Abstract
Along with the increasing availability of electronic medical record (EMR) data, phenome-wide association studies (PheWAS) and phenome-disease association studies (PheDAS) have become a prominent, first-line method of analysis for uncovering the secrets of EMR. Despite this recent growth, there is a lack of approachable software tools for conducting these analyses on large-scale EMR cohorts. In this article, we introduce pyPheWAS, an open-source python package for conducting PheDAS and related analyses. This toolkit includes 1) data preparation, such as cohort censoring and age-matching; 2) traditional PheDAS analysis of ICD-9 and ICD-10 billing codes; 3) PheDAS analysis applied to a novel EMR phenotype mapping: current procedural terminology (CPT) codes; and 4) novelty analysis of significant disease-phenotype associations found through PheDAS. The pyPheWAS toolkit is approachable and comprehensive, encapsulating data prep through result visualization all within a simple command-line interface. The toolkit is designed for the ever-growing scale of available EMR data, with the ability to analyze cohorts of 100,000 + patients in less than 2 h. Through a case study of Down Syndrome and other intellectual developmental disabilities, we demonstrate the ability of pyPheWAS to discover both known and potentially novel disease-phenotype associations across different experiment designs and disease groups. The software and user documentation are available in open source at https://github.com/MASILab/pyPheWAS.
Keywords: PheWAS, PheDAS, Electronic Medical Records, Phenotype, ICD
Introduction
Since the early 2000s, the introduction of computers in healthcare has led to the adoption of Electronic Medical Records (EMR) in healthcare systems across the globe. Initiatives such as the National Institutes of Health’s Clinical and Translational Science Awards have advanced this electronic healthcare landscape by providing funding for institutions to generate, store, and share healthcare information with the ultimate goal of improving patient care (MacKenzie et al., 2012). Many institutions, such as Intermountain Healthcare and Vanderbilt University Medical Center (VUMC), have risen to the challenge, building large EMR repositories that encompass patient demographics, insurance billing data, genetic sequences, medication records, laboratory testing, and more (Evans et al., 2012; Danciu et al., 2014). These rich EMR repositories create opportunities for “secondary use” of health data, meaning the utilization of health data outside of direct patient care. In medical research, this translates to opportunities for investigators to study disease progression and comorbidities, treatment efficacy, genetic factors, systemic problems, and biases in the medical system, among other goals (Safran et al., 2007). Yet, taking advantage of these complex databases is not a simple task; the EMR is often biased, incomplete, and inaccurate (Hripcsak & Albers, 2013). Consequently, rapid increases in the size and availability of EMR resources have led to a surge in the development of EMR analysis methods, particularly in the area of deriving and studying EMR phenotypes (Ahmad et al., 2002; Hripcsak & Albers, 2013; Kirby et al., 2016).
A particularly successful type of EMR phenotype analysis is the phenome-wide association study (PheWAS). This analysis is closely related to the genome-wide association study (GWAS), a framework in which a single phenotype is tested for associations with many genotypes (Hindorff et al., 2009). In contrast, a PheWAS tests the association between a single genotype and many EMR-derived phenotypes. This method was pioneered by Denny et al. (2010) with a proof of concept study that examined the associations between five single nucleotide polymorphisms (SNPs) and 776 EMR phenotypes; this PheWAS both replicated five previously reported SNP-disease associations and identified nineteen potentially novel associations, presenting PheWAS’ potential for supporting often-underpowered GWAS investigations. Three years later, the same group performed a large-scale trans-institutional validation of PheWAS, confirming its use as an unbiased phenotype interrogation technique and hypothesis generation tool (Denny et al., 2013). The 776 phenotypes used in the proof-of-concept study were derived from International Classification of Disease (ICD) version 9 billing codes; these phenotypes were designated PheWAS Codes, or PheCodes, and have since been publicly released and expanded to a cover a total of 1,866 EMR phenotypes (Denny et al., 2013; Wei et al., 2017a).
Since its conception, this groundbreaking technique has inspired many investigations of different sections in the genome. In a similar vein as its initial proof-of-concept, PheWAS has been used to examine the phenotype signature of the HLA-DRB1*1501 haplotype (a genetic variant linked with Multiple Sclerosis) (Hebbring et al., 2013), the major histocompatibility complex region of chromosome 6 (Liu et al., 2016), 31 SNPs associated with serum uric acid (Li et al., 2018), and other genome regions of interest revealed via GWAS (Denny et al., 2011). Other interesting applications of this technique include examining the contribution of Neanderthal genetic variants to the phenotypes of modern humans (Simonti et al., 2016), and evaluating self-reported ICD-9 records in a large-scale 23andMe database for the purpose of genetic drug targeting (Ehm et al., 2017).
Inspired by PheWAS, an alternative approach has emerged which scans the phenome for associations with non-genetic targets. This extension of PheWAS is advantageous due to the costly nature of genotyping, and therefore, the huge amount of EMR data available when linked genetic data are no longer necessary (Hebbring, 2014). This framework has been used to examine linked dental and medical records to identify ICD-9 phenotypes related to periodontitis (Boland et al., 2014). In a federated query task, it was used to retrieve records of patients who had a rare condition (multiple myeloma) across multiple institutions, and then further delineate specific subgroups that experienced serious complications (Warner et al., 2013). Other examples include scans of ICD-9 phenotype associations with white blood cell count (Warner & Alterovitz, 2012) and non-Hodgkin lymphoma in Medicare claims (Engels et al., 2016). Recently, we observed the potential for confusion of study designs with genetic and non-genetic phenome association studies. After consultation with the PheWAS team, we now refer to studies that do not include genetic markers but still use mass univariate regression as Phenome-Disease Association Studies (PheDAS) (Chaganti et al., 2019a), an example of which is shown in Fig. 1.
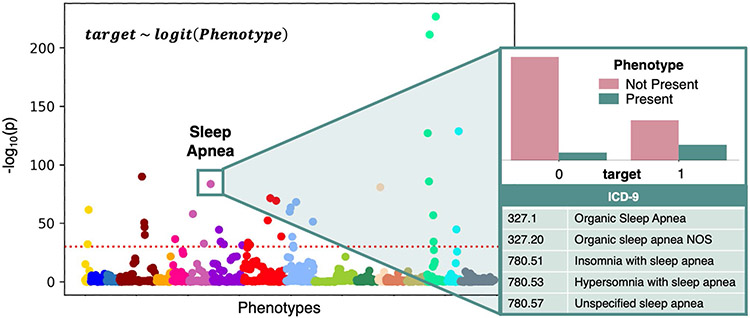
Fig. 1.
Overview of PheDAS. In the background, a Manhattan plot shows the statistical significance of many phenotypes in relation to a single target variable (target). Phenotypes are sorted into and colored by category, and the significance threshold for multiple comparisons correction is marked with a dashed horizontal line. These relationships were estimated by individually modeling the target variable as a function of each phenotype using a logistic regression. For a closer look, the significant phenotype Sleep Apnea is highlighted. The distribution of subjects from each target group that do (not) present the Sleep Apnea phenotype is shown, along with the ICD-9 codes that map to this this phenotype
In light of the pervasiveness of this EMR analysis technique, we present pyPheWAS: a comprehensive toolkit for performing PheWAS and PheDAS analyses. The original PheWAS software, written by the team that developed the PheWAS method, is implemented in R and includes core PheWAS functions (Carroll et al., 2014). The pyPheWAS package reimplements that core functionality in Python, a language that has become more widespread in the machine learning community and adds a collection of easy-to-use command line tools that covers everything from preprocessing EMR data to visualizing results. It includes analysis of ICD-9 and ICD-10 phenotypes, as well as a novel analysis for Current Procedural Terminology (CPT) code phenotypes. It is important to note that pyPheWAS is not a neuro-centric toolkit, although its methods allow investigators to explore the clinical progression of many neurological conditions. Additionally, pyPheWAS is agnostic to the dependent variable, and therefore can be used to implement either PheWAS or PheDAS; for the remainder of this article, we will focus specifically on PheDAS analyses.
In the following sections, we first describe the technical details of the pyPheWAS toolkit, including installation instructions, EMR data acquisition, data preprocessing, and analysis methods. Following this, we demonstrate the toolkit in action by performing a PheDAS analysis on a custom synthetic EMR dataset. We then perform a case study on real EMR data, comparing the EMR of Down Syndrome patients to patient with other Intellectual and Developmental Disabilities. Finally, we discuss PheDAS result interpretation and several limitations of the pyPheWAS package.
Methods
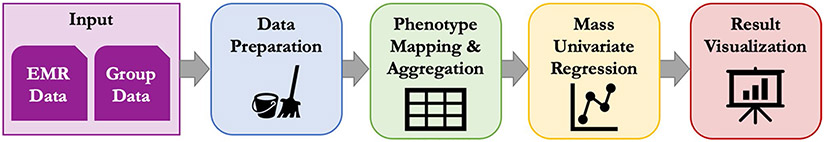
The overall workflow of a PheDAS analysis is shown in Fig. 2. EMR events and group demographic data are preprocessed, mapped to meaningful phenotypes, used to model a target variable (such as a disease group), and then visualized for interpretation. Figure 3 presents the pyPheWAS toolkit, a collection of command line scripts that aims to make PheDAS-style analysis highly approachable, as this process can quickly become intractable given the sheer scale of EMR data coupled with a lack of easy-to-use software. This section describes the form and function of each tool in detail. Source code for pyPheWAS may be found on GitHub (https://github.com/MASILab/pyPheWAS). The full user documentation may be found at https://pyphewas.readthedocs.io/en/latest/.
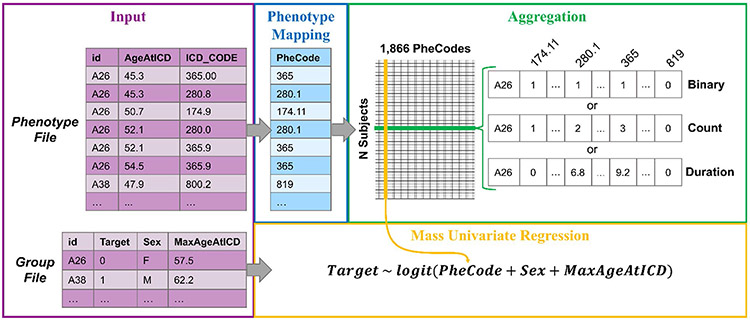
Fig. 2.
