Abstract
As in other Diptera, the telomeres of Chironomus thummi lack canonical short telomerase-specified repeats and instead contain complex sequences. They react to heat shock and other stress treatments by forming giant puffs at some chromosome termini, which are visible in polytene cells. All telomeres, except the telocentric end of chromosome four (4L), consist of large blocks of repeats, 176 bp in length. Three subfamilies of telomeric sequences have been found to show different distribution patterns between chromosome ends. TsA and TsC are characteristic of telomeres 3R and 4R, respectively, whereas TsB is present in the other non-telocentric telomeres. Heat shock transcription regulatory elements have been identified in the telomeric sequences, appearing differentially represented in the three subfamilies, but otherwise rather similar in size and sequence. Interestingly, TsA and TsB repeats share the well-conserved heat shock element (HSE) and GAGA motif, while the TATA box is only present in the former. Neither a HSE nor a TATA box appear in TsC repeats. Moreover, experimental data indicate that the HSE is functionally active in binding heat shock transcription factor (HSF). These results provide, for the first time, a molecular basis for the effect of heat shock on C.thummi telomeres and might also explain the different behaviour they show. A positive correlation between the presence of HSE and telomeric puffing and transcription under heat shock was demonstrated. This was also confirmed in the sibling species Chironomus piger. The significance of heat shock activation of telomeric repeats in relation to telomeric function is unknown at present, but it might be compared to the behaviour of other non-heat shock protein coding sequences, such as SINE-like and LINE-like retroelements, which have been reported to be activated by stress.
INTRODUCTION
In most organisms studied telomeric DNA consists of short (5–26 bp) tandem repeats maintained by telomerase, a special cellular reverse transcriptase responsible for RNA-templated regeneration of telomeres to counteract the loss of sequences due to incomplete replication at the DNA termini (reviewed in 1,2). However, there is increasing evidence for alternative telomere structures and different mechanisms for telomere maintenance. Telomeres in Drosophila melanogaster and its close relatives are elongated by retrotransposition of the terminus-specific HeT-A and TART non-LRT transposons (reviewed in 3). Telomeres with long complex tandem repeats have been found in other Diptera such as Chironomus (4,5), Anopheles (6) and, more recently, in Drosophila virilis and its sibling species (7). The same has also been found in some plants, for example members of the genera Allium and Aloe (8,9), and in the ciliate linear mitochondrial chromosome (10). These organisms may somehow represent an intermediate situation between short telomeric repeats and the long D.melanogaster retrotransposons, and deserve more detailed analysis. Chironomus telomeres offer an interesting model since different species have been extensively studied. In Chironomus pallidivitatus, 340 bp repeats reach the very end of the chromosome (11). Similar sequences, likely to be terminal, have been found in Chironomus thummi (176 bp), Chironomus tentans (350 bp) and Chironomus dilutus (341 bp) (4,12). Chironomid telomeres might regenerate DNA by a process different to that already characterised. In chironomids, telomeric repeats are transcribed (4,5) and large telomeric transcripts have been found in salivary glands and other larval tissues (13). Support for RNA-templated regeneration is found in the presence of reverse transcriptase-like activity in telomeric areas (14).
Chironomus thummi telomeres are particularly interesting since they may react to heat shock and other stress treatments by inducing terminal puffs. At telomere 3R, a prominent Balbiani ring-shaped puff named TBR is formed in polytene chromosomes. In cytological studies, TBRs show signs of transcriptional activation involving telomeric repeats. Upon induction, TBR binds specific factors such as Ku (15), Hsp90 (16), heat shock transcription factor (HSF) (13) and reverse transcriptase (14), an indication of the complex processes at this telomeric structure. However, five non-telocentric telomeres react weakly to heat shock. Finally, the 4R telomere and the 4L telocentromere seem to be refractory to heat shock. The significance of this particular behaviour of C.thummi telomeres, in which the heat shock response appears to be associated with telomeric organisation and/or function, is still to be elucidated. Recently, we described three different subfamilies of telomeric repeats (17). In the present investigation, the telomeric organisation of C.thummi was further characterised and a sequence showing high homology to the heat shock element (HSE) in some subfamilies of repeats was found. This element, highly represented in the telomeric blocks, was able to bind specifically to HSF in vitro. The existence of telomeric HSE, along with other regulatory elements such as TATA and GAGA boxes, may explain why C.thummi telomeres react to heat shock and other stress treatments.
MATERIALS AND METHODS
Animals
The experimental animals were fourth instar larvae of C.thummi, Chironomus piger and C.thummi × C.piger hybrids. These were raised under standard conditions at 18°C. Larvae were heat shocked at 35°C for 1 or 2 h in a preheated and aerated culture medium.
