Abstract
Purpose:
Metaplastic breast cancer (MpBC) is a rare subtype of breast cancer that is commonly triple-negative and poorly responsive to neoadjuvant therapy (NAT) in retrospective studies.
Experimental Design:
To better define clinical outcomes and correlates of response, we analyzed the rate of pathological complete response (pCR) to NAT, survival outcomes, and genomic and transcriptomic profiles of the pre-treatment tumors in a prospective clinical trial (NCT02276443). 211 patients with triple-negative breast cancer (TNBC), including 39 with MpBC, received doxorubicin-cyclophosphamide-based NAT.
Results:
Although not meeting the threshold for statistical significance, patients with MpBCs were less likely to experience a pCR (23% vs 40%; p=0.07), had shorter event-free survival (29.4 vs 32.2 months, p=0.15), metastasis-free survival (30.3 vs 32.4 months, p=0.22) and overall survival (32.6 vs 34.3 months, p=0.21). This heterogeneity is mirrored in the molecular profiling. Mutations in PI3KCA (23% vs 9%, p=0.07) and its pathway (41% vs 18%, p=0.02), were frequently observed and enriched in MpBCs. The gene expression profiles of each histologically defined subtype were distinguishable, and characterized by distinctive gene signatures. Among non-Mp TNBCs, 10% possessed a metaplastic-like gene expression signature and had pCR rates and survival outcomes similar to MpBC.
Conclusions:
Further investigations will determine if metaplastic-like tumors should be treated more similarly to MpBC in the clinic. The 23% pCR rate in this study suggests that patients with MpBC should be considered for NAT. To improve this rate, a pathway analysis predicted enrichment of HDAC and RTK/MAPK pathways in MpBC, which may serve as new targetable vulnerabilities.
INTRODUCTION
Metaplastic breast cancer (MpBC) is a rare and aggressive subset of breast cancer that is often characterized by an admixture of histologically distinct morphological components (1). MpBC comprises less than 1% of invasive breast cancers and is frequently triple-negative (>90%) (2). The National Comprehensive Cancer Network (NCCN) guidelines recognize metaplastic histology as an independent, negative prognostic indicator, as patients with MpBC usually have worse clinical outcomes compared to those with non-Mp triple-negative breast cancer (TNBC) (3,4).
The optimal treatment strategy for patients with localized MpBC remains controversial. Retrospective analyses of patients with MpBC treated with neoadjuvant therapy (NAT) have demonstrated pathological complete response (pCR) rates of ~10%, which is substantially lower than the 30-54% reported for non-Mp TNBC (5-7), prompting some to suggest that patients with MpBC should not receive NAT (7,8) or even chemotherapy as standard of care (9). Furthermore, survival outcomes associated with pCR are unknown in MpBC. Notably, most reported analyses of clinical outcomes in patients with MpBC are performed on retrospectively assembled datasets from larger, tertiary cancer centers where referral bias may enrich for more aggressive cases (10) or datasets generated through searches of national claims databases where the diagnosis of MpBC cannot be histologically confirmed (11).
To identify more effective treatments, genomic profiling has been done on MpBC. Like TNBCs, MpBCs commonly have mutations in TP53 (~50-75% of cases). However, mutations in PIK3CA (~20-50%), PTEN (~5-25%), and other members of the PI3K/AKT/mTOR pathway are enriched in MpBC relative to TNBC (12-16), suggesting that this pathway may be a rational therapeutic target for MpBC. Indeed, the combination of liposomal doxorubicin, bevacizumab, and mTOR inhibition with either temsirolimus or everolimus (DAT or DAE) demonstrate promising activity in patients with metastatic MpBC (17). Additionally, genomic profiling studies have sometimes identified frequent mutations in beta-catenin (18), although this finding has been controversial (13,19,20). Another frequent target of mutations or copy number changes are receptor tyrosine kinases (RTKs), including ERBB4 (15), FLT3 (15), and EGFR (21-24). However, a neoadjuvant strategy to target these aberrations has not yet been established.
As another approach to identify vulnerabilities, transcriptomic and proteomic studies have been performed in MpBC, demonstrating that epithelial-mesenchymal transition (EMT) and stem cell pathways are frequently up-regulated (19,25-28), potentially explaining the increased resistance to systemic therapy observed in these tumors (29). Further complicating the search for targets, considerable heterogeneity has been seen to exist across the subtypes of MpBC. For example, the spindle subtype of MpBC were claudin-low and enriched with mesenchymal stem-like characteristics, while other MpBC subtypes were more diverse (30). However, thus far, a gene- or pathway-based analysis of the subtypes of MpBC has not yet been reported, potentially due to limitations in sample number from this rare tumor type.
To better understand the heterogeneity in the biology, responses to NAT, and survival outcomes, we performed a prospective study of the patients and the genomic and transcriptomic profiles of the tumors from the ARTEMIS trial (NCT02276443).
MATERIALS AND METHODS
METHODS - PATIENTS
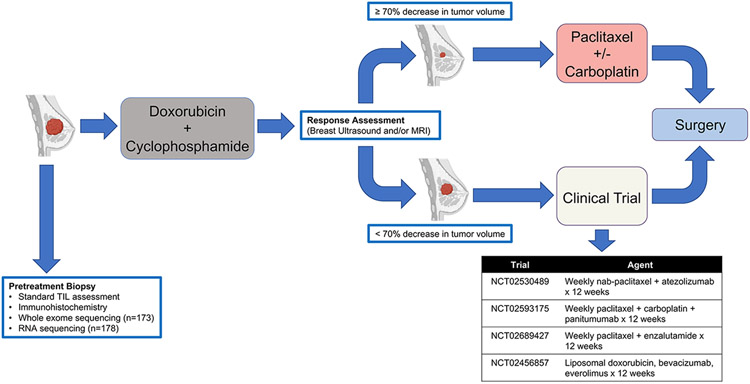
Written informed consent was obtained for all patients included in this study. All patients were enrolled in our prospective, IRB-approved clinical trial, “A Robust TNBC Evaluation FraMework to Improve Survival” (ARTEMIS, NCT02276443), which was conducted in accordance with the ethical standards of the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Treatment-naïve patients with localized TNBC (stage I-III) underwent a biopsy and breast ultrasound prior to starting a planned four cycles of doxorubicin-cyclophosphamide (AC) chemotherapy regimen (Fig 1). Patients deemed to have chemotherapy-sensitive disease after four cycles of AC (≥70% volumetric reduction by breast ultrasound) were recommended to receive standard paclitaxel-based chemotherapy as the second phase of NAT. Patients with chemotherapy-resistant TNBC (disease progression during AC or <70% volumetric reduction by breast ultrasound after four cycles of AC) were offered therapy on molecularly targeted “bucket” clinical trials using targeted therapy or immunotherapy in combination with chemotherapy based on the specific molecular characteristics of their tumor as the second phase of NAT (Fig 1). Upon completion of NAT, patients underwent surgical resection and residual cancer burden (RCB) status (31) was determined by dedicated breast pathologists. Patients who experienced disease progression precluding curative surgical resection while receiving NAT were classified as having RCB-III disease.
FIGURE 1. ARTEMIS (NCT02276443) Trial Schema.
Patients with stage I-III TNBC underwent a pre-treatment core needle biopsy prior to initiating standard of care NAT with doxorubicin and cyclophosphamide (AC). Clinical response by breast imaging (ultrasound and/or MRI) was performed after AC to determine if patients should receive standard taxane-based NAT or be offered enrollment on a clinical trial as part of therapeutic escalation.
METHODS - MATERIALS
TNBC was defined as breast cancer that is estrogen receptor (ER)-negative (<1%) or low positive (1 to <10%), progesterone receptor (PgR)-negative (<1%) or low positive (1 to <10%), and human epidermal growth factor receptor 2 (HER2)-negative according to HER2 testing guidelines from the American Society of Clinical Oncology/College of American Pathologists (32). Clinical anatomic stage was determined at the time of diagnosis using the American Joint Committee on Cancer Staging Manual (7th edition). The diagnosis of MpBC was made according to World Health Organization criteria using histologic evaluation of hematoxylin and eosin (H&E) stained slides (33). Tumors that exhibited metaplastic features in either the pre-treatment core needle biopsy or post-NAT surgical specimens were designated as MpBC and classified into squamous, spindle, and mesenchymal/matrix-producing (referred to as matrix-producing hereafter) subtypes. Stromal tumor-infiltrating lymphocytes (sTIL) were quantified according to guidelines set by the International TILs Working Group (34).
