Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) undergoes mutations at a high rate and with frequent genetic reassortment (antigenic drift/shift), leading to variability in targets. The receptor-binding domain (RBD) of the spike (S) protein has a major role in the binding of SARS-CoV-2 with human angiotensin-converting enzyme 2 (ACE2). Mutations at the RBD influence the binding interaction at the SARS-CoV-2 S–ACE2 interface and impact viral pathogenicity. Here, we discuss different reported mutations of concern in RBD, physicochemical characteristic changes resulting from mutated amino acids and their effect on binding between the RBD and ACE2. Along with mutation informatics, we highlight recently developed small-molecule inhibitors of RBD and the ACE2 interface. This information provides a rational basis for the design of inhibitors against the multivariant strains of SARS-CoV-2.
Keywords: SARS-CoV-2 variants, RBD, Mutation informatics, Entry inhibitors, Spike protein, COVID-19
Introduction
The novel coronavirus 2019 (COVID-19 or SARS-CoV-2) remains a global health emergency. Traditional/rational drug discovery approaches have so far failed to provide effective clinical treatments against multivariant (mutational) strains of coronavirus.1 The fast-emerging mutant strains highlight an urgent need to develop new drugs against wild-type and mutant strains of SARS-CoV-2.
The genome of all organisms serves as a source of biological targets. In drug research and development, identification of an appropriate biological target takes precedence over the design/discovery of its modulator.2 The complete genome (29, 903 nucleotides) of SARS-CoV-2 comprises 11 genes, encoding various proteins. A nucleotide similarity search revealed that SARS-CoV-2 is highly related (89.1% nucleotide similarity) to a group of SARS-like coronaviruses infecting in bats in China.3
The life cycle of SARS-CoV-2 involves various stages.4 The entry of virus into the host cell is the most important stage and is mediated by the S protein. The S protein is ∼ 180 kDa, comprises 1273 amino acids, and has two divisions: S1 (14–685) and S2 (686–1273).5 The S1 subunit is responsible for binding to the host cell receptor, ACE2, and S2 for the fusion of viral and host cell membranes. The S1 and S2 units are proteolytically cleaved at S2′ cleavage sites, which remain noncovalently linked and exist in a prefusion conformation. The S1 unit is further divided into the N-terminal domain (NTD), RBD, and C-terminal domain (CTD), whereas the S2 unit is subdivided in the fusion peptide (FP), heptad repeat, (HR1/2), transmembrane domain (TM), and cytoplasmic tail (CT). The RBD of the distal S1 subunit contributes to stabilization of the prefusion state of the membrane-anchored S2 subunit, which contains the fusion machinery. The S protein exists as a trimer, the three RBDs of which are positioned at its apex. In the ‘closed’ prefusion state, all three RBDs of the S trimer are packed tightly together and are not able to bind to ACE2. During viral entry, the RBDs adopt an ‘open-up’ conformation for binding to ACE2. The resulting post-fusion conformation of the S trimer is obtained following the loss of the NTD. Subsequently, the postfusion state undergoes conformational changes with respect to its prefusion conformation. Stabilization of the postfusion conformational state depends on the re-orientation of the C terminus.6, 7, 8, 9
The SARS-CoV-2 S protein complexed with human ACE2 has been resolved experimentally in various states (closed/prefusion/open/postfusion ACE2-bound/unbound S trimer, double ACE2-bound S trimer, dissociated monomeric S1 bound to ACE2, etc.). The SARS-CoV-2 S protein trimer bound to monoclonal antibodies has also been reported. The RBDs show a closed (‘lying down’) and open (‘standing up’) conformation during binding with ACE2. The 3-D arrangement of the RBDs is highly dynamic and undergoes continuous conformational rearrangement.10 Recent research showed that the rate of spontaneous mutation in SARS-CoV-2 is 1.3 × 10–6 ± 0.2 × 10–6 per base per infection cycle.11 The repeatedly mutating residues can alter the transmissibility (viral spread) and virulence (viral ability to enter and replicate inside host cell) of SARS-CoV-2 and decrease the efficacy of vaccines/drugs, thus posing serious challenges to the discovery of drugs against multivariant strains of SARS-CoV-2.
Given the importance of SARS-CoV-2, attempts have been made to use mutation informatics of the binding pocket of the RBD S protein to advance understanding of its transmissibility/virulence in terms of its ongoing mutations. Here, we use different bioinformatics databases/tools to determine the analytics of the physicochemical properties of the RBD from the S protein from wild-type and mutant strains of SARS-CoV-2. We also highlight the importance of small molecules against the SARS-CoV-2 S–ACE2 interface. Such information provides a rational basis for the development of new vaccines/drugs against this deadly virus.
