Abstract
Living biological systems, ranging from single cells to whole organisms, can sense, process information, and actuate in response to changing environmental conditions. Inspired by living biological systems, engineered living cells and non-living matrices are brought together, which gives rise to the technology of engineered living materials. By designing the functionalities of living cells and the structures of non-living matrices, engineered living materials can be created to detect variability in the surrounding environment and to adjust their functions accordingly, thereby enabling applications in health monitoring, disease treatment, and environmental remediation. Hydrogels, a class of soft, wet, and biocompatible materials, have been widely used as matrices for engineered living cells, leading to the nascent field of engineered living hydrogels. Here, we discuss the interactions between hydrogel matrices and engineered living cells, focusing on how hydrogels influence cell behaviours and how cells affect hydrogel properties. We also discuss the interactions between engineered living hydrogels and their environments, and how these interactions enable versatile applications. Finally, we highlight current challenges facing the field of engineered living hydrogels for their applications in clinical and environmental settings.
Keywords: engineered living hydrogels, synthetic biology, microbe-material interaction, real-world applications
Graphical Abstract
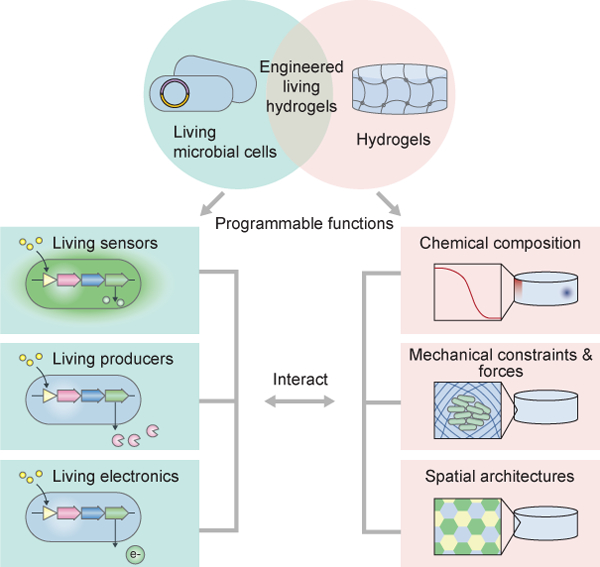
The convergence of engineering, biology, and materials science is providing unprecedented opportunities to integrate living microbes into hydrogel matrices. This integration constructs engineered living hydrogels with the capability of performing tasks associated with living microbes such as self-replication, self-adaption, and environmental responsiveness.
1. Introduction
Engineered living hydrogels are a new class of living systems that are generated by encapsulating living microbial cells in hydrogel matrices. Both components in engineered living hydrogels are programmable for specific goals: The living cells can be engineered with diverse capabilities, including sensing, chemical production, and electricity generation; whereas the hydrogels can be fabricated with various functions to create chemical gradients, mechanical confinement, and spatial distribution for the living cells (Figure 1). Compared with traditional liquid cultures of living cells, three-dimensional (3D) living hydrogels exhibit definite macroscale shape and microscale organization[1–4]. In a typical engineered living hydrogel, the hydrogel matrix provides living cells a variety of chemical and physical cues to modulate the cell behaviors; conversely, the living cells have versatile functions that can change the mechanical and functional properties of the hydrogel (Figure 1). In this review, cells or microbial cells refer to microorganisms, in particular, bacteria and fungi. Since the interactions between hydrogels and animal or plant cells have been systematically discussed in other reviews[5], they are not discussed in this review.
Figure 1.
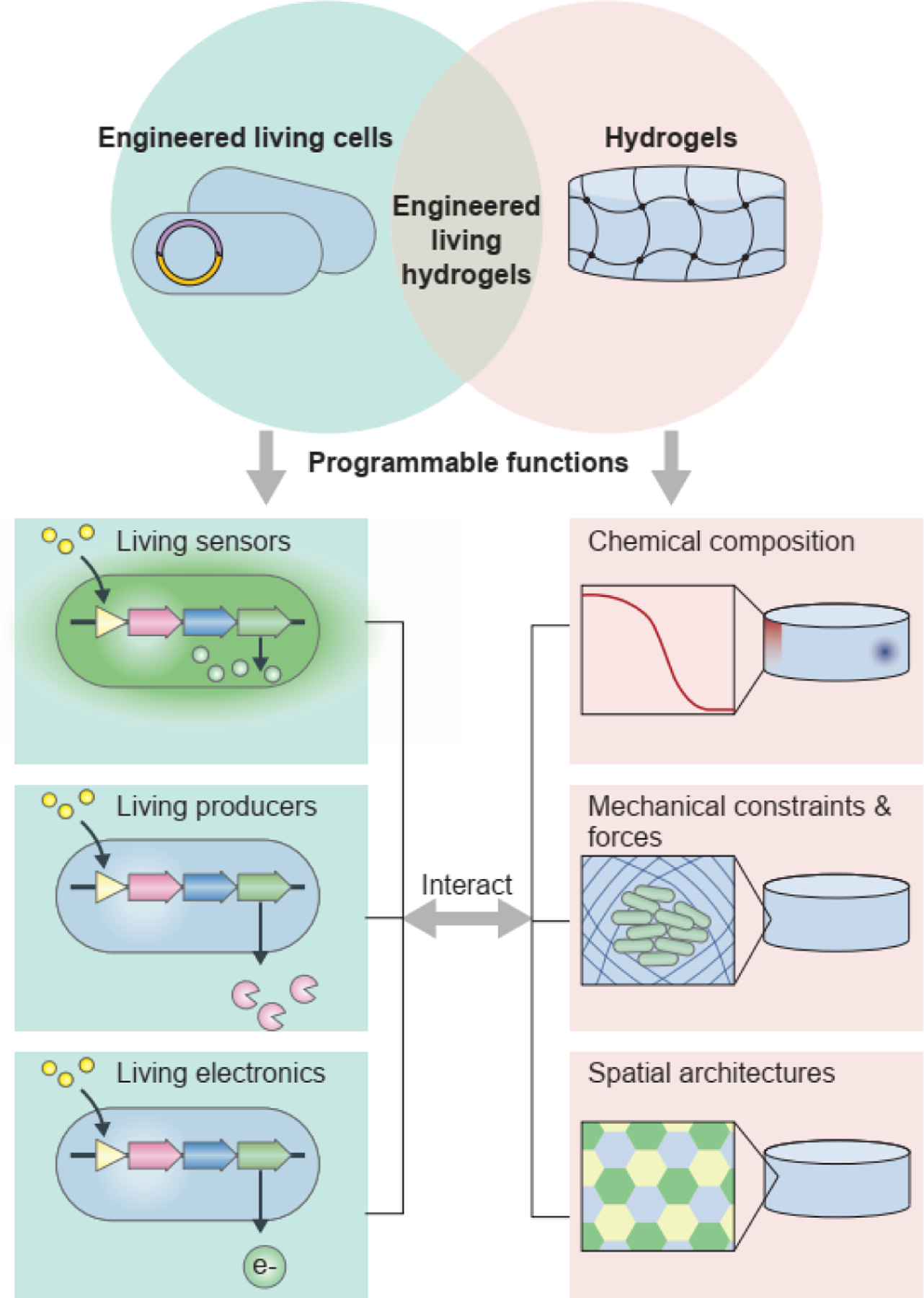
The convergence of engineered living cells and hydrogels gives rise to the technology of engineered living hydrogels. In the engineered living hydrogels, cells and hydrogels interact with each other. The living cells can be programmed with diverse functions, including sensing, chemical production, and electricity generation. The hydrogels can also be programmed with various functions, which create chemical gradients, mechanical confinement and forces, and spatial distribution for the engineered living cells.
The development of engineered living hydrogels is enabled by technological advances in the engineering of microbial cells. Microbial cells have been known for their critical role in the development and maintenance of our bodies and our planet. For example, microbiota in the digestive tract break down protein, carbohydrates, and fats into forms the body can use[6]. In the soil, rhizobia provide ammonium and amino acids to plants, thereby stimulating plant growth[7]. These microbial cell functions are determined by genes and molecular regulators (e.g., RNA, proteins), which govern gene expression levels[8]. Recent breakthroughs in genetic sequencing, DNA synthesis, and gene editing, as well as the emergence of synthetic biology, have made it possible to customize regulatory parts and to construct genetic circuits in microbial cells for many user-defined functions[9]. For example, microbial cells have been programmed by synthetic biologists to produce biofuels from renewable sources[10], to sense the presence of toxins and biomarkers[11], and to release interleukin inside the gut for treatment in situ[12]. To alleviate cross-reactivity and host-cell dependence, biologists can improve the reliability, standardization, modularity, and automated design of these circuits. Through rigorous characterization of libraries of genetic parts[13], predictable and scalable assembly of genetic parts into genetic circuits[14], and mathematical modelling of cell behaviors[15], it is possible to avoid unexpected interference between different genetic parts in a single genetic circuit and between host cells and the genetic circuits they harbor[16].
Material and manufacturing innovations are also indispensable for the development of engineered living hydrogels. Hydrogels, consisting of polymer networks infiltrated with water, have been adopted to form the matrices for microbial cells because of their unique material properties, such as biocompatibility, chemical permeability, and mechanical compliance[17, 18]. The high water content (e.g., 70–99 vol%) of the hydrogel provides sufficient hydration to the encapsulated living cells, while the crosslinked polymer network confers a solid form and structural integrity[17, 19]. Hydrogel matrices can be either produced by living cells or synthesized from naturally occurring polymers and synthetic polymers[18, 20]. Researchers are able to engineer the chemical composition of polymer networks and aqueous solutions, which provide biochemical cues for the living microbial cells. The hydrogel structures can be further designed so that they are formulated at different length scales (nanometer to millimeter) through versatile manufacturing techniques[21]. The structural features (e.g., geometry, porosity, dimension) of the hydrogels exert spatial constraints and mechanical forces on the living cells.
When the technologies of living microbial cells and hydrogel matrices converge, a diverse range of devices have been developed based on engineered living hydrogels, such as wearable biosensors[22, 23], water quality sensors[24], tissue adhesives[25, 26], drug-producing implants[27], and pollutant-degrading scaffolds[28]. Researchers have used engineered living hydrogels as a platform to study the fundamental aspects of microbial cells and hydrogel materials as well as their interactions. For example, scientists can monitor real-time gene network dynamics in single microbes and quorum sensing in microbial populations, when they are trapped in a hydrogel matrix[29–32]. This knowledge, in turn, can be applied to improve the performances of engineered living hydrogels and to extend the scope of their applications to biomedicine, industry, and environmental protection.
Here, we first discuss two pressing questions regarding engineered living hydrogels: how do hydrogel matrices alter cell behaviours, and how do living cells affect the matrix properties? Several reviews have thoroughly described existing engineered living materials[3, 4, 33–35]. Yet, the interplay between the hydrogel matrix and the microbial community in these materials has not been comprehensively discussed. We then review the ways that the engineered living hydrogels interact with the environment, focusing on the applications of sensing, treatment, and energy conversion that are enabled by engineered living hydrogels. Finally, we highlight the challenges that need to be overcome to unleash the full potential of engineered living hydrogels in real-world applications.
2. Types of hydrogels and hydrogel matrices
Depending on the configuration of the hydrogel matrix, engineered living hydrogels can be classified into three types: cell-generated hydrogels, synthetic hydrogels, and hydrogel chambers (Figure 2). In cell-generated and synthetic hydrogels, living microbial cells are dispersed throughout bulk hydrogel structures (Figure 2a,d), whereas in hydrogel chambers, the cells are contained in a hollow space inside (Figure 2g).
Figure 2.
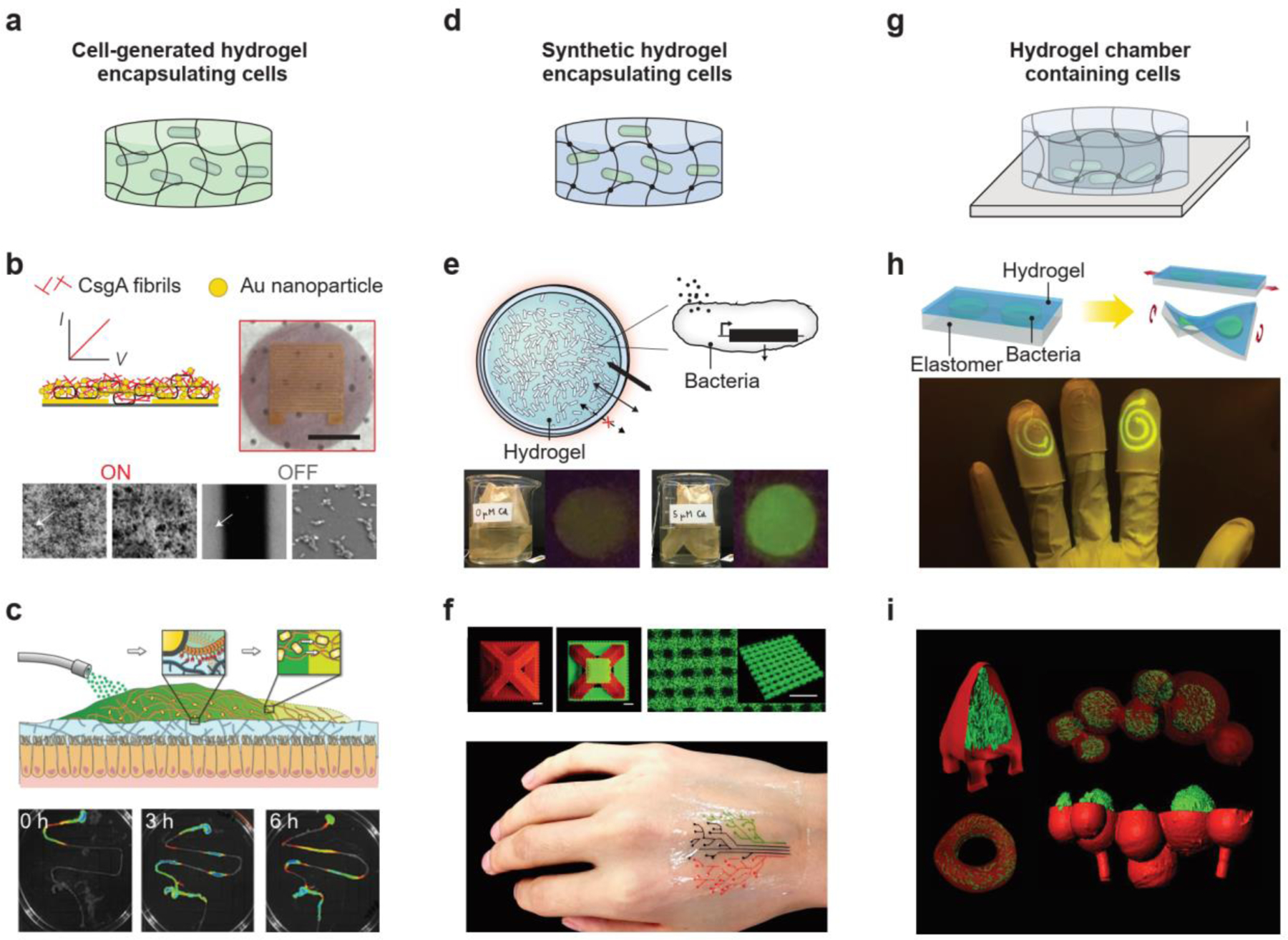
Representative hydrogel matrices in engineered living hydrogels. a-c) Living microbial cells dispersed in cell-generated hydrogels. Examples include an E. coli-produced curli fibril biofilm used as an electrical switch (b) and an E. coli-generated curli hydrogel used as a mucoadhesive patch in the gut (c). (b) Reproduced with permission[41]. Copyright 2014, Springer Nature. (c) Reproduced with permission[42]. Copyright 2019, Wiley-VCH. d-f) Living microbial cells dispersed in synthetic hydrogels. Examples include microbial cell-laden hydrogel beads used as a heavy-metal detector in the environment (e) and a 3D-printed, cell-laden hydrogel pattern used as a biosensor on the skin (f). (e) Reproduced with permission[24]. Copyright 2021, Springer Nature. (f) Reproduced with permission[22]. Copyright 2018, Wiley-VCH. g-i) Living microbial cells enclosed in hydrogel chambers. Examples include a stretchable hydrogel-elastomer hybrid containing microbial cells in channels (h) and a 3D-printed, core-shell hydrogel structure containing microbial cells in cavities (i). (h) Reproduced with permission[46]. Copyright 2017, National Academy of Sciences. (i) Reproduced with permission[53]. Copyright 2013, National Academy of Sciences.
Designs of cell-generated hydrogels are inspired by observations of microbial cells growing in liquid culture[1], which show that individual planktonic cells may spontaneously aggregate into micro-colonies surrounded with hydrated extracellular polymeric substances (EPS) called biofilms (Figure 2a)[2]. The ability to form biofilms is an important attribute of living microbes. The EPS in biofilms, which are generated by the microbial cells[2], are composed of polysaccharides, proteins, and nucleic acids, but their chemical compositions and architectures vary greatly, depending on the microbial cells present, shear forces, temperature, and available nutrients[1, 2, 36, 37]. For example, the Bacillus subtilis biofilm matrix consists of an exopolysaccharide and secreted proteins TasA and BslA, leading to the formation of fruiting body structures[38]. Gluconacetobacter xylinus can produce bacterial cellulose nanofibers (20–100 nm in diameter), which constitute the fibrous network of a biofilm matrix[39]. Microbial cells that are sequestered in a biofilm matrix behave differently from cells suspended in a liquid culture, at both the single-cell and population level[1]. For single cells, the biofilm matrix enables bridging and recognition between cells; for cell populations, the solid matrix temporarily immobilizes the cell populations and increases local cell densities[1, 2]. When cell-generated biofilms are adopted as a hydrogel matrix, the engineered living hydrogels have unique capabilities of self-replication, self-replenishing and self-healing[1]. As a result, cell-generated hydrogels can be applied for wound healing, building construction, and material patterning[40]. For example, E. coli-generated curli hydrogels can act as a mucoadhesive wound patch in the gut (Figure 2c)[41]. The hydrogels are persistent in the gastrointestinal (GI) tract for several days through autonomous self-regeneration[42]. Because biofilm formation can be influenced by environmental factors, the engineered living hydrogels can exhibit externally controlled patterning and environmentally switchable conductivity (Figure 2b)[42].
To encapsulate living microbial cells in synthetic hydrogels, the cells are first dispersed in an uncrosslinked solution, and the cell-containing solution then undergoes crosslinking during manufacturing[22]. Diverse fabrication approaches, including molding, emulsion, electrospinning, light-mediated patterning, and 3D printing, are used to manufacture synthetic hydrogels that encapsulate living cells (Figure 2d)[22, 24, 43, 44]. Alternatively, living microbial cells can be introduced into the pores or channels of synthetic hydrogels post-manufacturing[45]. In synthetic hydrogel matrices, the polymer networks can be composed of either naturally occurring polymers or synthetic polymers. Synthetic hydrogel matrices have more controllable chemical compositions and microstructures than cell-generated biofilm matrices. For example, by varying the mesh size of the polymer network, the diffusion of chemical compounds in synthetic hydrogels can be accelerated or decelerated[19]. The pore size in synthetic hydrogel matrices determines the degree of motility of the microbial cells. The structures and properties of synthetic hydrogels can be optimized for a variety of applications, such as disease detection and environmental monitoring. For example, microbial cell-laden hydrogel beads were used as a heavy-metal detector in the environment (Figure 2e)[24], and a 3D-printed, cell-laden hydrogel pattern was able to sense the biomarkers on the skin (Figure 2f)[22].
