Graphical abstract
Keywords: Lipid nanoparticles, LNP, Ionizable lipid, PEGylated lipid, Helper lipid, Nucleic acid, Physicochemical properties, pKa, Drug delivery, Targeting
Abstract
Lipid nanoparticles (LNPs) play an important role in mRNA vaccines against COVID-19. In addition, many preclinical and clinical studies, including the siRNA-LNP product, Onpattro®, highlight that LNPs unlock the potential of nucleic acid-based therapies and vaccines. To understand what is key to the success of LNPs, we need to understand the role of the building blocks that constitute them.
In this Review, we discuss what each lipid component adds to the LNP delivery platform in terms of size, structure, stability, apparent pKa, nucleic acid encapsulation efficiency, cellular uptake, and endosomal escape. To explore this, we present findings from the liposome field as well as from landmark and recent articles in the LNP literature. We also discuss challenges and strategies related to in vitro/in vivo studies of LNPs based on fluorescence readouts, immunogenicity/reactogenicity, and LNP delivery beyond the liver. How these fundamental challenges are pursued, including what lipid components are added and combined, will likely determine the scope of LNP-based gene therapies and vaccines for treating various diseases.
1. Introduction
The tremendous success of the vaccines from Pfizer-BioNTech (BNT162b2, also known as Comirnaty®) [1] and Moderna (mRNA-1273, also known as Spikevax®) [2] in combatting COVID-19 has demonstrated the value and rapid translational potential of lipid nanoparticles (LNPs). For the purposes of this Review, we define LNPs as sub-micron particles containing ionizable cationic lipids in addition to other types of lipids and encapsulated nucleic acid cargo. The LNP delivery system is vital to the success of these vaccines. LNPs act as a protective capsule for the nucleic acid cargo and prevent enzymatic degradation until nucleic acid delivery to the cytosol of the target cell. In the COVID-19 vaccines, the mRNA encodes an antigen, specifically a modified version of the SARS-CoV-2 spike surface protein, that triggers an immune response including the production of neutralizing antibodies.
Many years of research have culminated in modern LNP technology, which is now the most clinically advanced non-viral gene delivery system. Following in the footsteps of Onpattro®, the first RNA interference therapeutic approved by the FDA in 2018, LNP technology now enables gene editing, protein replacement, and vaccines [3]. In Onpattro®, LNP delivers short interfering RNA (siRNA) to the liver to silence the expression of the protein transthyretin, which causes transthyretin amyloidosis (ATTR). Developing and scaling-up Onpattro® provided a path for the LNP-mRNA vaccines, the fastest vaccines to ever be produced, that are central to the fight against COVID-19.
Preclinical studies have used LNPs to deliver nucleic acids beyond siRNA and mRNA, such as antisense oligonucleotides (ASOs) [4], microRNA [5], and DNA [6]. Several clinical trials are currently assessing LNP delivery of a broad range of payloads, including the first in vivo CRISPR/Cas9 treatment delivered intravenously (i.v.) to treat patients with ATTR [7]. Hence, LNPs seem to be a versatile nucleic acid delivery platform that overcomes major hurdles in gene therapy, namely nucleic acid degradation and limited cellular uptake. Here we define gene therapy broadly to include nucleic acid modalities that alter specific protein expression in cells to treat disease. These modalities include siRNA and ASOs to e.g., reduce the production of disease-causing proteins, mRNA and DNA to enable the generation of essential proteins that are missing or impaired in genetic diseases, and CRISPR/Cas9 to edit or inactivate defective genes. Cas9 enzyme and guide RNA can be delivered at the level of DNA and RNA. LNPs loaded with these new therapeutic modalities could hugely impact immuno-oncology and treatments for rare genetic and undruggable diseases, in addition to building on the recent success in rapid vaccine development.
Many informative reviews about employing LNPs for vaccines and gene therapy [8], [9] have largely focused on how nucleic acid cargo elicits biological changes and enacts therapeutic effects, with less attention to the LNP. In this Review, by contrast, we assess the liposome and the LNP literature with a focus on the various lipid-based building blocks that constitute LNPs in order to explicate what each lipid component adds to the LNP delivery platform in terms of size, structure, stability, nucleic acid encapsulation efficiency, cellular uptake, and endosomal escape. We also present considerations about the pKa properties of ionizable lipids as this parameter is key for efficient transfection [10], [11], [12]. Finally, we discuss challenges and strategies related to immunogenicity and LNP targeting beyond the liver. How these fundamental challenges are solved will likely determine the breadth of diseases LNP-based gene therapies can cover. To make a concise Review, we decided to focus mainly on the four specific lipid types currently used in the clinic [1], [2], [3] (Fig. 1 ). How preparation methods impact the physiochemical properties of LNPs lies beyond our current scope. Note that by lipid components we refer to amphipathic constituents and thus not solely biological lipids.
