Abstract
Purpose:
We designed a comprehensive multiple myeloma targeted sequencing panel to identify common genomic abnormalities in a single assay and validated it against known standards.
Experimental Design:
The panel comprised 228 genes/exons for mutations, 6 regions for translocations, and 56 regions for copy number abnormalities (CNA). Toward panel validation, targeted sequencing was conducted on 233 patient samples and further validated using clinical FISH (translocations), multiplex ligation probe analysis (MLPA; CNAs), whole-genome sequencing (WGS; CNAs, mutations, translocations), or droplet digital PCR (ddPCR) of known standards (mutations).
Results:
Canonical immunoglobulin heavy chain translocations were detected in 43.2% of patients by sequencing, and aligned with FISH except for 1 patient. CNAs determined by sequencing and MLPA for 22 regions were comparable in 103 samples and concordance between platforms was R2 = 0.969. Variant allele frequency (VAF) for 74 mutations were compared between sequencing and ddPCR with concordance of R2 = 0.9849.
Conclusions:
In summary, we have developed a targeted sequencing panel that is as robust or superior to FISH and WGS. This molecular panel is cost-effective, comprehensive, clinically actionable, and can be routinely deployed to assist risk stratification at diagnosis or posttreatment to guide sequencing of therapies.
Translational Relevance.
Here we provide a validated panel for targeted sequencing and analysis of myeloma and other plasma cell dyscrasias to identify the common genomic abnormalities that are diagnostic, prognostic, and clinically actionable. This panel can identify the common immunoglobulin translocations and copy number abnormalities currently detected by FISH, as well as less common translocations, MYC rearrangements, and mutations that are not currently tested for in a standard manner. We hope that adoption of a common sequencing panel will improve patient diagnostics and can be used to assist in risk stratification at diagnosis or posttreatment to guide therapeutic decision making.
Introduction
While personalized medicine in multiple myeloma is still in its infancy (1–11), next-generation sequencing (NGS) technologies have proven useful in identifying mutations, gene expression differences, and other key genetic events in multiple myeloma [refs. 12, 17; such as translocations and copy number abnormalities (CNA)] but so far their clinical utility has been limited (18). Despite efforts to use genomics to improve identification of patients with high-risk multiple myeloma, the detection of key translocations and CNA by FISH remains the standard in the clinic. Although FISH is the most frequently used technique across clinical diagnostic laboratories, there is a vast difference in the methodologies used including whether or not CD138+ cell selection is performed, regions of the genome probed, and limited interrogation of immunoglobulin heavy chain (IgH) locus rearrangements (19).
Other technologies, such as copy number arrays (20), and multiplex ligation-dependent probe amplification (MLPA; ref. 21) have also been used diagnostically to detect CNAs such as del(CDKN2C) on 1p, del(TP53) on 17p, and gain/amplification of CKS1B on 1q, which are associated with poor outcome. Together, the high-risk IgH translocations and del(TP53) are used to stratify high-risk patients according to the revised-ISS (R-ISS) criteria (22, 23). The addition of 1q gain or amplification, and TP53 mutation have also been used to further stratify patients as high risk (24, 25). MYC rearrangements are associated with poor outcome in multiple myeloma but the presence of the rearrangements is not easy to detect, due to the complexity of rearrangements and the high number of partner loci (26, 27). FISH can be used to detect the t(8;14) IgH-MYC rearrangement, but this only accounts for a minority of the cases (27, 28). A more unbiased methodology is required to detect all possible rearrangements.
Recently, additional high-risk markers have been reported, including biallelic alterations in TP53 or DIS3, arising from deletion or mutation of the remaining allele (25, 29–32). Newer patient segments such as Myeloma Genome Project (MGP) Double-Hit and Mayo Clinic (Rochester, MN) double- or triple-hit multiple myeloma identify patients with significant adverse prognosis (21, 24, 25) but their assessment is not widespread due to lack of availability of diagnostic tests.
Genomic risk stratification may also be extended to asymptomatic disease states of monoclonal gammopathy of undetermined significance and smoldering multiple myeloma (SMM). We and others have shown that IgH translocations, mutations in NRAS, KRAS, and FAM46C, as well as MYC translocations or abnormalities at 8q24 can define a high-risk group of patients with SMM who are likely to progress to multiple myeloma quickly, independent of current International Myeloma Working Group (IMWG) risk factors (33–35).
Here, we describe a comprehensive, cost-effective, hybridization capture-based, NGS assay panel for targeted sequencing of recurrently mutated key genes in newly diagnosed and relapsed multiple myeloma, genomic regions of CNA, translocations involving immunoglobulin (Ig) heavy and light chain loci, and MYC translocations. Previous versions of this panel have been used extensively in the research setting (1, 26, 27, 29, 30, 33, 36–39). We evaluated the current expanded and updated panel on 233 patient samples and extensively validated results using multiple comparative assays. A complete guidance document from laboratory methodology, capture design, bioinformatic pipeline, and analysis visualization tool has been made publicly available for others to utilize. The assay technology was transferred to a clinical diagnostic laboratory and its performance was compared with existing clinical diagnostic data. This newly developed, highly validated assay and platform enables rapid and reliable detection of patients with high-risk or therapeutically-targetable biomarkers and has the potential to guide risk-adapted treatment selection and sequencing as a personalized medicine strategy. Finally, we propose an engagement with the multiple myeloma community to consider available molecular profiling approaches including this panel to adopt an actionable strategy for diagnosis and treatment of patients with multiple myeloma.
Materials and Methods
Patients and samples
Patient material was obtained after written informed consent in accordance with the U.S. Common Rule and were approved by the Institutional Review Board. CD138+ plasma cells were magnetically sorted from bone marrow aspirates using the AutoMACS Pro (Miltenyi Biotec GmbH) or RoboSep (STEMCELL Technologies). The postselection plasma cell purity was determined by flow cytometry using anti-CD45-ECD (Beckman Coulter), anti-CD138 (Becton Dickinson), and only samples with more than 85% purity were used in this study. DNA was isolated from CD138+ plasma cells using the AllPrep DNA/RNA or Puregene kits (Qiagen). DNA from peripheral blood, saliva, or CD34+ stem cells was isolated and used as a matched nontumor control where available. For 39 samples, the CD138− fraction was used as the control sample. All DNA were eluted in low EDTA buffer.
