomatic (n = 1) infection; all but one had received 2 or 3 doses of vaccine. The patient P6 was over 70 years old, unvaccinated and required hospitalisation for hypoxemic respiratory failure 9 days after diagnosis infection (Supplementary Table 1). Follow-up samples collected 11 (D11) and 7 days (D7) after the initial diagnosis (D1 samples) were obtained for patients P1 and P5, respectively and were analysed by the same methods.
Confirmation of co-infections by whole genome sequencing
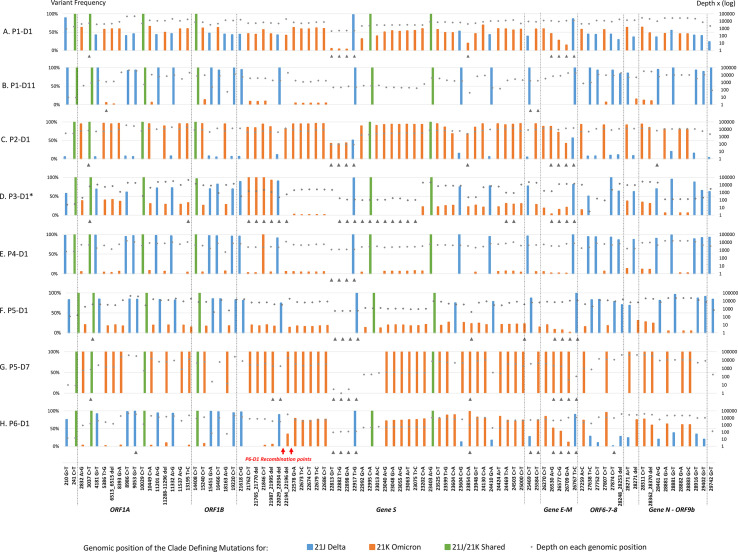
Co-infection was confirmed for 6/7 D1 samples, with both Delta and Omicron being detected in varying proportions (Supplementary Table 1). The distribution of VAF defining mutations of both variants was homogenous over the whole genome except for P6, which had predominant VAF Omicron mutations in the 5'part of the genome (Fig. 2 ). WGS results for patients P1, P5 and P6 were confirmed with another WGS method using independent RNA extraction and xGen™ SARS-CoV-2-Midnight-1200-Amplicon (IDT) sequencing on an Illumina Platform (Supplementary Figure S2).
Fig. 2.
Representation of variant allele frequency clade-defining mutations for 21J Delta and 21K Omicron along the SARS-CoV-2 genome. All 21J and 21K clade defining mutations along the genome are presented for P1-D1 (A) and P1-D7 (B), P5-D1 (C) and P5-D7 (D) and P6-D1 (E). Mutations are represented with the variant allele frequency on the first Y axis and the depth of each genomic position on the secondary Y axis. If the mutation was not present in the vcf file, the value was set at zero. The black arrows indicate clade defining mutations or deletions excluded for the proportion’s estimation of co-infection (no more than 13 per sample for the artic v4). P6-D1 recombination points were confirmed by Single Molecule Real Time sequencing.
Detection of a delta/omicron recombinant
IGV analysis of read pairs in the S gene of P6 sample showed 70 individual DNA fragments carrying both Delta and Omicron clade-defining mutations suggesting a recombination event between the genomic deletions 22029_22034 (Delta) and 22194_22196 (Omicron) (Supplementary Figure S3). Additionally, P6 was sequenced in the spike-region using the Pacific-Biosciences Single Molecule Real Time (SMRT) as previously described [9]. Three haplotypes were detected including two Delta-Omicron recombinant variants (R1 and R2) with distinct breaking points: R1 at positions 22035-22193 (51%), R2 at positions 22218-22586 (29%) and one Delta variant (20%) (Supplementary Figure S4).