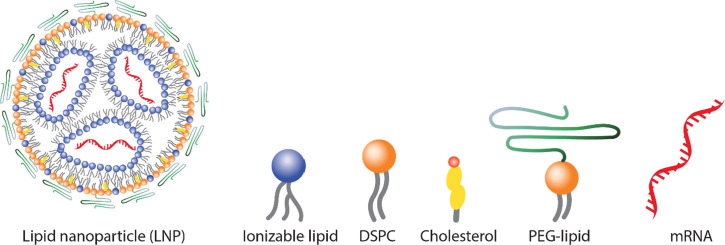
Fig. 1.
Simplistic illustration of LNP and its individual components, the focus of this Review. The LNP can encapsulate many nucleic acid cargo types including but not limited to DNA, ASOs, siRNA, microRNA, and mRNA (shown in this figure).
The aim of this Review is to help researchers understand the fundamental construction of the LNP delivery platform, and highlight challenges and opportunities with the technology. We hope to provide researchers with the pre-requisite knowledge to better design the next generation of LNPs.
2. The lipid components of clinically approved LNPs
All the current FDA-approved LNP formulations contain four lipids (depicted in Fig. 1) [13], [14], [15]: (1) an ionizable cationic lipid, helper lipids which include (2) 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) and (3) cholesterol, and (4) a polyethylene glycol (PEG)-lipid conjugate. These constituents facilitate monodisperse nanoparticle formation, improve nanoparticle stability, enable efficient nucleic acid encapsulation, aid cellular uptake, and promote endosomal escape of nucleic acid cargo.
The composition of modern nucleic acid delivery systems derives from traditional liposomal systems for small-molecule therapeutics. One example is liposomal doxorubicin, also called Doxil®, the first FDA-approved nanomedicine [16], which contains hydrogenated soy phosphatidylcholine (mainly DSPC), cholesterol, and PEG-lipid at a 56:38:5 molar ratio [17]). This formulation is the result of nearly two decades of optimization to ensure robust manufacturing methods in order to achieve the desired particle properties: higher encapsulation efficiencies using remote-loading processes, fewer unfavorable interactions with serum proteins, longer circulation lifetimes to take advantage of the enhanced permeation and retention (EPR) effect [18], better drug accumulation at the target site, and fewer dose-limiting toxicities. Applying such systems to nucleic acid delivery revealed that the large size and high negative charge density of nucleic acids required additional lipid functionalities, including active encapsulation methods that typical lipid components did not provide. Iteratively improved formulation design, manufacturing processes, and ionizable lipid efficacy and tolerability resulted in the LNP formulation typically used today. This formulation contains ionizable lipid, phospholipid, cholesterol, and PEG-lipid (about 50:10:38.5:1.5 mol%), the roles of which will be discussed below.
3. Role of different lipids in LNPs
Many of the structural and biological properties of LNPs are not solely ascribed to a single lipid component but rather the combination of lipids. With this in mind, we will describe how different lipids promote key LNP features.
3.1. Role of ionizable cationic lipids
Ionizable cationic lipids are defined (typically) by a tertiary amine that is deprotonated under neutral conditions and is positively charged in pH conditions below the acid-dissociation constant (pKa) of the lipid. They serve two key functions including facilitating nucleic acid encapsulation in LNPs and mediating endosomal membrane disruption to enable nucleic acid release to the cytosol. Further, the ionizable lipids may also play an important role in endosomal uptake, either directly through interaction between the positive charge on some ionizable lipids and the negatively charged cell membranes or via their binding to plasma proteins that support cellular uptake.
Over the past 20 years, ionizable lipid design has undergone numerous improvements leading to much higher transfection potency. The first ionizable lipid reported AL1 [19], also known as DODAP, was synthesized to assess the role of asymmetric lipid distribution in a lipid bilayer (inner vs outer leaflet). In the application of such lipids for nucleic acid delivery, therapeutic levels of delivery were not obtained due to the high doses required. Improvements to the lipid design resulted in nearly 8000-fold improvements in the therapeutic index. First, systematic study of lipid tail saturation has demonstrated that the linoleyl-derivative of 1,2-dioleyloxy-3-dimethylaminopropane (DODMA)), known as 1,2-dilinoleyloxy-n,n-dimethyl-3-aminopropane (DLinDMA), dramatically improved gene silencing, and additional lipid tail unsaturation did not provide additional benefits [20]. Interestingly, the ester-containing analogue, 1,2-bis(linoleoyloxy)-3-(dimethylamino)propane (DLinDAP), was found to be ineffective and required substantially higher doses because of labile bonds [21], [22]. Using the Factor VII murine model, the rational-design process next identified 2-[2,2-bis[(9Z,12Z)-octadeca-9,12-dienyl]-1,3-dioxolan-4-yl]-N,N-dimethylethanamine (DLin-KC2-DMA) or KC2 [22], a ketal-containing compound with 10-fold more activity than DLinDMA; the effective dose required to achieve 50% gene silencing was 0.1 mg siRNA per kg body weight with KC2, but DLinDMA required 1.0 mg/kg.