Immunocytochemistry
Salivary glands were isolated, fixed and stained with carmine:orcein as previously described (17). Squashes fixed in ethanol:acetic acid (3:1) were used for DNA–RNA detection. These were rehydrated and denatured for 10 min at 40°C in 50% acetic acid. Slides were then blocked for 10 min at room temperature in phosphate-buffered saline (PBS), 0.1% Tween-20 (PT) and 2% low fat powdered milk and incubated for 1 h at room temperature with anti-DNA–RNA hybrid IgG antibodies. After two washes in PT they were then incubated with anti-goat IgGs conjugated with fluorescein isothiocyanate (FITC) (Sigma). Squashes fixed in ethanol:acetic acid (3:1) containing 3.7% formaldehyde were used for HSF detection. These were rehydrated and incubated with anti-HSF IgG (18) diluted in PT with 1% blocking reagent (Roche) for 1 h at room temperature. After washing in PT, the secondary antibody used was anti-rabbit IgG conjugated with FITC (Sigma). Slides were finally mounted in 9:1 glycerol:10× PBS with 1 mg/ml p-phenylene diamine and observed under a Zeiss photomicroscope equipped with a CCD camera. Images were captured using a 1P-lab spectrum program and processed with Adobe PhotoShop 4.0.
DNA extraction and Southern blotting
Genomic DNA was extracted from 100 frozen larvae as described (17). Genomic and plasmid DNA digested with RsaI was electrophoresed in 1.5% agarose gels in 1× TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA) for 3 h at 60 V. Alkaline transfer onto positively charged nylon was performed following the manufacturer’s instructions (Boehringer). An RsaI fragment obtained by digestion of the TsA dimer was randomly primed with DIG High Prime (Boehringer), whereas the HSE probe was obtained by annealing oligonucleotides HSE29A (5′-CCCCT ATGGA AAATT CTAGA AAAAT CG-3′) and HSE29B (5′-CTCGA TTTTT CTAGA ATTTT CCATA GG-3′). It was then labelled with 11-Dig-ddUTP by terminal transferase (Roche) following the manufacturer’s instructions. Hybridisation and detection was performed using CDP-Star (Boehringer) diluted 1:1000 in 1% diethanolamine.
Electrophoretic mobility shift assay (EMSA)
The probe for the EMSAs was prepared by annealing complementary synthetic oligonucleotides followed by end-labelling with [32P]ATP (Amersham) and polynucleotide kinase (Roche). Chironomus thummi protein extracts were obtained from 10 control or treated larvae and human extracts were obtained from HeLa cells infected with either adenovirus or recombinant adenovirus carrying human Hsf1d202–-316 (19). Extracts were quantified using the Bradford method (Bio-Rad). The binding reaction was performed by preincubating either 10 mg C.thummi proteins or 2.5 mg human proteins with 2 mg poly(dI·dC) in CB buffer (0.4 mM EDTA, 80 mM Tris–HCl, pH 8, 20 mM MgCl2, 2 mM DTT, 2 mM CaCl2) on ice for 20 min. Two nanograms of end-labelled double-stranded probe (50 000–100 000 c.p.m.) were added to the reaction mixture and incubated for 30 min at 4°C. Samples were electrophoresed in 4% polyacrylamide gels in 0.5× TBE (89 mM Tris–borate and 2 mM EDTA), 10% glycerol buffer, 175 V, at room temperature for 2.5 h. In competition experiments, a 50-fold excess of unlabelled oligonucleotides was incubated with the reaction mixture for 15 min at 4°C before addition of the radiolabelled probe. Supershift assays were performed by incubating an antibody against HSF-1 (Stressgen) overnight at 4°C with the protein extracts before the radiolabelled probe was added to the reaction mixture. HSE29 (5′-CCCCT ATGGA AAATTCTAGA AAAAT CGAG-3′) and HSE29C (5′-CCCCT ATGAA AATTT GTAGA AAAAT CGAG-3′) were used as competitors.
In situ hybridisation
Salivary glands fixed in ethanol:acetic acid (3:1) and squashed in 50% acetic acid were denatured. Each slide was treated with 100 nmol telomeric HSE oligonucleotide (the same as that used in the Southern blot experiments) 3′-labelled by terminal transferase with 11-Dig-ddUTP (Roche) or TsA monomer labelled by random priming with 11-dUTP-biotin (Enzo) and incubated overnight at 37°C in hybridisation buffer (50% deionised formamide and 4× SSC, 0.4% SDS). Slides were washed in 2× SSC for 5 min and in PT. Detection of the probe was achieved with anti-digoxigenin IgGs conjugated with rhodamine (Roche) or FITC-conjugated EstrAvidin (Sigma). After washing, the slides were stained with Hoechst, mounted in 9:1 glycerol:10× PBS with 1 mg/ml p-phenylene diamine and observed with a Zeiss photomicroscope equipped with a CCD camera. Images were captured using the 1P-lab spectrum program and processed with Adobe PhotoShop 4.0.