PheDAS analysis pipeline. Inputs to the pipeline include EMR data (ICD-9, ICD-10, or CPT codes) and group data (disease group, sex, race, etc.). The data is first prepared for analysis via case–control matching and censoring. Next, the EMR data is mapped to a set of predefined phenotypes (PheWAS or ProWAS Codes) and aggregated across each subject’s record. Mass univariate regression is then performed across all phenotypes, where a target variable is modeled as a function of the phenotype plus any relevant covariates (such as sex or race) to determine the relationship between the target variable and each phenotype. Finally, the results are visualized to facilitate interpretation of target variable-phenotype relationship significance and effect size
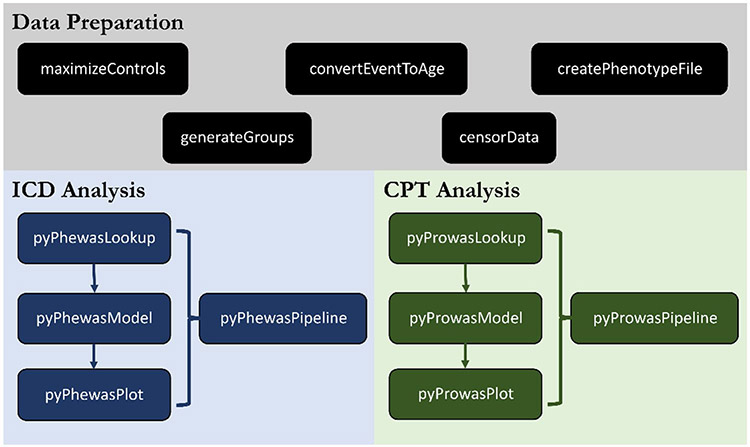
Fig. 3.
pyPheWAS package tools. The package is composed of three main tool sets: data preparation, ICD analysis, and CPT analysis. Data preparation tools focus on preprocessing EMR data, e.g., case/control matching (maximizeControls) and censoring events (censor-Data). The ICD analysis tools run PheDAS on ICD code data, while the CPT analysis tools run PheDAS on CPT code data. The function and usage of all tools are described in the Methods section
Requirements and Installation
pyPheWAS is a Python (version 3.6 +) package hosted on pypi.org, making installation quick and easy. On any computer which has Python 3 and the popular package manager pip already installed, the user must simply enter pip install pyPheWAS in a terminal or command line to install the software. All tools are accessed via command line. Note that there are no explicit hardware requirements for the pyPheWAS package, but the amount of memory available on the user’s system will limit the size of experiment that can be performed.
Beyond software, the only requirements for using pyPheWAS is the format of the input data. Two primary files are expected by pyPheWAS tools: the phenotype file (EMR data) and the group file (demographic data). The phenotype file contains EMR events for all subjects in the group file, with a single line for each event. Events include an ICD or CPT code and the subject’s age at the event. The group file contains demographic information, such as sex, and the target response variable which will be used in the logistic regression. The response variable may be pre-defined (such as a diagnosis), or it may be determined based on EMR data using the pyPheWAS data preparation tools. The phenotype and group files are linked by a column labeled ‘id’ which contains a unique identifier for each subject in the cohort.
EMR Data Acquisition
Many institutions have spent large amounts of time and resources to build multi-faceted data repositories that include genetic data, clinical records, and demographic information across large swaths of patient populations. A few prominent repositories include the Healthcare Cost and Utilization Project’s (HCUP) National Inpatient Sample (2021a), the eMERGE Network (eMERGE Consortium, 2021), VUMC’s Synthetic Derivative (VUMC-SD) (Danciu et al., 2014), Intermountain Healthcare’s Enterprise Data Warehouse (Evans et al., 2012), the Utah Population Database (2021b), and the Rochester Epidemiology Project (Rocca et al., 2012). Due to the sensitive nature of EMR and protections set forth by the Health Insurance Portability and Accountability Act (HIPAA), an approval process is generally required to obtain access to these repositories. For example, in order to obtain the ICD and CPT records used for this article’s Down Syndrome case study from VUMC-SD, we first were required to obtain study approval from Vanderbilt University’s Institutional Review Board, sign a data use agreement, and pay a fee for repository use. We then worked with analysts at VUMC-SD to identify our target population using specific ICD codes and other diagnosis information. With our population identified, the VUMC-SD then pulled the requested ICD, CPT, and demographic records. Such processes are common across many EMR repositories. Though these procedures were designed to protect patient information, they also present steep entry barriers for aspiring EMR researchers. Therefore, we have made the synthetic dataset developed for this article publicly available through pyPheWAS’s GitHub repository, allowing users to familiarize themselves more quickly with both EMR data and PheDAS methods (see the Results section for details). We hope that this resource will inspire similar accessibility efforts and enthusiasm for large-scale EMR analysis.
Data Preparation
The pyPheWAS package provides several useful data preparation functions so that users do not have to directly manipulate the very large data files often used for PheDAS studies.
Defining Case and Control Groups
The first step in a PheDAS study is defining which subjects are cases and which are controls. In the absence of externally defined group assignments (such as genetic markers (Denny et al., 2011) or white blood cell count (Warner & Alterovitz, 2012)), ICD codes themselves may be used as a proxy for diagnosis (Bastarache & Denny, 2011; Wei et al., 2017a) (although sources of error for this are well known (O’Malley et al., 2005)). The ICD-9 code 758.0 – Down’s syndrome, for example, may be used as a proxy for the actual clinical diagnosis of Down Syndrome. Due to the noisy nature of EMR, however, a minimum frequency threshold is applied to codes used for this proxy diagnosis based on the notion that the more frequently a subject is assigned a certain ICD code, the more likely it is that they legitimately have the target condition.
To address this need, the createPhenotypeFile function sorts subjects into case and control groups based on the presence or absence of ICD codes in subjects’ records. At a minimum, createPhenotypeFile requires a phenotype file, a list of ICD-9 and ICD-10 codes that define the case group, and the minimum frequency of those codes in a subject’s record to be considered part of the case group. Users may specify whether this frequency threshold is a daily threshold (code frequency is calculated based on the number of unique days over which a code is recorded; ignores multiple records of a code within a single day) or an absolute threshold (code frequency is calculated based on the absolute number of code events; includes multiple records of a code within a single day). All subjects listed in the phenotype file who have at least the minimum frequency of provided codes in their record are assigned to the case group (target = 1). Subjects who have the provided codes in their record but fall below the specified frequency are considered ambiguous and, consequently, excluded. All remaining subjects are assigned to the control group (target = 0). These group assignments are saved to a comma-separated values (CSV) file containing A) only subject IDs and target variable assignments, or B) the target variable assignment added to an existing group file specified by the user.
In the basic configuration described above, the control group is comprised of all non-case and non-ambiguous subjects. In some experiments, however, it may be desirable to enforce stricter control group inclusion criteria; create-PhenotypeFile provides two commonly used practices for narrowing the scope of PheDAS control groups. The first method excludes subjects from the control group based on both the provided case codes and codes related to those case codes; this prevents the control group from becoming contaminated by conditions similar to the target condition. The list of related codes may be supplied by the user or pulled from the ICD phenotype map (see the pyPhewasLookup section for details on the ICD phenotype map used by pyPheWAS). The second method allows users to target a specific condition for the control group. For example, a PheDAS could be performed comparing Alzheimer’s disease patients (case) to Vascular Dementia patients (controls). In this case, the user would supply createPhenotypeFile with lists of ICD-9 and ICD-10 codes for both the case group and the control group. The control group is then composed of subjects not in the case group that have at least the minimum frequency of provided control group codes in their record. Optionally, a second argument may be provided to the code frequency input; if this is specified, the second frequency value is applied to the control group.
Converting Dates to Ages
EMR event data is usually tagged with dates. In certain cases, a researcher may choose to study EMR records only within a specific period of time, or they may want to use age as a covariate. For convenience, the convertEventToAge script allows users to quickly convert dates associated with CPT and ICD events to subject ages at the events. This function requires the phenotype file for which event dates are to be converted and a corresponding group file that contains each subjects’ date of birth. Optionally, the user may specify the level of precision with which ages are saved in the output phenotype file.
Censoring Event Data
A common aim of medical studies is to examine specific periods of time in patients’ lives. For example, one may be interested in the EMR signature for the five years leading up to an Alzheimer’s Disease diagnosis or for children ages 10 to 18 who have Autism/Autism Spectrum Disorder. Data censoring such as this is incorporated into the pyPheWAS toolkit with the censorData function. Similar to other tools, this function requires a phenotype file containing the events to be censored and a group file containing subject information, along with user-specified censoring start and/or end years. Censoring can be applied to the data in two distinct ways. The first method censors the absolute value of event ages (e.g. the age at CPT or ICD code events) to only those that fall within the user-defined start and end years, such that all preserved events fulfill the equation
| (1) |
The second method instead censors event ages relative to an external event, such as subject age at diagnosis or surgery. In this case, the interval between the events is considered such that all preserved events fulfill the equation
| (2) |
The censored events are saved to a new phenotype file, and all subjects with event data remaining after censoring are written to a new group file.
Case–Control Matching
Another common practice in case–control studies such as PheDAS is matching a certain number of control subjects to each case subject based on specified group variables. The pyPheWAS toolkit includes case–control mapping through its maximizeControls tool. This tool requires a group file containing group variables and case/control assignments, a list of variables to match on, tolerance intervals for each of those matching variables, and the desired ratio of controls to cases. It constructs a bipartite graph from the cohort in which subjects are the vertices, matching variables are edges, and the case and control groups are two disjoint independent vertex sets. To find a first set of matches, it uses the Hopcroft-Karp algorithm (Hopcroft & Karp, 1973) to find a mapping between the case and control sets that results in maximal cardinality (i.e., matches). If the desired matching ratio is larger than 1:1, the first set of matched controls are removed from the graph, and the Hopcroft-Karp algorithm is applied again to find a second set; this repeats until either the desired matching ratio is satisfied or there are no more possible matches. A new group file is saved containing all matched subjects, along with a separate matched pairs file containing the explicit mapping between each individual case and its control(s).