The apparent pKa value (pKa of the ionizable lipids likely at the LNP surface (see discussion in Section 3.1.2)) has proven critical for efficient LNP delivery and transfection potency [10], [11], [12]. The landmark study by Jayaraman et al. [10], shows a strong correlation between hepatic gene-silencing activity in the Factor VII model and the apparent pKa of LNPs based on different ionizable lipid components. The correlation follows a bell-shaped curve with an optimum between 6.2 and 6.5. The ionizable lipid (6Z,9Z,28Z,31Z)-Heptatriaconta-6,9,28,31-tetraen-19-yl 4-(dimethylamino)butanoate (DLin-MC3-DMA) or MC3 with an apparent pKa 6.44 used in Onpattro® was identified from a library of 56 ionizable lipids consisting of a dilinoleyl-based hydrophobic tail with varying headgroups [10]. Subsequently, Whitehead et al. [11] prepared 1,400 lipid-like ionizable compounds, termed lipidoids, for siRNA delivery to develop robust molecular structure–function relationships. The full library was screened using an in vitro gene-silencing assay, and the 82 lipids that silenced more than 50% of firefly luciferase were further evaluated for in vivo activity. In total, 96 lipidoids were tested for their ability to knockdown the liver-expressed Factor VII protein in mice. This process revealed four knockdown efficacy criteria. Lipidoids containing at least one tertiary amine, at least three alkyl chains, and 13-carbon long alkyl chains are optimal; however, pKa appeared to be the most influential factor regarding in vivo gene-silencing efficacy in hepatocytes upon i.v. administration to mice. Indeed, pKa was the only criterion in this study [11] that, when not met, abolishes the ability of an LNP to facilitate high levels of gene silencing.
A team from Moderna synthesized 30 ionizable lipids and determined that ‘Lipid H’ (SM-102) (pKa 6.68) is the best ionizable lipid for mRNA vaccine delivery. Its attractive properties include good biodegradability, tolerability, protein expression, and immunogenicity [23]. This study showed that Lipid H performed better than MC3. Several studies have highlighted that LNP formulations optimized for siRNA delivery are not ideal for other payloads like mRNA and DNA [6], [24].
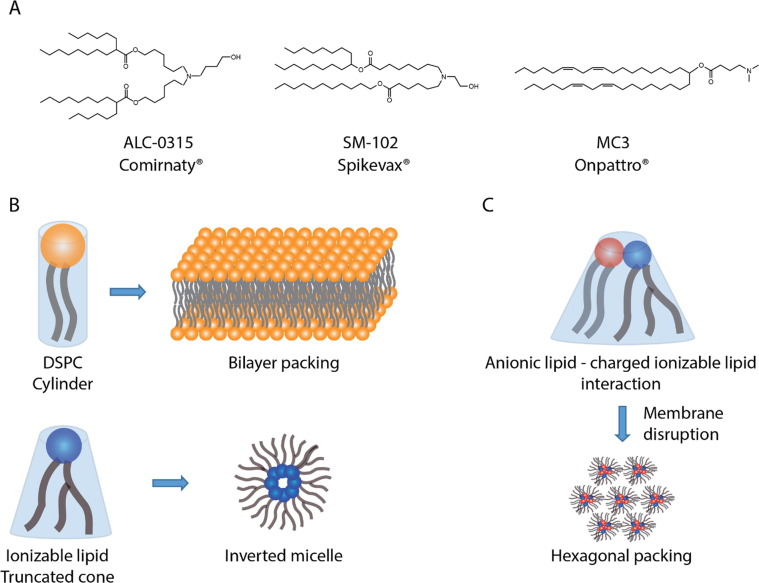
The three different ionizable lipids used in Comirnaty®, Spikevax®, and Onpattro® (current LNP-based clinical products) are shown in Fig. 2 A.
Fig. 2.