RESULTS
Presence of a HSE in telomeric sequences
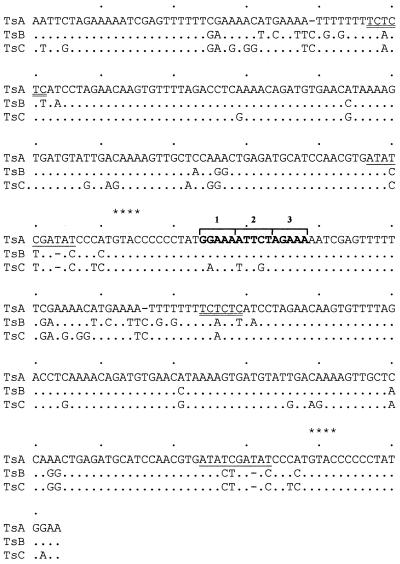
Three subfamilies of repeats, TsA, TsB and TsC, were characterised previously (17) by sequence analysis of single repeats (monomers). In the present investigation, a head-to-tail dimer of TsA was isolated from a genomic minilibrary (4) and TsB dimers by PCR. A closer inspection of the TsA dimer sequence led to the identification, in the junction of the two repeats, of the sequence gGAAaaTTCtaGAAa, which is highly homologous to the consensus sequence for the HSE, a transcription regulatory sequence shared by all heat shock coding genes. The HSE consensus sequence shows a repeating array of the 5 bp sequence nGAAn, where each module is inverted relative to the one immediately adjacent. The minimum size of an HSE is three modules (nGAAnnTTCnnGAAn) (20). The sequence containing the telomeric HSE shows great similarity to an HSE from the D.melanogaster hsp22 promoter (21). The interest of this finding in the context of telomeric activation by heat shock prompted a search for this element in dimers of the other types of repeats (TsB and TsC). Figure 1 shows the dimer sequence alignment. Homology with the HSE consensus sequence was observed in TsB, although less similarity was seen with TsC, due to the presence of three base substitutions.
Figure 1.
Sequence comparison of the dimers of telomeric repeats TsA, TsB and TsC. The TsA dimer was isolated from a C.thummi IIIR minilibrary (4). Six clones with a dimer were identified by restriction enzyme analysis, Southern blotting using as probe the previously described monomer and sequencing. The TsB dimer was obtained by PCR of whole genomic DNA with two primers, primer 1 (5′-ccggaattccgaAATTCTAGAAAAATGC-3′) and primer 2 (5′-ccggaattccggTTCCATAGGGGGG-3′), which contain the flanking TsA telomeric sequences (upper case) and EcoRI site (lower case) to facilitate cloning. The PCR products were cloned in pBluescript and sequenced. The location of putative regulatory regions in head-to-tail dimers are showed: bold indicates sequences homologous to the HSE consensus (nGAAnnTTCnnGAAn); double underlined shows homology to the GAGA factor consensus sequence (GAGAGAG); underlined indicates the putative TATA box with a central GC pair. Dots indicate homology, dashes insertion/deletion and asterisks RsaI sites. Sequence accession nos: TsA, M33211; TsB, AJ295633; TsC, AJ295634.
A detailed analysis of the sequences revealed other regulatory elements, including a TATA box with an internal GC pair only found in TsA (Fig. 1) and also a GAGA motif running from position 46 to 52 in TsA which was present in all three subfamilies of repeats. The telomeric GAGA motif is homologous to the consensus sequence of the GAGA transcription factor, (GA·TC)n, which is found in the promoters of several Drosophila genes (22).
Abundance and distribution of HSE in telomeres
An attempt was made to ascertain the abundance of the identified HSE in telomeric DNA. In Southern blots, genomic DNA was digested with RsaI to maintain the HSE within one monomer. Identical samples of RsaI-digested DNA were electrophoresed and hybridised separately with either the whole telomeric repeat or an oligonucleotide containing the telomeric HSE (Fig. 2). The signals obtained with both probes were comparable, suggesting HSE to be highly represented in the telomeric blocks of repeats.
Figure 2.