Evolution of the co-infection
For P1-D11 (collected 11 days after the first detection), SNP screening suggested the presence of Delta only. WGS showed a proportion of Delta-defining mutations at 96% with a low proportion of Omicron-defining mutations while at D1 proportions of Delta and Omicron were equivalent. For this patient, Delta outcompeted Omicron at D11. For P5-D7, SNP screening and WGS showed the presence only of Omicron (L452L/K471K) while at D1, Delta accounted for 80% of the co-infection. These two follow-up samples showed no divergence neither in the number or the percentage of defining mutations all along the genome, suggesting that no recombination occurred.
Discussion
This work shows that co-infection with two SARS-CoV-2 can occur, particularly during a major epidemic wave marked by a co-circulation of two highly transmissible variants in the same region as in December 2021 in France [10]. Few cases of SARS-CoV-2 co-infections have been reported [[2], [3], [4], [5]]. A recent study performed on 15,253 samples during the co-circulation of Delta and Omicron estimated the prevalence of such co-infection at 0.18% (28/15,523), similarly to the rate we observed [11]. The frequency of co-infections is rare but probably underestimated, as dual infection is difficult to detect. The use of specific RT-PCR assays targeting SNP is an efficient strategy for rapid detection of circulating SARS-CoV-2 but can detect co-infection only if SNPs of different variants are simultaneously screened and if the allele discrimination plots are carefully analysed. In our study, Nextclade assigned a clade from consensus sequences for all sequenced D1 samples but reported the presence of additional mutations as shown by high private mutation scores. Although this score lacks specificity to detect co-infections, it could be a warning to prompt a careful analysis of WGS data. The number and prevalence of low-frequency variants for clade-defining mutations can be used to detect co-infection but their determination is not systematically implemented in pipelines currently used in diagnostic laboratories [11].
The clinical presentation and vaccination status of co-infected patients were unremarkable although this has been previously reported that co-infections could lead to a more severe or longer disease [3]. Analysis of follow-up samples showed that Delta or Omicron can outcompete each other, as previously reported but only in immunocompromised patients [12]. Complementary analyses, such as neutralizing antibody levels against both variants, could help to better understand if the ability of variants to evade immunity after vaccination could explain why one variant outcompetes the other after co-infection.
Our study is monocentric which could be a limitation. However, it relied on prospective, systematic and homogeneous screening of positive samples for SNPs, independently of the immune status of patients. The detection of co-infections through multicentre studies might have been more difficult because of the heterogeneity in VOC screening strategies and methods between laboratories and countries.
The evidence of Delta/Omicron co-infections makes it possible for recombination events to occur. The presence of recombination for P6 was confirmed by SMRT which identified two breaking points in the spike-region. A native Delta haplotype was also detected. The presence of native Delta and recombinant fragments suggests that the recombination was under selection at the sampling date. Within host recombination of Delta and Omicron genomes was reported in a recent study [13]. Similarly, the breakpoint was localised at the 5′ end of the spike-region between positions 22,036 and 22,193 suggesting that this region could be a hot spot for recombination events. Genetic recombination may generate new variants with unpredictable epidemic or pathogenic properties, especially in the case of Delta-Omicron recombinants as these VOC are each characterised by the capacity to cause severe disease or by a higher transmissibility and immune escape, respectively. This underlines the need for continuous efforts to maintain an effective genomic surveillance of SARS-CoV2 infections. Moreover, a re-enforced attention to whole genome data beyond the consensus genome sequence is essential to improve detection of emerging variants co-infection and recombinants.
Transparency declaration
The authors declare that they have no conflicts of interest.
Funding statement
No special funding was received for the study.
Author's contributions
PC, MB and PM were involved in the screening of single nucleotide polymorphism of Delta and Omicron variant and in the whole genome sequencing analysis. CH, CA, AB, HC, CR, AM and JL participated in the analyses of clinical samples. LJ, AB, GD and BS were involved in the confirmation of the co-infection with the second WGS method (xGen™ SARS-CoV-2-Midnight-1200 Amplicon (IDT)) and the analysis of the data sequencing. JI and JL were involved in the confirmation of the recombination event for the P6 sample using the Single Molecule Real Time sequencing method (Pacific Biosciences). PC coordinated the data collection and analysis. PC, AM and CH wrote the first draft of the manuscript. All authors supervised the study progress and contributed to the manuscript and approved the final version.
