Summary
Post-translational modifications (PTMs), such as glycosylation and palmitoylation, are critical to protein folding, stability, intracellular trafficking, and function. Understanding regulation of PTMs of SARS-CoV-2 spike (S) protein could help the therapeutic drug design. Herein, the VSV vector was used to produce SARS-CoV-2 S pseudoviruses to examine the roles of the 611LYQD614 and cysteine-rich motifs in S protein maturation and virus infectivity. Our results show that 611LY612 mutation alters S protein intracellular trafficking and reduces cell surface expression level. It also changes S protein glycosylation pattern and decreases pseudovirus infectivity. The S protein contains four cysteine-rich clusters with clusters I and II as the main palmitoylation sites. Mutations of clusters I and II disrupt S protein trafficking from ER-to-Golgi, suppress pseudovirus production, and reduce spike-mediated membrane fusion activity. Taken together, glycosylation and palmitoylation orchestrate the S protein maturation processing and are critical for S protein-mediated membrane fusion and infection.
Subject areas: Biochemistry, Virology, Cell biology
Graphical abstract
Highlights
-
•
611LY612 mutation alters the glycosylation pattern of the SARS-CoV-2 S protein
-
•
611LY612 mutation reduces S protein surface expression level
-
•
Palmitoylation targets mature S protein to the Golgi and plasma membrane
-
•
Palmitoylation is required for pseudovirus and SARS-CoV-2 production
Biochemistry; Virology; Cell biology.
Introduction
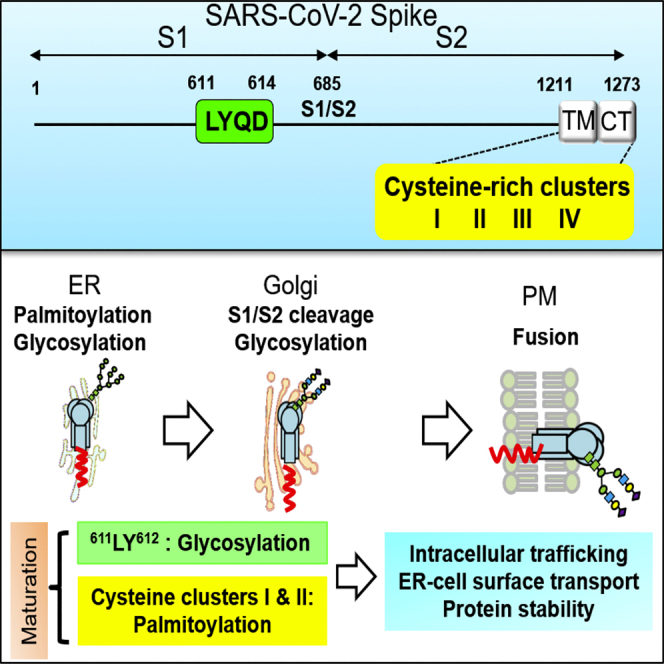
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged in Wuhan, China, in December 2019 and caused a coronavirus disease (COVID-19) outbreak. SARS-CoV-2 belongs to Betacoronavirus, which contains Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV-1, and mouse hepatitis virus (MHV) and has a high similarity to SARS-CoV-1 (Korner et al., 2020; Woo et al., 2010). The SARS-CoV-2 spike (S) glycoprotein interacts with angiotensin-converting enzyme 2 (ACE2) on the cell surface during virus entry (Hoffmann et al., 2020b) to mediate virus-host membrane fusion (Huang et al., 2020). After translation, the coronavirus S protein is processed by host proteases into S1 and S2 subunits, and the S2 protein is further cleaved at the S2′ site to facilitate virus entry (Peng et al., 2021). The SARS-CoV-1 and SARS-CoV-2 S proteins have ∼76% amino acid identity (Lan et al., 2020; Zhou et al., 2020). In the SARS-CoV-2 S protein, an additional furin-like recognition sequence (RRAR685↓S) is present at the S1/S2 cleavage site, which may contribute to the high transmissibility of SARS-CoV-2 (Coutard et al., 2020; Hoffmann et al., 2020a; Papa et al., 2021; Peacock et al., 2021; Xia et al., 2020).
The S glycoprotein is a critical target for pathogenic coronavirus vaccine development, and current COVID-19 vaccines employ full-length or portions of S protein as the antigen to induce neutralizing antibodies against SARS-CoV-2 entry (Li et al., 2020; Tregoning et al., 2020). A process by which suboptimal antibodies against viral glycoproteins enhance viral infection through the Fcγ receptor (Lee et al., 2020), antibody-dependent enhancement (ADE) has been a concern in vaccine development against pathogens such as dengue virus (Katzelnick et al., 2017; Ulrich et al., 2020), SARS-CoV-1, and MERS-CoV (Wan et al., 2020; Wang et al., 2016). Several studies show that ADE of SARS-CoV-2 is mediated by Fcγ receptor IIA or complement component C1q (Maemura et al., 2021; Okuya et al., 2022; Wang et al., 2022). Monoclonal antibodies specific for the 597LYQD600 motif of the SARS-CoV-1 S protein are shown to have ADE activity (Wang et al., 2016). An LYQD motif is also present in the SARS-CoV-2 S protein, but whether eliminating the ADE-associated sequence in the S antigen is beneficial for an effective COVID-19 vaccine design remains unknown.
Maturation of the S glycoprotein is critical for coronavirus infection and transmission and can also be one of the antiviral targets. The S protein undergoes several post-translational modifications (PTMs), including N-linked glycosylation, palmitoylation, and proteolytic processing as part of its maturation process (Fung and Liu, 2018). 12 out of 23 asparagine residues in the SARS-CoV-1 S protein are glycosylated (Krokhin et al., 2003). SARS-CoV-1 S proteins are glycosylated in the endoplasmic reticulum (ER) with high-mannose glycans, which are then further modified as complex N-glycans in the Golgi (Duan et al., 2020; Nal et al., 2005). Glycosylation can influence viral glycoprotein folding, function, immune evasion, and virus infection (Huang et al., 2021; Watanabe et al., 2019, 2020). The endodomains of SARS-CoV-1 and SARS-CoV-2 S proteins contain a cysteine-rich motif for palmitoylation, which might participate in membrane fusion and infectivity (Petit et al., 2007; Wu et al., 2021). It has been shown that zinc finger DHHC domain palmitoyltransferase 5 (zDHHC5) and Golgin subfamily A membrane 7 (GOLGA7) interact with S protein and induce its palmitoylation (Gordon et al., 2020; Wu et al., 2021; Zeng et al., 2021). How these PTMs affect SARS-CoV-2 S protein stability, intracellular trafficking, and function will need to be addressed more thoroughly.
In this study, the vesicular stomatitis virus (VSV) vector was used to generate the SARS-CoV-2 S pseudoviruses for the functional characterization of the LYQD and cysteine-rich motifs on S protein maturation and virus infectivity. Our results suggest that the LYQD motif was involved in the S protein glycosylation process, and the palmitoylation of the cysteine-rich motif participated in the S protein trafficking and maturation process.
Results
Mutations in the LYQD motif change the glycosylation pattern of the SARS-CoV-2 S protein
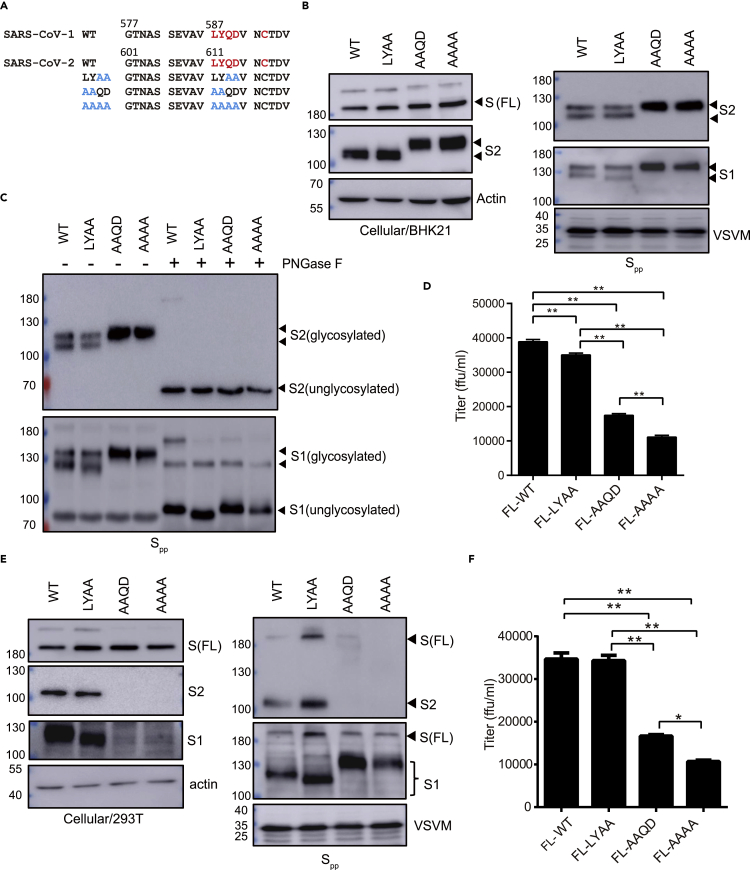
Antibodies recognizing the 587LYQD590 and C593 amino acid residues in the SARS-CoV-1 S protein have ADE activity (Wang et al., 2016). To examine whether the 611LYQD614 sequence in SARS-CoV-2 S protein maturation can be removed from the S protein, alanine substitution mutations in the LYQD sequence (611LYAA614, 611AAQD614, and 611AAAA614) were generated in the mammalian expression plasmid (Figure 1A). As the following cysteine residue might be involved in inter- or intra-molecule disulfide bond formation, C617 of SARS-CoV-2 S protein was not mutated. Wild type (WT) and mutant S protein expression in BHK21 cells was achieved through plasmid DNA transfection, followed by infection with the VSVΔG-GFP/G virus to secrete the S pseudotyped virus particles (Spp) into the culture medium. When the S protein expression in transfected cells was examined by immunoblotting with an anti-S2 antibody, full-length S protein (∼180 kDa) and S2 (∼100 kDa) were both detected in the cell lysate (Figure 1B, left panel). However, the S2 proteins of the AAQD and AAAA mutants had different electrophoretic mobility in contrast to those from the WT and LYAA mutants. Spp secreted into the culture supernatant was then examined by immunoblotting with antibodies specific for the S1 and S2 proteins (Figure 1B, right panel). Very little unprocessed FL S protein was detected in the Spp culture medium. The S1 and S2 proteins from WT and LYAA Spp each appeared as a doublet on an immunoblot, indicating two different protein sizes, but appeared as a single high molecular weight band in the culture medium containing AAQD and AAAA Spp.
