Abstract
The development of methods to reduce costs associated with the solubilization of cellulose is essential for the utilization of lignocellulose as a renewable feedstock for fuels and chemicals. One promising approach is the genetic engineering of ethanol-producing microorganisms that also produce cellulase enzymes during fermentation. By starting with an ethanologenic derivative (strain P2) of Klebsiella oxytoca M5A1 with the native ability to metabolize cellobiose, the need for supplemental β-glucosidase was previously eliminated. In the current study, this approach has been extended by adding genes encoding endoglucanase activities. Genes celY and celZ from Erwinia chrysanthemi have been functionally integrated into the chromosome of P2 using surrogate promoters from Zymomonas mobilis for expression. Both were secreted into the extracellular milieu, producing more than 20,000 endoglucanase units (carboxymethyl cellulase activity) per liter of fermentation broth. During the fermentation of crystalline cellulose with low levels of commercial cellulases of fungal origin, these new strains produced up to 22% more ethanol than unmodified P2. Most of the beneficial contribution was attributed to CelY rather than to CelZ. These results suggest that fungal enzymes with substrate profiles resembling CelY (preference for long-chain polymers and lack of activity on soluble cello-oligosaccharides of two to five glucosyl residues) may be limiting in commercial cellulase preparations.
The production of fuels and chemicals from cellulosic substrates using microbial biocatalysts offers the potential to reduce the use of fossil fuels and improve the environment (11, 18, 22, 29–31). However, the low activity of cellulase enzymes (15, 30) and the resulting cost of hydrolysis represent major barriers for the use of lignocellulosic feedstocks for fuels, bulk chemicals, and plastics (15, 29, 30, 41). The enzymatic hydrolysis of cellulose has been extensively studied but remains poorly understood (2, 3, 5, 21, 32). Hydrolysis results from the combined action of at least three classes of β-1,4-glucanase activities (2, 5, 26, 28): endoglucanases, exoglucanases, and cellobiases that complete the hydrolysis of soluble products (from two to six glucosyl residues) to monomeric glucose. Cellobiose and soluble cellobiosides are potent competitive inhibitors of endo- and exoglucanases that must be removed to prevent autoinhibition. Fungi such as Trichoderma reesei (19, 27, 39) secrete soluble β-1,4-glucosidases (cellobiase) to complete the hydrolysis process. In some bacteria, however, hydrolysis of soluble cellobiosides is completed intracellularly. Cellobiose and cellotriose are actively transported by a β-glucoside-specific phosphoenolpyruvate-dependent phosphotransferase system (PTS) (18, 20, 40). Intracellular cellobiose-phosphate is subsequently hydrolyzed to glucose and glucose-6-phosphate by a cytoplasmic phospho-β-glucosidase for immediate entry into glycolysis.
One approach to reduce the costs of cellulase enzymes for bioprocessing is to develop ethanologenic biocatalysts that provide a portion of the cellulolytic activity. This approach is being pursued with several naturally ethanologenic microorganisms such as Saccharomyces cerevisiae (8, 9, 33, 34), Zymomonas mobilis (7, 24, 31), and cellulolytic bacteria such as Clostridium thermocellum and C. thermosaccharolyticum (16, 35). Our laboratory is pursuing an alternative approach: the addition of cellulase activities to enteric bacteria (Escherichia coli B and Klebsiella oxytoca M5A1) that have been previously engineered to produce ethanol by adding the Z. mobilis pdc and adhB genes encoding the pyruvate-to-ethanol pathway (18). K. oxytoca M5A1 contains a native cellobiose-specific PTS and phospho-β-glucosidase (20, 25, 40), eliminating the need for extracellular β-glucosidase. The K. oxytoca operon containing the cellobiose utilization genes (casAB) has been cloned (20) and expressed in ethanologenic E. coli KO11 (18, 25) to construct a second, analogous biocatalyst. Subsequent studies have overexpressed the celZ endoglucanase (CelZ; formerly EGZ) from the plant saprophyte, Erwinia chrysanthemi (6, 10), in both K. oxytoca M5A1 (42) and E. coli B (44). By adding accessory genes (out genes) from E. chrysanthemi, high levels of endoglucanase activity were effectively secreted into the extracellular milieu by both organisms (13, 42, 44).
CelZ represents 95% of the total carboxymethyl cellulase (CMCase) activity in E. chrysanthemi (6). The remaining 5% is attributed to a second endoglucanase, CelY (formerly EGY). The celY gene has also been cloned and sequenced (13). Recent investigations have shown that CelY and CelZ differ in substrate preference (43). When mixed at ratios similar to those produced by cultures of E. chrysanthemi, these two enzymes function synergistically, indicating a potential need for both enzymes for optimal cellulose hydrolysis during fermentation.
In the current study, we have integrated the celY and celZ genes from E. chrysanthemi into the chromosome of K. oxytoca P2, an ethanologenic derivative of M5A1 (40). The secreted cellulase enzymes functioned together with commercial fungal cellulase to increase the production of ethanol during the simultaneous saccharification and fermentation (SSF) of crystalline cellulose.
MATERIALS AND METHODS
Bacteria, plasmids, and culture conditions.