Tissue Processing and Immunohistochemistry
Pre-treatment core needle biopsies of tumors were immediately placed in RNAlater (1-2 cores) or embedded in paraffin (1-2 cores). Slides from paraffin-embedded cores underwent IHC staining for Ki-67 (if not recorded in pathology reports from the referring center), androgen receptor (AR), vimentin, PTEN, and PD-L1. IHC staining for Ki-67, androgen receptor (AR), vimentin, and PTEN, were performed on unstained 4-μm thick tissue sections that had been prepared from a representative paraffin block of tumor, using the polymeric biotin-free horseradish peroxidase method on the Leica Microsystems Bond III autostainer (Leica Microsystems, Buffalo Grove, IL, USA). Slides were incubated at 60°C for 25 minutes. Heat-induced epitope retrieval was performed with TRIS-EDTA buffer for 20 minutes at 100°C (for Ki-67, PTEN), citrate buffer for 25 minutes at 100°C (for AR), or citrate buffer for 5 minutes at 100°C (for vimentin). Slides were incubated with mouse monoclonal antibodies to Ki-67 (clone MIB-1, Dako; 1:100), AR (clone AR441, Dako, Carpinteria, CA, USA; 1:30), vimentin (clone V9, 1:900, Dako), or PTEN (clone 6H2.1, 1:100, Dako North America Inc., Carpinteria, CA). The Refine Polymer Detection kit was used to detect bound antibody, with 3,3-diaminobenzidine serving as the chromogen (Leica Microsystems). IHC staining for programmed death-ligand 1 (PD-L1) was performed using the PD-L1 IHC 22C3 pharmDx kit (Dako) on the Dako AutostainerLink 48 according to the manufacturer’s instructions. Slides were counterstained with Mayer’s hematoxylin. Results were evaluated with known positive and negative tissue controls.
For Ki-67, the percentage of tumor cells with any nuclear staining of any intensity was recorded (low/moderate: ≤35% and high: >35%). For AR, the percentage and intensity of any nuclear staining in tumor cells were recorded (AR-negative: <10% and AR-positive: ≥10%). The threshold for AR-positivity was selected based on data from an earlier study suggesting that ≥ 10% AR-positivity was associated with response to enzalutamide in TNBC (35). For vimentin, positive staining was defined as moderate or strong cytoplasmic staining in ≥50% invasive tumor cells. For PTEN, unequivocal nuclear or cytoplasmic staining of any intensity in any proportion of invasive tumor cells was considered positive, and the stain was considered negative if there was no staining in invasive tumor cells. PD-L1 expression was quantified as the percentage of tumor area occupied by PD-L1-expressing tumor-infiltrating immune cells (PD-L1 negative: <1% and PD-L1-positive: ≥1%).
Whole exome sequencing
Genomic DNA was extracted and quantified (PicoGreen dsDNA quantitation assay; Invitrogen) and quality assessed (Genomic DNA ScreenTape; Agilent). 100-500 ng of genomic DNA was sheared (Covaris E220evolution Focused-ultrasonicator), and fragment size was assessed (Agilent 2200 TapeStation, High Sensitivity DNA Kit). Library preparation was performed using the optimized “with-bead” protocol (KAPA). PCR primers were removed (AMPure PCR Purification kit; Agencourt Bioscience) and samples quantified (KAPA Library Quantification Kit). Equimolar quantities of DNA were pooled (2-6 samples per pool) and captured (NimbleGen SeqCap EZ v3.0 Enrichment Kit). Captured libraries were sequenced on a HiSeq 4000 Illumina Sequencer on a version 3 TruSeq paired end flowcell for 2 x 100 paired end reads. BCL files were demultiplexed using CASAVA 1.8.2 with no mismatches.
We called variants using the BETSY system using a consensus-based pipeline previously described (36,37). In short, reads were aligned to hg19 using BWA, pre-processed using the GATK Best Practices workflow, and variants called with six callers, accepting ones found by at least 2 callers not seen in high discrepancy regions (38), using custom software. We annotated variants with Annovar and found copy number alterations using FACETS.
Whole transcriptomic sequencing
RNA was extracted (NORGEN Total RNA Purification Kit; Cat. 37500), treated with DNase I and purified (AMPure XP beads; Beckman Coulter Life Sciences). cDNA was prepared (Ovation RNA-Seq System V2; NuGEN). Up to 200 ng of cDNA was sheared (Covaris E220evolution Focused-ultrasonicator). Library preparation and hybridization was performed (Sciclone G3 NGSx Workstation; PerkinElmer, Inc.) using Agilent SureSelect XT Low Input Reagent Kit with indexes 1-96 and Agilent SureSelect Human All Exon v.4 probes. 500-1000 ng of library were hybridized as single sample reactions and sequenced on the Illumina NovaSeq 6000 platform for 2 x 150 paired end reads using Cycle Sequencing v3 reagents (Illumina). Data was pre-processed using the STAR aligner, HTSeqCount, RSEM, and FastQC.
Bioinformatics methods
The tumor samples were sequenced for gene expression across seven different batches. We noticed, via a principal components analysis (PCA) that two of the batches had a distinct expression profile from the other ones, despite having no discernible difference in biological covariates. Therefore, we believed this difference to be a technical artifact, and to correct for this, we applied the ComBat algorithm (39) and verified its ability to remove the batch effects using PCA.
Using the batch corrected data, we looked for genes that were differentially expressed between the metaplastic and non-metaplastic tumors using an empirical Bayes method (40). We controlled for multiple hypothesis using a false discovery rate (FDR) calculated using the Benjamini and Hochberg method, accepting as significant genes with at least a 2-fold change and less than 5% FDR.
For pathway analysis, we scored pathways using the ssGSEA algorithm implemented in the GSVA package (41). We calculated the scores for the Hallmarks signature set (42) as well as the Canonical pathways from the C2 set from the Molecular Signatures Database (43). We compared the ssGSEA scores of the metaplastic and non-metaplastic samples using a two-tailed unpaired Student’s t-test.
We calculated the Vanderbilt TNBC signatures using the TNBCtype web server. We performed the analysis on the data after removing batch effects with ComBat as described above. One sample had high gene expression of ESR1, despite being an ER-negative tumor by immunohistochemistry. We set the expression value to 0 so that it could be processed by TNBCtype.
To perform hierarchical clustering on gene expression profiles, we compared gene expression using a Euclidean distance with complete linkage agglomeration. We calculated the clusters using Cluster 3.0 .
We downloaded the processed gene expression data and cell type annotations for data set GSE57544 from the GEO database. We filled missing values with zeroes, removed duplicate genes by selecting the highest variance, log2 transformed, and then quantile normalized the data. We then merged this data set with our metaplastic samples by matching gene names, and then removed batch effects using ComBat, as above. We verified the batch effect removal using PCA plots.
To find the differentially expressed genes for each metaplastic subtype, we used the corrected gene expression data, and then compared the expression of genes from samples of each subtype against those from all other subtypes using a Student’s t test. We accepted as significant genes that had at least a 2-fold change in expression and a 5% FDR.
Statistical methods
Patient demographic and clinical characteristics were summarized by their count and frequency across patients with metaplastic and non-metaplastic TNBC (by histology). We compared these frequencies using the nonparametric Fisher’s exact test. We excluded subjects with missing data and did not perform any imputation.
Event-free survival (EFS) was defined as the time from diagnosis to disease progression during NAT to an extent that precludes curative surgical resection, local and/or distant recurrence after curative surgical resection, or death from any cause, whichever occurred first. Metastasis-free survival (MFS) was defined as the time from diagnosis to the development of distant metastatic disease or death from any cause, whichever occurred first. Overall survival (OS) was defined as the time from diagnosis to death from any cause. All time-to-event endpoints were censored at the time of last known follow-up.