Scanning the ICD Phenome
As outlined in Fig. 2, the core of PheDAS analysis may be broken up into three distinct phases: 1) mapping EMR data to phenotypes, 2) mass univariate regression of phenotypes, and 3) result visualization. The ICD analysis tools in the pyPheWAS package focuses on processing ICD-9-CM and ICD-10 codes, with individual functions devoted to each of the three phases: pyPhewasLookup, pyPhewasModel, and pyPhewasPlot, respectively. This section describes each of those functions in detail.
pyPhewasLookup
The pyPhewasLookup function transforms individual ICD code records into feature matrices ready to be processed by the pyPhewasModel function; Fig. 4 provides a detailed view of this function. It requires as input a phenotype file containing the ICD records of each subject and a group file containing the target and covariate variables. The feature matrices are constructed in two phases: 1) mapping and 2) aggregation. In the mapping phase, each ICD code in the phenotype file is mapped to its corresponding phenotype. The phenotype mapping used by pyPhewasLookup includes 1,866 hierarchical phenotype codes (PheCodes); it was originally constructed solely for ICD-9 codes by Denny et al. (2013), with later improvements to the ICD-9 mapping (Wei et al., 2017a) and the addition of an ICD-10 code mapping (Wu et al., 2019b). It should be noted that these mappings are not complete. They do not cover the full range of ICD-9 and ICD-10 codes, so ICD events in a subject’s record which are not included in the mapping are removed from the study. When these removals occur, pyPhewasLookup notifies the user regarding the number of removed events; optionally, the user may choose to export the list of removed events for further inspection.
Fig. 4.
Detailed look at phenotype mapping, aggregation, and regression in pyPhewasLookup. On the far left, excerpts from input phenotype and group files containing data from subjects A26 and A38 are shown. ICD codes from the phenotype file are mapped to corresponding PheCodes. These codes are then aggregated via one of three possible methods for each subject; binary, count, and duration aggregations for subject A26 are shown. Finally, the aggregated EMR data is combined with group data (in this case, the target variable Target, and covariates Sex and MaxAgeAtICD), and univariate regressions are computed for each PheCode
The aggregation phase next reformats the mapped data from longitudinal events to subject-by-PheCode feature matrices. Three types of feature matrices are created, in which the columns are PheCodes and the rows are subjects from the group file. The first matrix is the core of the PheWAS analysis; denoted the aggregate measure matrix, it contains a single aggregate measure for each PheCode across all subjects. To allow researchers to investigate different aspects of the EMR, three distinct types of aggregation may be performed: binary, count, and duration. Binary aggregation investigates the relationship between the target variable and the presence or absence of a PheCode. Its feature matrix contains only zeros (the PheCode was absent in the subject’s record) and ones (the PheCode was present in the subject’s record). Count aggregation investigates the relationship between the target variable and the number of occurrences of a PheCode. Its feature matrix contains positive integers that correspond to the total number of times each PheCode occurred in a subject’s record. Duration aggregation investigates the relationship between the target variable and the interval of time over which a PheCode is experienced. Its feature matrix contains the time in years between the first and last occurrences of each PheCode in a subject’s record.
The second and third feature matrices are independent of aggregation type and are created as optional covariates for pyPhewasModel. The ICD age feature matrix contains the maximum age recorded for each PheCode in a subject’s record; if the subject has no records of that PheCode, the subject’s overall maximum recorded age is reported. The PheWAS covariate matrix allows researchers to use the presence/absence of a specified PheCode as a covariate in the regression. Across all columns, it records a one if the specified PheCode is present in a subject’s record or zero if the specified PheCode is absent. All three feature matrices are saved as CSV files in preparation for the pyPhewasModel step.
pyPhewasModel
The pyPhewasModel function performs the mass logistic regression which is the focal point of PheDAS analyses. It requires the feature matrix files generated by pyPhewasLookup in addition to the group file. For each PheCode, pyPhewasModel computes a univariate logistic regression of the form
| (3) |
where the target variable and covariates are specified by the user, and Aphe is the aggregate measure vector for a particular PheCode phe taken from the aggregate measure matrix.
These regressions are only computed on PheCodes for which Aphe is non-zero in at least X subjects, where X is a user-defined threshold that defaults to 5. This requirement cuts out PheCodes which lack sufficient statistical power. The model is fit to the data via regularized maximum likelihood optimization. The Python library statsmodels is used to generate and fit the logit model to the PheCode data (Seabold & Perktold, 2010). Regression results are again saved in a CSV file for the user to review and visualize. This file reports the log odds ratio, confidence interval, standard error, and uncorrected p-value estimated from Aphe for each PheCode phe.
pyPhewasPlot
Visualization of the PheDAS mass regression is performed by the pyPhewasPlot function. It requires the regression file produced by pyPhewasModel and the user’s desired multiple comparisons correction method; both False Discovery Rate (FDR) and Bonferroni are available. From these inputs, it creates three complementary views of the PheDAS analysis using the Python matplotlib library (Hunter, 2007). The first is a Manhattan plot, a classic GWAS plot which compares statistical significance across PheCodes. This view presents PheCodes across the horizontal axis, with negative log10(p-value) along the vertical axis; PheCode markers on the plot are colored and sorted according to 18 general categories (mostly organ systems and disease groups, e.g. “circulatory system” and “mental disorders”), allowing users to distinguish related PheCodes. To enhance legibility, the plot only labels PheCodes which are significant after the chosen multiple comparisons correction is applied.
The second view is a Log Odds plot, which compares effect size across PheCodes. In this plot, the log odds of each PheCode and its confidence interval are plotted on the horizontal axis, with PheCodes plotted along the vertical axis. Similar to the Manhattan plot, PheCode markers are sorted and colored by category; only PheCodes which are significant after multiple comparisons correction are shown.
The final view is a Volcano plot. This view combines the previous two, presenting an overview of the entire experiment. In the Volcano plot, significance, negative log10(p-value), is represented by the vertical axis, and effect size, log odds, is represented by the horizontal. All Phe-Codes in the regression file are included on this plot, with marker color corresponding to each PheCodes’s level of significance (none, FDR, Bonferroni). To ensure legibility, only PheCodes that are significant after FDR or Bonferroni correction are labeled.
These three views together provide a comprehensive visualization of the PheWAS analysis. The Volcano plot allows the user to see an overview of the entire experiment, with the Manhattan and Log Odds plots then providing a detailed view for closer examination of significant results. The user has the option of either opening the plots in an interactive window or immediately saving them as image files.
pyPhewasPipeline
pyPhewasPipeline is a streamlined combination of pyPhewasLookup, pyPhewasModel, and pyPhewasPlot created for convenience. Its required inputs are the phenotype file, group file, and the regression type. All intermediate results (feature matrices, regressions) are saved. In addition to the Volcano plot, Manhattan and Log Odds plots are created for both FDR and Bonferroni corrections by default. Optional arguments allow users to modify every step of the pipeline (adding covariates, specifying significance level, etc.).
Scanning the CPT Phenome
Procedure wide association studies (ProWAS) are nearly identical to PheDAS, with one critical difference: the EMR data. While PheDAS investigates ICD code phenotypes, ProWAS investigates CPT code phenotypes. Examining ICD codes may provide insight into patient diagnoses; in a similar vein, examining CPT codes may reveal patterns in how patients are treated. As such, these tools are identical to their PheDAS counterparts, with the exception of the EMR-phenotype mapping. As with PheDAS, ProWAS consists of three main stages: 1) mapping EMR data to phenotypes, 2) mass univariate regression of phenotypes, and 3) result visualization. The CPT analysis tools for each of these stages are analogous to the ICD analysis tools: pyProwasLookup, pyProwasModel, and pyProwasPlot.
ProWAS employs a custom procedural phenotype map, linking 10,396 CPT codes to 1,681 ProWAS Codes (ProCodes) (Chaganti et al., 2017). This map is based on the Clinical Classification System for CPT codes provided by the Healthcare Cost and Utilization Project (HCUP) Agency for Healthcare Research and Quality (2018). Starting with 236 of the HCUP clinically meaningful CPT categories, additional granularity was added to the mapping with guidance from medical experts, until 1,681 ProCodes were defined. For example, the HCUP category 66 (Procedures on spleen) was split into ProCodes 66.1 (Splenectomy), 66.2 (Splenorrhaphy), and 66.3 (Laparoscopy). The full CPT-ProCode map may be found at https://github.com/MASILab/pyPheWAS.
Results
In this section, we demonstrate the utility of the pyPheWAS package via two example PheDAS experiments. In Experiment 1, we evaluate the package by analyzing a synthetic EMR dataset which contains several hand-crafted PheCode associations. In Experiment 2, we perform a case study on real EMR data, in which we compare subjects with Down Syndrome (DS) to controls with other Intellectual or Developmental Disabilities (IDD). A listing of all pyPheWAS commands used to implement these experiments are included in Appendix A.
Experiment 1: Synthetic Dataset
Dataset Construction
Our synthetic dataset consists of 10,000 individuals, split evenly into 5,000 case (Dx = 1) and 5,000 control (Dx = 0) subjects, where Dx is the target variable. Other demographic variables include biological sex and maximum age at visit (MAV). Sex was intentionally made a confounding variable by skewing the female:male ratios between the case and control groups. MAV was calculated as the maximum age recorded from ICD records generated for each individual. These synthetic demographic variables are summarized in Table 1.
Table 1.
Synthetic dataset demographic summary
| Subjects | Sex [% Female] |
Max Age At Visit [mean (std.)] |
|
|---|---|---|---|
| Case (Dx = 1) | 5,000 | 70% | 59.946 (9.563) |
| Control (Dx = 0) | 5,000 | 40% | 60.802 (9.448) |
While curating ICD code events for each individual, three types of PheCode associations were created. Primary PheCode associations were true associations between Dx and the PheCode. ICD events were generated such that each of these PheCodes would have a unique pre-specified effect size (log odds ratio) across the full cohort; individuals’ ages for each event were randomly generated using a uniform distribution over the range [30, 50]. pyPheWAS should accurately estimate each primary association’s effect size and determine that the association is statistically significant. We generated nine primary PheCode associations, including six positive associations and three negative associations (Table 2). In contrast, background PheCode associations were insignificant associations between Dx and the Phe-Code. ICD events were generated such that each background PheCode would have a small pre-specified effect size, randomly generated via a uniform distribution over the range [−0.1, 0.1]; again, individuals’ ages for each event were randomly generated using a uniform distribution over the range [30, 50]. pyPheWAS should accurately estimate each background association’s effect size but determine that the association is insignificant. Twenty background PheCode associations were generated for the synthetic dataset.
Table 2.