Strains and plasmids used in this study are listed in Table 1. E. coli DH5α and TOPO10F′ were used as hosts during plasmid constructions. The celZ gene was cloned from E. chrysanthemi P86021 (42, 44). The celY gene was cloned by Guiseppi et al. (13) from E. chrysanthemi 3937. The out genes were cloned by He et al. (14) from E. chrysanthemi EC16.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | lacZΔM15 recA | Bethesda Research Laboratory |
| TOP10F′ | hsdR mcrA lacZΔM15 endA recA; F′ tet lacI | Invitrogen |
| HB101 | recA lacY | ATCC 37159 |
| S17-1 | thi pro recA hsdR RP4-2-tet::Mu aphA::Tn7 λpir | 12 |
| Z. mobilis CP4 | Prototrophic | 18 |
| K. oxytoca | ||
| M5A1 | Prototrophic | 40 |
| P2 | pfl::pdc adhB cat | 40 |
| SZ6 | pfl::pdc adhB cat; integrated celZ tet | 42 |
| SZ12 | pfl::pdc adhB cat; integrated celZ celY kan | This study |
| SZ21 | pfl::pdc adhB cat; integrated celZ celY | This study |
| SZ22 | pfl::pdc adhB cat; integrated celY celZ::aac | This study |
| Plasmids | ||
| pUC18 | bla cloning vector | New England Biolabs |
| pUC19 | bla cloning vector | New England Biolabs |
| pCR2.1-TOPO | TA cloning vector, bla kan | Invitrogen |
| pMH18 | bla celY from E. chrysanthemi 3937 | 13 |
| pHPΩ45aac | bla aac source of apramycin gene | 4 |
| pBR322 | bla tet cloning vector | New England Biolabs |
| pRK2013 | kan, mobilizing plasmid | American Type Culture Collection |
| pCPP2006 | spm, ∼40-kbp fragment containing out genes from E. chrysanthemi EC16 | 14 |
| pFT-A | bla flp low-copy vector containing recombinase and temperature-conditional pSC101 replicon | 23 |
| pLOI2224 | kan integration vector containing conditional R6K replicon and two FRT sites | 23 |
| pLOI2302 | pUC19 containing AscI linkers inserted into blunt NdeI and SapI sites | 42 |
| pLOI2307 | bla celZ gene and a surrogate promoter from Z. mobilis DNA | 44 |
| pLOI2311 | PCR fragment containing celY gene cloned into pCR2.1-TOPO, expressed from lac promoter | 43 |
| pLOI2316 | pUC18 containing the celY gene on a Klenow-treated EcoRI fragment from pLOI2311 inserted into a HincII site; celY expressed from the lac promoter | This study |
| pLOI2317 | EcoRI-HindIII fragment from pLOI2316 inserted into the corresponding sites of pLOI2302 | This study |
| pLOI2318 | Sau3A1 fragment of Z. mobilis DNA exhibiting promoter activity inserted into the BamHI site of pLOI2317 | This study |
| pLOI2319 | Sau3A1 fragment of Z. mobilis DNA exhibiting promoter activity inserted into the BamHI site of pLOI2317 | This study |
| pLOI2320 | Sau3A1 fragment of Z. mobilis DNA exhibiting promoter activity inserted into the BamHI site of pLOI2317 | This study |
| pLOI2323 | Sau3AI fragment of Z. mobilis DNA exhibiting promoter activity inserted into the BamHI site of pLOI2317 | This study |
| pLOI2342 | Sau3A1 fragment of Z. mobilis DNA exhibiting promoter activity inserted into the BamHI site of pLOI2317 | This study |
| pLOI2348 | Random EcoRI fragment of K. oxytoca M5A1 DNA cloned into EcoRI site of pLOI2323 | This study |
| pLOI2349 | EcoRI linker inserted into the Klenow-treated SphI site of pLOI2307 | This study |
| pLOI2350 | EcoRI fragment (celZ and surrogate promoter) from pLOI2349 inserted into the EcoRI site of pLOI2224 | This study |
| pLOI2352 | AscI fragment (K. oxytoca fragment, Z. mobilis promoter fragment and celY) from pLOI2348 inserted into the AscI site of pLOI2350 | This study |
| pLOI2353 | EcoRI-AvaI fragment (tet gene) from pBR322 inserted into the ClaI site of pFT-A | This study |
| pLOI2354 | pUC19 derivative in which the multiple cloning sites from HindIII to SmaI were deleted by digestion, Klenow treatment, and self-ligation | This study |
| pLOI2355 | EcoRI fragment (celZ gene) from pLOI2349 inserted into the EcoRI site of pLOI2354 | This study |
| pLOI2356 | SmaI fragment containing the apramycin resistance gene (aac gene) from pHPΩ45aac inserted into the T4 DNA polymerase-treated PstI site of pLOI2355, disrupting the celZ gene | This study |
| pLOI2357 | EcoRI fragment (aac and disrupted celZ) from pLOI2356 inserted into the EcoRI site of pLOI2224 | This study |
| pLOI2358 | Subclone of pLOI2323 in which the internal PstI fragment was deleted, used for sequencing | This study |
| pLOI2359 | Subclone of pLOI2323 in which the ClaI-HindIII fragment was deleted, used for sequencing | This study |
E. coli cultures were grown at 37°C in Luria-Bertani (LB) broth (1) containing (per liter) 10 g of Difco tryptone, 5 g of Difco yeast extract, and 5 g of sodium chloride or on solid LB medium containing agar (1.5%). Sugar was always included in broth (5% glucose or sorbitol) and solid media (2% glucose) used for the growth of ethanologenic strains. Clones were screened for endoglucanase production using the Congo red method (38, 40). Endoglucanase indicator plates were prepared by supplementing LB agar with 0.3% low-viscosity carboxymethyl cellulose (CMC). Ampicillin (50 μg/ml), apramycin (100 μg/ml), kanamycin (50 μg/ml), chloramphenicol (40 μg/ml), and spectinomycin (100 μg/ml) were used for selection. Ethanologenic strains of K. oxytoca were maintained at 30°C on solid LB medium containing glucose (2%) and chloramphenicol (600 μg/ml).
Genetic methods.
Standard methods were used for plasmid construction, analyses, and sequencing (1). The ribosome-binding site and promoterless coding region of celY were amplified by the PCR using pMH18 as the template with the following primer pair: N terminus, 5′-CTGTTCCGTTACCAACAC-3′, and C terminus, 5′-GTGAATGGGATCACGAGT-3′. The E. chrysanthemi out genes (pCPP2006) were transferred by conjugation using pRK2013 for mobilization (42). Constructions were confirmed by sequencing using the dideoxy method and a LI-COR Model 4000-L DNA sequencer with fluorescent primers. The E. chrysanthemi celY and celZ genes were introduced into K. oxytoca P2 by electroporation using a Bio-Rad Gene Pulser. Recombinants were selected on solid medium containing kanamycin (50 mg/liter) as previously described (23, 42).
Primer extension analysis.
Promoter regions were identified by mapping the transcriptional start sites using IRD41-labeled fluorescent primers within the coding regions (44): 5′-ACCATCAGCATCAACGCCCAACAACG-3′ for celY and 5′-GACTGGATGGTTATCCGAATAAGAGAGAGG-3′ for celZ. Extension products were dissolved in loading buffer and compared to parallel dideoxy sequences (42) using the LI-COR Model 4000-L DNA sequencer (LI-COR, Inc., Lincoln, Nebr.).
Enzyme assay.
Endoglucanase activity was determined in vitro using 2% CMC as the substrate. Appropriate dilutions of cell-free culture broth (extracellular activity) or broth containing cells that had been disrupted by ultrasound (total activity) were assayed at 35°C in 50 mM citrate buffer (pH 5.2). Reactions were terminated by heating to 100°C for 10 min. Reducing sugars were measured using 3,5-dinitrosalicylic acid reagent with glucose as a standard (42, 44). Endoglucanase activity (CMCase) is expressed as micromoles of reducing sugar per minute (IU). Results are an average of two or more determinations.
Fermentation.
SSF tests were conducted in unbaffled, 500-ml flasks containing 200 ml of broth. Flasks were fitted with a rubber stopper and vented with an 18-gauge needle. Fermentations were conducted at 35°C (120 rpm) in LB medium containing 10% Sigmacell 50 (crystalline cellulose). Inocula were grown for 12 h in LB medium containing 5% glucose. Cells were harvested by centrifugation and resuspended in LB medium. Each flask was inoculated with approximately 64 mg of cells (dry weight).
Materials and chemicals.
Tryptone and yeast extract were products of Difco (Detroit, Mich.). Antibiotics, low-viscosity CMC, and Sigmacell 50 were obtained from the Sigma Chemical Company (St. Louis, Mo.). The IRD41-labeled fluorescent primers were purchased from LI-COR, Inc.
RESULTS
Construction of a promoter-probe vector for celY.
