Abstract
Although type 2C protein phosphatases (PP2Cs) have been demonstrated to play important roles in regulating plant development and various stress responses, their specific roles in rice abiotic stress tolerance are still largely unknown. In this study, the functions of OsPP65 in rice osmotic and salt stress tolerance were investigated. Here, we report that OsPP65 is responsive to multiple stresses and is remarkably induced by osmotic and salt stress treatments. OsPP65 was highly expressed in rice seedlings and leaves and localized in the nucleus and cytoplasm. OsPP65 knockout rice plants showed enhanced tolerance to osmotic and salt stresses. Significantly higher induction of genes involved in jasmonic acid (JA) and abscisic acid (ABA) biosynthesis or signaling, as well as higher contents of endogenous JA and ABA, were observed in the OsPP65 knockout plants compared with the wild-type plants after osmotic stress treatment. Further analysis indicated that JA and ABA function independently in osmotic stress tolerance conferred by loss of OsPP65. Moreover, metabolomics analysis revealed higher endogenous levels of galactose and galactinol but a lower content of raffinose in the OsPP65 knockout plants than in the wild-type plants after osmotic stress treatment. These results together suggest that OsPP65 negatively regulates osmotic and salt stress tolerance through regulation of the JA and ABA signaling pathways and modulation of the raffinose family oligosaccharide metabolism pathway in rice. OsPP65 is a promising target for improvement of rice stress tolerance using gene editing.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12284-022-00581-5.
Keywords: PP2C, Osmotic stress tolerance, ABA, JA, Raffinose family oligosaccharide, Rice
Background
As sessile organisms, plants are always faced with various environmental stresses such as high salinity, drought, and low temperature (Sharma et al. 2013). These stresses have adverse effects on plant growth and seed production. To survive, plants have developed intricate mechanisms to efficiently perceive external signals and tailor their responses to the precise environmental conditions encountered (Shinozaki et al. 2003; Sharma et al. 2013). Reversible protein phosphorylation mediated by protein kinases and phosphatases is one such adaptive cellular activity that allows plants to sense and transduce a broad range of environmental signals (Luan 2003; Fuchs et al. 2013; Singh et al. 2015). Plant protein phosphatases predominantly belong to three classes of protein serine⁄threonine phosphatases, of which the major class is the type 2C protein phosphatase (PP2C) class (Fuchs et al. 2013).
PP2C-type protein phosphatases, which require Mn2+ or Mg2+ for their activities, are the largest protein phosphatase family in plants and have been reported to play important roles in signal transduction pathways associated with stress tolerance, innate immunity, stomatal closure, seed dormancy, apical hook development, and plant yield (Schweighofer et al. 2004, 2007; Singh et al. 2016; Lu et al. 2017; Nishimura et al. 2018; Wang et al. 2020; Yu et al. 2020; Chen et al. 2021; Wong et al. 2021). For instance, PP2C49 negatively regulates Arabidopsis salt tolerance by inhibiting the activity of AtHKT1;1, whereas ZmPP2C55 positively regulates maize drought tolerance (Chu et al. 2021; Zhang et al. 2022). AP2C1, which inactivates the mitogen-activated protein kinases MPK4 and MPK6, regulates plant defense responses to both the necrotrophic pathogen Botrytis cinerea and hemibiotrophic pathogen Pseudomonas syringae in Arabidopsis (Schweighofer et al. 2007; Shubchynskyy et al. 2017). ABA-HYPERSENSITIVE GERMINATION 1 controls seed dormancy and germination by interacting with DELAY OF GERMINATION 1 in Arabidopsis (Née et al. 2017; Nishimura et al. 2018). A PP2C-1 allele from wild soybean ZYD7 enhances 100-seed weight by enlarging the size of integument cells and activating a series of genes related to seed traits (Lu et al. 2017).
In rice, 90 PP2C genes have been predicted, but the functions of only few members have been reported (Fujii and Toriyama 2008; Singh et al. 2010; Ni et al. 2019). OsPP18, OsPP2C09, OsPP108, and OsBIPP2C1 have been demonstrated to be involved in rice tolerance to abiotic stresses, such as drought, osmotic, salt, mannitol, and oxidative stresses (Hu et al. 2006; You et al. 2014; Singh et al. 2015; Miao et al. 2020; Min et al. 2021). The PP2C XA21 BINDING PROTEIN15 negatively regulates XA21-mediated innate immunity in rice (Park et al. 2008), and OsBIPP2C1/2 was found to significantly enhance resistance to tobacco mosaic virus when expressed in tobacco (Hu et al. 2006, 2009). A recent report indicated that OsPP95, which is negatively regulated by the ubiquitin E2 conjugase PHOSPHATE2, positively regulates phosphate homeostasis and remobilization by dephosphorylating the phosphate transporters (PTs) OsPT2 and OsPT8 and influencing their trafficking from the endoplasmic reticulum to plasma membrane in rice (Yang et al. 2020). Despite these intriguing results, the specific roles of most of the PP2C genes in rice are still largely unknown.
In our previous study to screen for proteins interacting with OsGF14b that are involved in rice drought response (Liu et al. 2019) through yeast two-hybrid assay, we identified that one PP2C protein, OsPP65, was among the candidate interaction partners. But its specific function in rice stress response has not been reported. In this study, we integrated transgenics, transcriptomics and metabolomics analyses to investigate the biological role of OsPP65 in rice abiotic stress tolerance. We found that the transcription of OsPP65 (LOC_Os04g37660) is remarkably induced by various stresses and that knockout of OsPP65 enhances tolerance to osmotic and salt stresses in rice. Further analysis showed that OsPP65 regulates rice responses to abiotic stresses by regulating the abscisic acid (ABA) and jasmonic acid (JA) signaling pathways independently. Moreover, analysis of the primary metabolomes of the wild-type cultivar Nipponbare and OsPP65 knockout plants indicated that the raffinose family oligosaccharide (RFO) metabolic pathway might play important roles in stress tolerance conferred by loss of OsPP65 in rice.
Results
Expression Profile of OsPP65 Under Different Stress Treatments
To analyze the responsiveness of OsPP65 to various abiotic stresses and hormones in rice, wild-type Nipponbare plants were subjected to cold, NaCl, PEG, H2O2, ABA, or JA treatment, and gene expression analysis was conducted by quantitative RT-PCR (qRT-PCR) at multiple time points from 0 to 24 h post treatment (hpt). The results showed that the transcription level of OsPP65 was significantly induced at almost all the time points after the NaCl and PEG treatments (Fig. 1). In response to H2O2 and the hormone treatments, the expression of OsPP65 first remarkably increased but then decreased to the normal level by 24 hpt. In cold-treated plants, significant induction of OsPP65 was observed at 3 and 6 hpt, whereas significant reduction of expression was detected at 24 hpt (Fig. 1). These results together indicate that OsPP65 is a stress-responsive PP2C gene that may play important roles in stress tolerance in rice.
Fig. 1.
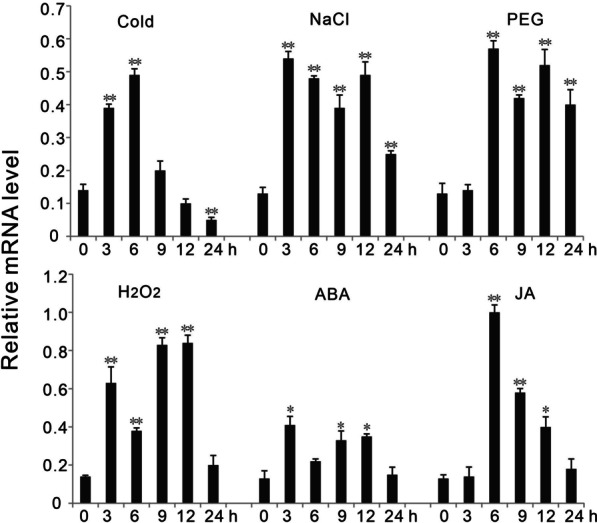
The change in expression of OsPP65 in response to various stress and chemical treatments. Cold, 8 ℃; NaCl, 150 mM NaCl; PEG, 20% PEG6000; H2O2, 1% H2O2, ABA, 100 μM ABA; JA, 100 μM JA. Values represent the means ± SD of three biological replicates (5 plants for each replicate), and “Relative expression level” indicates the expression relative to EF1α, which was used as an internal control. The asterisks indicate significant differences compared with the 0 h time point at *P < 0.05 and **P < 0.01 (Dunnett's test)
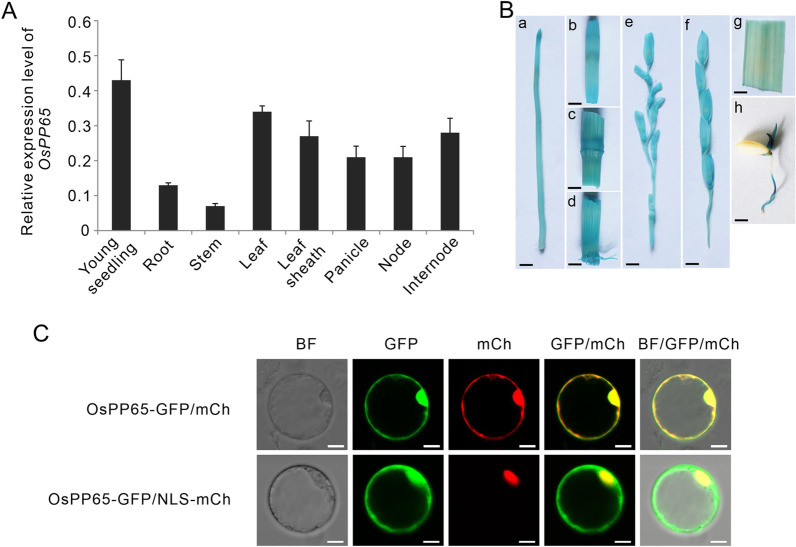
OsPP65 is Highly Expressed in Rice Seedlings and Leaves and Localized in the Nucleus and Cytoplasm
To investigate the biological role of OsPP65 in rice, we first analyzed the spatial and temporal expression of OsPP65 in different rice tissues and found that this gene was ubiquitously expressed during the entire rice life cycle, with a relatively higher transcript level in the seedlings and leaves (Fig. 2A). To validate this result, we generated transgenic plants in which expression of β-glucuronidase (GUS) was driven by the promoter of OsPP65. Histochemical analysis revealed GUS activity in all the tissues examined (Fig. 2B), which was consistent with the results of qRT-PCR. The subcellular localization of OsPP65 was determined through transient expression of an OsPP65-GFP fusion protein in rice protoplasts co-expressing a mCherry protein (as a marker for nucleus and cytoplasm localization) or a nuclear localization signal-tagged mCherry protein. At 24 h after transformation, fluorescence microscopy analysis showed that the OsPP65-GFP fusion protein was localized in the nucleus and cytoplasm of rice cells (Fig. 2C).