Panel design
Based on the findings of the MGP(1) and other multiple myeloma genome sequencing studies (2–4, 7, 8, 17, 40, 41), prognostically and biologically relevant genes and genomic regions were identified (Supplementary Fig. S1). By utilizing this information, two capture panels were designed: one for common multiple myeloma translocations, and another for mutation and CNA information (Supplementary Tables S1–S3). The mutation and CNA probe set covers approximately 1.19 Mb of the genome. Probes capture exonic regions (including flanking 10 bp) of 228 key multiple myeloma genes for mutation detection. An additional 471 SNPs were captured to aid in copy number variation detection either within or surrounding key genes or in other areas of the genome. For example, in addition to the 10 exons captured for TP53, an extra 40 SNPs around TP53 were included in the design for increased sensitivity to detect loss of the region. These SNPs were chosen with a population minor allele frequency > 0.35, and the change in B allele frequency between control and tumor samples was used in combination with read depth ratio to infer both deletions and gains. To avoid hybridization artifacts and low depth problems, SNPs in guanine cytosine (GC)-rich regions were excluded. For the mutation panel, 4,785 total regions were captured.
The translocation panel covers about 4.32 Mb of the genome. Tiling capture probes were designed to cover the V, D, and J segments as well as the entire constant region to identify Ig translocations. To detect MYC translocations and rearrangements, tiling probes were designed upstream and downstream of MYC (from NSMCE2 to GSDMC). Some sequences were omitted due to mappability problems in repetitive regions which prevent sequence-specific probe design, meaning that the capture regions are not contiguous. The specifics of the captured region can be found in the annotation files at https://github.com/bwalker2/Targeted-Panel-Analysis.
For both the mutation and translocation panels, the probes were empirically balanced by testing on a set of eight saliva DNA samples using the HyperCap (KAPA Biosystems) reagents. Any over- or undercapture of regions on the panels were balanced out by modifying the amounts of probes for each region until a roughly uniform coverage of the regions of interest was observed. The catalog numbers of the mutation and translocation panels (v2.1) are IRN 1000008523 and IRN 1000008533 (KAPA Biosystems), respectively. Future updates to panel designs will be documented at https://github.com/bwalker2.
Samples were processed using HyperCap reagents as described in Supplementary Methods and validated accordingly (Supplementary Figs. S2–S10). Libraries were sequenced using 75 bp paired end reads, to a mean total depth of 344× (mutation panel 867×, translocation panel 252×).
Targeted panel data analysis
For all samples the same informatics pipeline was used. bcl2fastq was used for demultiplexing and Burrows-Wheeler Aligner (BWA) mem (v. 0.7.12) for alignment to University of California Santa Cruz's (GRCh37/hg19) human reference genome. Strelka (v.2.9.2) was used for variant calling and single-nucleotide variants (SNV) were filtered using fpfilter (https://github.com/ckandoth/variant-filter) with a 5% variant allele frequency (VAF) cut-off. Indels were filtered using a 10% VAF cut-off. Variants were annotated using Variant Effect Predictor (v.101). To determine copy number, a normalized depth comparison between tumor and control samples was used and segments of SNP variance were utilized to identify regions of chromosomal deletion and gain. A Python library and command-line software toolkit, CNVKit (v 0.9.7) was used for copy number calling pipeline. Quality control (QC) metrics were calculated using Picard's (v 2.10.0) “CollectHsMetrics” command. Intra- and interchromosomal rearrangements were called using Manta (v1.6.0) with default settings and the exome flag specified. An SQLite database was generated using somatic variants by Strelka2, structural variants by Manta, copy number depth metrics by CNVKit, and QC metrics by Picard. Data were visualized using a custom built “RShiny” application, TarPan (42) showing the mutations, translocations, copy number, QC metrics, and cross-sample contamination estimations. In TarPan, copy number can be manually normalized based on the ratio and SNP allele calls using the best fitting chromosomes with the least variance (usually chromosome 2 or 10). A full pipeline is available at https://github.com/bwalker2/Targeted-Panel-Analysis.
Orthogonal technologies for validation
Orthogonal technologies were used to validate the results of the panel, including FISH, MLPA, and whole-genome sequencing (WGS). Details are provided in Supplementary Methods.
Data availability
The analytical methods generated in this study are available at https://github.com/bwalker2/Targeted-Panel-Analysis. Data have been submitted to the European Genome-Phenome Archive under accession numbers EGAD00001008689 and EGAD00001008735.
Results
Detection of key prognostic markers and risk stratification of patients
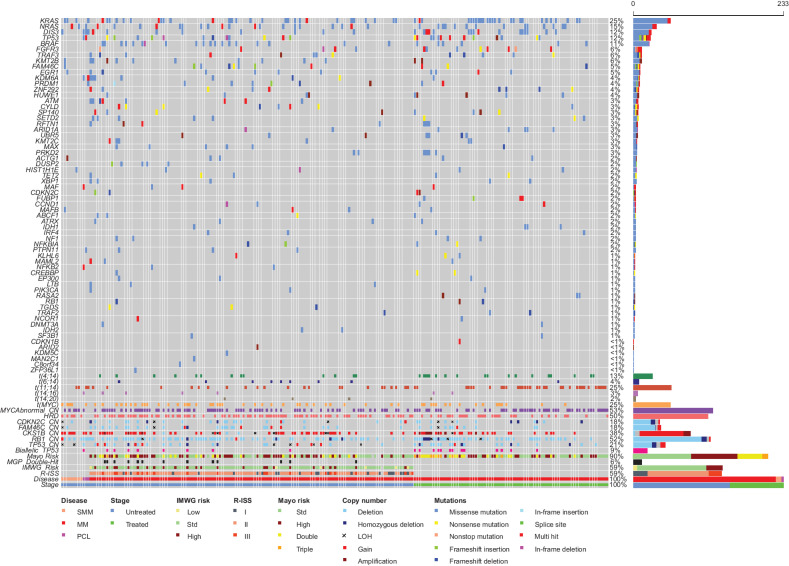
The targeted capture panel was tested on 233 samples from 190 patients with SMM (n = 9), multiple myeloma [n = 221, of which 138 were newly diagnosed multiple myeloma (NDMM)], and plasma cell leukemia (PCL; n = 3). Mutations, translocations, and CNAs were determined using a standard computational pipeline. In agreement with previous studies, we identified key mutations including KRAS (25%), NRAS (15%), DIS3 (12%), FAM46C (5%), BRAF (11%), and TP53 (12%). The frequency of 63 previously identified driver genes from MGP(1) in our dataset are shown in Fig. 1, along with identified key cytogenetic groups and CNAs. Notably, with the exception of SAMHD1, all other driver gene mutations were detected. Thus, the current panel is able to detect most of the driver genes identified thus far in NDMM, including in six genes commonly mutated in relapse refractory multiple myeloma (N. Ansari-Pour; unpublished data).