Hydrogel chambers are created through either soft lithography or light-based 3D printing[46, 47] (Figure 2g). Hydrogel adhesion technologies help ensure that the chambers that enclose the living microbial cells are completely sealed[48]. The chamber size imposes physical constraints on microbial cell growth. Depending on the desired cell population, the chamber dimension ranges from 1 μm for single cells to 1 mm for cell colonies[49–53]. When the microbial cells are enclosed inside the chamber, the hydrogel wall determines the chemical exchange between microbial cells and the environment. Wall thickness ranges from 2 μm to 1 mm[46, 47]. In addition to geometries, the chemistry of the hydrogel wall also plays a role in chemical exchange. However, the enclosed microbial cells are less influenced by the bulk properties (e.g., chemical and mechanical properties) of the hydrogel, compared with microbial cells encapsulated in bulk hydrogels, since they grow in a liquid medium within the hydrogel chamber. The hydrogel chambers are especially useful for fundamental research on gene network dynamics, cell growth, and intercellular interactions in a confined niche. For example, a 3D-printed hydrogel structure that traps microbial cells in its sealed cavities has been used to study the pathogenicity caused by multiple microbial species (Figure 2i)[46]. Hydrogel chambers are being used not only for basic research but also for biomedical applications, such as wearable devices that can detect environmental toxins (Figure 2h)[53].
3. Influence of hydrogels on living cells
The study of microbial dynamics is of great interest to researchers in the fields of soil microbiology, water purification, and biomedical engineering[54]. Microbial cells display a variety of dynamic behaviors at the levels of single cells, single-species populations, or mixed-species populations. The viability, motility, reproduction, and sporulation of planktonic cells have been intensively investigated[55, 56]. Yet, only a few studies have investigated the effect on microbial cell morphology and growth when the cells are physically confined in a solid matrix[51, 57]. Moreover, collective behaviors of microbial cell communities in a solid matrix, such as biofilm formation, quorum sensing, gene transfer, and nutrient competition, are highly dependent on the microbial cell organization within the hydrogel matrix[2, 36, 58]. The features of hydrogel matrices (e.g., chemical composition, microscale structure, and mechanical properties) define the local environment of microbial cells and affect cell dynamics and further determine whether the cells divide, move, secrete chemicals, exert forces, or express extracellular polymers.
3.1. Influence of chemical composition of polymer networks in hydrogels
One requirement for the polymer networks is to maintain the viability of encapsulated microbial cells. To ensure the biocompatibility of polymer networks, hydrogels have been made of polysaccharides or polyamides, which are abundant low-cost biomaterials that can support cell growth[18, 59], such as agar[60], agarose[61], alginate[62], cellulose[63], hyaluronate[47], mucin[64], curli protein[26], silk protein[65], gelatin[53], and gelatin methacrylate[30]. However, the chemical bonds in natural polymers are susceptible to hydrolytic or enzymatic degradation[66]. To maintain hydrogel stability, several kinds of synthetic polymers have been widely adopted, including polyacrylamide (PAAm)[67], polyvinyl alcohol (PVA)[43, 68, 69], and polyethylene glycol (PEG)[22, 70]. These polymers constitute crosslinked networks and support hydrogel matrices by virtue of their robust C-C and C-O-C linkages[18]. The chemical compositions of these synthetic hydrogels not only guarantee the long-term viability of the microbial cells, but also help protect them from harsh environmental conditions, including low temperature[71], low pH[24], and antibiotics[24]. For example, the number of viable microbial cells in alginate hydrogel beads remains unchanged when the beads are exposed to pH 4 and to kanamycin for 2 h, possibly because the diffusion of highly charged molecules is restricted by the anionic polymer network of alginate hydrogels[24]. Hydrogels with cationic groups in polymer networks, on the other hand, are usually used as antimicrobial materials rather than as scaffolds for engineered living hydrogels[72], because the cationic charges in antimicrobial peptides or quaternary ammonium compounds cause microbial lysis (Figure 3a, left)[73, 74].
Figure 3.
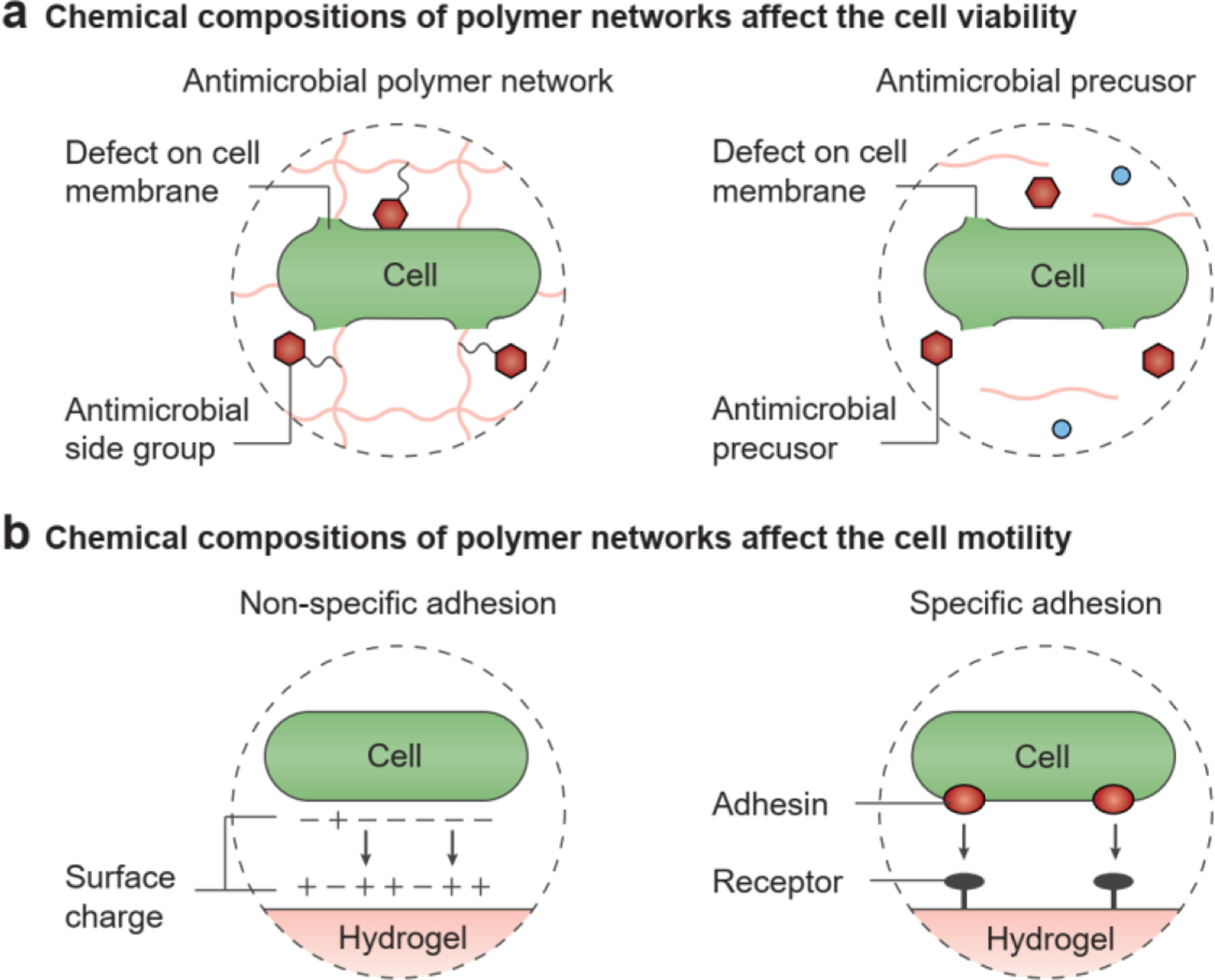
The chemical composition of polymer networks in hydrogels affects cell dynamics. a) Effects on cell viability: the antimicrobial side groups in the polymer network and antimicrobial precursors to prepare the polymer network can induce defects in the cell membrane and cause microbial cell death. b) Effects on cell motility: microbial cells can adhere to the hydrogel surface through non-specific or specific adhesion, which reduces cell motility.
Besides the final compositions, the process of fabricating the hydrogel may also affect its compatibility with living microbial cells. In general, two fabrication approaches are used to seed the microbial cells in the hydrogel matrices, seeding to and seeding from. The seeding to method involves introducing living microbial cells into a hydrogel matrix post-fabrication; thus, the final composition of the hydrogel determines its chemical interactions and biocompatibility. For example, growing microbial cells on an agar hydrogel plate is a routine way to observe microbial colony development in the laboratory[75, 76]. A growth medium, such as Lysogeny broth, and selective compounds, such as antibiotics, are usually added to the agar hydrogel, and then the cell culture will be spread across it[76]. In microfluidic devices, the microbial cells are seeded to the chambers after fabrication[32, 46]. In these two cases, cell viability is independent of the fabrication process. In contrast, the seeding from method involves establishing the living microbial cells at the beginning stage, before hydrogel crosslinking. The chemical precursors for hydrogel synthesis should be biologically compatible with the microbes, neither penetrating the cell membranes nor inhibiting intracellular processes. Microbial cells stay viable when they are dispersed in uncrosslinked polymer solutions, such as polyvinyl alcohol and alginate solutions[24, 43]. However, some monomers (e.g., acrylamide), oligomers (e.g., cationic peptides), and crosslinkers (e.g., glutaraldehyde) are efficient in microbial killing, so that they cannot be mixed with microbial cells in a precursor solution (Figure 3a, right)[73, 77].
In addition to affecting viability, hydrogels with diverse chemical properties can regulate cell attachment and adhesion (Figure 3b, left)[78]. Charge and hydrophilicity are two key factors that affect the microbe-hydrogel interfaces[78]. Because microbial cells are negatively charged in general, hydrogels with positive or neutral charges are more readily colonized than hydrogels with negative surface charges, owing to electrostatic forces[79]. In spite of facilitated adhesion, positively charged hydrogels will lead to lysis of colonized microbial cells, as mentioned above[72]. On the other hand, hydrophilic surfaces of hydrogels are generally more resistant to microbial adhesion, compared with hydrophobic materials (e.g., Teflon and natural rubbers)[80]. However, the hydrogel-microbe adhesion also depends on the particular degree of hydrophilicity of the microbial cell envelope. Still, a moderate number of microbial species with a hydrophilic cell surface preferentially colonize hydrophilic surfaces[78, 81].
Moreover, some natural polymers have exhibited specific chemical interactions with microbial cells (Figure 3b, right). Serum proteins are recognized for their ability to specifically bind some microbial strains. For example, fibronectin and fibrinogen bind Staphylococcus aureus[82], and laminin binds Streptococcus pyogenes[83] via ligand-receptor interactions. Microbial adhesion to hydrogels can be promoted by either grafting these proteins to existing hydrogels or constructing hydrogels with these proteins as backbones[84]. Selective adhesion has also been observed between microbial cells and polysaccharides. The binding between Clostridium thermocellum and cellulose is achieved through the cellulose-binding factor on the microbial cell surface[85]. In natural biofilms, the EPS can adhere to microbial cells and play a vital role in cell clustering and microcolony development[86]. The adhesion between microbial cells and hydrogel matrices limits the motility of microbial cells in the engineered living hydrogels (Figure 3b)[87, 88]. For example, in a living hydrogel system built by Guo et al., there is a high affinity between E. coli and the dextran-based hydrogels, and cell motility is reduced by 100-fold[88].
3.2. Influence of chemical composition of aqueous solution in hydrogels
The hydrogel’s molecular architecture (e.g., network meshes on the nanoscale) and microstructures (e.g., pores, channels, and chambers on the microscale) generate the spatial heterogeneity of nutrients, metabolites, oxygen, and signaling compounds[19, 36]. The heterogeneous distribution of these chemical species in the hydrogel matrix determines the microbial cell organization and regulates the growth, motility, signaling, metabolism, and responsiveness of the embedded microbial cells[36].
Hydrogels can be either non-porous or porous. For non-porous hydrogels, the mesh size of the polymer network, defined as the linear distance between two adjacent crosslinks, regulates the chemical diffusion of compounds (Figure 4a)[19, 89]. Depending on the crosslinking density of hydrogels, the mesh size of non-porous hydrogels is around 10 nm[19, 90]. Most small molecules involved in microbial metabolism (e.g., oxygen, glucose, lactose, acetate, and ammonium) have dimensions of 0.1–1 nm, so they can freely diffuse through a hydrogel (Figure 4a, left)[91]. The spatiotemporal distribution of small-molecule species in hydrogel matrices is governed by Fick’s laws of diffusion[92]. The hydrodynamic diameters of extracellular biopolymers, such as polysaccharides and proteins, are comparable to the mesh size (1–10 nm), thus posing a significant steric barrier for chemical diffusion (Figure 4a, right)[19, 89, 90]. Besides the size effect, chemical-polymer and chemical-cell interactions also play a role in establishing the chemical gradients in the hydrogels (Figure 4b). For example, alginate hydrogels can sequester cationic ions due to electrostatic attraction (Figure 4b, left)[93]. In addition, cells themselves can act as either a sink or a source of chemical species and tune the spatiotemporal profile of the chemical species in the hydrogel matrix (Figure 4b, right)[94, 95]. Therefore, in analyzing chemical heterogeneity, the simultaneous production, consumption, and transportation of chemical species should be considered, in alignment with the reaction-diffusion theory[95, 96].
Figure 4.
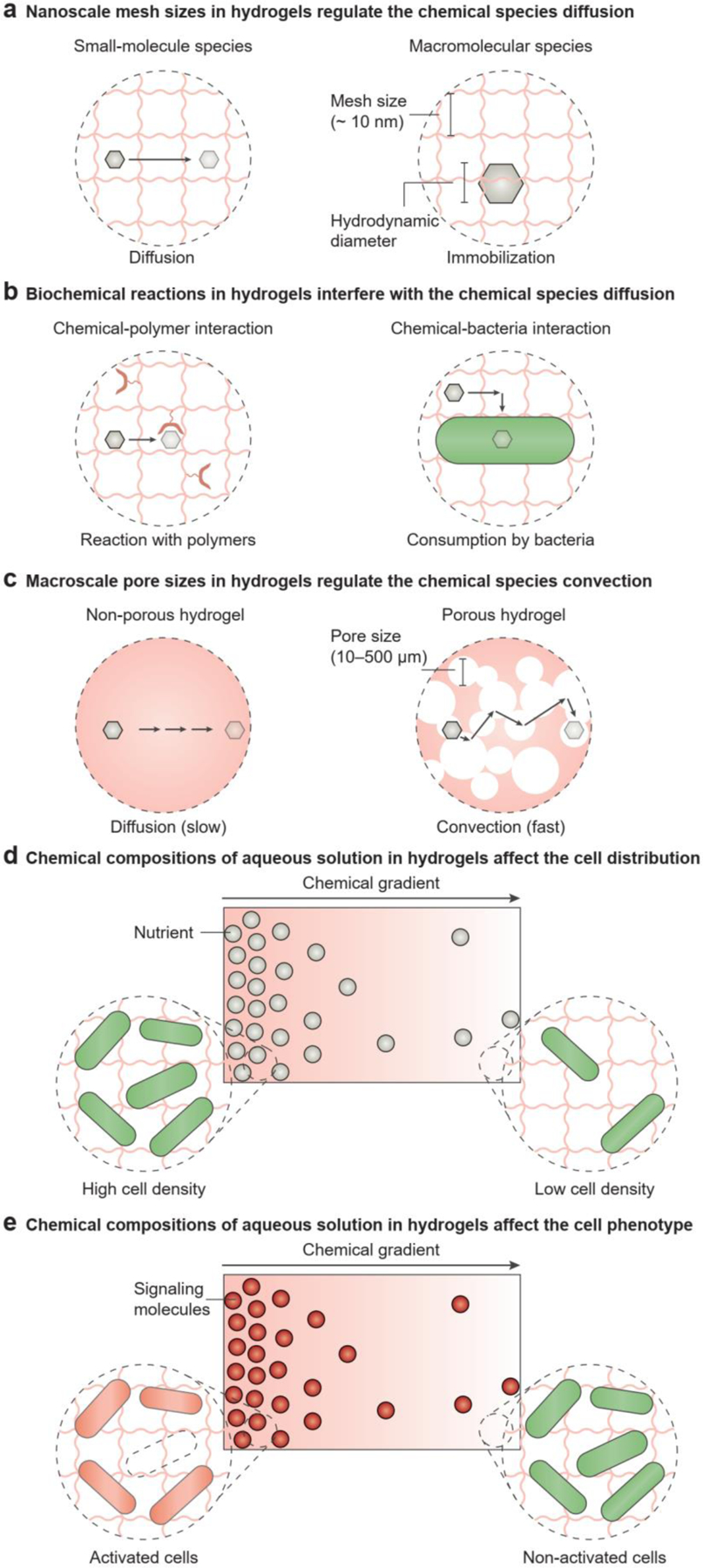
The chemical composition of aqueous solutions in hydrogels affects cell dynamics. a) In non-porous hydrogels the diffusion of chemical species is regulated by the nanoscale mesh of polymer networks. Non-porous hydrogels allow the diffusion of small molecules, but macromolecules are immobilized. b) Biochemical reactions in hydrogels may interfere with the diffusion of chemical species. For example, the chemicals may be consumed if they interact with the polymer network or cells. c) In porous hydrogels, the convection of chemical species is regulated by the macroscale pores. In contrast to the slow diffusion observed in non-porous hydrogels, porous hydrogels allow fast convection of chemicals. d) The chemical composition of aqueous solutions in hydrogels affects cell distribution. For example, the chemical gradient of nutrients sets up the gradient of cell-population densities: A sufficient nutrient supply leads to a high cell density, while an insufficient nutrient supply leads to a low cell density. e) The chemical composition of aqueous solutions in hydrogels affects the cell phenotype. For example, the chemical gradient of signaling molecules causes the population-density gradient of activated cells: A high concentration of signaling molecules leads to cell activation, while a low concentration does not affect the cell phenotypes.
In porous hydrogels, the macroscopic interconnected pores (pore size: 10 – 500 μm) allow the convective transport of chemical compounds (Figure 4c, right). Fabrication techniques used to create porous hydrogels include electrospinning[97], freeze-drying[69], and particle packing[21, 45]. Porous hydrogels also exist in nature as biofilms, which have large and non-uniform pore sizes[1, 2]. Wilking et al. discovered that the channels (~200 μm) formed in Bacillus subtilis biofilms can effectively enhance the liquid flow and facilitate the transport of nutrients and waste for microbial cells[98]. By injecting a fluorescent solution into the channel, these authors visualized the rapid flow across the whole biofilm[98]. The convection-diffusion equation can depict the transport of chemical species in porous hydrogels (Figure 4c, right).
Microbial cells can grow and move through the soft and porous hydrogels[45], and their growth and movement are regulated by the concentration gradients of metabolic substrates and products in the hydrogel matrix (Figure 4d). For example, oxygen is highly concentrated at the air-solid interface and depleted in the center of non-porous hydrogels, as confirmed by microelectrode measurement[36, 99, 100]. Different cells exhibit varied oxygen requirements, so they tend to grow non-uniformly at different locations of the hydrogel matrix[101]: Obligate aerobes and microaerophiles thrive near the hydrogel surface, where they obtain a sufficient supply of oxygen, whereas obligate anaerobes grow in the deep regions of the hydrogel, where oxygen is present at very low concentrations. On the other hand, facultative and aerotolerant anaerobes can be found throughout the hydrogel. Similarly, the distribution of nutrients leads to the collective movement of the cell population in the hydrogel matrix to maximize nutrient availability and cell survival (Figure 4d)[1, 36]. In contrast to oxygen and nutrients, localized high concentrations of antimicrobials and metabolic wastes gradually diminish microbial cell activity[1, 36].
The heterogeneous distribution of microbial cells within the hydrogel can be established not only through spontaneous cell growth or migration but also through programmed cell allocation to different regions. For example, the symbiotic growth of two microorganisms (e.g., Acetobacter and photosynthetic microalgae) generates a biological hydrogel made of bacterial cellulose, in which the two microorganisms are randomly distributed[63, 102, 103]. On the other hand, the microfabrication (e.g., multi-material 3D printing, soft lithography) of cell-laden hydrogels can compartmentalize different types of microbial cells to build spatially segregated microbial consortia[22, 31, 53, 71]. In the compartmentalized microbial consortia, the communication between microbial communities is usually enabled by the diffusion of signaling molecules through the hydrogel matrix[46].