PheDAS regression results for the primary and confounded PheCodes in the synthetic dataset
| PheCode | Phenotype | Actual LORa | Reg A |
Reg B |
|||
|---|---|---|---|---|---|---|---|
| LORa | p-valb | LORa | p-valb | ||||
| Primary | 338.2 | Chronic pain | 1.50 | 1.500 | ** | 1.490 | ** |
| 340 | Migraine | 1.10 | 1.099 | ** | 1.128 | ** | |
| 1011 | Complications of surgical and medical procedures | 0.70 | 0.700 | ** | 0.700 | ** | |
| 296.22 | Major depressive disorder | 0.60 | 0.600 | ** | 0.579 | ** | |
| 530.11 | GERD | 0.30 | 0.300 | ** | 0.302 | ** | |
| 401 | Hypertension | 0.25 | 0.249 | ** | 0.257 | ** | |
| 041 | Bacterial infection NOS | −0.20 | −0.200 | ** | −0.194 | ** | |
| 1009 | Injury, NOS | −0.60 | −0.599 | ** | −0.604 | ** | |
| 495 | Asthma | −1.00 | −1.000 | ** | −0.991 | ** | |
| Confounded | 174.1 | Breast cancer [female] | 0.66 / 0.00c | 0.662 | ** | 0.004 | - |
| 292.2 | Mild cognitive impairment | −0.2 | −0.199 | ** | −0.500 | - | |
log odds ratio
significant after Bonferroni correction (**), insignificant (−)
male + female log odds ratio / female-only log odds ratio
Finally, confounded PheCode associations were false positives caused by the confounding effect of either sex or age. Without controlling for the confounding variable, pyPheWAS should identify a significant association with these confounded PheCodes; including the confounding variable as a covariate, however, should reduce (or eliminate) the confounded association. PheCode 174.1 (Breast cancer [female]) was used as a sex-confounded PheCode (Table 2). To produce the confounding effect, ICD events were generated such that all females in the dataset had equal odds of having PheCode 174.1 in their record; event ages were generated in the same way as primary PheCodes. Because females were disproportionally represented across the case and control groups, however, the PheCode’s cohort-wide effect size is positively skewed to a 0.6 log odds ratio. Additionally, PheCode 292.2 (Mild cognitive impairment) was used as an age-confounded PheCode (Table 2). ICD events were generated such that PheCode 292.2 would have a −0.2 log odds ratio; however, event ages were randomly generated using a uniform distribution over the higher age range [65,70]. This resulted in PheCode 292.2 being highly associated with larger values of MAV. This synthetic EMR dataset has been made freely available on pyPheWAS’s GitHub.
PheDAS Analysis
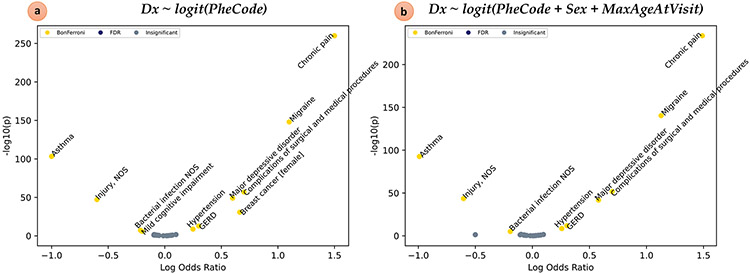
The synthetic EMR dataset was analyzed in a single command via pyPhewasPipeline. We first ran Reg A, a minimal PheDAS with no covariates (Fig. 5a, Table 2). Reg A successfully estimated the log odds ratios of all nine primary PheCodes and determined that they were statistically significant after Bonferroni multiple comparisons correction. The twenty background codes were accurately identified as insignificant. Reg A also correctly estimated the apparent effect sizes and significance of the two confounded PheCodes, 174.1 and 292.2; this was expected since Reg A did not properly control for the confounding variables. To remedy this, we next ran Reg B, a PheDAS that included both sex and MAV as covariates (Fig. 5b, Table 2). With this modification, pyPheWAS recognizes the confounded PheCodes and now correctly determines that they are insignificant.
Fig. 5.
PheDAS applied to a synthetic dataset. a) Volcano plot resulting from a PheDAS without covariates. pyPheWAS successfully identified the nine primary PheCode associations in the synthetic dataset and ignored the twenty background associations. The confounded PheCodes (Breast cancer [female] and Mild cognitive impairment) were also identified as significant. b) Volcano plot resulting from a PheDAS with the Sex and MaxAgeAtVisit covariates. Controlling for sex and age effects successfully repressed findings from confounded PheCodes (Breast cancer [female] and Mild cognitive impairment)
Experiment 2: Down Syndrome Case Study
Dataset Acquisition
This case study and its procedures were carried out in accordance with the Institutional Review Board of Vanderbilt University and VUMC. Our EMR dataset was obtained from the Synthetic Derivative at Vanderbilt University Medical Center as a fully deidentified collection of clinical data via the Vanderbilt Institute for Clinical and Translational Research. All researchers working with this data received proper Human Subjects training. Our initial cohort consisted of 901,883 subjects, each having records of sex, race, and date of birth. Collectively, these subjects had 20,519,770 ICD event records and 19,555,593 CPT event records.
Cohort Preparation
We first identified all DS cases and IDD controls in our cohort using the createPhenotypeFile tool. For this case study, we defined DS and IDD subjects based on ICD-9 and ICD-10 codes, which are listed in Appendix B. For both the DS and IDD groups, we required that a subject have at least 2 records of the codes listed in the Appendix to be included. From these criteria, we found 2,315 DS subjects and 106,059 IDD subjects. This control group was intentionally designed to cover a broad range of IDDs in order to elucidate phenotypic patterns that are unique to DS. Future investigations with more specific hypotheses, however, may benefit from curating a more targeted comparison group; for example, using PheDAS to compare autism spectrum disorder with DS could reveal more about the absence of psychiatric comorbid conditions in DS (Tables 3 and 4).
After obtaining subject event ages via the convertEvent-ToAge tool, we next used the censorData tool to restrict both the ICD and CPT data to only those events occurring previous to age 10. After this censoring, we were left with 1,830 DS and 52,138 IDD subjects that had both ICD and CPT events previous to age 10. Finally, due to the highly unbalanced nature of our cohort, we used the maximize-Controls tool to match our DS cases to IDD controls with a 1:2 ratio. Matching was performed based on sex (exact match), race (exact match), and minimum ICD/CPT event age (± 0.3 years). One DS subject was dropped at this point, as there did not exist a single suitable match in the IDD cohort (even after varying the tolerance for the minimum age matching criterion), leaving us with 1,829 DS subjects and 3,658 IDD subjects.
ICD Record Analysis
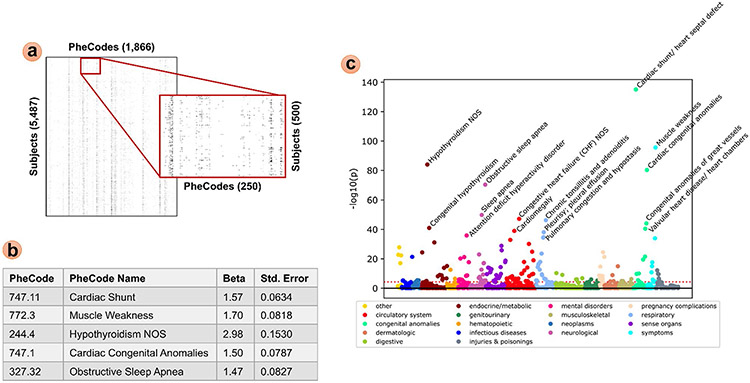
To analyze the ICD signature of DS subjects compared to IDD controls, we performed a binary pyPheWAS analysis. We constructed a binary feature matrix via pyPhewasLookup, then performed mass logistic regression across all PheCodes with the maximum ICD age feature matrix as a covariate using pyPhewasModel. Applying Bonferroni multiple comparisons correction resulted in 177 PheCodes that were statistically significant; the top five most significant PheCodes in this experiment were found to be Cardiac shunt/heart septal defect (747.11), Muscle weakness (772.30), Hypothyroidism NOS (244.40), Cardiac congenital anomalies (747.10), and Obstructive sleep apnea (327.32). All regression results were plotted via pyPhewasPlot with the Bonferroni threshold. This analysis and the resulting Manhattan plot are presented in Fig. 6. All three plots produced by pyPhewasPlot are included in the supplementary material, along with a subset of the tabular regression results.
Fig. 6.
Sample PheDAS of ICD records in DS vs. IDD subjects. (a) A binary feature matrix with PheCodes as columns and subjects as rows was constructed from the ICD event records mapped to PheCodes in pyPhewasLookup. (b) Mass univariate logistic regression was performed across PheCodes in the feature matrix using pyPhewasModel; regression results are listed for the top 5 most significant PheCodes (p < < < 0.001 after Bonferroni multiple comparisons correction). (c) Manhattan plot of all results is shown, with the top 14 most significant PheCodes labeled (p < < < 0.001 after Bonferroni multiple comparisons correction). The Bonferroni threshold is shown as a dotted red line
CPT Record Analysis
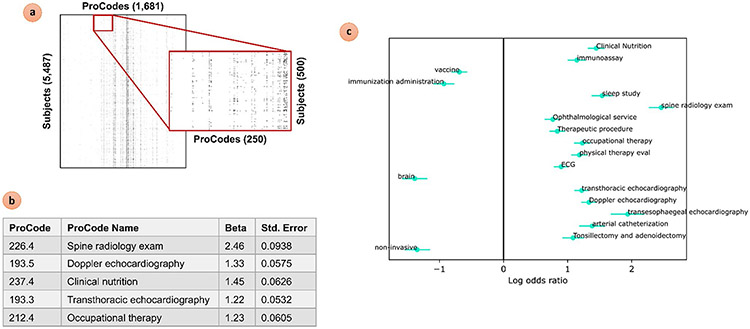
The CPT signature of DS subjects compared to IDD controls was analyzed in a similar manner. We first constructed a binary ProWAS feature matrix via pyProwasLookup. We then performed mass logistic regression across all ProCodes with the maximum CPT age feature matrix as a covariate using pyProwasModel. Applying Bonferroni multiple comparisons correction resulted in 109 ProCodes that were statistically significant, of which Spine radiology exam (226.4), Doppler echocardiography (193.5), Clinical nutrition (237.4), Transthoracic echocardiography (193.3), and Occupational therapy (212.4) were found to be the most significant. Due to the large number of significant ProCodes, the results were plotted via pyProwasPlot with a much stricter custom threshold (puncorrected < 1e-30) in order to pare down results for discussion. This ProWAS analysis and its Log Odds plot of significant results are shown in Fig. 7. All three plots produced by pyProwasPlot are also included in the supplementary material, along with a subset of the tabular regression results.