Figure 1.
Frequency of mutations in 63 key driver genes, translocations, hyperdiploidy, and key CNAs detected by targeted sequencing. Risk stratification of patients was determined from genomic and biochemical makers. MM, multiple myeloma; Std, standard.
Poor prognostic CNA markers in multiple myeloma include del1p (CDKN2C), gain/amp 1q (CKS1B), and del17p (TP53). In this dataset, deletion of CDKN2C was identified in 30 samples (12.8%) including homozygous deletion in 7 samples. Copy number–neutral LOH (CNN-LOH) was detected in an additional 5 samples. There was no significant difference in frequency of deletion of CDKN2C among the disease states. Gain (3 copies) or amplification (4+ copies) of 1q (CKS1B) was detected in 81 samples (34.8%), of which 11 were amplifications. Gain/amp 1q was detected in 62.5% SMM, 31.9% NDMM, 33.7% previously treated multiple myeloma, and 66.6% PCL with no significant difference between groups. Deletion of TP53 was detected in 36 samples (15.5%), including homozygous deletion in 6 samples. CNN-LOH was detected in an additional six samples. There was a significant increase in frequency of TP53 deletion between NDMM and previously treated multiple myeloma (P = 0.026): 11.6% versus 22.9%.
We applied genomic risk stratification criteria to the samples, exploring MGP Double-Hit, biallelic TP53, and Mayo Clinic risk classification (Fig. 1), which requires TP53 mutation status in addition to deletion (24, 25). MGP Double-Hit (biallelic TP53 abnormalities or gain 1q with ISS III) was applied to NDMM samples and identified 10.9% (15/138) of patients. Biallelic TP53 abnormalities were detected in 9.9% of samples; 9 of 138 (6.5%) were NDMM, 13 of 83 (15.7%) were previously treated, and 1 of 3 (33.3%) was PCL, and none in SMM. There was a significant increase in biallelic TP53 events from diagnosis to those previously treated (P = 0.015).
The Mayo Clinic risk classification, where t(4;14), t(14;16), t(14;20), gain 1q, del(17p), or mutation of TP53 are considered high-risk markers and are additive, was applied to all multiple myeloma samples identifying 90 of 221 standard risk, 72 of 221 high risk, 38 of 221 double hit, and 9 triple-hit multiple myeloma. Of these, the split between NDMM and previously treated multiple myeloma was 42.7% versus 37.3% standard, 34.8% versus 28.9% high, 15.2% versus 20.4% double, and 0.7% versus 9.6% triple hit. There was a significant increase in triple-hit multiple myeloma in previously treated patients (P = 0.0011) and all but one of the triple-hit patients had biallelic TP53 abnormalities.
Mutation detection and validation
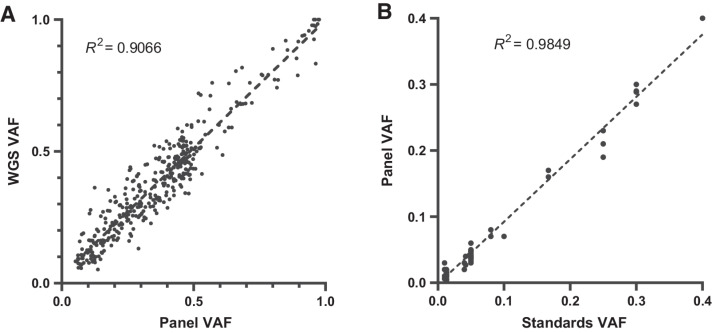
Of the 233 samples, WGS mutation data were available for 113. For this analysis, WGS data were considered only for regions captured by the mutation panel and further filtered for those with a protein coding effect. There were 379 variants detected that passed filtering by both sequencing methods. A comparison of VAFs between sequencing methods showed a correlation of R2 = 0.9006 (Fig. 2A).
Figure 2.
Validation of mutation VAF against matched WGS data (A) and DNA standards (B).
Mutation detection validation was performed using samples with known VAF for common mutations. Five DNA standards (Horizon Discovery; Supplementary Table S4) were used which had mutations at frequencies from 1.3% to 40% VAF engineered into them in key genes important in cancer. The VAF of the DNA standards is commercially determined by ddPCR and can be used to show that the mutations are detected at the correct frequency and that the bioinformatics pipeline is able to annotate them correctly. From these five standards, 74 mutations were assayed on the panel. The expected and observed VAF for each mutation were plotted giving a correlation coefficient of R2 = 0.9849 (Fig. 2B), indicating high concordance of results.
CNA validation
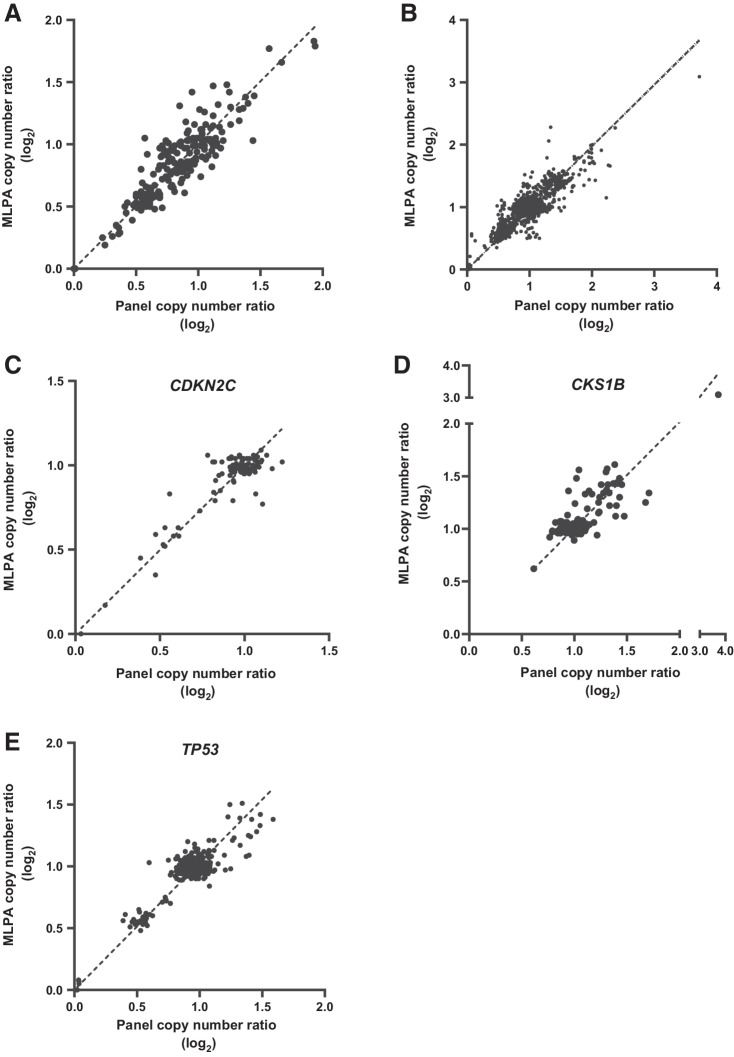
CNA was determined by targeted sequencing and by MLPA for 22 regions that were directly comparable. Initial validation of MLPA and sequencing was performed in a panel of 13 multiple myeloma cell lines. For all the 22 regions combined, a concordance of 99.61% was observed between MLPA and sequencing in the 13 cell lines (Fig. 3A; Supplementary Table S5; Supplementary Fig. S4). In 101 patient samples the concordance between the technologies was R2 = 0.987 (Fig. 3B). For the important prognostic regions, the concordance was R2 = 0.962 (CDKN2C), R2 = 0.986 (CKS1B), and R2 = 0.973 (TP53; Fig. 3C–E).