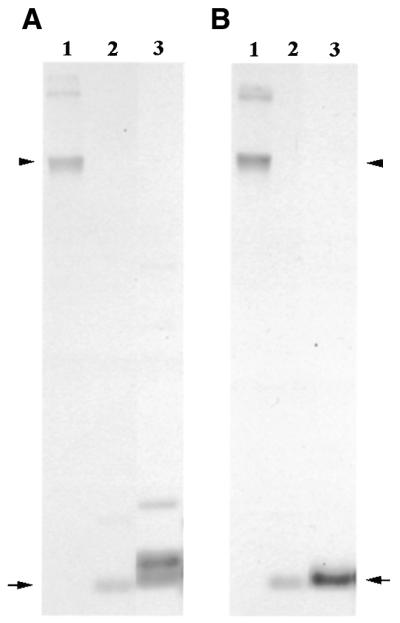
Southern blots of genomic DNA hybridised with the telomeric TsA monomer (A) or an oligonucleotide containing the HSE (B) as probes. Equal amounts of non-digested genomic DNA (lanes 1) and RsaI-digested genomic DNA (lanes 2) were loaded. As a control, RsaI-digested plasmid DNA containing a TsA dimer was used (lanes 3). Arrowheads indicate non-digested DNA and arrows indicate the 176 bp monomer band in RsaI-digested DNA.
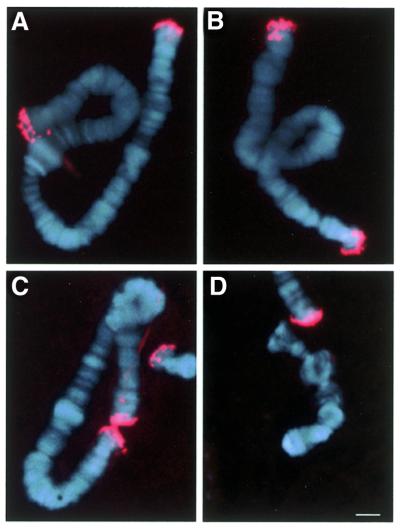
To determine the location of HSE at the level of individual telomeres, FISH experiments were performed on polytene chromosomes using an oligonucleotide covering the HSE as probe. As shown in Figure 3, a clear and distinct signal at all non-telocentric telomeres was obtained except for 4R. These results are in good agreement with the prediction made from the dimer sequence analysis, since TsC sequences (which lack HSE) are specific for the 4R telomere (17). Further, the strong fluorescent signal confirmed the Southern blot data (Fig. 2), indicating that HSE is abundant in telomeres of C.thummi.
Figure 3.
In situ hybridisation experiments with a 29 bp oligonucleotide containing the HSE (see Materials and Methods) detected with an anti-digoxigenin–TRIC conjugate antibody. A compiled image with Hoechst H-33528 staining shows the signal obtained for (A) chromosome I, (B) chromosome II, (C) chromosome III and (D) chromosome IV. The bar represents 10 µm.
In vitro binding of HSF by telomeric HSE
To learn whether HSE binds HSF, EMSAs were performed with an oligonucleotide that runs from position 166 to 194 (Fig. 1). This includes three modules with homology to the HSE consensus and an additional 7 nt in each direction, forming a total size of 29 nt in all. Incubation of labelled HSE29 with extracts from control C.thummi larvae showed no shift in mobility (Fig. 4A, lane 1). When protein extracts from heat shocked larvae were used, a band was seen (Fig. 4A, lane 2) which disappeared in experiments using a 50-fold excess of cold oligonucleotide as competitor (Fig. 4A, lane 3). Nevertheless, using a 50-fold excess of HSE29C, an equivalent oligonucleotide covering the same zone from TsC telomeric sequences, as competitor the band was not abolished (Fig. 4A, lane 4), suggesting that the three base difference between the oligonucleotides alters HSE binding. These data show that the telomeric HSE, present in the HSE29 oligonucleotide, binds a soluble protein induced, or activated, by heat shock, most probably HSF. This hypothesis was confirmed using heterologous extracts from human cells infected with adenovirus containing the HSF-1 gene, which constitutively express HSF-1, and a commercial antibody specific to human HSF. When the labelled HSE29 probe was incubated with protein extracts from human control cells infected only with vector, no bands were seen (Fig. 4B, lane 1). Incubation with protein extracts from human HSF-expressing cells gave three bands (Fig. 4B, lane 2), which probably correspond to different forms of HSF-1. No band was observed when the protein extract was preincubated with a specific antibody against HSF-1 (Fig. 4B, lane 3), whereas the band remained unaltered when preincubation was undertaken with preimmune serum (Fig. 4B, lane 4). These results show that the HSE sequences identified in the dimers of TsA and TsB telomeric units are able to bind HSF-1 in vitro, suggesting that this element may be functional in vivo, as later confirmed by in situ localisation of HSF (see Fig. 5), while TsC units are unable to bind HSF under the same conditions.
