Abstract
Phylogenetic analysis of tetracycline resistance genes encoding the ribosomal protection proteins (RPPs) revealed the monophyletic origin of these genes. The most deeply branching class, exemplified by tet and otrA, consisted of genes from the antibiotic-producing organisms Streptomyces rimosus and Streptomyces lividans. With a high degree of confidence, the corresponding genes of the other seven classes (Tet M, Tet S, Tet O, Tet W, Tet Q, Tet T, and TetB P) formed phylogenetically distinct separate clusters. Based on this phylogenetic analysis, a set of PCR primers for detection, retrieval, and sequence analysis of the corresponding gene fragments from a variety of bacterial and environmental sources was developed and characterized. A pair of degenerate primers targeted all tetracycline resistance genes encoding RPPs except otrA and tet, and seven other primer pairs were designed to target the specific classes. The primers were used to detect the circulation of these genes in the rumina of cows, in swine feed and feces, and in swine fecal streptococci. Classes Tet O and Tet W were found in the intestinal contents of both animals, while Tet M was confined to pigs and Tet Q was confined to the rumen. The tet(O) and tet(W) genes circulating in the microbiota of the rumen and the gastrointestinal tract of pigs were identical despite the differences in animal hosts and antibiotic use regimens. Swine fecal streptococci uniformly possessed the tet(O) gene, and 22% of them also carried tet(M). This population could be considered one of the main reservoirs of these two resistance genes in the pig gastrointestinal tract. All classes of RPPs except Tet T and TetB P were found in the commercial components of swine feed. This is the first demonstration of the applicability of molecular ecology techniques to estimation of the gene pool and the flux of antibiotic resistance genes in production animals.
Antibiotic resistance research has been and still is confined primarily to the study of cultivable bacterial isolates of mostly clinical origin. However, the cultivable isolates may represent only a fraction of the actual microbiota (1) where the antibiotic resistance genes reside. For example, no bacteria can be grown from more than 80% of all clinical samples sent to clinical microbiology laboratories (4), and certainly no antibiotic resistance profile can be determined if cultivation fails. Another important issue with antibiotic resistance is the fact that the wide use of antibiotics not only selects for drug-resistant pathogenic bacteria but also exerts selective pressure on the normal commensal microbiota. In light of the ubiquitously demonstrated phenomenon of horizontal antibiotic resistance gene transfer in the microbial world, the presence of such reservoirs may explain the rapid dissemination of antibiotic resistance from commensal organisms to the pathogenic microbiota. However, information regarding the antibiotic resistance pool in commensal microbiotas is very scarce, and the data are mostly phenotypical. Therefore, development of genotyping tools for detection and tracking of antibiotic resistance genes in a variety of commensal and pathogenic bacteria, as well as in the environment, is essential for understanding the ecology of antibiotic resistance.
One of the attractive models for studying the ecology of antibiotic resistance could be the genes conferring resistance to tetracyclines. Tetracyclines belong to a family of broad-spectrum antibiotics that includes tetracycline, chlortetracycline, doxycycline, and minocycline. These antibiotics inhibit protein synthesis in gram-positive and gram-negative bacteria by preventing the binding of aminoacyl-tRNA molecules to the 30S ribosomal subunit (36). Bacterial resistance to tetracycline is mediated mainly by two mechanisms, protection of ribosomes by large cytoplasmic proteins (5, 6, 23, 33, 43) and energy-dependent efflux of tetracycline (18, 33, 36). A third mechanism, enzymatic inactivation of tetracycline, is relatively uncommon and has been described in only one species (41). The first nomenclature for tetracycline resistance determinants was proposed in 1989 (17), and a recent update appeared in 1999 (19). The ribosomal protection mechanisms identified so far fall into six classes: Tet M, Tet O, TetB P, Tet Q, Tet S, and otrA (43). Almost all representatives of these classes have been sequenced and have been shown to encode proteins with N-terminal amino acid sequence similarity to translation elongation factors EF-Tu and EF-G (6, 22, 35, 43).
Since their introduction in the 1950s, tetracyclines have been widely used in human and veterinary medicine, as growth promoters in animal industry, and for prophylaxis in plant agriculture and aquaculture. At present, resistance to tetracyclines has spread to almost all bacterial genera, and this situation perhaps is the consequence of previous overuse. Among the ribosomal protection determinants, Tet M was described originally in streptococci (5, 24) and subsequently in a broad variety of gram-positive and gram-negative bacteria (32). The Tet S determinant was encountered first on a plasmid in the food pathogen Listeria monocytogenes (7), later in a number of Enterococcus faecalis strains (8), and recently on a plasmid of Lactococcus lactis isolated from raw milk (31). Tet O-related sequences were determined first in plasmids from campylobacteria (40, 42), then in streptococci (16, 45), and recently in a rumen bacterium, Butyrivibrio fibrisolvens (2). Finding nearly identical tet(Q) sequences in Prevotella ruminicola (a typical inhabitant of the rumen) and in Bacteroides (a typical inhabitant of the human gastrointestinal tract) (29) suggested that bacteria normally found in the guts of different species can exchange DNA, presumably during transient colonization of the animal intestine by human-associated bacteria or vice versa. This scenario is supported by recent work which described the occurrence of the tet(W) gene in the rumen and in human and pig intestinal microbiotas (37). Sequence analysis of this gene from taxonomically divergent rumen and human bacterial isolates showed that there was no or just one nucleotide substitution in a 1.25-kb amplified internal fragment. Misincorporation errors during amplification could not be ruled out, suggesting that the sequences may actually be identical in ruminal B. fibrisolvens, Selenomonas ruminantium, and Mitsuokella multiacidus isolates and in human isolates of Fusobacterium prausnitzii and Bifidobacterium longum (37). These findings demand that there be further research targeted at development of genotyping tools for tracking the movement of antibiotic resistance genes in the environment.
In this study, we initiated research to examine the molecular ecology of antibiotic resistance. As a model, we used tetracycline resistance genes encoding the ribosomal protection proteins (RPPs). Phylogenetic analysis revealed the monophyletic origin of these genes, which allowed us to design a set of PCR primers suitable for detection of RPP genes in general, as well as different classes. After validation, this set was used to detect the corresponding genes in the total DNA of swine fecal and rumen samples and in swine feed, as well as in fecal streptococcal isolates from pigs. The primers were also used in a PCR-denaturing gradient gel electrophoresis (DGGE) analysis to demonstrate the overall diversity and similarity of RPP genes in different ecosystems. The methods used in this work can be applied to study other phylogenetically coherent antibiotic resistance gene families.
MATERIALS AND METHODS
Phylogenetic analysis and primer design.
All currently available nucleotide sequences encoding RPPs, as well as the phylogenetically most closely related elongation factors, EF-Gs, were downloaded from the GenBank database (3). These included the sequences of the following RPP genes (the numbers in parentheses are GenBank accession numbers): E. faecalis DS16 tet(M) (M85225), E. faecalis Tn916 tet(M) (X56353), E. faecalis Tn1545 tet(M) (X04388), Neisseria meningitidis tet(M) (X75073), Ureaplasma urealyticum tet(M) (U08812), Gardnerella vaginalis tet(M) (U58986), Neisseria gonorrhoeae 6418 tet(M) (L12241), N. gonorrhoeae 2903 tet(M) (L12242), Staphylococcus aureus tet(M) (M21136), Streptococcus pneumoniae Tn5251 tet(M) (X90939), L. monocytogenes BM4210/pIP811 tet(S) (L09756), L. lactis K214/pK214 tet(S) (X92946), S. pneumoniae tet(O) (Y07780), Streptococcus mutans DL5 tet(O) (M20925), Campylobacter jejuni tet(O) (M18896), B. fibrisolvens tet(W) (AJ222769), Bacteroides fragilis tet(Q) (Z21523), Prevotella intermedia PDRC-11 tet(Q) (U73497), P. ruminicola tet(Q) (L33696), B. fragilis BF-2 tet(Q) (Y08615), Bacteroides thetaiotaomicron tet(Q) (X58717), Streptococcus pyogenes A498 tet(T) (L42544), Clostridium perfringens CW92 tetB(P) (L20800), Streptomyces lividans 1326 tet (M74049), and Streptomyces rimosus otrA (X53401). Elongation factor EF-G-encoding genes were obtained from Bacillus subtilis (D64127), E. faecalis (retrieved from www.tigr.org), Escherichia coli (X00415), Helicobacter pylori (AE001539), Thermus thermophilus (X16278), and Aquifex aeolicus (AE000669).