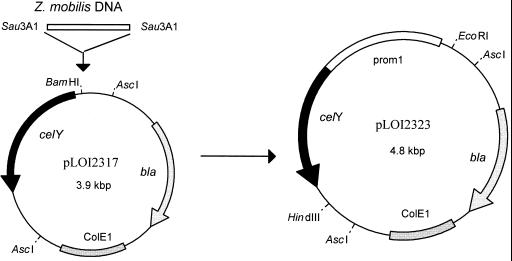
Although the celY gene from E. chrysanthemi was cloned previously, it was poorly expressed in E. coli from the original plasmid (13). To increase expression, a promoter-probe vector was constructed as follows using celY as the reporter (Fig. 1). The promoterless celY coding region with a native ribosome-binding site (1.2 kbp) was amplified by PCR using pMH18 as the template and inserted into the topoisomerase vector, PCR2.1-TOPO. A clone oriented to express celY from the lac promoter was selected and designated pLOI2311 (5.2 kbp). An EcoRI fragment containing the promoterless celY gene was isolated from pLOI2311. The ends of this fragment were blunted using Klenow polymerase prior to ligation into the HincII site of pUC18. A clone oriented to express celY from the lac promoter was selected (3.9 kbp), and expression was confirmed using endoglucanase indicator plates (pLOI2316). The promoterless celY gene was isolated from pLOI2316 as a 1.2-kbp fragment using EcoRI and HindIII and inserted into the corresponding sites of pLOI2302 (pUC19 derivative) to reverse the direction of the celY gene. As expected, the resulting construct DH5α(pLOI2317) was inactive on endoglucanase indicator plates due to the lack of a promoter. To facilitate the insertion of DNA fragments containing promoter regions, plasmid pLOI2317 (3.9 kbp) contains a BamHI site in the polylinker region, immediately upstream from the celY gene (Fig. 1).
FIG. 1.
Construction of promoter-probe vector for celY. Sau3A1 fragments of Z. mobilis chromosomal DNA were ligated into the BamHI site of pLOI2317 to provide a strong, surrogate promoter for the celY coding region (solid segments). The Z. mobilis DNA fragment (promoter 1) is shown as an open segment. Replicons and antibiotic resistance genes are stippled; other vector DNA is shown as thin connecting lines. The arrows indicate the direction of transcription.
Construction of plasmids with increased expression of celY in E. coli DH5α.
Sau3A1 fragments of Z. mobilis chromosomal DNA were used to provide a heterologous promoter that would not be subject to native regulatory mechanisms in K. oxytoca or interfere with subsequent integration into the K. oxytoca chromosome (42). Fragments of 0.5 to 1.5 kbp were isolated and randomly ligated into the BamHI site of pLOI2317 to generate a library of surrogate promoters (Fig. 1). Approximately 7,500 colonies were screened on endoglucanase indicator plates. One-third of the clones actively produced celY. The most active 100 colonies were identified by zone size, purified, and retested. The 30 clones with the largest zones of activity were grown overnight in LB medium and assayed for CMCase activity. The five most active clones are listed in Table 2 and exhibited approximately sevenfold-higher activity than the original clone, pMH18. Plasmid pLOI2323 was selected for further investigation.
TABLE 2.
Expression of celY in E. coli DH5α using Sau3A1 fragments of Z. mobilis chromosomal DNA as surrogate promotersa
| Plasmids expressing celY or celZ | Endoglucanase activity
|
||
|---|---|---|---|
| Extracellular (IU/liter) | Total (IU/liter) | % Extracellular | |
| pMH18 (native celY promoter) | 151 | 184 | 82 |
| pLOI2317 (promoterless celY vector) | 0 | 0 | 0 |
| celY expressed from surrogate promoters | |||
| pLOI2318 | 1,123 | 1,257 | 89 |
| pLOI2319 | 888 | 1,023 | 87 |
| pLOI2320 | 1,023 | 1,056 | 97 |
| pLOI2323 | 1,257 | 1,291 | 97 |
| pLOI2342 | 1,224 | 1,257 | 97 |
| pLOI2349 (celZ) | 3,414 | 16,234 | 21 |
All plasmids are pUC derivatives. Endoglucanase activity was measured using cultures grown at 37°C for approximately 16 h.
The DNA serving as a surrogate promoter in pLOI2323 was sequenced in both directions (GenBank accession no. AF305919). Based on database comparisons, this fragment appears to be derived from two pieces, an 890-bp fragment from the Z. mobilis chromosome that corresponds to a previously sequenced region plus a stray 41-bp fragment from pLOI2311 which was inadvertently ligated during the construction of pLOI2316. This 41-bp fragment alone did not serve as a functional promoter since no activity was observed prior to addition of the Z. mobilis DNA fragment. A BLAST search of the translated Z. mobilis sequence did not reveal identity to known genes. Four putative sites of transcriptional initiation were identified in DH5α(pLOI2323) by primer extension analysis. Upstream sequences share some similarity with three different sigma factors: ς32, ς38, and ς70 (Table 3). Although the differences in intensity were <2-fold, the sequence resembling the ς32 (rpoH) consensus appeared to be the most intense (36, 37).
TABLE 3.
Transcriptional initiation sites and putative promoter regions for the celY promoter in DH5α(pLOI2323)a
| Sequenceb | RNA start | Proposed ς factors | ς Factor consensus sequence
|
|
|---|---|---|---|---|
| −35 | −10 | |||
| ATATTTTTGATTTTTCAAGAAAAGCCTGATATCTTCCAACATCTT | T | ς70 | TTGACA | TATAAT |
| GATTTGATCCTCTAGAGTCAACCTGCTTGTTACTCGTGATCCCAT | A | ς70 | TTGACA | TATAAT |
| GAGTCAACCTGCTTGTTACTCGTGATCCCATTCACAAGGGCGAA | C | ς32 | CTTGAAA | CCCCAT |
| TTACTCGTGATCCCATTCACAAGGGCGAATTAATTCGCCCTT | C | ς38 | CCGCCT | TATACT |
Transcriptional starts for celY were identified by primer extension analysis. Four putative promoters are shown.
Upstream regions with similarity to E. coli −35 (left) and −10 (right) regions are underlined. RNA start sites are shown in boldface.
Construction of a vector for the integration celY and celZ into the chromosome of K. oxytoca P2.
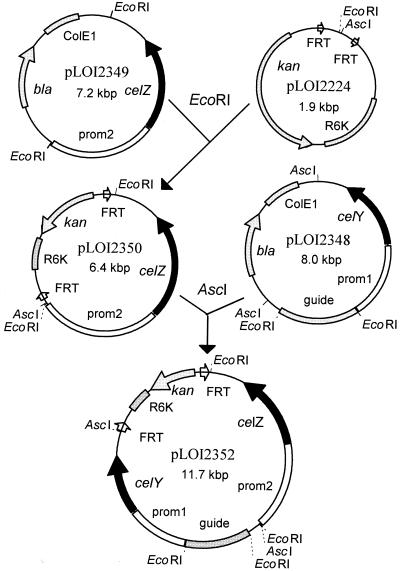
Plasmid pLOI2307 (7.2 kbp) was previously constructed and expressed celZ from a surrogate Z. mobilis promoter at high levels in recombinant E. coli DH5α (44) and K. oxytoca M5A1 (42). To facilitate subcloning of this hybrid celZ gene and promoter (4.5 kbp), an EcoRI linker was inserted into the T4 DNA polymerase-treated SphI site of pLOI2307 to provide flanking EcoRI sites for convenient excision (pLOI2349). Prior to constructing a plasmid containing celY and celZ, a random 3-kbp fragment of EcoRI-digested K. oxytoca M5A1 chromosomal DNA was inserted into pLOI2323 containing celY (and surrogate promoter) to serve as a guide for homologous recombination (pLOI2348; 8 kbp). This 3-kbp M5A1 fragment was partially sequenced and appears to encode the complete M5A1 glgP gene. In pLOI2348 (8 kbp), flanking AscI sites allowed the excision of a single 5.5-kbp fragment containing the M5A1 glgP gene, the Z. mobilis surrogate promoter, and E. chrysanthemi celY.