Figure 1.
The 611LY612 mutation in the antibody-dependent enhancement (ADE) domain leads to aberrant glycosylation of the SARS-CoV-2 spike (S) protein
(A) Amino acid sequences of the wild type (WT) and mutants close to the putative ADE domain of the SARS-CoV-2 spike protein. Red: ADE domain; Blue: mutation sites.
(B) BHK21 cells were transfected with plasmid DNA to introduce full-length (FL) spike protein expression and then infected with the recombinant VSV defective in glycoprotein expression (VSVΔG-GFP/G) for pseudovirus production. Spike protein expression in cells at 24 h post-transfection and in S pseudovirus (Spp) in the culture supernatant at 16 h post-VSVΔG-GFP/G infection was examined by immunoblotting.
(C) Assessment of S protein glycosylation in Spp via immunoblot after treatment with peptide N-glycosidase F (PNGase F).
(D) Quantification of the Spp titer via counting the GFP-positive BHK21-hACE2 cells at 16 h post-infection.
(E and F) 293T cells were used to express the LYQD-related S mutant protein and generate the mutant Spp. The protein expression in cell and in Spp was detected by immunoblotting (E), and the Spp titer was measured in BHK21-hACE2 cells (F). ∗∗p < 0.01. Error bars represent SEM and n = 3.
Because the SARS-CoV-2 S protein has 22 predicted N-linked glycosylation sites, with the N603 and N616 residues located close to the LYQD motif (Watanabe et al., 2020), we postulated that the differential electrophoretic mobility observed for the S1 and S2 mutants could be due to changes in protein glycosylation. To test this, WT and mutant Spp were treated with peptide N-glycosidase F (PNGase F) to remove N-linked oligosaccharides from the S protein. The protein size of S1 and S2 proteins from WT, AAQD, and AAAA Spp became the same after PNGase F treatment (Figure 1C), suggesting that the 611LY612 mutation altered the glycosylation pattern of S protein. When the Spp titer was measured by assessing the GFP signal in infected BHK21-hACE2 cells, the AAQD and AAAA Spp had significantly lower viral titer compared to the WT and LYAA Spp (Figure 1D). Taken together, these data suggest that mutation of LY residues affects the glycosylation pattern of S protein.
611LY612 sequence is required for high titer of Spp production in HEK293T
To test whether the LYQD sequence is also required for Spp production in other cell types, HEK293T was used for Spp packaging (Figures 1E and 1F). While the WT and LYAA mutant proteins show normal S protein processing and Spp production, only one dominant S1 and S2 form were present in Spp from HEK293T (Figure 1E), which was different from the Spp generated from BHK-21 cells. The S2 subunit of the AAQD and AAAA mutants might be unstable and could not be detected in HEK293T cell and medium. Surprisingly, a large amount of S1 subunit was detected in the culture medium. When the virus particles were precipitated with PEG (Figure S1), S1 and S2 subunits were present in the WT and LYAA Spp but not in the AAQD and AAAA Spp. This suggests that the S1 subunit in AAQD and AAAA mutants was present in the culture medium but not associated with virus particles. Viral titers of AAQD and AAAA Spp were also consistently reduced (Figure 1F).
The SARS-CoV-2 S protein contains 22 asparagine (N) residues, 16 of which have N-linked glycosylation (Shajahan et al., 2020; Walls et al., 2020; Watanabe et al., 2020). The N603 and N616 residues are close to 611LYQD614 sequence. Whether N603 and N616 mutation to alanine in S protein might show a similar phenotype as LY mutation was further tested. As shown in Figure S2A, the protein expression level and glycosylation pattern of the N603A and N616A mutants were very similar to the WT protein, suggesting that the phenotype of the 611LY612 mutation is not derived from the dysregulation of the neighboring glycosylation sites. Interestingly, the infectivity of the N603A and N616A Spp was significantly reduced (Figure S2B), indicating that appropriate glycosylation is critical for S protein-mediated infection.
The C-terminal tail of the S protein contains an ER-retention motif, and deletion of the tail facilitates S protein targeting to the plasma membrane for pseudovirus production (Lontok et al., 2004; McBride et al., 2007; Xiong et al., 2020). The S mutants with truncation of the C-terminal 19 amino acids (SΔ19) and LYQD-related mutations were generated to evaluate Spp production efficiency. The WT and LYAA SΔ19pp were produced with high efficiency, as indicated by immunoblotting and virus titration (Figure S3). The AAQD and AAAA SΔ19pp production were much lower than the WT and LYAA SΔ19pp. These data indicate that the LY sequence was required for a high titer of Spp and SΔ19pp production.
611LY612 sequence is required for S protein maturation and trafficking
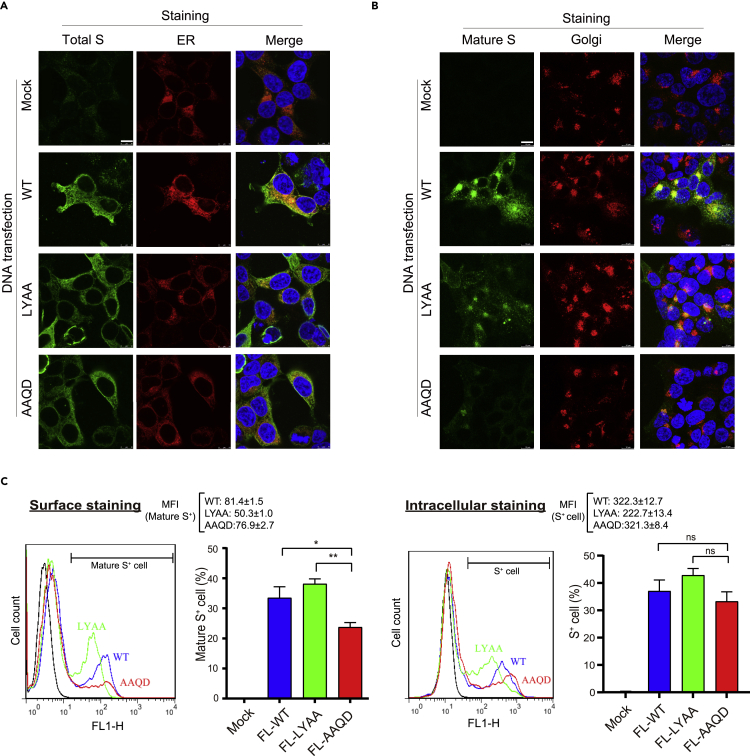
Protein glycosylation with diverse glycan occurs in the ER and Golgi. As 611LY612 mutation altered the glycosylation profile on both S1 and S2 subunits, we next examined whether 611LY612 sequence affects S protein subcellular distribution. When the expression pattern of S protein was examined by immunostaining with antibodies recognizing the S2 epitope (1A9), WT, LYAA, and AAQD S proteins were present in the cytoplasm and partially colocalized with the ER marker, which might correlate with the nascent S protein localization (Figure 2A). When an anti-S2 antibody (ECD45) with a high affinity to the prefusion state of trimeric S (mature S) was used for immunostaining, the mature WT and LYAA S protein expression were concentrated in the Golgi, and some speckles in the cytoplasm (Figure 2B). LY mutation showed a slightly reduced mature S expression compared to WT by immunostaining. In contrast, the signals for mature S of the AAQD mutant were very weak, and rarely present in the Golgi, suggesting that the LY mutation downregulates the S protein trafficking and maturation process. Some S proteins might further transport from Golgi to the cell surface, which may contribute to cell-cell fusion and spreading of SARS-CoV-2. Hence, whether the LYQD mutation affects S protein expression on the cell surface with anti-S2 antibodies (1A9 and ECD45) was examined by flow cytometry. As shown in Figure 2C, a higher percentage of cells had mature S protein expression on the cell surface in the WT and LYAA plasmid-transfected cells than in the AAQD-transfected group. The percentages of intracellular S protein expression of the WT, LYAA, and AAQD plasmid-transfected cells did not show any significant difference. The mean fluorescence intensity (MFI) of the LYAA mutant on the cell surface or intracellular was lower compared to the WT, suggesting that the protein stability of the LYAA mutant might also be slightly affected. Taken together, the LYQD sequence might sustain S protein stability and facilitate S protein moving from ER to Golgi for glycan modification and further to the plasma membrane during its maturation.
