Abstract
Strains of Streptococcus mutans produce at least three mutacins, I, II, and III. Mutacin II is a member of subgroup AII in the lantibiotic family of bacteriocins, and mutacins I and III belong to subgroup AI in the lantibiotic family. In this report, we characterize two mutacins produced by UA140, a group I strain of S. mutans. One is identical to the lantibiotic mutacin I produced by strain CH43 (F. Qi et al., Appl. Environ. Microbiol. 66:3221–3229, 2000); the other is a nonlantibiotic bacteriocin, which we named mutacin IV. Mutacin IV belongs to the two-peptide, nonlantibiotic family of bacteriocins produced by gram-positive bacteria. Peptide A, encoded by gene nlmA, is 44 amino acids (aa) in size and has a molecular mass of 4,169 Da; peptide B, encoded by nlmB, is 49 aa in size and has a molecular mass of 4,826 Da. Both peptides derive from prepeptides with glycines at positions −2 and −1 relative to the processing site. Production of mutacins I and IV by UA140 appears to be regulated by different mechanisms under different physiological conditions. The significance of producing two mutacins by one strain under different conditions and the implication of this property in terms of the ecology of S. mutans in the oral cavity are discussed.
Bacteriocins are a family of ribosomally synthesized peptide antibiotics that are produced by bacteria (8, 10, 11, 13, 22). Based on posttranslational modification, bacteriocins from gram-positive bacteria can be classified into two groups: class I, the modified bacteriocins (the lantibiotics), and class II, the unmodified bacteriocins (the nonlantibiotics). The lantibiotics are lanthionine-containing small-peptide antibiotics (11, 23) that contain dehydrated amino acid residues and thioether bridges resulting from posttranslational modifications. The nonlantibiotic bacteriocins can be divided into two groups (18): the one-peptide bacteriocins, represented by pediocin AcH and pediocin PA-1 (5, 9), and the two-peptide bacteriocins such as lactococcin G, plantaricin E/F, lactacin F, and thermophilin 13 (1, 6, 15, 17). Biosynthesis of the nonlantibiotic bacteriocins involves synthesis of a prepeptide, which consists of a leader peptide with two glycines at the processing site and a mature peptide moiety. At maturation, the leader peptide is cleaved off after the double-glycine region by a dedicated protease, releasing the mature peptide to the outside medium. The mature peptide may or may not be modified by intra- or intermolecular disulfide bond formation (10).
Mutans streptococci are considered major contributors to human caries (14). Some strains of Streptococcus mutans produce antimicrobial substances called mutacins (3, 7). Mutacins are active against closely related species as well as other gram-positive bacteria. Our laboratory divided mutacin-producing strains into three groups based on the presence or absence of a 5.6-kb residential plasmid and the antagonistic activity of the strains against each other (3). We isolated and then biochemically and genetically characterized mutacins I, II, and III, elaborated by group I, II, and III strains, respectively (19–21). Mutacins I, II, and III all belong to the lantibiotic family. During our initial attempt to isolate mutacin I from the prototype group I strain, UA140 (3), we found two active peaks in the high-pressure liquid chromatography (HPLC) profile of the crude mutacin extract. Peak 1 was eluted at the same fraction as mutacin I produced by CH43 (20), and peak 2 was eluted at a later fraction. Here we report characterization of peptides from the two active peaks. DNA and peptide sequence analyses revealed that the peptide in peak 1 is identical to mutacin I, while there are two peptides in peak 2. These peptides form a two-peptide bacteriocin belonging to the family of nonlantibiotic bacteriocins produced by gram-positive bacteria.
MATERIALS AND METHODS
Bacterial strains, media, and mutacin activity assays.