Figure 3.
Validation of copy number against MLPA. Copy number ratio (log2) was determined for 13 multiple myeloma cell lines by targeted panel sequencing and MLPA (A). Comparison of copy number ratio for multiple myeloma 101 patient samples for 22 common regions (B), with emphasis on regions associated with poor prognosis including CDKN2C (C), CKS1B (D), and TP53 (E).
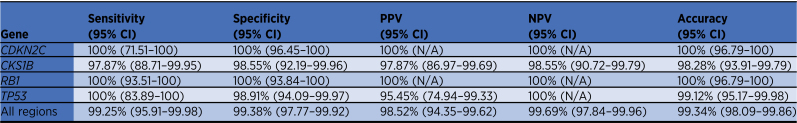
We compared the copy number determination between WGS and panel sequencing methods for the common prognostic regions, CDKN2C, CKS1B, TP53, and RB1 (Supplementary Tables S6–S10). At CDKN2C, a deletion (0 or 1 copies) was detected in 11 of 113 samples on the panel and matched with WGS data. For CKS1B, gain/amplification (≥3 copies) was detected in 46 of 113 samples, of which one was not detected by WGS. WGS did detect gain of CKS1B in one sample that was not detected by the panel. For RB1, deletion was detected in 55 of 113 samples by the panel and agreed with WGS data. For TP53, deletions were detected in 21 of 113 samples by both the panel and WGS. The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy for each region are shown in Table 1. All PPVs and NPVs were above 95%.
Table 1.
Detection rates of CNAs by targeted panel compared with WGS.
By combining all the data from these four loci, the overall performance of the assay for CNA detection, compared with WGS, was calculated: sensitivity (99.25%), specificity (99.38%), PPV (98.52%), NPV (99.69%), and accuracy (99.34%).
Detection of small homozygous deletions in CDKN2C, RB1, and TP53
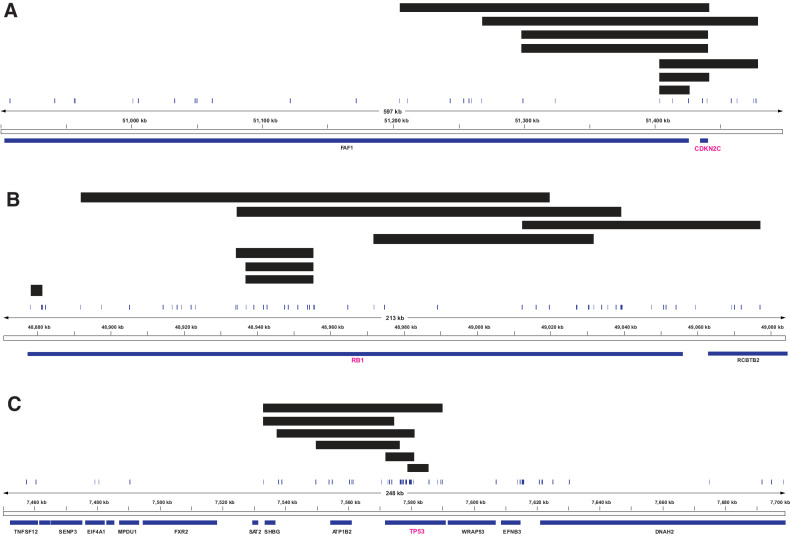
To further explore the utility of the panel, we examined homozygous deletions of the key tumor suppressor genes, CDKN2C, RB1, and TP53 (Fig. 4). For CDKN2C, the panel detected homozygous deletions in seven of 233 samples which ranged in size from 21.9 to 235.4 kb and affected both coding exons of the gene. Of these seven samples, four also had WGS and the homozygous deletions were detected only in one of four.
Figure 4.
Detection of homozygous deletions in the key tumor suppressor genes CDKN2C, RB1, and TP53. Samples with homozygous deletions plotted at the CDKN2C (A), RB1 (B), or TP53 (C) loci. Black bars indicate homozygous deletion events in samples. Gene/exon locations are shown below each plot and vertical lines indicate capture regions on the panel.
For RB1, homozygous deletions were detected in eight of 233 samples and ranged in size from 3.5 to 105.3 kb. WGS was available for four of eight, and a homozygous deletion was detected in one sample. The remaining three samples, where WGS did not detect the deletion, were from the same patient and the deletion was 18.6 kb. None of the homozygous deletions spanned the entire gene with most deleting several exons within the gene. As such, these deletions would be unlikely to be detected by FISH.
For TP53, six samples with a homozygous deletion were identified. Of these homozygous deletions, none covered the entire gene. The homozygous deletions ranged in size from 6.1 to 56.8 kb. Three of the six samples also had WGS, of which only one detected the homozygous deletion. The deletions that were not detected by WGS were 6.1, 8.1, and 27.6 kb in size. Given the small nature of all six homozygous deletions, they are unlikely to be detectable by FISH.