Figure 2 summarizes the construction of the celY, celZ integration vector from pLOI2349, pLOI2224, and pLOI2348. The recombinant celY and celZ genes containing surrogate promoters and the guide fragment were sequentially inserted into the core integration vector, pLOI2224 (23) using E. coli S17-1 as the host, to produce pLOI2352 (11.7 kbp). The 4.5-kbp EcoRI fragment from pLOI2349 containing celZ and promoter 2 was inserted into pLOI2224 using an EcoRI site to make pLOI2350 (6.4 kbp). The 5.5-kbp AscI fragment from pLOI2348 containing celY, promoter 1, and the guide fragment for integration was inserted into the AscI site of pLOI2350 to make pLOI2352 (11.7 kbp). The fragments containing cel genes were oriented such that expression from the surrogate promoters was divergent. The resulting vector contained an R6K replicon that does not function in DH5α or M5A1. The two FRT sites in pLOI2352 facilitate removal of the kanamycin gene and replicon after integration (23).
FIG. 2.
Construction of pLOI2352 for the functional integration of celY and celZ into the chromosome of ethanologenic K. oxytoca P2. Coding regions for celY and celZ are shown as solid segments. Fragments of Z. mobilis DNA that serve as promoters (prom1 and prom2) are shown as open segments. The K. oxytoca DNA fragment which serves as a guide for chromosomal integration (guide) is stippled. Replicons and antibiotic resistance genes are stippled; other vector DNA is shown as a thin connecting line. The arrows on segments indicate the direction of transcription. The small open arrows represent the FRT sites which are recognized by the flp recombinase. FRT sequences are asymmetrical and are arranged to allow the deletion of plasmid DNA (replicon and selectable marker) after chromosomal integration.
Functional integration of celY and celZ into the K. oxytoca P2 chromosome.
Plasmid pLOI2352 was introduced into P2 by electroporation, followed by selection for kanamycin resistance. Approximately 150 colonies were recovered, and all were positive on endoglucanase indicator plates. Ten clones with the largest zones of activity were purified, grown in broth, and assayed for endoglucanase activity. These produced 5 to 6 IU of endoglucanase activity per ml. One clone was selected for further study and designated SZ12.
Due to the natural resistance of K. oxytoca to ampicillin, an additional antibiotic resistance marker (tet) was added to plasmid pFT-A containing the flp recombinase gene to facilitate selection. The tetracycline gene was isolated as a 1.4-kbp EcoRI-to-AvaI fragment from pBR322. After treatment with Klenow polymerase, this fragment was ligated in the Klenow-treated ClaI site of pFT-A to produce pLOI2353 (7.0 kbp). This plasmid encodes resistance to both ampicillin and tetracycline, the FLP recombinase (flp) under the control of the tetracycline promoter, and a temperature-conditional pSC101 replicon.
Plasmid pLOI2353 was transformed into SZ12 and plated at 30°C with selection for tetracycline resistance. The presence of tetracycline also induced flp expression, resulting in a deletion of the kanamycin gene and the R6K replicon from the chromosomally integrated pLOI2352. Of 307 tetracycline-resistant colonies tested, >99% retained expression of the endoglucanase genes and were sensitive to kanamycin. Clones were purified, grown in broth, and assayed for endoglucanase activity. All were similar, and one was designated SZ21(pLOI2353). The helper plasmid was eliminated from SZ21 by overnight growth at 37°C.
Results from primer extension analysis of celY and celZ in SZ21 were similar to those observed in DH5α. A single major transcriptional start was identified for celZ that corresponded precisely to the most prominent start site in DH5α(pLOI2183) which contains the same promoter fragment (42, 44). DNA immediately upstream from this site resembles the recognition sequence for a ς70 promoter (36, 37). As observed with DH5α(pLOI2323) (Table 3), primer extension analysis of celY indicated the presence of multiple putative transcriptional start sites in SZ21. Although localized in the same regions as the start sites in DH5α(pLOI2323), all bands were of near equal intensities.
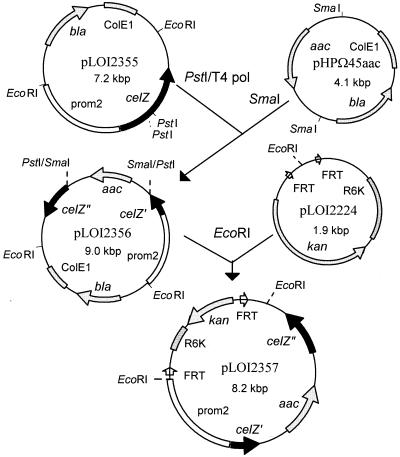
Construction of a celZ knockout mutation.
To confirm the presence of a functional celY in SZ21, a knockout mutation of the chromosomally integrated celZ was constructed by double, homologous recombination using plasmid pLOI2357 (Fig. 3). Plasmid pUC19 was digested with SmaI and HindIII, treated with Klenow polymerase, and self-ligated to eliminate many of the polylinker sites (pLOI2354). The remaining unique EcoRI site was used to insert a 4.5-kbp EcoRI fragment containing the promoter and celZ gene from pLOI2349 to make pLOI2355 (7.2 kbp). The 1.8-kbp SmaI fragment from pHPΩ45aac containing the apramycin resistance gene (aac) was then ligated into the central region of celZ, replacing a small internal PstI fragment (after treatment with T4 DNA polymerase) to produce pLOI2356 (9 kbp). The 6.3-kbp EcoRI fragment from this plasmid was isolated and inserted into the core integration vector, pLOI2224, to produce pLOI2357 (8.2 kbp). This plasmid contains a conditional R6K replicon and kanamycin resistance gene in addition to a celZ gene that is interrupted by an apramycin resistance gene.
FIG. 3.
Construction of pLOI2357 for the inactivation of celZ by double homologous recombination. Coding regions for celY are shown as solid segments. After disruption, the N-terminal and C-terminal segments are indicated by celZ′ and celZ", respectively. The fragment of Z. mobilis DNA that serves as a promoter (prom1) is shown as an open segment. Replicons and antibiotic resistance genes are stippled; other vector DNA is shown as a thin connecting line. The arrows on segments indicate the direction of transcription. The small open arrows represent the FRT sites which are recognized by the flp recombinase.
Plasmid pLOI2357 was electroporated into SZ21 with selection for apramycin. Approximately 10% of the recombinants were apramycin resistant and kanamycin sensitive, indicating a double homologous recombination event. These clones exhibited low levels of endoglucanase production on indicator plates (Table 4). One was selected and designated SZ22. Loss of CelZ and retention of CelY in SZ22 was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using the Pharmacia Phast Gel system (data not shown).