The group I S. mutans strain UA140 was isolated from a caries-active dental patient at the University of Alabama at Birmingham dental clinic. Streptococcus sanguinis strain NY101 was used as the indicator for routine mutacin activity assays. Other strains tested for sensitivity to mutacin include Streptococcus sobrinus OMZ176, S. sanguinis ATCC 10556, Streptococcus oralis ATCC 10557, Streptococcus gordonii ATCC 10558, Streptococcus mitis ATCC 903, S. mitis ATCC 33399, Streptococcus parasanguinis ATCC 15911, Streptococcus crista ATCC 49999, Actinomycetes odontolyticus ATCC 17929, A. naeslundii ATCC 12104, A. naeslundii ATCC 19039, A. naeslundii ATCC 19246, A. naeslundii ATCC 27044, and A. naeslundii ATCC 49340. All strains were grown in Trypticase soy broth or on Trypticase soy agar plates (Becton Dickinson and Company, Cockeysville, Md.). For mutacin activity assays, the indicator strains were grown in Trypticase soy broth at 37°C overnight anaerobically; 0.3 ml of the overnight culture was then added to 4 ml of Trypticase soy soft agar melted and cooled to 50°C. The mixture was overlaid on top of the plate spotted with purified mutacins. The zone of inhibition was inspected after overnight incubation of the plate at 37°C.
Isolation and purification of mutacin I and mutacin IV.
Isolation and purification of mutacin I from UA140 were performed using a membrane transfer technique and reverse-phase HPLC as described previously (20). For isolation of mutacin IV, UA140 was inoculated into 10 ml of Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) and incubated at 37°C aerobically in a shaker incubator. The overnight culture was diluted 1:50 into 1 liter of fresh Todd-Hewitt broth, and incubation continued for 24 h at 37 or 39°C in a floor shaker at 150 rpm. To each flask, 150 ml of chemically defined medium (60%; JRH Biosciences, Lenexa, Kans.) was then added, and incubation continued for 24 h. Mutacin IV was isolated from the culture supernatant by a chloroform extraction technique (19, 21). For purification, the crude extract of mutacin IV was applied to a Source 15RPC column and eluted with a gradient of buffers A (0.1% trifluoroacetic acid [TFA]) and B (0.085% TFA in 60% acetonitrile) using an LKB Purifier (Amersham Pharmacia Biotech, Piscataway, N.J.). The active fractions were pooled and dried in a lyophilizer. The pellet was redissolved in 0.25% TFA and subjected to a second round of purification as above. The active fractions were collected, dried in a lyophilizer, and used for sequence analysis and electrospray ionization mass spectrometry (EIMS).
Amino acid sequence analysis and database searching.
N-terminal peptide sequencing of mutacin IV was performed by Edman degradation. Since mutacin IV comprised two peptides which were inseparable by reverse-phase HPLC, each cycle of Edman degradation yielded two amino acid peaks. Based on different intensities of the two peaks, four tentative sequences of the first 10 amino acids (aa) were constructed. The sequences were then searched against the S. mutans sequence database at the University of Oklahoma (http://www.genome.ou.edu/smutans.html) via BLAST, and the DNA contig that contained the homologous region (contig 450) was obtained.
Nucleic acid accession numbers.
The sequence of the mutacin I biosynthesis gene from UA140 has been submitted to GenBank with accession no. AF238860.
RESULTS
Mutacin profiles of strain UA140.
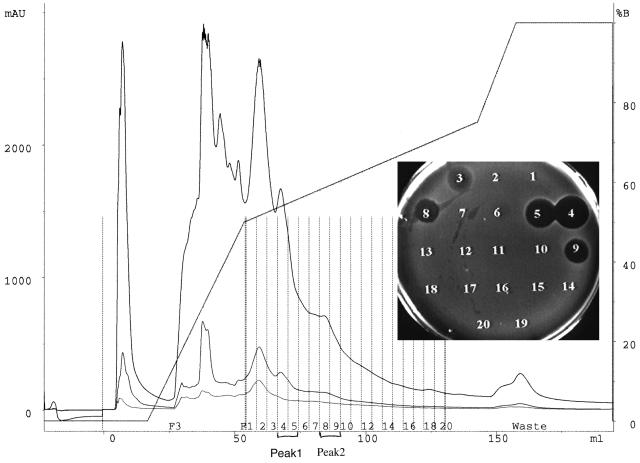
S. mutans strain UA140 is the prototype group I strain that has been used in the laboratory for many years (3). However, we found different profiles for mutacin isolated from UA140 and that from another group I strain, CH43. In CH43, only one active peak was detected in reverse-phase HPLC analysis (20), while two active peaks were apparent in UA140 (Fig. 1). Peak 1 (fractions 4 and 5) was eluted at approximately 31.8% acetonitrile, the same as for mutacin I produced by strain CH43 (20), while Peak 2 (fractions 8 and 9) was eluted at a higher acetonitrile concentration (∼34.8%). This result raised the interesting question as to whether UA140 produced two mutacins, or mutacin I was modified differently in UA140.