The Kaplan-Meier method was used to visualize and estimate survival curves for time-to-event outcomes, including EFS, MFS, and OS. The log-rank test was used to compare survival times between groups. Univariate Cox proportional hazards models were fit to evaluate the effects of histology on the time-to-event outcomes. Restricted mean survival times were also calculated to perform inference on survival times when evidence of non-proportional hazards was observed. To jointly model the incidence of local and distant recurrences, competing risk regression analyses were employed. Differences in binary outcomes across treatment groups were compared with Fisher’s exact test. All p-values were two-sided, and those less than 0.05 were considered statistically significant. We acknowledge the multiple testing burden, and we emphasize that the primary motivation for this study is a descriptive analysis as opposed to hypothesis testing.
DATA AVAILABILITY STATEMENT
The genomic and transcriptomic data supporting this study were generated in the ARTEMIS trial (NCT02276443). A superset of this data is being made publicly available with a manuscript describing the trial and molecular profiling. Before this occurs, the data used for this trial will be made available upon reasonable request from the corresponding authors.
RESULTS
Patient Characteristics and Clinical Outcomes
A total of 211 patients with TNBC were included in this study (Table 1, STable 1). Of these, 18% (39/211) had MpBC. Of the 39 patients with MpBC, 37 were diagnosed as having MpBC based on the pre-treatment tumor biospecimens and the remaining 2 patients were diagnosed as having MpBC based on the post-NAT surgical specimen. All patients received NAT: 148 (70%) received standard AC plus paclitaxel with or without carboplatin chemotherapy, and 63 (30%) received AC followed by therapy on a clinical trial; of the latter patients, 44 (21%) received chemotherapy and targeted therapy, and 19 (9%) received chemotherapy and atezolizumab. Of the 39 patients with MpBC, eight (21%) received less than four cycles of standard AC during the first phase of NAT. Among these eight patients, seven (18%) stopped AC early due to disease progression or lack of clinical response on breast imaging and were enrolled on a clinical trial. The last patient stopped AC early because of infectious complications. Of the 172 patients with non-metaplastic TNBC, eight (5%) received less than four cycles of standard AC during the first phase of NAT. All eight patients discontinued AC due to disease progression or lack of clinical response. Seven of these enrolled on a clinical trial, and the last patient received standard of care chemotherapy with carboplatin and paclitaxel for the second phase of NAT. Of the 31 patients with MpBC who received four cycles of AC, 16 had suboptimal clinical response as assessed by breast ultrasound after four cycles (<70% reduction in tumor volume). Of these 16 patients, 10 were enrolled on clinical trials and the remaining six received standard taxane-based chemotherapy (ineligible for clinical trial enrollment [n=2]; patient preference [n=2]; physician recommendation [n=2]). Of the 164 patients with non-metaplastic TNBC who received four cycles of AC, 161 had a breast ultrasound done after four cycles of AC. Among these 161 patients, 62 had suboptimal clinical response (assessed as above). Of these 62 patients, 35 were enrolled on clinical trials and the remaining 27 received standard taxane-based chemotherapy (ineligible for clinical trial enrollment [n=1]; insurance denial [n=3]; patient preference [n=6]; physician recommendation [n=17]). Three patients (one MpBC and two non-Mp) developed metastatic disease during NAT, which precluded curative surgical resection and one (non-Mp) experienced disease recurrence within one month of surgery. Among the remaining 207 patients, the MpBC and non-Mp patients received adjuvant systemic therapy (p=0.45) or radiation (p=0.26) at similar rates.
Table 1.
Clinicopathological characteristics and neoadjuvant therapy received.
| Characteristic | Metaplastic n=39 |
Non-metaplastic n=172 |
Total n=211 |
p value | |
|---|---|---|---|---|---|
| Median age at diagnosis – years (interquartile range) | 56.2 (45.6-63.5) | 54.9 (45.1-61.1) | 55.1 (45.4-61.6) | 0.47 | |
| Race - n (%) | White | 27 (69.2) | 100 (58.1) | 127 (60.2) | 0.64 |
| Black | 4 (10.3) | 34 (19.8) | 38 (18.0) | ||
| Hispanic | 5 (12.8) | 24 (14.0) | 29 (13.7) | ||
| Asian | 3 (7.7) | 12 (7.0) | 15 (7.1) | ||
| Other | 0 | 2 (1.2) | 2 (1.0) | ||
| Median tumor size – cm (interquartile range) | 3.4 (2.2-5.2) | 2.7 (2.1-3.9) | 2.8 (2.1-4.0) | 0.09 | |
| T category - n (%) | T1 | 7 (18.0) | 27 (15.7) | 34 (16.1) | 0.37 |
| T2 | 22 (56.4) | 118 (68.6) | 140 (66.4) | ||
| T3 | 7 (18.0) | 20 (11.6) | 27 (12.8) | ||
| T4 | 3 (7.7) | 7 (4.1) | 10 (4.7) | ||
| Nodal Status - n (%) | Negative | 33 (84.6) | 90 (52.3) | 123 (58.3) | <0.001 |
| Positive | 6 (15.4) | 82 (47.7) | 88 (41.7) | ||
| TNM Stage - n (%) | I | 5 (12.8) | 15 (8.7) | 20 (9.5) | 0.35 |
| II | 28 (71.8) | 114 (66.3) | 142 (67.3) | ||
| III | 6 (15.4) | 43 (25.0) | 49 (23.2) | ||
| Histologic grade - n (%) | 1 | 1 (2.6) | 0 | 1 (0.5) | 0.04 |
| 2 | 9 (23.1) | 22 (12.8) | 31 (14.7) | ||
| 3 | 29 (74.4) | 150 (87.2) | 179 (84.8) | ||
| Ki-67 - n (%) | Low | 3 (7.7) | 8 (4.7) | 11 (5.2) | 0.03 |
| Moderate | 12 (30.8) | 28 (16.3) | 40 (19.0) | ||
| High | 21 (53.9) | 133 (77.3) | 154 (73.0) | ||
| Missing | 3 (7.7) | 3 (1.7) | 6 (2.8) | ||
| Metaplastic Subtype - n (%) | Matrix producing | 18 (46.2)* | |||
| Spindle | 14 (35.9) | ||||
| Squamous | 5 (12.8) | ||||
| Mixed spindle / matrix producing | 2 (5.1) |
Two of the 18 tumors were diagnosed as MpBC based on the post-NAT surgical specimen.
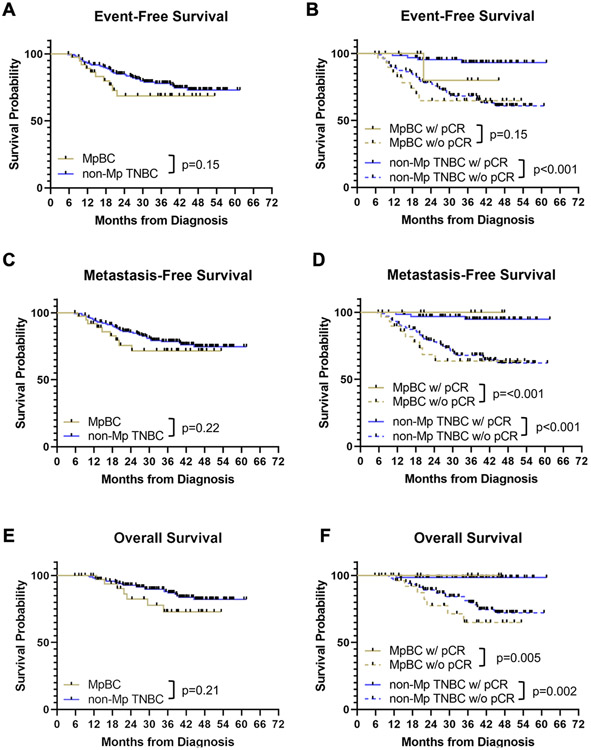
Compared to patients with non-Mp TNBCs, features strongly associated with MpBCs included node negativity (p<0.001), lower histologic grade (p=0.04), lower Ki67 (p=0.03), high vimentin (p<0.001), and lower rates of PD-L1 expression (p=0.04). There was a trend towards lower TIL infiltration (p=0.08) (Table 1), which has previously been associated with poor prognosis in hormone receptor-negative breast cancers (44). Patients with MpBCs were less likely to experience a pCR (23% vs 40%; p=0.07), had shorter EFS (restricted mean EFS: 29.4 months vs 32.2 months, p=0.15), MFS (restricted mean MFS: 30.3 months vs 32.4 months, p=0.22), and OS (restricted mean OS: 32.6 months vs 34.3 months, p=0.21) (Fig 2), although these differences did not achieve statistical significance likely due to small sample size. Among the patients with pCR, metaplastic histology had no impact on the favorable MFS and OS (Fig 2). One patient with MpBC with pCR who underwent breast-conserving surgery followed by radiation developed local disease recurrence that was treated with additional chemotherapy (gemcitabine and carboplatin) without response. She underwent a mastectomy, which showed residual disease, and has remained disease-free for 29 months. Of note, among the 38 patients with metaplastic breast cancer who underwent surgical resection with curative intent, 19 underwent a segmental mastectomy and the remaining 19 underwent a mastectomy. We estimated the risk of local recurrence by fitting a competing risk regression model and found no difference in the rates of local recurrence at 18 months (8% [segmental mastectomy] vs 6% [mastectomy], p=0.98).