Translocation breakpoint detection and validation
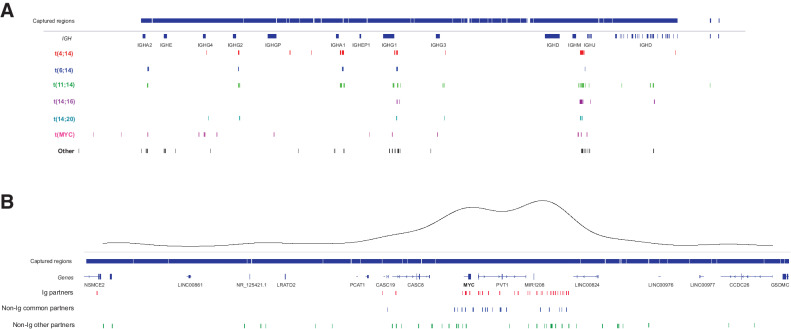
In the 233 patient samples, canonical Ig translocations were detected in 47% of samples, encompassing t(4;14), t(6;14), t(11;14), t(14;16), and t(14;20) in 13%, 4%, 25%, 3%, and 2%, respectively. This is consistent with the expected frequencies of these translocations, with some enrichment of t(4;14) and t(11;14) due to sample selection bias. The distribution of the translocation breakpoints at the IgH locus is shown in Fig. 5 and it aligns with previously published data (38).
Figure 5.
Translocation breakpoints. A,IGH@ locus breakpoints broken down by partner chromosome. V regions not shown for clarity. Captured regions extend to each V region. B,MYC region breakpoints broken down by Ig, non-Ig common (FOXO3, TXNDC5, FAM46C), and other partners. A kernel density plot shows the two main translocation hotspots centromeric of MYC and telomeric of PVT1.
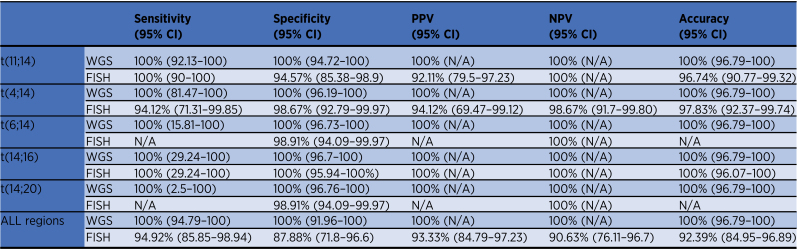
One hundred and sixteen samples had WGS data available to validate the capture panel results and to ensure that no translocations were missed. There was complete agreement between targeted panel and WGS calls for the canonical translocations: t(4;14) (n = 19), t(6;14) (n = 2), t(11;14) (n = 46), t(14;16) (n = 3), t(14;20) (n = 1), and no translocation detected (n = 45). As the results were completely consistent between the platforms, the sensitivity, specificity, PPV, NPV, and accuracy were all 100% (Table 2).
Table 2.
Detection rates of IGH translocations by targeted panel compared with WGS and FISH.
In addition, clinical FISH data were available for 92 samples. A total of 85 samples gave concordant results between technologies with translocations detected in 56 samples and not observed in 29 samples (Supplementary Table S11). FISH did not detect four translocations that were detected by targeted sequencing and WGS [one t(4;14), one t(14;20), and two t(11;14)], and in three additional samples a rearrangement at the IgH locus was detected by FISH but the partner chromosome was not identified [t(8;14), t(6;14), and t(11;14) detected by targeted panel and WGS]. In one sample a variant t(4;14) was detected by FISH but not by targeted sequencing or WGS. Therefore, targeted sequencing only failed to detect one variant translocation that was detected by FISH but gave more information on six samples than was given by FISH highlighting the superiority of sequencing approach over FISH methods. The statistical comparisons between the targeted sequencing panel (and also WGS as they were identical) and FISH are shown in Table 2 and Supplementary Table S11.
In addition, since immunoglobulin lambda chain (IgL) rearrangements have been shown to be prognostic in multiple myeloma (43), we examined the detection of these between panel and WGS data. The panel detected IgL translocations in 10 samples, including the most common rearrangement IgL:MYC in eight of the samples. Of these 10 samples, seven also had WGS data and were confirmed by that method. WGS sequencing identified nine samples with translocations involving the IgL locus, of which seven were detected by the panel. Of the two discordant samples, one was a t(8;22) and was resolved with realignment to hg38. The other discordant sample had a complex event involving five chromosomes (chr 5, 7, 14, 19, and 22) by WGS, of which three of the breakpoints (chr 7, 14, and 19) were detected by the panel.
Novel translocation partners detected by targeted sequencing
An advantage of capture panels is that novel events can also be detected. We have previously identified novel translocations to the Ig loci affecting partner proto-oncogenes (39, 44). In this study from 185 samples, we identified novel Ig translocations in 20 samples (10.8%). The partner loci included some known oncogenes such as CCND2, KMT2B, PAX5, MYCN, MAP3K14, BCL2, and TNFAIP8, but also identified some potentially novel oncogenes such as UST, TNFSF12, DEFB1, and LRRK2. KMT2B is also frequently mutated indicating multiple mechanisms of disrupting the gene in oncogenesis. The prognostic significance of these infrequent translocation partners is difficult to ascertain, but they may lead to better understanding of disease biology through identification of new driver genes.
MYC rearrangements and CNAs
We previously performed a comprehensive analysis of MYC translocations and CNAs in multiple myeloma using an identical panel design (27). The location of MYC translocation breakpoints in this dataset are shown in Fig. 5. The frequency of MYC translocations was 24.0% with 49.4% of samples having a CNA within 2 Mb of MYC, which we have shown can affect expression of MYC (27). Many samples with a translocation also had CNAs and so the total frequency of samples with MYC abnormalities was 66.9.1% (156/233; Supplementary Fig. S5).
Validation of MYC translocations detected by the panel against WGS data (n = 116) showed agreement in 91.4% of samples (106/116). Of the discordant samples (n = 10/116), MYC translocations were detected by the panel and not by WGS in four samples and were judged to be subclonal translocations with insufficient depth of coverage in the WGS. The remaining six translocations that were only detected by WGS had been filtered out due to mapping quality issues with hg19 alignments and were resolved with realignment to hg38.