A) The ionizable lipids used in the COVID-19 LNP vaccines Comirnaty® (ALC-0315) and Spikevax® (SM-102) and in Onpattro® (MC3). B) Lipid packing theory explaining the relation between the molecular shape of amphipathic compounds (in this case DSPC and an arbitrary ionizable lipid) and the geometry of their self-assembled structures (a detailed description of the critical packing parameters of surfactants can be found in [25], [26]). C) The proposed mechanism by which charged ionizable lipids mediate endosomal disruption [22].
3.1.1. Ionizable lipids enable efficient encapsulation of nucleic acids in LNPs
Ionizable lipids enable significant nucleic acid cargo encapsulation in LNPs, typically > 90% across nucleic acid modalities [27]. During particle formation, the ionizable lipids become positively charged, and, through electrostatic interactions with the negatively charged phosphate backbone of nucleic acid polymers, promote nucleic acid incorporation into the emerging nanoparticle. To ensure proper ionizable lipid protonation, the LNPs are prepared at an acidic pH (∼4) significantly lower than the apparent pKa value of the ionizable lipids (typically ∼ 6.5), followed by a buffer/pH exchange step. In 2012, a proposed mechanism [28] for this process hypothesized that ionizable lipid binding around the nucleic acid cargo in LNPs formed an inverted micellar arrangement of ionizable lipids in the core (Fig. 1), as suggested by in silico modelling. Since ionizable lipids are typically designed so that a cross-sectional area of the tail group is larger than that of the hydrophilic head groups, these lipids exhibit an inverted cone geometry (Fig. 2B). As per the molecular shape hypothesis [25], [26] this favors inverted micellular-like packing of the ionizable lipids around the nucleic acids in the core (Fig. 1). Recent studies suggest alternative LNP structures, which we elaborate on in Section 4.
3.1.2. The apparent pKaof ionizable lipids
As the apparent pKa of ionizable lipids regulates efficient LNP mediated transfection [10], [11], [12], we will outline some parameters that determine the apparent pKa within an LNP. The standard 2-(p-toluidino)-6-napthalene sulfonic acid (TNS) binding assay can assess the apparent pKa of LNPs. In aqueous solution, TNS does not fluoresce. Upon binding to a positively charged membrane through ionic interactions, the water molecules are excluded from the environment, thereby resulting in a fluorescent signal. Hence, the TNS-derived pKa values likely indicate the pKa properties of the easy accessible ionizable lipids at the LNP surface and are thus referred to as the apparent pKa [29]. However, it should be stated there is, thus far, no reported evidence that the TNS only measures ionizable lipids at the LNP surface. A recent study [30] showed that the difference between the apparent pKa derived from the TNS binding assay and the pKa derived from H-NMR measurements of the bare headgroup moiety, where the tertiary amine is present in aqueous solution, is around 2–3 pH units. The authors proposed a thermodynamical model that accounts for the measured pKa differences between NMR and TNS binding assays including the proton solvation free energy associated with the proton energy transfer from water to LNP and proton electrostatic repulsion or attraction to the LNP due to its electric potential. That said, the pKa values of the ionizable lipids in the LNP core, where most of the ionizable lipids are expected to be present due to their association with the nucleic acid cargo, could likely be higher than 9–10 due to the favorable electrostatic interactions between the protonated ionizable lipid and the negatively charged backbone of the encapsulated nucleic acids. Along these lines, monomeric tertiary amines are typically used as the ionizable functionality in ionizable lipids and have pKa values around 9–11 [31]. The alkyl groups should have a minor direct stabilizing or destabilizing effect on the protonated amine. In contrast heteroatoms including oxygen close to the nitrogen atom can lower the pka significantly like in case triethanolamine (pka 7.7). Further, the proximity between two amino groups within the same molecule greatly impacts their pKa values. For example, the two pKa values of 1,2-ethanediamine are ∼ 9.9 and ∼ 7.1 [31]. The different pKa values are due to an unfavorable charge repulsion between the two protonated amine groups in proximity, and thus, a lower pH is needed to protonate half of the second amino group. With this in mind, including the H-NMR study above [30], we need to emphasize that the LNP surface local environment is composed of ionizable lipids, the zwitterionic DSPC, and cholesterol, each of which contributes a different chemical structure including a tertiary amine, a quaternary amino group, a negatively charged phosphate, and a hydroxyl group from cholesterol. All these components can alter the pKa of the ionizable lipid due to proximity between the headgroups. The proximity between the charged moieties depends on the lipid geometrical packing which is determined by carbon tail saturation and length as well as lipid headgroup polarity (and effective charge density). Regarding the latter, an ionizable library based on a dilinoleyl hydrophobic tail and varying headgroups exhibited a wide range of apparent pKa values (4.17–8.12) [10]. Hence, the pKa of the ionizable lipid measured in LNPs likely depends on the molecular structure of its headgroup [10], its hydrophobic tail groups [32], and neighboring lipid packing, including LNP lipid composition [33]. Another study [29] showed that the measured pKa of ionizable lipids when dissolved in micelles (consisting of neutral surfactants) decreased when the number of ionizable lipids per micelle increased. This correlation supports the notion that repulsive interactions between closely packed charged ionizable lipids lower the apparent pKa of the ionizable lipid.