FIGURE 2. Survival outcomes in patients with metaplastic breast cancer (MpBC) and non-metaplastic (non-Mp) TNBC receiving neoadjuvant therapy (NAT).
(A) Kaplan-Meier plot of event-free survival for patients with MpBC (yellow) and non-Mp TNBC (blue); (B) Kaplan-Meier plot of event-free survival for patients with MpBC experiencing a pathological complete response (pCR) (solid yellow), patients with MpBC with residual disease (dashed yellow), patients with non-Mp TNBC experiencing a pCR (solid blue), and patients with non-Mp TNBC with residual disease (dashed blue); (C) Kaplan-Meier plot of metastasis-free survival for patients with MpBC (yellow) and non-Mp TNBC (blue); (D) Kaplan-Meier plot of metastasis-free survival for patients with MpBC experiencing a pCR (solid yellow), patients with MpBC with residual disease (dashed yellow), patients with non-Mp TNBC experiencing a pCR (solid blue), and patients with non-Mp TNBC with residual disease (dashed blue); (E) Kaplan-Meier plot of overall survival for patients with MpBC (yellow) and non-Mp TNBC (blue); (F) Kaplan-Meier plot of overall survival for patients with MpBC experiencing a pCR (solid yellow), patients with MpBC with residual disease (dashed yellow), patients with non-Mp TNBC experiencing a pCR (solid blue), and patients with non-Mp TNBC with residual disease (dashed blue).
MpBCs are histologically heterogeneous and categorized into spindle, squamous, or matrix-producing subtypes (1) based on WHO classifications, although some tumors can have ambiguous features in histology. We found that there was a trend towards higher rates of pCR rates in patients with MpBC of squamous histology (3/5, 60%) compared to those with spindle (3/14, 21%), matrix-producing (3/16, 19%), or mixed (0/4, 0%, including the 2 tumors diagnosed from the surgical specimen) histologies (p=0.07, squamous vs all others), although the numbers are not large enough to achieve statistical significance.
Metaplastic TNBC is enriched in PI3K pathway mutations
WES was completed in pre-treatment tumor biospecimens from 22 patients with MpBC and 151 patients with non-Mp TNBC. TP53 was the most frequently mutated gene in both MpBCs (59%) and non-MpTNBCs (51%). However, PIK3CA was more frequently mutated in MpBCs (23% vs 9%, p=0.07) (SFig 1A). Mutations in other members of the PI3K pathway were found in 41% of MpBCs compared to 18% of non-Mp TNBCs (SFig 1B, p=0.02). Mutations frequently occurred in established hotspots, with the PIK3CA H1047R mutation, previously reported to be the most common somatic PIK3CA mutation (45), seen in eight of thirteen (62%) non-Mp TNBCs with PIK3CA mutations, and two of five (40%) MpBCs. The next two most common mutations, E545K, was present in one MpBC and one non-Mp TNBC, and E542R was seen in two non-MpTNBCs. Other previously reported somatic mutations were seen in one non-MpTNBCs each: AKT1 E17K (46) and MTOR E1799K (47). Thus, there is an enrichment of pathogenic alterations among the detected PI3K pathway mutations. In this cohort of MpBCs, there were no mutations in CTNNB1 or enrichment in Wnt pathway genes (data not shown). EGFR amplifications were seen in 17% of MpBCs and in one tumor, a frameshift mutation was seen in the C2 loop of PTEN in a codon previously deemed to be necessary for proper PTEN localization (48). Additionally, we saw no differences in the domains frequently mutated in either TP53 or PIK3CA (SFig 1C). Collectively, these data show that both MpBCs and non-Mp TNBCs have similar genetic alterations, although mutations in the PI3K pathway are more frequently observed in MpBCs. Neither the presence of TP53 or PIK3CA mutations was associated with pathological response to NAT (data not shown).
Metaplastic and non-metaplastic TNBCs express distinct genes
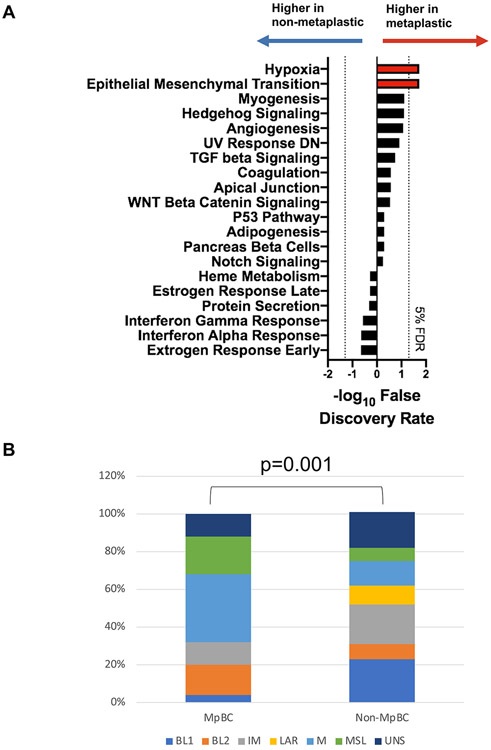
We next examined differences in gene expression using 178 tumors (including 25 MpBCs) with high-quality samples by RNA-Seq. 115 genes were differentially expressed (≥ 2 fold change, FDR <5%) (SFig 2A-C) in MpBCs vs non-MpTNBCs and many of the differentially expressed genes were associated with cell-type identity. The non-Mp TNBCs were enriched with markers for epithelial differentiation, such as EPCAM, CDH1, ESRP1, as well as classical markers for luminal epithelial cells, including cytokeratins, KRT8, KRT18, and KRT19. The MpBCs had significantly higher expression of SNAI2, a potent inducer of epithelial to mesenchymal transition (EMT) (49) as well as a driver of the mammary stem cell state (50). Further, many components of the extracellular matrix, including COL5A3, COL11A2, COL2A1, and FBN2, were enriched in MpBC. To determine whether the differences in transcriptomic profiles observed between MpBCs and non-Mp TNBCs can be grouped into specific pathways, we used ssGSEA to predict the activation of pathways in each individual sample, and then compared the scores seen in MpBC and non-Mp samples using a Student’s t-test. At a 5% FDR cutoff, two Hallmarks pathways were significant: hypoxia and EMT (Fig 3A). Further, we tested whether the MpBCs were associated with the mesenchymal TNBC subtype using Vanderbilt classification (51) and found enrichment for mesenchymal (M) and mesenchymal stem-like (MSL) subtypes (Fig 3B). These data suggest that MpBCs exhibit a strong mesenchymal gene expression signature that indicates a divergence from an epithelial phenotype. These findings are consistent with other studies showing enrichment of EMT in MpBC (25,29,52). Notably, enrichment in EMT has correlated with poor survival and lack of pCR to neoadjuvant therapy (NAT) in breast cancer (29).
FIGURE 3. Metaplastic breast cancers (MpBCs) and non-metaplastic (non-Mp) TNBCs have distinct gene expression profiles.
(A) Bar plot showing the 10 Hallmarks pathways predicted to be most significantly activated in MpBC or non-MP TNBC. The x-axis shows the −log10 of the false discovery rate (FDR), where the values for the pathways associated with non-Mp TNBCs are inversed so that the most significant ones go to the left. The pathways most significantly associated with MpBCs go to the right and are shown in red. The dotted lines indicate a 5% FDR. (B) Stacked bar graph showing the distribution of TNBC subtypes (legend) within MpBCs (left) and non-Mp TNBCs (right).