Figure 2.
611LY612 mutation blocks S protein maturation
(A and B) 293T/17 cells were transfected with plasmid DNA encoding WT and mutant S proteins, and the subcellular expression pattern was examined by immunofluorescent staining with the antibodies specific for S2 (1A9 and ECD45 clones), PDI (ER marker), and RCAS1 (Golgi marker). The nascent S protein was recognized by an anti-S2 (1A9) antibody (A). Mature S protein was stained with an anti-S2 (ECD45) antibody (B). Scale bars: 10 μm.
(C) The 293T/17 cells expressing S protein were subjected to surface staining with anti-S2 (ECD45) antibody and intracellular staining with anti-S2 (1A9) antibody for flow cytometry. MFI: mean fluorescence intensity of the positive cells. ∗p < 0.05, ∗∗p <0.01. Error bars represent SEM and n=3.
Palmitoylation is required for efficient Spp and SARS-CoV-2 production
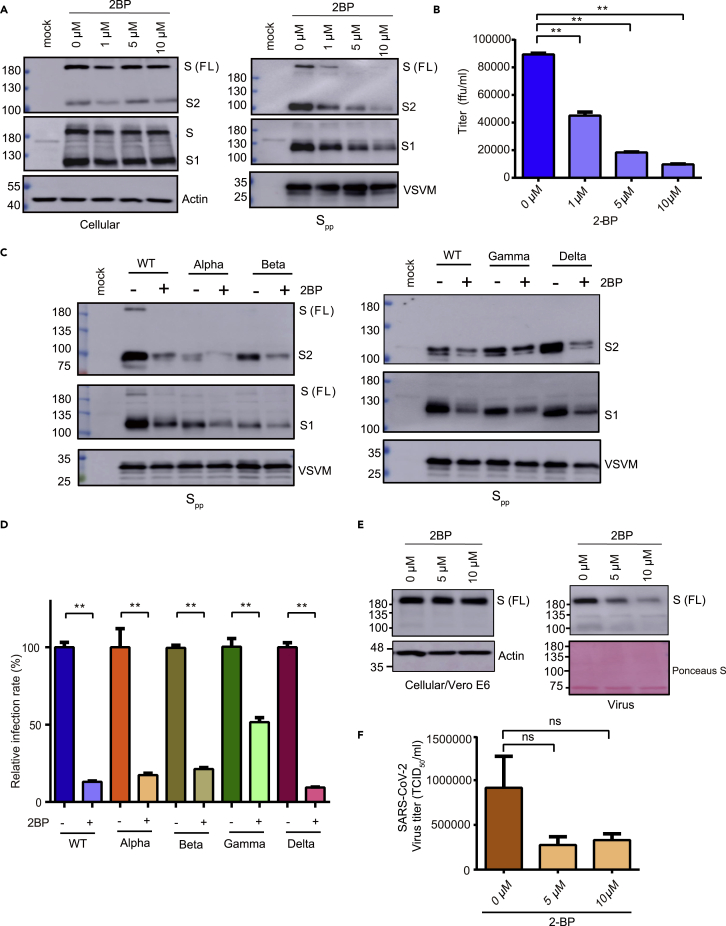
Protein palmitoylation primarily functions in protein subcellular trafficking. The endodomain of SARS-CoV-2 S protein contains a cysteine-rich motif for palmitoylation. To evaluate the function of palmitoylation in S protein maturation and virus infectivity, full-length S protein expression and Spp packaging in HEK293T cells were examined in the presence of a general palmitoylation inhibitor, 2-bromopalmitate (2BP). While S, S1, and S2 protein intracellular expression was not altered by the 2BP treatment at non-toxic concentrations (1–10 μM; Figures 3A and S4), the Spp packaging and titer in the culture medium were reduced (Figures 3A and 3B). As the cysteine-rich motif remains intact in the SΔ19 protein, the SΔ19pp production was also inhibited by the 2BP treatment (Figure S5). Genetic lineages of SARS-CoV-2 continue to evolve some variants, such as alpha, beta, gamma, and delta variants, which may have a higher transmission rate and decrease the effectiveness of COVID-19 vaccines (Chakraborty et al., 2021). Palmitoylation inhibition by 2BP reduced variant Spp packaging and virus titer (Figures 3C and 3D) but did not affect the S, S1, and S2 intracellular expression (Figure S6). To examine the importance of S protein palmitoylation in SARS-CoV-2 virus replication, Vero E6 cells were infected with SARS-CoV-2 virus (moi = 1) in the presence of 2BP treatment for 48 h. As shown in Figure 3E, palmitoylation inhibition did not affect intracellular S protein expression but blocked SARS-CoV-2 virus production in the culture medium. The virus titer was slightly lower in the presence of 2BP (Figure 3F). In summary, palmitoylation inhibition suppresses Spp and SARS-CoV-2 virus production.
Figure 3.
2-Bromopalmitate (2BP) inhibits Spp and SARS-CoV-2 production
(A) Expression of S protein in 293T/17 cells and Spp in the presence of 2BP treatment (1–10 μM) were examined by immunoblotting.
(B) The Spp titer was evaluated in BHK21-hACE2 cells.
(C) The WT and variant (alpha, beta, gamma, and delta strains) Spp generated from 293T/17 cells in the presence of 2BP treatment (10 μM) was examined by immunoblotting.
(D) The WT and variant Spp titers were evaluated.
(E and F) Vero E6 cells were infected by SARS-CoV-2 (hCoV-19/Taiwan/4/2020, moi = 1) in the presence of 2BP. The cell lysate and supernatant were collected at 48 h post-infection and subjected to immunoblotting (E) and virus titration (F). ∗∗p < 0.01. Error bars represent SEM and n = 3.
SARS-CoV-2 S protein is palmitoylated at multiple cysteine residues
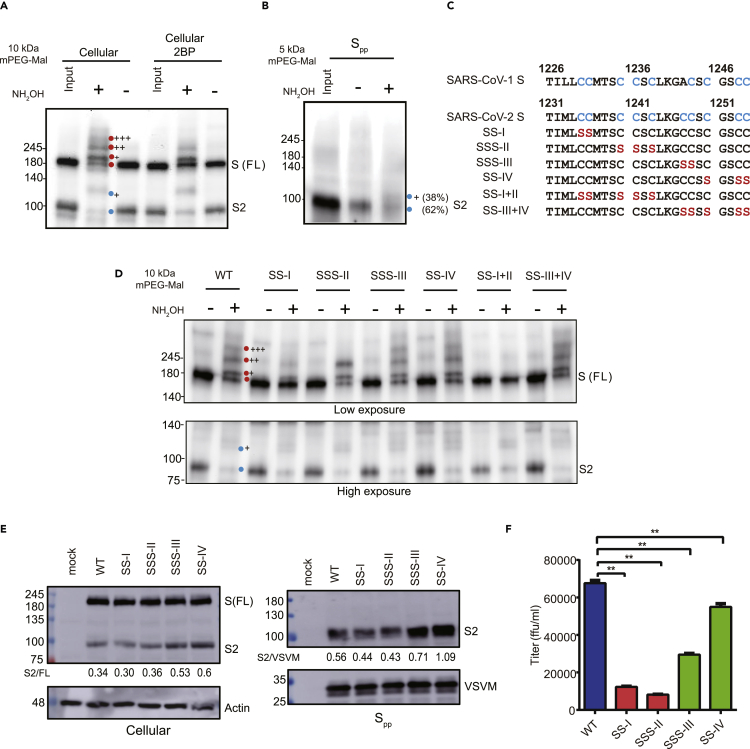
To confirm S protein palmitoylation was inhibited by 2BP, the palmitoylated S protein was monitored by using a modified protocol of the acyl-PEG exchange gel (Percher et al., 2016). As shown in Figure 4A, there were at least three palmitoylated species of the S protein as observed by the slower migrating bands on the Western blot after acyl-PEG exchange. The palmitoylation level of S protein was inhibited by the 2BP treatment. We also detected a single major palmitoylated species of the S2 protein with or without 2BP treatment. Similarly, the palmitoylation level of SΔ19 was also suppressed by the 2BP treatment (Figure S7). Furthermore, the palmitoylated S2 protein was also incorporated in the Spp virus (Figure 4B), suggesting that S protein palmitoylation might directly participate in virus packaging.