In addition to molecular structure and packing, which partly determine the electrostatic interactions that impact the pKa, environmental parameters, such as ionic strength and the dielectric constant, also influence the measured pKa. Therefore, a lower local dielectric constant at the lipid-water interface compared to the bulk aqueous solution may elevate the charge effects on the pKa value.
The research summarized above highlights the complexity of the pKa value of ionizable lipids. Such complexity contributes to the intricacy of the LNP structure, the mechanisms by which LNPs are endocytosed, and the subsequent endosomal disruption mediated by the ionizable lipids.
3.1.3. Ionizable lipids may be essential for cellular internalization of LNPs
Whether the ionizable lipids play a direct role in endosomal uptake has not been thoroughly investigated. Studies on LNP-siRNA accumulation in the liver show that the low-density lipoprotein receptor (LDLR) on hepatocytes mediates the LNP uptake; in apoE-deficient or LDLR-deficient mice, LNP-siRNA do not display the same gene silencing potency [34]. ApoE is an exchangeable apolipoprotein and a ligand for LDLR-mediated uptake of remnant chylomicrons [35]. Consequently, the apoE adsorption on LNPs turn them into biomimicking remanent chylomicrons that are taken up by hepatocytes. This mechanism clearly demonstrates how, rather than being an obstacle, protein (apoE) corona formation on nanomedicines introduced into blood can be exploited for targeting strategies. ApoE adsorption to liposomes does not drive hepatocyte uptake of negatively charged liposomes [36], and hepatocyte uptake of neutral liposomes is >20-fold lower in apoE-deficient mice. Surface charge, including the composition of the protein adsorbed to lipid-based particles, thus plays a role in the clearance mechanism. When using ionizable lipids/LNPs with an apparent pKa of ∼6.4, about 10% of the ionizable lipids are positively charged in blood (pH 7.4) according to the Henderson–Hasselbalch equation. The surface charge may not only affect the protein corona composition on nanoparticles but also influence the conformation of the adsorbed proteins, including apoE. Taken together, these inquiries show the interplay between nanomedicines, including LNPs and biomolecules such as apoE in biological fluids, is affected by surface composition, surface charge, and particle size [37]. Such interplay determines the in vivo fate of many nanomedicines. More detailed discussions and recent literature about how size and surface charge affect LNP biodistribution will be presented in Section 5 .
The internalization mechanism of the mRNA-LNP COVID-19 vaccines following intramuscular (i.m.) injection remains poorly characterized. Further, the types of cells that are transfected by the vaccines have not been fully detailed. The B cell mediated antibody responses may likely be triggered by delivery and expression of the antigen in antigen-presenting cells (APCs). Dendritic cells, the most potent APC, have been reported to elicit T cell mediated responses, including the induction of T follicular helper cells which are a subset of CD4 + T cells that regulate germinal center B cell maturation to high-affinity memory B cells, and long-lived plasma cells [38]. It is likely that dendritic cells at the point of injection are transfected by the LNPs. However, other types of APCs may also be mobilized by the expression of the spike protein.
3.1.4. Proposed mechanism by which LNPs mediate endosomal disruption
Generally, cells are expected to take up LNPs via endocytosis [21]. As endosomes mature, pH decreases to values below the apparent pKa value of the ionizable lipids, a high proportion of which are consequently protonated. It has been suggested that the electrostatic interactions between the cationic ionizable lipids in the LNPs and the anionic lipids in the endosomal membrane lead to a disruption of the endosomal membrane [22]. More specifically, the oppositely charged lipids may help transform the planar bilayer structure within the endosomal membrane to a more hexagonal-like structure (inverted-like micellular structure, Fig. 2), thereby disrupting the membrane [22], [39], [40]. The interplay between cationic and anionic lipids, both drive electrostatic interaction between the charged headgroups (Fig. 2C) and through their tail/head geometry promote an inverted cone structure (Fig. 2B). That said, due to endocytic recycling, the proportion of nucleic acid cargo that escapes the endosomes is reportedly less than 5% [41]. Another study [42] estimated that only 1–2% of gold-labeled siRNAs loaded into LNPs escape from the endosomes into the cytosol, and escape only occurs within a limited window of time when the LNPs reside in a specific compartment sharing early and late endosomal characteristics. The ionizable lipids in fact must dissociate from the nucleic acids in the cytosol to ensure that the mRNA or siRNA fits into the translational or silencing machinery respectively, a necessity that can be challenging. Hence, new ionizable lipids and a better understanding of endosomal uptake and escape should enable the development of LNPs that even more efficiently deliver nucleic acids to target cells.