Figure 4.
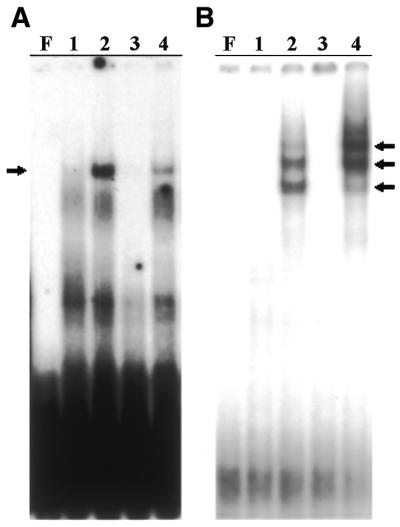
EMSA with extracts of (A) C.thummi and (B) HeLa cells using an oligonucleotide containing telomeric HSE as probe. The C.thummi extracts used were from non-treated larvae (lane 1) and from larvae heat shocked for 2 h at 35°C (lanes 2–4). As competitors, the same non-labelled probes were used (lane 3) as well as an unlabelled oligonucleotide (HSE29C) covering the same region as the TsC repeat (lane 4), in 50-fold excess. HeLa extracts are from control cells (lane 1), infected with adenovirus, and from cells constitutively expressing HSF (lanes 2–4), infected with the adenovirus containing the HSF-1 gene. In (B) the extract in lane 3 was preincubated with a commercial antibody against HSF-1 and that in lane 4 was previously incubated with a preimmune serum from rabbit. F indicates free probe and arrows show HSF bands.
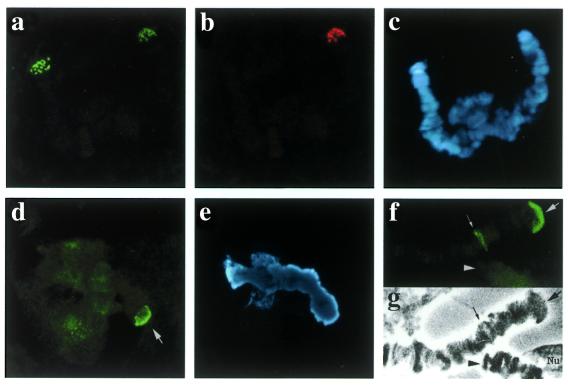
Figure 5.
Double in situ hybridisation in chromosome IV from C.piger × C.thummi hybrids with the telomeric 176 repeat labelled with 11-dUTP-biotin and detected with FITC (green) (a) and with a telomeric HSE oligonucleotide labelled with 11-dUTP-DIG and detected with TRIC (red) (b). In (c) Hoechst H-33258 staining shows the two chromatids unpaired, as frequently occurs. Immunohistochemical visualisation of HSF in heat shocked (1 h, 35°C) cells from C.piger: (d) Chromosome IV telomere 4R (arrow); (e) the corresponding Hoechst H-33258 image. HSF immunolocalization in heat shocked (2 h, 35°C) C.thummi larva: (f) positive signal at TBR III (3R telomere) (arrow) and heat shock puff III-A3 (small arrow) (note the absence of signal at the 4R end; arrowhead); (g) the corresponding phase contrast image.
Involvement of HSE in heat shock-induced puffing
Patterns of puffing, transcription and HSF binding in individual telomeres were analysed in polytene cells of C.thummi under heat shock conditions (Table 1). The results show a positive correlation between the presence of heat shock regulatory elements and the ability of specific telomeres to react to heat shock. Particularly evident was the case of telomere 3R, characterised by prominent puff formation (TBR III), HSF binding and transcriptional activation upon heat shock (Fig. 5e and f). TsA telomeric sequences containing HSE, GAGA and TATA boxes are specifically found in this telomere. In contrast, at telomere 4R, characterised by TsC sequences lacking HSE, neither puffing nor any other sign of activation was found (Fig. 5e and f). Other non-telocentric telomeres containing HSE and characterised by the presence of TsB sequences showed rather irregular behaviour upon heat shock. While slight signs of transcriptional activation were detected at these telomeres, puff formation and HSF binding were only occasionally observed.