Fig. 2.
The expression patterns of OsPP65 in different rice tissues and its subcellular localization. A Transcription analysis of OsPP65 in different rice tissues by quantitative RT-PCR. Values represent the means ± SD of three biological replicates. B GUS staining analysis of OsPP65 promoter-GUS expression in different rice tissues. a, root; b, the third node; c, the second node; d, the first node; e, panicle of the booting stage; f, panicle of the heading stage; g, leaf; h, 3 day-old seedling. Scale bar = 5 mm. C, Subcellular localization of OsPP65 in rice protoplasts. BF indicates bright field, mCh indicates mCherry, and NLS-mCh indicates nuclear localization signal-tagged mCherry. Scale bar = 10 μm
Knockout of OsPP65 Enhances Tolerance to Osmotic and Salt Stresses
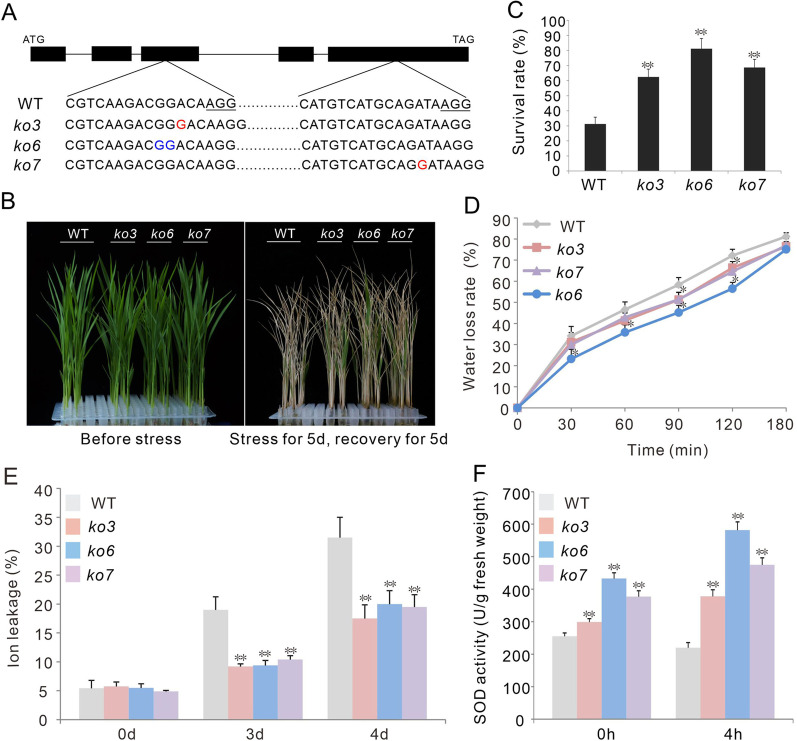
To explore the regulatory roles of OsPP65 in rice responses to abiotic stress, we generated OsPP65 knockout (ko) plants using CRISPR/Cas9 gene editing technology. Two target sites were selected for gene editing and 20 independent transgenic lines were obtained. Through PCR amplification and sequencing analysis, three homozygous lines with different frameshift mutations (ko3, ko6, and ko7) were chosen for stress tolerance evaluation (Fig. 3A, Additional file 1: Fig. S1).
Fig. 3.
OsPP65 knockout rice plants show enhanced osmotic stress tolerance. Data represent means ± SD of three biological replicates (16 plants for each replicate), and the asterisks indicate significant differences compared with the wild-type (WT) plants at *P < 0.05 and **P < 0.01 (Dunnett's test). A Three homozygous transgenic lines used for phenotype analysis. Inserted nucleotides are shown in red, while missing nucleotides are shown in blue. B Phenotypes of the WT and OsPP65 knockout plants before and after osmotic stress treatment. Scale bars = 5 cm. C Survival rates of the osmotic stress-treated plants after 5 days of recovery. D, Relative water loss rate of 2-week-old leaves of WT, ko3, ko6, and ko7. E, Relative ion leakage in rice leaves after osmotic stress for 3 days and 4 days. F, SOD activities in the seedlings of WT and OsPP65 knockout plants before (0 h) and after (4 h) osmotic stress treatment
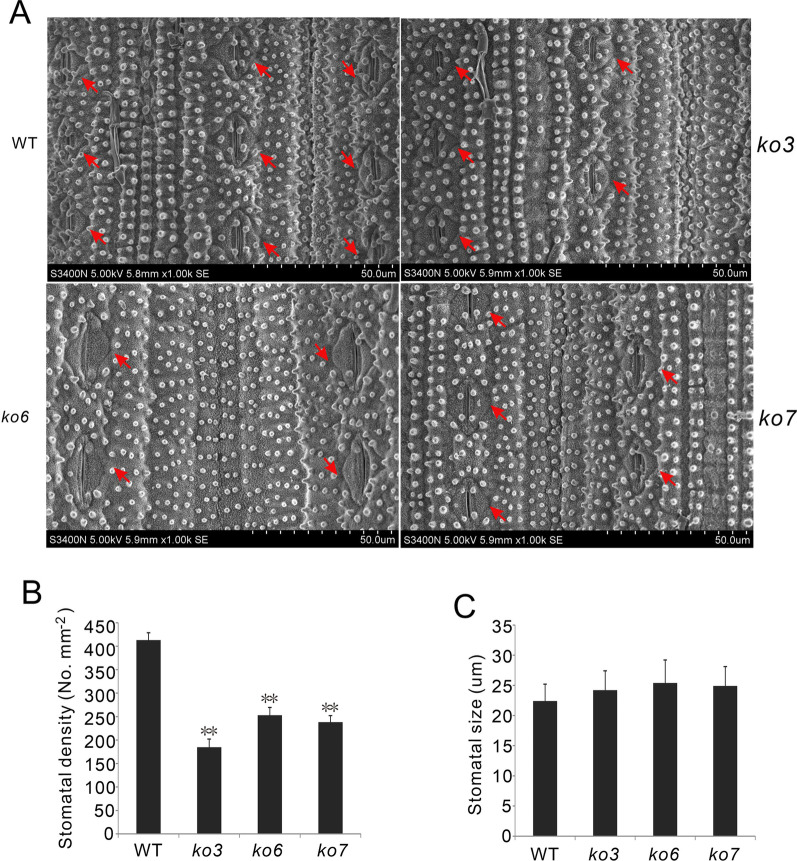
To assess the role of OsPP65 in osmotic stress tolerance, 2-week-old rice plants were subjected to 20% PEG 6000 for 5 days and allowed to recover under normal water supply levels (Fig. 3B). After recovery growth for 5 days, the survival rates of the three transgenic lines were all greater than 62%, which was significantly higher than that of the wild-type plants (31%) (P < 0.01, Fig. 3C). Ion leakage analysis revealed that the osmotic stress-induced increase in ion leakage was significantly lower in OsPP65 knockout plants than that in the wild-type plants (P < 0.01) (Fig. 3E). Moreover, enzymatic assays showed that the activities of superoxide dismutase (SOD), a reactive oxygen species scavenging enzyme, were much stronger in OsPP65 knockout plants than in the wild-type plants both before and after PEG treatment (Fig. 3F). To observe the water loss rate in vitro, leaves were detached and exposed to the open air at room temperature and weighed at different times. The water loss rate of the wild-type leaves was higher than that of the leaves of the three OsPP65 knockout plants at all time points after air-drying (Fig. 3D). Since the water loss rate is closely related to the number and size of stomata, we next quantified the density and size of stomata in leaves. As shown in Fig. 4, the stomata densities were 55%, 39%, and 43% lower in ko3, ko6, and ko7, respectively, as compared with those in the wild-type plants. However, no significant differences in stomata size were observed between the wild-type and transgenic plants (Fig. 4C), indicating that OsPP65 affects stomata density instead of stomata size in rice. The decrease of stomata density in the OsPP65 knockout plants is more conductive to maintain the appropriate temperature and water status of leaves than in the wild-type plants when subjected to osmotic stress. Taken together, these results suggest that knockout of OsPP65 enhances osmotic stress tolerance in rice plants.
Fig. 4.
Stomatal density and size in the leaves of WT and OsPP65 knockout rice plants. A Photographs showing the stomatal density in WT and OsPP65 knockout plant leaves. Red arrows indicate the stomates in leaves. B Quantification of the stomatal density in leaves of WT and OsPP65 knockout plants. C Quantification of stomatal size in leaves of WT and OsPP65 knockout plants. Data are presented as means ± SD of three biological replicates (5 plants for each replicate) and **P < 0.01 (Dunnett’s test)
To test the function of OsPP65 in tolerance to salt stress, seeds of the wild-type and OsPP65 knockout plants were germinated on 1/2 MS medium supplemented with or without NaCl. As shown in Figure S2, no significant differences were identified between the wild-type and OsPP65 knockout plants in terms of seedling growth in the absence of NaCl. In the presence of 150 mM NaCl, the shoot and root growth of both the wild-type and OsPP65 knockout plants were severely inhibited at 6 days after germination, but the OsPP65 knockout plants were markedly less inhibited by NaCl treatment than the wild-type plants (Additional file 1: Fig. S2). Similar results were also observed for 200 mM NaCl, indicating that knockout of OsPP65 enhances salt tolerance in rice plants.