Sequences were aligned with the multiple-sequence alignment program CLUSTAL W (44). The two-parameter model of Kimura (14) was used for construction of neighbor-joining trees (34). The statistical significance of branching was evaluated by bootstrap analysis (11) involving the construction of 1,000 trees from resampled data. Sequences within clusters were separately aligned and compared with each other. PCR primers were designed to satisfy specificity and so that they could potentially be used in multiplex PCR with simultaneous coamplification of the V3 region of 16S ribosomal DNA (rDNA) (26) and in PCR-DGGE analysis (27). The nine sets of primers and the expected amplicon sizes are shown in Table 1. A GC clamp (CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGG) was added to the reverse primers for use in DGGE analysis (26). The primers used for amplification of the bacterial V3 region of 16S rDNA were the primers described previously (27).
TABLE 1.
PCR primers targeting the RPP classes
| Primer | Class targeted | Sequence | PCR annealing temp (°C) | Amplicon size (bp) |
|---|---|---|---|---|
| Ribo2-FWa | All except Otr A | GGMCAYRTGGATTTYWTIGC | Touchdownb | 1,187 |
| Ribo2-RV | TCIGMIGGIGTRCTIRCIGGRC | |||
| TetB/P-FW | TetB P | AAAACTTATTATATTATAGTG | 46 | 169 |
| TetB/P-RV | TGGAGTATCAATAATATTCAC | |||
| TetM-FW | Tet M | ACAGAAAGCTTATTATATAAC | 55 | 171 |
| TetM-RV | TGGCGTGTCTATGATGTTCAC | |||
| TetO-FW | Tet O | ACGGARAGTTTATTGTATACC | 60 | 171 |
| TetO-RV | TGGCGTATCTATAATGTTGAC | |||
| OTR-FW | Otr A | GGCATYCTGGCCCACGT | 66 | 212 |
| OTR-RV | CCCGGGGTGTCGTASAGG | |||
| TetQ-FW | Tet Q | AGAATCTGCTGTTTGCCAGTG | 63 | 169 |
| TetQ-RV | CGGAGTGTCAATGATATTGCA | |||
| TetS-FW | Tet S | GAAAGCTTACTATACAGTAGC | 50 | 169 |
| TetS-RV | AGGAGTATCTACAATATTTAC | |||
| TetT-FW | Tet T | AAGGTTTATTATATAAAAGTG | 46 | 169 |
| TetT-RV | AGGTGTATCTATGATATTTAC | |||
| TetW-FW | Tet W | GAGAGCCTGCTATATGCCAGC | 64 | 168 |
| TetW-RV | GGGCGTATCCACAATGTTAAC |
FW, forward; RV, reverse.
PCR conditions are described in Materials and Methods.
Environmental samples and DNA extraction.
Samples of whole rumen contents were obtained from eight fistulated steers maintained at the Research Farm of the University of Illinois at Urbana-Champaign. The animals used are treated with antibiotics only in case of disease, and no antibiotics are given for prophylaxis or growth promotion. No tetracyclines had been used for disease treatment in this group of animals, and the animals were considered free of tetracycline selective pressure. Fecal samples from six sows were collected at the Swine Research Farm of the University of Illinois at Urbana-Champaign. In this facility, in addition to therapeutic use, antibiotics are added to feed for prophylactic and growth-promoting purposes. The sows were routinely fed Tylan (Elanco Animal Health, Indianapolis, Ind.) at a concentration of 40 mg per kg of feed. The antibiotic was switched to chlortetracycline (400 mg per kg of feed) for 2 weeks and then back to Tylan. Fecal samples were collected 3 weeks after the switch back to Tylan. Rumen and fecal samples were frozen at −20°C for future isolation of total DNA. Samples of pig feed were stored at room temperature before DNA isolation. Total DNA was isolated from the fecal, rumen, and feed samples by using a Soil DNA Purification Kit (Mo Bio, Solana Beach, Calif.) according to the manufacturer's protocol.
Organisms, plasmids, and culture techniques.
The organisms and plasmids used in this study for validation and control are listed in Table 2. C. jejuni subsp. jejuni ATCC 43503 was grown at 37°C under microaerophilic conditions on ATCC medium 1115. C. perfringens JIR4202 was grown anaerobically at 37°C on BHIB medium (Difco Laboratories, Detroit, Mich.). S. pyogenes CIP105079 was grown aerobically on BHIB medium (Difco) at 37°C. E. coli strains were grown on Luria-Bertani medium at 37°C with aeration. Media were solidified when necessary with 1.8% (wt/vol) agar (Difco). Tetracycline (Sigma Chemical Co., St. Louis, Mo.) was added at a concentration of 10 μg/ml to cultures of C. jejuni, C. perfringens, and S. pyogenes. The cloned tet genes were maintained in E. coli by selection of plasmid antibiotic markers (ampicillin or kanamycin at a concentration of 50 μg/ml or chloramphenicol at a concentration of 20 μg/ml).
TABLE 2.
Characteristics of bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| C. jejuni subsp. jejuni | Tcr, tet(O) | ATCC 43503 |
| S. pyogenes A498 | Tcr, tet(T) | 9 |
| C. perfringens JIR4202 | Tcr, tetA(P) tetB(P) | 38 |
| Plasmids | ||
| pBT-1 | pJRD215 carrying a 2.5-kb SstI fragment containing the tet(Q) gene of TcrEmrDOT | 28 |
| pJIR667 | pPR328 carrying a 1.1-kb PstI-EcoRI internal fragment of the tetB(P) gene | 20 |
| pFD310 | Carries the tet(M) gene from S. agalactiae | 39 |
| pGEM-tetW | pGEM carrying a 2.4-kb PCR product with the tet(W) gene from B. fibrisolvens | 2 |
| pGEM-tetO | pGEM carrying the tet(O) gene from B. fibrisolvens | 2 |
| pVP2 | pUC18 carrying a 5.88-kb ClaI fragment of pK214 encompassing the tet(S) gene | 31 |
| pAT451 | pUC18 carrying a 4.5-kb ClaI fragment of pIP811 with the tet(S) gene | 7 |
| pCT10 | Contains the tet gene from S. lividans 1326 | 10 |
For isolation of fecal streptococci, fresh fecal samples from six pigs were resuspended in phosphate-buffered saline (pH 7.0), and dilutions were plated on MRS agar (Difco) plates. For detection of tetracycline resistance, colonies were transferred to the same medium with 10 μg of tetracycline per ml. The taxonomic affiliations of the resulting isolates were confirmed by sequencing 1.4-kb fragments of 16S rDNA amplified with bacterial primers (15). DNA similarity matrixes were calculated by using the DNADIST program in the PHYLIP package (12). Restriction fragment length polymorphism (RFLP) analysis was accomplished by AluI restriction digestion of amplified 1.4-kb fragments, followed by electrophoresis on a 3.0% agarose gel.