Figure 4.
Palmitoylation of WT and cysteine-rich cluster mutants S protein
(A) Palmitoylated proteins in 293T/17 cells were treated with 10 kDa methoxy polyethylene glycol maleimide (mPEG-mal) and immunoblotted with an anti-S2 antibody.
(B) Palmitoylated S protein in Spp was labeled with 5 kDa mPEG-mal and detected by immunoblotting.
(C) Sequence of cysteine-rich motif of the WT and mutant S proteins.
(D) Palmitoylation of cysteine-rich cluster mutants as determined by acyl-PEG exchange.
(E) The S protein expression of WT and cysteine-rich cluster mutants in 293T/17 cells and Spp was examined by immunoblotting with an anti-S2 antibody.
(F) The titers for WT and cysteine mutants Spp were evaluated. Error bars represent SEM and n = 3. Red circle: S protein; blue circle: S2 subunit. +: number of different palmitoylated species.
The cysteine-rich motifs of SARS-CoV-1 and SARS-CoV-2 S protein each contain four clusters known to be involved in protein palmitoylation. To examine the functional importance of the cysteine-rich clusters in SARS-CoV-2 S protein palmitoylation and Spp production, we generated cysteine-to-serine mutations in the cysteine-rich motif of S protein (SS-I, SSS-II, SSS-III, SS-IV, SS-I + II, and SS-III + IV) (Figure 4C). Consistent with previous reports (Petit et al., 2007; Wu et al., 2021), clusters I and II are crucial for SARS-CoV-2 S protein palmitoylation. SS-I and SSS-II mutants showed drastically diminished palmitoylation levels and palmitoylated forms S protein were barely detectable for the SS-I + II mutant (Figure 4D). In contrast, the major palmitoylated species of the SARS-CoV-2 S protein were still observed for the SSS-III, SS-IV, and SS-III + IV mutants. S protein palmitoylation is important for Spp production because S2 expression level in Spp and Spp titers was reduced in the SS-I, SSS-II, and SS-I + II mutants (Figures 4E, 4F, and S8). Interestingly, the SSS-III, SS-IV, and SS-III + IV mutants showed higher Spp production but reduced virus titers (Figures 4E and S8), suggesting that cysteine clusters III and IV might have additional roles in S protein processing and Spp production that are independent of protein palmitoylation. Collectively, these results demonstrate that cysteine clusters I and II are important for S protein palmitoylation and Spp production.
Palmitoylation targets mature S protein to the Golgi and plasma membrane
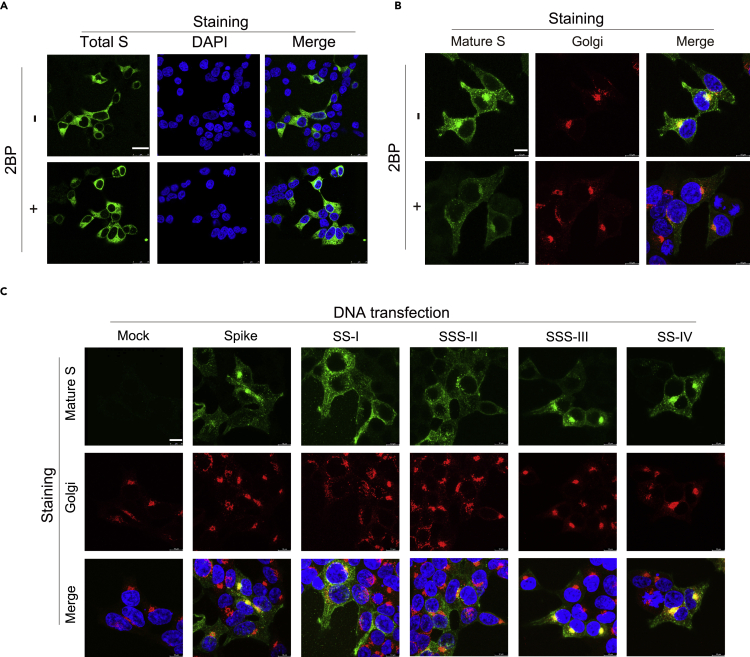
As palmitoylation may influence protein localization, trafficking, and stability (Aicart-Ramos et al., 2011; Yang et al., 2020), we evaluated whether palmitoylation modulates S protein subcellular localization. The localization of nascent S protein was unaffected with 2BP treatment when the S protein was stained with anti-S1 antibody (clone HL263) (Figure 5A). In contrast, the localization of mature S protein stained by ECD45 antibody was concentrated in the Golgi, plasma membrane, and speckles in the cytoplasm. Upon 2BP treatment, the fluorescent intensity of mature S protein was reduced and dispersed as an ER-like distribution (Figure 5B). The S protein subcellular localization of the cysteine cluster mutants was also examined. Similarly, consistent with palmitoylation targeting mature S protein to the Golgi, the palmitoylation-defective SS-I and SSS-II mutant S proteins were distributed outside the Golgi (Figure 5C). On the other hand, the mature SSS-III and SS-IV S proteins remained predominantly concentrated in the Golgi and plasma membrane. Furthermore, the mature SS-I + II mutant showed a more dispersed compartment outside the Golgi, and the SS-III + IV combination mutant was even more condensed in the Golgi and plasma membrane despite similar localization of the nascent S proteins (Figure S9). In summary, palmitoylation is important for the subcellular localization and trafficking of the S protein.
Figure 5.
Palmitoylation inhibition and cysteine-rich clusters I & II mutation blocks S protein maturation
(A and B) The mature S protein in the DNA transfected 293T/17 cells with or without 2BP treatment for 24 h was fixed and stained with antibodies specific for (A) S1 and (B) S2 & RCAS1 (Golgi marker). Scale bars: 25 μm (A) or 10 μm (B).
(C) The S protein carrying cysteine-rich cluster mutations in 293T/17 cells (24 h) were fixed and stained with antibodies specific for S2 and RCAS1 (Golgi marker). Nuclear DNA was counterstained with DAPI dye. Scale bars: 10 μm.
S protein palmitoylation is required for the ACE2-mediated cell fusion
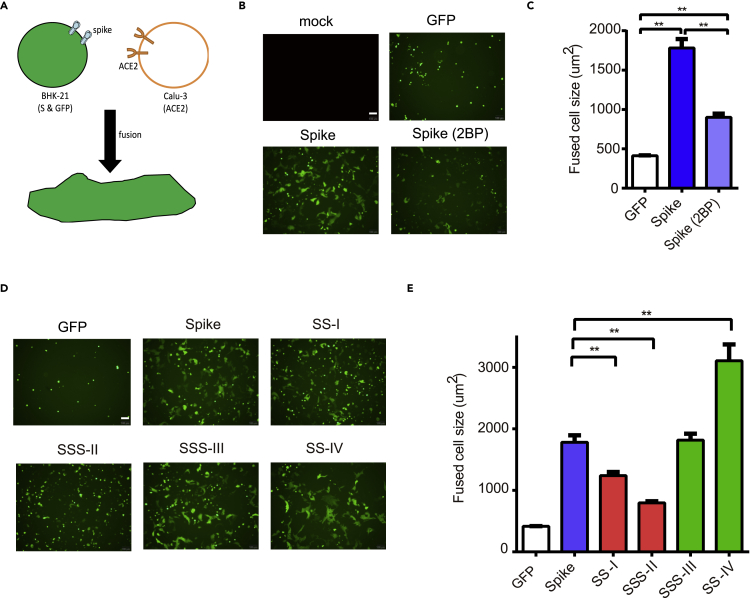
Other than assembling into the virion, mature S proteins may be expressed on host cell surfaces and induce membrane fusion with neighboring cells (Duan et al., 2020; Nishima and Kulik, 2021). As some palmitoylated S proteins observed at the plasma membrane (Figure 5), we examined whether palmitoylation is required for S protein-mediated cell fusion. GFP- and S-expressing BHK-21 cells were overlaid on the calu-3 cell film at 4°C for 45 min. After washing with PBS, the S protein-ACE2-mediated cell fusion between BHK-21 and Calu3 was monitored by tracing the GFP+ multinucleated cells after 4 h incubation (Figure 6A). The GFP+ cells without S protein expression remained as single cells (300–400 μm2) while those with S protein expression became large multinucleate cells (1500–2000 μm2) upon cell fusion (Figures 6B and 6C). Upon palmitoylation inhibition by 2BP, the GFP+ multinucleate cells had reduced cell-cell fusion (1000 μm2 on average). Consistent with S palmitoylation being important for spike-mediated membrane fusion, significantly reduced cell fusion was observed for palmitoylation-defective SS-I and SSS-II mutants (Figures 6D and 6E). Further highlighting the different functions of clusters III and IV, cell-cell fusion was not reduced when these clusters were mutated. Interestingly, SS-IV mutation enhanced the cell-cell fusion by about 50%, although likely through palmitoylation-independent mechanisms. Taken together, these data indicate that the palmitoylation of S protein in cysteine clusters I and II is involved in the mature S protein trafficking to the Golgi and cell surface to facilitate S-ACE2-mediated cell fusion.