Subtypes of MpBC have divergent, yet overlapping, gene expression profiles
To identify distinctions in the gene expression profiles across the MpBC subtypes, we categorized our cohort of 25 MpBCs with RNA-Seq profiles into squamous, spindle, or matrix-producing subtypes based upon tumor histology. Three of these tumors did not fit these categories cleanly and were classified as Other. Then, to increase the number of samples, we integrated publicly available data from another MpBC study (30). This increased the number of samples with both transcriptome profiling and subtype information from 22 to 41 (SFig 3A).
Following batch effect correction (SFig 3B), we performed an unbiased principal component analysis across the samples and found that the squamous and spindle subtypes of MpBC have the most visually distinct gene expression profiles; however, not every tumor clustered cleanly with others of its subtype, reminiscent of the ambiguities seen in the histology. For each of the principal components, we associated the values of the tumors from each subtype against the values of the tumors from the other subtypes using a two-tailed non-parametric Wilcoxon rank-sum test (SFig 3C). The first principal component (20% of variance) distinguished the squamous subtype tumors from the others (0.4% FDR), while the second (18% of variance) distinguished the spindle subtype (0.3% FDR) and matrix-producing subtypes (2% FDR) (SFig 3D). No other principal components were significantly associated with any of the subtypes. We interpreted this finding to mean that each of the subtypes have identifiable gene expression profiles, although there are some similarities. There was one matrix-producing tumor that clustered closely among the spindle tumors, and upon further examination of the histology, it was confirmed to be sarcomatoid with mesenchymal differentiation. Although this tumor did not resemble a typical spindle cell carcinoma histologically, it nevertheless appeared to share a similar gene expression profile as the spindle-type tumors.
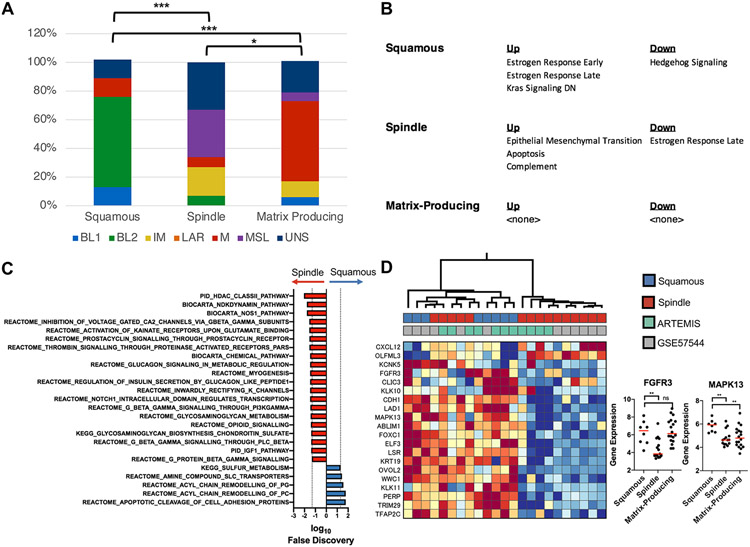
Noting expression differences across the subtypes, we next tried to understand the nature of these differences. Using the Vanderbilt TNBC signatures (53), we found that the squamous subtype was enriched for BL2 tumors, whereas the spindle and matrix-producing subtypes were enriched for the MSL and M tumors, respectively (Fig 4A). We also evaluated genes that were specifically expressed (either higher or lower) in each of the three subtypes (SFig 4). Each subtype expressed a distinctive set of genes.
FIGURE 4. Histologic subtypes of metaplastic breast cancer (MpBC) have heterogeneous gene expression profiles.
(A) Stacked bar graphs showing the distribution of TNBC subtypes within each histologic subtype of MpBC. The distribution of TNBC subtypes across the columns were compared, and the statistical significance is indicated according to: * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001. (B) The Hallmarks pathways that are predicted to be up- or down- regulated (false discovery rate [FDR] < 5%) in each of the histologic subtypes of MpBC are shown. (C) The Canonical pathways that are enriched in the spindle (red bars, towards the left) and squamous (blue bars, towards the right) at a 5% FDR cutoff are shown. (D) Heatmap showing the expression levels of the significant genes (rows) from the Hallmarks Estrogen Response pathways across the MpBCs (columns). The colors indicate the normalized expression values of the genes, where warm and cold colors indicate higher and lower expression, respectively. The first row of boxes under the dendrogram show the subtype of the metaplastic tumors, and the second row indicates the data set. Beeswarm plots (right panel) show the expression level of FGFR3 and MAPK13 across each of the histologic subtypes of MpBCs (x-axis). The statistical significance is denoted using the same notation as in Fig 4A.
Pathway analyses using the Hallmark and Canonical gene sets revealed that the spindle subtype was increased in the EMT gene expression signature (Fig 4B), consistent with the histology. The Canonical gene sets predicted that these tumors had an increased HDAC activity (Fig 4C) suggesting that epigenetic reprogramming maintained through HDACs may influence tumor biology, potentially indicating subtype sensitivity to HDAC inhibition.
Squamous MpBCs were enriched in Hallmark estrogen receptor (ER) response signaling compared to spindle MpBCs (Fig 4B) and, given the lack of ER expression by IHC, we further explored the expression patterns of individual genes within these ER response signatures (Fig 4D). The result revealed a set of genes that were up-regulated in all squamous, as well as a subset of the spindle type MpBCs. Three of these genes were previously found to be linked to an RTK-activated MAPK signaling pathway: FGFR3, MAPK13 (a p38 MAPK), and ELF3 (54), suggesting that RTK/MAPK signaling may be an important driver of squamous MpBCs.
Some non-metaplastic tumors exhibit a metaplastic gene expression profile and response
Given the heterogeneity of gene expression profiles observed across different histologic subtypes of MpBC, we next investigated whether any non-Mp TNBC tumors exhibit the metaplastic signature, or share components of the metaplastic phenotype. To do this, we evaluated the expression data for the entire cohort using unbiased clustering based on the metaplastic signature, the genes whose expression distinguishes MpBCs from non-Mp TNBCs (SFig 2C). This revealed three major clusters (SFig 5A). Clusters 2 and 3 were enriched in MpBCs, while cluster 1 was comprised mainly of non-Mp TNBCs (SFig 5B). We then compared the subtypes of the MpBCs across clusters 2 and 3, which showed squamous MpBCs were enriched in cluster 2 and spindle MpBCs in cluster 3 (SFig 5B).
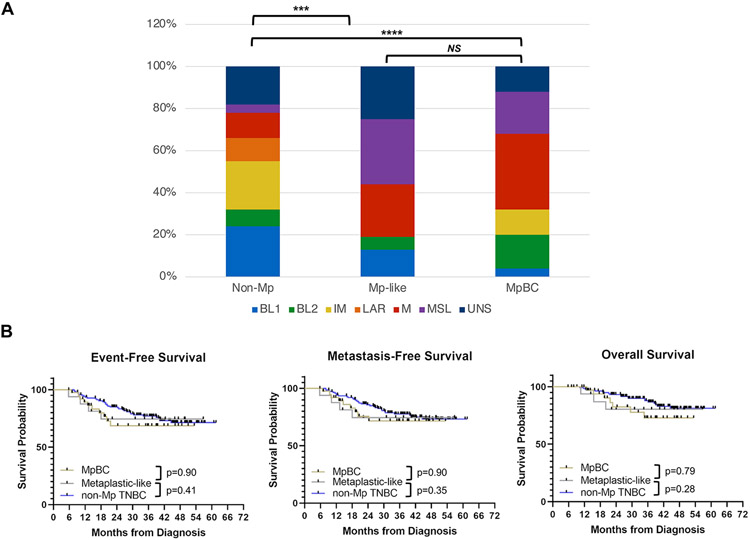
Notably, in cluster 2, several non-Mp TNBCs had gene expression profiles that resembled MpBCs, though they displayed no metaplastic features on histology, thus deemed metaplastic-like based on gene signature. Both the metaplastic-like TNBCs and MpBCs were enriched in the Vanderbilt M and MSL subtypes (Fig 5A). Using Hallmark gene sets, hypoxia and EMT gene signatures were upregulated in MpBCs and metaplastic-like TNBCs (SFig 5C). Interestingly, the expression of several gene signatures such as angiogenesis and TGF-β signaling was significantly higher in metaplastic-like TNBCs but not MpBCs. The pCR rate observed in patients with metaplastic-like TNBC was 31% (5/16) compared with 39% (54/137) in patients with non-Mp TNBC without metaplastic-like gene expression profiles and 20% (5/25) in patients with MpBC. Despite the apparent difference in pCR rates observed between patients with metaplastic-like TNBC and MpBC, patients with metaplastic-like TNBCs and MpBCs appeared to have similar survival outcomes (Fig 5B), suggesting that gene expression profiling may identify a subset of aggressive non-Mp TNBCs that are phenotypically similar to MpBCs.