Mutations in the target site of the S protein RBD
The WHO has reported varied transmissibility and virulence of different mutated strains (alpha, beta, gamma, delta, lambda, and omicron) of SARS-CoV-2.12 Various number of mutations (e.g., T19R, A67V, D80A, D80G, T95I, G142D, Y145D, R158G, L212I, D215G, A222V, N234Q, W258L, N331Q, G339D, N343Q, R346K S371L, S373P, S375F, etc.) have been reported in the S1 protein of these strains since 2019.13, 14 The binding pocket of the RBD is located between S338 to Q506 and the mutations reported in this domain are responsible for altered binding interactions with human ACE2.15 Although D614G is located outside the binding pocket of RBD, it modulates viral transmissibility and infectivity and is thought to result in resistance to COVID-19 vaccines.13, 14
Mutation informatics
A WHO assessment of circulating variants of SARS-CoV-2 states that a clear understanding of the physicochemical characteristics of the mutated amino acid residues of each variant is needed.12 The recent successful application of target-based drug design/protein informatics investigations of mutational effect in drug targets inspired researchers to use these techniques for the development of potential new antiviral agents against wild-type and mutant SARS-CoV-2 strains.2, 16, 17
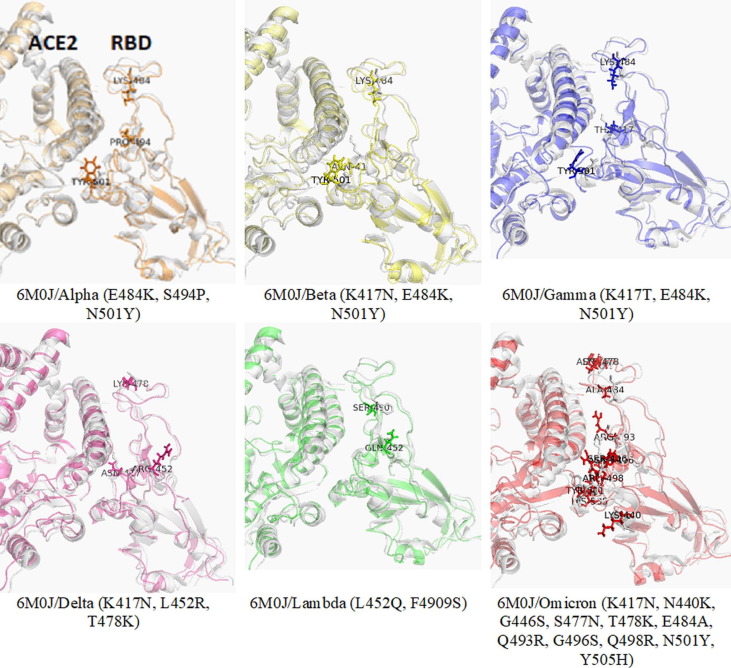
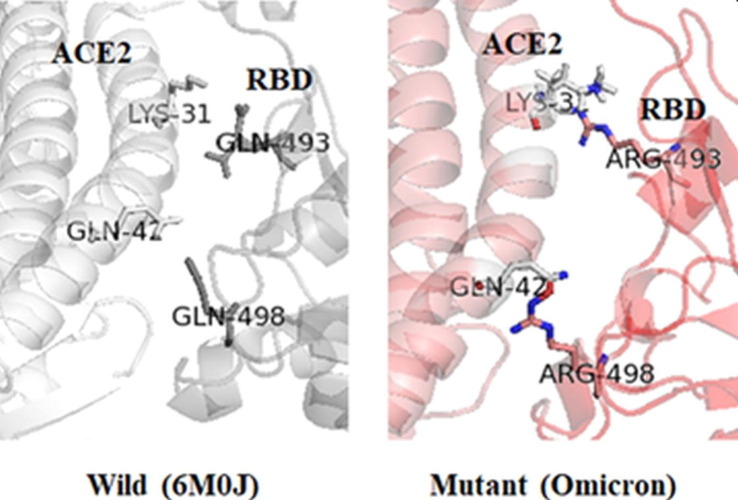
The sequences/3-D structures of relevant wild-type and mutated targets are analyzed with different bioinformatics tools to unravel their changing geometry and physicochemical characteristics.2, 16 Such assessment of physicochemical properties resulting from mutations in the binding pocket of target proteins will help to design potent molecules against multiple strains of SARS-CoV-2. Primary mutation informatics data are shown in Table 1 to understand the effect of mutation on the RBD-ACE2 binding mechanism in the alpha, beta, gamma, delta, lambda, and omicron variants. The point mutation informatics data predicted N501Y, T478K, Q493R, and Q498R as noticeable mutations of SARS-CoV-2, which might be responsible for stabilizing the binding interaction between RBD and ACE2. The mutations significantly impacted the polarity, isoelectric point, and hydropathy index of mutated residues (L452R, Q493R, and Q498R) of the delta and omicron variants. The Q493R and Q498R mutations in omicron resulted in a higher affinity between RBD and ACE2 (Figure 1, Figure 2 ). The results also indicate arginine as a major contributor to the increased affinity between RBD and ACE2 and that it might be responsible for the altered pathogenicity of the virus. These two mutations might also be responsible for the high transmissibility rate in omicron. The replacement of amino acids in the target protein with amino acids with dissimilar physicochemical properties caused changes in the mechanistic interaction between RBD and ACE2.17 Zahradnik et al. predicted the severity of Q498R in the virus even before the emergence of omicron.18 The potential energy (PE: –16 446 kcal/mol) of the RBD–ACE2 structural model of the delta (K417N, L452R, and T478K) variant was lower compared with the alpha, beta, gamma, and lambda structural models (Table 1). However, PE (–11 293 kcal/mol) was higher in the omicron structural model. Thus, the PE indicated that the delta model is more stable than the other models.
Table 1.