Figure 6.
Palmitoylation of S protein facilitates the ACE2-mediated cell fusion
(A) S protein-ACE2-mediated cell fusion assay.
(B and C) The S and GFP coexpression plasmids were introduced to BHK21 cells by DNA transfection for 24 h. The transfected cells were collected and overlaid on Calu-3 cells at 4°C for 1 h, and the unbound cells were removed with PBS wash. After further 4 h incubation, the images of the GFP+ cells were captured by fluorescent microscope. GFP+ cell sizes in five randomly selected fields were quantified using ImageJ (C).
(D and E) The cysteine mutants were subjected to the cell fusion assays (D). GFP+ cell sizes in five randomly selected fields were quantified using ImageJ (E). Scale bars: 100 μm ∗∗p < 0.01. Error bars represent SEM and n = 3.
Discussion
The VSVΔG-based pseudovirus was applied to characterize SARS-CoV-2 S protein maturation and virus infection. The 611LY612 sequence in the putative ADE motif of S protein is required for correct glycosylation and maturation, which is essential for the production of infectious Spp particles. In contrast, the S protein can tolerate 613QD614 mutation in the putative ADE motif without affecting glycosylation and virus infection. The 613QD614 could be further evaluated in a vaccination setting. Palmitoylation targets the mature S protein trafficking to the Golgi and plasma membrane, which subsequently facilitates pseudovirus packaging and cell-cell membrane fusion. The four cysteine clusters of S protein have distinct roles in these processes. The study reveals that glycosylation and palmitoylation coordinate the S protein maturation processes, which are critical for the protein to function.
Glycosylation on the newly translated protein facilitates correct protein folding. Glycans could influence the conformation of S protein (Sztain et al., 2021), affect the binding affinity between S protein and ACE2 receptor (Casalino et al., 2020; Koehler et al., 2020), and avoid S protein from antibody recognition (Wang, 2020; Watanabe et al., 2020; Zhao et al., 2021). Based on our results, the S protein in Spp showed two different glycosylation forms in BHK21 cells but only one form (lower molecular weight) in 293T cells. The difference could be due to the diversity of glycosylation machinery in various cell types or fast degradation of the S protein with alternative glycosylation in 293T cells. The 611LY612 S protein with an altered glycosylation profile (higher molecular weight) could be incorporated in Spp in BHK21 cells, and the 611LY612 mutant Spp showed lower infectivity. The SARS-CoV-2 S protein contains multiple N-linked glycosylation sites, and N603 & N616 residues are close to 611LYQD614 sequence. The glycosylation levels of the N603A and N616A mutants are different from the 611LY612 mutant, suggesting that the low maturation process of the 611LY612 mutant is not due to the changes in the glycosylation of the neighboring amino acid sequence. Although the LY sequence is located in the S1 subunit, the S2 glycosylation profile was also affected, suggesting that the LY mutation keeps the S protein from trafficking to the Golgi, where glycan modification on both S1 and S2 subunits can be further processed. In 293T/17 cells, the S1 and S2 subunits of 611LY612 mutant might be unstable and subjected to protein degradation. It is also possible that the 611LY612 might impact protein folding, leading to ER quality control retention (Benyair et al., 2011).
Palmitoylation is important for SARS-CoV-1 and SARS-CoV-2 S protein maturation in order to function during virus infection and membrane fusion. Palmitoylation might facilitate S protein trafficking from ER to the cell surface during its maturation. The cysteine cluster I and II of SARS-CoV-1 and SARS-CoV-2 S proteins are the main palmitoylation site, as shown in our study and several studies (Mesquita et al., 2021; Petit et al., 2007; Puthenveetil et al., 2021). Host proteins zDHHC5 and GOLGA7 are involved in the palmitoylation of S protein (Gordon et al., 2020; Wu et al., 2021; Zeng et al., 2021). zDHHC enzymes such as zDHHC2, zDHHC3, zDHHC8, zDHHC9, and zDHHC20 may also contribute to the palmitoylation process of S protein (Mesquita et al., 2021; Puthenveetil et al., 2021; Ramadan et al., 2022). zDHHC enzymes distribute at ER, Golgi, and plasma membranes throughout the cellular secretory pathway (Malgapo and Linder, 2021). Zeng et al. showed that the intracellular distribution of S protein was not altered in the zDHHC5 or GOLGA7 knockout condition (Zeng et al., 2021), but our data show that mutations of the palmitoylation sites reduce the Golgi localization of the mature S protein. The discrepancy may be due to the difference in antibody selection for immunostaining. The other possibility was that the zDHHC family enzymes might compensate for the deficiency of the zDHHC5 or GOLGA7 and complete the palmitoylation of S protein. Mutations in cysteine clusters I and II also lead to a deficient ER-to-cell surface trafficking of the S protein, S1/S2 cleavage, virus packaging, and S-mediated membrane fusion. The cluster I and II mutants carry two palmitoylation sites of S proteins, whereas cluster III and IV mutants might contain more than two palmitoylation sites of S proteins. Moreover, the mutations in the cysteine clusters III and IV seem further enhance Golgi localization, S1/S2 cleavage, and virus packaging, suggesting that they might have a regulatory role in these processes. Based on the S protein structure prediction (Zheng et al., 2021), the cysteine-rich motif is located in the cytoplasmic tail proximally to the transmembrane domain. The cysteine-rich clusters I and II are in a stable alpha-helix structure and might be more accessible for zDHHC enzyme, which is known to interact with SARS-CoV-2 S protein. As the cysteine-rich motif in SARS-CoV-2 S is highly conserved in all the variants, S palmitoylation might be a potential target for antiviral drug development.
S protein trafficking is via the secretory pathway (Bracquemond and Muriaux, 2021; Mendonca et al., 2020). After synthesized, S protein is modified in ER and Golgi for post-translational modifications. It has been shown that the S protein has ER export and ER retrieval signals at the cytoplasmic tail (Cattin-Ortola et al., 2021). How the LYQD motif-associated glycosylation regulation and palmitoylation of S protein coordinate with these ER export and ER retrieval signals in the maturation process will be worth further investigation.
Limitations of the study
First, we have utilized vesicular stomatitis virus pseudoviruses to study glycosylation and palmitoylation of S protein. However, it is difficult to develop SARS-CoV-2 infectious cDNA clone to understand glycosylation and palmitoylation during virus replication and package. Second, detecting ADE activity from AAQD and AAAA Spp is not very easy because viral titers of AAQD and AAAA Spp were lower than that of LYAA Spp. Further studies will be required to evaluate the glycosylation mechanisms of LYQD motif and palmitoylation of S protein coordinate with these ER signal peptides in the maturation process.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CoV-2 S2 (1A9) | Genetex, Hsinchu, Taiwan | GTX632604 |
| Anti-VSV-M | Absolute, Boston, MA | Ab01404–2.0 |
| Anti-beta-actin | Genetex, Hsinchu, Taiwan | GTX109639 |
| Anti-CoV-2 Spike S1 | Genetex, Hsinchu, Taiwan | GTX635672 |
| Anti-CoV-2 mature S2 (ECD 45) | Antaimmu BioMed, Hsinchu, Taiwan | ATBM-A-08 |
| Anti-PDI | Cell signaling, Danvers, MA | 45596 |
| Anti-RCAS1 | Cell signaling, Danvers, MA | 12290 |
| Bacterial and virus strains | ||
| DH5α | Yeastern Biotech, Taipei, Taiwan | FYE678 |
| SARS-CoV-2 | Taiwan Centers for Disease Control | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| PNGase F | NEB, Ipswich, MA | P0704S |
| Lipofectamine 2000 | ThermoFisher, Waltham, MA | 11668 |
| DAPI | ThermoFisher, Waltham, MA | D21490 |
| 2-Bromohexadecanoic acid (2BP) | Sigma-Aldrich, Darmstadt, Germany | 238422 |
| M199 medium | Gibco; ThermoFisher, Waltham, MA | 11150–059 |
| TPCK-trypsin | Sigma-Aldrich, Darmstadt, Germany | T8802 |
| MTT solution | Sigma-Aldrich, Darmstadt, Germany | M-0283 |
| Versene solution | Gibco, Waltham, MA | 15040066 |
| Protease inhibitor cocktail | ThermoFisher, Waltham, MA | 78437 |
| Phenylmethylsulfonyl fluoride | Sigma-Aldrich, Darmstadt, Germany | 329-98-6 |
| Transcription Factor Buffer Set | BD Pharmingen™; ThermoFisher, Waltham, MA | AB_2869424 |
| SuperNuclease | SinoBiological,Beijing,China | SSNP01 |
| TCEP solution | ThermoFisher, Waltham, MA | 77720 |
| N-Ethylmaleimide | Sigma-Aldrich, Darmstadt, Germany | 04259 |
| mPEG-Mal | Sigma-Aldrich, Darmstadt, Germany | 99126-64-4 |
| MEM alpha medium | Cytiva; Hyclone, Marlborough, MA | SH30265 |
| FBS | Cytiva; Hyclone, Marlborough, MA | SH30396 |
| Pen Strep solution | Corning, New York | 20-003 |
| HEPES buffer | Biological Industries, Israel | 03-025 |
| DMEM/High glucose medium | Cytiva; Hyclone, Marlborough, MA | SH30243 |
| MEM medium | ThermoFisher, Waltham, MA | 11095080 |
| Experimental models: Cell lines | ||
| Vero E6 | provided by Dr. Shiow-Ju Lee, NHRI | N/A |
| HEK293T/17 | provided by Dr. Chia-Yi Yu, NHRI | N/A |
| BHK21 | provided by Dr. Chia-Yi Yu, NHRI | N/A |
| Calu-3 | provided by Dr. Yueh-Hsin Ping, NYCU | N/A |
| Software and algorithms | ||
| GraphPad PRISM 6.2 | https://www.graphpad.com | N/A |
| ImageJ | https://imagej.nih.gov/ij/ | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Guann-Yi Yu (guannyiy@nhri.edu.tw).