TABLE 4.
Effect of out genes (pCPP2006) on endoglucanase production by derivatives of K. oxytoca P2
| Strain or Spezyme additive | CMC zonea (mm) | OD550b | CMCase activityc
|
||
|---|---|---|---|---|---|
| Extracellular (IU/liter) | Total (IU/liter) | Secretion (%) | |||
| P2 | 0 | 10.5 | 0 | 0 | 0 |
| SZ6 | 8.5 | 11.0 | 1,920 | 8,800 | 22 |
| SZ21 | 6.7 | 11.0 | 1,620 | 7,800 | 21 |
| SZ22 | 2.0 | 10.0 | 480 | 879 | 55 |
| P2(pCPP2006) | 0 | 10.0 | 0 | 0 | 0 |
| SZ6(pCPP2006) | 10.8 | 9.6 | 13,800 | 22,300 | 62 |
| SZ21(pCPP2006) | 11.5 | 10.2 | 20,100 | 26,900 | 75 |
| SZ22(pCPP2006) | 2.0 | 9.7 | 449 | 833 | 54 |
| Spezyme CE (10 ml/liter)d | 27,000 | ||||
| Spezyme CP (10 ml/liter)d | 33,400 | ||||
Diameter of cleared zone on CMC indicator plates.
Culture density after 24 h of incubation (30°C in LB medium containing 5% sorbitol).
Endoglucanase activity was measured using cultures grown for 24 h.
Dilution equivalent to the highest Spezyme level used in fermentation experiments (Table 5).
It is interesting that cell clumping in liquid culture, typical of M5A1 and P2, was eliminated by the functional expression of celZ from integrated genes or from plasmids. Clumping was not affected by the functional expression of celY alone and provides a convenient marker for expression.
Effect of the E. chrysanthemi out genes (pCPP2006) on the extracellular secretion of CelY and CelZ in derivatives of K. oxytoca P2.
Table 4 summarizes the endoglucanase activities exhibited by cellulolytic derivatives of ethanologenic K. oxytoca P2. Strain SZ6 (42) contains a chromosomally integrated hybrid celZ gene with the same promoter fragment used to construct SZ21. Despite the presence of two endoglucanase genes in SZ21, extracellular and total endoglucanase activities were 13% lower in this strain than in SZ6. Most of the endoglucanase activity produced by SZ21 can be attributed to celZ. SZ22, a celZ mutant of SZ21, expressed only 11% of the endoglucanase produced by the parent containing functional celY and celZ genes. In strains SZ6 and SZ21 containing a functional celZ, most of the endoglucanase activity (primarily CelZ) was cell associated. In strain SZ22, containing a functional celY alone, half of the endoglucanase activity was extracellular.
Previous studies have shown that the addition of the out genes (pCPP2006) to recombinant E. coli and K. oxytoca M5A1 harboring celZ caused a dramatic increase in the functional expression of celZ and in the fraction of CelZ that was secreted into the extracellular milieu (42, 44). The same effects were observed for ethanologenic K. oxytoca SZ21 containing celZ and celY (Table 4). Addition of the out genes to SZ22 (inactive celZ) had no effect on the functional expression of celY or the extent of CelY secretion. This celZ mutant, SZ22(pCPP2006), produced only 3% of the total endoglucanase activity (CelY) produced by SZ21(pCPP2006) containing functional celY and celZ genes. It is interesting that the secreted endoglucanase produced by SZ21(pCPP2006) with the out genes was substantially higher than the sum of the individual activities expressed from the same respective promoters in SZ6 (CelZ) and SZ22 (CelY), a finding consistent with synergism between these two enzymes (43). In this assay, synergy is estimated to be 1.4-fold the arithmetic sum of the individual activities [SZ6(pCPP2006) and SZ22(pCPP2006)] for the combination of extracellular enzymes produced by SZ21(pCPP2006).
Synergism between recombinant E. chrysanthemi endoglucanase (CelZ and CelY) and fungal cellulase (Spezyme CE) during the fermentation of cellulose to ethanol.
SSF experiments were performed with flasks without pH control to evaluate the combined effects of fungal cellulase (Spezyme) and cellulase enzymes produced by the biocatalysts on ethanol production from Sigmacell 50, a highly crystalline substrate (Table 5). Although very low levels of ethanol were produced by all strains in the absence of Spezyme, strains SZ6(pCPP2006) and SZ21(pCPP2006) containing functional celZ genes produced higher levels of ethanol (P ≤ 0.05) than strain SZ22(pCPP2006) containing only a functional celY gene and strain P2(pCPP2006) lacking both endoglucanase genes. In the absence of both Spezyme and Sigmacell 50, all strains produced 0.22 g of ethanol per liter. The additional increment of ethanol produced by SZ6(pCPP2006) and SZ21(pCPP2006) during incubation with Sigmacell 50 is attributed to hydrolysis of the small fraction of amorphous cellulose in the substrate by CelZ (42–44). Digestion of amorphous cellulose by CelY alone produces saccharides that are too large to be transported and metabolized without further hydrolysis (43).
TABLE 5.
Ethanol production from Sigmacell 50 (100 g/liter)
| Strain | Genencor Spezyme
|
Fermentationa
|
|||
|---|---|---|---|---|---|
| Type | Addition (ml/liter) | n | Ethanol concn ± SD (g/liter)b | % Control ± SD | |
| P2(pCPP2006) | None | 0 | 3 | 0.23 ± 0.01 | 100 ± 2.0 |
| SZ6(pCPP2006) | None | 0 | 3 | 0.28 ± 0.02* | 124 ± 8.5 |
| SZ21(pCPP2006) | None | 0 | 3 | 0.26 ± 0.02* | 116 ± 8.5 |
| SZ22(pCPP2006) | None | 0 | 3 | 0.24 ± 0.01 | 107 ± 1.0 |
| P2(pCPP2006) | CE | 5.0 | 6 | 13.7 ± 0.3 | 100 ± 2.0 |
| SZ6(pCPP2006) | CE | 5.0 | 6 | 13.8 ± 0.3 | 101 ± 2.4 |
| SZ21(pCPP2006) | CE | 5.0 | 6 | 16.0 ± 0.5** | 117 ± 3.5 |
| SZ22(pCPP2006) | CE | 5.0 | 6 | 15.2 ± 0.3** | 112 ± 1.5 |
| P2(pCPP2006) | CE | 10.0 | 6 | 20.7 ± 0.5 | 100 ± 2.1 |
| SZ6(pCPP2006) | CE | 10.0 | 6 | 21.2 ± 0.1 | 103.4 ± 0.4 |
| SZ21(pCPP2006) | CE | 10.0 | 6 | 24.6 ± 0.5** | 121 ± 2.3 |
| SZ22(pCPP2006) | CE | 10.0 | 6 | 25.2 ± 1.1** | 122 ± 5.0 |
| P2(pCPP2006) | CP | 5.0 | 6 | 15.2 ± 0.3 | 100 ± 1.7 |
| SZ21(pCPP2006) | CP | 5.0 | 6 | 17.8 ± 0.4** | 116 ± 2.2 |
| P2(pCPP2006) | CP | 10.0 | 6 | 25.3 ± 0.7 | 100 ± 2.6 |
| SZ21(pCPP2006) | CP | 10.0 | 6 | 27.2 ± 0.3** | 107 ± 1.2 |
Cultures grown without added cellulose or Spezyme produced 0.22 ± 0.01 g of ethanol per liter. Spezyme contained approximately 100 FPU/ml. The addition of 5 or 10 ml of Spezyme corresponds to 5 and 10 FPU/g of cellulose, respectively.