Fig. 7.
Sample PheDAS of CPT records in DS vs. IDD subjects. (a) A binary feature matrix with ProCodes as columns and subjects as rows was constructed from the CPT event records mapped to ProCodes in pyProwasLookup. (b) Mass univariate logistic regression was performed across ProCodes in the feature matrix using pyProwas-Model; regression results are listed for the top 5 most significant ProCodes (p < < < 0.001 after Bonferroni multiple comparisons correction). (c) The Log Odds plot of top 18 most significant PheCodes (p < < < 0.001 after Bonferroni multiple comparisons correction) is shown, created via pyProwasPlot
Discussion
This article presents the pyPheWAS comprehensive toolkit for performing PheDAS analyses on EMR data. We have described the PheDAS process, wherein EMR data, specifically ICD or CPT codes, are first mapped to meaningful phenotypes and aggregated across each patient’s record. These aggregate measures are then used along with specified covariates to perform mass univariate regression of a target variable on each phenotype. The results of this mass univariate regression are visualized in several ways to facilitate interpretation. We verified the pyPheWAS package by analyzing a synthetic dataset and then further illustrated its function in a real-world setting via a case study comparing DS subjects with non-DS IDD controls. With the analysis complete, our final consideration focuses on how to interpret PheDAS experiments.
The first question we must ask of a PheDAS is how do we verify its correctness? Since PheDAS is primarily a hypothesis generation method, there is no “correct” set of values we can test the strength, significance, or number of associations against. Despite this, PheDAS has a built-in verification test: expected associations. For practically any disease being tested via PheDAS, there are several previously known phenotype associations. These expected associations may be used as reassuring results in a study; a sanity check that establishes baseline credibility for all regression results (Pendergrass et al., 2011). Several such reassuring results are present in the ICD and CPT analyses of our case study. The Manhattan plot in Fig. 6 shows that the PheCodes for Cardiac Congenital Anomalies, Hypothyroidism, and Obstructive Sleep Apnea were found to have positive associations with DS, all of which are known co-morbidities of DS (Bull et al., 2011) (Davidson, 2008). Similarly, the Log Odds plot in Fig. 7 shows that the ProCodes for Echocardiography (ECG), Clinical Nutrition, Sleep Studies, and Physical Therapy were found to be significantly positively associated with DS; again, these ProCodes would be expected as they are procedures which could be used to diagnose and treat known co-morbidities of Down Syndrome (Bull et al., 2011).
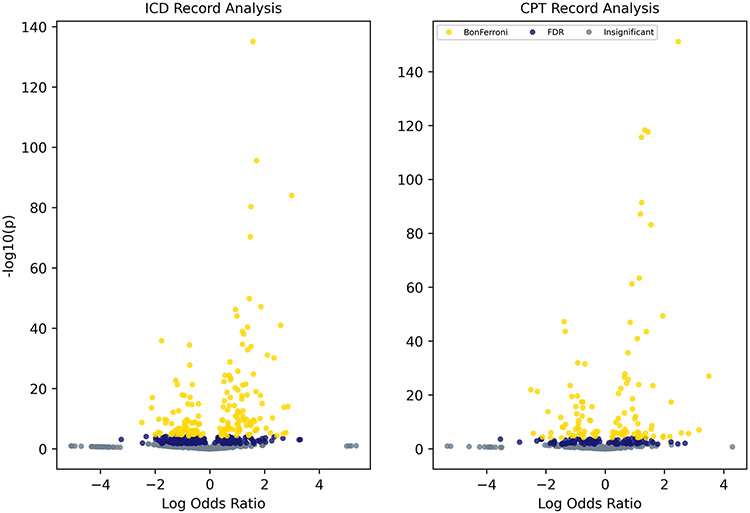
With our expected associations established, the next task is identifying unknown or interesting associations in the PheDAS. The volcano plot may serve as a helpful guide in this step, since it provides an overview of all results and directly links statistical significance with effect size. When viewed via pyPhewasPlot and pyProwasPlot, zooming and panning functions allow users interactively identify results of interest. Figure 8 shows the volcano plots for both the ICD and CPT analyses described in the Results section; it should be noted that phenotype labels have been removed in this figure for legibility.
Fig. 8.
Sample volcano plots. Phenotype labels have been removed for legibility. Users may directly interact with these plots via pyPhewas-Plot and pyProwasPlot. Zooming and panning across the plot enable users to explore phenotypes with regard to both significance and effect size. Thresholds for multiple comparisons correction are presented visually via color (Bonferroni in yellow, FDR in dark blue, and no significance in gray)
An alternative approach for interpreting PheDAS results is assessing the novelty of disease-phenotype associations in terms of existing literature. Previous work has presented a formal method for assessing this type of novelty in Phe-DAS (Chaganti et al., 2019b). In brief, a novelty score is calculated for each disease-phenotype association in a Phe-DAS that measures the degree to which it is already known based on data mined from PubMed abstracts. If a disease-phenotype pairing is present in a large number of PubMed abstracts, the association is assigned a low novelty score and considered well known. In contrast, if a disease-phenotype pairing is present in only a few PubMed abstracts, the association is assigned a high novelty score and considered unknown. This framework is advantageous for exploratory studies in particular, as it does not require a clinical expert to manually review all results and filters the number of potentially novel or interesting PheDAS results down to a manageable amount. This novelty score framework is also available as part of the pyPheWAS package, though not covered in depth here.
We have shown that PheDAS methods are powerful in isolation, but several studies have also demonstrated their utility as support for other types of analyses. Warner et al. performed a proof-of-concept study which employed the PheDAS framework in order to identify subjects for a trans-institutional cohort of multiple myeloma patients (Warner et al., 2013). Li et al. used PheWAS for hypothesis generation in the context of phenotypes related to the genetic components that drive serum uric acid level, then performed a conventional analysis to investigate causal relevance for the identified phenotypes (Li et al., 2018). In the realm of medical imaging, PheDAS has been used successfully to study diseases of the eye and optic nerve. In one such study, PheCode and ProCode feature matrices were used alongside imaging-derived features in a model of visual function for subjects with glaucoma and thyroid eye disease; inclusion of the EMR data was found to improve the explained variance of disease outcomes (Chaganti et al., 2017). Another study used PheDAS to identify PheCodes associated with several optic nerve diseases, then used the identified phenotypes combined with optic nerve imaging features to classify disease subjects and controls. Again, combining the PheCode feature vectors with imaging-derived features produced the most accurate classifiers (Chaganti et al., 2019a). This framework could be extended to the domain of neuroimaging, allowing researchers to support their models of neurological disease with EMR context.
There are several limitations to keep in mind when working with EMR data and the pyPheWAS package. Inherent variability in EMR data is well documented (O’Malley et al., 2005). For example, the ICD coding system’s primary function is to bill insurance companies, not to serve as a proxy for diagnosis. ICD codes are generated by a coding specialist who translates clinician notes into insurance billing codes; this process has many opportunities for noise to enter the system, including at the patient-physician interface (patient-physician communication, physician training, expertise, and attention to detail), at the physician-coder interface (variations in clinical practices, coder training and expertise, facility quality assurance), and from simple human errors (O’Malley et al., 2005). Additionally, EMRs suffer from broader issues of record fragmentation (such as when a patient moves between institutions) and a bias toward sicker populations (EMR events are usually recorded during illness) (Hripcsak & Albers, 2013). Some of this error may be mitigated while creating case and control groups with the createPhenotypeFile tool. Users may specify a code frequency threshold which must be met for a subject to be considered a “true” case or control; enforcing higher temporal thresholds on ICD code events reduces the possibility that mis-coded subjects are mistakenly included in the case or control groups. Additionally, the mapping from ICD codes to PheCodes further reduces EMR variability by consolidating large groups of highly-related ICD codes into a single PheCode (Wei et al., 2017b).
Another common challenge with large-scale association methods such as GWAS and PheDAS is confounding. Users have several options for addressing this issue within the pyPheWAS toolkit. The case–control matching tool, maximizeControls, allows users to match the distributions of potentially confounding variables, such as sex or age, between the case and control populations. Confounding variables may also be added as covariates in the mass univariate regression step; users may specify both primary variables (height or weight) and combined terms (height divided by weight) via the group file to control for various confounding effects. Furthermore, after completing a PheDAS experiment, users should carefully consider the verification of their results by identifying plausible biological links for identified associations and replicating their analysis in an independent population (Smith & Ebrahim, 2002).
These strategies may be used to control for common confounding factors, but investigators should also carefully consider more subtle confounders that might influence their group composition. Individuals suffering from chronic diseases, for example, tend to have more hospital visits and therefore higher numbers of secondary medical diagnoses than individuals with acute ailments; because of this, comparing a chronic disease case group to an acute disease control group may result in false positive phenotype associations unrelated to the chronic disease of interest. This common but challenging scenario could be mitigated in several ways, such as including visit frequency as a matching criterion or redefining the control as a comparable chronic disease. Ultimately, it falls to the investigators using pyPheWAS to precisely select case and control group populations so that their study design properly addresses their specific research question.
A few additional limitations are related directly to the pyPheWAS toolkit. As was previously stated, the ICD-phenotype maps do not cover the full range of possible ICD codes; specifically, the map includes 15,558 ICD-9 codes and 9,505 ICD-10 codes (Denny et al., 2013; Wei et al., 2017a; Wu et al., 2019a). Users are notified when their datasets contain ICD-9 and ICD-10 codes which are not in the mapping and may choose to save the excluded ICD events for inspection. Relatedly, the pyPheWAS map is limited to processing only ICD-9 and ICD-10 codes; newer coding systems such as ICD-11 are not yet supported. To work with an expanded set of ICD-9 and ICD-10 codes or to incorporate ICD-11, users may wish to use a custom phenotype map with pyPheWAS. Though this feature is currently not supported, pyPheWAS is an open source tool, allowing researchers to customize its functionality. To incorporate a custom phenotype map, users may clone the pyPheWAS project from GitHub and replace the default map within the source code. This modification would require that the user first edit their custom map’s headings to match the default map’s headings, and then point the map loading function in the source code to their local custom map. In a similar vein, the pyPheWAS package currently performs only mass logistic regression. Other regression methods have proven interesting in PheDAS analyses, however; for example, one study used of a linear regression to study phenotypic associations with white blood cell count (Warner & Alterovitz, 2012). Again, though this feature is not currently supported, the open source nature of the pyPheWAS toolkit provides the opportunity for other researchers to build in new capabilities. The key modification required for a custom regression type would involve replacing the logistic regression in pyPhewasModel with an alternate regression model from the statsmodels python package (Seabold & Perktold, 2010) and specifying which output values to pull from the fitted model. An alternative statistical python package such as scikitlearn (Pedregosa et al., 2011) may also be used, but would require more modifications to the modeling input and output structure. The pyPheWAS website contains more detailed directions for users wishing to implement either a custom phenotype map or regression modifications.