FIG. 1.
First-pass HPLC profile of crude mutacin extract from strain UA140 grown on a membrane. Fractions of 1 ml (dotted vertical lines) were collected during elution using buffers A (0.1% TFA) and B (0.085% TFA in 60% acetonitrile) and then tested for antimicrobial activity using NY101 as the indicator. The two active peaks, 1 (fractions 4 and 5) and 2 (fractions 8 and 9), are labeled. mAU, milli-absorption units.
To answer this question, peptides from the two active peaks of UA140 were purified and characterized. EIMS of the peptide in peak 1 revealed a molecular mass of 2,364 Da, identical to that of mutacin I (20). To further confirm that the peptide in peak 1 was indeed mutacin I, we derivatized the peptide with ethanethiol (16) and determined the sequence of the six N-terminal amino acids by Edman degradation. Analysis of the N-terminal region revealed the sequence F1-SEC (S-ethylcysteine)2-SEC3-L4-SEC5-L6, identical to that determined for mutacin I (20). SEC was the product of ethanethiol insertion into the double bond of dehydrated serine or the thioether bridge in lanthionine. These results confirmed that the peptide in peak 1 was mutacin I.
Production of peptides in peak 2.
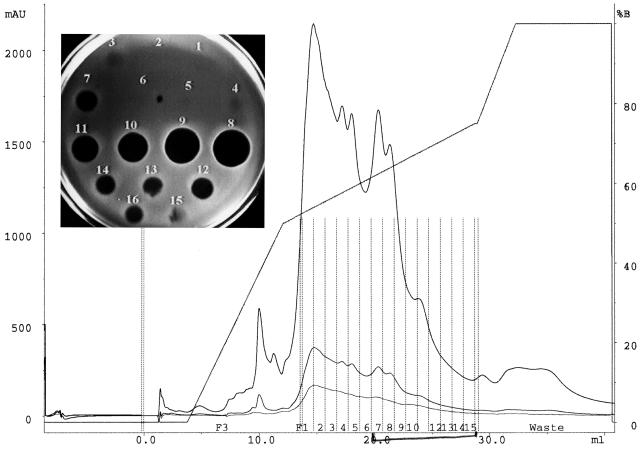
With the membrane transfer technique that was used in the initial isolation of mutacin from UA140, the peptide in peak 2 was produced at a much lower level than that of mutacin I (Fig. 1). To produce enough peptide in peak 2 for biochemical analysis, we tested different growth conditions. To our surprise, the peptide in peak 2 was produced nearly exclusively in a liquid culture grown aerobically at 37 or 39°C (see Materials and Methods), while under the same conditions, mutacin I was nearly undetectable (Fig. 2). This result suggested that production of mutacin I and production of the second active peptide were controlled by different mechanisms.
FIG. 2.
Product profile of UA140 grown in liquid culture under aerobic conditions. Crude mutacin extract was isolated and analyzed using the HPLC buffers and programs used for Fig. 1. Unlike the HPLC profile in Fig. 1, which showed two active peaks, only the second peak (fractions 7 to 14) was detected; the first peak (fraction 3) was barely detectable. mAU, milli-absorption units.
Characterization of the peptides in peak 2.