Besides heterogeneous cell distribution, chemical gradients in the hydrogel matrix also regulate the metabolic pathways and physiological activities of microbial cells are (Figure 4e). Microbial cells can sense their chemical environment locally (e.g., oxidative/osmotic/pH stress, and electron donors/acceptors) and adjust their gene expression and physiological activities accordingly. For example, cells may respond to antibiotics, which can reduce metabolic activity or even cause death[104]. Cell populations produce quorum-sensing molecules and change their behaviors according to cell density[105]. In addition, yeast cells immobilized in hydrogel matrices exhibit a higher metabolic rate and increased ethanol and protein production, compared to planktonic yeast cells in suspension cultures[44, 100, 106, 107]. A possible explanation is that the chemical concentrations (e.g., hydrogen ion, oxygen) and the osmotic stress which the hydrogels present are different from those in the liquid culture[106].
3.3. Influence of mechanical constraints in hydrogels
At the single-cell level, the spatial constriction imposed by the hydrogel structure affects microbial cell motility. Cell motility in a porous hydrogel matrix is similar to the conventional run-and-tumble mode in liquid media[49, 108]. As reported by Bhattacharjee et al., in jammed packings of hydrogel particles with pore sizes of 1.5–3.6 μm, the microbial cells follow the hop-and-trap dynamics: during the hopping phase, microbial cells move through extended, directed paths in the pore space; whereas during the trapping phase, cells are confined for extended periods of time (Figure 5a, top)[45]. The hop length decreases as the pore size in the hydrogel matrix becomes smaller[109]. For smaller structures in microfluidic devices, Mannik et al. observed that flagellated microbial cells still retain their motility if the channel width is 1.3 times the cell diameter (i.e., width: 1.2 μm)[49]. However, when the feature size of the hydrogel microstructures is comparable to or even smaller than cell dimensions (≤ 1 μm), microbial cell movement is prevented. Microbial cells then elongate and divide, rather than move through very narrow microchannels (width: 0.4 μm) (Figure 5a, bottom)[49].
Figure 5.

The mechanical constraints imposed by hydrogel structures affect cells dynamics. a) Effects on cell motility when cells pass through a spatially constrained structure. For example, in a porous matrix with the pore dimension larger than the cells, the cells exhibit hop-and-trap dynamics (a, top); in a narrow channel with the channel diameter smaller than the cells, the cells “move” via cell growth with shape change (a, bottom). Note that the microfluidic channels shown here are made of silicon and polydimethylsiloxane (PDMS) instead of hydrogels. b) Effects on cell growth when the hydrogel structure constitutes a confined space. For example, in a closed chamber, cell growth is limited by the chamber size (b, top); in a narrow chamber, filamentous microbial cells grow into the structure of the chamber (b, bottom).
In addition to motility, the morphology of microbial cells can be altered by the hydrogel. Cell shape is altered after they exit narrow microchannels, possibly because the mechanical stress applied by the microchannels deforms the microbial cell walls (Figure 5a, bottom)[49–51]. Takeuchi et al. reported an embossing technique of microchambers (feature size: 2.0 μm) in agarose hydrogels, in which filamentous microbial cells grow into defined shapes (Figure 5b, bottom)[110]. In their experiments, the cells can bend during elongation and adapt their shapes according to the microchamber structures (e.g., crescents, zigzags, sinusoids, and spirals) in the hydrogel[110]. When yeast cells are encapsulated in hydrogel inks for 3D printing, the viscoelastic property of hydrogels appear to affect the proliferation patterns of yeast colonies and alter the sizes of yeast cells[111].
At the population level, the microstructures are physical barriers that limit cell division and population growth. With a sufficient nutrient supply and space, the cell population undergoes fast, exponential growth, because the microbial cells divide by binary fission and double in numbers after each generation time (i.e., 20–60 min)[56, 86]. On the contrary, when microbial cells are embedded in a microscale chamber made of hydrogels (feature size: 10–20 μm), the rapid population expansion is retarded after microbial cells occupy all available spaces in the chamber (Figure 5b, top)[52, 53, 112]. For example, in one study microbial cells in the microfabricated chambers of protein or gelatin-based hydrogel chambers were observed to be densely packed after ten to twelve hours of incubation, and the expansion rate of the cell population dramatically decreased due to the limited space[52, 53].
3.4. Influence of mechanical forces in hydrogels
Although the effects of mechanical forces on eukaryotic cells have been investigated[113], only a few studies have addressed how microbial cells are regulated by their mechanical environment[111, 114, 115]. In fact, various mechanical forces in the hydrogel matrix can actively modulate cell phenotypes, including morphology, growth, adhesion, motility, and biofilm formation and dispersion.
Cell growth in a non-porous hydrogel leads to chain stretching of the polymer network. Stretching of polymer chains at the molecule level then builds up the elastic stress around the microbial cells. When encapsulated in a hydrogel matrix, the cells experience mechanical compression as they grow within the solid environment[116]. Compared to cells in soft hydrogels, the cell growth rate decreases in stiff hydrogels with a high Young’s modulus[43, 116, 117]. The elongation of rod-shaped microbial cells is inhibited by the hydrogel matrix, although the cells can remain metabolically active (Figure 6a, left)[116]. At a higher cell density, the repulsive forces between microbial cells and EPS-based biopolymers lead to spontaneous cell aggregation in the hydrogels owing to the effect of macromolecular crowding[96, 118]. The initial cell aggregates expand by recruiting neighboring microbial cells and grow into densely populated microcolonies[86]. The growth of these microcolonies in the hydrogels generates additional mechanical stress by deforming the matrix, which in turn suppresses the continued expansion of the microcolonies (Figure 6a, right)[112, 115].
Figure 6.
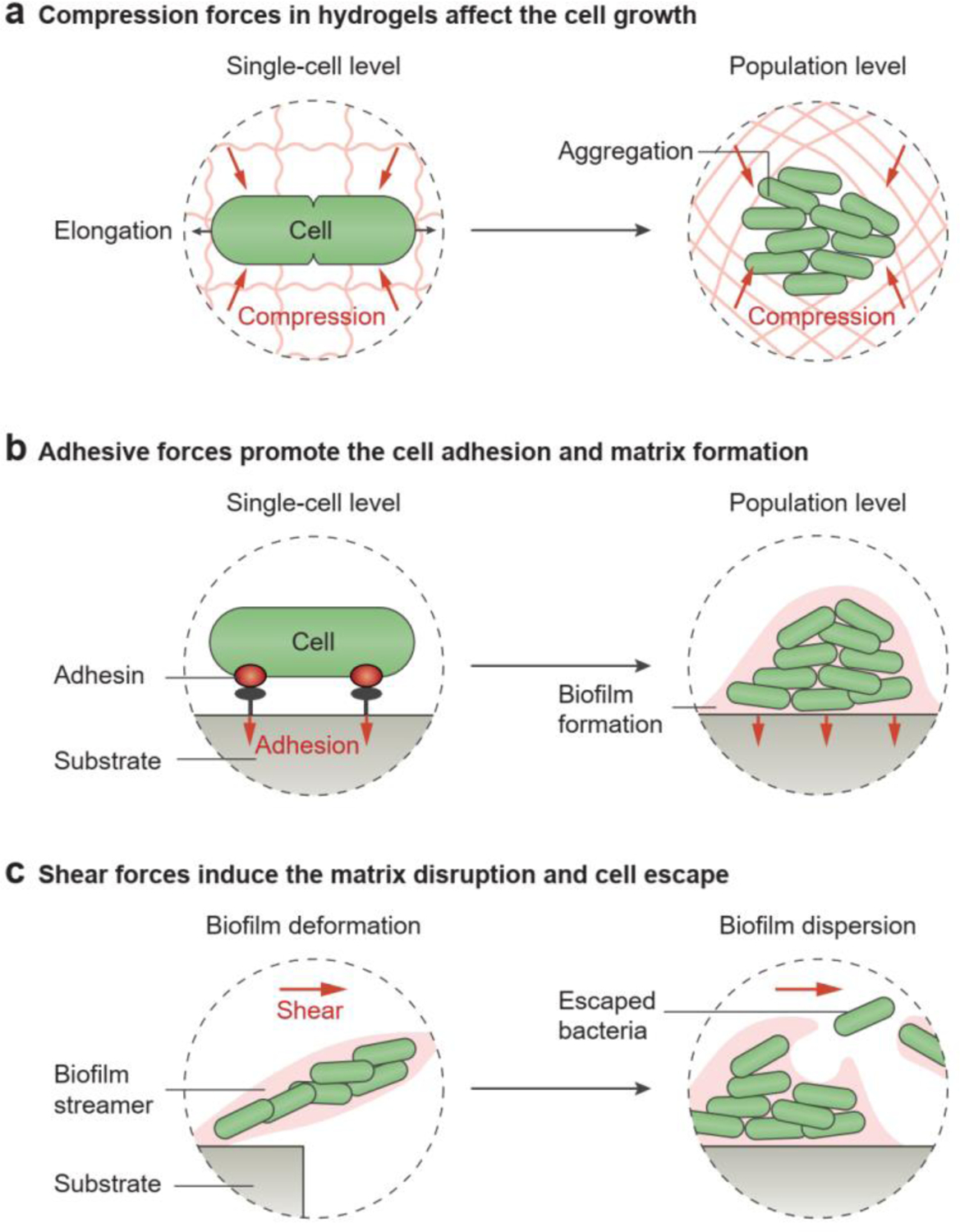
The mechanical forces in hydrogels affect cell growth, adhesion, and escape. a) The compression forces limit cell growth at the single-cell level and population level. b) The adhesion forces between the cell and substrate surface promote cell adhesion and biofilm formation. c) The shear forces caused by the fluid flow induce the deformation and dispersion of the natural hydrogel matrix, which allow the encapsulated microbial cells to escape.
As biofilms are usually generated at air-solid or air-liquid interfaces, they are subject to mechanical interactions with solid and liquid substrates[115, 119]. Typical mechanical interactions between biofilms and substrates include surface adhesion and shear stress (Figure 6b,c). As a result of receptor-ligand interactions between the microbial cell surface and its attached substrate, cell adhesion is the initial step in biofilm formation (Figure 6b, left)[2]. Microbial cells have the ability to strengthen this adhesion under a tensile load[120]. As reported in a few papers, mechanical stress can promote the adhesion of S. aureus and Staphylococcus epidermidis, and trigger E. coli biofilm generation (Figure 6b, right)[121, 122].
In the presence of fluid motion, shear stress applied to the microbial biofilm can deform biofilm architecture by promoting the formation of filamentous structures (i.e., streamers, Figure 6c, left)[114, 123]. Moreover, vigorous fluid flow may lead to the dispersion or detachment of the cohesive cell community (Figure 6c, right)[124]; formerly biofilm-embedded microbial cells can be transferred to and colonize new sections of a microfluidic channel[86, 125]. To ensure mechanical stability and structural integrity, living hydrogels are engineered such that the matrices resist fracture and fatigue[20, 126]. For example, in a hybrid scaffold made of hydrogel and elastomer, both bulk properties and hydrogel-elastomer interfaces were designed to be tough enough to prevent undesirable leakage of the encapsulated cells[46].
4. Influence of living cells on hydrogels
Conventional materials and devices that have been deployed in factories are generally unresponsive to dynamic environments. In contrast, living organisms can navigate environments, communicate, and build complex materials by initiating changes in gene expression in response to specific signals[127]. In engineered living hydrogels, living cells can generate, regenerate, reinforce, or degrade hydrogel materials and also form patterns on hydrogel surfaces[33, 40]. In addition, living cells, equipped with natural or synthetic genetic circuits, are designed to tune the hydrogel properties in response to environmental variations; the ability to interact with their surroundings considerably expands the applications of hydrogels[128].
4.1. Influence of living cells on hydrogel generation
Microorganisms can be viewed as biological factories that can efficiently convert carbon sources to a wide range of extracellular biopolymers, including polysaccharides, polyamides, and polyesters[129, 130]. The biopolymers they produce can be used to create new structures for numerous industrial and medical applications. Engineering the biosynthetic pathways of microbial cells via genes and culture environments provides opportunities to regulate the structures and properties of the material produced[35, 131].
The ability to engineer microbial cells to produce biopolymers and generate hydrogel materials depends on understanding biopolymer synthesis and secretion pathways. For metabolic engineering, the first step is to quantify metabolic pathways, enzyme kinetics, and cell growth rates (Figure 7a). Researchers can then modify existing pathways, by making reactions more efficient or removing undesirable side reactions (Figure 7a)[132]. Strategies such as directed evolution and environmental selection have been used to optimize production[132, 133]. As a bottom-up approach, genetic engineering can be used for versatile biopolymer production. Sequencing and cloning of biosynthetic genes and the selection of proper genetic parts (e.g., promoters, ribosome binding sites, genes of interest, and origin of replication) have expanded the available genetic toolkit and improved production efficiency (Figure 7b). For example, the biosynthesis of highly ordered cellulose fibrils is dictated by the bacterial cellulose synthase operon[134]. The genes of bacterial cellulose synthase in Gluconacetobacter xylinus were placed under the control of a stronger promoter, which increased the cellulose production rate 10-fold[135, 136]. By using inducible promoters, synthase expression could be regulated by an external chemical signal (e.g., arabinose) (Figure 7b)[136, 137]. Besides the engineering of genetic parts, biosynthetic gene expression can be controlled by other transcriptional (e.g., CRISPR-dCas9-based systems[138]) and post-transcriptional (e.g., engineered riboregulators[139, 140]) approaches. Researchers also engineered B. subtilis to secrete a protein-based hydrogel matrix that allows cell attachment and silica biomineralization[141].
Figure 7.
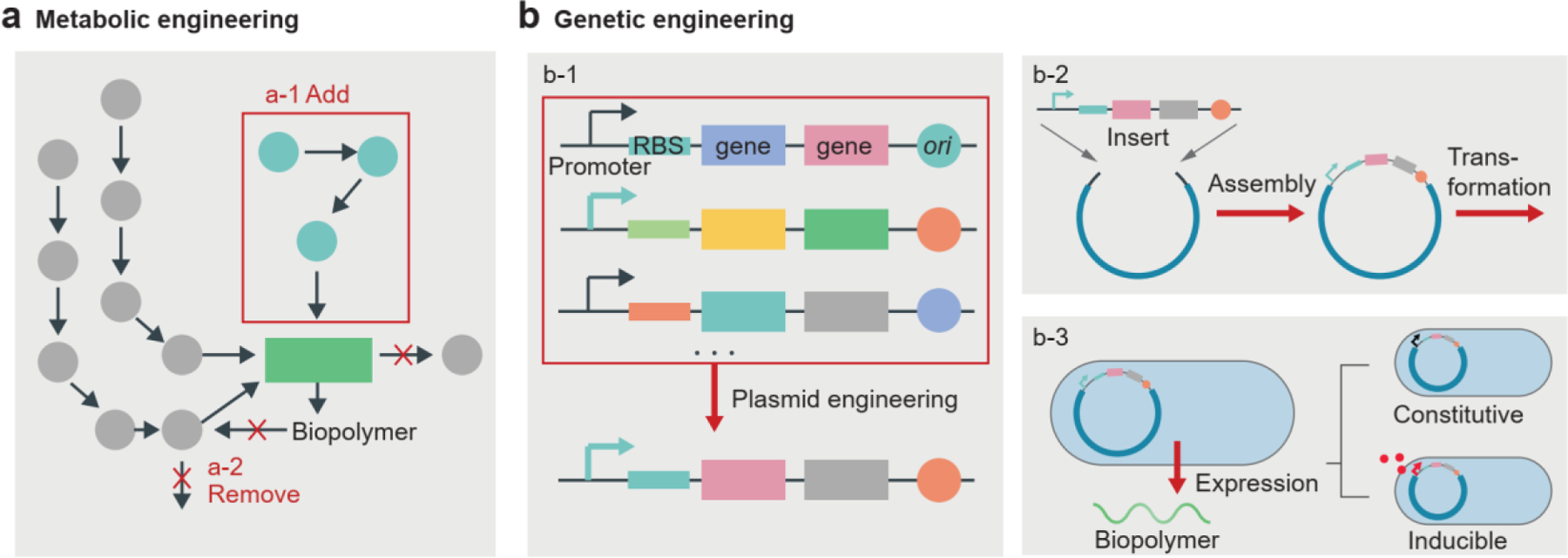
Cell engineering to regulate the generation of biopolymers. a) Metabolic engineering involves adding (a-1) or removing (a-2) biochemical pathways to optimize the production of the desired biopolymers. b) Genetic engineering involves plasmid engineering (b-1) and transformation (b-2), resulting in the expression of desired genes in microbial cells (b-3). Genetic engineering allows the constitutive and inducible gene expression of biopolymer production by microbial cells. RBS, ribosome binding site; ori, origin of replication.
The microbial biofilm is a ubiquitous form of aggregated biopolymers generated by microbial cells and composed of exopolysaccharides (for example, cellulose, alginate, and hyaluronate) (Figure 8a)[1, 2]. However, natural biofilms are usually weak and tend to lose their structural integrity under shear loading[142]. Bacterial cellulose is an exceptional material with high mechanical robustness. Gluconacetobacter xylinus in a liquid culture produces cellulose nanofibrils at the liquid-air interface, thereby generating a pellicle of bacterial cellulose (Figure 8a, b)[143]. Owing to its highly crystalline cellulose nanofibrils, the bacterial cellulose is ultra-stiff (Young’s modulus: 7–12 GPa) and strong (tensile strength: 50–200 MPa) in the hydrated state[144]. Besides bacterial cellulose, protein-based amyloid fibers and polysaccharide-based mycelia are two other biopolymers produced by microorganisms (Figure 8a). Amyloid fibers are readily generated by enterobacteria (e.g., Escherichia coli) and develop into any functional 2D architectures[41, 145], while mycelia are produced by fungi (e.g., Ganoderma lucidum) and can be grown as 3D shapes determined by molds[146]. The biosynthesis circuits can be transplanted to non-polymer-producing microbial cells for the production of tailor-made biopolymers, e.g., polyhydroxyalkanoate, hyaluronate, and poly-γ-glutamate[147]. Other than biopolymers, synthetic polymers such as poly(methyl methacrylate) and polystyrene can be mediated by microbial metabolism[148]. Shewanella oneidensis can control the radical polymerization of monomers and crosslinking of polymers by first consuming dissolved oxygen via aerobic respiration, and then directing extracellular electron flux to a metal catalyst[148, 149].
Figure 8.