Materials availability
Plasmids in this paper will be shared by the lead contact upon request.
Experimental model and subject details
Cell, virus, and reagent
BHK21 cells were maintained in MEM alpha medium (Cytiva; Hyclone, Marlborough, MA) containing 10% FBS (Cytiva; Hyclone, Marlborough, MA), 1× Penicillin Streptomycin (PS) solution (100 IU/mL Penicillin and 100 μg/mL Streptomycin, Corning, New York), and 1× HEPES buffer (Biological Industries, Israel). HEK293T/17 cells and Vero E6 cells were maintained in 10% FBS in DMEM/High glucose medium (Cytiva; Hyclone, Marlborough, MA) with 1× PS solution. Calu-3 cells were maintained in 10% FBS in MEM medium (ThermoFisher, Waltham, MA) with 1× PS solution. Human ACE2 (hACE2) overexpression in BHK21 cells was introduced by the lentiviral vector carrying the hACE2 gene as described previously (Su et al., 2020). SARS-CoV-2 virus (hCoV-19/Taiwan/4/2020) was kindly provided by the Taiwan Centers for Disease Control and was amplified in Vero E6 cells in M199 medium with 2 μg/mL TPCK-trypsin. All SARS-CoV-2 experiments were performed in a biosafety level 3 (BSL-3) laboratory. 2-Bromohexadecanoic acid (2BP) was purchased from Sigma-Aldrich (Darmstadt, Germany).
VSV based-SARS-CoV-2 S pseudovirus (Spp)
Full-length S (Wuhan-Hu-1 strain, MN908947.3) or C-terminal 19 amino acid deletion mutant (SΔ19) were PCR-amplified and inserted between NheI and NotI restriction enzyme sites in the pVax1 vector (ThermoFisher). Mutations of the 611LYQD614 sequence (LYAA, AAQD, and AAAA), N603A, and N616A were generated by site-directed mutagenesis. The VSVΔG-GFP/G virus was recovered from the rVSV-ΔG-GFP-2.6 plasmid and helper plasmids (G, L, N, and P; Kerafast, Boston, MA) using methods described previously (Hsu et al., 2021; Lin et al., 2022; Whitt, 2010). To generate VSV based-SARS-CoV-2 S pseudovirus (Spp) or related mutant viruses, BHK21 cells were transfected with pVax1-SARS-CoV-2 S or mutant plasmids using Lipofectamine 2000 (ThermoFisher, Waltham, MA) and infected with the VSVΔG-GFP/G virus (multiplicity of infection [moi] = 5) the next day. After incubation for 24 h, the supernatant was clarified by centrifugation at 1,320 g for 10 min and stored at −80°C. For titration, the BHK21-hACE2 cells were infected with serially diluted pseudoviruses, and the virus titer (ffu/mL) was measured by counting GFP-positive cells.
Method details
Immunoblotting and PNGase F treatment
Cells were lysed in lysis buffer (50 mM Tris, 250 mM NaCl, 3 mM EDTA, 10% Triton X-100, 0.5% NP-40, and 10% glycerol). Cell lysate (10–50 μg) or viral supernatant (equal volume) were subjected to immunoblotting. Primary antibodies used in the study were anti-SARS-CoV-2 S2 (Genetex, Hsinchu, Taiwan; GTX632604), anti-VSV-M (Absolute, Boston, MA), and anti-beta-actin (Genetex, Hsinchu, Taiwan). The RBD antiserum was collected from rabbits immunized with RBD recombinant protein purified from E. coli. To remove N-linked oligosaccharides, viral supernatants were treated with peptide N-glycosidase F (PNGase F; NEB, Ipswich, MA) at 37°C for 1 h and then subjected to immunoblotting.
Immunofluorescence assay
293T/17 cells were transfected with plasmid DNA to express full-length or mutant S protein on coverslips, and the cells were fixed in 4% paraformaldehyde and permeabilized in 0.1% Triton X-100 of PBS buffer at 24 h post-transfection. Primary antibodies used in the study were: Spike S1 (Genetex; GTX635672), Spike S2 (clone 1A9, Genetex, GTX632604), mature S2 (clone ECD45, Antaimmu BioMed, Hsinchu, Taiwan; ATBM-A-08), PDI (Cell signaling, Danvers, MA), RCAS1 (Cell Signaling). Fluorescent secondary antibodies were purchased from Genetex or ThermoFisher. The cells were counterstained with DAPI (ThermoFisher) and examined by a confocal microscope (Leica TCS SP5).
Flow cytometry
The surface and intracellular staining of S protein for flow cytometry was performed with the protocol modified from the literature (Carter-Timofte et al., 2021; Cattin-Ortola et al., 2021; Chiuppesi et al., 2020). Briefly, HEK293T/17 cells were transfected with plasmid DNA to induce S protein expression for 24 h and dissociated from the culture dish with Versene solution (Gibco, Waltham, MA). For surface staining, cells were washed twice in FACS buffer (0.5% FBS in PBS), incubated with anti-mature S2 (1:500, ECD45) antibodies in FACS buffer for 30 min at 4°C, and staining with an Alexa 488-labeled secondary antibody for 30 min at 4°C. For intracellular staining, cells were fixed and permeabilized with Fix/Perm buffer (BD biosciences; ThermoFisher) for 5 min and washed three times in Perm wash buffer. The cells were then stained with anti-spike S2 (1:1000, clone 1A9) and fluorescent secondary antibodies for 30 min at 4°C. The stained cells were analyzed by flow cytometry (BD FACScalibur).
S protein-mediated cell-cell fusion assay
Calu-3 cells were seeded in a 12-well plate (1 × 106 cells/well) and incubated for 48 h to form cell films. BHK-21 cells were co-transfected with Spike and GFP plasmids (5:1) for 24 h. The Spike/GFP expressing BHK-21 cells were harvested and added into the Calu-3 cell film at 4°C for 45 min. After PBS washing twice, the mixed cells were incubated in a D-MEM medium for cell-cell fusion at 37°C for 4 h. The fusion level was evaluated by examining GFP-expressing cells with an inverted fluorescence microscope (Olympus IX73). The GFP-expressing cell images were used to quantify cell size using the ImageJ software.