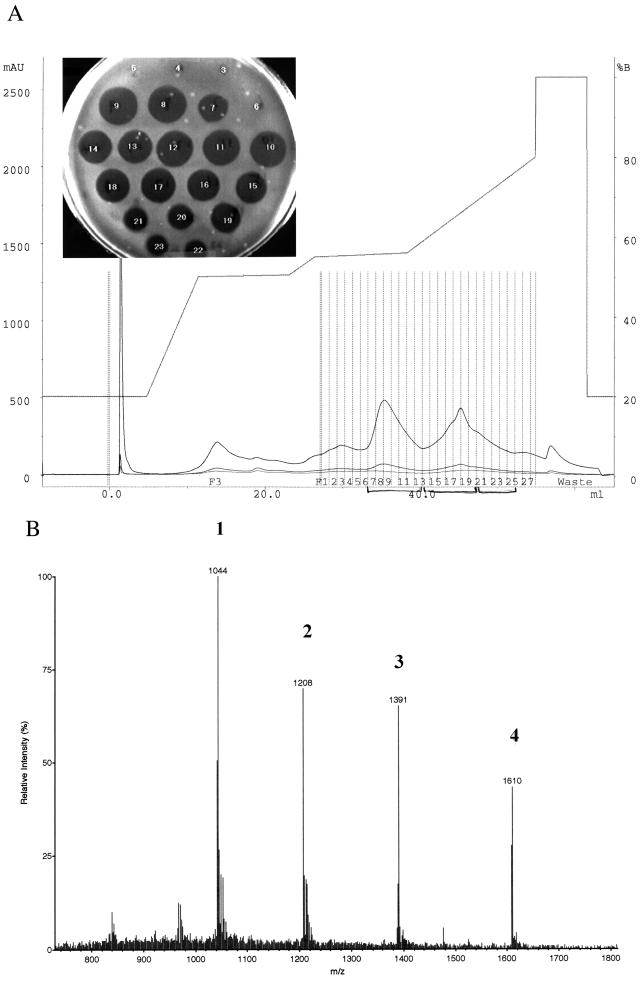
The peptides in peak 2 were purified by reverse-phase HPLC using a program different from that used for mutacin I (20). After the first pass, the active fractions were pooled and subjected to a second round of purification (Fig. 3A). In contrast to the HPLC profile of mutacin I, the active fractions of the peptides in peak 2 were not confined to one peak; instead, they trailed the main peak (fractions 7 to 13) to later fractions (fractions 14 to 25). To identify the materials in these fractions, the active fractions were collected as three segments; segment 1 contained fractions 7 to 13, segment 2 contained fractions 8 to 20, and segment 3 contained fractions 21 to 25. The three segments were then analyzed by EIMS. For segment 1, which appeared to be a single peak, two peptides were revealed; one had a molecular mass of 4,169 Da, the other had a molecular mass of 4,826 Da. (Fig. 3B). The EIMS profiles of segments 2 and 3 were more complex, but they all contained the two peptides found in segment 1 (data not shown). Thus, we concluded that either one or both peptides in segment 1 were responsible for the observed antimicrobial activity.
FIG. 3.
Purification and EIMS analysis of mutacin IV. (A) HPLC profile of the second pass for mutacin IV purification, using a different gradient to obtain better separation of different components. The active fractions were divided into three segments (fractions 7 to 13, 14 to 20, and 21 to 25) for EIMS analysis. (B) EIMS analysis of mutacin IV from fractions 7 to 13 in panel A. Peaks 1 and 3 correspond to quadruply and triply charged molecules of 4,169 Da, respectively; peaks 2 and 4 correspond to quadruply and triply charged molecules of 4,826 Da, respectively. mAU, milli-absorption units.
N-terminal sequences of the two peptides in segment 1.
The measured molecular mass of the two peptides in segment 1 was about twice the molecular mass of mutacin I (2,364 Da), suggesting that the two peptides could be either some kind of dimer of mutacin I or entirely different peptides. To solve this puzzle, we attempted to sequence these peptides. However, with every buffer tested, we could not separate the peptides by HPLC. Finally, using HPLC fraction 8 (Fig. 3A), in which the 4,169-Da peptide represented a higher proportion than the 4,826-Da peptide, we obtained the first 10-aa sequence with two peaks at each cycle. Using the relative intensity of each peak as a reference and some random matching we constructed four tentative sequences (data not shown). By searching the S. mutans genome database, we found DNA segments encoding peptides homologous to sequence 2 (KVSGGEAVAA) and sequence 3 (DKQAADTFLS) in the same contig (contig 450).
Sequence analysis of contig 450.