PCR and DGGE.
A typical PCR mixture (total volume, 20 μl) contained 25 pmol of each primer (except for the degenerate universal primers, which were used at a concentration of 100 pmol per mixture), 1× ExTaq reaction buffer (Takara Shuzo, Orsu, Japan), each deoxynucleoside triphosphate at a concentration of 100 μM, and 1.0 U of ExTaq DNA polymerase (Takara Shuzo). A 200-ng portion of purified DNA or one-half of the biomass of a 1- to 2-mm-diameter colony was used as a template. PCR amplification (25 cycles) was performed with a GeneAmp 2400 PCR system (Perkin-Elmer, Norwalk, Conn.) as follows: initial denaturation at 94°C for 5 min, followed by 25 cycles of 94°C for 30 s, 30 s of annealing at the annealing temperatures shown in Table 1, and 30 s of extension at 72°C, and a final extension step at 72°C for 7 min. A touchdown PCR with the degenerate Ribo2 primers (Table 1) was performed as follows: initial denaturation at 94°C for 5 min; 22 cycles of denaturation at 94°C for 30 s, annealing for 30 s with 1°C decrements at temperatures of 72 to 50°C, and extension at 72°C for 30 s; 20 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s; and final extension at 72°C for 7 min. If unidentified substances in a DNA preparation inhibited the PCR, 1 μl of the reaction mixture was used as a template for further amplification. Aliquots (5 μl) were analyzed by electrophoresis on a 2.5% (wt/vol) agarose gel (NuSieve; FMC Bioproducts, Rockland, Maine) containing the fluorescent dye GelStar (FMC Bioproducts). Gels with amplicons generated by the Ribo2 primers were stained with ethidium bromide.
For DGGE, polyacrylamide gels with urea-formamide gradients (8% acrylamide, 15 to 60% urea-formamide, 0.5× TAE buffer; pH 7.4) were polymerized on Gel-Bond support sheets (FMC Bioproducts). Electrophoresis was performed at 60°C and 150 V for 2 h and then at 200 V for 1 h by using the D-Gene System (Bio-Rad Laboratories, Richmond, Calif.). After electrophoresis, the gels were rinsed in double-distilled H2O, fixed in a solution containing 10% ethanol and 0.5% acetic acid, and silver stained. Gel images were captured and digitized with a Bio-Rad system that included a GS-710 calibrated imaging densitometer connected to a G3 Macintosh computer with the Diversity Database fingerprinting software. For cloning and sequencing of DGGE bands, the corresponding amplicons were excised from the gels and equilibrated in TE buffer at room temperature for 30 min, and 1 μl of the buffer with diffused DNA was used for reamplification.
For estimation of the proportion of antibiotic resistance-carrying microbiota, total DNA from the rumen and the standard strain [C. jejuni with tet(O)] were subjected to multiplex PCR [using amplification conditions for tet(O)] with primer sets TetO and V3 (targeting the V3 region of 16S rDNA). In this multiplex PCR, a C. jejuni DNA template was used at various dilutions. The gel images were digitized, and densitograms were generated with the NIH Image program (http://rsb.info.nih.gov/nih-image). Then the densitogram of the rumen multiplex PCR was compared to the range of C. jejuni amplification data. In lines having the same density of the TetO signal, the V3 signal intensities were compared for the total rumen DNA (all bacterial sequences amplified) and C. jejuni DNA. From this comparison, the approximate proportion of the suspected tet(O)-carrying bacteria was calculated.
Cloning and sequencing of PCR amplicons.
PCR products were cloned by using a TA Cloning kit (Invitrogen, Carlsbad, Calif.). White colonies of ampicillin-resistant transformants were screened for the presence of tet fragments by PCR by using the same primer set that was used for amplification. DNA sequence analysis of recombinant plasmids was performed for both strands (primers M13F and M13R) at the University of Illinois Biotechnology Center. On-line similarity searching was performed by using the BLAST (Basic Local Alignment Search Tool) family of programs in GenBank (21).
RESULTS
Phylogenetic analysis.
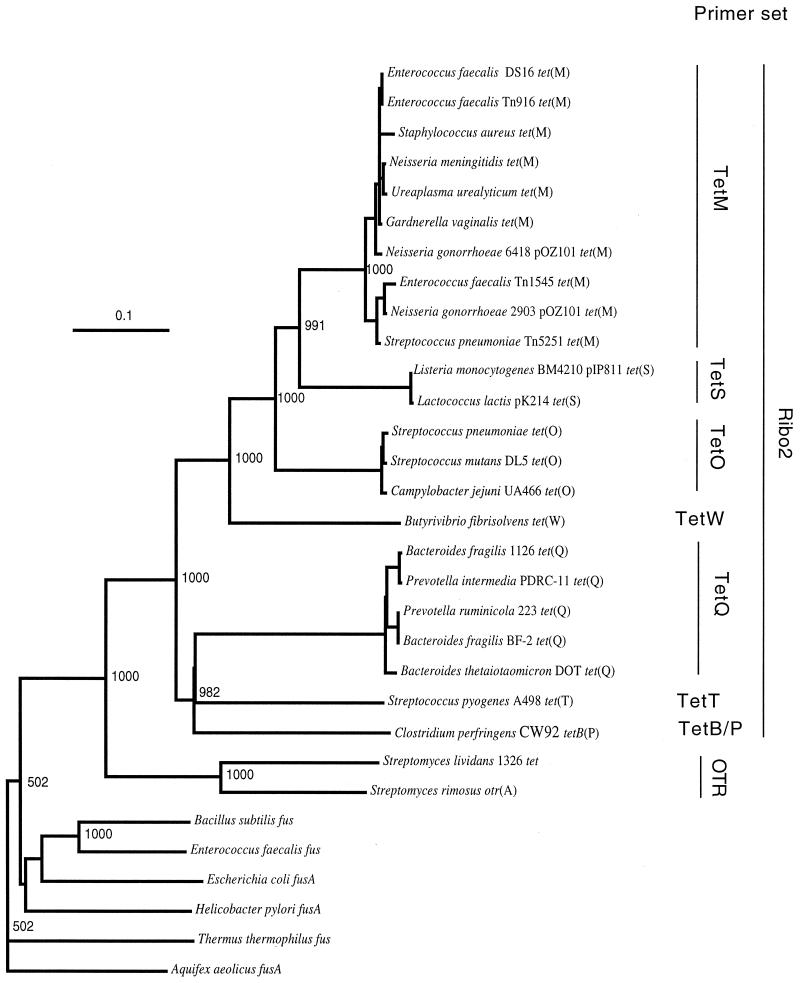
Phylogenetic analysis was performed with 25 complete nucleotide sequences encoding RPPs and with six complete sequences encoding phylogenetically closely related translation elongation factors belonging to family G (EF-G). With 100% bootstrap support, this analysis confirmed the monophyletic origin of RPP genes and the early branching from the other group of elongation factors, EF-G (Fig. 1). The number of substitutions per base pair was approximately 2.4 times higher in the RPP cluster than in the EF-G cluster. Within the RPP supercluster, there are eight clusters corresponding to the recently revised classes Tet M, Tet S, Tet O, Tet W, Tet Q, Tet T, TetB P, and otrA. Three of these clusters, Tet W, Tet T, and TetB P, are represented by only a single sequence at present.
FIG. 1.