In this work, we have presented pyPheWAS, a command line toolkit for implementing PheDAS analyses. We have demonstrated a typical PheDAS analysis of children with Down Syndrome compared to children with other intellectual and developmental disorders, complete with suggestions for verifying and interpreting the large amount of statistically significant results. Whether on its own or in combination with other analyses, the pyPheWAS toolkit provides an approachable method for taking advantage of the EMR and integrating this rich resource into our studies of neurological disease.
Information Sharing Statement
The source code for the pyPheWAS software package and the synthetic dataset described in this article are both available at https://github.com/MASILab/pyPheWAS. Software documentation, including instructions for installing the pyPheWAS package, are available at https://pyphewas.readthedocs.io/en/latest/. The dataset used for Experiment 2 (Down Syndrome Case Study) were obtained under license from the Synthetic Derivative at Vanderbilt University Medical Center and are not available to the general public.
Supplementary Material
Acknowledgements
The dataset used for the analyses described were obtained from Vanderbilt University Medical Center’s Synthetic Derivative which is supported by institutional funding and by the Vanderbilt CTSA grants from the National Center for Research Resources, Grant 1UL1RR024975-01, and now at the National Center for Advancing Translational Sciences, Grant 2UL1TR000445-06. Research in this publication was supported by the EKS NICHD of the NIH under Awards P50HD103537, U54HD083211, and U54HD083211-S1. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This research was also supported in part by NSF CAREER 1452485 and NIH grants 5R21EY024036. This project was supported in part by ViSE/VICTR. This research was conducted with the support from Intramural Research Program, National Institute on Aging, NIH. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN. Thank you to Kunal P. Nabar for his work in the early stages of development for pyPheWAS.
Key Terms
- GWAS
Genome-wide association study; mass logistic regression comparing many genotypes to one phenotype.
- PheWAS
Phenome-wide association study; mass logistic regression comparing many phenotypes to one genotype
- PheDAS
Phenome-disease association study; mass logistic regression comparing many ICD phenotypes to one non-genetic target variable
- ProWAS
Procedure-wide association study; mass logistic regression comparing many CPT-phenotypes to one non-genetic target variable
- PheWAS Code
ICD phenotype code used in PheWAS and PheDAS analyses (abbreviated PheCode)
- ProWAS Code
CPT phenotype code used in ProWAS analyses (abbreviated ProCode)
- ICD Code
International Classification of Disease billing code
- CPT Code
Current Procedural Terminology code
Appendix
Appendix A: listing of Case study commands
Experiment 1
pyPhewasPipeline –phenotype = icds.csv –-group = group. csv –reg_type = log –response = Dx –postfix = RegA –legacy = True.
pyPhewasPipeline –-phenotype = icds.csv –group = group.csv –reg_type = log –response = Dx –post-fix = RegB –legacy = True –covariates = MaxAge + Sex.
Experiment 2: Cohort Preparation
createPhenotypeFile –phenotype = master_ICDs.csv –group = master_group.csv –code_freq = 2 –group-out = group.csv.
–case_codes = DS_codes.txt –ctrl_codes = IDD_codes. txt.
convertEventToAge –phenotype = master_ICDs.csv –group = group.csv –etype = ICD –phenotypeout = ICDs_age.csv.
–eventcolumn = ICD_DATE.
convertEventToAge –phenotype = master_CPTs.csv –group = group.csv –etype = CPT –phenotypeout = CPTs_age.csv.
–eventcolumn = CPT_DATE.
censorData –phenotype = ICDs_age.csv –group = group. csv –efield = AgeAtICD –end = 10 –phenotypeout = ICDs_age_cen.csv.
–groupout = group_icd_cen.csv.
censorData –phenotype=CPTs_age.csv –group = group_icd_cen.csv –efield = AgeAtCPT –end = 10.
–phenotypeout = CPTs_age_cen.csv –groupout = group_icd_cpt_cen.csv.
maximizeControls –input = group_icd_cpt_cen.csv –keys = SEX,RACE,MinAgeAtVisit –deltas = ",,0.3" –goal = 2.
–output = group_icd_cpt_cen_matched.csv.
Experiment 2: ICD Record Analysis
pyPhewasLookup –reg_type = log –group = group_icd_cpt_cen_matched.csv –phenotype = ICDs_age_cen.csv.
–outfile = fm_phewas.csv.
pyPhewasModel –reg_type = log –covariates=MaxAge-AtICD –feature_matrix = fm_phewas.csv.
–group=group_icd_cpt_cen_matched.csv –outfile=reg_phewas.csv.
pyPhewasPlot –statfile = reg_phewas.csv –thresh_type = custom –custom_thresh = 1e-30 –outfil = custom_prowas_plots.png.
Experiment 2: CPT Record Analysis
pyProwasLookup –reg_type = log –group = group_icd_cpt_cen_matched.csv –phenotype = CPTs_age_cen.csv.
–outfile = fm_prowas.csv.
pyProwasModel –reg_type = log –covariates = MaxA-geAtCPT –feature_matrix = fm_prowas.csv.
–group=group_icd_cpt_cen_matched.csv –outfile = reg_prowas.csv.
pyProwasPlot –statfile = reg_prowas.csv –thresh_type = custom –custom_thresh = 1e-30 –outfile = custom_prowas_plots.png.
Appendix B: ICD codes used to define case study groups
Table 3.
Down Syndrome Group
| ICD Version | ICD Code | ICD Name |
|---|---|---|
| 9 | 758.0 | Down’s syndrome |
| 10 | Q90.0 | Trisomy 21; nonmosaicism (meiotic nondisjunction) |
| Q90.1 | Trisomy 21, mosaicism (mitotic nondisjunction) | |
| Q90.2 | Trisomy 21, translocation | |
| Q90.9 | Down syndrome, unspecified |
Table 4.
Other Intellectual and Developmental Disabilities Group
| ICD Version | ICD Code | ICD Name |
|---|---|---|
| 9 | 314.00 | Attention deficit disorder without mention of hyperactivity |
| 314.01 | Attention deficit disorder with hyperactivity | |
| 314.2 | Hyperkinetic conduct disorder | |
| 317 | Mild intellectual disabilities | |
| 318 | Other specified intellectual disabilities | |
| 318.0 | Moderate intellectual disabilities | |
| 318.1 | Severe intellectual disabilities | |
| 318.2 | Profound intellectual disabilities | |
| 319 | Unspecified intellectual disabilities | |
| 315.39 | Other developmental speech or language disorder | |
| 315.31 | Expressive language disorder | |
| 315.32 | Mixed receptive-expressive language disorder | |
| 315.34 | Speech and language developmental delay due to hearing loss | |
| 315.35 | Childhood onset fluency disorder | |
| 315.02 | Developmental dyslexia | |
| 315 | Specific delays in development | |
| 315.0 | Developmental reading disorder | |
| 315.00 | Developmental reading disorder; unspecified | |
| 315.09 | Other specific developmental reading disorder | |
| 315.2 | Other specific developmental learning difficulties | |
| 315.4 | Developmental coordination disorder | |
| 315.8 | Other specified delays in development | |
| 315.9 | Unspecified delay in development | |
| 299 | Pervasive developmental disorders | |
| 299.0 | Autistic disorder | |
| 299.00 | Autistic disorder; current or active state | |
| 299.01 | Autistic disorder; residual state | |
| 299.1 | Childhood disintegrative disorder | |
| 299.10 | Childhood disintegrative disorder; current or active state | |
| 299.8 | Other specified pervasive developmental disorders | |
| 299.80 | Other specified pervasive developmental disorders; current or active state | |
| 299.81 | Other specified pervasive developmental disorders; residual state | |
| 299.9 | Unspecified pervasive developmental disorder | |
| 299.90 | Unspecified pervasive developmental disorder; current or active state | |
| 330.8 | Other specified cerebral degenerations in childhood | |
| 307.21 | Transient tic disorder | |
| 307.22 | Chronic motor or vocal tic disorder | |
| 307.23 | Tourette's disorder | |
| 307.2 | Tics | |
| 307.3 | Stereotypic movement disorder | |
| 333.71 | Athetoid cerebral palsy | |
| 9 | 343.8 | Other specified infantile cerebral palsy |
| 343.9 | Infantile cerebral palsy; unspecified | |
| 759.83 | Fragile X syndrome | |
| 759.81 | Prader-Willi syndrome | |
| 799.51 | Attention or concentration deficit | |
| 799.52 | Cognitive communication deficit | |
| 799.53 | Visuospatial deficit | |
| 799.54 | Psychomotor deficit | |
| 799.55 | Frontal lobe and executive function deficit | |
| 784.52 | Fluency disorder in conditions classified elsewhere | |
| 784.59 | Other speech disturbance | |
| 784.61 | Alexia and dyslexia | |
| 315.01 | Alexia | |
| 784.69 | Other symbolic dysfunction | |
| 784.6 | Other symbolic dysfunction | |
| 784.60 | Symbolic dysfunction; unspecified | |
| F70 | Mild intellectual disabilities | |
| F71 | Moderate intellectual disabilities | |
| F72 | Severe intellectual disabilities | |
| F73 | Profound intellectual disabilities | |
| 10 | F78 | Other intellectual disabilities |
| F79 | Unspecified intellectual disabilities | |
| F80.0 | Phonological disorder | |
| F80.1 | Expressive language disorder | |
| F80.2 | Mixed receptive-expressive language disorder | |
| F80.4 | Speech and language development delay due to hearing loss | |