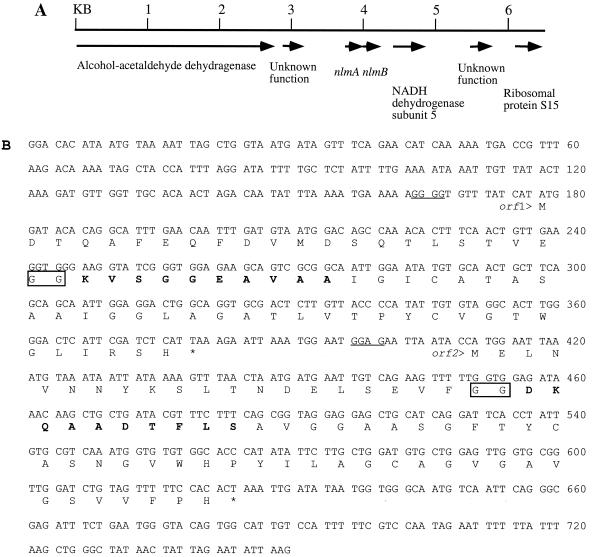
Analysis of the region encoding the homologous peptides revealed two open reading frames (ORFs), encoding two peptides of 67 and 71 aa, respectively (Fig. 4B). The above-mentioned sequence 2 corresponded to K24 to A33 of Orf1, and sequence 3 corresponded to D23 to S32 of Orf2. This result suggested that the two peptides in peak 2 probably derived from peptide precursors encoded by orf1 and orf2, respectively. Adding further support to this notion, the calculated molecular masses of peptide K24–H67 of Orf1 and peptide D23–H71 of Orf2 were 4,168 and 4,826 Da, respectively. These values matched perfectly the measured molecular masses (4,169 and 4,826 Da, respectively) of the two peptides in peak 2. Furthermore, close inspection of Orf1 and Orf2 revealed the presence of glycines at positions −1 and −2 relative to the cleavage site in both peptides (Fig. 4B, boxed letters). Glycines at positions −1 and −2 are well conserved among nonlantibiotic as well as lantibiotic subgroup AII bacteriocins and are thought to be involved with proteolytic cleavage of the prepeptides (10). Searching the DNA sequence surrounding orf1 and orf2 revealed that the two genes are surrounded by either housekeeping genes or genes with unknown functions (Fig. 4A). No protease or ABC transporter genes were found in the vicinity.
FIG. 4.
(A) Genomic organization of the mutacin IV gene locus. The genes encoding proteins with similarity to known enzymes or proteins are labeled. nlmA and nlmB are structural genes for the mutacin IV prepeptides. (B) DNA and deduced amino acid sequences of the two ORFs in contig 450. Bold letters in Orf1 and Orf2 correspond to peptide sequences obtained by N-terminal sequencing of the two peptides in mutacin IV. Boxed letters are the double glycines at the prepeptide cleavage site. The putative ribosomal binding sites for orf1 and orf2 are underlined.
Similarity of Orf1 and Orf2 with other peptides in GenBank.
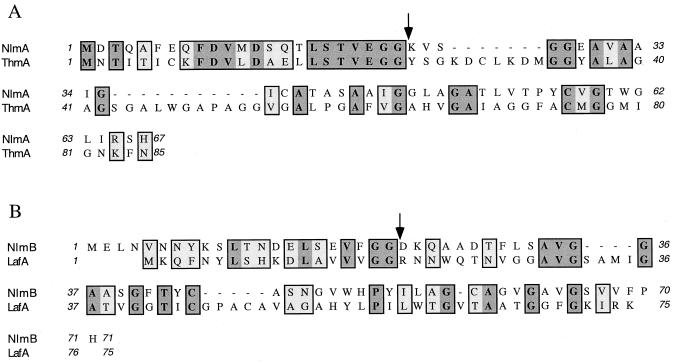
A GenBank search for similar peptides revealed that Orf1 was similar to ThmA and that Orf2 was similar to LafA (Fig. 5). ThmA is the active component in the two-peptide nonlantibiotic bacteriocin, thermophilin 13, produced by Streptococcus thermophilus (15). LafA is one of the peptides in another two-peptide nonlantibiotic bacteriocin, lactacin F, produced by Lactococcus johnsonii (6). These results indicate that the two peptides in peak 2 of mutacin extract from UA140 belong to a two-peptide nonlantibiotic bacteriocin, which we designate mutacin IV. Accordingly, orf1 and orf2 are named nlmA and nlmB, for nonlantibiotic mutacin gene A and nonlantibiotic mutacin gene B, respectively. It is noteworthy that a recently characterized mutacin, mutacin N, from group I-like S. mutans strain N (2), showed high degree of similarity to the mature peptide of NlmB of mutacin IV.