OsPP65 Mediated Stress Tolerance is Involved in Activation of the ABA and JA Signaling Pathways
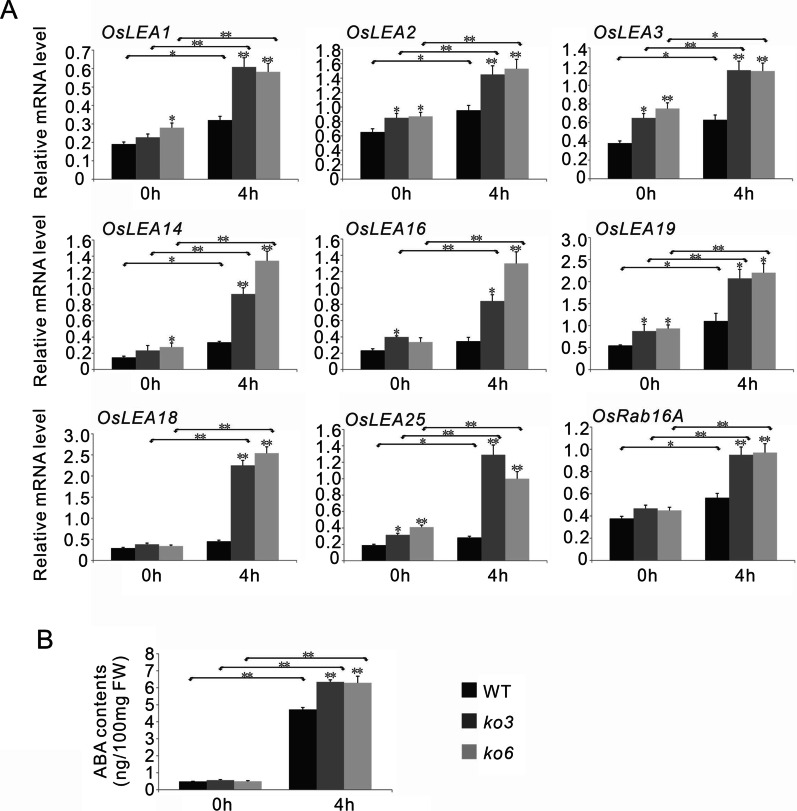
To identify the possible downstream signaling pathways regulated by OsPP65 in response to stress, RNA sequencing (RNA-seq) analysis of global gene expression was conducted in the wild-type and ko6 plants under normal conditions and 4 h after osmotic stress treatment. Under normal conditions, only 36 up-regulated genes and 20 down-regulated genes in ko6 plants were observed to have transcript levels more than twofold higher and lower, respectively, than those in the wild-type plants (false discovery rate ≤ 0.05; Additional file 2: Table S1). However, when subjected to osmotic stress treatment, 139 up-regulated genes and 88 down-regulated genes were identified in OsPP65 knockout plants compared with wild-type plants. Interestingly, among the 139 up-regulated genes, we found nine genes encoding late embryogenesis abundant (LEA) proteins, which are involved in ABA signaling, implying a role of OsPP65 in the control of the ABA signaling pathway. In addition, two allene oxide synthase genes (OsAOS2 and OsAOS3), which function in JA biosynthesis, were up-regulatedin ko6 plants compared with the wild-type plants, suggesting that the stress tolerance conferred by loss of OsPP65 might also be associated with the JA signaling pathway.
To test these inferences, we first analyzed the expression levels of the differentially expressed genes in wild-type, ko3, and ko6 plants under normal and osmotic stress conditions by qRT-PCR. The transcripts of two lipoxygenase genes (OsLOX2 and OsLOX9), which are involved in JA biosynthesis, were also analyzed in these plants. The results showed that seven OsLEA genes (OsLEA1/2/3/14/19/25/29) were strongly induced by osmotic stress in both wild-type and OsPP65 knockout plants, but their expression levels were significantly higher in the OsPP65 knockout plants than in wild-type plants under osmotic stress conditions (Fig. 6A). Similarly, the transcription levels of OsLEA16/18, OsLOX2, OsAOS2, and OsAOS3 were also remarkably higher in the OsPP65 knockout plants than in wild-type plants after stress treatment, but induction of their expression by osmotic stress was only observed in the OsPP65 knockout plants (Figs. 5A, 6A). Consistent with the results of qRT-PCR, the endogenous ABA contents were significantly increased in both wild-type and OsPP65 knockout plants under osmotic stress, and the ABA concentrations were higher in OsPP65 knockout plants than in wild-type plants (Fig. 5B). Significant accumulation of JA was also observed in the OsPP65 knockout plants under osmotic stress conditions, and the JA contents were remarkably higher in the OsPP65 knockout plants than in the wild-type plants (Fig. 6B). These results indicate that the stress tolerance conferred by loss of OsPP65 involves activation of the ABA and JA signaling pathways.
Fig. 6.
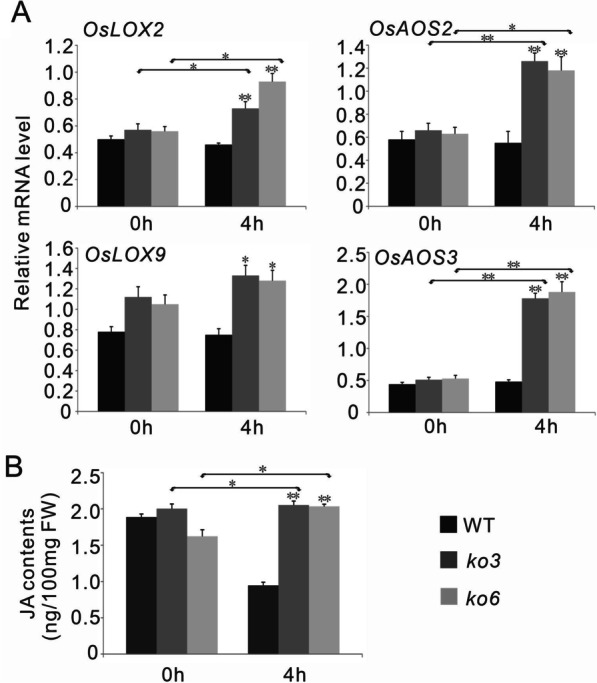
OsPP65 modulates the expression of genes involved in JA signaling and the accumulation of JA in rice. A Expression analysis of genes associated with JA signaling in WT and OsPP65 knockout plants before (0 h) and after (4 h) osmotic stress treatment. Data represent means ± SD of three biological replicates (5 plants for each replicate). B Quantification of endogenous JA levels in the WT and OsPP65 knockout plants. FW, fresh weight. Data represent means ± SD of three biological replicates (15 plants for each replicate). The asterisks indicate significant differences compared with the WT plants or 0 h treatment at **P < 0.01 and *P < 0.05 (Dunnett's test)
Fig. 5.
OsPP65 modulates the expression of genes involved in ABA signaling and the accumulation of ABA in rice. A Transcription analysis of the genes involved in ABA signaling in WT and OsPP65 knockout plants before (0 h) and after (4 h) osmotic stress treatment. B Quantification of endogenous ABA levels in the WT and OsPP65 knockout plants. FW, fresh weight. Data represent means ± SD of three biological replicates (5 plants for each replicate), and the asterisks indicate significant differences compared with the WT plants or 0 h treatment at **P < 0.01 and *P < 0.05 (Dunnett's test)
To further validate the involvement of ABA and JA signaling pathways in OsPP65-mediated stress tolerance, seed germination assays were conducted to analyze the effects of exogenous ABA and methyl jasmonate (MeJA) on the growth of OsPP65 knockout plants. The results showed that elongation of the roots of OsPP65 knockout seedlings was less sensitive to exogenous ABA and MeJA treatments than that of the wild-type seedlings (Additional file 1: Figs. S3 and S4). Intriguingly, two ABA-responsive elements and three JA-responsive CGTCA motifs were identified in the promoter of OsPP65 through bioinformatics analysis (Qin et al. 2020; Additional file 3: Table S2). All these results together demonstrated that stress tolerance conferred by loss of OsPP65 is at least partially dependent on the activation of the ABA and JA signaling pathways in rice.
The simultaneous accumulation of ABA and JA prompted us to test whether ABA and JA interact synergistically in OsPP65-mediated osmotic stress tolerance in rice. Toward this end, we analyzed the transcription of four JA synthesis-related genes (OsAOS2, OsAOS3, OsLOX2, and OsLOX9) in the wild-type and OsPP65 knockout plants after ABA treatment, as well as the transcription of two ABA synthesis-related genes (OsNCED3 and OsNCED4) and five OsLEA genes after JA treatment. ABA treatment did not affect the transcription of the four JA synthesis-related genes, and the endogenous contents of JA were not remarkably affected by ABA treatment in both the wild-type and OsPP65 knockout plants (Additional file 1: Fig. S5). Similar results were also observed for JA treatment (Additional file 1: Fig. S6). These observations suggest that ABA and JA function independently in osmotic stress tolerance conferred by loss of OsPP65 in rice.