3.1.5. Ionizable lipids may play a role in LNP tolerability and immunogenicity
In mRNA vaccines, LNPs may likely exert an adjuvant effect in addition to stabilizing mRNA and facilitating intracellular delivery. Nanoparticle size, shape, and rigidity are important factors in immunological activation (reviewed in [43]) but their mechanisms are still not fully revealed. That said, it has been shown that some cationic lipids activate toll-like receptors and therefore help induce proinflammatory cytokines and co-stimulatory molecules [44]. Recently, a study [38] investigated the adjuvant effect of a proprietary ionizable lipid by administering similar-sized spherical mRNA-LNPs either with or without the proprietary ionizable lipid to mice: the ionizable lipid produced the adjuvant effect by activating interleukin-6 via the mRNA-LNP, but toll-like receptors did not seem to be part of the mechanism. Comparison with immunological responses caused by mRNA-LNPs comprising the cationic lipid, DOTAP, instead of the proprietary ionizable lipid suggested that not all lipids used for nucleic acid vaccine delivery are potent adjuvants when incorporated in an LNP formulation. Hassett et al. [23] drew similar conclusions and, using 30 different LNPs in mice, illustrated that mRNA-LNP immunogenicity depended highly on the ionizable lipid structure. Interestingly, an optimal lipid pKa for immunogenicity was between 6.6 and 6.9 (slightly higher than the typical optimal pKa (6.2–6.5) for transfection [10]), indicating that, independent of cytosolic mRNA delivery, lipid pKa may play a role in formulation interactions with the immune system. In non-human primates, however, five LNP formulations with different ionizable lipids did not result in varied immunogenicity, even though one lipid resulted in 3-fold higher protein expression. This further indicates that protein expression is not the only factor determining vaccine immunogenicity. Yet the data also showed that LNP-driven immune stimulation did not equate to increased immunogenicity. Indeed, there is still much to learn about LNP mechanisms of immune stimulation, tolerability, and immunogenicity and the correlations between these.
While LNP-based immunological activation processes can support the efficacy of vaccines, LNP-mediated immune responses may be less attractive for therapeutic nucleic acid treatments. The toxicity of permanently positive charged lipids [45], [46], [47] and the immunogenicity of certain ionizable lipids are driving the development of degradable ionizable lipids in LNPs for nucleic acid delivery beyond vaccines. Cui et al. [46] emphasize that such toxicity stems almost exclusively from the headgroup. The ionizable lipids used in Comirnaty®, Spikevax®, and Onpattro®, shown in Fig. 2A, all contain esters that make them suitable for hydrolysis degradation. Interestingly, while MC3 is not completely biodegradable and metabolites persist in rats and non-human primates [48], the doses required for clinical application show no evidence of toxicity [3]. That said, pre-dosing of an immunosuppressive cocktail consisting of acetaminophen, a glucocorticoid, and an H1/H2-blocker [3] is required before administering Onpattro® to deal with potential infusion-related reactions. In another approach, replacing double bonds with ester linkages generates hydrolytic cleavage products that are readily incorporated into catabolic pathways, without losing potency [32]. Such biodegradable ionizable lipids containing ester bonds have shown rapid elimination and excretion as well as substantial tolerability in rodents and non-human primates after i.v. [32] and i.m. [23] administration.
3.2. Role of PEG-lipids
Even though PEG-lipids constitute the smallest molar percentage of the lipid components in LNPs (typically ∼ 1.5 mol%), they influence several key properties thereof: population size and dispersity [24], [49], [50], [51], [52], [53]; LNP aggregation prevention [49], [50]; and particle stability during both preparation and storage. Furthermore, PEG-lipids also affect factors such as nucleic acid encapsulation efficiency [49]; circulation half-life [49]; in vivo distribution [49], [54]; transfection efficiency [49]; and immune response [55]. All these properties are somewhat related to the molar ratio of the PEG-lipid as well as the structure and length of both the PEG chain and the lipid tail (alkyl/dialkyl chain(s)). The molecular structures of different PEG-lipids discussed in this Review are illustrated in Fig. 3 A (PEG-lipids used in the clinical approved LNPs) and 3B. Unless otherwise stated, the PEG-lipid referred to herein is the linear PEG with a molecular weight of 2000 Da, the PEG molecular mass (length) that is also used in Doxil®.