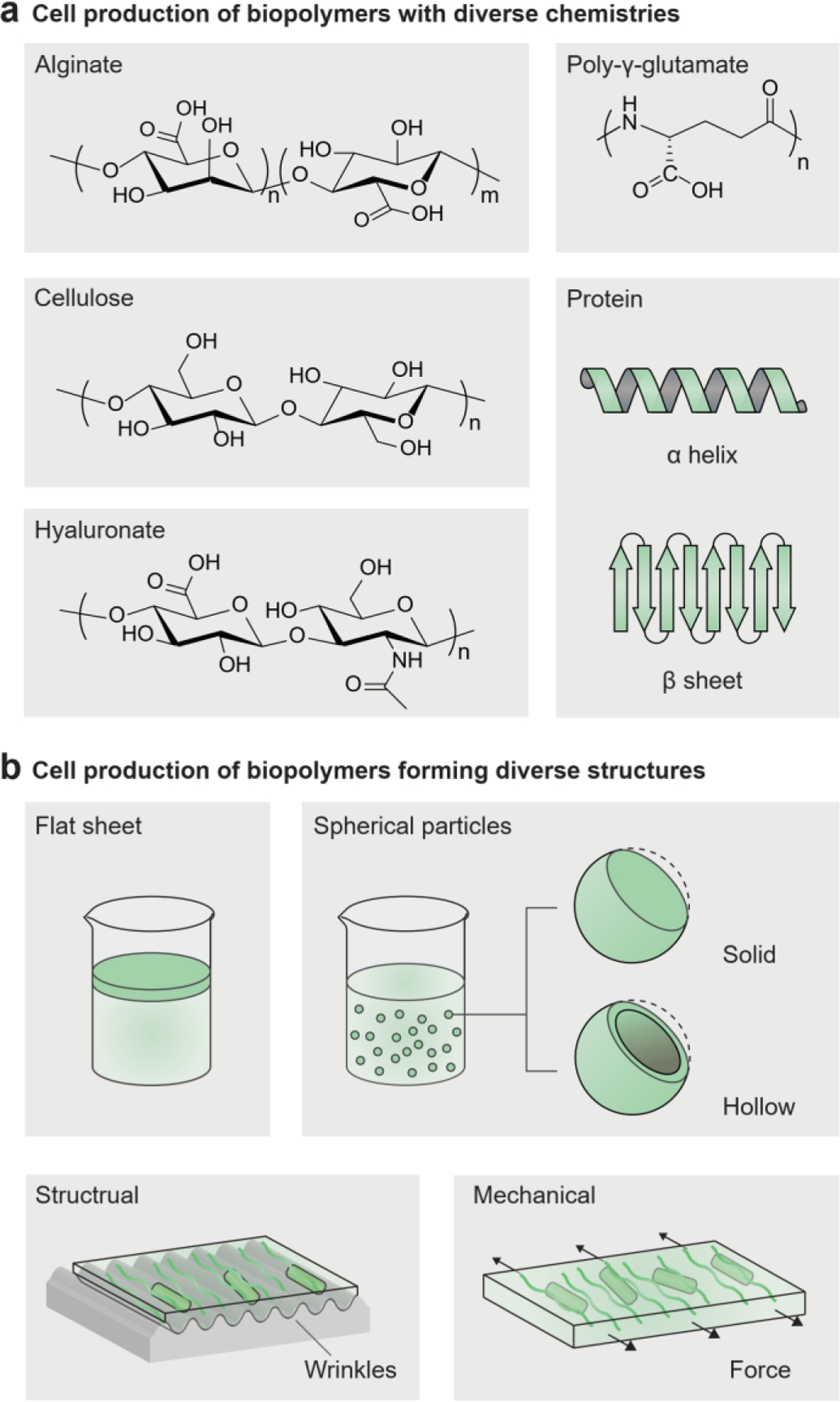
Living cells generate biopolymers with diverse chemistries and forming diverse structures. a) Microbial cells can produce diverse biopolymers, including polysaccharides (alginate, cellulose, hyaluronate) and polyamides (poly-γ-glutamate and protein). b) Microbial cells can produce biopolymers that form diverse structures and microstructures, including flat sheets, solid particles, hollow particles, and aligned fibrils.
Cell culture conditions can be used to adjust the structures of cell-generated materials[131]. The common bacterial cellulose pellicle is a flat-sheet structure floating at the interface between the culture medium and air (Figure 8b)[39]. Spherical particles of bacterial cellulose can be generated in an agitated culture (diameter: 0.5–8 mm), in contrast to the flat sheet grown on a static culture[39, 150]. The higher agitation frequency leads to a higher mass transfer rate and a smaller particles size (Figure 8b)[39, 151]. Moreover, researchers can fabricate bacterial cellulose with various geometries and curvatures, including solid spheres, hollow capsules, and customized 3D shapes, by surrounding the culture media with hydrophobic particles or hydrophobic liquids (Figure 8b)[152]. The hydrophobic materials stabilize the curved air-water interface, allowing for macroscopic shape control during bacterial cellulose production.
The mechanical and physical properties of cell-generated materials can further be modulated by adjusting the cell culture parameters. The addition of materials such as chitosan, gelatin, clay, or silica to the cell culture media alters the molecular composition of the bacterial cellulose as well as its mechanical properties[153, 154]. Cell-generated nanofibrils can be aligned by directing anisotropic cell growth in situ. For example, electrical fields[155], physical structures[156, 157], and mechanical stresses[154, 158] can guide nanofibril orientation during or after material production (Figure 8b). As a result, mechanical strength and toughness can be simultaneously enhanced compared to what is obtained with randomly oriented bacterial cellulose[154, 156].
4.2. Influence of living cells on hydrogel repair and reinforcement
There is a growing demand for self-regenerating materials for industrial and biomedical applications. Cell-generated living hydrogels are regenerative, which means, they can be reused and expanded to additional living materials[42, 141, 159, 160]. The autonomous expansion of engineered living hydrogels indicates their potential applications in cost-effective manufacturing and scalable building materials[160, 161].
Another critical capability of cell-produced materials is to repair damaged materials[26, 141, 160, 162–166]. The metabolites involved in microbial cells can heal cracks in matrix materials[162]. For example, cracks in concrete can be repaired when the incorporated microbial spores recover their metabolic activity and induce calcium carbonate precipitation in the concrete, as reported by Ehrlich et al. in 1996[167]. Sequentially, the precipitates build up and form a cohesive seal in the crack, repairing the concrete (Figure 9b)[167, 168]. Sustainable replenishment of materials is particularly crucial in dynamic environments. For example, peristalsis in the intestine washes away materials attached to the intestinal wall. When continuously replenished by microbial cells, hydrogels are able to maintain their structure and therapeutic effects[25, 42, 169]. Newly generated materials adhere to substrates (e.g., crack surface or intestinal mucus) through surface-binding proteins, such as mussel foot proteins[170] and trefoil factors[25].
Figure 9.
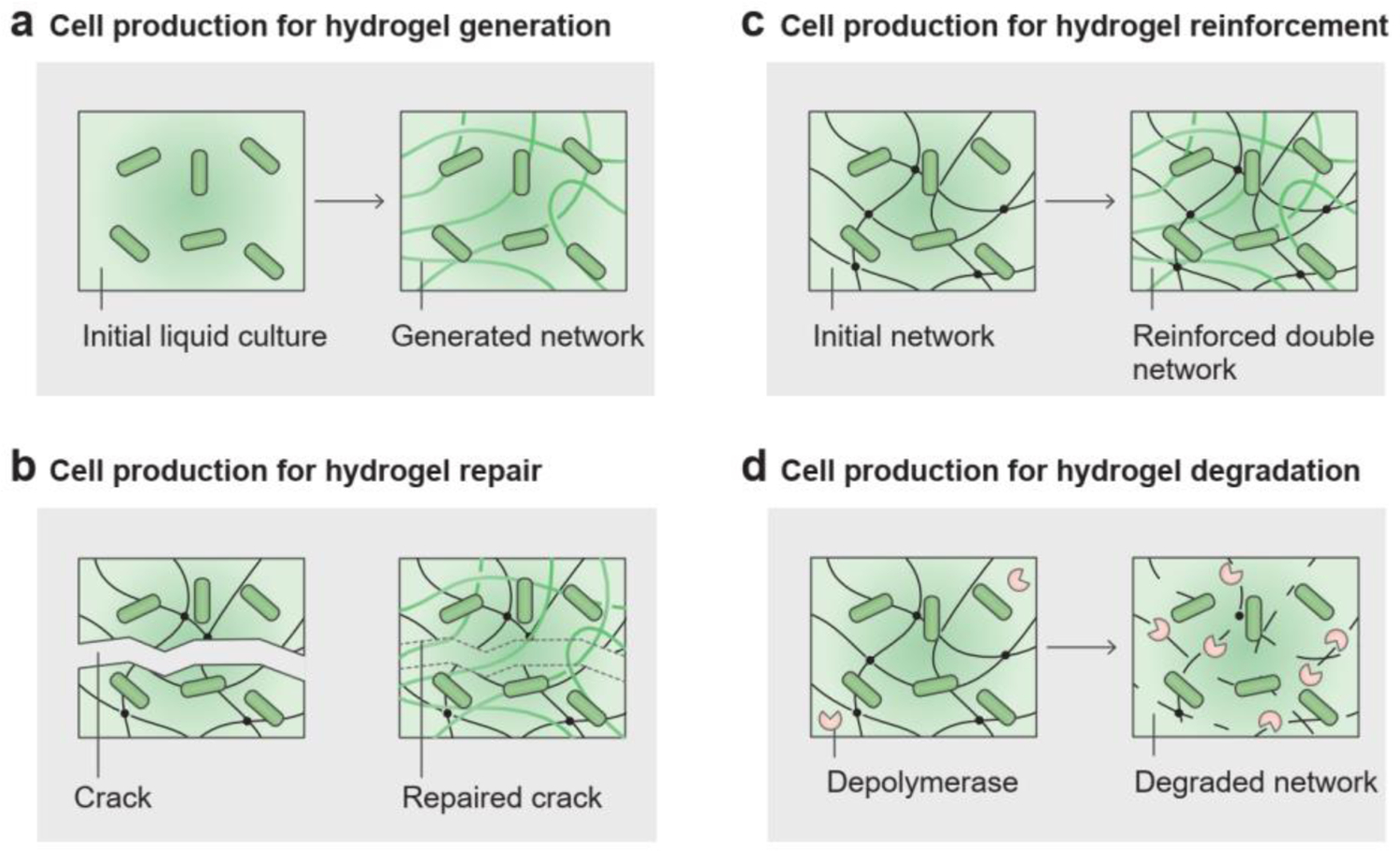
Living cells produce biomass for hydrogel generation, repair, reinforcement, and degradation. a) Microbial cells in liquid culture can generate new solid matrices by producing biopolymers. b) Microbial cells can repair damaged materials by sealing cracks in the matrix. c) Microbial cells can reinforce existing architectures by introducing another polymer network into the matrix. d) Microbial cells can induce degradation of a polymer network by secreting depolymerases in the matrix.
In addition to self-regeneration and self-repair, the existing hydrogel matrix can also be reinforced by living cells (Figure 9c)[37, 159, 171]. Monomers released by microbial cells can react and form a polymer network in the existing polymer network of a hydrogel matrix, leading to increased mechanical strength[166]. Bacterially induced calcium carbonate biomineralization can efficiently increase the toughness of sand-gelatin hydrogels by 1.5-fold[160]. Glucose, a small-molecule product of photosynthesis in living cells, can crosslink with isocyanate groups in synthetic hydrogels. Yu et al. used localized light exposure to adjust the glucose production of chloroplasts, thereby controlling the local reinforcement and repair of hydrogels (Figure 9c)[172].
4.3. Influence of living cells on hydrogel degradation
Microbial cells can induce the mechanical and chemical degradation of hydrogel matrices. On the one hand, population growth of microbial cells can deform the hydrogel, followed by the mechanical fracture of the hydrogel chambers[52, 53]. Localized cell growth and death may initiate mechanical instabilities, such as wrinkling and buckling, during biofilm development[122, 173].
On the other hand, microbial species found in nature degrade a wide range of natural polymers; thus, microbes can trigger the chemical degradation of hydrogel matrices (Figure 9d). For example, hydrolytic exoenzymes produced by enterobacteria are effective in decomposing lipid- and protein-based polymers[174]; while cellulase-producing microbial cells (i.e., Pseudomonas fluorescens, B. subtilis) isolated from the soil can be used to degrade cellulose-based polymers[175]. Through a series of metabolic activities, extracellular depolymerases and hydrolases secreted by cells can break down the long-chain polymers of the hydrogel matrices into low-molecular-weight oligomers or monomers (Figure 9d). The low-molecular-weight degradation products can then be taken up by the cells and used as carbon and energy sources[130].
In contrast to natural polymers, synthetic polymers (in particular, -CH2-CHR-) are less vulnerable to enzymatic or hydrolytic attack, because of the stable carbon-carbon bonds on their synthetic polymer backbone[176]. The great natural source of diverse microorganisms remains unexploited for efficient degradation of PAAm, PVA, polyacrylic acid (PAA), and other compounds. PEG was found to be degraded by a strain of Pseudomonas aerugirosa in 1975[177]: the microbial cells excrete an enzyme which converts low- and high-molecular-weight PEG to a product the cells can utilize[177, 178]. The recent discovery of polyester-degrading enzymes produced by Ideonella sakaiensis has been put to use to decompose polyethylene terephthalate (PET) bottles[179]. Compared to dry plastics, hydrogels have a higher water content and more surface area that cells can colonize; therefore, they are more likely to undergo enzymatic degradation.
4.4. Influence of living cells on hydrogel pattern formation
Microbial cell populations and cell phenotypes are being used to produce dynamic patterns on materials at different length and time scales that can represent complex tasks, including computing, image processing, and display[96, 180]. The patterns can be produced through either applying external cues or engineering genetic circuits. The heterogeneity of single-strain cell phenotypes in hydrogel matrices usually can be set up by morphogen gradients (Figure 10a, left)[22, 36]. The morphogens can be nutrients or signaling molecules, which act as spatial cues to activate or inhibit localized microbial cell responses. Besides chemical signals, optical signals can precisely regulate pattern formation at high spatial resolution. When microbial cells were engineered to produce pigments in response to light, they could display greyscale or color photographs in hydrogels (Figure 10a, right)[76, 181].
Figure 10.
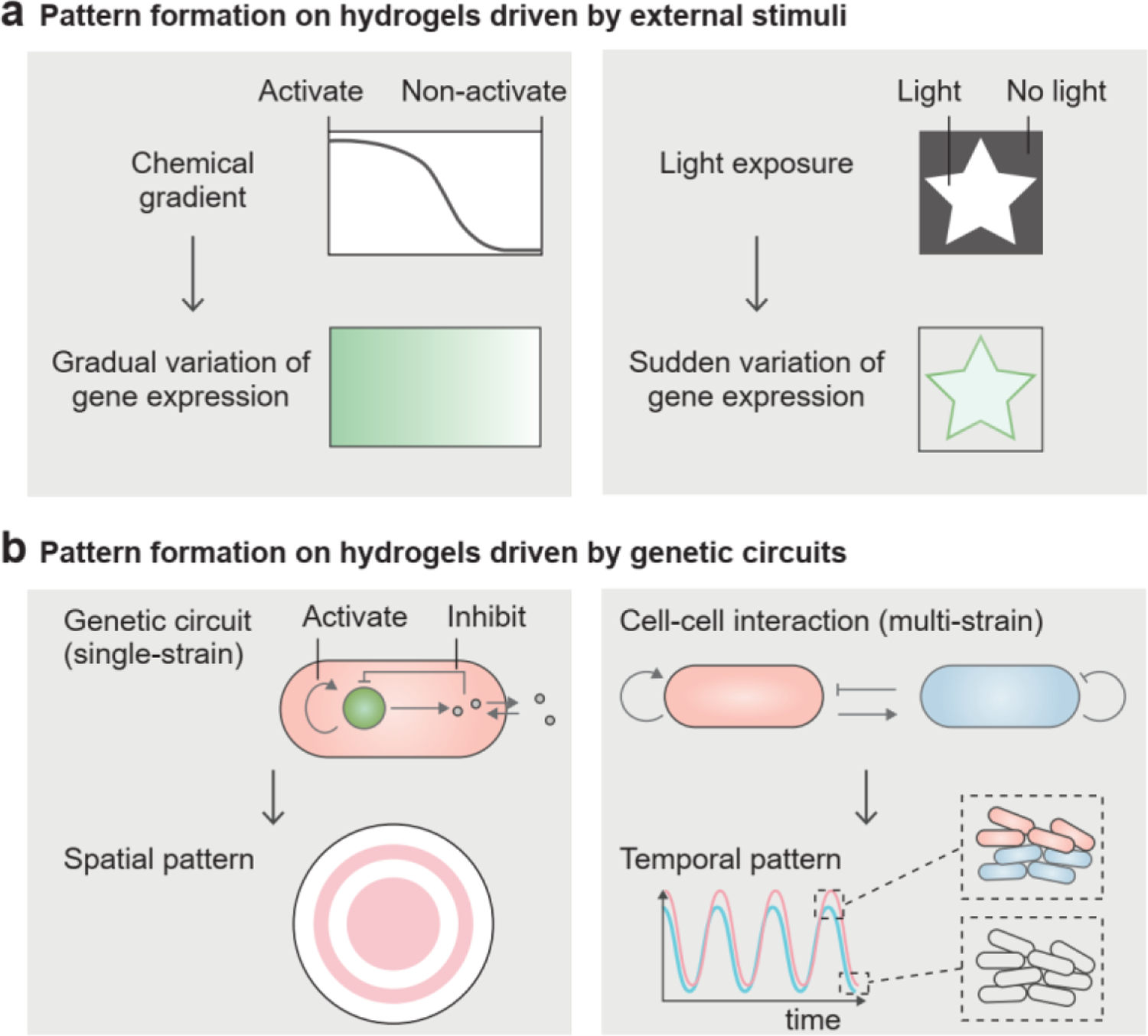
Living cells generate patterns on hydrogels. a) Non-uniform distribution of external stimuli (e.g., chemical gradient and light exposure) triggers the variation of gene expression of microbial cells, so that gradually or suddenly varied patterns are displayed on the material. b) Synthetic genetic circuits in single-strain and multi-strain systems can produce spatially or temporally varied patterns on the material.
Self-organized patterns of gene expression can also be engineered in the absence of any external stimuli (Figure 10b, left)[182, 183]. For example, cells harboring both locally activating and globally inhibitory genetic modules exhibited a ring-shaped fluorescent population pattern on an agar gel (Figure 10b, left)[182, 184]. A similar design of genetic circuits applied in a microfluidic cell culture produced a dynamic pattern with temporal oscillation in a microfluidic chamber and spatial variation in different chambers[185]. Eventually, the synchronized oscillation in the whole microfluidic device could be achieved through the global modulation of cell-released, quorum-sensing molecules[185]. Gene expression is more complicated for multiple microbial strains than for single strains due to cell-cell interactions[186, 187]. The population of each microbial strain can exhibit stable and oscillating dynamics over time (Figure 10b, right). For example, in a cooperative cell system, subpopulations of different microbial strains showed in-phase oscillations (Figure 10b, right)[186]. Moreover, the coordination among multiple microbial strains could produce delicate patterns and versatile functions, such as band detection and XOR logic gates[188, 189]. Spatial patterns for the band detection and XOR logic gates consisted of two and four microbial biofilms, respectively[188, 189]. The microbial strains in different biofilms carried different genetic circuits, and their inputs and outputs were wired to each other for cell-cell communication[22].
5. Applications of engineered living hydrogels
Besides the reciprocal influences between living microbial cells and hydrogel matrices within it, the engineered living hydrogel can also interact with external environments, and these interactions enable versatile applications in industry and biomedicine[3, 4]. In an engineered living hydrogel, the microbial cells usually perform active functions in response to environmental variations[11, 127], whereas the hydrogel matrix usually acts as a passive scaffold for maintaining cell viability, chemical diffusion, and light transmission. In most cases, the hydrogels do not significantly interfere with active functions of cells, though in some cases, the hydrogels are involved in signal transduction and amplification, as well as readout[17]. The bidirectional communication between engineered living hydrogels and the environment facilitates their use as sensors for health or environmental conditions, therapies for diseases, treatments for environmental pollution, actuators for mechanical energy conversion, and batteries for electrical energy conversion.
5.1. Engineered living hydrogels for sensing
In a typical sensing process, external signals (e.g., heat, light or chemicals) are first delivered through the hydrogel matrix to the living microbial cells within it (Figure 11a). Upon receiving the signals, cells report on these external signals through natural or engineered genetic modules. The sensing outputs are usually fluorescence, bioluminescence, or conductivity in cells (Figure 11a)[11, 46, 190, 191], and the collective cell expression in the hydrogel matrix can be further quantified in terms of light intensity or electrical resistance[22, 43, 46]. During the sensing process, signal transportation is coupled to biochemical reactions. The pathways of signal transportation include heat conduction, light transmission, and chemical diffusion. The overall sensing kinetics is usually dependent on the slow biochemical reactions in cells, rather than the fast signal transportation in hydrogels[46].