FIGURE 5. Metaplastic-like TNBCs are a subset of non-metaplastic (non-Mp) TNBCs with gene expression profiles resembling metaplastic breast cancers (MpBC) and are associated with poor long-term outcomes.
(A) The distribution of TNBC subtypes for non-Mp TNBC, metaplastic-like TNBC, and bona-fide MpBCs are shown in the columns. The statistical significance is annotated as in Fig 4A. (B) Kaplan-Meier plots of event-free, metastasis-free, and overall survival for patients with MpBC (yellow line), non-Mp TNBC (blue line), and metaplastic-like TNBC (gray line).
DISCUSSION
This analysis represents the largest prospective evaluation of NAT in patients with triple-negative MpBC. We report a pCR rate of 23%, which is higher compared to historical data from retrospective studies (~10%) (5,7,8,55-58). Notably, through frequent monitoring of disease response to NAT by imaging, we were able to identify disease progression or lack of response resulting in early discontinuation of AC in 18% (7/39) of patients with MpBC. A further 26% (10/39) of patients with MpBC were enrolled on clinical trials of chemotherapy plus novel therapeutic agents due to suboptimal clinical response after completing 4 cycles of AC, for a total of 44% (17/39).
Several reasons could potentially explain the higher pCR rate observed in our study. First, the ARTEMIS trial only enrolled patients with TNBC. Thus, our study was restricted to patients with triple-negative MpBC. Although >90% of MpBCs are of the triple-negative phenotype (1), earlier retrospective studies of NAT in MpBC included small numbers of patients with ER-positive MpBC (4,14,55). Thus, the lower rates of pCR observed in these earlier retrospective studies may be due, in part, to the heterogeneity of the patient population being studied. Second, patients in our study uniformly received anthracycline-based NAT while treatment regimens received by patients included in those earlier studies were more heterogeneous, and possibly less effective (5,7).
The molecular profiles of MpBC elucidated in our study propose novel treatment strategies for MpBC. First, consistent with earlier studies (12-16), we observed an enrichment of PI3K pathway alterations which suggests an opportunity for augmenting responses by targeting this pathway in patients with MpBC. To date, the success of mTOR-based systemic therapy regimens such as DAT/DAE have not been replicated in the neoadjuvant setting and chemotherapy remains the only FDA-approved treatment in this setting (17). In this cohort of MpBCs, there were no mutations in CTNNB1 or enrichment in Wnt pathway genes consistent with some prior studies (13,19), but contradicting others (18,20). Second, the lack of PD-L1 expression and trend towards lower TIL infiltration in previously untreated MpBC compared with non-Mp TNBC suggests that immune checkpoint blockade may only be effective in a small subset of patients with MpBC. There have been two case reports of patients with advanced, MpBC experiencing dramatic responses to the combination of chemotherapy and PD-L1 blockade (59,60). However, both these patients had tumors demonstrating significant PD-L1 expression, which was uncommon in our cohort of patients. Finally, we observed subtype-specific differences in the molecular profile of MpBC, with squamous-type and spindle-type MpBCs enriched for RTK/MAPK and HDAC signaling, respectively. This finding underscores the fact that MpBC, while rare, is highly heretogeneous. Thus, there is an urgent need for personalized therapeutic strategies in order to maximize cure rates in this disease.
From an unbiased transcriptomic analysis, we identified a cohort of patients with non-Mp TNBC with gene expression profiles resembling that of patients with MpBC (metaplastic-like). Although metaplastic-like TNBCs did not have metaplastic features on histology, the clinical outcomes of patients with metaplastic-like TNBCs more closely resembled those of patients with MpBC. These data suggest that the poor outcomes associated with metaplastic histology may be a consequence of an altered biological process reflected in a gene expression signature that can serve as a prognostic biomarker in patients with non-Mp TNBC.
Our study has several limitations. First, although our cohort is large for this rare breast cancer type, the reported 95% confidence intervals for pCR is wide, suggesting that further validation in larger, prospective studies is required. In addition, the modest sample size precluded formal hypothesis testing in this study and clinically significant differences in outcomes did not cross the threshold for statistical significance. Second, because NAT is considered standard of care for patients with TNBC, pre-treatment molecular profiling in our study had to be performed on core needle biopsies. This limited our ability to obtain a more representative sample of the entire tumor. Notably, in two patients with MpBC, there were no metaplastic features noted on the pre-treatment core needle biopsy and the diagnosis of MpBC was made in retrospect on the post-NAT surgical specimen. In our analysis, this sampling bias might be reflected in the deviations between the histological subtypes and those seen in the gene expression profiling, as the histological classification was performed on paraffin-embedded tissue and the transcriptomics on the fresh tissue from the biopsy. Third, all seven patients (18%) with metaplastic TNBC who stopped AC early due to progression in our study were subsequently enrolled on a targeted therapy clinical trial for the second phase of NAT. Thus, based on our data alone, it is not possible to determine if early progression on AC in metaplastic TNBC implies universal resistance to chemotherapy and ineffectiveness of standard taxane-based therapy. Finally, because patients were offered enrollment on clinical trials of experimental therapies only if there was demonstrable evidence of resistance to initial AC chemotherapy, direct comparisons of pCR rates between patients receiving experimental therapies and those receiving standard taxane-based chemotherapy as the second phase of NAT could not be made since such an analysis would be biased by the enrichment of patients with chemotherapy-resistant disease among those receiving experimental therapies. Although beyond the scope of this study, there is a critical need for randomized trials for patients with suboptimal response to initial standard NAT that reveal the effectiveness of novel experimental therapies compared to standard NAT.
In summary, the deep molecular characterization of MpBC tumors in this study has revealed the molecular heterogeneity in this disease and proposed new avenues for the development of treatments. While these novel therapeutic strategies are investigated, our data suggest that patients with MpBC should be considered for sequential anthracycline- and taxane-based NAT. Further, given the heterogeneity in the biology and response to NAT, biomarkers are needed to ensure appropriate patient selection, and close monitoring by serial breast imaging can assess for early disease progression during therapy.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Treatment for stage I-III metaplastic breast cancer (MpBC) is controversial as neoadjuvant therapy (NAT) has low reported rates of pathological complete response (pCR). To identify more effective strategies, we have performed a large prospective study of clinical outcomes and molecular profiles of MpBC. Our observed pCR rate of 23% is higher than previously reported in retrospective studies, suggesting that NAT may be a viable treatment strategy in this setting. Further, we observe that histologic subtypes of MpBC have divergent transcriptomic profiles, underscoring the need to consider subtype-specific strategies. Finally, we identify a subset of non-metaplastic TNBCs with metaplastic-like gene expression signatures and poor clinical outcomes, suggesting that tumor transcriptomics can refine the definition of the metaplastic phenotype and improve clinical prognostication in TNBC.