FIG. 5.
Similarity of NlmA and NlmB with other peptides in GenBank. (A) Sequence alignment of NlmA and ThmA; (B) sequence alignment of NlmB and LafA. ThmA and LafA are each one of two components in the two-peptide nonlantibiotic bacteriocin thermophilin 13 and lactacin F, respectively. Dark gray boxes represent identical amino acids; light gray boxes denote conserved changes; arrows indicate cleavage sites for the prepeptide.
Antimicrobial spectrum of mutacin IV.
To test the antimicrobial spectrum of mutacin IV, purified mutacin IV was spotted on agar plate at 100 μg per spot and tested for antimicrobial activity against a panel of selected oral streptococci and actinomycetes. As shown in Table 1, mutacin IV was active against all members in the mitis group of oral streptococci that we tested (S. sanguinis, S. parasanguinis, S. oralis, S. mitis, and S. gordonii) and less active against S. sobrinus, a member of the mutans group of oral streptococci. In contrast, mutacin IV did not show any activity against species of actinomycetes. This result suggests that mutacin IV may play an important role in the interspecies competition between mutans and the mitis group of oral streptococci.
TABLE 1.
Antimicrobial spectrum of mutacin IV against oral streptococci and actinomycetes
| Strain | Species | Zone of inhibition (diam [mm]) |
|---|---|---|
| OMZ176 | S. sobrinus | 3 |
| NY101 | S. sanguinis | 18 |
| ATCC 10556 | S. sanguinis | 17 |
| ATCC 10557 | S. oralis | 6 |
| ATCC 10558 | S. gordonii | 21 |
| ATCC 903 | S. mitis | 13 |
| ATCC 33399 | S. mitis | 7 |
| ATCC 15911 | S. parasanguinis | 16 |
| ATCC 49999 | S. crista | 19 |
| ATCC 12104 | A. naeslundii genospecies 1 | NMa |
| ATCC 19039 | A. naeslundii | NM |
| ATCC 17929 | A. odontolyticus | NM |
| ATCC 19246 | A. naeslundii genospecies 2 | NM |
| ATCC 27044 | A. naeslundii genospecies 2 | NM |
| ATCC 49340 | A. naeslundii | NM |
NM, not measurable.
DISCUSSION
In this study, we determined the nature of the two antimicrobial substances produced by S. mutans strain UA140; one is identical to mutacin I produced by strain CH43, and the other is a two-peptide nonlantibiotic bacteriocin, which we named mutacin IV. The two peptides of mutacin IV, NlmA and NlmB, are encoded by two genes in a single operon located on contig 450 in the S. mutans genome database. nlmA encodes a prepeptide of 67 aa, consisting of a 23-aa leader peptide with a double-glycine cleavage site and a 44-aa mature peptide. nlmB encodes a prepeptide of 71 aa consisting of a 22-aa leader peptide, also with a double-glycine processing signal, and a 49-aa mature peptide. The fact that the calculated molecular mass of the mature NlmA and NlmB peptides matches the measured molecular mass of the purified mutacin IV peptides suggests that neither peptide is posttranslationally modified.