Figure 11.
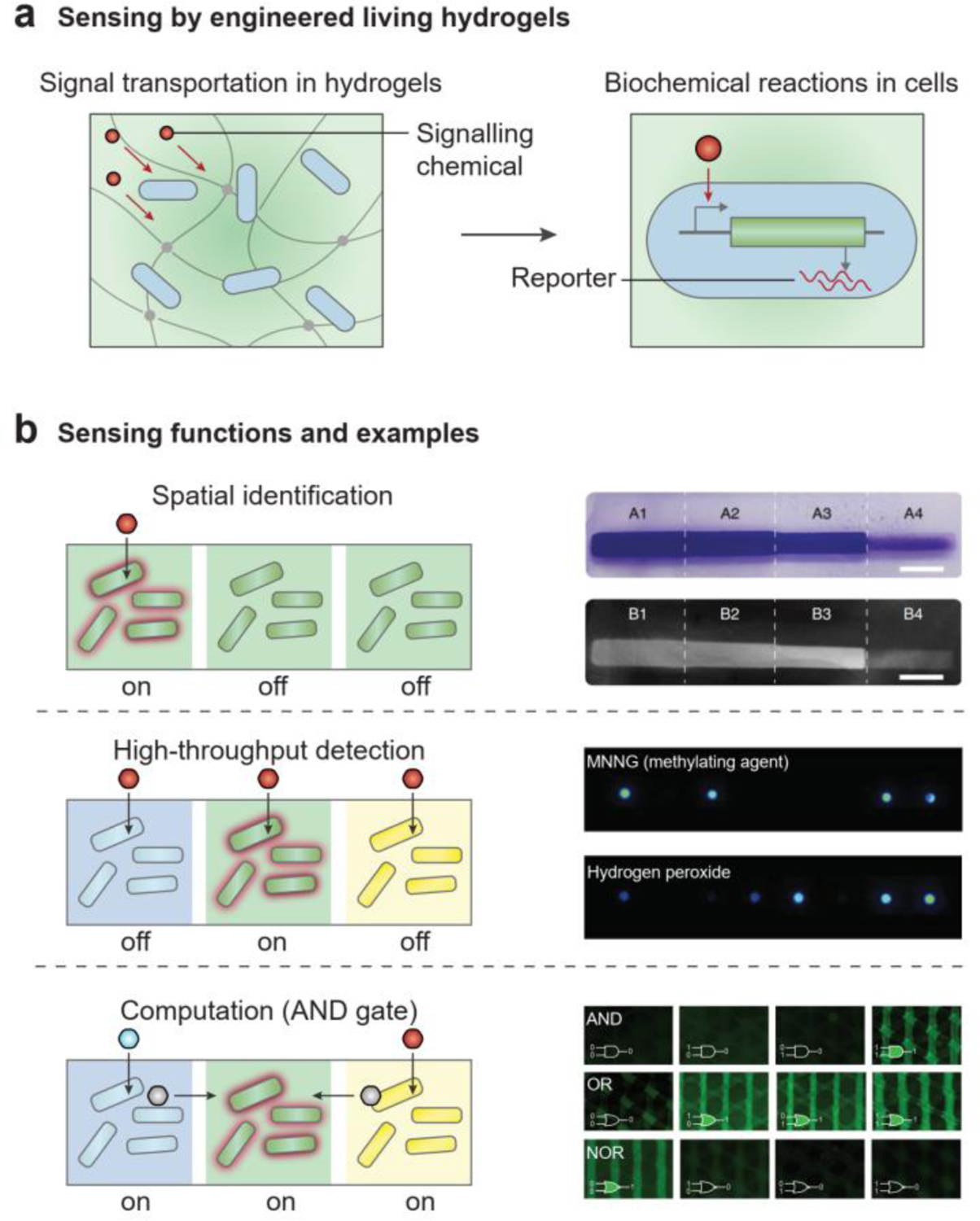
Engineered living hydrogels can sense environmental signals. a) Sensing by engineered living hydrogels in two coupled steps: signal transportation in hydrogels and biochemical reactions in cells. b) Sensing functions and examples by engineered living hydrogels. (b, top) Spatial identification of light intensity. Reproduced with permission[170]. Copyright 2021 Springer Nature. (b, middle) High-throughput detection of toxic chemicals. Reproduced with permission[193]. Copyright 2014 Royal Society of Chemistry. (b, bottom) Logic gate sensing and computation. Reproduced with permission[22]. Copyright 2018 Wiley-VCH.
Because cells can be distributed at different regions in the hydrogel matrix, living sensors can have multiple functions. First, the increased spatial complexity provides more detailed information about the signal source, such as where and how many signals are produced in the environment (Figure 11b, top)[170]. For example, both the location and the degree of mineralization in an E. coli biofilm can be regulated by light intensity. As shown in Figure 11b (top), the part of the biofilm that was exposed to the highest light intensity[170] had the highest mineral density. Second, when multiple types of cells are incorporated in a single hydrogel matrix and each cell type is exclusively sensitive to a specific signal, various signals can be sensed in an efficient manner (Figure 11b, middle)[22, 46, 192, 193]. For example, a dip-stick type living sensor that incorporates eight bacterial strains can detect eight different toxins simultaneously (Figure 11b, middle)[193]. Third, the distribution of different microbial cells and their connectivity allow engineered living hydrogels to perform advanced computing functions, such as signal digitization, signal amplification, Boolean logic gates, and data storage (Figure 11b, bottom)[22, 188, 189, 194]. In a logic gate sensor, the engineered living hydrogel can simultaneously receive a multitude of signals as inputs, and it can return an output after a series of Boolean operations (Figure 11b, bottom)[22, 188].
The sensing ability of engineered living hydrogels enables numerous applications in personal health and environmental sustainability[190, 195], such as monitoring metabolite production, disease signals, and environmental hazards. For example, living sensors to detect pathological glycosuria in urine from diabetic patients were created by encapsulating genetically engineered E. coli and B. subtilis in PVA-alginate hydrogel beads[68]. As another example, the fluorescent protein expression of living microbial cells in a PAAm-alginate hydrogel is proportionate to the concentration of heavy metal in the surroundings[24], indicating that heavy-metal ions in water samples can be quantified by fluorescence in the hydrogels for environmental evaluation[24].
There are still several technical challenges in applying living sensors to practical problems regarding response time, detectable limit, sensitivity, and reliability. These challenges can be addressed by optimizing the genetic circuits in the microbial cells and the cell density in the hydrogel matrix. For example, researchers optimized the transcriptional and translational levels of sensing elements to enhance the sensitivity of living sensors[196]. The distribution and population control of different cell types in the engineered living hydrogels ensure the reliability and ultra-sensitivity of the living sensors[197]. In addition, decreased path length for signal transportation in hydrogels can increase the accelerate the sensing process[19, 46]. Compared to planktonic cells, chemical sensing by cells encapsulated in hydrogels exhibits higher signal-to-noise and increased linearity[198].
5.2. Engineered living hydrogels for treatment
Engineered living hydrogels can produce a wide range of soluble biomolecules, including metabolites, antibiotics, and enzymes for disease treatment and environmental remediation[3, 164, 199]. Biomolecules produced by living microbial cells can be transported through the hydrogel matrix and delivered to the environment (Figure 12a). For example, antimicrobial agents (e.g., penicillin[60], lysostaphin[61], thiocillin[61], and deoxyviolacein[200]) and extracellular proteins (e.g., laccases[63], lactamase[201], organophosphate hydrolase[164]) can be readily released from microorganism-containing hydrogels; these molecules can be used to treat diseases (e.g., therapeutics) or to reduce environmental pollution (e.g., pollutant-degrading enzymes). Toxin and pollutants in the environment can also be absorbed by the hydrogel matrix and metabolized by the cells (Figure 12b)[47, 103, 164, 202]. For example, genetically engineered E. coli immobilized in alginate hydrogels can effectively remove urea and ammonia in uremic rats with renal failure[203]. Ralstonia metallidurans in alginate hydrogels can convert the toxic Hg(II) to non-toxic Hg(0)[204]. The phenol degradation capability of Pseudomonas putida has been demonstrated in a 3D-printed hydrogel matrix[47]. Thus, engineered living hydrogels can help break down body wastes, disease by-products and environmental hazards.
Figure 12.
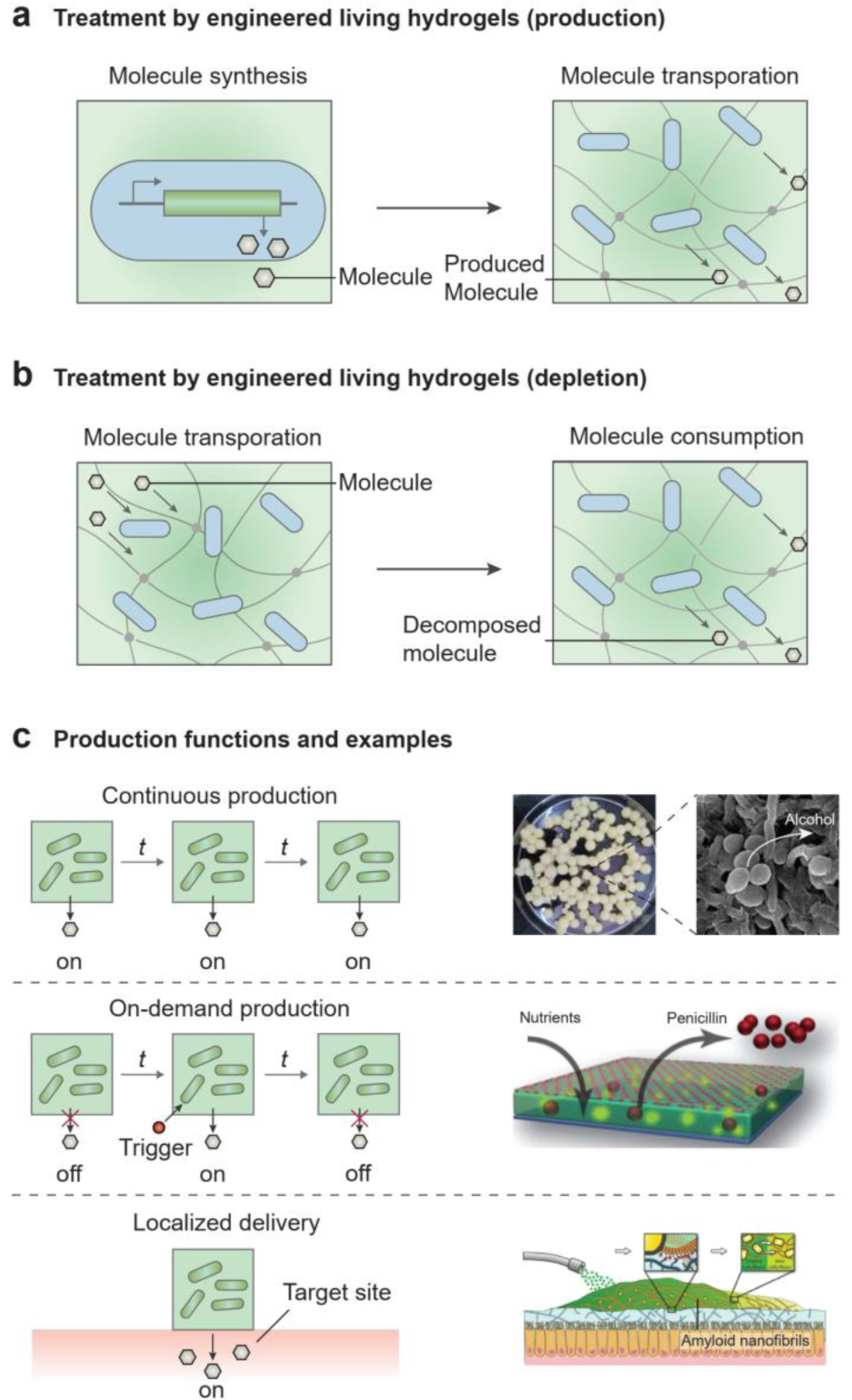
Engineered living hydrogels can treat diseases or alleviate environmental pollution. a) Molecular production: synthesis of the molecules in cells and transportation of the synthesized molecules in hydrogels, which can be used to treat diseases (e.g., therapeutics) or to remediate the environment (e.g., pollutant-degrading enzymes). b) Molecular depletion: transportation of the molecules in hydrogels and consumption of the molecules by cells, which can be used to treat diseases (e.g., by removing body wastes or disease by-products) or to remediate the environment (i.e., by removing pollutants). c) Production functions and examples by engineered living hydrogels. (c, top) Continuous production of alcohol. Reproduced with permission[206]. Copyright 2018 Frontiers Media. (c, middle) On-demand production of penicillin. Reproduced with permission[60]. Copyright 2012 Wiley-VCH. (c, bottom) Localized generation of therapeutics. Reproduced with permission[42]. Copyright 2019 Wiley-VCH.
Furthermore, the production of biomolecules by engineered living hydrogels can be controlled temporally and spatially. For the treatment of chronic diseases and prolonged environmental issues, continuous production of therapeutics is needed, thus necessitating robust, constitutive gene expression in microbial cells (Figure 12c, top)[63, 205]. As an example of sustained production, yeast immobilized in hydrogel beads can constantly produce alcohol (Figure 12c, top)[206]. On the other hand, temporally controlled, on-demand production of molecules may be required for acute diseases or incidents of environmental pollution. In this case, biomolecules can be produced in response to environmental stimuli, such as pollutants or pathogenic biomarkers via the inducible synthesis or the controllable release of biomolecules (Figure 12c, middle)[61, 107, 207]. For example, fungi in a living hydrogel can produce penicillin once nutrients are provided (Figure 12c, middle)[60]. The spatial confinement of microbial cells within hydrogel matrices can alleviate biosafety concerns of genetically engineered cells. Whereas direct administration of free drug-producing microbial cells to the body may alter the original microbiota and cause severe immune responses[25, 208], encapsulating such cells in a hydrogel matrix leaves the microbiota and host unaffected[25, 43, 209]. When the engineered living hydrogels are localized at target sites, therapeutic molecules can be produced and delivered locally to overcome the side effects of systemic delivery (Figure 12c, bottom)[25, 43]. For example, self-generated hydrogels can be programmed to adhere to specific tissues of the GI tract selectively for therapy (Figure 12c, bottom)[42].
In addition to soluble biomolecules, insoluble nanomaterials, such as protein-based or polysaccharide-based nanofibrils, can be generated by engineered living hydrogels; these may eventually be used to treat skin or gut injuries. Microbial biofilms composed of curli amyloid nanofibrils can be grown from engineered E. coli and used as wound dressing and tissue adhesives[25, 165, 169, 210]. When applied on the intestinal walls of colitis-induced mice, these biofilms promote tissue regeneration (Figure 12c, bottom). Furthermore, Lactococcus lactis can produce non-soluble matrix materials (e.g., fibronectin) and soluble proteins (e.g., growth factors) at the same time[27, 211]. The growth factor-containing living hydrogels trigger the adhesion and differentiation of mammalian cells cultured on top of it.
5.3. Engineered living hydrogels for energy conversion
Engineered living hydrogels can convert one type of energy into another, providing an alternative and sustainable way of producing energy and fuel[212, 213]. As a result of their metabolic activities, microbial cells generate chemical, electrical, and mechanical energy. Hydrogels that encapsulate the microbial cells can either serve as an inert matrix or facilitate the energy conversion of the cells by improving light absorption for solar harvesting, promoting electron transfer for electricity generation, and lowering the energy barrier for mechanical actuation.
Photosynthetic microalgae or cyanobacteria incorporated into the hydrogels can convert light energy into chemical energy through cellular respiration (Figure 13a)[102, 214, 215]. Photosynthesis allows sustainable production of metabolites (e.g., glucose, hydrogen) and high-value chemical compounds (e.g., biofuels, pharmaceuticals) (Figure 13a)[103, 216]. The cyanobacteria exhibit a similar hydrogen production rate whether or not they are encapsulated in a hydrogel matrix (Figure 13d, top)[215]. By genetically engineering metabolic pathways, metabolic yield can be improved and advanced biofuels can be produced[217]. Besides metabolic engineering, interfacing non-photosynthetic microbial cells (e.g., Moorella thermoacetica) with light-responsive nanoparticles can lead to the photosynthesis of high-value chemicals from sunlight and carbon dioxide[218]. In addition, photosynthetic microorganisms can harvest solar energy to produce electricity[219], and thus enable a series of optoelectronic devices such as phototransistors, photodetectors, and photovoltaics[220].
Figure 13.
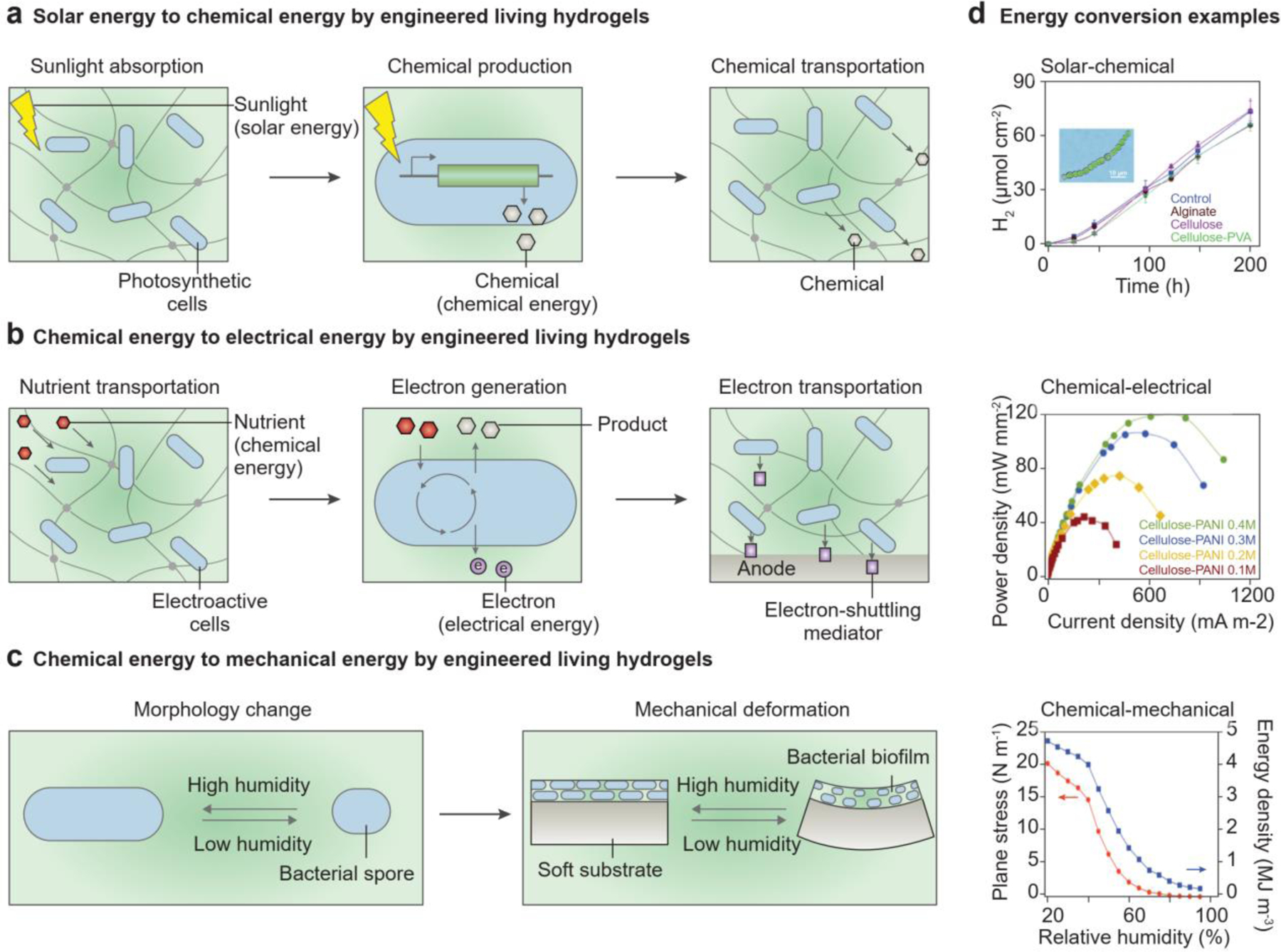
Engineered living hydrogels that can generate chemical, electrical, or mechanical energy. a) Chemical energy is generated by engineered living hydrogels in three coupled steps: sunlight absorption in hydrogels, chemical production in cells, and molecule transportation in hydrogels. b) Electrical energy is generated by engineered living hydrogels in three coupled steps: nutrient transportation in hydrogels, nutrient consumption and electron generation in cells, and electron transfer in hydrogels. c) Mechanical energy is generated by engineered living hydrogels in two coupled steps: morphological change of the cells and mechanical deformation of hydrogels. d) Representative applications of engineered living hydrogels used for energy conversion. (d, top) Solar to chemical energy conversion. Representative curves of hydrogen production by cyanobacteria entrapped in different hydrogels in the sunlight. Reproduced with permission[215]. Copyright 2018, Royal Society of Chemistry. (d, middle) Chemical to electrical energy conversion. Representative curves of power density when the cellulose-polyaniline hydrogel is used as an anode in microbial fuel cells. Reproduced with permission[225]. Copyright 2016, Elsevier. (d, bottom) Chemical to mechanial energy conversion. Representative curves of plane stress at the interface and energy density of the biofilm layer, when the bilayer of the biofilm and substrate are at different relative humidity. Reproduced with permission[229]. Copyright 2014, Springer Nature.