OsPP65 Mediates Stress Tolerance by Modulating the Metabolism of Raffinose Family Oligosaccharides
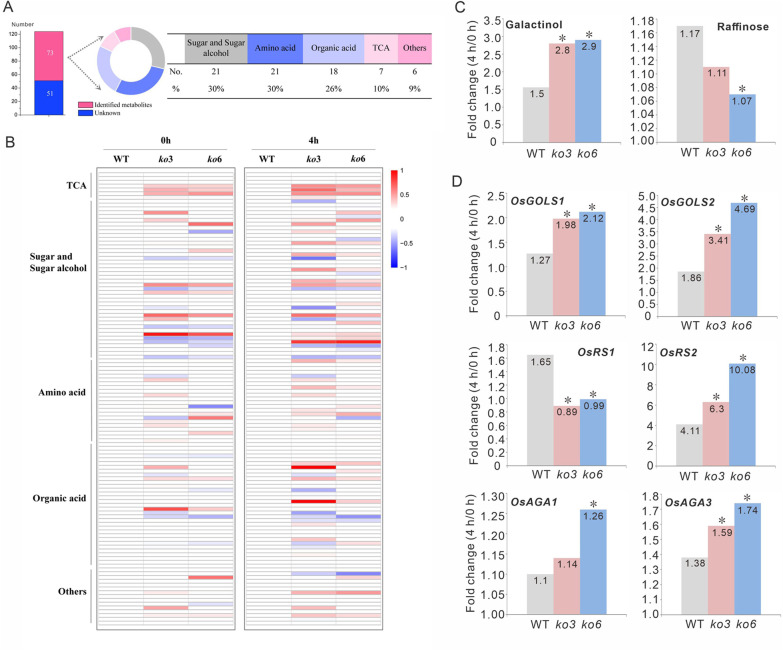
To further investigate the molecular mechanism underlying the stress tolerance conferred by loss of function of OsPP65, primary metabolomic studies were performed using the seedlings of wild-type and OsPP65 knockout plants (the transgenic lines ko3 and ko6) with (4 h) or without (0 h) osmotic stress treatment by gas chromatography tandem mass spectrometry (GC–MS). The omics data matrix consisted of retention time, an accurate mass-to-charge ratio (m/z), and the relative content of each metabolite in each sample (Additional file 4: Table S3). There were 124 metabolite peaks detected in the rice seedlings, and 73 metabolites were conclusively identified based on authentic standards, namely 21 amino acid-related compounds, 18 organic acids, 21 sugar-related metabolites, seven TCA cycle intermediates, and six other compounds (Fig. 7A, Additional file 4: Table S3). To find potential metabolomic variations between wild-type and OsPP65 knockout plants, orthogonal partial least squares-discriminant analysis was performed, and the results revealed that separation between the wild-type and OsPP65 knockout plants was more evident at 4 h under osmotic stress (Additional file 1: Fig. S7).
Fig. 7.
OsPP65-mediated stress response involves the regulation of RFO metabolism in rice. A The number of primary metabolites and unknown metabolites identified by GC–MS. B The number and proportion of identified metabolites in different classes. C The fold changes in galactinol and raffinose contents in WT and OsPP65 knockout plants at 4 h after osmotic stress treatment relative to the 0 h treatment. D The fold changes in the expression of key enzymes involved in RFO metabolism in WT and OsPP65 knockout plants at 4 h after osmotic stress treatment relative to the 0 h treatment. Data represent means ± SD of three biological replicates (15 plants for each replicate), and the asterisks indicate significant differences compared with the WT plants at *P < 0.05 (Dunnett's test)
We observed that the magnitudes of the fold changes of most metabolites were higher after 4 h of osmotic stress compared with 0 h both for the wild-type and OsPP65 knockout plants (Fig. 7B), especially for some TCA cycle intermediates, which were lower in the OsPP65 knockout lines (Fig. 7B). However, the contents of three metabolites (galactose, galactinol, and raffinose) belonging to the RFO metabolic pathway were higher in both the wild-type and OsPP65 knockout plants at 4 h after osmotic stress treatment relative to the 0 h treatment (Fig. 7C and Additional file 1: Fig. S8). The fold changes of galactose and galactinol were higher in the OsPP65 knockout plants than those in the wild-type plants, whereas the fold change of raffinose was lower in the OsPP65 knockout plants compared with that in the wild-type plants (Fig. 7C). To validate this result, the gene expression of key enzymes involved in RFO metabolism was analyzed. Consistent with the metabolomic results, stress treatment remarkably induced the expression of galactinol synthase 1 and 2 (OsGOLS1 and OsGOLS2), raffinose synthase 2 (OsRS2), and alkaline α-galactosidase 1 and 3 (OsAGA1 and OsAGA3) in both the wild-type and OsPP65 knockout plants, but the fold changes of these enzymes were higher in OsPP65 knockout plants compared with those in the wild-type plants (Fig. 7D). The higher expression levels of OsAGA genes, which function in raffinose catabolism, in the OsPP65 knockout plants could explain the lower content of raffinose in these plants relative to the wild-type plants. In addition to the products of the RFO metabolic pathway, stress treatment also significantly induced the accumulation of fructose, glucose, glucopyranose, sorbopyranose, rhamnopyranose, and lactulose in both the wild-type and OsPP65 knockout plants, and the fold changes of these metabolites were remarkably higher in the OsPP65 knockout plants than those in the wild-type plants (Additional file 1: Fig. S8). These observations together suggest the probably significant roles of these sugars in rice stress response mediated by OsPP65.
Discussion
OsPP65 Negatively Regulates Osmotic and Salt Stress Tolerance in Rice
PP2C-type protein phosphatases are a group of highly conserved regulatory proteins that are present in virtually all eukaryotic organisms (Singh et al. 2016). Many PP2C proteins have been reported to play important roles in disease resistance, abiotic stress response, and development in plants (Schweighofer et al. 2007; Lu et al. 2017; Nishimura et al. 2018; Wang et al. 2020; Chen et al. 2021). In rice, there are 90 PP2C proteins that are distributed on different chromosomes (Singh et al. 2010). However, only seven of these proteins have been confirmed to function in disease resistance or stress tolerance in previous studies (Hu et al. 2006, 2009; Park et al. 2008; You et al. 2014; Singh et al. 2015; Miao et al. 2020; Yang et al. 2020). The roles of the other members in stress responses are still unknown. In this study, we have demonstrated that OsPP65 is significantly induced by osmotic and salt stresses. The OsPP65 knockout rice plants exhibited enhanced tolerance to osmotic stress induced by PEG, as manifested by higher survival rates and SOD activities and lower water loss rates and ion leakage under stress conditions compared with the wild-type plants. The OsPP65 knockout plants also showed increased tolerance to salt stress at the germination stage, suggesting that OsPP65 functions as a negative regulator of osmotic and salt stress tolerance in rice. Intriguingly, we also found that OsPP65 is also involved in leaf blast and bacterial blight resistance in rice (unpublished data). Because of its multiple functions in both abiotic and biotic stresses, we believe that OsPP65 provides a promising target for improvement of stress tolerance and disease resistance in rice plants using a gene editing approach.
In this study, lower stomata density was identified in the OsPP65 knockout plants compared with the wild-type plants. OsSPCH2 and OsICE1 have been demonstrated to play pivotal roles in determining stomata density in rice. The osspch2 and osice1 mutant plants exhibited greatly reduced stomata densities when compared to the wild-type plants (Wu et al. 2019). To analyze that if OsPP65 could affect the expression of OsSPCH2 and OsICE1, we detected the expression levels of the two genes in wild-type and OsPP65 knockout plants. The result showed that transcription levels of OsSPCH2 and OsICE1 were all down-regulated in the knockout plants compared to the wild-type plants (Additional file 1: Fig. S9), indicating that reduced stomata density conferred by loss of OsPP65 may at least partially due to the decreased expression of the marker genes of stomata development in rice.
OsPP65 Mediated Abiotic Stress Response Involves the Regulation of ABA and JA Signaling in Rice
It has been well demonstrated that PP2C proteins are central components of the ABA signaling pathway and function as negative regulators of ABA signaling and responses in plants (Singh et al. 2015, 2016). Phytohormone ABA is a core regulator of plant responses to various abiotic stresses (Shen et al. 2017; Li et al. 2019, 2021; Ullah et al. 2020). Consistent with these previous reports, the gene expression, hormone quantification, and seed germination analyses performed here show that OsPP65-mediated abiotic stress response also involves regulation of the ABA signaling pathway in rice. Interestingly, we identified several pieces of evidence showing that there is also a tight association between the OsPP65 and the JA signaling pathway during rice abiotic stress response. First, the transcripts of four JA synthesis-related genes, OsLOX2, OsLOX9, OsAOS2, and OsAOS3, were significantly higher in the OsPP65 knockout plants compared with wild-type plants after osmotic stress treatment. Second, hormone quantification analysis indicated that the endogenous JA level was significantly higher in the OsPP65 knockout plants than in wild-type plants under osmotic stress. Third, three JA-responsive elements were identified in the promoter of OsPP65 and the transcription of OsPP65 was strongly induced by exogenous JA treatment. Lastly, seed germination experiments demonstrated that the OsPP65 knockout plants were less sensitive to exogenous JA than the wild-type plants. All these results together suggested that OsPP65 functions as an negative important regulator of the JA signaling pathway in the regulation of rice response to osmotic stress, consisting with the regulatory roles of JA in rice responses to various abiotic stresses, such as salt, drought and osmotic stresses (Hazman et al. 2015; Kurotani et al. 2015; Wu et al. 2015; Dhakarey et al. 2017; Tang et al. 2020). This result also provides new evidence of the relationship between PP2C proteins and the JA signaling pathway during plant stress response. Interestingly, in plants, the hormone JA is converted into bioactive form JA-isoleucine (JA-Ile) through conjugating isoleucine with JA, and this step is catalyzed by the enzyme JASMONATE RESISTANT 1 (JAR1) (Svyatyna et al. 2014). In the present study, the expression levels of OsJAR1 were analyzed in both the wild-type and OsPP65 knockout plants with or without osmotic stress. The results showed that osmotic stress repressed the expression of OsJAR1 and the expression levels of OsJAR1 were remarkably lower in the OsPP65 knockout plants than in the wild-type plants under osmotic stress (Additional file 1: Fig. S10), suggesting that the OsPP65 knockout plants could lead to less active OsJAR1 and lastly might resulted in the accumulation of JA when subjected to osmotic stress treatment.
Given that previous studies have indicated that dehydration stress-induced JA accumulation is dependent on ABA in Arabidopsis (Liu et al. 2016a), JA application can enhance foliar ABA concentration in barley (Bandurska et al. 2003), and JA deficiency (jar1) diminishes ABA accumulation in Arabidopsis (De Ollas et al., 2015), we performed a set of experiments to test whether the stress tolerance conferred by loss of OsPP65 involves JA acting in concert with ABA or acting independently. The results showed that ABA biosynthesis was not affected by JA treatment and that JA biosynthesis was not affected by ABA treatment, implying that the JA and ABA signaling pathways function independently in OsPP65-mediated abiotic stress response in rice. However, it should be pointed out that the effects of JA on other aspects of the ABA pathway (i.e., distribution, transport, and signaling) and the effects of ABA on other aspects of the JA signaling pathway were not investigated in our present study. Therefore, we cannot rule out the possibility that the JA and ABA signaling pathways act in concert with each other in relation to these other aspects.