Table 1. Telomeric activation at different chromosome ends in C.thummi after 2 h of 35°C heat shock.
| Il | Ir | IIl | IIr | IIIl | IIIr | IVr | |
|---|---|---|---|---|---|---|---|
| Puff size was measured as large (+++), medium (++), small (+) or not visible (–). Puffing frequency was considered high (H) when it reached >30%, medium (M) when between 10 and 30% and low when <10% (L). In telomere IVr no puffing was observed. Transcriptional activity was studied using an antibody against DNA–RNA hybrids. The frequency of telomeres with a positive signal (+, >50%; ++, >60%; +++, >80%) is indicated. HSF association indicates the presence (+) or absence (–) of a positive signal. At least five different larvae and 30–40 chromosomes per preparation were scored in each case. | |||||||
| Size of puff | + | ++ | – | – | – | +++ | – |
| Puffing frequency | M | M | L | L | L | H | N |
| Transcriptional activity | ++ | ++ | ++ | ++ | + | +++ | – |
| HSF association | – | + | – | – | – | + | – |
The correlation between HSE and telomeric activation was further confirmed by analysing the behaviour of C.piger telomeres under heat shock. In this species, a TBR was specifically induced by temperature at telomere 4R (23). A FISH experiment using HSE oligonucleotide as probe showed a clear positive signal at telomere 4R in C.piger polytene chromosomes but not in C.thummi telomeres when probed in C.thummi × C.piger hybrids (Fig. 5a–c). Further, HSF accumulated at the 4R C.piger telomere during heat shock (Fig. 5d and e). Thus, the presence of HSE, along with the capability of binding HSF, seem to be the basic features connected to full expression of telomeric puffing upon heat shock.
DISCUSSION
The existence of heat shock regulatory elements in the telomeric sequences reported in this paper provides, for the first time, a molecular basis for understanding the singular behaviour of C.thummi telomeres. These telomeres can be induced, by heat shock, to form giant, actively transcribing terminal puffs, named TBRs, which are most pronounced at the 3R chromosome end (24). Other chromosomal ends are only occasionally induced to puff, although signs of transcription could be detected under heat shock conditions. Finally, neither of the ends of chromosome 4 was found to respond to heat shock.
Telomeric HSE, found both in the TsA and TsB subfamilies of repeats, is highly homologous to the HSE consensus sequence established for heat shock protein coding genes. Telomeric HSE is able to bind HSF in vitro, as demonstrated by EMSA, and might be active in vivo, as suggested by HSF immunolocalisation in heat shock-induced telomeres.
The presence of HSE appears to be a necessary condition for telomeric activation by heat shock. The lack of response at the 4R end correlates with the lack of signal in FISH when probed with HSE oligonucleotides consistent with the TsC content of this telomere (17). Furthermore, in the related species C.piger, where telomere 4R always forms a puff after heat shock, HSE is also present and HSF localises at this site.
Other transcriptional regulatory elements such as TATA and GAGA motifs were also found in the telomeric sequences. They are differentially represented in TsA and TsB telomeric subfamilies of repeats, a possible explanation for the differences in heat shock activation observed among individual chromosome termini. Telomere 3R, which is always induced after heat shock, consists of large homogeneous blocks of tandem repeats of TsA sequences containing the minimal heat shock promoter elements (HSE, TATA and GAGA) considered capable of reacting to thermal stimuli (25). The lack of a TATA box in TsB sequences might be related to the more irregular reaction to heat shock of this group of telomeres. Other possibilities, such as chromatin structure constraints, cannot, however, be ruled out.
The alternative, that telomeric induction in C.thummi may be due to heat shock protein coding genes located at the chromosomal termini, is remote in view of the present results. The existence of heat shock regulatory elements integrated in the telomeric structure, possibly acting as internal promoters, seems more likely. In fact, RNA complementary to the telomeric repeats has been detected both locally and in northern blots after heat shock induction (26).
Heat shock activation of telomeres has only been observed in C.thummi and its sibling species C.piger, and not in several other Chironomus species investigated to date. All are similarly organised as large blocks of complex AT-rich sequences, but they show differences in the size of repeats as well as strong divergence at the sequence level (27). Nevertheless, constitutive transcription of telomeric repeats appears as a common feature (4,5). It is tempting to speculate that transcription of telomeric sequences might be directly involved in telomere maintenance through an RNA-templated mechanism, as suggested by the presence of retrotranscriptase detected in situ at telomeres (14).
The fact that heat shock responsiveness seems to be restricted to some telomeres in C.thummi and its sibling species C.piger make doubtful its requirement for essential telomeric maintenance functions. Nevertheless, it is remarkable that heat shock also activates transcription of the Zepp element of Chlorella telomeres (28). Zepp is a retrotransposable LINE-like element that accumulates at Chlorella subtelomeric regions, forming the actual terminus on chromosome I, which lacks the telomeric short repeats (29).