The production of lantibiotic mutacin I and nonlantibiotic mutacin IV in UA140 appeared to be regulated by different mechanisms. Mutacin I could be produced only on a membrane or on a plate with stab culture. This condition is reminiscent of a biofilm on the tooth surface, suggesting that mutacin I production may be triggered by dense colonization of the tooth surface by oral bacteria. In contrast, mutacin IV can be easily produced in liquid culture, e.g., by planktonic cells. This finding raised an interesting question as to what role the two mutacins may play in colonization of the tooth surface by S. mutans. Colonization studies have shown that in a newly exposed tooth surface, S. sanguinis and other members of the mitis group of oral streptococci are the initial colonizers and will remain predominant as long as conditions permit. When the pH on the tooth surface becomes low, due to consumption of fermentable sugars, the number of S. sanguinis decreases and that of S. mutans increases. If low-pH conditions persist, S. mutans will become predominant. Statistical data and in vitro competition studies also found a reverse relationship between S. mutans and S. sanguinis numbers (4, 12). While this reverse relationship may result from difference in acid tolerance between S. mutans and S. sanguinis, other factors may also be involved. For example, mutacin production may be used by S. mutans as a tool to gain advantage over the competitor, S. sanguinis. From our findings that mutacin IV is produced by planktonic cells while mutacin I is produced by biofilm-like cells, we speculate that production of the two mutacins may serve different purposes during the process of colonization by S. mutans. For instance, production of mutacin IV by planktonic cells in saliva may help S. mutans kill the primary colonizers on the tooth surface to make room for its own population. Once colonized on the tooth surface, the lantibiotic mutacin may be produced to inhibit potential competitors. In support of this hypothesis, the antimicrobial spectrum of mutacin IV is specifically against members of the mitis group of oral streptococci (Table 1), while that of mutacin I is much broader (data not shown). Since high levels of S. mutans are associated with caries whereas high levels of S. sanguinis are associated with caries-free sites, it is reasonable to speculate that mutacin production may play an important role in the colonization and pathogenesis of S. mutans.
By searching clinical isolates of S. mutans for the presence of mutacin IV genes by PCR, we found >50% positive results (data not shown). Unlike the lantibiotic mutacins I, II, and III, which are clustered within distinct ethnic groups (3), the distribution of mutacin IV genes was not restricted to the mutacin groups nor to the ethnic groups of people from whom the strains were isolated. We believe that understanding the physiology of mutacin production will provide valuable insight into the ecology of S. mutans in the oral cavity.
ACKNOWLEDGMENTS
We thank R. Krull for technical assistance, K. Morrison for assistance with the N-terminal sequencing of mutacins, and M. Kirk for assistance with EIMS.
This work was supported by NIH grant RO1 DE09082.
REFERENCES
- 1.Anderssen E L, Diep D B, Nes I F, Eijsink V G H, Nissen-Meyer J. Antagonistic activity of Lactobacillus plantarum C11: two new two-peptide bacteriocins, plantaricins EF and JK, and the induction factor plantaricin A. Appl Environ Microbiol. 1998;64:2269–2272. doi: 10.1128/aem.64.6.2269-2272.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balakrishnan M, Simmonds R S, Carne A, Tagg J R. Streptococcus mutans strain N produces a novel low molecular mass non-lantibiotic bacteriocin. FEMS Microbiol Lett. 2000;183:165–169. doi: 10.1111/j.1574-6968.2000.tb08952.x. [DOI] [PubMed] [Google Scholar]