The Important Roles of RFOs in the Osmotic Stress Tolerance Conferred by Loss of OsPP65 in Rice
The functions of galactinol and raffinose in plant abiotic stress tolerance have been well-documented by many studies. For example, rice plants overexpressing AtGolS2 exhibit improved galactinol content and increased grain yield in terms of panicle number and grain fertility compared with the control plants (Selvaraj et al. 2017). Ectopic expression of BhGolS1 leads to significant accumulation of galactinol and raffinose as well as elevated drought tolerance in tobacco (Wang et al. 2009). Consistent with previous results, we found that the contents of galactinol and raffinose were increased in both the wild-type and OsPP65 knockout plants under osmotic stress, and higher galactinol contents were identified in the OsPP65 knockout plants, which had higher stress tolerance compared with the wild-type plants. However, in contrast to galactinol, the raffinose level was slightly lower in the OsPP65 knockout plants. We hypothesized that the decreased raffinose level might be due to increased raffinose catabolism in the OsPP65 knockout plants. Just as expected, the expression levels of OsAGA1 and OsAGA3, two enzymes involved in raffinose catabolism, were remarkably higher in the OsPP65 knockout plants compared with the wild-type plants after stress treatment. These observations indicate that the rice stress tolerance conferred by OsPP65 involves in modulating the RFO metabolic pathway. This is the first report to dissect the associations between PP2C protein and the RFO metabolism pathway in rice. Li et al. (2020) revealed that the enhanced drought tolerance conferred by overexpression of ZmRS is due to the increased myo-inositol levels following galactinol hydrolysis, with the higher ratio of myo-inositol to raffinose positively regulating plant drought stress responses. Our results here also suggest that the molecular mechanisms by which RFOs regulate plant abiotic stress tolerance may be different between plant species.
The Potential Associations Between Sugars and the Phytohormone Signaling Pathways in OsPP65 Mediated Stress Tolerance
In addition to alterations in the RFO metabolic pathway, higher concentrations of fructose, glucose, glucopyranose, sorbopyranose, lactulose, and rhamnopyranose were also observed in the OsPP65 knockout plants compared with the wild-type plants after osmotic stress, indicating the important roles of these sugars in regulating the abiotic stress response conferred by OsPP65. Intriguingly, the associations between sugars and the phytohormone signaling pathways in regulating plant abiotic stresses have been demonstrated by many studies (Rook et al. 2006; Rodriguez et al. 2019; Saddhe et al. 2021). Cheng et al. (2002) reported that exogenous glucose significantly induced the expression of ABA biosynthesis-related genes and thus caused the accumulation of endogenous ABA in Arabidopsis. External application of JA leads to the accumulation of soluble sugars in many crop plants thereby improving the overall plant performance under different abiotic stresses (Ghoulam et al. 2002; Harpreet et al. 2013; Abdelgawad et al. 2014). The simultaneous accumulation of ABA, JA, and sugars observed in the present study prompted us to speculate that there might be cross-talk between these signals in OsPP65-mediated osmotic stress tolerance in rice. For instance, the increased levels of ABA and/or JA may lead to the accumulation of sugars, or the increased level of sugars may induce the accumulation of ABA and/or JA in the OsPP65 knockout plants. Further research will be needed to answer these questions.
Conclusion
In summary, our results demonstrated that a stress-responsive PP2C protein, OsPP65, negatively regulates osmotic and salt stress tolerance by modulating the ABA/JA signaling pathways and RFO metabolism pathway in rice. OsPP65 is a promising target for improvement of rice stress tolerance using a gene editing approach.
Materials and Methods
Stress Treatments
The japonica cultivar Nipponbare was used in the present study. Cold treatment was conducted by incubating 14-day-old rice plants at 8 ℃ in a plant growth chamber. For salt and osmotic stress treatments, 2-week old seedlings were cultured in Kimura B nutrient solution containing 150 mM NaCl and 20% PEG 6000, respectively. For SA, JA, and H2O2 treatment, 2-week-old rice seedlings were sprayed with a 100 μM hormone solution (SA or JA) or 1% H2O2. Seedlings were sampled at 3 h, 6 h, 9 h, 12 h, and 24 h after treatment for gene expression analysis.
Plasmid Construction and Rice Transformation
To generate the vector for gene editing of OsPP65 using CRISPR/Cas9 technology, two target sites (CGGGTTCGTCAAGACGGACA and CTCACCATGTCATGCAGATA) were selected and the expression cassettes were obtained by overlap PCR using the corresponding primers (OsPP65-crispr-6aF/6aR and OsPP65-crispr-6bF/6bR, Additional file 5: Table S4). Next, the expression cassettes were inserted into pYLCRISPR/Cas9Pubi-B according to Ma et al. (2015). For promoter analysis, an ~ 2.4-kb fragment upstream of the translational starting site of OsPP65 was amplified using primers OsPP65-GUS-F/R (Additional file 5: Table S4), and this fragment was then inserted into the pCAMBIA1381Z vector. Rice transformation was conducted using the same method as described previously (Toki et al. 2006).
Quantitative RT-PCR Analysis
Different rice samples which collected from five plants in each biological replicate were ground in liquid nitrogen, and total RNA was extracted using the Trizol reagent (Invitrogen, Carlsbad, CA, USA). Quantitative RT-PCR analysis was performed as described in our previous study (Liu et al. 2016b). Primers used for gene expression analysis are listed in Additional file 5: Table S4.
Subcellular Localization Analysis
For protein subcellular localization analysis, the coding sequence of OsPP65 was amplified using primers OsPP65-GFP-F/R (Additional file 5: Table S4) and inserted into the 35S-GFP vector to produce the OsPP65-GFP fusion construct. Subsequently, the fusion plasmids were transiently expressed in rice stem protoplasts or Arabidopsis leaf protoplasts as described previously (Zhang et al. 2011). After incubation in the dark for 24 h at 26 ℃, GFP fluorescence was examined by laser confocal microscopy (Zeiss LSM710, Germany).
Histochemical GUS Analysis
Different tissues of the transgenic plants transformed with the OsPP65 promoter-GUS construct were incubated in staining buffer at 37 °C for histochemical GUS analysis as described previously (Liu et al. 2016b). After 24 h in the dark, the tissues were decolored in 75% (v/v) ethanol and photographed by an ordinary Canon camera.
Determination of Tolerance to Osmotic Stress
For osmotic stress treatment, germinated seeds of the wild-type and OsPP65 knockout plants were cultured in Kimura B nutrient solution in 96-well plates in a plant growth chamber under 16 h light/8 h dark (26 ℃). The light intensity was 540 μmol m−2 s−1 and humidity was 75%. After two weeks, the seedlings were subjected to osmotic stress by transferring them into fresh Kimura B nutrient solution containing 20% PEG6000. Five days later, the treated seedlings were recovered in fresh Kimura B nutrient solution for another 5 days. To measure the ion leakage, the second and third leaves were excised from the seedling and incubated in 10 ml of distilled water overnight. The ratio of conductivity values measured by a DDS-11AT conductivity meter (Leici, Shanghai, China) before and after boiling was calculated as the ion leakage from the leaves. The water loss rates of the wild-type Nipponbare and OsPP65 knockout plants were measured using the same method as described by Liu et al. (2014).
For salt stress treatment, seeds of wild-type and OsPP65 knockout plants were sterilized with 75% ethanol and 2.5% NaClO and germinated on 1/2 MS medium with (150 and 200 mM) or without (0 mM) NaCl for 7 days. Next, the lengths of shoot and primary roots were measured.
Determination of Stomatal Number
Leaves of 2-week old wild-type Nipponbare and OsPP65 knockout plants were fixed with 2.5% glutaraldehyde, and pictures of stomata were acquired by scanning electron microscopy (S-3400N, Hitachi, Japan) as described previously (You et al. 2013).
RNA-Seq Analysis
Seedlings (15 plants for each replicate) of the wild-type Nipponbare and OsPP65 knockout plants (line ko6) with (4 h) or without (0 h) osmotic stress treatment were collected for RNA-seq analysis. The quality of the RNA-seq raw reads was assessed, and the reads were trimmed by the software trim_galore with the parameters “–stringency 3 –length 36 -e 0.1”. Sequencing reads that passed quality control were mapped onto the Nipponbare (MSU7, Oryza sativa japonica) reference genome using STAR (2.7.1a) software (Alexander et al. 2013), and an expression matrix was generated using RSEM (v1.3.1) software (Li and Colin 2011). Differential gene expression was calculated using the DESeq2 package in R with a filtering threshold of twofold or greater alteration in expression and an adjusted P value less than 0.05. Three biological replicates were performed for each treatment. The raw data has been submitted to NCBI (http://www.ncbi.nlm.nih.gov/) with accession number PRJNA808176.
Hormone Quantification and Untargeted Metabolomic Analysis
Seedlings (15 plants for each replicate) of the wild-type Nipponbare and OsPP65 knockout plants (lines ko3 and ko6) with (4 h) or without (0 h) osmotic stress treatments were collected for hormone quantification and metabolomic analysis. Hormones were measured using LC–MS while the metabolomic analysis was conducted using GC–MS as described in our previous studies (Liu et al. 2016c, 2018; Yan et al. 2018, 2021). The GC–MS raw data were processed using MassHunter Qualitative Analysis B.06.00 software (Agilent Technologies Inc., Santa Clara, CA, USA) and MassHunter Quantitative Analysis B.07.01 software (Agilent Technologies Inc., Santa Clara, CA, USA). Afterwards, the NIST mass spectral library and an in-house mass spectral database established using authentic standards were used together for metabolite identification.