A number of mobile and/or interspersed repetitive elements are activated by heat shock, such as certain SINEs (30–33) and retrotransposons (34,35). The activation of such elements has, until now, not been considered part of the cellular stress response. However, it has recently been suggested that transcriptional heat shock activation of SINEs could play a protective role (36). To our knowledge, C.thummi telomeric repeats are the first short repetitive DNA units in which heat shock regulatory elements have been identified. The physiological importance of telomeric heat shock activation remains to be determined.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Prof. Jan Erik Edström for critically reading the manuscript and valuable suggestions. We acknowledge Mrs A. Partearroyo for technical assistance. J.L.M. was the recipient of a fellowship from the MEC. This work was supported by a grant from the MCYT, PB98-0646.
REFERENCES
- 1.Lingner J. and Cech,T.R. (1998) Telomerase and chromosome end maintenance. Curr. Opin. Genet. Dev., 8, 226–232. [DOI] [PubMed] [Google Scholar]
- 2.McEachern M.J., Krauskopf,A. and Blackburn,E.H. (2000) Telomeres and their control. Annu. Rev. Genet., 34, 331–358. [DOI] [PubMed] [Google Scholar]
- 3.Pardue M.L. and DeBaryshe,P.G. (1999) Telomeres and telomerase: more than the end of the line. Chromosoma, 108, 73–82. [DOI] [PubMed] [Google Scholar]
- 4.Carmona M.J., Morcillo,G., Galler,R., Martinez-Salas,E., de la Campa,A.G., Diez,J.L. and Edström,J.-E. (1985) Cloning and molecular characterization of a telomeric sequence from a temperature-induced Balbiani ring. Chromosoma, 92, 108–115. [Google Scholar]
- 5.Saiga H. and Edstrom,J.E. (1985) Long tandem arrays of complex repeat units in Chironomus telomeres. EMBO J., 4, 799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biessmann H., Donath,J. and Walter,M.F. (1996) Molecular characterization of the Anopheles gambiae 2L telomeric region via an integrated transgene. Insect Mol. Biol., 5, 11–20. [DOI] [PubMed] [Google Scholar]
- 7.Biessmann H., Zurovcova,M., Yao,J.G., Lozovskaya,E. and Walter,M.F. (2000) A telomeric satellite in Drosophila virilis and its sibling species. Chromosoma, 109, 372–380. [DOI] [PubMed] [Google Scholar]
- 8.Pich U., Fritsch,R. and Schubert,I. (1996) Closely related Allium species (Alliaceae) share a very similar satellite sequence. Plant Syst. Evol., 202, 255–264. [Google Scholar]
- 9.Adams S.P., Leitch,I.J., Bennett,M.D. and Leitch,A.R. (2000) Aloe L.—a second plant family without (TTTAGGG)n telomeres. Chromosoma, 109, 201–205. [DOI] [PubMed] [Google Scholar]
- 10.Morin G.B. and Cech,T.R. (1986) The telomeres of the linear mitochondrial DNA of Tetrahymena thermophila consist of 53 bp tandem repeats. Cell, 46, 873–883. [DOI] [PubMed] [Google Scholar]
- 11.López C.C., Nielsen,L. and Edström,J.E. (1996) Terminal long tandem repeats in chromosomes from Chironomus pallidivitatus. Mol. Cell. Biol., 16, 3285–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosen M. and Edström,J. (2000) DNA structures common for chironomid telomeres terminating with complex repeats. Insect Mol. Biol., 9, 341–347. [DOI] [PubMed] [Google Scholar]
- 13.Morcillo G., Diez,J.L. and Botella,L.M. (1994) Heat shock activation of telomeric sequences in different tissues of Chironomus thummi. Exp. Cell Res., 211, 163–167. [DOI] [PubMed] [Google Scholar]
- 14.Lopez C.C., Rodriguez,E., Diez,J.L., Edstrom,J. and Morcillo,G. (1999) Histochemical localization of reverse transcriptase in polytene chromosomes of chironomids. Chromosoma, 108, 302–307. [DOI] [PubMed] [Google Scholar]
- 15.Gorab E., Botella,L.M., Quinn,J.P., Amabis,J.M. and Diez,J.L. (1996) Ku-related antigens are associated with transcriptionally active loci in Chironomus polythene chromosomes. Chromosoma, 105, 150–157. [DOI] [PubMed] [Google Scholar]
- 16.Morcillo G., Diez,J.L., Carbajal,M.E. and Tanguay,R.M. (1993) HSP90 associates with specific heat shock puffs (hsrω) in polytene chromosomes of Drosophila and Chironomus. Chromosoma, 103, 648–659. [DOI] [PubMed] [Google Scholar]