Phylogenetic placement of tetracycline resistance genes encoding RPPs. The sequence of the A. aeolicus fusA gene for translation elongation factor EF-G was used as the outgroup to root the tree. The number at each node is the number of times that that tree configuration occurred in 1,000 bootstrap trials. The scale bar indicates 0.1 fixed nucleotide substitution per sequence position. The sets of PCR primers (Table 1) targeting various classes of RPP genes are shown on the right.
This analysis suggests that there was early branching between the RPP genes of antibiotic-producing strains, tet and otrA, and the other RPP genes circulating in pathogenic and saprophytic bacteria (Fig. 1). Thus, based on available sequence data, no evidence of recent horizontal transfer of RPP genes from antibiotic-producing strains to other bacteria exists at present. However, there is a high level of similarity, as shown by the extremely short branch lengths, among the sequences in taxonomically distantly related bacteria for classes Tet M, Tet S, Tet O, and Tet Q (Fig. 1).
Design and validation of PCR primers targeting RPP genes.
Evidence of the monophyletic origin of the RPP genes opened the possibility of designing primers that target all genes in the cluster. However, early branching and further independent diversification, together with the high G+C contents of the tet and otrA genes, precluded incorporation of these genes into the alignment analysis. Thus, the design of the universal primer pair was based on the sequences belonging to seven classes of RPP genes. As mentioned above, the rate of nucleotide substitution in the RPP cluster is higher than that in other elongation factors, and therefore, the overall sequence structure is less conserved. Because of this, the design of the universal primer pair involved a substantial level of degeneracy (Table 1). Primer pairs specific for the individual classes, together with the expected amplicon sizes, are shown in Table 1.
This set of primers was rigorously tested in PCR performed with DNA and colony biomasses of control strains (Table 3). In all cases except the tet gene with OTR primers, amplicons of the expected size were produced with positive controls. We suspect that the failure to amplify tet with OTR primers was due to structural instability of this gene on a high-copy-number plasmid. The gene was also shown to be structurally unstable in its original host, S. lividans (10). As expected, no signal was produced with the pJIR667 template when the universal and class-specific primer sets were used (Table 3). During a previous gene cloning procedure, the upstream region of the gene was deleted (20). The forward primers of the Ribo2 and TetB/P sets target this lost region, and no amplification is expected because of this. In all other cases, sequence analysis of PCR-generated amplicons from control strains confirmed the specificity of the primers and the identity of the amplified product. To simplify the detection procedure, biomasses from both colonies and liquid cultures were used for PCR, and these amplifications were also successful (Table 3).
TABLE 3.
Validation of PCR primers with control templates
| Template | Amplification with PCR primer sets
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ribo2 | TetB/P | TetM | TetO | OTR | TetQ | TetS | TetT | TetW | |
| Genomic or plasmid DNA | |||||||||
| C. jejuni subsp. jejuni [tet(O)] | + | − | − | + | − | − | − | − | − |
| S. pyogenes A498 [tet(T)] | + | − | − | − | − | − | − | + | − |
| C. perfringens JIR4202 [tetA(P) tetB(P)] | + | + | − | − | − | − | − | − | − |
| pBT-1 [tet(Q)] | + | − | − | − | − | + | − | − | − |
| pJIR667 [ΔtetB(P)] | − | − | − | − | − | − | − | − | − |
| pFD310 [tet(M)] | + | − | + | − | − | − | − | − | − |
| pGEM-tetW [tet(W)] | + | − | − | − | − | − | − | − | + |
| pGEM-tetO [tet(O)] | + | − | − | + | − | − | − | − | − |
| pVP2 [tet(S)] | + | − | − | − | − | − | + | − | − |
| pAT451 [tet(S)] | + | − | − | − | − | − | + | − | − |
| pCT10 [tet] | − | − | − | − | − | − | − | − | − |
| Cell biomass | |||||||||
| C. jejuni subsp. jejuni [tet(O)] | + | − | − | + | − | − | − | − | − |
| S. pyogenes A498 [tet(T)] | + | − | − | − | − | − | − | + | − |
| C. perfringens JIR4202 [tetA(P) tetB(P)] | + | + | − | − | − | − | − | − | − |
| E. coli(pBT-1) [tet(Q)] | + | − | − | − | − | + | − | − | − |
| E. coli(pJIR667) [ΔtetB(P)] | − | − | − | − | − | − | − | − | − |
| E. coli(pFD310) [tet(M)] | + | − | + | − | − | − | − | − | − |
| E. coli(pGEM-tetW) [tet(W)] | + | − | − | − | − | − | − | − | + |
| E. coli(pGEM-tetO) [tet(O)] | + | − | − | + | − | − | − | − | − |
| E. coli(pVP2) [tet(S)] | + | − | − | − | − | − | + | − | − |
| E. coli(pAT451) [tet(S)] | + | − | − | − | − | − | + | − | − |
| E. coli(pCT10) [tet] | − | − | − | − | − | − | − | − | − |
Detection of RPP genes in the rumen.
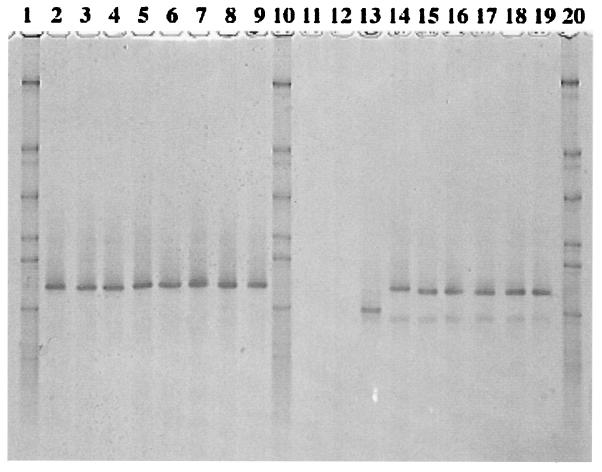
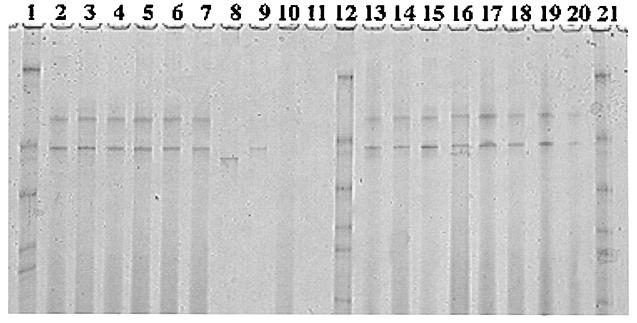
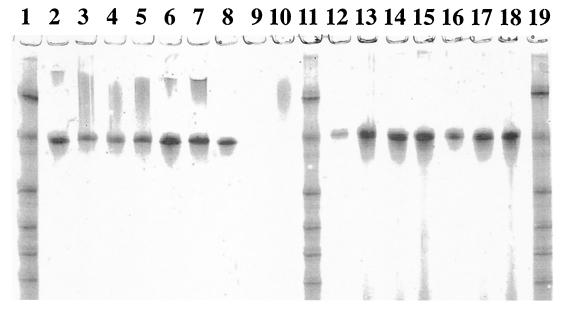
Total DNA preparations from the rumen samples were subjected to PCR amplification with the universal primer set, the Ribo2 set, followed by analysis with class-specific primers. Although the Ribo2 primer set performed well with pure cultures (Table 3), it was not sufficiently selective during amplification from total community DNA. According to the analysis with class-specific primers, the rumen microbiota appeared to bear the tet(O), tet(Q), and tet(W) genes (data not shown). The coamplified V3 region of 16S rDNA (data not shown) allowed a rough estimate of the proportion of the rumen bacterial microbiota carrying the corresponding resistance genes. Comparative analysis of densitograms by using biomass of C. jejuni as a standard suggested that up to 5% of ruminal bacteria may carry tet(O). The rumen samples were also subjected to PCR-DGGE analysis with the TetO, TetQ, and TetW primer sets, in which GC clamps were attached to the reverse primers. This analysis revealed the uniformity of the tet(W) and tet(O) genes circulating in the rumen microbiota of the cows (Fig. 2, lanes 2 through 9, and Fig. 3, lanes 13 through 20). DGGE bands from tet(W) samples were excised, reamplified, sequenced, and found to be identical to the tet(W) sequence (2). However, the DGGE band from this control template migrated farther than our samples (Fig. 2, lane 13), and careful inspection of the nucleotide sequence of the tet(W) control revealed a nucleotide substitution (T→C), which may have been incorporated during amplification from B. fibrisolvens (2). A DGGE analysis with the TetO primers produced similar results, with no variations among animals but with two bands (Fig. 3, lanes 13 through 20). Sequence analysis revealed that these two bands, which were in the region of tet(O), were actually identical, but for unknown reasons, the upper band also contained the reverse DGGE primer misincorporated at the 5′ end of the forward primer, thus increasing the melting temperature and creating artificial heterogeneity. A DGGE analysis with the TetQ primers revealed diversity among tet(Q) in the rumen, as well as animal-to-animal variation (Fig. 4). There were at least five bands, and sequence analysis confirmed the heterogeneity at the sequence level.