Statistical Analysis
Two independent experiments were conducted for stress treatment, hormone quantification and untargeted metabolomic analysis. Three or four biological replicates were performed for each experiment. Three independent experiments were conducted for stress tolerance analysis, stress-related gene expression analysis, and three biological replicates were performed for each experiment. All results are presented as means ± standard derivation (SD) of three biological replicates. Statistically significant difference analysis is conducted by “Dunnett's test” using IBM SPSS statistics software (version 23).
Supplementary Information
Additional file 1: Fig. S1. Phenotypes of the wild-type (WT) and OsPP65 knockout plants at 30 days after flowering under normal conditions. Fig. S2. OsPP65 knockout rice plants show enhanced salt stress tolerance at the germination stage. A, Seeds of the WT and OsPP65 knockout plants were germinated on 1/2 MS medium with (150 and 200 mM) or without (0 mM) NaCl for 7 days. B, Shoot lengths of the germinated seeds of WT and OsPP65 knockout plants. C, Root lengths of the germinated seeds of WT and OsPP65 knockout plants. Data represent means ± SD of three biological replicates (20 plants for each replicate), and the asterisks indicate significant differences compared with the WT plants at *P < 0.05 (Dunnett's test). Fig. S3. The OsPP65 knockout seeds were less sensitive to exogenous ABA relative to the wild-type seeds. A, Photograph showing the germinated seeds of WT and OsPP65 knockout plants with or without ABA treatment. B, Shoot lengths of the germinated seeds of WT and OsPP65 knockout plants. C, Root lengths of the germinated seeds of WT and OsPP65 knockout plants. Data represent means ± SD of three biological replicates (20 plants for each replicate) and the asterisks indicate significant differences compared to the WT plants at *P < 0.05 (Dunnett's test). Fig. S4. The OsPP65 knockout seeds were less sensitive to exogenous JA relative to the WT seeds. A, Phenotypes of the germinated seeds of WT and OsPP65 knockout plants with or without JA treatment. B, Shoot lengths of the germinated seeds of WT and OsPP65 knockout plants. C, Root lengths of the germinated seeds of WT and OsPP65 knockout plants. Data represent means ± SD of three biological replicates (20 plants for each replicate) and the asterisks indicate significant differences compared to the WT plants at *P < 0.05 (Dunnett's test). Fig. S5. Exogenous ABA treatment did not affect the JA signaling pathway in OsPP65 knockout plants. A, Transcription analysis of four JA biosynthesis genes in the WT and OsPP65 knockout plants before and after ABA treatment. B, Quantification of endogenous JA in the WT and OsPP65 knockout plants before and after ABA treatment. FW, fresh weight. Fig. S6. Exogenous JA treatment did not affect the JA signaling pathway in OsPP65 knockout plants. A, Transcription analysis of two ABA biosynthesis genes and five OsLEA genes in the WT and OsPP65 knockout plants before and after JA treatment. Data represent means ± SD of three biological replicates and the asterisks indicate significant differences compared to the WT plants at **P < 0.01 and *P < 0.05 (Dunnett's test). B, Quantification of endogenous JA in the WT and OsPP65 knockout plants before and after JA treatment. FW, fresh weight. Fig. S7. Score plot generated from the GC–MS metabonomics data using the crossvalidated OPLS-DA model. Fig. S8. The fold changes of other sugars or amino acids in WT and OsPP65 knockout plants at 4 h osmotic stress treatment relative to 0 h treatment. Data represent means ± SD of three biological replicates and the asterisks indicate significant differences compared to the WT plants at **P < 0.01 and *P < 0.05 (Dunnett's test). Fig. S9. Expression levels of OsSPCH2 and OsICE1 in WT and OsPP65 knockout plants under normal condition. Data represent means ± SD of three biological replicates and the asterisks indicate significant differences compared to the WT plants at *P < 0.05 and **P < 0.01 (Dunnett's test). Fig. S10. Expression levels of OsJAR1 in WT and OsPP65 knockout plants with or without osmotic stress treatment. Data represent means ± SD of three biological replicates and the asterisks indicate significant differences compared to the WT plants at *P < 0.05 (Dunnett's test).
Additional file 2: Table S1. Differentially expressed genes in ko6 plants by RNA-seq analysis before (0 h) and after (4 h) osmotic stress treatment. |log2 Ratio| ≥1 and FDR ≤ 0.05.
Additional file 3: Table S2. Cis-elements analysis of the OsPP65 promoter.
Additional file 4: Table S3. 120 metabolites identified by GC-MS in WT and the OsPP65 knockout plants. Number after the metabolite represents the isomer.
Additional file 5: Table S4. Primers used for vector construction and quantitative RT-PCR analysis.
Acknowledgements
Not applicable
Abbreviations
- PP2C
Type 2C protein phosphatases
- ABA
Abscisic acid
- JA
Jasmonic acid
- PT
Phosphate transporters
- Hpt
Hour post treatment
- GUS
β-Glucuronidase
- ko
Knockout
- RNA-seq
RNA sequencing
- SOD
Superoxide dismutase
- LEA
Late embryogenesis abundant protein
- AOS
Allene oxide synthase 2
- LOX2
Lipoxygenase 2
- MeJA
Methyl jasmonate
- GC–MS
Gas chromatography tandem mass spectrometry
- GOLS1
Galactinol synthase 1
- RS2
Raffinose synthase 2
- AGA1
Alkaline α-galactosidase 1
- RFOs
Raffinose family oligosaccharides
- JA-Ile
JA-isoleucine
- JAR1
JASMONATE RESISTANT 1
Author Contributions
Q L, SJ Y, and C L conceived and designed the experiment. Q L, JR D, WJ H conducted the experiments, performed data analysis and wrote the manuscript. H Y, SW W, WY L, XX M, WF C, JL X participated in material development, sample preparation and data analysis. Q L and SJ Y drafted proposals and corrected the manuscript. All authors read and approved the final manuscript.
Funding
This work was jointly funded by Special Program for Crop Germplasm Resources of Guangdong Province (Governor's Special Program 2018–2019), The investment project of Department of Agriculture and Rural Affairs, Development and Reform Commission, Guangdong province (No. 272 of 2021), the Natural Science Foundation of Guangdong Province (No. 2021A1515011107), the Science and Technology Program of Guangdong Province (No. 2021A0505030050), the Special Fund for Scientific Innovation Strategy-Construction of High Level Academy of Agriculture Science (Nos. R2020PY-JX019 and R2020PY-JX001), and Guangdong Key Laboratory of New Technology in Rice Breeding (2020B1212060047).
Availability of Data and Materials
The datasets supporting the conclusions of this article are provided within the article and its additional files.
Declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qing Liu, Email: liuqing198504@126.com.
Jierong Ding, Email: jierongdjr@163.com.
Wenjie Huang, Email: huangwenjie@agrogene.ac.cn.
Hang Yu, Email: yuhang5783@163.com.
Shaowen Wu, Email: wushaowen@agrogene.ac.cn.
Wenyan Li, Email: liwy1023@foxmail.com.
Xingxue Mao, Email: maoxingxue@qq.com.
Wenfeng Chen, Email: nsktfz@163.com.
Junlian Xing, Email: 834430993@qq.com.
Chen Li, Email: lic11111@sina.com.
Shijuan Yan, Email: shijuan@agrogene.ac.cn.