- 3.Caufield P W, Childers N K, Allen D N, Hansen J B. Distinct bacteriocin groups correlate with different groups of Streptococcus mutans plasmids. Infect Immun. 1985;48:51–56. doi: 10.1128/iai.48.1.51-56.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caufield P W, Dasanayake A P, Li Y, Pan Y, Hsu J, Hardin J M. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect Immun. 2000;68:4018–4023. doi: 10.1128/iai.68.7.4018-4023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ennahar S, Aoude-Werner D, Sorokine O, van orsselaer A, Bringel F, Hubert J C, Hasselmann C. Production of pediocin AcH by Lactobacillus plantarum WHE92 isolated from cheese. Appl Environ Microbiol. 1996;62:4381–4387. doi: 10.1128/aem.62.12.4381-4387.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fremaux C, Ahn C, Laenhammer T R. Molecular analysis of the lactacin F operon. Appl Environ Microbiol. 1993;59:3906–3915. doi: 10.1128/aem.59.11.3906-3915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamada S, Ooshima T. Production and properties of bacteriocins (mutacins) from Streptococcus mutans. Arch Oral Biol. 1975;20:641–648. doi: 10.1016/0003-9969(75)90131-4. [DOI] [PubMed] [Google Scholar]
- 8.Hansen J N. Antibiotics synthesized by posttranslational modifications. Annu Rev Microbiol. 1993;47:535–564. doi: 10.1146/annurev.mi.47.100193.002535. [DOI] [PubMed] [Google Scholar]
- 9.Henderson J T, Chopko A L, Dyck van Wassenaar P D. Purification and primary structure of pediocin PA-1, produced by Pediococcus acidilactici PAC-10. Arch Biochem Biophys. 1992;295:5–12. doi: 10.1016/0003-9861(92)90480-k. [DOI] [PubMed] [Google Scholar]
- 10.Jack R W, Tagg J R, Ray B. Bacteriocins of gram-positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung G. Lantibiotics: a survey. In: Jung G, Sahl H G, editors. Nisin and novel lantibiotics. Leiden, The Netherlands: ESCOM Science Publishers; 1991. pp. 1–34. [Google Scholar]
- 12.Kemp C W, Robrish S A, Curtis M A, Sharer S A, Bowen W H. Application of a competition model to the growth of Streptococcus mutans and Streptococcus sanguis in binary continuous culture. Appl Environ Microbiol. 1983;45:1277–1282. doi: 10.1128/aem.45.4.1277-1282.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolter R, Moreno F. Genetics of ribosomally synthesized peptide antibiotics. Annu Rev Microbiol. 1992;46:141–163. doi: 10.1146/annurev.mi.46.100192.001041. [DOI] [PubMed] [Google Scholar]
- 14.Loesche W J. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marciset O, Jeronimus-Stratingh M C, Mollet B, Poolman B. Thermophilin 13, a nontypical antilisterial poration complex bacteriocin, that functions without a receptor. J Biol Chem. 1997;272:14277–14284. doi: 10.1074/jbc.272.22.14277. [DOI] [PubMed] [Google Scholar]
- 16.Meyer H E, Heber M, Eisermann B, Korte H, Metzger J W, Jung G. Sequence analysis of lantibiotics: chemical derivatization procedures allow a fast access to complete Edman degradation. Anal Biochem. 1994;223:185–190. doi: 10.1006/abio.1994.1571. [DOI] [PubMed] [Google Scholar]
- 17.Moll G, Hildeng-Hauge H, Nissen-Meyer J, Nes I F, Konings W N, Driessen A J M. Mechanistic properties of the two-component bacteriocin lanctococcin G. J Bacteriol. 1998;180:96–99. doi: 10.1128/jb.180.1.96-99.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nissen-Meyer J, Nes I F. Ribosomally synthesized antimicrobial peptides: their function, structure, biosynthesis, and mechanism of action. Arch Microbiol. 1997;167:67–77. [PubMed] [Google Scholar]
- 19.Novak J, Caufield P W, Miller E J. Isolation and biochemical characterization of a novel lantibiotic mutacin from Streptococcus mutans. J Bacteriol. 1994;176:4316–4320. doi: 10.1128/jb.176.14.4316-4320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi F, Chen P, Caufield P W. Purification and biochemical characterization of mutacin I from the group I strain of Streptococcus mutans, CH43, and genetic analysis of the mutacin I biosynthesis genes. Appl Environ Microbiol. 2000;66:3221–3229. doi: 10.1128/aem.66.8.3221-3229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi F, Chen P, Caufield P W. Purification of mutacin III from the group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthetic genes. Appl Environ Microbiol. 1999;65:3880–3887. doi: 10.1128/aem.65.9.3880-3887.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahl H-G, Bierbaum G. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from Gram-positive bacteria. Annu Rev Microbiol. 1998;52:41–79. doi: 10.1146/annurev.micro.52.1.41. [DOI] [PubMed] [Google Scholar]
- 23.Sahl H-G, Jack R W, Bierbaum G. Biosynthesis and biological activities of lantibiotics with unique post-translational modifications. Eur J Biochem. 1995;230:827–853. doi: 10.1111/j.1432-1033.1995.tb20627.x. [DOI] [PubMed] [Google Scholar]