Primary mutation informatics of the RBD region of different variants in reference to wild-type protein (PDB code: 6M0J) of SARS-CoV-2a.
| Variants | Position | W/M | Side-chain polarityb | PIc | HId | ΔΔG (kcal/mol)e | Overall stability of modelf | Potential energy of modeled structure (kcal/mol)g |
|---|---|---|---|---|---|---|---|---|
| Alpha | 484 | E | AP | 3.22 | −3.5 | –0.34 | Destabilizing | –13 858 |
| K | BP | 9.73 | −3.9 | |||||
| 494 | S | P | 5.68 | −0.8 | 3.21 | Stabilizing | ||
| P | NP | 6.30 | 3.21 | |||||
| 501 | N | P | 5.41 | −3.5 | 0.78 | Stabilizing | ||
| Y | P | 5.66 | 0.78 | |||||
| Beta | 417 | K | BP | 9.73 | −3.9 | –2.08 | Destabilizing | –12 034 |
| N | P | 5.41 | –2.08 | |||||
| 484 | E | AP | 3.22 | −3.5 | –0.34 | Destabilizing | ||
| K | BP | 9.73 | −3.9 | |||||
| 501 | N | P | 5.41 | −3.5 | 0.78 | Stabilizing | ||
| Y | P | 5.66 | –1.3 | |||||
| Gamma | 417 | K | BP | 9.73 | −3.9 | –0.85 | Destabilizing | –11 779 |
| T | P | 5.60 | −0.7 | |||||
| 484 | E | AP | 3.22 | −3.5 | –0.34 | Destabilizing | ||
| K | BP | 9.73 | –0.34 | |||||
| 501 | N | P | 5.41 | −3.5 | 0.78 | Stabilizing | ||
| Y | P | 5.66 | –1.3 | |||||
| Delta | 417 | K | BP | 9.73 | −3.9 | –2.08 | Destabilizing | –16 446 |
| N | P | 5.41 | −3.5 | |||||
| 452 | L | NP | 5.96 | 3.8 | –0.48 | Destabilizing | ||
| R | BP | 10.76 | −4.5 | |||||
| 478 | T | P | 5.60 | −0.7 | 0.41 | Stabilizing | ||
| K | BP | 9.73 | −3.9 | |||||
| Lambda | 452 | L | NP | 5.96 | 3.8 | –3.5 | Destabilizing | –12 034 |
| Q | P | 5.65 | −3.5 | |||||
| 490 | F | NP | 5.48 | 2.8 | –0.66 | Destabilizing | ||
| S | P | 5.68 | −0.8 | |||||
| Omicron | 417 | K | BP | 9.73 | −3.9 | –2.08 | Destabilizing | –11 293 |
| N | P | 5.41 | −3.5 | |||||
| 440 | N | P | 5.41 | −3.5 | –7.31 | Destabilizing | ||
| K | BP | 9.73 | −3.9 | |||||
| 446 | G | NP | 5.97 | –0.4 | –1.94 | Destabilizing | ||
| S | P | 5.68 | −0.8 | |||||
| 477 | S | P | 5.68 | −0.8 | 0.69 | Stabilizing | ||
| N | P | 5.41 | −3.5 | |||||
| 478 | T | P | 5.60 | −0.7 | 0.41 | Stabilizing | ||
| K | BP | 9.73 | −3.9 | |||||
| 484 | E | AP | 3.22 | −3.5 | –0.43 | Destabilizing | ||
| A | NP | 6.00 | 1.8 | |||||
| 493 | Q | P | 5.65 | −3.5 | 2.89 | Stabilizing | ||
| R | BP | 10.76 | −4.5 | |||||
| 496 | G | NP | 5.97 | –0.4 | –1.96 | Destabilizing | ||
| S | P | 5.68 | −0.8 | |||||
| 498 | Q | P | 5.65 | −3.5 | 2.73 | Stabilizing | ||
| R | BP | 10.76 | −4.5 | |||||
| 501 | N | P | 5.41 | −3.5 | 0.78 | Stabilizing | ||
| Y | P | 5.66 | –1.3 | |||||
| 505 | Y | P | 5.66 | –1.3 | –3.18 | Destabilizing | ||
| H | BP | 7.59 | –3.2 | |||||
| R | BP | 10.76 | −4.5 |
Abbreviations: Ap, acidic polar; Bp, basic polar; M, mutant; N, neutral; Np, nonpolar; P, polar; W, wild type (PDB code: 6M0J).
Side-chain charge, pH 7.4.
Isoelectric point at pH 7.0.
Hydropathy index.
Difference in free energy of unfolding between wild-type and mutant proteins (CUPSAT webtool).
Overall stability of model (CUPSAT webtool).
Figure 1.
The superimposed structure of wild-type severe acute respiratory syndrome-coronavirus 2 (SARS-Cov-2) [Protein Data Bank (PDB): 6M0J] and homology models of the alpha/beta/gamma/delta/lambda/omicron variants highlight the variation in binding interaction between the receptor-binding domain (RBD) and angiotensin-converting enzyme 2(ACE2).
Figure 2.
Magnification of wild-type severe acute respiratory syndrome-coronavirus 2 (SARS-Cov-2) [Protein Data Bank (PDB): 6M0J] and omicron mutations (Q493R and Q498R) highlight major variations in the binding interaction between the receptor-binding domain (RBD) and angiotensin-converting enzyme 2(ACE2).