| F80.81 | Childhood onset fluency disorder | |
| F80.82 | Social pragmatic communication disorder | |
| F80.89 | Other developmental disorders of speech and language | |
| F80.9 | Developmental disorder of speech and language; unspecified | |
| F81.0 | Specific reading disorder | |
| F81.2 | Mathematics disorder | |
| F81.81 | Disorder of written expression | |
| F81.89 | Other developmental disorders of scholastic skills | |
| F82 | Specific developmental disorder of motor function | |
| F84.0 | Autistic disorder | |
| F84.2 | Rett's syndrome | |
| F84.3 | Other childhood disintegrative disorder | |
| F84.5 | Asperger's syndrome | |
| F84.8 | Other pervasive developmental disorders | |
| F84.9 | Pervasive developmental disorder; unspecified | |
| F88 | Other disorders of psychological development | |
| F89 | Unspecified disorder of psychological development | |
| F90.0 | Attention-deficit hyperactivity disorder; predominantly inattentive type | |
| F90.1 | Attention-deficit hyperactivity disorder; predominantly hyperactive type | |
| F90.2 | Attention-deficit hyperactivity disorder; combined type | |
| F90.8 | Attention-deficit hyperactivity disorder; other type | |
| F90.9 | Attention-deficit hyperactivity disorder; unspecified type | |
| F94.0 | Selective mutism | |
| F94.1 | Reactive attachment disorder of childhood | |
| F94.2 | Disinhibited attachment disorder of childhood | |
| F94.8 | Other childhood disorders of social functioning | |
| F94.9 | Childhood disorder of social functioning; unspecified | |
| 10 | F95.0 | Transient tic disorder |
| F95.1 | Chronic motor or vocal tic disorder | |
| F95.2 | Tourette's disorder | |
| F95.8 | Other tic disorders | |
| F95.9 | Tic disorder; unspecified | |
| F98.4 | Stereotyped movement disorders | |
| F98.8 | Other specified behavioral and emotional disorders with onset usually occurring in childhood and adolescence | |
| F98.9 | Unspecified behavioral and emotional disorders with onset usually occurring in childhood and adolescence | |
| G11.0 | Congenital nonprogressive ataxia | |
| G11.1 | Early-onset cerebellar ataxia | |
| G11.2 | Late-onset cerebellar ataxia | |
| G11.3 | Cerebellar ataxia with defective DNA repair | |
| G11.4 | Hereditary spastic paraplegia | |
| G11.8 | Other hereditary ataxias | |
| G11.9 | Hereditary ataxia; unspecified | |
| G80.0 | Spastic quadriplegic cerebral palsy | |
| G80.1 | Spastic diplegic cerebral palsy | |
| G80.3 | Athetoid cerebral palsy | |
| G80.4 | Ataxic cerebral palsy | |
| G80.8 | Other cerebral palsy | |
| G80.9 | Cerebral palsy; unspecified | |
| G93.0 | Cerebral cysts | |
| Q99.2 | Fragile X chromosome | |
| Q86.0 | Fetal alcohol syndrome (dysmorphic) | |
| Q86.8 | Other congenital malformation syndromes due to known exogenous causes | |
| Q87.1 | Congenital malformation syndromes predominantly associated with short stature | |
| Q93.81 | Velo-cardio-facial syndrome | |
| Q93.88 | Other microdeletions | |
| Q93.89 | Other deletions from the autosomes | |
| H53.10 | Unspecified subjective visual disturbances | |
| H53.121 | Transient visual loss; right eye | |
| H53.122 | Transient visual loss; left eye | |
| H53.123 | Transient visual loss; bilateral | |
| H53.129 | Transient visual loss; unspecified eye | |
| H53.131 | Sudden visual loss; right eye | |
| H53.132 | Sudden visual loss; left eye | |
| H53.133 | Sudden visual loss; bilateral | |
| H53.139 | Sudden visual loss; unspecified eye | |
| H53.141 | Visual discomfort; right eye | |
| H53.142 | Visual discomfort; left eye | |
| H53.143 | Visual discomfort; bilateral | |
| H53.149 | Visual discomfort; unspecified | |
| H53.15 | Visual distortions of shape and size | |
| H53.16 | Psychophysical visual disturbances | |
| H53.19 | Other subjective visual disturbances | |
| H53.30 | Unspecified disorder of binocular vision | |
| H53.31 | Abnormal retinal correspondence | |
| H53.32 | Fusion with defective stereopsis | |
| H53.33 | Simultaneous visual perception without fusion | |
| H53.34 | Suppression of binocular vision | |
| H53.40 | Unspecified visual field defects | |
| H53.451 | Other localized visual field defect; right eye | |
| 10 | H53.452 | Other localized visual field defect; left eye |
| H53.459 | Other localized visual field defect; unspecified eye | |
| H53.453 | Other localized visual field defect; bilateral | |
| H53.461 | Homonymous bilateral field defects; right side | |
| H53.462 | Homonymous bilateral field defects; left side | |
| H53.469 | Homonymous bilateral field defects; unspecified side | |
| H53.47 | Heteronymous bilateral field defects | |
| H53.481 | Generalized contraction of visual field; right eye | |
| H53.482 | Generalized contraction of visual field; left eye | |
| H53.483 | Generalized contraction of visual field; bilateral | |
| H53.489 | Generalized contraction of visual field; unspecified eye | |
| H53.50 | Unspecified color vision deficiencies | |
| H53.59 | Other color vision deficiencies | |
| H53.8 | Other visual disturbances | |
| H53.9 | Unspecified visual disturbance | |
| H90.0 | Conductive hearing loss; bilateral | |
| H90.2 | Conductive hearing loss; unspecified | |
| H90.3 | Sensorineural hearing loss; bilateral | |
| H90.41 | Sensorineural hearing loss; unilateral; right ear; with unrestricted hearing on the contralateral side | |
| H90.42 | Sensorineural hearing loss; unilateral; left ear; with unrestricted hearing on the contralateral side | |
| H90.5 | Unspecified sensorineural hearing loss | |
| H90.6 | Mixed conductive and sensorineural hearing loss; bilateral | |
| H90.71 | Mixed conductive and sensorineural hearing loss; unilateral; right ear; with unrestricted hearing on the contralateral side | |
| H90.72 | Mixed conductive and sensorineural hearing loss; unilateral; left ear; with unrestricted hearing on the contralateral side | |
| H90.8 | Mixed conductive and sensorineural hearing loss; unspecified | |
| H90.A11 | Conductive hearing loss; unilateral; right ear with restricted hearing on the contralateral side | |
| H90.A12 | Conductive hearing loss; unilateral; left ear with restricted hearing on the contralateral side | |
| H90.A21 | Sensorineural hearing loss; unilateral; right ear; with restricted hearing on the contralateral side | |
| H90.A22 | Sensorineural hearing loss; unilateral; left ear; with restricted hearing on the contralateral side | |
| H90.A31 | Mixed conductive and sensorineural hearing loss; unilateral; right ear with restricted hearing on the contralateral side | |
| H90.A32 | Mixed conductive and sensorineural hearing loss; unilateral; left ear with restricted hearing on the contralateral side | |
| H93.25 | Central auditory processing disorder | |
| F99 | Mental disorder; not otherwise specified | |
| R13.0 | Aphagia | |
| R13.1 | Dysphagia | |
| R13.11 | Dysphagia; oral phase | |
| R13.12 | Dysphagia; oropharyngeal phase | |
| R13.13 | Dysphagia; pharyngeal phase | |
| R13.14 | Dysphagia; pharyngoesophageal phase | |
| R13.19 | Other dysphagia | |
| R41.9 | Unspecified symptoms and signs involving cognitive functions and awareness | |
| R41.1 | Anterograde amnesia | |
| R41.2 | Retrograde amnesia | |
| R41.3 | Other amnesia | |
| 10 | R41.81 | Age-related cognitive decline |
| R41.82 | Altered mental status; unspecified | |
| R41.83 | Borderline intellectual functioning | |
| R41.840 | Attention and concentration deficit | |
| R41.841 | Cognitive communication deficit | |
| R41.842 | Visuospatial deficit | |
| R41.843 | Psychomotor deficit | |
| R41.844 | Frontal lobe and executive function deficit | |
| R41.89 | Other symptoms and signs involving cognitive functions and awareness | |
| R44.0 | Auditory hallucinations | |
| R44.1 | Visual hallucinations | |
| R44.2 | Other hallucinations | |
| R44.8 | Other symptoms and signs involving general sensations and perceptions | |
| R44.9 | Unspecified symptoms and signs involving general sensations and perceptions | |
| R47.82 | Fluency disorder in conditions classified elsewhere | |
| R47.89 | Other speech disturbances | |
| R47.9 | Unspecified speech disturbances | |
| R48.0 | Dyslexia and alexia | |
| R48.1 | Agnosia | |
| R48.2 | Apraxia | |
| R48.8 | Other symbolic dysfunctions | |
| R48.9 | Unspecified symbolic dysfunctions | |
| R62.0 | Delayed milestone in childhood | |
| R62.50 | Unspecified lack of expected normal physiological development in childhood | |
| R62.51 | Failure to thrive (child) | |
| R62.52 | Short stature (child) | |
| R62.59 | Other lack of expected normal physiological development in childhood |
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s12021-021-09553-4.
Data Availability Statement
The data that support the findings of this case study are available from the Synthetic Derivative at Vanderbilt University Medical Center, but restrictions apply to the availability of this data, which were used under license for the current study, and so are not publicly available.