- 17.Martinez J.L., Edström,J.E., Morcillo,G. and Diez,J.L. (2001) Telomeres in Chironomus thummi are characterised by different subfamilies of complex DNA repeats. Chromosoma, 110, 214–220. [DOI] [PubMed] [Google Scholar]
- 18.Westwood J.T., Clos,J. and Wu,T. (1991) Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature, 353, 822–827. [DOI] [PubMed] [Google Scholar]
- 19.Zuo J., Rungger,D. and Voellmy,R. (1995) Multiple layers of regulation of human heat shock transcription factor 1. Mol. Cell. Biol., 15, 4319–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandes M., O’Brien,T. and Lis,J.T. (1994) Structure and regulation of heat-shock promoters. In Morimoto,R.I., Tissieres,A. and Georgopoulos,C. (eds) The Biology of Heat-shock Proteins and Molecular Proteins. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Amin J., Ananthan,J. and Voellmy,R. (1988) Key features of heat shock regulatory elements. Mol. Cell. Biol., 8, 3761–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkins R.C. and Lis,J.T. (1997) Dynamics of potentiation and activation: GAGA factor and its role in heat shock gene regulation Nucleic Acids Res., 25, 3963–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morcillo G., Barettino,D., Carmona,M.J., Carretero,M.T. and Diez,J.L. (1988) Telomeric DNA sequences differentially activated by heat shock in two Chironomus subspecies. Chromosoma, 103, 648–659. [DOI] [PubMed] [Google Scholar]
- 24.Santa Cruz M.C., Morcillo,G. and Diez,J.L. (1984) Ultrastructure of a temperature-induced Balbiani ring in Chironomus thummi. Biol.Cell, 52, 205–212. [DOI] [PubMed] [Google Scholar]
- 25.Shopland L.S., Hirayoshi,K., Fernandes,M. and Lis,T.J. (1995) HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes Dev., 9, 2756–2769. [DOI] [PubMed] [Google Scholar]
- 26.Botella L.M., Morcillo,G., Barettino,D. and Diez,J.L. (1991) Heat-shock induction and cytoplasmic localization of transcripts from telomeric-associated sequences in Chironomus thummi. Exp. Cell Res., 196, 206–209. [DOI] [PubMed] [Google Scholar]
- 27.Kamnert I., Lopez,C.C., Rosen,M. and Edström,J.E. (1997) Telomeres terminating with long complex tandem repeats. Hereditas, 127, 175–180. [DOI] [PubMed] [Google Scholar]
- 28.Higashiyama T., Noutoshi,Y., Fujie,M. and Yamada,T. (1997) Zepp, a LINE-like retrotransposon accumulated in the Chlorella telomeric region. EMBO J., 16, 3715–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noutoshi Y., Arai,R., Fujie,M. and Yamada,T. (1998) Structure of the Chlorella Zepp retrotransposon: nested Zepp clusters in the genome. Mol. Gen. Genet., 259, 256–263. [DOI] [PubMed] [Google Scholar]
- 30.Liu W.M., Chu,W.M., Choudary,P.V. and Schmid,C.W. (1995) Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res., 25, 1758–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fornace A.J. and Mitchell,J.B. (1986) Induction of B2 RNA polymerase III transcription by heat shock: enrichment for heat shock induced sequences in rodent cells by hybridisation subtraction. Nucleic Acids Res., 14, 5793–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fornace A.J., Alamo,I., Hollander,M.C. and Lamoreaux,E. (1989) Induction of heat shock protein transcripts and B2 transcripts by various stresses in Chinese hamster cells. Exp. Cell Res., 182, 61–74. [DOI] [PubMed] [Google Scholar]
- 33.Kimura R.H., Choudary,P.V. and Schmid,C.W (1999) Silk worm Bm1 SINE RNA increases following cellular insults. Nucleic Acids Res., 27, 3380–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strand D.J. and McDonald,F. (1985) Copia is transcriptionally responsive to environmental stress. Nucleic Acids Res., 13, 4401–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuker C., Cappello,J., Lodish,H.F., George,P. and Chung,S. (1984) Dictyostelium transposable element DIRS-1 has 350-base-pair inverted terminal repeats that contain a heat shock promoter. Proc. Natl Acad. Sci. USA, 81, 2660–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li T., Spearow,J., Rubin,C.M. and Schmid,C.W. (1999) Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene, 239, 367–372. [DOI] [PubMed] [Google Scholar]