Comparison of multiple myeloma targeted sequencing panels
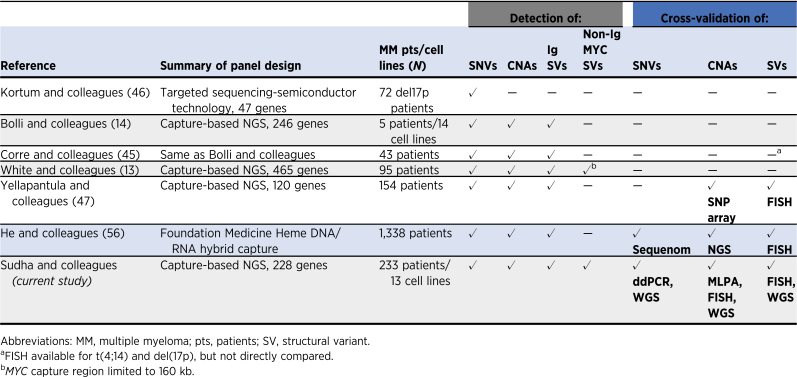
Several other multiple myeloma targeted sequencing panels have been described and are summarized in Table 3 (13, 14, 45–47). Most of those panels could detect mutations, CNAs, and IgH locus rearrangements, however, they were not universally validated using orthogonal technologies. Of these, the Yellapantula and colleagues (47) panel is the most characterized with validation by FISH for translocations and SNP array for CNAs. One key aspect missing from the Yellapantula and colleagues panel is that it only detects MYC abnormalities partnered with the IGH locus. The MGP panel also has the region surrounding MYC on 8q24 assayed, allowing for the detection of non-Ig partners which are more frequent than Ig partners. MYC rearrangements have been shown to be prognostic and associated with a shorter time to progression from SMM to symptomatic multiple myeloma (33, 35, 48). MGP is the only panel to be validated against WGS and show comparable identification of mutations, CNAs, and translocations between the methodologies. This comparison indicates that for those laboratories which cannot yet perform WGS on all multiple myeloma samples diagnostically, the MGP panel is a viable, cost-effective, and accurate alternative to generate prognostically meaningful data.
Table 3.
Targeted NGS-based assays in multiple myeloma.
Discussion
WGS of patient samples is increasingly popular in research laboratories and can also be utilized for clinical diagnostics (18). However, the cost, processing time, and high throughput computational expertise required to analyze data can be prohibitive for smaller nonacademic centers. We have developed and validated a sequencing panel that is relevant to prognosis, risk stratification, and treatment of patients with multiple myeloma and have described an end-to-end protocol for laboratory and bioinformatic processing of samples and data visualization. This panel has been utilized, in different forms, for the analysis of SMM, NDMM, previously treated multiple myeloma, and PCL patient samples (1, 25, 27, 29, 30, 33, 36, 49). Although not yet extensively used in relapsed refractory multiple myeloma (RRMM), the assay contains regions of interest for this setting, including the p53 pathway (TP53, ATM, ATR), PRDM1, and CRBN (41, 50). Other common abnormalities can also be detected including exonic deletions of KDM6A, deletion of FGFR3 in t(4;14) samples, and deletions of negative regulators of the NF-κB pathway, BIRC2/3, TRAF2/3, and CYLD, as well as NIK (MAP3K14) rearrangements (Supplementary Figs. S6–S9; refs. 51–53). We have formally validated the data generated here against WGS, MLPA, clinical FISH, and mutation standards for translocations, copy number, and mutation identification. In addition, the low input amount of genomic DNA (100 ng) used here allows for the profiling of samples with low disease burden where there are few cells to analyze. We have also successfully performed the assay with only 50 ng of DNA without loss of performance.
For translocation detection we report 100% concordance with WGS data, confirming that WGS is not required for accurate detection of these structural events in multiple myeloma. Furthermore, this assay can be utilized for the detection of translocations in other B-cell malignancies. In addition, we show that MYC structural alterations (interchromosomal translocations, CNAs) can be detected with this assay. The breadth and complexity of MYC abnormalities have resulted in the underestimation of this locus by FISH, where only 10% to 15% of NDMM samples have the abnormality, whereas targeted sequencing and WGS identifies up to 50% of patients with an abnormality (27, 54).
CNAs were validated against WGS and MLPA, which has been used in clinical trials (21). Compared with MLPA, the panel showed a correlation of R2 = 0.987 and compared with WGS the sensitivity and specificity, were 94.89% and 99.68%, respectively. The main advantage for the panel against WGS was in the detection of small homozygous deletions, where multiple algorithms were required to detect all homozygous deletions in the WGS data. The number of individually analyzed probes in exons and surrounding SNPs in the panel gave more confidence in detecting these small events.
Other targeted panels have been described for the examination of multiple myeloma patient samples (14, 47, 55), but none has been used as extensively or is as exhaustive as the MGP Panel encompassing the three main drivers of multiple myeloma: mutations, CNAs, and translocations (Table 3). We have used the translocation part of the panel as a bolt-on for exome studies (37), before incorporating it into a targeted design, which has now been used in over 550 tumor samples. Previously described targeted panels were not robustly tested nor cross-validated across platforms and laboratories. We have demonstrated the performance of our targeted panel against multiple well-established methods including ddPCR, MLPA, FISH, and WGS. We also provide a complete workflow including a graphical user interface (42) that can be adopted in any laboratory and modified to suit their needs.
The MGP Panel has been adopted for retrospective analysis of clinical trial samples (K.L. Yong; personal communication) and for use in clinical care. Our goal is to broadly share this panel with the multiple myeloma community to improve opportunities and parity across academic and community centers to quickly and easily identify patients with high-risk disease or targetable genetic mutations. Despite several efforts to construct a genomics-based molecular profiling platform in multiple myeloma, this approach has not been broadly adopted in clinical care nor for improved risk stratification of patients. We encourage the multiple myeloma community (guided by organizations such as IMWG and International Myeloma Society) to seriously consider a thorough examination of the different methods and potentially build consensus around adoption of a molecular-profiling strategy.