References
- Ahmad NA, Kochman ML, Long WB, Furth EE, & Ginsberg GG (2002). Efficacy, safety, and clinical outcomes of endoscopic mucosal resection: A study of 101 cases. Gastrointestinal Endoscopy, 55, 390–396. 10.1067/mge.2002.121881 [DOI] [PubMed] [Google Scholar]
- Bastarache L, Denny JC (2011). The Use of ICD-9 Codes in Genetic Association Studies. In: AMIA Annual Symposium Proceedings, p 1738 [Google Scholar]
- Boland MR, Hripcsak G, Albers DJ, Wei Y, Wilcox AB, Wei J, Li J, Lin S, Breene M, Myers R, Zimmerman J, Papapanou PN, & Weng C (2014). Discovering medical conditions associated with periodontitis using linked electronic health records. Journal of Clinical Periodontology, 40, 1–19. 10.1111/jcpe.12086.Discovering [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull MJ, Saal HM, Braddock SR, Enns GM, Gruen JR, Perrin JM, Saul RA, Tarini BA, Hersh JH, Mendelsohn NJ, Hanson JW, Lloyd-Puryear MA, Musci TJ, Rasmussen SA, Downs SM, & Spire P (2011). Clinical report - Health supervision for children with Down syndrome. Pediatrics, 128, 393–406. 10.1542/peds.2011-1605 [DOI] [PubMed] [Google Scholar]
- Carroll RJ, Bastarache L, & Denny JC (2014). R PheWAS: Data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics, 30, 2375–2376. 10.1093/bioinformatics/btu197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti S, Mawn LA, Kang H, Egan J, Resnick SM, Beason-Held LL, Landman BA, & Lasko TA (2019a). Electronic Medical Record Context Signatures Improve Diagnostic Classification Using Medical Image Computing. IEEE J Biomed Heal INFORMATICS, 23, 2052–2062. 10.1017/9781316671849.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti S, Robinson JR, Bermudez C, Lasko T, Mawn LA, Landman BA (2017). EMR-Radiological Phenotypes in Diseases of the Optic Nerve and their Association with Visual Function. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, pp 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti S, Welty VF, Taylor W, Albert K, Failla MD, Cascio C, et al. (2019). Discovering novel disease comorbidities using electronic medical records. PLoS One, 14, 1–14. 10.1371/journal.pone.0225495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danciu I, Cowan JD, Basford M, Wang X, Saip A, Osgood S, Shirey-Rice J, Kirby J, & Harris PA (2014). Secondary use of clinical data: The Vanderbilt approach. Journal of Biomedical Informatics, 52, 28–35. 10.1016/j.jbi.2014.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson MA (2008). Primary Care for Children and Adolescents with Down Syndrome. Pediatric Clinics of North America, 55, 1099–1111. 10.1016/j.pcl.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, Field JR, Pulley JM, Ramirez AH, Bowton E, Basford MA, Carrell DS, Peissig PL, Kho AN, Pacheco JA, Rasmussen LV, Crosslin DR, Crane PK, Pathak J, … Roden DM (2013). Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nature Biotechnology, 31, 1102–1110. 10.1038/nbt.2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y, Chai HS, Bastarache L, Zuvich R, Peissig P, Carrell D, Ramirez AH, Pathak J, Wilke RA, Rasmussen L, Wang X, Pacheco JA, Kho AN, Hayes MG, … De Andrade M (2011). Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: Using electronic medical records for genome- and phenome-wide studies. American Journal of Human Genetics, 89, 529–542. 10.1016/j.ajhg.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, & Crawford DC (2010). PheWAS: Demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics, 26, 1205–1210. 10.1093/bioinformatics/btq126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehm MG, Aponte JL, Chiano MN, Yerges-Armstrong LM, Johnson T, Barker JN, et al. (2017). Phenome-wide association study using research participants’ self-reported data provides insight into the Th17 and IL-17 pathway. PLoS One, 12, 1–14. 10.1371/journal.pone.0186405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- eMERGE Consortium. (2021). Lessons learned from the eMERGE Network: Balancing genomics in discovery and practice. Hum Genet Genomics Adv, 2, 100018. 10.1016/j.xhgg.2020.100018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels EA, Parsons R, Besson C, Morton LM, Enewold L, Ricker W, Yanik EL, Arem H, Austin AA, & Pfeiffer RM (2016). Comprehensive evaluation of medical conditions associated with risk of non-Hodgkin lymphoma using medicare claims (“MedWAS”). Cancer Epidemiology, Biomarkers & Prevention, 25, 1105–1113. 10.1158/1055-9965.EPI-16-0212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RS, Lloyd JF, & Pierce LA (2012). Clinical use of an enterprise data warehouse. American Medical Informatics Association Annual Symposium Proceedings, 2012, 189–198. [PMC free article] [PubMed] [Google Scholar]
- HCUP CCS-Services and Procedures. (2018). Healthcare Cost and Utilization Project. [Google Scholar]
- Healthcare Cost and Utilization Project Overview of the National (Nationwide) Inpatient Sample (NIS). (2021a). https://www.hcup-us.ahrq.gov/nisoverview.jsp [Google Scholar]
- Hebbring SJ (2014). The challenges, advantages and future of phenome-wide association studies. Immunology, 141, 157–165. 10.1111/imm.12195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbring SJ, Schrodi SJ, Ye Z, Zhou Z, Page D, & Brilliant MH (2013). A PheWAS approach in studying HLA-DRB1*1501. Genes and Immunity, 14, 187–191. 10.1038/gene.2013.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, & Manolio TA (2009). Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A, 106, 9362–9367. 10.1073/pnas.0903103106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopcroft JE, & Karp RM (1973). An n5/2 Algorithm for Maximum Matchings in Bipartite Graphs. SIAM Journal on Computing, 2, 225–231. 10.1137/0202019 [DOI] [Google Scholar]
- Hripcsak G, & Albers DJ (2013). Next-generation phenotyping of electronic health records. J Am Med Informatics Assoc, 20, 117–121. 10.1136/amiajnl-2012-001145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JD (2007). Matplotlib : A 2D Graphics Environment. Comput Sci Eng, 9, 90–95. [Google Scholar]
- Kirby JC, Speltz P, Rasmussen LV, Basford M, Gottesman O, Peissig PL, Pacheco JA, Tromp G, Pathak J, Carrell DS, Ellis SB, Lingren T, Thompson WK, Savova G, Haines J, Roden DM, Harris PA, & Denny JC (2016). PheKB: A catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Informatics Assoc, 23, 1046–1052. 10.1093/jamia/ocv202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Meng X, Spiliopoulou A, Timofeeva M, Wei WQ, Gifford A, Shen X, He Y, Varley T, McKeigue P, Tzoulaki I, Wright AF, Joshi P, Denny JC, Campbell H, & Theodoratou E (2018). MR-PheWAS: Exploring the causal effect of SUA level on multiple disease outcomes by using genetic instruments in UK biobank. Annals of the Rheumatic Diseases, 77, 1039–1047. 10.1136/annrheumdis-2017-212534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ye Z, Mayer JG, Hoch BA, Green C, Rolak L, Cold C, Khor SS, Zheng X, Miyagawa T, Tokunaga K, Brilliant MH, & Hebbring SJ (2016). Phenome-wide association study maps new diseases to the human major histocompatibility complex region. Journal of Medical Genetics, 53, 681–689. 10.1136/jmedgenet-2016-103867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie SL, Wyatt MC, Schuff R, Tenenbaum JD, & Anderson N (2012). Practices and perspectives on building integrated data repositories: Results from a 2010 CTSA survey. J Am Med Informatics Assoc, 19, e119–e124. 10.1136/amiajnl-2011-000508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley KJ, Cook KF, Price MD, Wildes KR, Hurdle JF, & Ashton CM (2005). Measuring diagnoses: ICD code accuracy. Health Services Research, 40, 1620–1639. 10.1111/j.1475-6773.2005.00444.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Alexandre P, Cournapeau D, Brucher M, Perrot M, & Duchesnay E (2011). Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research, 12, 2825–2830. [Google Scholar]
- Pendergrass SA, Brown-Gentry K, Dudek SM, Torstenson ES, Ambite JL, Avery CL, Buyske S, Cai C, Fesinmeyer MD, Haiman C, Heiss G, Hindorff LA, Hsu CN, Jackson RD, Kooperberg C, Le Marchand L, Lin Y, Matise TC, Moreland L, … Ritchie MD (2011). The use of phenome-wide association studies (PheWAS) for exploration of novel genotype-phenotype relationships and pleiotropy discovery. Genetic Epidemiology, 35, 410–422. 10.1002/gepi.20589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca WA, Yawn BP, & Sauver JL, Grossardt BR, Melton LJ,. (2012). History of the Rochester epidemiology project: Half a century of medical records linkage in a US population. Mayo Clinic Proceedings, 87, 1202–1213. 10.1016/j.mayocp.2012.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran C, Bloomrosen M, Hammond WE, Labkoff S, Markel-Fox S, Tang PC, & Detmer DE (2007). Toward a National Framework for the Secondary Use of Health Data: An American Medical Informatics Association White Paper. J Am Med Informatics Assoc, 14, 1–9. 10.1197/jamia.M2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabold S, Perktold J (2010). Statsmodels: Econometric and Statistical Modeling with Python. In: PROC. OF THE 9th PYTHON IN SCIENCE CONF. pp 92–96 [Google Scholar]
- Simonti CN, Vernot B, Bastarache L, Bottinger E, Carrell DS, Chisholm RL, Crosslin DR, Hebbring SJ, Jarvik GP, Kullo IJ, Li R, Pathak J, Ritchie MD, Roden DM, Verma SS, Tromp G, Prato JD, Bush WS, Akey JM, Denny JC, Capra JA (2016). The phenotypic legacy of admixture between modern humans and Neandertals. Science (80- ) 351:737–741. 10.1126/science.aad2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GD, & Ebrahim S (2002). Data dredging, bias, or confounding. British Medical Journal, 325, 1437–1438. 10.1136/bmj.325.7378.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utah Population Database. (2021b). https://uofuhealth.utah.edu/huntsman/utah-population-database/ [Google Scholar]
- Warner JL, & Alterovitz G (2012). Phenome based analysis as a means for discovering context dependent clinical reference ranges. American Medical Informatics Association Annual Symposium Proceedings, 2012, 1441–1449. [PMC free article] [PubMed] [Google Scholar]
- Warner JL, Alterovitz G, Bodio K, & Joyce RM (2013). External phenome analysis enables a rational federated query strategy to detect changing rates of treatment-related complications associated with multiple myeloma. J Am Med Informatics Assoc, 20, 696–699. 10.1136/amiajnl-2012-001355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W-Q, Bastarache LA, Carroll RJ, Marlo JE, Osterman TJ, Gamazon ER, et al. (2017a). Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One, 12, 1–16. 10.1371/journal.pone.0175508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W-Q, Bastarache LA, Carroll RJ, Marlo JE, Osterman TJ, Gamazon ER, Cox NJ, Roden DM, & Denny JC (2017b). Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One, 12, e0175508. 10.1371/journal.pone.0175508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Gifford A, Meng X, Li X, Campbell H, Varley T, Zhao J, Carroll R, Bastarache L, Denny JC, Theodoratou E, & Wei W-Q (2019a). Mapping ICD-10 and ICD-10-CM Codes to Phecodes: Workflow Development and Initial Evaluation. JMIR Med Informatics, 7, e14325. 10.2196/14325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Gifford A, Meng X, Li X, Campbell H, Varley T, Zhao J, Carroll R, Bastarache L, Denny JC, Theodoratou E, & Wei WQ (2019b). Mapping ICD-10 and ICD-10-CM codes to phecodes: Workflow development and initial evaluation. Journal of Medical Internet Research, 21, 1–13. 10.2196/14325 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this case study are available from the Synthetic Derivative at Vanderbilt University Medical Center, but restrictions apply to the availability of this data, which were used under license for the current study, and so are not publicly available.