These preliminary results go some way to explain the mutational effect on RBD–ACE2 binding. High-end molecular dynamic (MD) simulations and experimental studies of generated models will help to further understand the virulence mechanism of different SARS-CoV-2 variants.
Since January 2022, experimental results have been published concerning the delta and omicron SARS-CoV-2 variants.19, 20 These results are comparable to the bioinformatic findings explained herein. Thus, we highlight bioinformatics results that are comparable to experimental findings. The T478K mutation has a key role in stabilizing and reshaping the RBM loop473–490, enhancing the interaction with ACE2.19 The structure of the omicron S trimer with ACE2 revealed that omicron has a six- to ninefold increased affinity for ACE2 because of the additional mutations. The Q498R mutation also increased the affinity of RBD for ACE2. In addition, N501Y and T478K enhanced the RBD–ACE2 interactions. The study also reported that omicron RBD is thermodynamically less stable and more dynamic compared with wild-type RBD.20
Considering the scope of MD simulations in mimicking biological milieu, some research groups have explored the entry and fusion of SARS-CoV-2 into the human environment. Ching et al. discussed the key interacting residues between RBD and ACE2 based on MD simulation results. They found K417, Y449, F486, N487, L455, F456, Y489, Q493, Y495, Q498, T500, N501, and Y505 to be hotspot residues in RBD, and K353, K31, D30, D355, H34, D38, Q24, T27, Y83, Y41, and E35 to be hotspots in ACE2. In the alpha and beta variants of RBD, the binding interactions are slightly different because of the N501Y mutation. The alpha variant binds more tightly with ACE2 compared with the beta variant and wild-type RBD. In the beta variant, K417N reduces the binding affinity with ACE2, whereas E484K slightly increases it. In both the alpha and beta variants, N501Y increases the binding affinity between RBD and ACE2.21 Ma et al. used μs-scale coarse-grained MD simulations of RBD–ACE2 complexes and reported that, in the binding pocket, F486, Q498, and Y505 contribute optimally to the binding free energy between RBD and ACE2.22
Alaofi et al. investigated the effect of N501Y, E484K L452R, S477N, and N439K mutations on the RBD-ACE2 binding mechanism through MD simulation. They revealed that N501Y, N439K, and E484K increased the rigidity of RBD and the flexibility of the receptor-binding motif (RBM). Furthermore, S477N and L452R increased the flexibility of the RBM and maintained the flexibility of RBD compared with the wild-type RBD.23 Rezaei et al. compared the binding behavior of SARS-CoV-2 and SARS-CoV through MD simulations. They also investigated the effect of 42 reported mutations on the thermodynamic stability, free energy, solvation, electrostatic potential, and hydrophobicity of RBD. The study highlighted the criticality of the K417N and N439K mutations and that they might be responsible for the altered binding affinity of RBD for ACE2.24
Dehury et al. validated the effect of mutational residues (Y449A, N487A, Y489A, N501A, and Y505A) on the structure, function, and dynamics of RBD and ACE2 through MD simulation (150 ns) studies. Analysis of trajectories of mutated protein structure models indicated that the loss of important hydrogen bonds and nonbonded contacts was critical for molecular recognition of RBD–ACE2, highlighting the effect of RBD mutations on viral flexibility and, thus, its affinity for ACE2. These results are largely in agreement with previously reported experimental research.25 Veeramachaneni et al. performed MD simulations and analyzed the RBD hotspot amino acid residue interactions with ACE2. Binding interactions, root-mean-square deviation (RMSD), root mean square fluctuation (RMSF), and potential/total energy profile revealed the rigidity and stability of nCoV and ACE2 models.26 Chakraborty et al. also reported the effect of mutations on the binding affinity between SARS-CoV-2 S protein and human ACE2.27
Research is ongoing for the development and application of in silico techniques to learn more about the transmissibility and virulence of different SARS-CoV-2 variants.28, 29, 30 Singh et al. established a web tool by profiling 28 SARS-CoV-2 and coronavirus-associated receptors and factors (SCARFs) using single cell RNA-sequencing data from a range of healthy human tissues to predict the virulence of the virus.28 Greaney et al. mapped all mutations to the RBD that escape binding by different antibodies.29 Recently, Tragni et al. developed a web tool for predicting the RBD/ACE2 binding affinity to advance our understanding of the transmissibility and virulence of different variants of SARS-CoV-2, given that this affinity helps to predict the interaction energy between the protein–protein interface.30
Mutations in the binding pocket of RBD cause many changes in its critical properties, including, shape, size, isoelectric point, surface area, polar/nonpolar area, deep surface area, volume, stability/flexibility of the pocket, and other physiochemical properties. Many of these parameters have not yet been adequately explored. MD simulations work as ‘computational’ microscopes and can provide information about the inter/intramolecular interactions at atomistic level. MD simulations along with an in-depth analysis of the physicochemical characteristics of wild-type and different mutants of SARS-CoV-2 will increase our understanding of the transmissibility and virulence mechanisms. This will provide an opportunity to explore how mutations affect the binding and stability of RBD-ACE2 and help to design broad-spectrum drug candidates against this virus.