Supplementary Material
Acknowledgments
Research support was received from Bristol Myers Squibb. B.A. Walker was partially supported by a grant from the Leukemia and Lymphoma Society and the Daniel and Lori Efroymson Chair. The Indiana Myeloma Registry is funded in part by support from the Indiana University Precision Health Initiative, Miles for Myeloma, the Harry and Edith Gladstein Chair, and the Omar Barham Fighting Cancer Fund. Computational infrastructure at Indiana University (Indianapolis, IN) was funded in part by Lilly Endowment Inc. through the Indiana University Pervasive Technology Institute. Authors are grateful to Gail Vance and Mirian Salazar for helpful discussions.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
M. Czader reports grants and personal fees from Beckman Coulter outside the submitted work. N. Ansari-Pour reports other support from BMS during the conduct of the study. K.L. Yong reports grants from Amgen during the conduct of the study. W.E. Pierceall reports other support from Bristol Myers Squibb during the conduct of the study. E. Flynt reports other support from Bristol Myers Squibb outside the submitted work. S. Gooding reports grants from Bristol Myers Squibb during the conduct of the study. K. Ramasamy reports grants from Bristol Myers Squibb outside the submitted work. B.A. Walker reports grants from Bristol Myers Squibb and Leukemia and Lymphoma Society during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
P. Sudha: Data curation, formal analysis, validation, investigation, methodology, writing–review and editing. A. Ahsan: Data curation, formal analysis, supervision, validation, investigation, methodology, writing–original draft. C. Ashby: Data curation, formal analysis, investigation, methodology, writing–review and editing. T. Kausar: Data curation, investigation, methodology, writing–review and editing. A. Khera: Data curation, formal analysis, writing–review and editing. M.H. Kazeroun: Data curation, formal analysis, methodology, writing–review and editing. C.-C. Hsu: Investigation, methodology, writing–review and editing. L. Wang: Investigation, methodology, writing–review and editing. E. Fitzsimons: Data curation, investigation, methodology, writing–review and editing. O. Salminen: Data curation, investigation, methodology, writing–review and editing. P. Blaney: Methodology, writing–review and editing. M. Czader: Data curation, investigation, methodology, writing–review and editing. J. Williams: Supervision, methodology, writing–review and editing. M.I. Abu Zaid: Resources, investigation, methodology, writing–review and editing. N. Ansari-Pour: Formal analysis, supervision, methodology, writing–review and editing. K.L. Yong: Resources, supervision, writing–review and editing. F. van Rhee: Resources, writing–review and editing. W.E. Pierceall: Resources, writing–original draft, project administration. G.J. Morgan: Resources, supervision, writing–review and editing. E. Flynt: Conceptualization, resources, supervision, funding acquisition, investigation, methodology, writing–original draft. S. Gooding: Resources, data curation, supervision, investigation, writing–review and editing. R. Abonour: Resources, supervision, writing–review and editing. K. Ramasamy: Resources, supervision, writing–review and editing. A. Thakurta: Conceptualization, resources, supervision, funding acquisition, investigation, methodology, writing–original draft. B.A. Walker: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, methodology, writing–original draft.
References
- 1. Walker BA, Mavrommatis K, Wardell CP, Ashby TC, Bauer M, Davies FE, et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 2018;132:587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Szalat R, Munshi NC. Genomic heterogeneity in multiple myeloma. Curr Opin Genet Dev 2015;30:56–65. [DOI] [PubMed] [Google Scholar]
- 3. Manier S, Salem KZ, Park J, Landau DA, Getz G, Ghobrial IM. Genomic complexity of multiple myeloma and its clinical implications. Nat Rev Clin Oncol 2017;14:100–13. [DOI] [PubMed] [Google Scholar]
- 4. Manier S, Salem K, Glavey SV, Roccaro AM, Ghobrial IM. Genomic aberrations in multiple myeloma. Cancer Treat Res 2016;169:23–34. [DOI] [PubMed] [Google Scholar]
- 5. Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer 2012;12:335–48. [DOI] [PubMed] [Google Scholar]
- 6. Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun 2014;5:2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 2014;25:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature 2011;471:467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harding T, Baughn L, Kumar S, Van Ness B. The future of myeloma precision medicine: integrating the compendium of known drug resistance mechanisms with emerging tumor profiling technologies. Leukemia 2019;33:863–83. [DOI] [PubMed] [Google Scholar]
- 10. Gonzalez-Calle V, Fonseca R. [Towards precision medicine in myeloma: new evidence and challenges]. Medicina (B Aires) 2017;77:222–6. [PubMed] [Google Scholar]
- 11. Erin Flynt AT. Shadow of the Future: Precision medicine in multiple myeloma. J Precis Med 2019;5. [Google Scholar]
- 12. Lagana A, Beno I, Melnekoff D, Leshchenko V, Madduri D, Ramdas D, et al. Precision medicine for relapsed multiple myeloma on the basis of an integrative multiomics approach. JCO Precis Oncol 2018;2:PO.18.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. White BS, Lanc I, O'Neal J, Gupta H, Fulton RS, Schmidt H, et al. A multiple myeloma-specific capture sequencing platform discovers novel translocations and frequent, risk-associated point mutations in IGLL5. Blood Cancer J 2018;8:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bolli N, Li Y, Sathiaseelan V, Raine K, Jones D, Ganly P, et al. A DNA target-enrichment approach to detect mutations, copy number changes and immunoglobulin translocations in multiple myeloma. Blood Cancer J 2016;6:e467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bolli N, Biancon G, Moarii M, Gimondi S, Li Y, de Philippis C, et al. Analysis of the genomic landscape of multiple myeloma highlights novel prognostic markers and disease subgroups. Leukemia 2018;32:2604–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kortum KM, Mai EK, Hanafiah NH, Shi CX, Zhu YX, Bruins L, et al. Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes. Blood 2016;128:1226–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walker BA. Whole exome sequencing in multiple myeloma to identify somatic single nucleotide variants and key translocations involving immunoglobulin Loci and MYC. Methods Mol Biol 2018;1792:71–95. [DOI] [PubMed] [Google Scholar]
- 18. Hollein A, Twardziok SO, Walter W, Hutter S, Baer C, Hernandez-Sanchez JM, et al. The combination of WGS and RNA-Seq is superior to conventional diagnostic tests in multiple myeloma: Ready for prime time? Cancer Genet 2020;242:15–24. [DOI] [PubMed] [Google Scholar]
- 19. Yu Y, Brown Wade N, Hwang AE, Nooka AK, Fiala MA, Mohrbacher A, et al. Variability in cytogenetic testing for multiple myeloma: A comprehensive analysis from across the United States. JCO Oncol Pract 2020;16:e1169–e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leeksma AC, Baliakas P, Moysiadis T, Puiggros A, Plevova K, Van der Kevie-Kersemaekers AM, et al. Genomic arrays identify high-risk chronic lymphocytic leukemia with genomic complexity: a multi-center study. Haematologica 2021;106:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shah V, Johnson DC, Sherborne AL, Ellis S, Aldridge FM, Howard-Reeves J, et al. Subclonal TP53 copy number is associated with prognosis in multiple myeloma. Blood 2018;132:2465–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Palumbo A, Avet-Loiseau H, Oliva S, Lokhorst HM, Goldschmidt H, Rosinol L, et al. Revised International Staging System for multiple myeloma: A report from International Myeloma Working Group. J Clin Oncol 2015;33:2863–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abe Y, Ishida T. Immunomodulatory drugs in the treatment of multiple myeloma. Jpn J Clin Oncol 2019;49:695–702. [DOI] [PubMed] [Google Scholar]