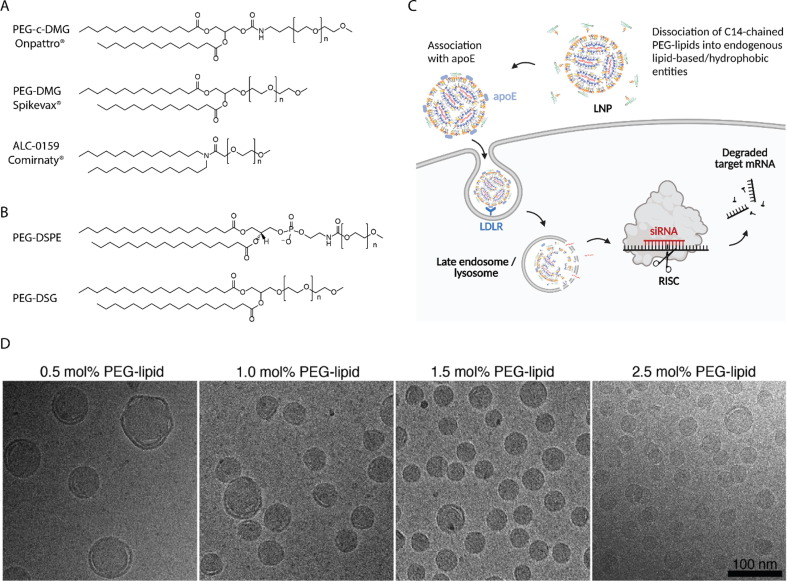
Fig. 3.
A) The PEG-lipids used in Onpattro® (PEG-c-DMG) and the COVID-19 vaccine LNP products Spikevax® (PEG-DMG) and Comirnaty® (ALC-0159). B) Additional PEG-lipids discussed in this Review. C) The proposed uptake mechanism for Onpattro® [56]. The figure was created with BioRender.com. D) Cryo-TEM images of LNPs with varying molar ratios of PEG-lipid. Reproduced from [52] with permission from the Royal Society of Chemistry.
3.2.1. PEG-lipids control LNP size and stability
PEG-lipids contribute to LNP self-assembly via the hydrophilic steric barrier that PEG chains form at the LNP surface [50]. During LNP formation, the PEG chain extends away from the surface of the emerging particle, and sufficient PEG-lipid accumulation per particle prevents heterogenous formulations. The steric PEG barriers also support particle stability by preventing aggregation [49], [50], [52]. Lokugamage et al. [57] demonstrated that formulations completely lacking PEG-lipids produced unstable, polydisperse LNPs exceeding 200 nm in diameter. Other studies have shown that including as little as 0.5 mol% of PEG-lipid resulted in stable, homogenous LNPs smaller than 80 nm [52]. The stability that PEG provides during formulation also endure long-term. LNPs containing mRNA can be stable for up to 3 weeks at 4 °C in buffer, as quantified by consistent size, PDI, zeta-potential and encapsulation efficiencies during storage [49], [58]. The current European Medicines Agency (EMA)-approved shelf lives for mRNA COVID-19 vaccines are 9 months between −25 °C and −15 °C or 30 days at 2–8 °C for Moderna (Spikevax®) and 9 months between −90 °C and −60 °C or 1 month at 2–8 °C for Pfizer-BioNTech (Comirnaty®) [59]. The different storage requirements may be due to disparate LNP formulations, including excipients and mRNA, or the companies’ particular development approaches [60]. Comparatively, Onpattro® has a clinical shelf life of 36 months, refrigerated, according to an EMA report [61]. Research has determined that LNPs formulated with different PEG-lipids have similar long-term stability, indicating that different PEG chain compositions can sufficiently provide the steric repulsive forces necessary during particle formation [49]. PEG-lipids also increase stability during subsequent LNP processing. Lokugamage et al. [57] nebulized LNPs for pulmonary delivery and observed that LNPs with high molar ratios of PEG-lipids resulted in higher pulmonary transfection efficiency than LNPs with lower molar ratios of PEG-lipids. LNPs damaged during nebulization are expected to aggregate, suggesting that higher PEG-density on LNPs limits aggregation and subsequently promotes cellular uptake.