FIG. 2.
DGGE analysis of tet(W) amplicons from steer rumen and pig fecal samples. Lanes 1, 10, and 20, synthetic marker composed of known 16S rDNA sequences with various G+C contents; lanes 2 through 9, rumen samples from steers 277, 279, 280, 281, J277, J279, J280, and J281, respectively; lanes 11 and 12, negative controls pBT-1 [tet(Q)] and pJIR667 [ΔtetB(P)], respectively; lane 13, positive control pGEM-tetW [tet(W)]; lanes 14 through 19, fecal samples from pigs 1 through 6, respectively.
FIG. 3.
DGGE analysis of tet(O) amplicons from pig fecal and steer rumen samples. Lanes 1, 12, and 21, synthetic marker composed of known 16S rDNA sequences with various G+C contents; lanes 2 through 7, fecal samples from pigs 1 through 6, respectively; lanes 8 and 9, S. alactolyticus O19 and O31, respectively; lanes 10 and 11, negative controls pBT-1 [tet(Q)] and pJIR667 [ΔtetB(P)], respectively; lanes 13 through 20, rumen samples from steers 277, 279, 280, 281, J277, J279, J280, and J281, respectively.
FIG. 4.
DGGE analysis of tet(Q) amplicons from steer rumen samples. Lanes 1 and 11, synthetic marker composed of known 16S rDNA sequences with various G+C contents; lanes 2 through 9, rumen samples from steers 277, 279, 280, 281, J277, J279, J280, and J281, respectively; lane 10, positive control pBT-1 [tet(Q)].
Detection of RPP genes in swine feces.
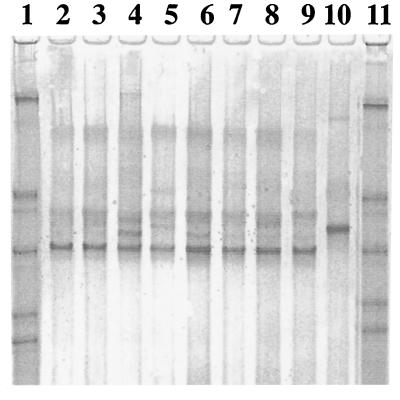
Total DNA preparations from swine fecal samples were subjected to PCR amplification with the universal primer set, the Ribo2 set, and also with class-specific primers. Tet M, Tet O, and Tet W determinants were detected in the swine intestinal microbiota (data not shown). Further PCR-DGGE analysis (Fig. 5) and sequencing demonstrated that swine tet(M) is identical to the control template, tet(M) cloned from Streptococcus agalactiae. Interestingly, the tet(O) and tet(W) genes circulating in the pig herd had the same mobility on DGGE gels as the corresponding genes from the rumina of steers (Fig. 2 and 3). Sequence analysis of the excised and cloned major DGGE bands confirmed that these two classes of genes were identical in the two types of animals. The difference was that swine fecal samples produced an additional minor tet(W) band migrating farther than the major band (Fig. 2, lanes 14 through 19). Several attempts to clone this minor band were unsuccessful, and sequence information for this band is not available.
FIG. 5.
DGGE analysis of tet(M) amplicons from pig fecal samples and streptococcal isolates. Lanes 1, 11, and 19, synthetic marker composed of known 16S rDNA sequences with various G+C contents; lanes 2 through 7, fecal samples from pigs 1 through 6, respectively; lane 8, positive control pFD310 [tet(M)]; lanes 9 and 10, negative controls pBT-1 [tet (Q)] and PCR mixture without a template, respectively; lanes 12 through 18, S. alactolyticus M15, M113, M118, M33, M35, M30, and M32, respectively.
Detection of RPP genes in fecal streptococci from swine.
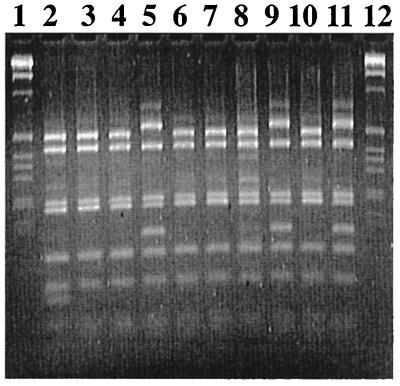
Fecal streptococcal isolates from swine (n = 150; 25 isolates from each of six animals) were characterized by RFLP and 16S rDNA sequence analyses. As determined by the RFLP analysis, these strains could be divided into at least three groups (Fig. 6). Sequence analysis allowed identification (sequence similarity, >99%) as strains of Streptococcus alactolyticus (Table 4). A majority (94.7%) of the isolates were resistant to tetracycline at a concentration of 10 μg/ml (Table 5). PCR analysis with our set of primers revealed that all of the resistant isolates carried the tet(O) gene (Table 5).Approximately 22% of the strains carried tet(M) in addition to tet(O). No other tetracycline resistance determinants conferring ribosomal protection were detected in these isolates (Table 5). Thus, the tetracycline-resistant swine S. alactolyticus populations were characterized by the invariable presence of tet(O), and 22% of the strains carried both tet(O) and tet(M).
FIG. 6.
RFLP analysis of swine S. alactolyticus isolates. Lanes 1 and 12, 1-kb ladder (Gibco BRL); lanes 2 through 11, isolates M15, M19, M113, M118, M33, M35, M310, M312, M321, and O31, respectively. The first group includes only M15; the second group includes M19, M113, M33, M35, M310, and M321; and the third group consists of M118, M312, and O31.
TABLE 4.
16S rDNA similarity matrix for swine streptococcal isolates
| rDNA source | % Similarity to rDNA of:
|
||||
|---|---|---|---|---|---|
| M15 | O31 | M118 | M19 | S. alactolyticus | |
| M15 | 100 | ||||
| O31 | 99.54 | 100 | |||
| M118 | 99.82 | 99.54 | 100 | ||
| M19 | 99.82 | 99.56 | 99.82 | 100 | |
| S. alactolyticus | 99.82 | 99.56 | 99.82 | 99.82 | 100 |
TABLE 5.