ACKNOWLEDGMENTS
We are grateful to the patients who provided tumor biopsies for these studies. This work was funded by a Conquer Cancer Career Development Award supported by Fleur Fairman (to C.Y.), the 2018 Gianni Bonadonna Breast Cancer Research Fellowship (Conquer Cancer Foundation to C.Y.), the Winterhof fund (to S.L.M.), and generous philanthropic contributions to the Moon Shots Program of The University of Texas MD Anderson Cancer Center. Pre-sequencing processing work was completed by the Cancer Genomics Laboratory Moon Shots Platform and the Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy at The University of Texas MD Anderson Cancer Center. Sequencing, data generation, and biostatistics analysis were supported in part by the NIH/NCI Cancer Center Support Grant (award number P30 CA016672) and used the Biostatistics Resource Group. Funding and drug support were provided (in part) by Amgen Inc.; Astellas Pharma Global Development, Inc.; Genentech, USA Inc.; Novartis AG; Pfizer Inc. C.Y. was additionally supported by the Allison and Brian Grove Endowed Fellowship for Breast Medical Oncology and the Susan Papizan Dolan Fellowship in Breast Oncology. J.T.C. was supported by the Cancer Prevention and Research Institute of Texas (RP170668, RP160710) and the National Institutes of Health’s National Cancer Institute (U54CA209978). Any opinions, findings, and conclusions expressed in this material are those of the author(s) and do not necessarily reflect those of the sponsors.
Footnotes
Conflict of interest statement:
C.Y. has received research support (to the institution) from Amgen, Merck, Genentech, and GSK. D.T. has received research support (to the institution) and serves on the steering committee and advisory board for research strategies for Novartis. J.K.L. has received research support from Novartis, Medivation/Pfizer, Genentech, GSK, EMD-Serono, Astra-Zeneca, Medimmune, and Zenith; participated in a Speaker’s Bureau for MedLearning, Physician’s Education Resource, Prime Oncology, Medscape, Clinical Care Options, and Medpage; received Honoraria from UpToDate; is a member on advisory committees and/or boards for Astra-Zeneca, Ayala, and Pfizer (all uncompensated); and serves on review panels for NCCN, ASCO, NIH, PDQ, and SITC. N.T.U. has received research support (to the institution) from Amgen and Pfizer. B.L. has received research support from Takeda Oncology, Genentech, Merck, Puma Biotechnology, Celcuity, Amgen, and Novartis. E.A.M. has received compensation for participation in the scientific advisory board for Exact Sciences (previously Genomic Health), Merck, and Roche; and participated in steering committees (uncompensated) for Bristol Myers Squibb, Eli Lilly, and Roche. S.L.M. is an employee of Eli Lilly. All other authors have no relevant conflict of interest disclosures.
REFERENCES
- 1.Tse GM, Tan PH, Putti TC, Lui PC, Chaiwun B, Law BK. Metaplastic carcinoma of the breast: a clinicopathological review. J Clin Pathol 2006;59(10):1079–83 doi 10.1136/jcp.2005.030536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abouharb S, Moulder S. Metaplastic breast cancer: clinical overview and molecular aberrations for potential targeted therapy. Curr Oncol Rep 2015;17(3):431 doi 10.1007/s11912-014-0431-z. [DOI] [PubMed] [Google Scholar]
- 3.National Comprehensive Cancer Network. October/13/2020. Breast Cancer (Version 6.2020). <https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf>. 10/13/2020. [Google Scholar]
- 4.Bae SY, Lee SK, Koo MY, Hur SM, Choi MY, Cho DH, et al. The prognoses of metaplastic breast cancer patients compared to those of triple-negative breast cancer patients. Breast Cancer Res Treat 2011;126(2):471–8 doi 10.1007/s10549-011-1359-8. [DOI] [PubMed] [Google Scholar]
- 5.Hennessy BT, Giordano S, Broglio K, Duan Z, Trent J, Buchholz TA, et al. Biphasic metaplastic sarcomatoid carcinoma of the breast. Ann Oncol 2006;17(4):605–13 doi 10.1093/annonc/mdl006. [DOI] [PubMed] [Google Scholar]
- 6.Sikov WM, Berry DA, Perou CM, Singh B, Cirrincione CT, Tolaney SM, et al. Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple-negative breast cancer: CALGB 40603 (Alliance). J Clin Oncol 2015;33(1):13–21 doi 10.1200/JCO.2014.57.0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Hilli Z, Choong G, Keeney MG, Visscher DW, Ingle JN, Goetz MP, et al. Metaplastic breast cancer has a poor response to neoadjuvant systemic therapy. Breast Cancer Res Treat 2019;176(3):709–16 doi 10.1007/s10549-019-05264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corso G, Frassoni S, Girardi A, De Camilli E, Montagna E, Intra M, et al. Metaplastic breast cancer: Prognostic and therapeutic considerations. J Surg Oncol 2021;123(1):61–70 doi 10.1002/jso.26248. [DOI] [PubMed] [Google Scholar]
- 9.He X, Ji J, Dong R, Liu H, Dai X, Wang C, et al. Prognosis in different subtypes of metaplastic breast cancer: a population-based analysis. Breast Cancer Res Treat 2019;173(2):329–41 doi 10.1007/s10549-018-5005-6. [DOI] [PubMed] [Google Scholar]
- 10.Melton LJ 3rd. Selection bias in the referral of patients and the natural history of surgical conditions. Mayo Clin Proc 1985;60(12):880–5 doi 10.1016/s0025-6196(12)64794-6. [DOI] [PubMed] [Google Scholar]
- 11.Motheral BR, Fairman KA. The use of claims databases for outcomes research: rationale, challenges, and strategies. Clin Ther 1997;19(2):346–66 doi 10.1016/s0149-2918(97)80122-1. [DOI] [PubMed] [Google Scholar]
- 12.Ross JS, Badve S, Wang K, Sheehan CE, Boguniewicz AB, Otto GA, et al. Genomic profiling of advanced-stage, metaplastic breast carcinoma by next-generation sequencing reveals frequent, targetable genomic abnormalities and potential new treatment options. Arch Pathol Lab Med 2015;139(5):642–9 doi 10.5858/arpa.2014-0200-OA. [DOI] [PubMed] [Google Scholar]
- 13.Ng CKY, Piscuoglio S, Geyer FC, Burke KA, Pareja F, Eberle CA, et al. The Landscape of Somatic Genetic Alterations in Metaplastic Breast Carcinomas. Clin Cancer Res 2017;23(14):3859–70 doi 10.1158/1078-0432.CCR-16-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joneja U, Vranic S, Swensen J, Feldman R, Chen W, Kimbrough J, et al. Comprehensive profiling of metaplastic breast carcinomas reveals frequent overexpression of programmed death-ligand 1. J Clin Pathol 2017;70(3):255–9 doi 10.1136/jclinpath-2016-203874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edenfield J, Schammel C, Collins J, Schammel D, Edenfield WJ. Metaplastic Breast Cancer: Molecular Typing and Identification of Potential Targeted Therapies at a Single Institution. Clin Breast Cancer 2017;17(1):e1–e10 doi 10.1016/j.clbc.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Zhai J, Giannini G, Ewalt MD, Zhang EY, Invernizzi M, Niland J, et al. Molecular characterization of metaplastic breast carcinoma via next-generation sequencing. Hum Pathol 2019;86:85–92 doi 10.1016/j.humpath.2018.11.023. [DOI] [PubMed] [Google Scholar]
- 17.Basho RK, Yam C, Gilcrease M, Murthy RK, Helgason T, Karp DD, et al. Comparative Effectiveness of an mTOR-Based Systemic Therapy Regimen in Advanced, Metaplastic and Nonmetaplastic Triple-Negative Breast Cancer. Oncologist 2018;23(11):1300–9 doi 10.1634/theoncologist.2017-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes MJ, Thomas D, Emmons A, Giordano TJ, Kleer CG. Genetic changes of Wnt pathway genes are common events in metaplastic carcinomas of the breast. Clin Cancer Res 2008;14(13):4038–44 doi 10.1158/1078-0432.CCR-07-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 2009;69(10):4116–24 doi 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geyer FC, Lacroix-Triki M, Savage K, Arnedos M, Lambros MB, MacKay A, et al. beta-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol 2011;24(2):209–31 doi 10.1038/modpathol.2010.205. [DOI] [PubMed] [Google Scholar]
- 21.Reis-Filho JS, Pinheiro C, Lambros MB, Milanezi F, Carvalho S, Savage K, et al. EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. J Pathol 2006;209(4):445–53 doi 10.1002/path.2004. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert JA, Goetz MP, Reynolds CA, Ingle JN, Giordano KF, Suman VJ, et al. Molecular analysis of metaplastic breast carcinoma: high EGFR copy number via aneusomy. Mol Cancer Ther 2008;7(4):944–51 doi 10.1158/1535-7163.MCT-07-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horlings HM, Weigelt B, Anderson EM, Lambros MB, Mackay A, Natrajan R, et al. Genomic profiling of histological special types of breast cancer. Breast Cancer Res Treat 2013;142(2):257–69 doi 10.1007/s10549-013-2740-6. [DOI] [PubMed] [Google Scholar]