RBD–ACE2 interface as a small-molecule target
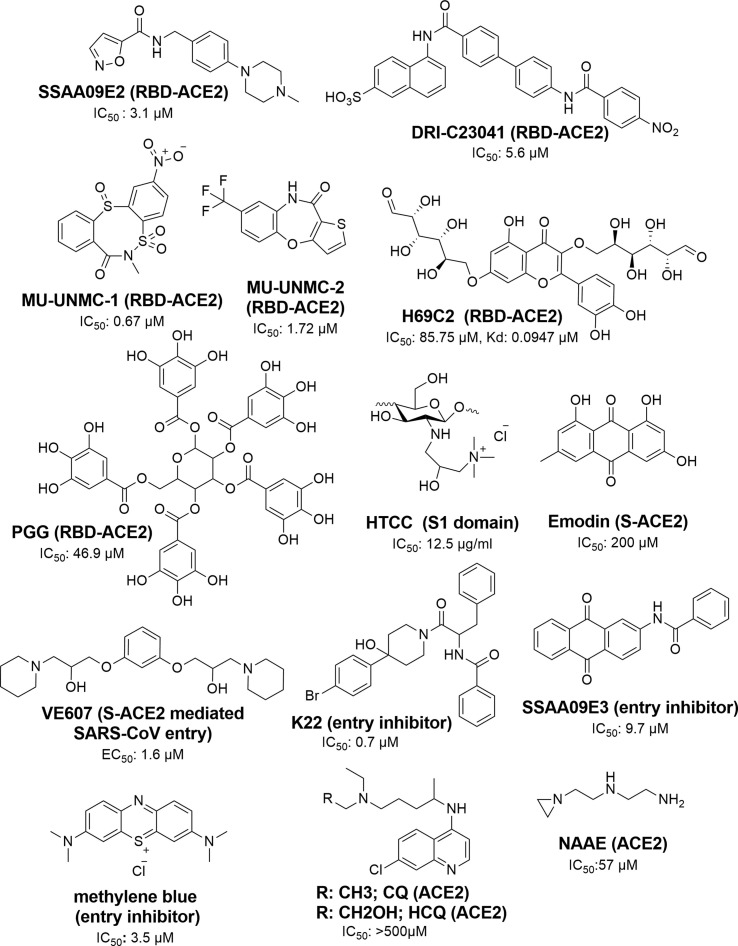
To date, COVID-19 management has relied mainly on the use of vaccines. The US Food and Drug Administration (FDA)-approved remdesivir and monoclonal antibodies (mAbs) are used for the emergency and conditional treatment of patients with COVID-19.31 Current research into drugs to treat COVID-19 includes peptides, antibodies, natural products, drug repurposing, as well as the synthesis of new molecules.32, 33, 34 In drug discovery programs, small molecules as therapeutics agents have an important role because of economic limitations. Various small molecules are reported as inhibitors of SARS-CoV-2 entry [i.e., of S proteins (S1/S2/RBD) and ACE2 (RBD/ACE2 interface)].35, 36 Important scaffolds in the identified molecules include oxazole-carboxamides (e.g., SSAA09E2), benzamides (e.g., DRI-C23041, K22, and SSAA09E3), anthraquinone (e.g., SSAA09E3 and Emodin), piperidine (e.g., VE607), alkylamines (NAEE), carbasugars (e.g., MCCCS-B, GTCP, CDCP, and H69D1), carbohydrate polymers (chitosan derivatives), Methylene Blue derivatives, phenolic derivatives (brazilin, theaflavin-3,3′-digallate, and curcumin), flavanone (e.g., Luteolin) and pentagalloyl glucose (e.g., PGG) (Fig. 3 ).35, 36, 37, 38, 39 Furthermore, a few new classes of compound, namely dithiazocin (MU-UNMC-1) and oxazepin (MU-UNMC-2), were recently discovered as SARS-CoV-2 RBD/ACE2 interface inhibitors.40
Figure 3.
Small-molecule inhibitors of Spike (S) protein receptor-binding domain (RBD) and angiotensin-converting enzyme 2 (ACE2) entry and their targets. HYPERLINK "SPS:refid::bib35_bib36_bib37_bib38_bib39_bib40_bib41_bib42_bib43_bib44_bib45" 35–45.
Among the S protein inhibitors, SSAA09E2, an attachment inhibitor, blocks the binding of SARS-CoV-2 S protein RBD with ACE2 (IC50 = 3.1 μM).35 Razizadeh et al. studied MD simulations of SSAA09E2 and the RBD–ACE2 complex [Protein Data Bank (PDB): 6M0J] and suggested that SSAA09E2 can interfere with the bonded and nonbonded interactions as well as the conformational flexibility of the interaction. The oxygen of the carboxamide group of SSAA09E2 forms a hydrogen bond with RBD by interacting with Gly496, whereas the rest of the molecule shows nonbonded interactions with Arg403, Tyr449, Tyr453, Ser494, Tyr495, Gly496, and Tyr505.41 DRI-C23041 is a newly discovered molecule that acts at the interface of the S proteins (S1 and RBD) and ACE2. The results indicated the strong interaction of this compound with RBD [binding affinity constant (Kd) = 3.7 nM] compared with the S1 protein (Kd = 14.7 nM).36 K22 is a benzamide derivative showing strong inhibitory activity (IC50 = 0.7 μM) against HCoV-229E. It could block the entry port of the virus and is active against range of viruses, including SARS-CoV.35 SSAA09E3 is also a potential inhibitor of SARS-CoV. It contains anthraquinone and benzamide scaffolds and inhibits (IC50 = 9.7 μM) SARS-CoV entry into humans by interfering with the virus–host cell membrane fusion mechanism.35 Emodin, an anthraquinone derivative from plants in the genera Rheum and Polygonum, inhibited S protein (IC50 = 200 μM) and showed dose-dependent interactions with ACE2.35, 42.