- 24. Rajkumar SV, Kumar S. Multiple myeloma current treatment algorithms. Blood Cancer J 2020;10:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walker BA, Mavrommatis K, Wardell CP, Ashby TC, Bauer M, Davies F, et al. A high-risk, double-hit, group of newly diagnosed myeloma identified by genomic analysis. Leukemia 2019;33:159–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walker BA, Wardell CP, Brioli A, Boyle E, Kaiser MF, Begum DB, et al. Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J 2014;4:e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mikulasova A, Ashby C, Tytarenko RG, Qu P, Rosenthal A, Dent JA, et al. Microhomology-mediated end joining drives complex rearrangements and over expression of MYC and PVT1 in multiple myeloma. Haematologica 2019;105:1055–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abdallah N, Baughn LB, Rajkumar SV, Kapoor P, Gertz MA, Dispenzieri A, et al. Implications of MYC rearrangements in newly diagnosed multiple myeloma. Clin Cancer Res 2020;26:6581–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boyle EM, Ashby C, Tytarenko RG, Deshpande S, Wang H, Wang Y, et al. BRAF and DIS3 mutations associate with adverse outcome in a long-term follow-up of patients with multiple myeloma. Clin Cancer Res 2020;26:2422–32. [DOI] [PubMed] [Google Scholar]
- 30. Ashby C, Tytarenko RG, Wang Y, Weinhold N, Johnson SK, Bauer M, et al. Poor overall survival in hyperhaploid multiple myeloma is defined by double-hit bi-allelic inactivation of TP53. Oncotarget 2019;10:732–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weinhold N, Ashby C, Rasche L, Chavan SS, Stein C, Stephens OW, et al. Clonal selection and double-hit events involving tumor suppressor genes underlie relapse in myeloma. Blood 2016;128:1735–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Flynt E, Bisht K, Sridharan V, Ortiz M, Towfic F, Thakurta A. Prognosis, biology, and targeting of TP53 dysregulation in multiple myeloma. Cells 2020;9:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boyle EM, Deshpande S, Tytarenko R, Ashby C, Wang Y, Bauer MA, et al. The molecular make up of smoldering myeloma highlights the evolutionary pathways leading to multiple myeloma. Nat Commun 2021;12:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bustoros M, Sklavenitis-Pistofidis R, Park J, Redd R, Zhitomirsky B, Dunford AJ, et al. Genomic profiling of smoldering multiple myeloma identifies patients at a high risk of disease progression. J Clin Oncol 2020;38:2380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Misund K, Keane N, Stein CK, Asmann YW, Day G, Welsh S, et al. MYC dysregulation in the progression of multiple myeloma. Leukemia 2020;34:322–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thanendrarajan S, Tian E, Qu P, Mathur P, Schinke C, van Rhee F, et al. The level of deletion 17p and bi-allelic inactivation of TP53 has a significant impact on clinical outcome in multiple myeloma. Haematologica 2017;102:e364–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, et al. Mutational spectrum, copy number changes, and outcome: Results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol 2015;33:3911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Walker BA, Wardell CP, Johnson DC, Kaiser MF, Begum DB, Dahir NB, et al. Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells. Blood 2013;121:3413–9. [DOI] [PubMed] [Google Scholar]
- 39. Walker BA, Wardell CP, Ross FM, Morgan GJ. Identification of a novel t(7;14) translocation in multiple myeloma resulting in overexpression of EGFR. Genes Chromosomes Cancer 2013;52:817–22. [DOI] [PubMed] [Google Scholar]
- 40. Walker BA, Morgan GJ. The genomic features associated with high-risk multiple myeloma. Oncotarget 2018;9:35478–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gooding S, Ansari-Pour N, Towfic F, Estevez MO, Chamberlain PP, Tsai KT, et al. Multiple cereblon genetic changes are associated with acquired resistance to lenalidomide or pomalidomide in multiple myeloma. Blood 2021;137:232–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ashby C, Rutherford M, Bauer MA, Peterson EA, Wang Y, Boyle EM, et al. TarPan: an easily adaptable targeted sequencing panel viewer for research and clinical use. BMC Bioinf 2020;21:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Barwick BG, Neri P, Bahlis NJ, Nooka AK, Dhodapkar MV, Jaye DL, et al. Multiple myeloma immunoglobulin lambda translocations portend poor prognosis. Nat Commun 2019;10:1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morgan GJ, He J, Tytarenko R, Patel P, Stephens OW, Zhong S, et al. Kinase domain activation through gene rearrangement in multiple myeloma. Leukemia 2018;32:2435–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Corre J, Cleynen A, du Pont SR, Buisson L, Bolli N, Attal M, et al. Multiple myeloma clonal evolution in homogeneously treated patients. Leukemia 2018;32:2636–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kortum KM, Langer C, Monge J, Bruins L, Zhu YX, Shi CX, et al. Longitudinal analysis of 25 sequential sample-pairs using a custom multiple myeloma mutation sequencing panel (M(3)P). Ann Hematol 2015;94:1205–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yellapantula V, Hultcrantz M, Rustad EH, Wasserman E, Londono D, Cimera R, et al. Comprehensive detection of recurring genomic abnormalities: a targeted sequencing approach for multiple myeloma. Blood Cancer J 2019;9:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Walker BA, Wardell CP, Murison A, Boyle EM, Begum DB, Dahir NM, et al. APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat Commun 2015;6:6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schinke C, Boyle EM, Ashby C, Wang Y, Lyzogubov V, Wardell C, et al. Genomic analysis of primary plasma cell leukemia reveals complex structural alterations and high-risk mutational patterns. Blood Cancer J 2020;10:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ziccheddu B, Biancon G, Bagnoli F, De Philippis C., Maura F, Rustad EH, et al. Integrative analysis of the genomic and transcriptomic landscape of double-refractory multiple myeloma. Blood Adv 2020;4:830–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 2009;41:521–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Keats JJ, Maxwell CA, Taylor BJ, Hendzel MJ, Chesi M, Bergsagel PL, et al. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood 2005;105:4060–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Demchenko YN, Glebov OK, Zingone A, Keats JJ, Bergsagel PL, Kuehl WM. Classical and/or alternative NF-kappaB pathway activation in multiple myeloma. Blood 2010;115:3541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sharma N, Smadbeck JB, Abdallah N, Zepeda-Mendoza C, Binder M, Pearce KE, et al. The prognostic role of MYC structural variants identified by NGS and FISH in multiple myeloma. Clin Cancer Res 2021;27:5430–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kortum KM, Langer C, Monge J, Bruins L, Egan JB, Zhu YX, et al. Targeted sequencing using a 47 gene multiple myeloma mutation panel (M (3) P) in -17p high risk disease. Br J Haematol 2015;168:507–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. He J, Abdel-Wahab O, Nahas MK, Wang K, Rampal RK, Intlekofer AM, et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 2016;127:3004–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The analytical methods generated in this study are available at https://github.com/bwalker2/Targeted-Panel-Analysis. Data have been submitted to the European Genome-Phenome Archive under accession numbers EGAD00001008689 and EGAD00001008735.