Engineered living hydrogels with electroactive microbial cells (i.e., exoelectrogens) can generate electrical energy from chemical energy[221]. Exoelectrogens such as Shewanella oneidensis and Geobacter species consume chemical energy in organic waste and renewable biomass and produce electrons (Figure 13b)[213, 222, 223]. If cells are attached to an electrode surface, electrons can be transferred directly to the anode via the outer-membrane proteins (e.g., c-type cytochrome) or conductive pilis[213, 222, 224]. If the cells are distributed in a hydrogel electrolyte, electrons can be transferred from the microbial cells to the electrodes via indirect mechanisms, for example, metabolic products (e.g., hydrogen) or electron-shuttling mediators (e.g., neutral red) (Figure 13b)[213, 224]. The addition of extracellular mediators, such as carbon nanotubes, graphene, or polyaniline, to the hydrogel matrix can increase electron transfer efficiency[225, 226]. For example, increased amounts of polyaniline in bacterial cellulose hydrogels can enhance the output power density of microbial fuel cells (Figure 13d, middle)[225].
Engineered living hydrogels can also convert chemical energy to mechanical energy (Figure 13c). Individual microbial cells move freely in liquid by transforming chemical energy to mechanical energy. Cell motility, e.g., chemotaxis or galvanotaxis, is high-speed (i.e., >100 body lengths per second) and responsive to stimuli[227]. This motility is significantly retarded when the cells are encapsulated in hydrogels. On the other hand, macroscopic actuation of the overall engineered living hydrogel can be driven by reversible cell hydration[23] and irreversible cell growth[228]. Soft substrates or encapsulations made of hydrogels ensure the deformability of the biohybrid structures. When microbial biofilms are grown on the soft substrates, cell morphology changes in accordance with the environmental humidity, leading to the mechanical actuation of the engineered living hydrogel (Figure 13c; Figure 13d, bottom)[23, 229]. Furthermore, spatially controlled cell proliferation can program 3D shape transformation of engineered living hydrogels[228].
6. Challenges and perspectives
Since the 2000s, the development of engineered living hydrogels has been facilitated by technological advances in genetic engineering, metabolic engineering, and fabrication tools. Advances in the fundamental understanding of material chemistry, polymer physics, and cell biology have also contributed to research on engineered living hydrogels. A search in PubMed for “engineered living materials,” a broad category that includes engineered living hydrogels, yielded more than 6,000 published papers since 2010, which indicates a booming topic in academia. However, maximizing the applicability of engineered living hydrogels to biomedical, industrial and environmental fields will require a deeper examination of the design principles used in their production.
One of the biggest hurdles for translation is that the undefined, changing environment surrounding the engineered living hydrogel can lead to high variability in its behavior. A variety of synthesis and fabrication techniques have been adopted to expand the experimental scope for gaining external control of living cell phenotypes and hydrogel architectures. When engineered living hydrogels are intended to be deployed in environments that are nutrient-deficient (e.g., tap water) or toxic (e.g., wastewater), the hydrogel matrices need to be designed to support the long-term functioning of microbial cells. For example, engineered living hydrogels that will be used in wastewater can be programmed for nutrient diffusion and toxin sequestration. In this case, hydrogel chemistry and microfabrication can be applied so that the transport of these molecules is controllable. For topical drug delivery, localized environmental remediation, and other functions adapted to particular environments, hydrogel matrices can be engineered through polymer physics and lab-on-a-chip technology to provide precise spatiotemporal modulation of biochemical and biophysical cues and to accommodate living cells to dynamic environments. One of these fabrication techniques, 3D printing, is an emerging technology for additive manufacturing that allows precise control over matrix geometry and cell populations[22, 47, 53, 62, 230]. High-resolution extrusion-based 3D printing can be achieved by optimizing printing parameters (e.g., nozzle diameter, nozzle movement speed, extrusion flow rate) and ink properties (e.g., rheological behavior and crosslinking mechanism). Furthermore, multi-material 3D printing enables different microbes to be immobilized at designated locations within the hydrogel matrix[22, 53]. With precisely allocated microbial cells in it, the 3D-printed engineered living hydrogels can respond to the changing environment in a synergistic mode.
At a more fundamental level, the functionality of engineered living hydrogels can be expanded by genetically engineering the behavior of living cells, for example, by tailoring responses to specific stimuli or enhancing recombinant protein yields. An enlarged genetic toolkit could be used to engineer more microbial species with versatile functions, for example, gut microbes that can treat GI disorders[231] or soil bacteria that can promote plant growth. In addition, the high expression levels of recombinant proteins for sensing and treatment usually represent a metabolic burden for microbial cells, which may reduce protein yields in harsh environments[232]. For the cells to work as efficient factories delivering the intended products at high yields, greater consistency in protein production is required. Finally, under the actual working conditions, living cells respond not only to the stimuli in the environment, but also to the hydrogel matrices. The precise characterization of genetic circuits and the physiology of the cells when they are inside the hydrogels remains rather underexplored. Understanding the ways that microbial cells respond to mechanical forces and constraints, as well as other changing conditions, will help improve their fitness.
Advances in computational techniques are helping to increase the programmability and predictability of engineered living hydrogels. However, theoretical modelling of engineered living hydrogels is still a challenge in the field due to a lack of tools to measure dynamic conditions in cells and hydrogels. The design of genetic circuits in microbial cells is traditionally supported by mechanistic mathematical modeling, which involves analysis of complex biochemical reactions and quantification of reaction kinetics[233]. In addition, many parameters in existing models remain to be quantified. The important tools of computational biology and bioinformatics, particularly, machine learning and artificial intelligence, are gaining attention for genetic engineering of microbial cells and deciphering of genetic networks and biological activities; furthermore, these tools can be applied to large and complex biological datasets[234]. Material modeling and simulation have been used to guide choices not only about the biotic components but also about hydrogel chemistry and the manufacturing of hydrogel structures[235]. Last but not least, multiscale and multiphysics modeling of the processes (e.g., sensing, producing) is highlighting new opportunities to predict the spatiotemporal responses of engineered living hydrogels under various conditions[22, 46].
For environmental and clinical applications, some engineered living hydrogels do not yet meet the requirements of biosafety and biocompatibility, which depend on specific regulatory frameworks. Concerns about the biosafety of genetically modified cells[236] can be addressed by introducing genetic kill switches in the genetically engineered microbial cells or adopting auxotrophic cells, to effectively impede cell escape and survival in the environment[140, 236, 237]. In addition to biological approaches, the physical encapsulation of microbial cells in a non-porous, highly-crosslinked hydrogel matrix can also reduce the chance of inadvertent escape[24]. When the engineered living hydrogels are intended to be used for biomedical applications, the biocompatibility and immunogenicity of the foreign materials (e.g., hydrogel matrices and living cells) with the body should be considered[238]. Several types of microbial cells, including Lactococcus, Lactobacillus, and Bifidobacterium, have been classified as Generally Recognized as Safe (GRAS status) for human consumption[239]. Furthermore, hydrogel encapsulation can provide better integration between the implanted microbial cells and the host mammalian cells[240]. Hydrogels’ superior biocompatibility and biofunctionality enable bridging interfaces that closely mimic the mechanical and chemical properties of extracellular matrices[18]. For example, several hydrogel components made of PVA and hyaluronate have been used in devices approved by U.S. Food and Drug Administration, due to their extraordinary biocompatibility[241]. Once implanted, the hydrogels can even promote cell adhesion, proliferation, and tissue in-growth, providing biocompatible interfaces between microbial cells and surrounding tissues[25].
7. Conclusion
The convergence of engineering, biology, and materials science is providing unprecedented opportunities to integrate living cells (e.g., microbes or microbial consortia) into soft materials (e.g., hydrogels)[18, 242]. This integration yields engineered living hydrogels with the capabilities of self-replication, self-regulation, and environmental responsiveness. The combinations of microbial cells and hydrogel matrices are often selected in an empirical manner. In this review, we have summarized the interactions between hydrogels and microbial cells, as well as the interactions between engineered living hydrogels and the environment. Our understanding of these fundamental interactions provides a foundation for the rational design and fabrication of living hydrogels and can spur technological innovations in designing new living hydrogels. Looking forward, accelerating the future testing and application of engineered living hydrogels in the real world will require collaboration among engineers, biologists, clinicians, regulatory agencies, and others. Once the associated ethical, legal, and social implications have been thoroughly explored, engineered living hydrogels can be built to address a variety of societal needs, ranging from health management to environmental remediation to infrastructure construction.
Acknowledgements
M.E.I. and Y.L. contributed equally to this work. The authors acknowledge Dr. Karen Pepper for editing the manuscript. This work is supported by the National Institutes of Health (1-R01-HL153857–01, X.Z.), National Science Foundation (EFMA-1935291, X.Z.), Pew Charitable Trusts (00030623, M.E.I.), Leona M. and Harry B. Helmsley Charitable Trust (3239, M.E.I, Y.L, T.K.L.), and the US Army Research Office through the Institute for Soldier Nanotechnologies at MIT (W911NF-13-D-0001, X.Z.).
Biographies

Xinyue Liu received her Bachelor’s degree in Polymer Science from Sichuan University in 2015 and Ph.D. in Mechanical Engineering from Massachusetts Institute of Technology (MIT) in 2021. She is currently a postdoctoral associate at MIT. During her Ph.D., she developed biomedical devices based on engineered living materials. Her current research interest is developing intelligent soft materials and systems for both human health and environmental sustainability.

Maria Eugenia Inda is currently a Pew Postdoctoral Fellow in the Synthetic Biology Center at MIT. She received her Ph.D. from Institute of Molecular and Cell Biology of Rosario. She is building biosensors to diagnose and treat inflammatory disorders in the gut, such as inflammatory bowel disease and celiac disease. Specifically, she is equipping the bacteria dwelling in the intestines with sensors to recognize the molecular markers of inflammation and command them as sentinels to patrol the gut and secrete therapeutic molecules in situ.

Yong Lai received his Ph.D. in Microbiology from the University of Hong Kong in 2016, where he studied polymicrobial interactions mediated by secondary metabolites. He is currently a postdoctoral research associate in the Synthetic Biology Center at MIT, focusing on engineering smart living diagnostic and therapeutic devices. He is using synthetic biology to develop technologies that can decipher complex biological processes in infectious diseases, such as changes in gene expression in infected cells, and tackle important problems that threaten public health.

Timothy K. Lu is an Associate Professor in the Department of Electrical Engineering and Computer Science and an Associate Member of the Broad Institute of MIT and Harvard. He received his undergraduate and M.Eng. degrees from MIT in Electrical Engineering and Computer Science. He obtained an M.D. from Harvard Medical School and Ph.D. from the Harvard-MIT Health Sciences and Technology Medical Engineering and Medical Physics Program. His group’s research focuses on engineering fundamental technologies to enable the scalable design of biological systems and on applying synthetic biology to solve medical and industrial problems.

Xuanhe Zhao is a Professor in Department of Mechanical Engineering and Department of Civil and Environmental Engineering at MIT. He received his Ph.D. in Engineering Science from Harvard University. The mission of his research group is to advance science and technology on the interfaces between humans and machines for addressing grand societal challenges in health and sustainability with integrated expertise in mechanics, materials, and biotechnology. A major focus of Zhao Lab’s current research is the study and development of soft materials and systems.
Contributor Information
Xinyue Liu, Department of Mechanical Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Maria Eugenia Inda, Synthetic Biology Group, Research Laboratory of Electronics, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Yong Lai, Synthetic Biology Group, Research Laboratory of Electronics, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Timothy K. Lu, Synthetic Biology Group, Research Laboratory of Electronics, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Xuanhe Zhao, Department of Mechanical Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Department of Civil and Environmental Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
References
- [1].Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S, Nature Reviews Microbiology 2016, 14, 563. [DOI] [PubMed] [Google Scholar]
- [2].Flemming H-C, Wingender J, Nature Reviews Microbiology 2010, 8, 623. [DOI] [PubMed] [Google Scholar]
- [3].Tang T-C, An B, Huang Y, Vasikaran S, Wang Y, Jiang X, Lu TK, Zhong C, Nature Reviews Materials 2021, 6, 332. [Google Scholar]
- [4].Rodrigo-Navarro A, Sankaran S, Dalby MJ, del Campo A, Salmeron-Sanchez M, Nature Reviews Materials 2021, 6, 1175. [Google Scholar]
- [5].Wang H, Heilshorn SC, Advanced Materials 2015, 27, 3717; K. Zhang, Q. Feng, Z. Fang, L. Gu, L. J. C. R. Bian, Chemical Reviews 2021, 121, 11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Oliphant K, Allen-Vercoe E, Microbiome 2019, 7, 91; A. E. Kaoutari, F. Armougom, J. I. Gordon, D. Raoult, B. Henrissat, Nature Reviews Microbiology 2013, 11, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Prell J, Poole P, Trends in Microbiology 2006, 14, 161; E. M. Lodwig, A. H. F. Hosie, A. Bourdès, K. Findlay, D. Allaway, R. Karunakaran, J. A. Downie, P. S. Poole, Nature 2003, 422, 722. [DOI] [PubMed] [Google Scholar]
- [8].Lazdunski AM, Ventre I, Sturgis JN, Nature Reviews Microbiology 2004, 2, 581; L. S. Waters, G. Storz, Cell 2009, 136, 615. [DOI] [PubMed] [Google Scholar]
- [9].Kobayashi H, Kærn M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ, Proceedings of the National Academy of Sciences 2004, 101, 8414; D. E. Cameron, C. J. Bashor, J. J. Collins, Nature Reviews Microbiology 2014, 12, 381; W. Weber, M. Fussenegger, Nature Reviews Genetics 2012, 13, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, Khandurina J, Trawick JD, Osterhout RE, Stephen R, Estadilla J, Teisan S, Schreyer HB, Andrae S, Yang TH, Lee SY, Burk MJ, Van Dien S, Nature Chemical Biology 2011, 7, 445. [DOI] [PubMed] [Google Scholar]
- [11].Inda ME, Lu TK, Annual Review of Microbiology 2020, 74, 337. [DOI] [PubMed] [Google Scholar]
- [12].Braat H, Rottiers P, Hommes DW, Huyghebaert N, Remaut E, Remon JP, van Deventer SJH, Neirynck S, Peppelenbosch MP, Steidler L, Clinical Gastroenterology and Hepatology 2006, 4, 754; L. Steidler, W. Hans, L. Schotte, S. Neirynck, F. Obermeier, W. Falk, W. Fiers, E. Remaut, Science 2000, 289, 1352. [DOI] [PubMed] [Google Scholar]
- [13].Adames NR, Wilson ML, Fang G, Lux MW, Glick BS, Peccoud J, Nucleic Acids Research 2015, 43, 4823; A. Arkin, Nature Biotechnology 2008, 26, 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nielsen Alec AK, Der Bryan S, Shin J, Vaidyanathan P, Paralanov V, Strychalski Elizabeth A, Ross D, Densmore D, Voigt Christopher A, Science 2016, 352, aac7341; B. Wang, R. I. Kitney, N. Joly, M. Buck, Nature Communications 2011, 2, 508. [DOI] [PubMed] [Google Scholar]