The piperidine derivative VE607 was discovered using a phenotype-based high-throughput screening (HTS) method from a set of structurally diverse small molecules. In a pseudotype virus entry assay, VE607 precisely blocked virus entry with an EC50 of 3 μM and inhibited SARS-CoV plaque formation with an IC50 of 1.6 μM. An alkylamine derivative, N-(2-aminoethyl)-1 aziridine-ethanamine (NAAE) was discovered by using a virtual screening approach targeting the ACE2 binding site. NAAE is not only an original ACE2 inhibitor, but also active against the cell fusion mechanism, which is mediated by the S protein of SARS. The compound showed IC50 = 57 μM and Ki = 459 μM against the virus.35
Wang et al. discovered carbasugar molecules (MCCS-B, GTCP, CDCP, and H69D1) and H69C2 as RBD–ACE2 interface inhibitors. Among these compounds, H69C2 showed strong binding affinity (Kd = 0.0947 mM) and antiviral activity (IC50 = 85.75 mM), as well as hydrogen bond interactions with D405, E406, and R408 and a nonbonded interaction with Y505, Y495, S494, and L455.43 Chitosan is a glycan obtained from marine organisms. HTCC, a chitosan derivative, is a potential inhibitor (IC50 = 12.5 mg/ml) of coronaviruses. In vitro and ex vivo studies in human airway epithelia models confirmed that HTCC can effectively block SARS-CoV-2 infection by interacting with the S protein S1 domain.44 In a docking experiment, N-benzyl-O-acetyl-chitosan, imino-chitosan, and sulfated-chitosan showed high binding affinity (–6.0 to –6.6 kcal/mol) with wild-type and mutant variants (alpha/B.1.1.7, gamma/P.1) of the SARS-CoV-2 S protein RBD. These compounds bind with Tyr 449, Asn 501, Tyr 501, Gln 493, and Gln 498. The hydroxyl and amine groups of chitosan have essential role in binding with the RBD.45
In experimental studies, phenolic compounds, including brazilin, theaflavin-3,3′-digallate, curcumin, and flavonoids, were also shown to be RBD inhibitors, with brazilin, TF-3, and curcumin affecting the cellular entry of SARS-CoV-2.38 In silico studies predicted that the inhibitory activity of flavonoids results from its binding with ACE2. In this class, luteolin gained attention as a SARS-CoV-2 inhibitor.44 Methylene Blue, a tricyclic phenothiazine, inhibited the entry of a pseudovirus S protein into ACE2-expressing cells (IC50 3.5 μM).37 Although, chloroquine (CQ) and hydroxychloroquine (HCQ) inhibited SARS-CoV-2 during in vitro experiment, CQ, siramesine, and suramin failed to exhibit inhibitory activity in this assay. The mode of action of this class has indicated that they modulate the restriction of pH-mediated S protein cleavage at the ACE2 binding site.37
A natural product, PGG, showed binding with RBD in a docking and biolayer interferometry (BLI) binding assay. The compound has low binding energy (–8 kcal/mol) and resides in-between binding residues from Glu 340 to Lys 356.39 Recently, Acharya et al. identified MU-UNMC-1 (IC50 = 0.67 mM) and MU-UNMC-2 (IC50 = 1.72 mM) binding to the interface of RBD–ACE2 through virtual screening/docking of 8 million compounds. These compounds were also active against SARS-CoV-2 variant beta/B.1.351 (IC50 = 9.27 and 3.00 mM, respectively) and the Scotland variant B.1.222 (IC50 = 2.64 mM). MU-UNMC-1 showed binding interactions with K417, R403, Q409, and Y505, and MU-UNMC-2 with R403, Y453, and Y505.40
Fig. 3 details the chemical structures of reported inhibitors against the RBD–ACE2 interface. These inhibitors have considerable substructural/functional group similarity. The functional group similarities (benzamide, piperidine, naphthalene, anthraquinone, phenolic groups, and alkylamines) in reported inhibitors indicate the importance of these groups in the design of potential molecules against SARS-CoV-2. Details of the functional groups, physicochemical properties of reported inhibitors, and binding pocket (surface area, polar or nonpolar side-chain polarities, electrostatic interactions, etc.) of wild-type and mutant virus will help to design novel and potential molecules against SARS-CoV-2. It also provides scope to develop potential hybrid drug molecules to treat the infection. Mutations cause changes in residues, whereas backbone atoms remain unchanged. Thus, one could design molecules to provide strong interactions with the backbone atoms of the target proteins. In another approach, molecules can be designed to interact stereo-selectively with the conserved residues of the binding pocket. Some studies have reported that mutations will not occur in conserved residues. Thus, another strategy is to design molecules against dual/multiple targets of the organism, which would increase the chance of the molecules being effective against multiple strains of the virus.2
Concluding remarks
Combating SARS-CoV-2 variants is a global challenge. This review has highlighted new ideas for the design and development of inhibitors against wild-type and mutant SARS-CoV-2 involving mutation informatics. Investigation of the physicochemical characteristics of each mutation is crucial to understand their transmissibility and virulence. The information obtained can be used rationally to design drugs against multivariant strains. Different physicochemical properties indicated that mutations reported in the delta (L452R) and omicron (Q493R, Q498R) variants have predicted major variation in terms of the RBD–ACE2 interaction. In a model of the RBD–ACE2 interaction in omicron, Q493R and Q498R of the RBD showed stronger interactions with ACE2 compared with the wild type (PDB: 6M0J). The reported small-molecule inhibitors are active against the RBD–ACE2 interface and show significant structural (benzamide, piperidine, naphthalene, anthraquinone, phenolic group and alkylamines) similarities, which offers scope to develop new potent hybrid molecules. Inhibitors rationally designed to bind to the backbone residues/conserved residues/multiple targets of SARS-CoV-2 provide significant scope to develop effective treatments against this deadly virus.