Tetracycline resistance phenotypes and genotypes of swine fecal streptococci
| Animal no. | No. of isolates | No. of resistant phenotypes | No. of strains analyzed | No. of resistant genotypes
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| tetB(P) | tet(M) | tet(Q) | tet(O) | tet(S) | tet(T) | tet(W) | ||||
| 1 | 25 | 20 (80)a | 19 | NAb | 4 (21.1) | 0 | 19 (100) | NA | NA | NA |
| 2 | 25 | 25 (100) | 18 | NA | NA | NA | 18 (100) | NA | NA | NA |
| 3 | 25 | 23 (92) | 22 | 0 | 5 (22.7) | NA | 22 (100) | NA | NA | 0 |
| 4 | 25 | 25 (100) | NA | NA | NA | NA | NA | NA | NA | NA |
| 5 | 25 | 24 (96) | 22 | NA | NA | NA | NA | NA | 0 | NA |
| 6 | 25 | 25 (100) | 22 | NA | NA | NA | NA | 0 | NA | NA |
| Total | 150 | 142 (94.7) | 103 | 0 | 9 (21.9) | 0 | 59 (100) | 0 | 0 | 0 |
The values in parentheses are percentages.
NA, not analyzed.
DGGE and sequence analyses of the amplified tet(M) fragments from streptococcal strains demonstrated that these fragments were identical (Fig. 5, lanes 12 through 18). Moreover, they were identical to the fragments amplified directly from the total DNA, as well as to the control template (Fig. 5). The control template, tet(M), was originally cloned from S. agalactiae, and its 16S rDNA sequence was 97% similar to those of our streptococcal isolates. Apparently, in our herd S. alactolyticus populations could be considered one of the main reservoirs of the tet(M) gene in the swine intestinal microbiota.
DGGE analysis of streptococcal tet(O) revealed some degree of heterogeneity. In particular, the amplicons from two isolates, O19 and O35, migrated farther through DGGE gels than the amplicons from nine other streptococci migrated (the results for O19 are shown in lane 8 of Fig. 3). Sequence analysis of these two amplicons revealed a single A→G substitution (but the locations were different). However, as with tet(M), the majority of tet(O) amplicons (as exemplified by S. alactolyticus O31 in Fig. 3) had the same melting characteristics as the amplicons amplified from the total swine fecal DNA (Fig. 3). Because of the universal presence of tet(O), S. alactolyticus populations could be considered one of the main reservoirs of the tet(O) gene in the swine intestinal microbiota. In addition, the TetO-generated DGGE bands from swine streptococcal isolates had the same mobility on DGGE gels as the bands from rumen samples (Fig. 3). The occurrence of tet(O) in cultivable rumen bacteria was not studied, and it is not clear in which part of the rumen microbiota the gene resides. As in pig samples, the organisms containing the gene may be the ruminal streptococci, which have been shown to possess transferable tetracycline resistance (13).
Detection of RPP genes in swine feed.
Because of the presence of unidentified inhibitory substances, a second round of PCR was necessary in the experiments performed with swine feed, and therefore, the detection limit of this assay was lower than that of the assay performed with the fecal and rumen samples. The presence of bacterial DNA in all premix and mixed samples was confirmed by amplification of the V3 region of bacterial 16S rDNA (Table 6). The presence of RPP genes in these samples was confirmed first with the Ribo2 primer set and then with class-specific primers (Table 6). First, the feed components were sampled before the corresponding diet mixes were prepared for three different age groups. These groups were the starters (ages, 3 to 6 weeks), growers (6 weeks to 6 months), and finishers (antibiotics were withdrawn before slaughtering). The corn component used to prepare the mixes for all age groups contained tet(W), tet(O), tet(Q), and tet(M), while the soybean component also contained the tet(S) gene (Table 6). The resistance gene profiles of the commercial whey preparation and the protein plasma product were similar to that of the soybean component. Interestingly, the commercial preparation of Tylan (a macrolide which was used in the grower diet) also contained tetracycline resistance genes with a profile similar to that of the corn component (Table 6). The antibiotic mixture used for the starter group (chlortetracycline, sulfonamide, and penicillin) contained DNA of tetracycline resistance genes, particularly that of tet(W), tet(O), tet(Q), tet(M), and tet(S), and had a profile similar to those of the soybean, whey, and plasma product components (Table 6).
TABLE 6.
Detection of RPP genes in swine feed and feed components
| Sample | Detection with primer sets
|
|||||||
|---|---|---|---|---|---|---|---|---|
| V3 region | TetB/P | TetM | TetQ | TetO | TetS | TetT | TetW | |
| Corn | + | − | + | + | + | − | − | + |
| Soybean | + | − | + | + | + | + | − | + |
| Whey | + | − | + | + | + | + | − | + |
| Plasma protein | + | − | + | + | + | + | − | + |
| CSP | + | − | + | + | + | + | − | + |
| Tylan | + | − | + | + | + | − | − | + |
| Starter mix (freshly prepared) | + | − | + | + | + | + | − | + |
| Starter mix (inside barn) | + | − | + | + | + | + | − | + |
| Grower mix (freshly prepared) | + | − | + | + | + | − | − | + |
| Grower mix (inside barn) | + | − | + | + | + | − | − | + |
| Finisher mix (freshly prepared) | + | − | + | + | + | − | − | + |
| Finisher mix (inside barn) | + | − | + | + | + | − | − | + |
A second set of samples was taken from fresh mixes and the feed mixes inside the barns to test the possibility that there was cross-contamination of the feed inside the barns. However, since the food components and the mixes already contained the resistance genes circulating in the pig gut microbiota [tet(M), tet(O), and tet(W)], it was not possible to test this contamination effect, and there was no difference between the antibiotic resistance profiles of the freshly prepared feed mix and the mix obtained inside the barns (Table 6). Interestingly, the tet(S) signal, which was detected in the soybean component, disappeared in the mixes used for the grower and finisher stages. Also, the finisher diet, which was free of any antibiotics, contained the resistance genes that supposedly came from the corn and soybean components (Table 6).
DISCUSSION
This work was the first attempt to use the molecular ecology approach to study antibiotic resistance and, in particular, to estimate the gene pool and flux of antibiotic resistance genes in production animals. With this approach, the first step is elucidation of the evolutionary history of the genes of interest. From the phylogenetic analysis, it is evident that the elongation factors conferring resistance to tetracycline form, with a high degree of confidence, a phylogenetically coherent group separated from other elongation factors. Within this group, there are eight clusters, which correspond to the eight currently defined classes of RPPs (Tet M, Tet S, Tet O, Tet W, Tet Q, Tet T, TetB P, and otrA).
The most deeply branching class, exemplified by tet and otrA, is the class obtained from the antibiotic-producing organisms S. lividans and S. rimosus. Based on the sequence information available, there is no evidence of recent horizontal transfer of RPP genes from antibiotic-producing strains to commensal or pathogenic microbiotas. Hybridization data indicate, however, that some mycobacteria may actually carry the resistance genes originally described in streptomycetes (30). Additional sequence information concerning the mycobacterial RPP genes is required to decide whether there was a potential horizontal transfer event from antibiotic-producing strains. Another interesting aspect of the two available gene sequences of antibiotic-producing streptomycetes is that they are quite divergent. The length of the branch between the two genes is actually comparable to the length of the branch separating the Tet M, Tet S, and Tet O classes (Fig. 1). If more sequence data from this class of genes were available, perhaps definition of at least two new classes would be necessary.