Student t test shows that there is significant difference in ethanol production compared to the respective P2 controls at each Spezyme dilution. A P value of ≤0.001 is indicated by two asterisks; a P value of ≤0.05 is indicated by one asterisk.
Spezyme CE and Spezyme CP contain a commercially optimized combination of endoglucanase, exoglucanase, and cellobiase activities (3, 5, 27, 28). Despite this optimization, Spezyme-supplemented fermentations with two of the endoglucanase-producing biocatalysts, SZ21(pCPP2006) and SZ22(pCPP2006), produced significantly higher levels of ethanol than the control P2(pCPP2006), which lacks endoglucanase genes. The combinations of Spezyme and SZ21(pCPP2006) and SZ22(pCPP2006) were synergistic in terms of ethanol production, which was up to 20% higher than the sum of ethanol produced by each individually (P ≤ 0.001). Synergy was observed for both dilutions of Spezyme CE and for Spezyme CP. This synergistic effect can be attributed primarily to CelY since this is the only endoglucanase produced by SZ22(pCPP2006). No synergy was observed for SZ6(pCPP2006) that produces CelZ alone.
DISCUSSION
Although the development of a recombinant biocatalyst that can supply all of the enzymes needed for the hydrolysis of cellulose and ethanol production has yet to be achieved, incremental progress has been made with both bacterial and yeast systems. Plasmid-based strains of Z. mobilis have been developed that express low levels (estimated to be <100 U/liter) of both an endoglucanase and a cellobiase from Xanthomonas albilineans (31). Higher levels of activity (600 to 1,000 U/liter) were produced by plasmid-based strains of Z. mobilis expressing an endoglucanase gene from Cellulomonas uda (24) and the celZ gene (CelZ) from E. chrysanthemi (7). Although the C. uda endoglucanase remained cell associated in recombinant Z. mobilis, up to 40% of the CelZ was released from the periplasm in late stationary phase without apparent lysis (7). No data were presented concerning the utilization of cellobiose or β-1-4-linked glucosides for growth and ethanol production by this strain.
Cellobiase and three different cellulase genes have been coexpressed from a single plasmid in recombinant Saccharomyces cerevisiae (33): cellobiase (bgl1) from Endomyces fibuliger, endoglucanase (end1) from Butyrivibrio fibrisolvens, cellobiohydrolase (cbh1) from Phanerochaete chrysosporium, and cellodextrinase (cel1) from Ruminococcus flavefaciens. Secretion of the gene products was facilitated by adding various leader sequences. URA3-based autoselection in an auxotrophic host was used to ensure plasmid retention. The resulting strain expressed higher levels of cellobiase activity than cellulase activity, sufficient to allow slow growth of the recombinant on cellobiose. An analogous construct expressing two cellulase genes (34) produced higher levels of endoglucanase (approximately 1,200 U/liter). No data were presented regarding ethanol production by these strains or growth on polymeric β-1-4-linked glucosides.
A recent study has shown that substantial levels of cellulases can be produced and secreted in recombinant S. cerevisiae by integrating up to 50 copies of the β-glucosidase gene from Bacillus circulans and an endo- or exocellulase gene from Bacillus sp. strain DO4 into multiple chromosomal δ sites of Ty1 retrotransposons (8, 9). Both genes contained added leader sequences to facilitate efficient secretion. In batch fermentations, 300 to 500 U of both activities were produced per liter. Recombinant strains achieved approximately 30% more cell mass and ethanol than the parent strain during growth on a mixture of glucose and soluble cellobiosides (9). A novel two-step process was developed using the recombinant in which high levels of cell mass and cellulase were initially produced aerobically from glucose (9). After supplementing this broth with additional commercial cellulase, crystalline cellulose substrate was added and fermented to ethanol. Although broth from the first step contained up to 700 filter paper units (FPU) of cellulase activity per liter (Whatman filter paper hydrolysis), no ethanol was produced without the further addition of commercial cellulase. Production of these enzymes by recombinant S. cerevisiae allowed a 40% reduction in the amount of commercial enzyme required, from 30 to 18 FPU/g of cellulose.
Our approach to reduce the requirement for fungal cellulase has been to functionally express genes encoding the Z. mobilis pathway in K. oxytoca, an abundant organism in pulp mill waste that has the natural ability to transport and metabolize cellobiose (20, 40). This recombinant can rapidly and efficiently produce 40 to 50 g/liter of ethanol from cellobiose (40). We have now added two endoglucanase genes from a closely related organism that macerates plant cell walls and tissues in nature, i.e., E. chrysanthemi (6, 10). High levels of endoglucanase activity were produced (over 20,000 U/liter), equivalent to 1% of the endoglucanase present in concentrated commercial cellulase products (Table 4 and reference 27). These activities are more than an order of magnitude higher than was previously reported for engineered strains of S. cerevisiae or Z. mobilis. To minimize some of the problems associated with the expression of heterologous genes in industrial strains, unregulated promoters were isolated from random fragments of Z. mobilis DNA using functional assays, hybrid genes were integrated into the chromosome to ensure stability, and the antibiotic resistance markers used in construction were deleted using the FLP recombinase system (23) to facilitate future genetic modifications. Approximately 95% of the CMCase activity produced by this strain was attributed to the celZ gene product, with the balance attributed to the celY product.
CelY and CelZ appear to be secreted by different mechanisms. Approximately 70% of the CelZ was secreted as an extracellular product when the E. chrysanthemi out genes were added on a plasmid (pCPP2006), consistent with a type II secretion system (16). Half of the CelY activity was secreted in the presence or absence of the out genes, consistent with a type IV secretion system (17). The integrated genes have been maintained without selection for over 6 months. However, pCPP2006 containing the out genes is less stable and requires antibiotic selection to ensure maintenance. In SSF experiments with added commercial cellulase, two of the new K. oxytoca recombinants, SZ21 and SZ22, produced more ethanol than the parent strain lacking E. chrysanthemi cellulases. Both of these strains also produced ethanol levels equivalent to the best yeast SSF experiments (9) using approximately one-third of the amount of added commercial cellulase (5 PFU/g of cellulose versus 18 FPU/g of cellulose for recombinant yeast [9]).
Previous studies have shown that endoglucanases CelY and CelZ function synergistically to affect the hydrolysis of CMC and amorphous cellulose (43). This synergy is based on differences in substrate range and mechanism of action. CelZ hydrolyzes cellotriose, cellotetraose, cellopentaose, amorphous cellulose, and CMC. CelY hydrolyzes polymeric substrates to products of approximately 10 glucosyl residues. Optimum synergy was observed with a high ratio of CelZ to CelY, similar to that produced in nature by E. chrysanthemi and by SZ21. Strains SZ6(pCPP2006) and SZ21(pCPP2006) expressing CelZ and CelY+CelZ, respectively, produced small amounts of ethanol from Sigmacell 50 even without the addition of fungal enzymes (Table 5). Both ethanol values were significantly higher than strains grown in broth without added cellulose and strain P2(pCPP2006) with cellulose (no endoglucanase genes). This small amount of ethanol is attributed to the hydrolysis of a minor amorphous component of Sigmacell 50.