- 24.McCart Reed AE, Kalaw E, Nones K, Bettington M, Lim M, Bennett J, et al. Phenotypic and molecular dissection of metaplastic breast cancer and the prognostic implications. J Pathol 2019;247(2):214–27 doi 10.1002/path.5184. [DOI] [PubMed] [Google Scholar]
- 25.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A 2010;107(35):15449–54 doi 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Beca FF, Caetano P, Gerhard R, Alvarenga CA, Gomes M, Paredes J, et al. Cancer stem cells markers CD44, CD24 and ALDH1 in breast cancer special histological types. J Clin Pathol 2013;66(3):187–91 doi 10.1136/jclinpath-2012-201169. [DOI] [PubMed] [Google Scholar]
- 27.McQuerry JA, Jenkins DF, Yost SE, Zhang Y, Schmolze D, Johnson WE, et al. Pathway activity profiling of growth factor receptor network and stemness pathways differentiates metaplastic breast cancer histological subtypes. BMC Cancer 2019;19(1):881 doi 10.1186/s12885-019-6052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Djomehri SI, Gonzalez ME, da Veiga Leprevost F, Tekula SR, Chang HY, White MJ, et al. Quantitative proteomic landscape of metaplastic breast carcinoma pathological subtypes and their relationship to triple-negative tumors. Nat Commun 2020;11(1):1723 doi 10.1038/s41467-020-15283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oon ML, Thike AA, Tan SY, Tan PH. Cancer stem cell and epithelial-mesenchymal transition markers predict worse outcome in metaplastic carcinoma of the breast. Breast Cancer Res Treat 2015;150(1):31–41 doi 10.1007/s10549-015-3299-1. [DOI] [PubMed] [Google Scholar]
- 30.Weigelt B, Ng CK, Shen R, Popova T, Schizas M, Natrajan R, et al. Metaplastic breast carcinomas display genomic and transcriptomic heterogeneity [corrected]. Mod Pathol 2015;28(3):340–51 doi 10.1038/modpathol.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Symmans WF, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol 2007;25(28):4414–22 doi 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 32.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013;31(31):3997–4013 doi 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 33.Lakhani SR, International Agency for Research on Cancer., World Health Organization. WHO classification of tumours of the breast. Lyon: International Agency for Research on Cancer; 2012. 240 p. p. [Google Scholar]
- 34.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol 2015;26(2):259–71 doi 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar V, Yu J, Phan V, Tudor IC, Peterson A, Uppal H. Androgen Receptor Immunohistochemistry as a Companion Diagnostic Approach to Predict Clinical Response to Enzalutamide in Triple-Negative Breast Cancer. JCO Precision Oncology 2017(1):1–19 doi 10.1200/PO.17.00075. [DOI] [PubMed] [Google Scholar]
- 36.Chen X, Chang JT. Planning bioinformatics workflows using an expert system. Bioinformatics 2017;33(8):1210–5 doi 10.1093/bioinformatics/btw817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Echeverria GV, Powell E, Seth S, Ge Z, Carugo A, Bristow C, et al. High-resolution clonal mapping of multi-organ metastasis in triple negative breast cancer. Nat Commun 2018;9(1):5079 doi 10.1038/s41467-018-07406-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnell EK, Ronning P, Campbell KM, Krysiak K, Ainscough BJ, Sheta LM, et al. Standard operating procedure for somatic variant refinement of sequencing data with paired tumor and normal samples. Genet Med 2019;21(4):972–81 doi 10.1038/s41436-018-0278-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8(1):118–27 doi 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 40.Efron B, Tibshirani R. Empirical bayes methods and false discovery rates for microarrays. Genet Epidemiol 2002;23(1):70–86 doi 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- 41.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7 doi 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1(6):417–25 doi 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27(12):1739–40 doi 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurozumi S, Matsumoto H, Kurosumi M, Inoue K, Fujii T, Horiguchi J, et al. Prognostic significance of tumour-infiltrating lymphocytes for oestrogen receptor-negative breast cancer without lymph node metastasis. Oncol Lett 2019;17(3):2647–56 doi 10.3892/ol.2019.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez-Saez O, Chic N, Pascual T, Adamo B, Vidal M, Gonzalez-Farre B, et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res 2020;22(1):45 doi 10.1186/s13058-020-01284-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yesiloz U, Kirches E, Hartmann C, Scholz J, Kropf S, Sahm F, et al. Frequent AKT1E17K mutations in skull base meningiomas are associated with mTOR and ERK1/2 activation and reduced time to tumor recurrence. Neuro Oncol 2017;19(8):1088–96 doi 10.1093/neuonc/nox018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov 2014;4(5):554–63 doi 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh G, Odriozola L, Guan H, Kennedy CR, Chan AM. Characterization of a novel PTEN mutation in MDA-MB-453 breast carcinoma cell line. BMC Cancer 2011;11:490 doi 10.1186/1471-2407-11-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet 2005;37(10):1047–54 doi 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 2012;148(5):1015–28 doi 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X, Li J, Gray WH, Lehmann BD, Bauer JA, Shyr Y, et al. TNBCtype: A Subtyping Tool for Triple-Negative Breast Cancer. Cancer Inform 2012;11:147–56 doi 10.4137/CIN.S9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lien HC, Hsiao YH, Lin YS, Yao YT, Juan HF, Kuo WH, et al. Molecular signatures of metaplastic carcinoma of the breast by large-scale transcriptional profiling: identification of genes potentially related to epithelial-mesenchymal transition. Oncogene 2007;26(57):7859–71 doi 10.1038/sj.onc.1210593. [DOI] [PubMed] [Google Scholar]
- 53.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121(7):2750–67 doi 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H, Yu Z, Huo S, Chen Z, Ou Z, Mai J, et al. Overexpression of ELF3 facilitates cell growth and metastasis through PI3K/Akt and ERK signaling pathways in non-small cell lung cancer. Int J Biochem Cell Biol 2018;94:98–106 doi 10.1016/j.biocel.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 55.Ong CT, Campbell BM, Thomas SM, Greenup RA, Plichta JK, Rosenberger LH, et al. Metaplastic Breast Cancer Treatment and Outcomes in 2500 Patients: A Retrospective Analysis of a National Oncology Database. Ann Surg Oncol 2018;25(8):2249–60 doi 10.1245/s10434-018-6533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dave G, Cosmatos H, Do T, Lodin K, Varshney D. Metaplastic carcinoma of the breast: a retrospective review. Int J Radiat Oncol Biol Phys 2006;64(3):771–5 doi 10.1016/j.ijrobp.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 57.Cimino-Mathews A, Verma S, Figueroa-Magalhaes MC, Jeter SC, Zhang Z, Argani P, et al. A Clinicopathologic Analysis of 45 Patients With Metaplastic Breast Carcinoma. Am J Clin Pathol 2016;145(3):365–72 doi 10.1093/ajcp/aqv097. [DOI] [PubMed] [Google Scholar]
- 58.Polamraju P, Haque W, Cao K, Verma V, Schwartz M, Klimberg VS, et al. Comparison of outcomes between metaplastic and triple-negative breast cancer patients. Breast 2020;49:8–16 doi 10.1016/j.breast.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Al Sayed AD, Elshenawy MA, Tulbah A, Al-Tweigeri T, Ghebeh H. Complete Response of Chemo-Refractory Metastatic Metaplastic Breast Cancer to Paclitaxel-Immunotherapy Combination. Am J Case Rep 2019;20:1630–5 doi 10.12659/AJCR.918770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams S Dramatic response of metaplastic breast cancer to chemo-immunotherapy. NPJ Breast Cancer 2017;3:8 doi 10.1038/s41523-017-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic and transcriptomic data supporting this study were generated in the ARTEMIS trial (NCT02276443). A superset of this data is being made publicly available with a manuscript describing the trial and molecular profiling. Before this occurs, the data used for this trial will be made available upon reasonable request from the corresponding authors.