Modified acyl-PEG exchange (APE) to monitor S protein palmitoylation
S protein palmitoylation was detected by a modified version of the acyl-PEG exchange method described previously (Percher et al., 2016, 2017). HEK293T/17 cells expressing the wild type and mutant S glycoproteins were washed thrice in ice-cold PBS and cell pellets were stored at −80°C until lysis. Cells were lysed in 4SB buffer [4% (w/v) SDS, 150 mM NaCl, 50 mM triethanolamine pH 7.4] containing 1× Halt™ protease inhibitor cocktail (Thermo Scientific 78437), 5 mM phenylmethylsulfonyl fluoride (Sigma 329-98-6) and 1 μL/mL SuperNuclease (SinoBiological SSNP01). Protein concentrations were measured using the BCA assay (Thermo Scientific 23225). 1.5 μL of 0.5 M EDTA pH 8 (final EDTA concentration 5 mM) was added to 200 μg of protein in 141.75 μL lysis buffer prior to treatment with 10 mM TCEP (3 μL 500 mM TCEP, Thermo Scientific 77720) for 30 min at room temperature with rocking. 3.75 μL of freshly prepared 1 M N-ethylmaleimide (NEM, Sigma 04259, final concentration 25 mM) in ethanol was added to the samples and further incubated at room temperature for 2 h. Since we observed significant degradation for the S protein with the multiple precipitation steps in the APE protocol described by Percher et al., we chose to terminate reductive alkylation and remove excess NEM from the samples by buffer exchange using the Microcon-10 kDa filter units (Millipore MRCPRT010). Samples were concentrated and washed thrice with 500 μL SDS-TEA buffer (2% SDS, 150 mM NaCl, 50 mM triethanolamine pH 7.4, 4 mM EDTA) by centrifuging the tubes at 14,000 g for 1.5 h after each addition of SDS-TEA buffer. Samples (∼150 μL) were recovered by inverting the filter device over a new tube and centrifuging at 1,000 g for 5 min. Each sample was then split equally into two tubes (30 μL each). One half of each sample was subjected to hydroxylamine cleavage by adding 90 μL of freshly prepared 1 M NH2OH (Sigma 5470-11-1) in TEA buffer/Triton X-100 buffer pH 7.4 (final NH2OH concentration 0.75 M). As controls, 90 μL of TEA buffer/Triton X-100 buffer pH 7.4 was added to the other half of each sample. After incubation for 1 h with mixing at room temperature, samples were chloroform-methanol precipitated to remove the hydroxylamine by sequential addition of 4, 1.5 and 3 volumes of ice-cold methanol, chloroform and water, respectively. After mixing and centrifuging for 5 min at 20,000 g, 4°C, the upper aqueous layer was removed and 4 volumes of chilled methanol was added prior to mixing by gentle inversion. Proteins were pelleted by centrifuging for 2 min at 20,000 g, 4°C, washed with 1 mL chilled methanol, centrifuged again and air-dried prior to resuspension in 30 μL of 4SB buffer containing 4 mM EDTA. Sonication may be necessary to fully resuspend the proteins. For mPEG alkylation, 90 μL of freshly prepared 1.33 mM 10 kDa mPEG-Mal (Sigma 99126-64-4, final concentration 1 mM) in TEA/Triton X-100 buffer was added to each tube and incubated for 2 h with mixing at room temperature. Samples were then methanol-chloroform precipitated as described above. Air-dried protein pellets were resuspended in 50 μL SDS buffer before the addition of 17.4 μL 4× SDS-loading buffer (20% glycerol, 125 mM Tris·HCl, pH 6.8, 4% SDS, 0.05% bromophenol blue) and 2.6 μL TCEP (Thermo Scientific, 77720). Samples were heated for 5 min at 95°C, separated by SDS-PAGE, and transferred to PVDF membranes for Western blot analysis using the anti-S antibody (Genetex GTX632604) and secondary antibody (Jackson ImmunoResearch 115-035-003).
Cell proliferation assay
Cell proliferation kit was following the manufacturer’s instructions and previously described methods (Tsai et al., 2016). In brief, 1×104 of 293T cells were cultured in 96-well plates. After 2BP treatment at 37°C for 24 h, the MTT labeling reagent (Sigma-Aldrich, Darmstadt, Germany) was added to each well at a final concentration of 0.5 mg/mL for additional 4 h incubation. Finally, the reaction was terminated by adding the stop solution, and the absorbance was measured at 570 nm.
Quantification and statistical analysis
All data are represented as the mean ± SEM and were analyzed using one-way ANOVA with Tukey’s test. A p-value < 0.05 indicates a statistically significant difference.
Acknowledgments
The SARS-CoV-2-related experiments were performed by the P3 research team at NHRI. The confocal images were obtained with assistance from the optical biology core facility of NHRI. The ECD45 antibody was kindly provided by AnTaimmu BioMed. The study was supported by the Ministry of Health and Welfare, Taiwan (MOHW 109-TDU-C-222-000010) and the Ministry of Science and Technology (MOST 109-2327-B-400-004).
Author contributions
C.-F.T., M.M.Z., Y.-H.P., C.-Y.Y., H.-W.C., and G.-Y.Y. designed the experiments. C.-F.T., W.-T.T., C.-H.C., H.-J.C., J.-J.L., Y.-P.K., S.-S.D., W-H.T., and E.-J.L. performed the experiments and analyzed the data. C.-F.T. and G.-Y.Y. prepared the manuscript. All authors participated in discussion, reviewed, and approved the manuscript.
Declaration of interests
All authors declare no competing interests.
Published: August 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104709.
Supplemental information
Data and code availability
-
•
Data reported in this paper will be shared by the lead contact upon request.
-
•
There is no original code associated with this work.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aicart-Ramos C., Valero R.A., Rodriguez-Crespo I. Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta. 2011;1808:2981–2994. doi: 10.1016/j.bbamem.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Benyair R., Ron E., Lederkremer G.Z. Protein quality control, retention, and degradation at the endoplasmic reticulum. Int. Rev. Cell Mol. Biol. 2011;292:197–280. doi: 10.1016/B978-0-12-386033-0.00005-0. [DOI] [PubMed] [Google Scholar]
- Bracquemond D., Muriaux D. Betacoronavirus assembly: clues and perspectives for elucidating SARS-CoV-2 particle formation and egress. mBio. 2021;12:e0237121. doi: 10.1128/mBio.02371-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-Timofte M.E., Arulanandam R., Kurmasheva N., Fu K., Laroche G., Taha Z., van der Horst D., Cassin L., van der Sluis R.M., Palermo E., et al. Antiviral potential of the antimicrobial drug atovaquone against SARS-CoV-2 and emerging variants of concern. ACS Infect. Dis. 2021;7:3034–3051. doi: 10.1021/acsinfecdis.1c00278. [DOI] [PubMed] [Google Scholar]
- Casalino L., Gaieb Z., Goldsmith J.A., Hjorth C.K., Dommer A.C., Harbison A.M., Fogarty C.A., Barros E.P., Taylor B.C., McLellan J.S., et al. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattin-Ortolá J., Welch L.G., Maslen S.L., Papa G., James L.C., Munro S. Sequences in the cytoplasmic tail of SARS-CoV-2 Spike facilitate expression at the cell surface and syncytia formation. Nat. Commun. 2021;12:5333. doi: 10.1038/s41467-021-25589-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A.R., Bhattacharya M., Agoramoorthy G., Lee S.S. Evolution, mode of transmission, and mutational landscape of newly emerging SARS-CoV-2 variants. mBio. 2021;12:e0114021. doi: 10.1128/mBio.01140-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiuppesi F., Salazar M.D., Contreras H., Nguyen V.H., Martinez J., Park Y., Nguyen J., Kha M., Iniguez A., Zhou Q., et al. Development of a multi-antigenic SARS-CoV-2 vaccine candidate using a synthetic poxvirus platform. Nat. Commun. 2020;11:6121. doi: 10.1038/s41467-020-19819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L., Zheng Q., Zhang H., Niu Y., Lou Y., Wang H. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front. Immunol. 2020;11:576622. doi: 10.3389/fimmu.2020.576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13:405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.T.-A., Tien C.-F., Yu G.-Y., Shen S., Lee Y.-H., Hsu P.-C., Wang Y., Chao P.-K., Tsay H.-J., Shie F.-S. The effects of aβ1-42 binding to the SARS-CoV-2 spike protein S1 subunit and angiotensin-converting enzyme 2. Int. J. Mol. Sci. 2021;22:8226. doi: 10.3390/ijms22158226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.C., Lai Y.J., Liao C.C., Yang W.F., Huang K.B., Lee I.J., Chou W.C., Wang S.H., Wang L.H., Hsu J.M., et al. Targeting conserved N-glycosylation blocks SARS-CoV-2 variant infection in vitro. EBioMedicine. 2021;74:103712. doi: 10.1016/j.ebiom.2021.103712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzelnick L.C., Gresh L., Halloran M.E., Mercado J.C., Kuan G., Gordon A., Balmaseda A., Harris E. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358:929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler M., Delguste M., Sieben C., Gillet L., Alsteens D. Initial step of virus entry: virion binding to cell-surface glycans. Annu. Rev. Virol. 2020;7:143–165. doi: 10.1146/annurev-virology-122019-070025. [DOI] [PubMed] [Google Scholar]
- Korner R.W., Majjouti M., Alcazar M.A.A., Mahabir E. Of mice and men: the coronavirus MHV and mouse models as a translational approach to understand SARS-CoV-2. Viruses. 2020;12:880. doi: 10.3390/v12080880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokhin O., Li Y., Andonov A., Feldmann H., Flick R., Jones S., Stroeher U., Bastien N., Dasuri K.V.N., Cheng K., et al. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol. Cell. Proteomics. 2003;2:346–356. doi: 10.1074/mcp.M300048-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]