SSF experiments (Table 5) demonstrated that bacterial cellulases produced by ethanologenic K. oxytoca functioned synergistically with added commercial cellulase to increase ethanol production (7 to 22%) from crystalline cellulose (Sigmacell 50). Surprisingly, the beneficial effect was attributed almost exclusively to CelY, despite the fact that CelY activities were quite low in comparison to CelZ. Strain SZ22 expressing CelY was nearly equivalent to strain SZ21 expressing both activities. Strain SZ6 expressing only CelZ showed little increase in ethanol despite the production of over 20,000 U of endoglucanase activity per liter. Differences in the effectiveness of CelY and CelZ in combination with Spezyme may result from differences in substrate specificities and modes of action. Fungi such as T. reesei produce multiple endoglucanase activities which are presumed to function together with exoglucanases during the hydrolysis of crystalline cellulose (26, 32, 39). It is possible that the fungal activity resembling CelZ is not limiting hydrolysis in dilutions of Spezyme. In contrast to CelZ, CelY does not hydrolyze soluble cellobiosides but preferentially acts on longer-chain substrates, producing ends which can function as new sites for exoglucanase activity. Thus, endoglucanases with an activity profile resembling that for CelY may be limiting in fungal preparations supplied commercially.
In the absence of fungal cellulase additions, previous studies have shown that CelY and CelZ function synergistically to degrade amorphous cellulose (43). In nature, lignocellulosic substrates are depolymerized by mixtures of extracellular enzymes produced by consortia of fungi and bacteria. Thus, it is not surprising that a mixture E. chrysanthemi enzymes and T. reesei enzymes can improve the digestion of lignocellulosic substrates during bioconversion to ethanol.
ACKNOWLEDGMENTS
We thank F. Barras for sharing plasmid pMH18 containing the celY gene from E. chrysanthemi 3937 and A. Collmer for sharing plasmid pCPP2006 containing the out genes from E. chrysanthemi EC16.
This research was supported in part by grants from the U.S. Department of Agriculture, National Research Initiative (98-35504-6177); the U.S. Department of Energy, Office of Basic Energy Science (FG02-96ER20222); and the Florida Agricultural Experiment Station, University of Florida.
Footnotes
Florida Agricultural Experiment Station journal series no. R-07702.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Deidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 2.Baker J O, Adney W S, Thomas S R, Nieves R A, Chou Y C, Vinzant T B, Tucker M P, Laymon R A, Himmel M E. Synergism between purified bacterial and fungal cellulases. In: Saddler J N, Himmel M E, editors. Enzymatic degradation of insoluble carbohydrates. ACS Symposium Series 618. Washington, D.C.: American Chemical Society; 1995. pp. 113–141. [Google Scholar]
- 3.Beguin P, Aubert J P. The biological degradation of cellulose. FEMS Microbiol Rev. 1994;13:25–58. doi: 10.1111/j.1574-6976.1994.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 4.Blondelet-Rouault M-H, Weiser J, Lebrihi A, Branny P, Pernodet J-L. Antibiotic resistance gene cassette derived from the Ω interposon for use in E. coli and Streptomyces. Gene. 1997;190:315–317. doi: 10.1016/s0378-1119(97)00014-0. [DOI] [PubMed] [Google Scholar]
- 5.Boisset C, Fraschini C, Schulein M, Henrissat B, Chanzy H. Imaging the enzymatic digestion of bacterial cellulose ribbons reveals the endo character of the cellobiohydrolase ce16A from Humicola insolens and its mode of synergy with cellobiohydrolase cel7A. Appl Environ Microbiol. 2000;66:1444–1452. doi: 10.1128/aem.66.4.1444-1452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer M H, Cami B, Chambost J P, Magnan M, Cattaneo J. Characterization of a new endoglucanase from Erwinia chrysanthemi. Eur J Biochem. 1987;162:311–316. doi: 10.1111/j.1432-1033.1987.tb10602.x. [DOI] [PubMed] [Google Scholar]
- 7.Brestic-Goachet N, Gunasekaran P, Cami B, Baratti J C. Transfer and expression of an Erwinia chrysanthemi cellulase gene in Zymomonas mobilis. J Gen Microbiol. 1989;135:893–902. [Google Scholar]
- 8.Cho K M, Yoo Y J, Kang H S. δ-Integration of endo/exo-glucanase and β-glucosidase genes into the yeast chromosomes for direct conversion of cellulose to ethanol. Enzyme Microb Technol. 1999;25:23–30. [Google Scholar]
- 9.Cho K M, Yoo Y J. Novel SSF process for ethanol production from microcrystalline cellulose using the δ-integrated recombinant yeast, Saccharomyces cerevisiae L2612δGC. J Microbiol Biotechnol. 1999;9:340–345. [Google Scholar]
- 10.Collmer A, Keen N T. The role of pectic enzymes in plant pathogenesis. Annu Rev Phytopathol. 1996;24:383–409. [Google Scholar]
- 11.Dale B E. Biobased industrial products: bioprocess engineering when cost really counts. Biotechnol Prog. 1999;15:775–776. doi: 10.1021/bp990286f. [DOI] [PubMed] [Google Scholar]
- 12.De Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guiseppi A, Aymeric J L, Cami B, Barras F, Creuzet N. Sequence analysis of the cellulase-encoding celY gene of Erwinia chrysanthemi: a possible case of interspecies gene transfer. Gene. 1991;106:109–114. doi: 10.1016/0378-1119(91)90573-t. [DOI] [PubMed] [Google Scholar]
- 14.He S Y, Lindeberg M, Chatterjee A K, Collmer A. Cloned Erwinia chrysanthemi out genes enable Escherichia coli to selectively secrete a diverse family of heterologous proteins to its milieu. Proc Natl Acad Sci USA. 1991;88:1079–1083. doi: 10.1073/pnas.88.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Himmel M E, Ruth M F, Wyman C E. Cellulase for commodity products from cellulosic biomass. Curr Opin Biotechnol. 1999;10:358–364. doi: 10.1016/S0958-1669(99)80065-2. [DOI] [PubMed] [Google Scholar]
- 16.Hogsett D A, Ahn H J, Bernardez T D, South C R, Lynd L R. Direct microbial conversion—prospects, progress, and obstacles. Appl Biochem Biotechnol. 1992;34:527–541. [Google Scholar]
- 17.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingram L O, Aldrich H C, Borges A C C, Causey T B, Martinez A, Morales F, Saleh A, Underwood S A, Yomano L P, York S W, Zaldivar J, Zhou S. Enteric bacterial catalysts for fuel ethanol production. Biotechnol Prog. 1999;15:855–866. doi: 10.1021/bp9901062. [DOI] [PubMed] [Google Scholar]