- [15].Del Vecchio D, Dy AJ, Qian Y, Journal of the Royal Society Interface 2016, 13, 20160380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Meng F, Ellis T, Nature Communications 2020, 11, 5174; R. Kwok, Nature 2010, 463, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu X, Liu J, Lin S, Zhao X, Materials Today 2020, 36, 102. [Google Scholar]
- [18].Lee KY, Mooney DJ, Chemical Reviews 2001, 101, 1869. [DOI] [PubMed] [Google Scholar]
- [19].Li J, Mooney DJ, Nature Reviews Materials 2016, 1, 16071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sun J-Y, Zhao X, Illeperuma WRK, Chaudhuri O, Oh KH, Mooney DJ, Vlassak JJ, Suo Z, Nature 2012, 489, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Daly AC, Riley L, Segura T, Burdick JA, Nature Reviews Materials 2020, 5, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liu X, Yuk H, Lin S, Parada GA, Tang T-C, Tham E, de la Fuente-Nunez C, Lu TK, Zhao X, Advanced Materials 2018, 30, 1704821. [DOI] [PubMed] [Google Scholar]
- [23].Wang W, Yao L, Cheng C-Y, Zhang T, Atsumi H, Wang L, Wang G, Anilionyte O, Steiner H, Ou J, Zhou K, Wawrousek C, Petrecca K, Belcher AM, Karnik R, Zhao X, Wang DIC, Ishii H, Science Advances 2017, 3, e1601984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tang T-C, Tham E, Liu X, Yehl K, Rovner AJ, Yuk H, de la Fuente-Nunez C, Isaacs FJ, Zhao X, Lu TK, Nature Chemical Biology 2021, 17, 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Praveschotinunt P, Duraj-Thatte AM, Gelfat I, Bahl F, Chou DB, Joshi NS, Nature Communications 2019, 10, 5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhong C, Gurry T, Cheng AA, Downey J, Deng Z, Stultz CM, Lu TK, Nature Nanotechnology 2014, 9, 858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hay JJ, Rodrigo‐Navarro A, Petaroudi M, Bryksin AV, García AJ, Barker TH, Dalby MJ, Salmeron‐Sanchez M, Advanced Materials 2018, 30, 1804310. [DOI] [PubMed] [Google Scholar]
- [28].Tay PKR, Nguyen PQ, Joshi NS, ACS Synthetic Biology 2017, 6, 1841. [DOI] [PubMed] [Google Scholar]
- [29].Bennett MR, Hasty J, Nature Reviews Genetics 2009, 10, 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Connell JL, Kim J, Shear JB, Bard AJ, Whiteley M, Proceedings of the National Academy of Sciences 2014, 111, 18255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ge Z, Girguis PR, Buie CR, Lab on a Chip 2016, 16, 480. [DOI] [PubMed] [Google Scholar]
- [32].Prindle A, Samayoa P, Razinkov I, Danino T, Tsimring LS, Hasty J, Nature 2012, 481, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nguyen PQ, Courchesne N-MD, Duraj-Thatte A, Praveschotinunt P, Joshi NS, Advanced Materials 2018, 30, 1704847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rivera-Tarazona LK, Campbell ZT, Ware TH, Soft Matter 2021, 17, 785. [DOI] [PubMed] [Google Scholar]
- [35].Molinari S, Tesoriero RF, Ajo-Franklin CM, Matter 2021, 4, 3095. [Google Scholar]
- [36].Stewart PS, Franklin MJ, Nature Reviews Microbiology 2008, 6, 199. [DOI] [PubMed] [Google Scholar]
- [37].Balasubramanian S, Aubin-Tam M-E, Meyer AS, ACS Synthetic Biology 2019, 8, 1564. [DOI] [PubMed] [Google Scholar]
- [38].Arnaouteli S, Bamford NC, Stanley-Wall NR, Kovács ÁT, Nature Reviews Microbiology 2021, 19, 600; S. S. Branda, J. E. González-Pastor, S. Ben-Yehuda, R. Losick, R. Kolter, Proceedings of the National Academy of Sciences 2001, 98, 11621. [DOI] [PubMed] [Google Scholar]
- [39].Wang J, Tavakoli J, Tang Y, Carbohydrate Polymers 2019, 219, 63. [DOI] [PubMed] [Google Scholar]
- [40].Kovelakuntla V, Meyer AS, Nature Chemical Biology 2021, 17, 630. [DOI] [PubMed] [Google Scholar]
- [41].Chen AY, Deng Z, Billings AN, Seker UOS, Michelle Y. Lu, Citorik RJ, Zakeri B, Lu TK, Nature Materials 2014, 13, 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Duraj-Thatte AM, Courchesne N-MD, Praveschotinunt P, Rutledge J, Lee Y, Karp JM, Joshi NS, Advanced Materials 2019, 31, 1901826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu X, Yang Y, Inda ME, Lin S, Wu J, Kim Y, Chen X, Ma D, Lu TK, Zhao X, Advanced Functional Materials 2021, 31, 2010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Saha A, Johnston TG, Shafranek RT, Goodman CJ, Zalatan JG, Storti DW, Ganter MA, Nelson A, ACS Applied Materials & Interfaces 2018, 10, 13373. [DOI] [PubMed] [Google Scholar]
- [45].Bhattacharjee T, Datta SS, Nature Communications 2019, 10, 2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liu X, Tang T-C, Tham E, Yuk H, Lin S, Lu TK, Zhao X, Proceedings of the National Academy of Sciences 2017, 114, 2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schaffner M, Rühs PA, Coulter F, Kilcher S, Studart AR, Science Advances 2017, 3, eaao6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yuk H, Zhang T, Lin S, Parada GA, Zhao X, Nature Materials 2016, 15, 190; H. Yuk, T. Zhang, G. A. Parada, X. Liu, X. Zhao, Nature Communications 2016, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Männik J, Driessen R, Galajda P, Keymer JE, Dekker C, Proceedings of the National Academy of Sciences 2009, 106, 14861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Amir A, Babaeipour F, McIntosh DB, Nelson DR, Jun S, Proceedings of the National Academy of Sciences 2014, 111, 5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hol FJH, Dekker C, Science 2014, 346, 1251821. [DOI] [PubMed] [Google Scholar]
- [52].Kaehr B, Shear JB, Proceedings of the National Academy of Sciences 2008, 105, 8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Connell JL, Ritschdorff ET, Whiteley M, Shear JB, Proceedings of the National Academy of Sciences 2013, 110, 18380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ranjard L, Richaume A, Research in Microbiology 2001, 152, 707; P. V. Laxma Reddy, B. Kavitha, P. A. Kumar Reddy, K.-H. Kim, Environmental Research 2017, 154, 296; I. Francolini, C. Vuotto, A. Piozzi, G. Donelli, APMIS 2017, 125, 392. [DOI] [PubMed] [Google Scholar]
- [55].Berg HC, Nature 1975, 254, 389; D. W. Adams, J. Errington, Nature Reviews Microbiology 2009, 7, 642; G. H. Wadhams, J. P. Armitage, Nature Reviews Molecular Cell Biology 2004, 5, 1024; E. I. Tocheva, D. R. Ortega, G. J. Jensen, Nature Reviews Microbiology 2016, 14, 535. [DOI] [PubMed] [Google Scholar]
- [56].Moger-Reischer RZ, Lennon JT, Nature Reviews Microbiology 2019, 17, 679. [DOI] [PubMed] [Google Scholar]
- [57].Wessel AK, Hmelo L, Parsek MR, Whiteley M, Nature Reviews Microbiology 2013, 11, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Mukherjee S, Bassler BL, Nature Reviews Microbiology 2019, 17, 371; M. E. Hibbing, C. Fuqua, M. R. Parsek, S. B. Peterson, Nature Reviews Microbiology 2010, 8, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Naahidi S, Jafari M, Logan M, Wang Y, Yuan Y, Bae H, Dixon B, Chen P, Biotechnology Advances 2017, 35, 530. [DOI] [PubMed] [Google Scholar]
- [60].Gerber LC, Koehler FM, Grass RN, Stark WJ, Angewandte Chemie 2012, 51, 11293. [DOI] [PubMed] [Google Scholar]
- [61].González LM, Mukhitov N, Voigt CA, Nature Chemical Biology 2020, 16, 126. [DOI] [PubMed] [Google Scholar]
- [62].Lehner BAE, Schmieden DT, Meyer AS, ACS Synthetic Biology 2017, 6, 1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Gilbert C, Tang TC, Ott W, Dorr BA, Shaw WM, Sun GL, Lu TK, Ellis T, Nature Materials 2021, 20, 691. [DOI] [PubMed] [Google Scholar]
- [64].McGuckin MA, Lindén SK, Sutton P, Florin TH, Nature Reviews Microbiology 2011, 9, 265. [DOI] [PubMed] [Google Scholar]
- [65].Wang C, Xia K, Zhang Y, Kaplan DL, Accounts of Chemical Research 2019, 52, 2916. [DOI] [PubMed] [Google Scholar]
- [66].Nair LS, Laurencin CT, Progress in Polymer Science 2007, 32, 762. [Google Scholar]
- [67].Tuson HH, Renner LD, Weibel DB, Chemical Communications 2012, 48, 1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Courbet A, Endy D, Renard E, Molina F, Bonnet J, Science Translational Medicine 2015, 7, 289ra83. [DOI] [PubMed] [Google Scholar]
- [69].Gutiérrez MC, García-Carvajal ZY, Jobbágy M, Yuste L, Rojo F, Abrusci C, Catalina F, del Monte F, Ferrer ML, Chemistry of Materials 2007, 19, 1968. [Google Scholar]
- [70].Akselrod GM, Timp W, Mirsaidov U, Zhao Q, Li C, Timp R, Timp K, Matsudaira P, Timp G, Biophysical Journal 2006, 91, 3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Johnston TG, Yuan S-F, Wagner JM, Yi X, Saha A, Smith P, Nelson A, Alper HS, Nature Communications 2020, 11, 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Li P, Poon YF, Li W, Zhu H-Y, Yeap SH, Cao Y, Qi X, Zhou C, Lamrani M, Beuerman RW, Kang E-T, Mu Y, Li CM, Chang MW, Jan Leong SS, Chan-Park MB, Nature Materials 2011, 10, 149. [DOI] [PubMed] [Google Scholar]
- [73].Reddy KVR, Yedery RD, Aranha C, International Journal of Antimicrobial Agents 2004, 24, 536. [DOI] [PubMed] [Google Scholar]
- [74].Jia Z, shen D, Xu W, Carbohydrate Research 2001, 333, 1. [DOI] [PubMed] [Google Scholar]
- [75].Jett BD, Hatter KL, Huycke MM, Gilmore MS, Biotechniques 1997, 23, 648. [DOI] [PubMed] [Google Scholar]
- [76].Fernandez-Rodriguez J, Moser F, Song M, Voigt CA, Nature Chemical Biology 2017, 13, 706. [DOI] [PubMed] [Google Scholar]
- [77].Charoenpanich J, Applied Bioremediation: Active and Passive Approaches 2013, 99; H.-W. Leung, Ecotoxicology environmental safety 2001, 49, 26. [Google Scholar]
- [78].An YH, Friedman RJ, Journal of Biomedical Materials Research 1998, 43, 338. [DOI] [PubMed] [Google Scholar]
- [79].Berne C, Ellison CK, Ducret A, Brun YV, Nature Reviews Microbiology 2018, 16, 616. [DOI] [PubMed] [Google Scholar]
- [80].Fletcher M, Loeb GI, Appl Environ Microbiol 1979, 37, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Hogt A, Dankert J, De Vries J, Feijen J, Microbiology 1983, 129, 2959. [DOI] [PubMed] [Google Scholar]
- [82].Kuusela P, Nature 1978, 276, 718; M. Herrmann, P. E. Vaudaux, D. Pittet, R. Auckenthaler, P. D. Lew, F. S. Perdreau, G. Peters, F. A. Waldvogel, The Journal of Infectious Diseases 1988, 158, 693. [DOI] [PubMed] [Google Scholar]
- [83].Switalski L, Speziale P, Höök M, Wadström T, Timpl R, Journal of Biological Chemistry 1984, 259, 3734. [PubMed] [Google Scholar]
- [84].Cook AD, Sagers RD, Pitt WG, Journal of Biomaterials Applications 1993, 8, 72. [DOI] [PubMed] [Google Scholar]
- [85].Bayer EA, Kenig R, Lamed R, Journal of Bacteriology 1983, 156, 818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Paula AJ, Hwang G, Koo H, Nature Communications 2020, 11, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Tuson HH, Weibel DB, Soft Matter 2013, 9, 4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Guo S, Dubuc E, Rave Y, Verhagen M, Twisk SA, van der Hek T, Oerlemans GJ, van den Oetelaar MC, van Hazendonk LS, M. Brüls, ACS Synthetic Biology 2020, 9, 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Amsden B, Macromolecules 1998, 31, 8382. [Google Scholar]
- [90].Rehmann MS, Skeens KM, Kharkar PM, Ford EM, Maverakis E, Lee KH, Kloxin AM, Biomacromolecules 2017, 18, 3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wu Y, Joseph S, Aluru NR, The Journal of Physical Chemistry B 2009, 113, 3512. [DOI] [PubMed] [Google Scholar]
- [92].Figueiredo L, Pace R, D’Arros C, Réthoré G, Guicheux J, Le Visage C, Weiss P, Journal of Tissue Engineering and Regenerative Medicine 2018, 12, 1238. [DOI] [PubMed] [Google Scholar]
- [93].Ouwerx C, Velings N, Mestdagh M, Axelos MA, Polymer Gels and Networks 1998, 6, 393. [Google Scholar]
- [94].Zhang TC, Fu YC, Bishop PL, Water Environment Research 1995, 67, 992; D. De Beer, A. Schramm, C. M. Santegoeds, M. Kuhl, Applied Environmental Microbiology 1997, 63, 973. [Google Scholar]
- [95].Stewart PS, Journal of Bacteriology 2003, 185, 1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Ghosh P, Mondal J, Ben-Jacob E, Levine H, Proceedings of the National Academy of Sciences 2015, 112, E2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Sarioglu OF, San Keskin NO, Celebioglu A, Tekinay T, Uyar T, Colloids Surfaces B: Biointerfaces 2017, 152, 245; Y. Liu, M. H. Rafailovich, R. Malal, D. Cohn, D. Chidambaram, Proceedings of the National Academy of Sciences 2009, 106, 14201. [DOI] [PubMed] [Google Scholar]
- [98].Wilking JN, Zaburdaev V, De Volder M, Losick R, Brenner MP, Weitz DA, Proceedings of the National Academy of Sciences 2013, 110, 848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Ramsing NB, Kühl M, Jørgensen B, Appl Environ Microbiol 1993, 59, 3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Butelmann T, Priks H, Parent Z, Johnston TG, Tamm T, Nelson A, Lahtvee P-J, Kumar R, ACS Applied Bio Materials 2021, 4, 7195. [DOI] [PubMed] [Google Scholar]
- [101].Borer B, Tecon R, Or D, Nature Communications 2018, 9, 1; Z. Lu, J. A. Imlay, Nature Reviews Microbiology 2021, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Das AAK, Bovill J, Ayesh M, Stoyanov SD, Paunov VN, Journal of Materials Chemistry B 2016, 4, 3685. [DOI] [PubMed] [Google Scholar]
- [103].He F, Ou Y, Liu J, Huang Q, Tang B, Xin F, Zhang J, Jiang M, Chen S, Yu Z, Advanced Materials 2022, 18, 2104820. [DOI] [PubMed] [Google Scholar]
- [104].McMahon MAS, Xu J, Moore JE, Blair IS, McDowell DA, Applied Environmental Microbiology 2007, 73, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Miller MB, Bassler BL, Annual Reviews in Microbiology 2001, 55, 165. [DOI] [PubMed] [Google Scholar]
- [106].Verbelen PJ, De Schutter DP, Delvaux F, Verstrepen KJ, Delvaux FR, Biotechnology Letters 2006, 28, 1515. [DOI] [PubMed] [Google Scholar]
- [107].Yuan S-F, Brooks SM, Nguyen AW, Lin W-L, Johnston TG, Maynard JA, Nelson A, Alper HS, Bioactive Materials 2021, 6, 2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Tailleur J, Cates M, Physical Review Letters 2008, 100, 218103; S. A. Biondi, J. A. Quinn, H. Goldfine, AIChE Journal 1998, 44, 1923. [DOI] [PubMed] [Google Scholar]
- [109].Bhattacharjee T, Datta SS, Soft Matter 2019, 15, 9920. [DOI] [PubMed] [Google Scholar]
- [110].Takeuchi S, DiLuzio WR, Weibel DB, Whitesides GM, Nano Letters 2005, 5, 1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Priks H, Butelmann T, Illarionov A, Johnston TG, Fellin C, Tamm T, Nelson A, Kumar R, Lahtvee P-J, ACS Applied Bio Materials 2020, 3, 4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Holt LJ, Hallatschek O, Delarue M, Methods Cell Biol 2018, 147, 215. [DOI] [PubMed] [Google Scholar]
- [113].Engler AJ, Sen S, Sweeney HL, Discher DE, Cell 2006, 126, 677. [DOI] [PubMed] [Google Scholar]
- [114].Persat A, Nadell Carey D., Kim Minyoung K., Ingremeau F, Siryaporn A, Drescher K, Wingreen Ned S., Bassler Bonnie L., Gitai Z, Stone Howard A., Cell 2015, 161, 988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Dufrêne YF, Persat A, Nature Reviews Microbiology 2020, 18, 227. [DOI] [PubMed] [Google Scholar]
- [116].Tuson HH, Auer GK, Renner LD, Hasebe M, Tropini C, Salick M, Crone WC, Gopinathan A, Huang KC, Weibel DB, Molecular Microbiology 2012, 84, 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Genova LA, Roberts MF, Wong Y-C, Harper CE, Santiago AG, Fu B, Srivastava A, Jung W, Wang LM, Krzemiński Ł, Mao X, Sun X, Hui C-Y, Chen P, Hernandez CJ, Proceedings of the National Academy of Sciences 2019, 116, 25462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Zhou H-X, Rivas G, Minton AP, Annual Review of Biophysics 2008, 37, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Qin B, Fei C, Bridges AA, Mashruwala AA, Stone HA, Wingreen NS, Bassler BL, Science 2020, 369, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Thomas W, Annual Review of Biomedical Engineering 2008, 10, 39. [DOI] [PubMed] [Google Scholar]
- [121].Kerrigan SW, Clarke N, Loughman A, Meade G, Foster TJ, Cox D, Arteriosclerosis, Thrombosis, and Vascular Biology 2008, 28, 335; E. K. Chu, O. Kilic, H. Cho, A. Groisman, A. Levchenko, Nature Communications 2018, 9, 1. [DOI] [PubMed] [Google Scholar]
- [122].Asally M, Kittisopikul M, Rué P, Du Y, Hu Z, Çağatay T, Robinson AB, Lu H, Garcia-Ojalvo J, Süel GM, Proceedings of the National Academy of Sciences 2012, 109, 18891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Das S, Kumar A, Scientific Reports 2014, 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Rumbaugh KP, Sauer K, Nature Reviews Microbiology 2020, 18, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Drescher K, Shen Y, Bassler BL, Stone HA, Proceedings of the National Academy of Sciences 2013, 110, 4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Zhao X, Soft Matter 2014, 10, 672; S. Lin, J. Liu, X. Liu, X. Zhao, Proceedings of the National Academy of Sciences 2019, 116, 10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Brophy JAN, Voigt CA, Nature Methods 2014, 11, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Gardner TS, Cantor CR, Collins JJ, Nature 2000, 403, 339. [DOI] [PubMed] [Google Scholar]
- [129].Moradali MF, Rehm BHA, Nature Reviews Microbiology 2020, 18, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Rehm BHA, Nature Reviews Microbiology 2010, 8, 578. [DOI] [PubMed] [Google Scholar]
- [131].Molinari S, Tesoriero RF, Li D, Sridhar S, Cai R, Soman J, Ryan KR, Ashby PD, Ajo-Franklin CM, bioRxiv 2022, 2021.11.12.468079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Mitra R, Xu T, Xiang H, Han J, Microbial Cell Factories 2020, 19, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Currin A, Swainston N, Day PJ, Kell DB, Chemical Society Reviews 2015, 44, 1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Wong HC, Fear AL, Calhoon RD, Eichinger GH, Mayer R, Amikam D, Benziman M, Gelfand DH, Meade JH, Emerick AW, Proceedings of the National Academy of Sciences 1990, 87, 8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Mangayil R, Rajala S, Pammo A, Sarlin E, Luo J, Santala V, Karp M, Tuukkanen S, ACS Synthetic Biology 2017, 9, 19048. [DOI] [PubMed] [Google Scholar]