- Li Y.D., Chi W.Y., Su J.H., Ferrall L., Hung C.F., Wu T.C. Coronavirus vaccine development: from SARS and MERS to COVID-19. J. Biomed. Sci. 2020;27:104. doi: 10.1186/s12929-020-00695-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.-J., Tien C.-F., Kuo Y.-P., Lin E.-J., Tsai W.-H., Chen M.-Y., Tsai P.-J., Su Y.-W., Pathak N., Yang J.-M., et al. Furin and TMPRSS2 resistant spike induces robust humoral and cellular immunity against SARS-CoV-2 lethal infection. Front. Immunol. 2022;13:872047. doi: 10.3389/fimmu.2022.872047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lontok E., Corse E., Machamer C.E. Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J. Virol. 2004;78:5913–5922. doi: 10.1128/JVI.78.11.5913-5922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maemura T., Kuroda M., Armbrust T., Yamayoshi S., Halfmann P.J., Kawaoka Y. Antibody-dependent enhancement of SARS-CoV-2 infection is mediated by the IgG receptors FcgammaRIIA and FcgammaRIIIA but does not contribute to aberrant cytokine production by macrophages. mBio. 2021;12:e0198721. doi: 10.1128/mBio.01987-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malgapo M.I.P., Linder M.E. Substrate recruitment by zDHHC protein acyltransferases. Open Biol. 2021;11:210026. doi: 10.1098/rsob.210026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride C.E., Li J., Machamer C.E. The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J. Virol. 2007;81:2418–2428. doi: 10.1128/JVI.02146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonca L., Howe A., Gilchrist J.B., Sun D., Knight M.L., Zanetti-Domingues L.C., Bateman B., Krebs A.S., Chen L., Radecke J., et al. SARS-CoV-2 assembly and egress pathway revealed by correlative multi-modal multi-scale cryo-imaging. bioRxiv. 2020 doi: 10.1101/2020.11.05.370239. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita F.S., Abrami L., Sergeeva O., Turelli P., Qing E., Kunz B., Raclot C., Paz Montoya J., Abriata L.A., Gallagher T., et al. S-acylation controls SARS-CoV-2 membrane lipid organization and enhances infectivity. Dev. Cell. 2021;56:2790–2807.e8. doi: 10.1016/j.devcel.2021.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nal B., Chan C., Kien F., Siu L., Tse J., Chu K., Kam J., Staropoli I., Crescenzo-Chaigne B., Escriou N., et al. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S. J. Gen. Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- Nishima W., Kulik M. Full-length computational model of the SARS-CoV-2 spike protein and its implications for a viral membrane fusion mechanism. Viruses. 2021;13:1126. doi: 10.3390/v13061126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuya K., Hattori T., Saito T., Takadate Y., Sasaki M., Furuyama W., Marzi A., Ohiro Y., Konno S., Hattori T., Takada A. Multiple routes of antibody-dependent enhancement of SARS-CoV-2 infection. Microbiol. Spectr. 2022;10:e0155321. doi: 10.1128/spectrum.01553-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa G., Mallery D.L., Albecka A., Welch L.G., Cattin-Ortolá J., Luptak J., Paul D., McMahon H.T., Goodfellow I.G., Carter A., et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog. 2021;17:e1009246. doi: 10.1371/journal.ppat.1009246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T.P., Goldhill D.H., Zhou J., Baillon L., Frise R., Swann O.C., Kugathasan R., Penn R., Brown J.C., Sanchez-David R.Y., et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021;6:899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- Peng R., Wu L.A., Wang Q., Qi J., Gao G.F. Cell entry by SARS-CoV-2. Trends Biochem. Sci. 2021;46:848–860. doi: 10.1016/j.tibs.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percher A., Ramakrishnan S., Thinon E., Yuan X., Yount J.S., Hang H.C. Mass-tag labeling reveals site-specific and endogenous levels of protein S-fatty acylation. Proc. Natl. Acad. Sci. USA. 2016;113:4302–4307. doi: 10.1073/pnas.1602244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percher A., Thinon E., Hang H. Mass-Tag labeling using acyl-PEG exchange for the determination of endogenous protein S-fatty acylation. Curr. Protoc. Protein Sci. 2017;89:14.17.1–14.17.11. doi: 10.1002/cpps.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C.M., Chouljenko V.N., Iyer A., Colgrove R., Farzan M., Knipe D.M., Kousoulas K.G. Palmitoylation of the cysteine-rich endodomain of the SARS-coronavirus spike glycoprotein is important for spike-mediated cell fusion. Virology. 2007;360:264–274. doi: 10.1016/j.virol.2006.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveetil R., Lun C.M., Murphy R.E., Healy L.B., Vilmen G., Christenson E.T., Freed E.O., Banerjee A. S-acylation of SARS-CoV-2 spike protein: mechanistic dissection, in vitro reconstitution and role in viral infectivity. J. Biol. Chem. 2021;297:101112. doi: 10.1016/j.jbc.2021.101112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan A.A., Mayilsamy K., McGill A.R., Ghosh A., Giulianotti M.A., Donow H.M., Mohapatra S.S., Mohapatra S., Chandran B., Deschenes R.J., et al. Identification of SARS-CoV-2 spike palmitoylation inhibitors that results in release of attenuated virus with reduced infectivity. Viruses. 2022;14:531. doi: 10.3390/v14030531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajahan A., Supekar N.T., Gleinich A.S., Azadi P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30:981–988. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C.I., Kao Y.T., Chang C.C., Chang Y., Ho T.S., Sun H.S., Lin Y.L., Lai M.M.C., Liu Y.H., Yu C.Y. DNA-induced 2'3'-cGAMP enhances haplotype-specific human STING cleavage by dengue protease. Proc. Natl. Acad. Sci. U S A. 2020;117:15947–15954. doi: 10.1073/pnas.1922243117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztain T., Ahn S.H., Bogetti A.T., Casalino L., Goldsmith J.A., Seitz E., McCool R.S., Kearns F.L., Acosta-Reyes F., Maji S., et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat. Chem. 2021;13:963–968. doi: 10.1038/s41557-021-00758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregoning J.S., Brown E.S., Cheeseman H.M., Flight K.E., Higham S.L., Lemm N.M., Pierce B.F., Stirling D.C., Wang Z., Pollock K.M. Vaccines for COVID-19. Clin. Exp. Immunol. 2020;202:162–192. doi: 10.1111/cei.13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W.T., Lo Y.C., Wu M.S., Li C.Y., Kuo Y.P., Lai Y.H., Tsai Y., Chen K.C., Chuang T.H., Yao C.H., et al. Mycotoxin patulin suppresses innate immune responses by mitochondrial dysfunction and p62/Sequestosome-1-dependent mitophagy. J. Biol. Chem. 2016;291:19299–19311. doi: 10.1074/jbc.M115.686683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H., Pillat M.M., Tárnok A. Dengue fever, COVID-19 (SARS-CoV-2), and antibody-dependent enhancement (ADE): a perspective. Cytometry A. 2020;97:662–667. doi: 10.1002/cyto.a.24047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;183:1735. doi: 10.1016/j.cell.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Sun S., Tai W., Chen J., Geng Q., He L., Chen Y., Wu J., Shi Z., et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J. Virol. 2020;94:e02015-19. doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. Coronaviruses' sugar shields as vaccine candidates. Curr. Trends Immunol. 2020;21:17–23. [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang L., Kuwahara K., Li L., Liu Z., Li T., Zhu H., Liu J., Xu Y., Xie J., et al. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates. ACS Infect. Dis. 2016;2:361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Deng T., Zhang Y., Niu W., Nie Q., Yang S., Liu P., Pei P., Chen L., Li H., Cao B. ACE2 can act as the secondary receptor in the FcgammaR-dependent ADE of SARS-CoV-2 infection. iScience. 2022;25:103720. doi: 10.1016/j.isci.2021.103720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Bowden T.A., Wilson I.A., Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta, Gen. Subj. 2019;1863:1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt M.A. Generation of VSV pseudotypes using recombinant DeltaG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J. Virol. Methods. 2010;169:365–374. doi: 10.1016/j.jviromet.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Huang Y., Lau S.K., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Zhang Z., Wang X., Zhang J., Ren C., Li Y., Gao L., Liang X., Wang P., Ma C. Palmitoylation of SARS-CoV-2 S protein is essential for viral infectivity. Signal Transduct. Targeted Ther. 2021;6:231. doi: 10.1038/s41392-021-00651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Lan Q., Su S., Wang X., Xu W., Liu Z., Zhu Y., Wang Q., Lu L., Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct. Targeted Ther. 2020;5:92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H.L., Wu Y.T., Cao J.L., Yang R., Liu Y.X., Ma J., Qiao X.Y., Yao X.Y., Zhang B.H., Zhang Y.L., et al. Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressing BHK21 cells. Emerg. Microbes Infect. 2020;9:2105–2113. doi: 10.1080/22221751.2020.1815589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Chatterjee V., Ma Y., Zheng E., Yuan S.Y. Protein palmitoylation in leukocyte signaling and function. Front. Cell Dev. Biol. 2020;8:600368. doi: 10.3389/fcell.2020.600368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X.T., Yu X.X., Cheng W. The interactions of ZDHHC5/GOLGA7 with SARS-CoV-2 spike (S) protein and their effects on S protein's subcellular localization, palmitoylation and pseudovirus entry. Virol. J. 2021;18:257. doi: 10.1186/s12985-021-01722-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Chen H., Wang H. Glycans of SARS-CoV-2 spike protein in virus infection and antibody production. Front. Mol. Biosci. 2021;8:629873. doi: 10.3389/fmolb.2021.629873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Zhang C., Li Y., Pearce R., Bell E.W., Zhang Y. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep. Methods. 2021;1:100014. doi: 10.1016/j.crmeth.2021.100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data reported in this paper will be shared by the lead contact upon request.
-
•
There is no original code associated with this work.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.