References
- Abdelgawad ZA, Khalafaallah AA, Abdallah MM. Impact of methyl jasmonate on antioxidant activity and some biochemical aspects of maize plant grown under water stress condition. Agric Sci. 2014;5(12):1077–1088. [Google Scholar]
- Alexander D, Carrie AD, Felix S, Jorg D, Chris Z, Sonali J, Philippe B, Mark C, Thomas RG. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandurska H, Stroin´ski A, Kubis´ J, The effect of jasmonic acid on the accumulation of ABA, proline and spermidine and its influence on membrane injury under water deficit in two barley genotypes. Acta Physiol Plant. 2003;25:279–285. doi: 10.1007/s11738-003-0009-0. [DOI] [Google Scholar]
- Chen Q, Bai L, Wang W, Shi H, Ramón Botella J, Zhan Q, Liu K, Yang HQ, Song CP. COP1 promotes ABA-induced stomatal closure by modulating the abundance of ABI/HAB and AHG3 phosphatases. New Phytol. 2021;229(4):2035–2049. doi: 10.1111/nph.17001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Endo A, Zhou L, Penney J, Chen H, Arroyo A, Leon P, Nambara E, Asami T, Seo M, Koshiba T, Sheen J. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002;14(11):2723–2743. doi: 10.1105/tpc.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu ML, Chen PW, Meng SF, Xu P, Lan WZ. The Arabidopsis phosphatase PP2C49 negatively regulates salt tolerance through inhibition of AtHKT1;1. J Integr Plant Biol. 2021;63(3):528–542. doi: 10.1111/jipb.13008. [DOI] [PubMed] [Google Scholar]
- De Ollas C, Arbona V, Gómez Cadenas A. Jasmonoyl isoleucine accumulation is needed for abscisic acid build-up in roots of Arabidopsis under water stress conditions. Plant Cell Environ. 2015;38(10):2157–70. doi: 10.1111/pce.12536. [DOI] [PubMed] [Google Scholar]
- Dhakarey R, Raorane ML, Treumann A, Peethambaran PK, Schendel RR, Sahi VP, Hause B, Bunzel M, Henry A, Kohli A, Riemann M. Physiological and proteomic analysis of the rice mutant cpm2 suggests a negative regulatory role of jasmonic acid in drought tolerance. Front Plant Sci. 2017;8:1903. doi: 10.3389/fpls.2017.01903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S, Grill E, Meskiene I, Schweighofer A. Type 2C protein phosphatases in plants. FEBS J. 2013;280(2):681–693. doi: 10.1111/j.1742-4658.2012.08670.x. [DOI] [PubMed] [Google Scholar]
- Fujii S, Toriyama K. DCW11, down-regulated gene 11 in CW-type cytoplasmic male sterile rice, encoding mitochondrial protein phosphatase 2c is related to cytoplasmic male sterility. Plant Cell Physiol. 2008;49(4):633–640. doi: 10.1093/pcp/pcn036. [DOI] [PubMed] [Google Scholar]
- Ghoulam C, Foursy A, Fares K. Effects of salt stress on growth, inorganic ions and proline accumulation in relation to osmotic adjustment in five sugar beet cultivars. Environ Exp Bot. 2002;47(1):39–50. doi: 10.1016/S0098-8472(01)00109-5. [DOI] [Google Scholar]
- Harpreet K, Poonam S, Geetika S. Sugar accumulation and its regulation by jasmonic acid in Brassica napus L. under salt stress. J Physiol Biochem. 2013;9(4):53–64. [Google Scholar]
- Hazman M, Hause B, Eiche E, Nick P, Riemann M. Increased tolerance to salt stress in OPDA-deficient rice ALLENE OXIDE CYCLASE mutants is linked to an increased ROS-scavenging activity. J Exp Bot. 2015;66(11):3339–3352. doi: 10.1093/jxb/erv142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XB, Song FM, Zheng Z. Molecular characterization and expression analysis of a rice protein phosphatase 2C gene, OsBIPP2C1, and overexpression in transgenic tobacco conferred enhanced disease resistance and abiotic tolerance. Physiol Plant. 2006;127(2):225–236. doi: 10.1111/j.1399-3054.2006.00671.x. [DOI] [Google Scholar]
- Hu XB, Zhang HJ, Li GJ, Yang YX, Zheng Z, Song FM. Ectopic expression of a rice protein phosphatase 2C gene OsBIPP2C2 in tobacco improves disease resistance. Plant Cell Rep. 2009;28:985–995. doi: 10.1007/s00299-009-0701-7. [DOI] [PubMed] [Google Scholar]
- Kurotani K, Hayashi K, Hatanaka S, Toda Y, Ogawa D, Ichikawa H, Ishimaru Y, Tashita R, Suzuki T, Ueda M, Hattori T, Takeda S. Elevated levels of CYP94 family gene expression alleviate the jasmonate response and enhance salt tolerance in rice. Plant Cell Physiol. 2015;56(4):779–789. doi: 10.1093/pcp/pcv006. [DOI] [PubMed] [Google Scholar]
- Li B, Colin ND. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Zhang Y, Liu Y, Li X, Hao G, Han Q, Dirk LMA, Downie AB, Ruan YL, Wang J, Wang G, Zhao TJ. Raffinose synthase enhances drought tolerance through raffinose synthesis or galactinol hydrolysis in maize and Arabidopsis plants. J Biol Chem. 2020;295(23):8064–8077. doi: 10.1074/jbc.RA120.013948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang Q, Yang X, Han J, Zhu Z. OsANN3, a calcium-dependent lipid binding annexin is a positive regulator of ABA-dependent stress tolerance in rice. Plant Sci. 2019;284:212–220. doi: 10.1016/j.plantsci.2019.04.019. [DOI] [PubMed] [Google Scholar]
- Li X, Yu B, Wu Q, Min Q, Zeng R, Xie Z, Huang J. OsMADS23 phosphorylated by SAPK9 confers drought and salt tolerance by regulating ABA biosynthesis in rice. PLoS Genet. 2021;17(8):e1009699. doi: 10.1371/journal.pgen.1009699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Ma Y, Chen N, Guo SY, Liu HL, Guo XY, Chong K, Xu YY. Overexpression of stress-inducible OsBURP16, the β subunit of polygalacturonase 1, decreases pectin content and cell adhesion and increases abiotic stress sensitivity in rice. Plant Cell Environ. 2014;37:1144–1158. doi: 10.1111/pce.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sun X, Liao W, Zhang J, Liang J, Xu W. Involvement of OsGF14b adaptation in the drought resistance of rice plants. Rice. 2019;12(1):82. doi: 10.1186/s12284-019-0346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Staswick PE, Avramova Z. Memory responses of jasmonic acid-associated Arabidopsis genes to a repeated dehydration stress. Plant Cell Environ. 2016;39(11):2515–2529. doi: 10.1111/pce.12806. [DOI] [PubMed] [Google Scholar]
- Liu Q, Yang JY, Zhang SH, Zhao JL, Feng AQ, Yang TF, Wang XF, Mao XX, Dong JF, Zhu XY, Leung H, Leach JE, Liu B. OsGF14b positively regulates panicle blast resistance, but negatively regulates leaf blast resistance in rice. Mol Plant Microbe Interact. 2016;29(1):46–56. doi: 10.1094/MPMI-03-15-0047-R. [DOI] [PubMed] [Google Scholar]
- Liu Q, Yang JY, Yan SJ, Zhang SH, Zhao JL, Wang WJ, Yang TF, Wang XF, Mao XX, Dong JF, Zhu XY, Liu B. The germin-like protein OsGLP2-1 enhances resistance to fungal blast and bacterial blight in rice. Plant Mol Biol. 2016;92(4–5):411–423. doi: 10.1007/s11103-016-0521-4. [DOI] [PubMed] [Google Scholar]
- Liu Q, Yan SJ, Huang WJ, Yang JY, Dong JF, Zhang SH, Zhao JL, Yang TF, Mao XX, Zhu XY, Liu B. NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice. Plant Mol Biol. 2018;98(4–5):289–302. doi: 10.1007/s11103-018-0768-z. [DOI] [PubMed] [Google Scholar]
- Lu X, Xiong Q, Cheng T, Li QT, Liu XL, Bi YD, Li W, Zhang WK, Ma B, Lai YC, Du WG, Man WQ, Chen SY, Zhang JS. A PP2C-1 allele underlying a quantitative trait locus enhances soybean 100-seed weight. Mol Plant. 2017;10(5):670–684. doi: 10.1016/j.molp.2017.03.006. [DOI] [PubMed] [Google Scholar]
- Luan S. Protein phosphatases in plants. Annu Rev Plant Biol. 2003;54:63–92. doi: 10.1146/annurev.arplant.54.031902.134743. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8(8):1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Miao J, Li XF, Li XB, Tan WC, You AQ, Wu SJ, Tao YJ, Chen C, Wang J, Zhang DP, Gong ZY, Yi CD, Yang ZF, Gu MH, Liang GH, Zhou Y. OsPP2C09, a negative regulatory factor in abscisic acid signalling, plays an essential role in balancing plant growth and drought tolerance in rice. New Phytol. 2020;227(5):1417–1433. doi: 10.1111/nph.16670. [DOI] [PubMed] [Google Scholar]
- Min MK, Kim R, Hong WJ, Jung KH, Lee JY, Kim BG. OsPP2C09 is a bifunctional regulator in both ABA-dependent and independent abiotic stress signaling pathways. Int J Mol Sci. 2021;22(1):393. doi: 10.3390/ijms22010393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Née G, Kramer K, Nakabayashi K, Yuan B, Xiang Y, Miatton E, Finkemeier I, Soppe WJJ. DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat Commun. 2017;8(1):72. doi: 10.1038/s41467-017-00113-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Fu X, Zhang H, Li X, Cai X, Zhang P, Liu L, Wang Q, Sun M, Wang QW, Zhang A, Zhang Z, Jiang M. Abscisic acid inhibits rice protein phosphatase PP45 via H2O2 and relieves repression of the Ca(2+)/CaM- dependent protein kinase DMI3. Plant Cell. 2019;31(1):128–152. doi: 10.1105/tpc.18.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Tsuchiya W, Moresco JJ, Hayashi Y, Satoh K, Kaiwa N, Irisa T, Kinoshita T, Schroeder JI, Yates JR, 3rd, Hirayama T, Yamazaki T. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat Commun. 2018;9(1):2132. doi: 10.1038/s41467-018-04437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ, Peng Y, Chen X, Dardick C, Ruan D, Bart R, Canlas PE, Ronald PC. Rice XB15, a protein phosphatase 2C, negatively regulates cell death and XA21-mediated innate immunity. PLoS Biol. 2008;6(9):e231. doi: 10.1371/journal.pbio.0060231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q, Wang YX, Huang LY, Du FP, Zhao XQ, Li ZK, Wang WX, Fu BY. A U-box E3 ubiquitin ligase OsPUB67 is positively involved in drought tolerance in rice. Plant Mol Biol. 2020;102:89–107. doi: 10.1007/s11103-019-00933-8. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Parola R, Andreola S, Pereyra C, Martínez-Noël G. TOR and SnRK1 signaling pathways in plant response to abiotic stresses: do they always act according to the “yin-yang” model? Plant Sci. 2019;288:110220. doi: 10.1016/j.plantsci.2019.110220. [DOI] [PubMed] [Google Scholar]
- Rook F, Hadingham SA, Li Y, Bevan MW. Sugar and ABA response pathways and the control of gene expression. Plant Cell Environ. 2006;29(3):426–434. doi: 10.1111/j.1365-3040.2005.01477.x. [DOI] [PubMed] [Google Scholar]