Acknowledgements
The authors thank the reviewers and Dr. Y.S. Prabhakar, Chief Scientist of CSIR-Central Drug Research Institute (India) for scientific input in the manuscript revision.
References
- 1.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verma S., Prabhakar Y.S. Target based drug design - a reality in virtual sphere. Curr Med Chem. 2015;22:1603–1630. doi: 10.2174/0929867322666150209151209. [DOI] [PubMed] [Google Scholar]
- 3.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercurio I., Tragni V., Busto F., De Grassi A., Pierri C.L. Protein structure analysis of the interactions between SARS-CoV-2 spike protein and the human ACE2 receptor: from conformational changes to novel neutralizing antibodies. Cell Mol Life Sci. 2021;78:1501–1522. doi: 10.1007/s00018-020-03580-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turonova B., Sikora M., Schürmann C., Hagen W.J.H., Welsch S., Blanc F.E.C., et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J., Xiao T., Cai Y., Chen B. Structure of SARS-CoV-2 spike protein. Curr Opin Virol. 2021;50:173–182. doi: 10.1016/j.coviro.2021.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amicone M., Borges V., Alves M.J., Isidro J., Ze-Ze L., Duarte S., et al. Mutation rate of SARS-CoV-2 and emergence of mutators during experimental evolution. Evol Med Public Health. 2022;10:142–155. doi: 10.1093/emph/eoac010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.WHO. Tracking SARS-CoV-2 Variants. Geneva; WHO; 2022.
- 13.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.CDC. SARS-CoV-2 Variant Classifications and Definitions. Atlanta; CDC; 2022uns.
- 15.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 16.Debnath U., Verma S., Jain S., Katti S.B., Prabhakar Y.S. Pyridones as NNRTIs against HIV-1 mutants: 3D-QSAR and protein informatics. J Comput Aided Mol Des. 2013;27:637–654. doi: 10.1007/s10822-013-9667-1. [DOI] [PubMed] [Google Scholar]
- 17.Iqbal S., Pérez-Palma E., Jespersen J.B., May P., Hoksza D., Heyne H.O., et al. Comprehensive characterization of amino acid positions in protein structures reveals molecular effect of missense variants. Proc Natl Acad Sci U S A. 2020;117:28201–28211. doi: 10.1073/pnas.2002660117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zahradník J., Marciano S., Shemesh M., Zoler E., Harari D., Chiaravalli J., et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat Microbiol. 2021;6:1188–1198. doi: 10.1038/s41564-021-00954-4. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y., Liu C., Zhang C., Wang Y., Hong Q., Xu S., et al. Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies. Nat Commun. 2022;13:871. doi: 10.1038/s41467-022-28528-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin W., Xu Y., Xu P., Cao X., Wu C., Gu C., et al. Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody. Science. 2022;375:1048–1053. doi: 10.1126/science.abn8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jawad B., Adhikari P., Podgornik R., Ching W.Y. Key interacting residues between RBD of SARS-CoV-2 and ACE2 receptor: combination of molecular dynamics simulation and density functional calculation. J Chem Inf Model. 2021;61:4425–4441. doi: 10.1021/acs.jcim.1c00560. [DOI] [PubMed] [Google Scholar]
- 22.Koehler M., Ray A., Moreira R.A., Juniku B., Poma A.B., Alsteens D. Molecular insights into receptor binding energetics and neutralization of SARS-CoV-2 variants. Nat Commun. 2021;12:6977. doi: 10.1038/s41467-021-27325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alaofi A.L., Shahid M. Mutations of SARS-CoV-2 RBD may alter its molecular structure to improve its infection efficiency. Biomolecules. 2021;11:1273. doi: 10.3390/biom11091273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rezaei S., Sefidbakht Y., Uskoković V. Comparative molecular dynamics study of the receptor-binding domains in SARS-CoV-2 and SARS-CoV and the effects of mutations on the binding affinity. J Biomol Struct Dyn. 2022;40:4662–4681. doi: 10.1080/07391102.2020.1860829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dehury B., Raina V., Misra N., Suar M. Effect of mutation on structure, function and dynamics of receptor binding domain of human SARS-CoV-2 with host cell receptor ACE2: a molecular dynamics simulations study. J Biomol Struct Dyn. 2021;39:7231–7245. doi: 10.1080/07391102.2020.1802348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veeramachaneni G.K., Thunuguntla V.B.S.C., Bobbillapati J., Bondili J.S. Structural and simulation analysis of hotspot residues interactions of SARS-CoV 2 with human ACE2 receptor. J Biomol Struct Dyn. 2021;39:4015–4025. doi: 10.1080/07391102.2020.1773318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakraborty S. Evolutionary and structural analysis elucidates mutations on SARS-CoV2 spike protein with altered human ACE2 binding affinity. Biochem Biophys Res Commun. 2021;534:374–380. doi: 10.1016/j.bbrc.2020.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh M., Bansal V., Feschotte C. A single-cell RNA expression map of human coronavirus entry factors. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greaney A.J., Starr T.N., Barnes C.O., Weisblum Y., Schmidt F., Caskey M., et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat Commun. 2021;12:4196. doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tragni V., Preziusi F., Laera L., Onofrio A., Mercurio I., Todisco S., et al. Modeling SARS-CoV-2 spike/ACE2 protein-protein interact-ions for predicting the binding affinity of new spike variants for ACE2, and novel ACE2 structurally related human protein targets, for COVID-19 handling in the 3PM context. EPMA J. 2022;13:149–175. doi: 10.1007/s13167-021-00267-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.FDA. Know Your Treatment Options for COVID-19. Silver Spring; FDA: 2022.