The available sequence data support the scenario that early branching and lengthy independent diversification of eight (or more) clusters of RPPs occurred well before the “antibiotic era.” While the functional role of these proteins in antibiotic-producing bacteria is evident (they provide protection against the synthesized antibiotics), it is more challenging to explain their presence and function in bacteria from other ecological niches that have no or limited contact with the soil microbiota (e.g., the gastrointestinal tract). The long evolutionary history of RPP genes supports the hypothesis that these genes might have served some metabolic functions other than providing antibiotic resistance. Protein synthesis is a vital cell process, and there should be mechanisms that support proper functioning of the translation machinery and buffer possible undesirable effects of low-molecular-weight metabolites of the cell. Thus, the alternative elongation factors may have been selected in this way in some bacteria and may have assumed a role in protecting ribosomes against tetracyclines only recently.
At the same time, the rapid movement of the tetracycline-resistant elongation factors to taxonomically divergent commensal and pathogenic bacteria is a very recent evolutionary event on the phylogenetic time scale and can most probably be attributed to horizontal transfers within the clusters in the antibiotic era. Some of the genes are located on plasmids (e.g., pOZ101, pIP811, or pK214) or conjugative transposons (e.g., Tn916, Tn5251, or Tn1545), thus facilitating transfer between species and genus boundaries.
Proof of the monophyletic origin of the RPP genes opened the possibility of designing primer sets targeting all classes, as well as class-specific primers. However, it appeared that the early branching and further independent diversification of the otrA genes, together with a high G+C content, precluded incorporation of these genes into the alignment. Also, the number of substitutions per base pair appeared to be higher in the RPP genes than in other elongation factors, and therefore, the overall sequence structure is less conserved. Thus, the overall design of the universal primer pair involves a substantial level of degeneracy and does not include the genes from the antibiotic-producing streptomycetes. Primers were validated in PCR that included crude bacterial biomass and fecal material, which is notorious for the presence of PCR-inhibiting substances. This fact could be useful for rapid screening for the presence of the RPP genes in bacteria without a DNA isolation step. The primers also were designed to amplify short sequences, thus allowing use in PCR-DGGE analysis. Therefore, such an analysis could be performed with total DNA preparations of environmental origin, thus allowing for the first time access to the pool and diversity of RPP genes in a given ecosystem.
The primers were used to detect the occurrence of RPP genes in the rumina of cows, in swine feed and feces, and in swine fecal streptococci. The Tet O and Tet W determinants were found in the intestinal contents of both types of animals, while Tet M was confined to pigs and Tet Q was confined to the rumen. Approximate estimates suggest that up to 5% of the bacteria in the rumen and swine intestine may carry the tet(O) gene. Another interesting observation is that tet(W) and the majority of the tet(O) genes circulating in the two different animal herds, which had very different antibiotic use regimens, were actually identical. The identity of the tet(W) genes obtained from bovine and ovine rumen and human intestinal isolates was demonstrated in a recent study (37). Obviously, this finding could be extended to include yet another animal model (pig) and another gene [tet(O)]. The occurrence of identical tetracycline resistance genes in different hosts provides additional evidence that there are extensive pools of antibiotic resistance genes that are actively exchanged at least between domestic animals. However, genetic transfer itself is not a guarantee that the transferred antibiotic resistance gene will be maintained in another host. The second observation concerning the persistence of antibiotic resistance in the apparent absence of antibiotic selective pressure [cattle that have no antibiotic in their feed but carry intestinal bacteria with tet(O), tet(Q), and tet(W) and swine feed containing a diverse group of resistance genes] raises the question of how resistance persists. The possession of an antibiotic resistance gene by a bacterium is certainly advantageous in the presence of the corresponding antibiotic. In the absence of the antibiotic, however, the cost of carrying of the resistance gene should reduce the bacterial fitness and the resistant phenotype should be replaced by the sensitive phenotype. However, a recent reexamination of this topic suggested that bacteria may have been able to adapt to the burden of resistance with little or no cost to their fitness (25). In this scenario, the antibiotic-resistant microbiota would successfully compete with the sensitive counterpart even in the absence of selection. Such adaptations would preclude resistant lineages from reverting to sensitivity and make control of antibiotic resistance even more difficult.
The significant outcome of the sequence analysis of tetracycline resistance genes in cultivable streptococcal isolates is that the nucleotide sequences of tet(M) and the majority of tet(O) genes are identical to those of the corresponding genes acquired directly from fecal DNA. This is yet another validation of the in vitro analysis approach and suggests that the pool of resistance genes, initially discovered in total DNA, could be tracked to specific bacterial populations in the gut. In our case, S. alactolyticus could be considered one of the main reservoirs of the tet(M) and tet(O) genes in the swine intestinal microbiota. Based on RFLP and sequence analyses of 16S rDNA of S. alactolyticus isolates, this is not a clonal population but is represented by at least three subpopulations. Therefore, circulation of identical tet(M) and tet(O) genes in this genetically diverse group of bacteria suggests that there is horizontal exchange of tetracycline resistance genes rather than coexistence of several tetracycline-resistant clones.
Compared with the rumen and fecal samples, the components of the swine feed appeared to be contaminated with a more diverse group of RPPs, and only two classes (Tet T and TetB P) were absent. No attempt to isolate resistant bacteria was made, but the ubiquitous presence of these genes, together with the bacterial V3 markers, suggests that the feed components may have been contaminated by bacteria carrying the corresponding resistance genes. It is not clear whether these bacteria were dead or viable; regardless, the feed was genetically contaminated. The experiments were designed to detect possible cross-contamination of the swine feed by on-farm dust and fecal material, but it appeared that the components of swine feed already carried more diverse markers of tetracycline resistance, including that in the swine gut microbiota. This suggests that the actual source of antibiotic resistance gene contamination of swine feed was something else and requires further independent research. At this time, we hypothesize that at least for the corn and soybean components the source may have been manure from farms on which antibiotics were used, which was applied to the land. Whey, a by-product of cheese manufacturing, may contain a residual biomass of tetracycline-resistant lactic acid bacteria and propionobacteria. However, we have no information concerning the source of antibiotic resistance gene contamination in other components of the swine feed, such as the plasma protein and especially the antibiotic preparations, which are supposedly the products of sterile fermentation.