- Saddhe AA, Manuka R, Penna S. Plant sugars: Homeostasis and transport under abiotic stress in plants. Physiol Plant. 2021;171(4):739–755. doi: 10.1111/ppl.13283. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 2004;9(5):236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Kant M, Schuurink R, Mauch F, Buchala A, Cardinale F, Meskiene I. The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell. 2007;19(7):2213–2224. doi: 10.1105/tpc.106.049585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj MG, Ishizaki T, Valencia M, Ogawa S, Dedicova B, Ogata T, Yoshiwara K, Maruyama K, Kusano M, Saito K, Takahashi F, Shinozaki K, Nakashima K, Ishitani M. Overexpression of an Arabidopsis thaliana galactinol synthase gene improves drought tolerance in transgenic rice and increased grain yield in the field. Plant Biotechnol J. 2017;15:1465–1477. doi: 10.1111/pbi.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Vleesschauwer DD, Sharma MK, Ronald PC. Recent advances in dissecting stress-regulatory crosstalk in rice. Mol Plant. 2013;6(2):250–260. doi: 10.1093/mp/sss147. [DOI] [PubMed] [Google Scholar]
- Shen J, Lv B, Luo L, He J, Mao C, Xi D, Ming F. The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci Rep. 2017;7:40641. doi: 10.1038/srep40641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6(5):4105–4107. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- Shubchynskyy V, Boniecka J, Schweighofer A, Simulis J, Kvederaviciute K, Stumpe M, Mauch F, Balazadeh S, Mueller-Roeber B, Boutrot F, Zipfel C, Meskiene I. Protein phosphatase AP2C1 negatively regulates basal resistance and defense responses to Pseudomonas syringae. J Exp Bot. 2017;68(5):1169–1183. doi: 10.1093/jxb/erw485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Giri J, Kapoor S, Tyagi AK, Pandey GK. Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics. 2010;11:435. doi: 10.1186/1471-2164-11-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Jha SK, Bagri J, Pandey GK. ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS ONE. 2015;10(4):e0125168. doi: 10.1371/journal.pone.0125168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Pandey A, Srivastava AK, Tran LS, Pandey GK. Plant protein phosphatases 2C: from genomic diversity to functional multiplicity and importance in stress management. Crit Rev Biotechnol. 2016;36(6):1023–1035. doi: 10.3109/07388551.2015.1083941. [DOI] [PubMed] [Google Scholar]
- Svyatyna K, Jikumaru Y, Brendel R, Reichelt M, Mithöfer A, Takano M, Kamiya Y, Nick P, Riemann M. Light induces jasmonate-isoleucine conjugation via OsJAR1-dependent and -independent pathways in rice. Plant Cell Environ. 2014;37(4):827–839. doi: 10.1111/pce.12201. [DOI] [PubMed] [Google Scholar]
- Tang G, Ma J, Hause B, Nick P, Riemann M. Jasmonate is required for the response to osmotic stress in rice. Environ Exp Bot. 2020;175:104047. doi: 10.1016/j.envexpbot.2020.104047. [DOI] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006;47(6):969–976. doi: 10.1111/j.1365-313X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- Ullah F, Xu Q, Zhao Y, Zhou DX. Histone deacetylase HDA710 controls salt tolerance by regulating ABA signaling in rice. J Integr Plant Biol. 2020 doi: 10.1111/jipb.13042. [DOI] [PubMed] [Google Scholar]
- Wang J, Sun N, Zhang F, Yu R, Chen H, Deng XW, Wei N. SAUR17 and SAUR50 differentially regulate PP2C-D1 during apical hook development and cotyledon opening in Arabidopsis. Plant Cell. 2020;32(12):3792–3811. doi: 10.1105/tpc.20.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhu Y, Wang L, Liu X, Liu Y, Phillips J, Deng X. A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta. 2009;230(6):1155–1166. doi: 10.1007/s00425-009-1014-3. [DOI] [PubMed] [Google Scholar]
- Wong JH, Klejchová M, Snipes SA, Nagpal P, Bak G, Wang B, Dunlap S, Park MY, Kunkel EN, Trinidad B, Reed JW, Blatt MR, Gray WM. SAUR proteins and PP2C.D phosphatases regulate H+-ATPases and K+ channels to control stomatal movements. Plant Physiol. 2021;185(1):256–273. doi: 10.1093/plphys/kiaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Ye H, Yao R, Zhang T, Xiong L. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 2015;232:1–12. doi: 10.1016/j.plantsci.2014.12.010. [DOI] [PubMed] [Google Scholar]
- Wu Z, Chen L, Yu Q, Zhou W, Gou X, Li J, Hou S. Multiple transcriptional factors control stomata development in rice. New Phytol. 2019;223(1):220–232. doi: 10.1111/nph.15766. [DOI] [PubMed] [Google Scholar]
- Yan SJ, Huang WJ, Gao JD, Fu H, Liu J. Comparative metabolomic analysis of seed metabolites associated with seed storability in rice (Oryza sativa L.) during natural aging. Plant Physiol Biochem. 2018;127:590–598. doi: 10.1016/j.plaphy.2018.04.020. [DOI] [PubMed] [Google Scholar]
- Yan SJ, Liu Q, Naake T, Huang WJ, Chen MY, Kong Q, Zhang S, Li WY, Li X, Liu QJ, Yang JW, Fernie AR, Liu B. OsGF14b modulates defense signaling pathways in rice panicle blast response. Crop J. 2021;9(4):725–738. doi: 10.1016/j.cj.2020.10.007. [DOI] [Google Scholar]
- Yang ZL, Yang J, Wang Y, Wang F, Mao WX, He QJ, Xu JM, Wu ZC, Mao CZ. PROTEIN PHOSPHATASE95 regulates phosphate homeostasis by affecting phosphate transporter trafficking in rice. Plant Cell. 2020;32(3):740–757. doi: 10.1105/tpc.19.00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Zong W, Hu HH, Li XH, Xiao JH, Xiong LZ. A STRESS-RESPONSIVE NAC1-regulated protein phosphatase gene rice Protein Phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice. Plant Physiol. 2014;166(4):2100–2114. doi: 10.1104/pp.114.251116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Zong W, Li XK, Ning J, Hu HH, Li XH, Xiao JH, Xiong LZ. The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J Exp Bot. 2013;64(2):569–583. doi: 10.1093/jxb/ers349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Han J, Li L, Zhang Q, Yang G, He G. Wheat PP2C-a10 regulates seed germination and drought tolerance in transgenic Arabidopsis. Plant Cell Rep. 2020;39(5):635–651. doi: 10.1007/s00299-020-02520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PY, Yuan Z, Wei L, Qiu X, Wang GR, Liu ZX, Fu JX, Cao LR, Wang TC. Overexpression of ZmPP2C55 positively enhances tolerance to drought stress in transgenic maize plants. Plant Sci. 2022;314:111127. doi: 10.1016/j.plantsci.2021.111127. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, Wang J, Wang H. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods. 2011;7(1):30. doi: 10.1186/1746-4811-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Phenotypes of the wild-type (WT) and OsPP65 knockout plants at 30 days after flowering under normal conditions. Fig. S2. OsPP65 knockout rice plants show enhanced salt stress tolerance at the germination stage. A, Seeds of the WT and OsPP65 knockout plants were germinated on 1/2 MS medium with (150 and 200 mM) or without (0 mM) NaCl for 7 days. B, Shoot lengths of the germinated seeds of WT and OsPP65 knockout plants. C, Root lengths of the germinated seeds of WT and OsPP65 knockout plants. Data represent means ± SD of three biological replicates (20 plants for each replicate), and the asterisks indicate significant differences compared with the WT plants at *P < 0.05 (Dunnett's test). Fig. S3. The OsPP65 knockout seeds were less sensitive to exogenous ABA relative to the wild-type seeds. A, Photograph showing the germinated seeds of WT and OsPP65 knockout plants with or without ABA treatment. B, Shoot lengths of the germinated seeds of WT and OsPP65 knockout plants. C, Root lengths of the germinated seeds of WT and OsPP65 knockout plants. Data represent means ± SD of three biological replicates (20 plants for each replicate) and the asterisks indicate significant differences compared to the WT plants at *P < 0.05 (Dunnett's test). Fig. S4. The OsPP65 knockout seeds were less sensitive to exogenous JA relative to the WT seeds. A, Phenotypes of the germinated seeds of WT and OsPP65 knockout plants with or without JA treatment. B, Shoot lengths of the germinated seeds of WT and OsPP65 knockout plants. C, Root lengths of the germinated seeds of WT and OsPP65 knockout plants. Data represent means ± SD of three biological replicates (20 plants for each replicate) and the asterisks indicate significant differences compared to the WT plants at *P < 0.05 (Dunnett's test). Fig. S5. Exogenous ABA treatment did not affect the JA signaling pathway in OsPP65 knockout plants. A, Transcription analysis of four JA biosynthesis genes in the WT and OsPP65 knockout plants before and after ABA treatment. B, Quantification of endogenous JA in the WT and OsPP65 knockout plants before and after ABA treatment. FW, fresh weight. Fig. S6. Exogenous JA treatment did not affect the JA signaling pathway in OsPP65 knockout plants. A, Transcription analysis of two ABA biosynthesis genes and five OsLEA genes in the WT and OsPP65 knockout plants before and after JA treatment. Data represent means ± SD of three biological replicates and the asterisks indicate significant differences compared to the WT plants at **P < 0.01 and *P < 0.05 (Dunnett's test). B, Quantification of endogenous JA in the WT and OsPP65 knockout plants before and after JA treatment. FW, fresh weight. Fig. S7. Score plot generated from the GC–MS metabonomics data using the crossvalidated OPLS-DA model. Fig. S8. The fold changes of other sugars or amino acids in WT and OsPP65 knockout plants at 4 h osmotic stress treatment relative to 0 h treatment. Data represent means ± SD of three biological replicates and the asterisks indicate significant differences compared to the WT plants at **P < 0.01 and *P < 0.05 (Dunnett's test). Fig. S9. Expression levels of OsSPCH2 and OsICE1 in WT and OsPP65 knockout plants under normal condition. Data represent means ± SD of three biological replicates and the asterisks indicate significant differences compared to the WT plants at *P < 0.05 and **P < 0.01 (Dunnett's test). Fig. S10. Expression levels of OsJAR1 in WT and OsPP65 knockout plants with or without osmotic stress treatment. Data represent means ± SD of three biological replicates and the asterisks indicate significant differences compared to the WT plants at *P < 0.05 (Dunnett's test).
Additional file 2: Table S1. Differentially expressed genes in ko6 plants by RNA-seq analysis before (0 h) and after (4 h) osmotic stress treatment. |log2 Ratio| ≥1 and FDR ≤ 0.05.
Additional file 3: Table S2. Cis-elements analysis of the OsPP65 promoter.
Additional file 4: Table S3. 120 metabolites identified by GC-MS in WT and the OsPP65 knockout plants. Number after the metabolite represents the isomer.
Additional file 5: Table S4. Primers used for vector construction and quantitative RT-PCR analysis.
Data Availability Statement
The datasets supporting the conclusions of this article are provided within the article and its additional files.