Acknowledgments
We thank all the technicians for excellent technical assistance. We are grateful to Jeff Watts for revision of the English manuscript.
Editor: L. Kaiser
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2022.06.030.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Assessment of the further spread and potential impact of the SARS-CoV-2 Omicron variant of concern in the EU/EEA. European Centre for Disease Prevention and Control; 2022. https://www.ecdc.europa.eu/en/publications-data/covid-19-omicron-risk-assessment-further-emergence-and-potential-impact 19th update [Internet] [cited 2022 Feb 17]. Available from: [Google Scholar]
- 2.Samoilov A.E., Kaptelova V.V., Bukharina A.Y., Shipulina O.Y., Korneenko E.V., Saenko S.S., et al. Case report: change of dominant strain during dual SARS-CoV-2 infection. BMC Infect Dis. 2021;21:959. doi: 10.1186/s12879-021-06664-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pedro N., Silva C.N., Magalhães A.C., Cavadas B., Rocha A.M., Moreira A.C., et al. Dynamics of a dual SARS-CoV-2 lineage Co-infection on a prolonged viral shedding COVID-19 case: insights into clinical severity and disease duration. Microorganisms. 2021;9:300. doi: 10.3390/microorganisms9020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosch S., Mpina M., Nyakurungu E., Borico N.S., Obama T.M.A., Ovona M.C., et al. Genomic surveillance enables the identification of Co-infections with multiple SARS-CoV-2 lineages in Equatorial Guinea. Front Public Health. 2021;9 doi: 10.3389/fpubh.2021.818401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francisco Junior R. da S., de Almeida L.G.P., Lamarca A.P., Cavalcante L., Martins Y., Gerber A.L., et al. Emergence of within-host SARS-CoV-2 recombinant genome after coinfection by gamma and Delta variants: a case report. Front Public Health. 2022;10 doi: 10.3389/fpubh.2022.849978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pérez-Losada M., Arenas M., Galán J.C., Palero F., González-Candelas F. Recombination in viruses: mechanisms, methods of study, and evolutionary consequences. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2015;30:296–307. doi: 10.1016/j.meegid.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson B., Boni M.F., Bull M.J., Colleran A., Colquhoun R.M., Darby A.C., et al. Generation and transmission of interlineage recombinants in the SARS-CoV-2 pandemic. Cell. 2021;184:5179–5188. doi: 10.1016/j.cell.2021.08.014. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vellas C., Del Bello A., Debard A., Steinmeyer Z., Tribaudeau L., Ranger N., et al. Influence of treatment with neutralizing monoclonal antibodies on the SARS-CoV-2 nasopharyngeal load and quasispecies. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2022;28:139.e5–139.e8. doi: 10.1016/j.cmi.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coronavirus : circulation des variants du SARS-CoV-2. https://www.santepubliquefrance.fr/dossiers/coronavirus-covid-19/coronavirus-circulation-des-variants-du-sars-cov-2 [Internet]. [cited 2022 Feb 28]. Available from:
- 11.Bal A., Simon B., Destras G., Chalvignac R., Semanas Q., Oblette A., et al. France; 2021. 2022. Detection and prevalence of SARS-CoV-2 co-infections during the Omicron variant circulation.https://www.medrxiv.org/content/10.1101/2022.03.24.22272871v1 [Internet]. medRxiv; 2022 [cited 2022 Apr 27]. p. 2022.03.24.22272871. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rockett R.J., Draper J., Gall M., Sim E.M., Arnott A., Agius J.E., et al. Co-Infection with SARS-COV-2 Omicron and Delta variants revealed by genomic surveillance. 2022. https://www.medrxiv.org/content/10.1101/2022.02.13.22270755v1 [Internet]. medRxiv; [cited 2022 Feb 18]. p. 2022.02.13.22270755. Available from: [DOI] [PMC free article] [PubMed]
- 13.Evidence for SARS-CoV-2 Delta and Omicron co-infections and recombination. https://www.medrxiv.org/content/10.1101/2022.03.09.22272113v2 medRxiv [Internet]. [cited 2022 Apr 14]. Available from: [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.