In this study molecular ecology tools were used to study the antibiotic resistance problem, and the results suggest that this approach has the potential of uncovering the reservoirs and determining the identities of antibiotic resistance genes in a variety of ecosystems. This approach could be easily extended to other classes of antibiotic resistance genes in order to understand the pathways leading to acquisition of drug resistance by human- and animal-pathogenic bacteria.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbosa T M, Scott K P, Flint H J. Evidence for recent intergenic transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O), in ruminal bacteria. Environ Microbiol. 1999;1:53–64. doi: 10.1046/j.1462-2920.1999.00004.x. [DOI] [PubMed] [Google Scholar]
- 3.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F, Rapp B A, Wheeler D L. GenBank. Nucleic Acids Res. 1999;27:12–17. doi: 10.1093/nar/27.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergeron M G, Quellette M. Diagnosing bacterial infectious diseases in one hour: an essential upcoming revolution. Infection. 1995;23:69–72. doi: 10.1007/BF01833867. [DOI] [PubMed] [Google Scholar]
- 5.Burdett V. Streptococcal tetracycline resistance mediated at the level of protein synthesis. J Bacteriol. 1986;165:564–569. doi: 10.1128/jb.165.2.564-569.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burdett V. Purification and characterization of Tet(M), a protein that renders ribosomes resistant to tetracycline. J Biol Chem. 1991;266:2872–2877. [PubMed] [Google Scholar]
- 7.Charpentier E, Gerbaud G, Courvalin P. Characterization of a new class of tetracycline-resistance gene tet(S) in Listeria monocytogenes BM4210. Gene. 1993;131:27–34. doi: 10.1016/0378-1119(93)90665-p. [DOI] [PubMed] [Google Scholar]
- 8.Charpentier E, Gerbaud G, Courvalin P. Presence of the Listeria tetracycline resistance gene tet(S) in Enterococcus faecalis. Antimicrob Agents Chemother. 1994;38:2330–2335. doi: 10.1128/aac.38.10.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clermont D, Chesneau O, De Cespedes G, Horaud T. New tetracycline resistance determinants coding for ribosomal protection in streptococci and nucleotide sequence of tet(T) isolated from Streptococcus pyogenes A498. Antimicrob Agents Chemother. 1997;41:112–116. doi: 10.1128/aac.41.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dittrich W, Schrempf H. The unstable tetracycline resistance gene of Streptomyces lividans 1326 encodes a putative protein with similarities to translational elongation factors and Tet(M) and Tet(O) proteins. Antimicrob Agents Chemother. 1992;36:1119–1124. doi: 10.1128/aac.36.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.53c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 13.Jonecova Z, Marekova M, Kmei V. Conjugative transfer of tetracycline resistance in rumen streptococcal strains. Folia Microbiol. 1994;39:83–86. doi: 10.1007/BF02814537. [DOI] [PubMed] [Google Scholar]
- 14.Kimura M. A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 15.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- 16.LeBlanc D J, Lee L N, Titmas B M, Smith C J, Tenover F C. Nucleotide sequence analysis of tetracycline resistance gene tet(O) from Streptococcus mutans DL5. J Bacteriol. 1998;170:3618–3626. doi: 10.1128/jb.170.8.3618-3626.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy S B, McMurry L M, Burdett V, Courvalin P, Hillen W, Roberts M C, Taylor D E. Nomenclature for tetracycline resistance determinants. Antimicrob Agents Chemother. 1989;33:1373–1374. doi: 10.1128/aac.33.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy S B. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992;36:695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy S B, McMurry L M, Barbosa T M, Burdett V, Courvalin P, Hillen W, Roberts M C, Rood J I, Taylor D E. Nomenclature for new tetracycline resistance determinants. Antimicrob Agents Chemother. 1999;43:1523–1524. doi: 10.1128/aac.43.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyras D, Rood J I. Genetic organization and distribution of tetracycline resistance determinants in Clostridium perfringens. Antimicrob Agents Chemother. 1996;40:2500–2504. doi: 10.1128/aac.40.11.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madden T L, Tatusov R L, Zhang J. Application of network BLAST server. Methods Enzymol. 1996;266:131–141. doi: 10.1016/s0076-6879(96)66011-x. [DOI] [PubMed] [Google Scholar]
- 22.Manavathu E K, Hiratsuka K, Taylor D E. Nucleotide sequence analysis and expression of a tetracycline resistance gene from Campylobacter jejuni. Gene. 1988;62:17–26. doi: 10.1016/0378-1119(88)90576-8. [DOI] [PubMed] [Google Scholar]
- 23.Manavathu E K, Fernandez C L, Cooperman B S, Taylor D E. Molecular studies on the mechanism of tetracycline resistance mediated by Tet(O) Antimicrob Agents Chemother. 1990;34:71–77. doi: 10.1128/aac.34.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin P, Trieu-Cuot P, Courvalin P. Nucleotide sequence of the Tet M tetracycline resistance determinant of the streptococcal conjugative shuttle transposon Tn1545. Nucleic Acids Res. 1986;14:7047–7058. doi: 10.1093/nar/14.17.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris A, Kellner J D, Low D E. The superbugs: evolution, dissemination and fitness. Curr Opin Microbiol. 1998;1:524–529. doi: 10.1016/s1369-5274(98)80084-2. [DOI] [PubMed] [Google Scholar]
- 26.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muyzer G, Hottentrager S, Teske A, Wawer C. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA—a new molecular approach to analyze the genetic diversity of mixed microbial communities. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 3.4.4.1–3.4.4.22. [Google Scholar]
- 28.Nikolich M P, Shoemaker N B, Salyers A A. A Bacteroides tetracycline resistance gene represents a new class of ribosome protection tetracycline resistance. Antimicrob Agents Chemother. 1992;36:1005–1012. doi: 10.1128/aac.36.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikolich M P, Hong G, Shoemaker N B, Salyers A A. Evidence for the horizontal transfer of tetQ between bacteria that normally colonize humans and bacteria that normally colonize livestock. Appl Environ Microbiol. 1994;60:3255–3260. doi: 10.1128/aem.60.9.3255-3260.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang Y, Brown B A, Steingrube V A, Wallace R J, Jr, Roberts M C. Tetracycline resistance determinants in Mycobacterium and Streptomyces species. Antimicrob Agents Chemother. 1994;38:1408–1412. doi: 10.1128/aac.38.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perreten V, Schwarz F, Cresta L, Boeglin M, Dasen G, Teuber M. Antibiotic resistance spread in food. Nature. 1997;389:801–802. doi: 10.1038/39767. [DOI] [PubMed] [Google Scholar]
- 32.Roberts M C. Epidemiology of tetracycline-resistance determinants. Trends Microbiol. 1994;2:353–357. doi: 10.1016/0966-842x(94)90610-6. [DOI] [PubMed] [Google Scholar]
- 33.Roberts M C. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev. 1996;19:1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 34.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez-Pescador R, Brown J T, Roberts M C, Urdea M S. Homology of the Tet(M) with translational elongation factors: implication for potential modes of tet(M)-conferred tetracycline resistance. Nucleic Acids Res. 1988;16:1218. doi: 10.1093/nar/16.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnappinger D, Hillen W. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch Microbiol. 1996;165:359–369. doi: 10.1007/s002030050339. [DOI] [PubMed] [Google Scholar]
- 37.Scott K P, Melville C M, Barbosa T M, Flint H J. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob Agents Chemother. 2000;44:775–777. doi: 10.1128/aac.44.3.775-777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sloan J, McMurray L M, Lyras D, Levy S B, Rood J I. The Clostridium perfringens Tet P determinant comprises two overlapping genes: tetA(P), which mediates active tetracycline efflux, and tetB(P), which is related to the ribosomal protection family of tetracycline-resistance determinants. Mol Microbiol. 1994;11:403–415. doi: 10.1111/j.1365-2958.1994.tb00320.x. [DOI] [PubMed] [Google Scholar]
- 39.Smith C J, Rogers M B, McKee M L. Heterologous gene expression in Bacteroides fragilis. Plasmid. 1992;27:141–154. doi: 10.1016/0147-619x(92)90014-2. [DOI] [PubMed] [Google Scholar]
- 40.Sougakoff W, Papadopoulou B, Nordman P, Courvalin P. Nucleotide sequence and distribution of tet(O) gene encoding tetracycline resistance in Campylobacter coli. FEMS Microbiol Lett. 1987;44:153–159. [Google Scholar]
- 41.Speer B S, Salyers A A. Novel aerobic tetracycline resistance gene that chemically modifies tetracycline. J Bacteriol. 1989;171:148–153. doi: 10.1128/jb.171.1.148-153.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor D E, Hiratsuka K, Ray H, Manavathu E K. Characterization and expression of a cloned tetracycline resistance determinant from Campylobacter jejuni plasmid pUA466. J Bacteriol. 1987;169:2984–2989. doi: 10.1128/jb.169.7.2984-2989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor D E, Chau A. Tetracycline resistance mediated by ribosomal protection. Antimicrob Agents Chemother. 1996;40:1–5. doi: 10.1128/aac.40.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Widdowson C A, Klugman K P, Hanslo D. Identification of the tetracycline resistance gene, tet(O), in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:2891–2893. doi: 10.1128/aac.40.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]