- 32.Aleem A., Akbar Samad A.B., Slenker A.K. StatPearls Publishing; 2022. Emerging variants of SARS-CoV-2 and novel therapeutics against coronavirus (COVID-19) [Google Scholar]
- 33.Shah J.N., Guo G.Q., Krishnan A., Ramesh M., Katari N.K., Shahbaaz M., et al. Peptides-based therapeutics: emerging potential therapeutic agents for COVID-19. Therapie. 2022;77:319–328. doi: 10.1016/j.therap.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singla R.K., He X., Chopra H., Tsagkaris C., Shen L., Kamal M.A., et al. Natural products for the prevention and control of the COVID-19 pandemic: sustainable bioresources. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.758159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiu S., Dick A., Ju H., Mirzaie S., Abdi F., Cocklin S., et al. Inhibitors of SARS-CoV-2 entry: current and future opportunities. J Med Chem. 2020;63:12256–12274. doi: 10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bojadzic D., Alcazar O., Chen J., Chuang S.T., Condor Capcha J.M., et al. Small-molecule inhibitors of the coronavirus spike: ACE2 protein-protein interaction as blockers of viral attachment and entry for SARS-CoV-2. ACS Infect Dis. 2021;7:1519–1534. doi: 10.1021/acsinfecdis.1c00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bojadzic D., Alcazar O., Buchwald P. Methylene Blue inhibits the SARS-CoV-2 spike-ACE2 protein-protein interaction-a mechanism that can contribute to its antiviral activity against COVID-19. Front Pharmacol. 2021;11 doi: 10.3389/fphar.2020.600372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goc A., Sumera W., Rath M., Niedzwiecki A. Phenolic compounds disrupt spike-mediated receptor-binding and entry of SARS-CoV-2 pseudo-virions. PLoS ONE. 2021;16 doi: 10.1371/journal.pone.0253489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen R.H., Yang L.J., Hamdoun S., Chung S.K., Lam C.W., Zhang K.X., et al. 1,2,3,4,6-Pentagalloyl glucose, a RBD-ACE2 binding inhibitor to prevent SARS-CoV-2 infection. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.634176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Acharya A., Pandey K., Thurman M., Klug E., Trivedi J., Sharma K., et al. Discovery and evaluation of entry inhibitors for SARS-CoV-2 and its emerging variants. J Virol. 2021;95 doi: 10.1128/JVI.01437-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Razizadeh M, Nikfar M, Liu Y. Small molecules to destabilize the ACE2-RBD complex: a molecular dynamics study for potential COVID-19 therapeutics. ChemRxiv. Published online December 16, 2020; http://dx.doi.org/10.26434/chemrxiv.13377119.v1.
- 42.Ho T.Y., Wu S.L., Chen J.C., Li C.C., Hsiang C.Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 2007;74:92–101. doi: 10.1016/j.antiviral.2006.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L., Wu Y., Yao S., Ge H., Zhu Y., Chen K., et al. Discovery of potential small molecular SARS-CoV-2 entry blockers targeting the spike protein. Acta Pharmacol Sin. 2021;1–9 doi: 10.1038/s41401-021-00735-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milewska A., Chi Y., Szczepanski A., Barreto-Duran E., Dabrowska A., Botwina P., et al. HTCC as a polymeric inhibitor of SARS-CoV-2 and MERS-CoV. J Virol. 2021;95:e01622–e1720. doi: 10.1128/JVI.01622-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Modak C., Jha A., Sharma N., Kumar A. Chitosan derivatives: a suggestive evaluation for novel inhibitor discovery against wild type and variants of SARS-CoV-2 virus. Int J Biol Macromol. 2021;187:492–512. doi: 10.1016/j.ijbiomac.2021.07.144. [DOI] [PMC free article] [PubMed] [Google Scholar]