- 19.Karlsson J, Medve J, Tjerneld F. Hydrolysis of steam-pretreated lignocellulose: Synergism and adsorption for cellobiohydrolase I and endoglucanase II of Trichoderma reesei. Appl Biochem Biotechnol. 1999;82:243–258. doi: 10.1385/abab:82:3:243. [DOI] [PubMed] [Google Scholar]
- 20.Lai X, Davis F C, Hespell R B, Ingram L O. Cloning of cellobiose phosphoenolpyruvate-dependent phosphotransferase gene: functional expression in recombinant Escherichia coli and identification of a putative binding region for disaccharides. Appl Environ Microbiol. 1997;63:355–363. doi: 10.1128/aem.63.2.355-363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee I, Evans B R, Woodward J. The mechanism of cellulase action on cotton fibers: evidence from atomic force microscopy. Ultramicroscopy. 2000;82:213–221. doi: 10.1016/s0304-3991(99)00158-8. [DOI] [PubMed] [Google Scholar]
- 22.Lynd L E, Wyman C E, Gerngross T U. Biocommodity engineering. Biotechnol Prog. 1999;15:777–793. doi: 10.1021/bp990109e. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Morales F, Borges A C, Martinez A, Shanmugam K T, Ingram L O. Chromosomal integration of heterologous DNA in Escherichia coli with precise removal of markers and replicons used during construction. J Bacteriol. 1999;181:7143–7148. doi: 10.1128/jb.181.22.7143-7148.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Misawa N, Okamoto T, Nakamura K. Expression of a cellulase gene in Zymomonas mobilis. J Biotechnol. 1988;7:167–178. [Google Scholar]
- 25.Moniruzamman M, Lai X, York S W, Ingram L O. Isolation and molecular characterization of high-performance cellobiose-fermenting spontaneous mutants of ethanologenic Escherichia coli KO11 containing the Klebsiella oxytoca casAB operon. Appl Environ Microbiol. 1997;63:4633–4637. doi: 10.1128/aem.63.12.4633-4637.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nidetzky B, Steiner W, Claeyssens M. Synergistic interaction of cellulases from Trichoderma reesei during cellulose degradation. In: Saddler J N, Himmel M E, editors. Enzymatic degradation of insoluble carbohydrates. ACS Symposium Series 618. Washington, D.C.: American Chemical Society; 1995. pp. 90–112. [Google Scholar]
- 27.Nieves R A, Ehrman C I, Adney W S, Elander R T, Himmel M E. Technical communication: survey and analysis of commercial cellulase preparations suitable for biomass conversion to ethanol. World J Microbiol Biotechnol. 1998;14:301–304. [Google Scholar]
- 28.Ohmiya K, Sakka K, Karita S, Kimura T. Structure of cellulases and their applications. Biotechnol Genet Eng Rev. 1997;14:365–414. doi: 10.1080/02648725.1997.10647949. [DOI] [PubMed] [Google Scholar]
- 29.Philippidis G P, Hatzis C. Biochemical engineering analysis of critical process factors in the biomass-to-ethanol technology. Biotechnol Prog. 1997;13:222–231. doi: 10.1021/bp970017u. [DOI] [PubMed] [Google Scholar]
- 30.Sheehan J, Himmel M. Enzymes, energy, and the environment: a strategic perspective on the U.S. Department of Energy's research and development activities for bioethanol. Biotechnol Prog. 1999;15:817–827. doi: 10.1021/bp990110d. [DOI] [PubMed] [Google Scholar]
- 31.Su P, Liu C Q, Lucas R J, Delaney S F, Dunn N W. Simultaneous expression of genes encoding endoglucanase and β-glucosidase in Zymomonas mobilis. Biotechnol Lett. 1993;15:979–984. [Google Scholar]
- 32.Tomme P, Warren R A J, Gilkes N R. Cellulose hydrolysis by bacteria and fungi. Adv Microbiol Physiol. 1995;37:1–81. doi: 10.1016/s0065-2911(08)60143-5. [DOI] [PubMed] [Google Scholar]
- 33.Van Rensburg P, Van Zyl W H, Pretorius I S. Engineering yeast for efficient cellulose degradation. Yeast. 1998;14:67–76. doi: 10.1002/(SICI)1097-0061(19980115)14:1<67::AID-YEA200>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 34.Van Rensburg P, Willem H V Z, Pretorius I S. Co-expression of a Phanerochaete chrysosporium cellobiohydrolase gene and a Butyrivibrio fibrisolvens endo-β-1, 4-glucanase gene in Saccharomyces cerevisiae. Curr Genet. 1996;30:246–250. doi: 10.1007/s002940050128. [DOI] [PubMed] [Google Scholar]
- 35.Van Walsum G P, Lynd L R. Allocation of ATP to synthesis of cells and hydrolytic enzymes in cellulolytic fermentative microorganisms: bioenergetics, kinetics, and bioprocessing. Biotechnol Bioeng. 1998;58:316–320. [PubMed] [Google Scholar]
- 36.Wang L, Gralla J D. Multiple in vivo roles for the −12 region elements of sigma 54 promoters. J Bacteriol. 1998;180:5626–5631. doi: 10.1128/jb.180.21.5626-5631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wise A, Brems R, Ramakrishnan V, Villarejo M. Sequences in the −35 region of Escherichia coli rpoS-dependent genes promote transcription by EςS. J Bacteriol. 1996;178:2785–2793. doi: 10.1128/jb.178.10.2785-2793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood T M, Bhat K M. Methods for measuring cellulase activities. Methods Enzymol. 1988;160:87–112. [Google Scholar]
- 39.Woodward J. Synergism in cellulase systems. Bioresource Technol. 1991;36:67–75. [Google Scholar]
- 40.Wood B E, Ingram L O. Ethanol production from cellobiose, amorphous cellulose and crystalline cellulose by recombinant Klebsiella oxytoca containing chromosomally integrated Zymomonas mobilis genes for ethanol production and plasmids expressing thermostable cellulase genes from Clostridium thermocellum. Appl Environ Microbiol. 1992;58:2103–2110. doi: 10.1128/aem.58.7.2103-2110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyman C E. Biomass ethanol: technical progress, opportunities, and commercial challenges. Annu Rev Energy Environ. 1999;24:189–226. [Google Scholar]
- 42.Zhou S, Ingram L O. Engineering endoglucanase-secreting strains of ethanologenic Klebsiella oxytoca P2. J Indust Microbiol Biotechnol. 1999;22:600–607. doi: 10.1038/sj.jim.2900666. [DOI] [PubMed] [Google Scholar]
- 43.Zhou S, Ingram L O. Synergistic hydrolysis of carboxymethyl cellulose and acid-swollen cellulose by two endoglucanases (CelZ and CelY) from Erwinia chrysanthemi. J Bacteriol. 2000;182:5676–5682. doi: 10.1128/jb.182.20.5676-5682.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou S, Yomano L P, Saleh A Z, Davis F C, Aldrich H C, Ingram L O. Enhancement of expression and apparent secretion of Erwinia chrysanthemi endoglucanase (encoded by celZ) in Escherichia coli B. Appl Environ Microbiol. 1999;65:2439–2445. doi: 10.1128/aem.65.6.2439-2445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]