- [136].Mangayil R, Rajala S, Pammo A, Sarlin E, Luo J, Santala V, Karp M, Tuukkanen S, ACS Applied Materials & Interfaces 2017, 9, 19048. [DOI] [PubMed] [Google Scholar]
- [137].Florea M, Hagemann H, Santosa G, Abbott J, Micklem CN, Spencer-Milnes X, de Arroyo Garcia L, Paschou D, Lazenbatt C, Kong D, Proceedings of the National Academy of Sciences 2016, 113, E3431; K. T. Walker, V. J. Goosens, A. Das, A. E. Graham, T. Ellis, Microbial Biotechnology 2019, 12, 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Nature Methods 2013, 10, 973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ, Nature Biotechnology 2004, 22, 841. [DOI] [PubMed] [Google Scholar]
- [140].Hayashi N, Lai Y, Mimee M, Lu TK, bioRxiv 2021, 2021.11.03.467106. [Google Scholar]
- [141].Kang S-Y, Pokhrel A, Bratsch S, Benson JJ, Seo S-O, Quin MB, Aksan A, Schmidt-Dannert C, Nature Communications 2021, 12, 7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Boudarel H, Mathias J-D, Blaysat B, Grédiac M, npj Biofilms and Microbiomes 2018, 4, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Mikkelsen D, Flanagan BM, Dykes G, Gidley M, Journal of Applied Microbiology 2009, 107, 576; F. Esa, S. M. Tasirin, N. A. Rahman, Agriculture and Agricultural Science Procedia 2014, 2, 113. [DOI] [PubMed] [Google Scholar]
- [144].Guhados G, Wan W, Hutter JL, Langmuir 2005, 21, 6642; H. Zhao, J. Li, K. Zhu, IOP Conference Series: Materials Science and Engineering 2018, 394, 022041. [DOI] [PubMed] [Google Scholar]
- [145].Knowles TP, Buehler MJ, Nature Nanotechnology 2011, 6, 469; M. M. Barnhart, M. R. Chapman, Annual Review of Microbiology 2006, 60, 131. [DOI] [PubMed] [Google Scholar]
- [146].Haneef M, Ceseracciu L, Canale C, Bayer IS, Heredia-Guerrero JA, Athanassiou A, Scientific Reports 2017, 7, 1; R. Abhijith, A. Ashok, C. Rejeesh, Materials Today: Proceedings 2018, 5, 2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Park SJ, Lee SY, Lee Y, Applied Biochemistry and Biotechnology 2004, 114, 373; L. J. Chien, C. K. Lee, Biotechnology Progress 2007, 23, 1017; H. Jiang, L. Shang, S. H. Yoon, S. Y. Lee, Z. Yu, Biotechnology Letters 2006, 28, 1241. [PubMed] [Google Scholar]
- [148].Fan G, Graham AJ, Kolli J, Lynd NA, Keitz BK, Nature Chemistry 2020, 12, 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Fan G, Dundas CM, Graham AJ, Lynd NA, Keitz BK, Proceedings of the National Academy of Sciences 2018, 115, 4559; A. J. Graham, C. M. Dundas, G. Partipilo, I. E. Miniel Mahfoud, T. FitzSimons, R. Rinehart, D. Chiu, A. E. Tyndall, A. M. Rosales, B. K. Keitz, bioRxiv 2021, 2021.10.17.464678; A. J. Graham, C. M. Dundas, A. Hillsley, D. S. Kasprak, A. M. Rosales, B. K. Keitz, ACS Biomaterials Science Engineering 2020, 6, 1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Singhsa P, Narain R, Manuspiya H, Cellulose 2018, 25, 1571. [Google Scholar]
- [151].Watanabe K, Tabuchi M, Morinaga Y, Yoshinaga F, Cellulose 1998, 5, 187. [DOI] [PubMed] [Google Scholar]
- [152].Greca LG, Lehtonen J, Tardy BL, Guo J, Rojas OJ, Materials Horizons 2018, 5, 408; T. Hoshi, K. Yamazaki, Y. Sato, T. Shida, T. Aoyagi, Heliyon 2018, 4, e00873; S. Shin, H. Kwak, D. Shin, J. Hyun, Nature Communications 2019, 10, 4650. [Google Scholar]
- [153].Gorgieva S, Trček J, Nanomaterials 2019, 9, 1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Wang S, Li T, Chen C, Kong W, Zhu S, Dai J, Diaz AJ, Hitz E, Solares SD, Li T, Advanced Functional Materials 2018, 28, 1707491. [Google Scholar]
- [155].Sano MB, Rojas AD, Gatenholm P, Davalos RV, Annals of Biomedical Engineering 2010, 38, 2475. [DOI] [PubMed] [Google Scholar]
- [156].Prathapan R, Ghosh AK, Knapp A, Vijayakumar A, Bogari NNJ, Abraham BD, Al-Ghabkari A, Fery A, Hu J, ACS Applied Bio Materials 2020, 3, 7898. [DOI] [PubMed] [Google Scholar]
- [157].Putra A, Kakugo A, Furukawa H, Gong JP, Osada Y, Uemura T, Yamamoto M, Polymer Journal 2008, 40, 137. [Google Scholar]
- [158].Wang S, Jiang F, Xu X, Kuang Y, Fu K, Hitz E, Hu L, Advanced Materials 2017, 29, 1702498. [DOI] [PubMed] [Google Scholar]
- [159].Balasubramanian S, Yu K, Meyer AS, Karana E, Aubin-Tam M-E, Advanced Functional Materials 2021, 31, 2011162. [Google Scholar]
- [160].Heveran CM, Williams SL, Qiu J, Artier J, Hubler MH, Cook SM, Cameron JC, Srubar WV, Matter 2020, 2, 481. [Google Scholar]
- [161].Qiu J, Artier J, Cook S, Srubar WV, Cameron JC, Hubler MH, iScience 2021, 24, 102083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Caro-Astorga J, Walker KT, Herrera N, Lee K-Y, Ellis T, Nature Communications 2021, 12, 5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Jimenez-Lopez C, Jroundi F, Pascolini C, Rodriguez-Navarro C, Piñar-Larrubia G, Rodriguez-Gallego M, González-Muñoz MT, International Biodeterioration & Biodegradation 2008, 62, 352. [Google Scholar]
- [164].Huang J, Liu S, Zhang C, Wang X, Pu J, Ba F, Xue S, Ye H, Zhao T, Li K, Wang Y, Zhang J, Wang L, Fan C, Lu TK, Zhong C, Nature Chemical Biology 2019, 15, 34. [DOI] [PubMed] [Google Scholar]
- [165].An B, Wang Y, Jiang X, Ma C, Mimee M, Moser F, Li K, Wang X, Tang T-C, Huang Y, Liu Y, Lu TK, Zhong C, Matter 2020, 3, 2080. [Google Scholar]
- [166].Dai Z, Yang X, Wu F, Wang L, Xiang K, Li P, Lv Q, Tang J, Dohlman A, Dai L, Shen X, You L, Nature Communications 2021, 12, 3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Ehrlich HL, Chemical Geology 1996, 132, 5. [Google Scholar]
- [168].Kua HW, Gupta S, Aday AN, Srubar WV, Cement and Concrete Composites 2019, 100, 35. [Google Scholar]
- [169].Wang X, Cao Z, Zhang M, Meng L, Ming Z, Liu J, Science Advances 2020, 6, eabb1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Wang Y, An B, Xue B, Pu J, Zhang X, Huang Y, Yu Y, Cao Y, Zhong C, Nature Chemical Biology 2021, 17, 351. [DOI] [PubMed] [Google Scholar]
- [171].Axpe E, Duraj-Thatte A, Chang Y, Kaimaki D-M, Sanchez-Sanchez A, Caliskan HB, Dorval Courchesne N-M, Joshi NS, ACS Biomaterials Science & Engineering 2018, 4, 2100. [DOI] [PubMed] [Google Scholar]
- [172].Yu K, Feng Z, Du H, Xin A, Lee KH, Li K, Su Y, Wang Q, Fang NX, Daraio C, Proceedings of the National Academy of Sciences 2021, 118, e2016524118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Fei C, Mao S, Yan J, Alert R, Stone HA, Bassler BL, Wingreen NS, Košmrlj A, Proceedings of the National Academy of Sciences 2020, 117, 7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Hossain TJ, Chowdhury SI, Mozumder HA, Chowdhury MNA, Ali F, Rahman N, Dey S, Frontiers in Microbiology 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Sadhu S, Maiti TK, Microbiology Research Journal International 2013, 235. [Google Scholar]
- [176].Dussud C, Ghiglione J-F, “Bacterial degradation of synthetic plastics”, presented at CIESM Workshop Monogr, 2014. [Google Scholar]
- [177].Haines J, Alexander M. J. A. m., Applied Microbiology 1975, 29, 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].White GF, Russell NJ, Tidswell E. C. J. M. r., Microbiological Reviews 1996, 60, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, Toyohara K, Miyamoto K, Kimura Y, Oda K, Science 2016, 351, 1196. [DOI] [PubMed] [Google Scholar]
- [180].Hoppensteadt F, Jäger W, Biological growth and spread, Springer, 1980, 68. [Google Scholar]
- [181].Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD, Cell 2009, 137, 1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Cao Y, Ryser MD, Payne S, Li B, Rao CV, You L, Cell 2016, 165, 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Payne S, Li B, Cao Y, Schaeffer D, Ryser MD, You L, Molecular Systems Biology 2013, 9, 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Liu C, Fu X, Liu L, Ren X, Chau CKL, Li S, Xiang L, Zeng H, Chen G, Tang L-H, Lenz P, Cui X, Huang W, Hwa T, Huang J-D, Science 2011, 334, 238. [DOI] [PubMed] [Google Scholar]
- [185].Danino T, Mondragón-Palomino O, Tsimring L, Hasty J, Nature 2010, 463, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Chen Y, Kim JK, Hirning AJ, Josić K, Bennett MR, Science 2015, 349, 986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Liao MJ, Din MO, Tsimring L, Hasty J, Science 2019, 365, 1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R, Nature 2005, 434, 1130. [DOI] [PubMed] [Google Scholar]
- [189].Tamsir A, Tabor JJ, Voigt CA, Nature 2011, 469, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Cao Y, Feng Y, Ryser MD, Zhu K, Herschlag G, Cao C, Marusak K, Zauscher S, You L, Nature biotechnology 2017, 35, 1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Chen B, Kang W, Sun J, Zhu R, Yu Y, Xia A, Yu M, Wang M, Han J, Chen Y, Teng L, Tian Q, Yu Y, Li G, You L, Liu Z, Dai Z, Nature Chemical Biology 2022, 18, 289. [DOI] [PubMed] [Google Scholar]
- [192].Smith RSH, Bader C, Sharma S, Kolb D, Tang T-C, Hosny A, Moser F, Weaver JC, Voigt CA, Oxman N, Advanced Functional Materials 2020, 30, 1907401. [Google Scholar]
- [193].Jung I, Seo HB, Lee J.-e., Kim B. Chan, Gu MB, Analyst 2014, 139, 4696. [DOI] [PubMed] [Google Scholar]
- [194].Farzadfard F, Lu TK, Science 2014, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Chen F, Warnock RL, Van der Meer JR, Wegner SV, ACS Sensors 2020, 5, 2205; C. A. Mora, A. F. Herzog, R. A. Raso, W. J. Stark, Biomaterials 2015, 61, 1; A. Y. Zhou, M. Baruch, C. M. Ajo-Franklin, M. M. Maharbiz, PLOS ONE 2017, 12, e0184994; M. Mimee, P. Nadeau, A. Hayward, S. Carim, S. Flanagan, L. Jerger, J. Collins, S. McDonnell, R. Swartwout, R. J. Citorik, V. Bulović, R. Langer, G. Traverso, A. P. Chandrakasan, T. K. Lu, Science 2018, 360, 915. [DOI] [PubMed] [Google Scholar]
- [196].Salis HM, Mirsky EA, Voigt CA, Nature Biotechnology 2009, 27, 946; Y. Chen, J. M. L. Ho, D. L. Shis, C. Gupta, J. Long, D. S. Wagner, W. Ott, K. Josić, M. R. Bennett, Nature Communications 2018, 9, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Xu C, Yu H, Chinese Journal of Chemical Engineering 2021, 30, 112; R. N. Alnahhas, M. Sadeghpour, Y. Chen, A. A. Frey, W. Ott, K. Josić, M. R. Bennett, Nature Communications 2020, 11, 3659; H. J. Kim, J. Q. Boedicker, J. W. Choi, R. F. Ismagilov, Proceedings of the National Academy of Sciences 2008, 105, 18188; W. M. Shaw, H. Yamauchi, J. Mead, G.-O. F. Gowers, D. J. Bell, D. Öling, N. Larsson, M. Wigglesworth, G. Ladds, T. Ellis, Cell 2019, 177, 782. [Google Scholar]
- [198].Atkinson JT, Su L, Zhang X, Bennett GN, Silberg JJ, Ajo-Franklin CM, bioRxiv 2021, 2021.06.04.447163. [Google Scholar]
- [199].Riglar DT, Silver PA, Nature Reviews Microbiology 2018, 16, 214. [DOI] [PubMed] [Google Scholar]
- [200].Sankaran S, Becker J, Wittmann C, del Campo A, Small 2019, 15, 1804717. [DOI] [PubMed] [Google Scholar]
- [201].Dai Z, Lee AJ, Roberts S, Sysoeva TA, Huang S, Dzuricky M, Yang X, Zhang X, Liu Z, Chilkoti A, You L, Nature Chemical Biology 2019, 15, 1017. [DOI] [PubMed] [Google Scholar]
- [202].Gerber LC, Koehler FM, Grass RN, Stark WJ, Proceedings of the National Academy of Sciences 2012, 109, 90; M. G. Nussbaumer, P. Q. Nguyen, P. K. R. Tay, A. Naydich, E. Hysi, Z. Botyanszki, N. S. Joshi, ChemCatChem 2017, 9, 4328; J. K. Sakkos, B. R. Mutlu, L. P. Wackett, A. Aksan, ACS Applied Materials & Interfaces 2017, 9, 26848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Prakash S, Chang TMS, Nature Medicine 1996, 2, 883; S. Prakash, T. Chang, Biotechnology and Bioengineering 1995, 46, 621. [DOI] [PubMed] [Google Scholar]
- [204].Kim HJ, Du W, Ismagilov RF, Integrative Biology 2010, 3, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Saltzman DA, Heise CP, Hasz DE, Zebede M, Kelly SM, Curtiss III R, Leonard AS, Anderson PM, Cancer Biotherapy & Radiopharmaceuticals 1996, 11, 145. [DOI] [PubMed] [Google Scholar]
- [206].Moreno-García J, García-Martínez T, Mauricio JC, Moreno J, Frontiers in Microbiology 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Ryan R, Green J, Williams P, Tazzyman S, Hunt S, Harmey J, Kehoe S, Lewis C, Gene Therapy 2009, 16, 329. [DOI] [PubMed] [Google Scholar]
- [208].Wang J, Wang Y, Zhang E, Zhou M, Lin J, Yang Q, Microbial Cell Factories 2019, 18, 1; M. Guo, S. Yi, Y. Guo, S. Zhang, J. Niu, K. Wang, G. Hu, Viral Immunology 2019, 32, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Cao Z, Wang X, Pang Y, Cheng S, Liu J, Nature Communications 2019, 10, 5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Li Y, Li K, Wang X, Cui M, Ge P, Zhang J, Qiu F, Zhong C, Science Advances 2020, 6, eaba1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Rodrigo-Navarro A, Rico P, Saadeddin A, Garcia AJ, Salmeron-Sanchez M, Scientific Reports 2014, 4, 5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Dovì VG, Friedler F, Huisingh D, Klemeš JJ, Journal of Cleaner Production 2009, 17, 889. [Google Scholar]
- [213].Lovley DR, Nature Reviews Microbiology 2006, 4, 497. [DOI] [PubMed] [Google Scholar]
- [214].David C, Karande R, Bühler K, Cyanobacteria Biotechnology, 2021, 477. [Google Scholar]
- [215].Jämsä M, Kosourov S, Rissanen V, Hakalahti M, Pere J, Ketoja JA, Tammelin T, Allahverdiyeva Y, Journal of Materials Chemistry A 2018, 6, 5825. [Google Scholar]
- [216].Zhang H, Liu H, Tian Z, Lu D, Yu Y, Cestellos-Blanco S, Sakimoto KK, Yang P, Nature nanotechnology 2018, 13, 900. [DOI] [PubMed] [Google Scholar]
- [217].Nozzi N, Oliver J, Atsumi S, Frontiers in Bioengineering and Biotechnology 2013, 1; S. Atsumi, W. Higashide, J. C. Liao, Nature Biotechnology 2009, 27, 1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Sakimoto KK, Wong AB, Yang P, Science 2016, 351, 74; S. Cestellos-Blanco, H. Zhang, J. M. Kim, Y.-x. Shen, P. Yang, Nature Catalysis 2020, 3, 245. [DOI] [PubMed] [Google Scholar]
- [219].Zou Y, Pisciotta J, Billmyre RB, Baskakov IV, Biotechnology and Bioengineering 2009, 104, 939; M. Rosenbaum, Z. He, L. T. Angenent, Current Opinion in Biotechnology 2010, 21, 259. [DOI] [PubMed] [Google Scholar]
- [220].Buscemi G, Vona D, Trotta M, Milano F, Farinola GM, Advanced Materials Technologies 2022, 7, 2100245. [Google Scholar]
- [221].Lin X, Nishio K, Konno T, Ishihara K, Biomaterials 2012, 33, 8221. [DOI] [PubMed] [Google Scholar]
- [222].Logan BE, Rossi R, Ragab A. a., Saikaly PE, Nature Reviews Microbiology 2019, 17, 307. [DOI] [PubMed] [Google Scholar]
- [223].Logan BE, Nature Reviews Microbiology 2009, 7, 375. [DOI] [PubMed] [Google Scholar]
- [224].McKinlay JB, Zeikus JG, Applied Environmental Microbiology 2004, 70, 3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Mashkour M, Rahimnejad M, Mashkour M, Journal of Power Sources 2016, 325, 322. [Google Scholar]
- [226].Liu X-W, Huang Y-X, Sun X-F, Sheng G-P, Zhao F, Wang S-G, Yu H-Q, ACS Applied Materials & Interfaces 2014, 6, 8158. [DOI] [PubMed] [Google Scholar]
- [227].Adler JJS, Science 1966, 153, 708; L. Ricotti, B. Trimmer, A. W. Feinberg, R. Raman, K. K. Parker, R. Bashir, M. Sitti, S. Martel, P. Dario, A. J. S. R. Menciassi, Science Robotics 2017, 2. [DOI] [PubMed] [Google Scholar]
- [228].Rivera-Tarazona L, Bhat V, Kim H, Campbell Z, Ware T, Science Advances 2020, 6, eaax8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Chen X, Mahadevan L, Driks A, Sahin O, Nature Nanotechnology 2014, 9, 137. [DOI] [PubMed] [Google Scholar]
- [230].Studart AR, Chemical Society Reviews 2016, 45, 359; Z. Wang, Z. Wang, Y. Zheng, Q. He, Y. Wang, S. Cai, Science Advances 2020, 6, eabc0034. [DOI] [PubMed] [Google Scholar]
- [231].Mimee M, Tucker AC, Voigt CA, Lu TK, Cell Systems 2015, 1, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Ceroni F, Boo A, Furini S, Gorochowski TE, Borkowski O, Ladak YN, Awan AR, Gilbert C, Stan G-B, Ellis T, Nature Methods 2018, 15, 387. [DOI] [PubMed] [Google Scholar]
- [233].Ellis T, Wang X, Collins JJ, Nature Biotechnology 2009, 27, 465; A. J. Lopatkin, J. J. Collins, Nature Reviews Microbiology 2020, 18, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Camacho DM, Collins KM, Powers RK, Costello JC, Collins JJ, Cell 2018, 173, 1581; M. Baek, F. DiMaio, I. Anishchenko, J. Dauparas, S. Ovchinnikov, G. R. Lee, J. Wang, Q. Cong, L. N. Kinch, R. D. Schaeffer, Science 2021, 373, 871. [DOI] [PubMed] [Google Scholar]
- [235].Butler KT, Davies DW, Cartwright H, Isayev O, Walsh A, Nature 2018, 559, 547; Y. Mao, Q. He, X. Zhao, Science Advances 2020, 6, eaaz4169. [DOI] [PubMed] [Google Scholar]
- [236].Lee JW, Chan CTY, Slomovic S, Collins JJ, Nature Chemical Biology 2018, 14, 530. [DOI] [PubMed] [Google Scholar]
- [237].Rovner AJ, Haimovich AD, Katz SR, Li Z, Grome MW, Gassaway BM, Amiram M, Patel JR, Gallagher RR, Rinehart J, Isaacs FJ, Nature 2015, 518, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Maxmen A, Nature Medicine 2017, 23, 5; J. M. Anderson, A. Rodriguez, D. T. Chang, Seminars in Immunology 2008, 20, 86. [DOI] [PubMed] [Google Scholar]
- [239].Hansen SJZ, Morovic W, DeMeules M, Stahl B, Sindelar CW, Frontiers in Microbiology 2018, 9; F. D. Administration, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Moya-Ramirez I, Kotidis P, Marbiah M, Kim J, Kontoravdi K, Polizzi K, ACS Synthetic Biology 2022; L. Goers, P. Freemont, K. M. Polizzi, 2014, 11, 20140065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Baker MI, Walsh SP, Schwartz Z, Boyan BD, Journal of Biomedical Materials Research Part B: Applied Biomaterials 2012, 100, 1451; H. Kim, H. Jeong, S. Han, S. Beack, B. W. Hwang, M. Shin, S. S. Oh, S. K. Hahn, Biomaterials 2017, 123, 155. [DOI] [PubMed] [Google Scholar]
- [242].Sharp PA, Langer R, Science 2011, 333, 527. [DOI] [PubMed] [Google Scholar]