LNP size must be controlled during preparation as it plays a decisive role in nanoparticle pharmacokinetics and biodistribution [62] and LNP biodistribution, delivery efficiency, and transfection potency [63]. Multiple studies have shown that increasing the molar ratio of the PEG-lipid coheres with significantly smaller LNPs, independent of other lipid components [24], [52], [64]. Kulkarni et al. [52] used cryo-transmission electron microscopy (Cryo-TEM) to visualize how varying the PEG molar ratio affects the lipid structure (Fig. 3D). It is hypothesized that the PEG-lipid is located only at the LNP surface [54], hence raising the mol% of the PEG-lipid (surface molecule) leads to a higher surface area:volume ratio and thus decrease in particle size. In these studies, varying the molar ratio of PEG-lipids requires concurrently varying the molar ratio of another lipid. Most studies choose to alter cholesterol content [51], [52]. Kulkarni et al. [52] investigated the impact of DSPC and cholesterol mol% on LNP size and observed no significant differences when varying DSPC-cholesterol (1:1 mol) within a 40–60 mol% range [15]. This indicates that the effect of concurrently modifying cholesterol ratios by up to 2 mol%, as in studies focused on changing the molar ratio of PEG, is presumably negligible.
The lipid tail structure in PEG-lipids also influences LNPs biological activity. Since the PEG-lipid is incorporated into the LNP membrane through the hydrophobic tail (the alkyl/acyl chains), PEG-lipids with longer tails are less likely to dissociate from the LNP. Studies have reported this correlation between desorption rate and lipid tail length both in vitro [49], [65] and in vivo [51]. Mui et al. [51] showed that PEG-lipid desorption from LNPs in circulation, measured one hour after in vivo administration, was 45% for PEG-lipids with C14 dialkyl chains (the length of the alkyl/acyl groups is reported as CX, where X is the number of carbon atoms in the alkyl/acyl chain) and only 1.3% and 0.2% for PEG-lipids with C16 and C18 dialkyl chains, respectively. Studies have generally found that longer alkyl chains improve LNP stability in biological fluids. That said, even short C12 and C14 dialkyl/diacyl chains provide sufficient stability during LNP self-assembly [49], [51]. However, in the presence of a lipid sink (e.g. plasma), which contains endogenous lipid-based particles including lipoproteins and extracellular vesicles plus high levels of albumin with its many hydrophobic pockets [66], the short-alkyl-chained PEG-lipids are prone to dissociating into the endogenous lipid-associating entities. In addition to biological lipid-based nanoparticles in blood, cell membranes can also function as a lipid sink.
3.2.1.1. PEG-lipids mediate indirect targeting capabilities of LNPs
In addition to impacting structural properties such as size and stability, different PEG-lipid molar ratios and compositions greatly influence in vivo distribution and degree of cellular interaction, thereby controlling the ability of LNPs and LNP-like particles to sufficiently deliver their nucleic acid cargo inside the cells of interest [49], [54], [65], [67] PEGylation is a widely used method for preventing rapid nanoparticle clearance and increasing in vivo circulation time [49]. For example, in Doxil®, which delivers the anti-cancer drug doxorubicin (Dox), including PEG-1,2-distearoyl-sn-glycero-3-phosphorylethanolamine (PEG-DSPE) extends circulation time [17]. By increasing circulation time, these well-anchored PEG-lipids on the surface of Doxil®-liposomes boost Dox accumulation at the tumor site [17] and decrease cardiotoxicity, as compared to free doxorubicin [68], [69]. However, the generally high stability and stealth-like properties of PEGylated liposomes limit the cellular interactions that can lead to low cellular uptake and poor endosomal escape [70], an outcome that is counter-productive for nucleic acid delivery. Therefore, while particle stabilization with persistent PEG-lipids is helpful during formation and for extending circulation times, it creates a paradox known as the PEG dilemma [14], [51], [65], [67], [71] wherein improved accumulation outside the liver corresponds with reduced particle activity [49].
The short chained diacyl PEG-lipid PEG-carbamate-1,2-dimyristoyl-sn-glycerol (PEG-c-DMG) is introduced in the Onpattro® formulation (Fig. 3A) to do ‘the exact opposite’ to what the PEG-DSPE is facilitating. While PEG-DSPE ensures long circulation [49], PEG-c-DMG tends to desorb from the LNPs upon i.v. administration, to exploit an already established route for specific cellular uptake in the liver. Blood contains significant apoE, especially on LDLs and remanent chylomicrons which are taken up in the liver via the LDLR. PEG-lipids with short lipid tails tend to desorb from LNPs [6], [51], [63], allowing the particles to adsorb apoE [34]. This allows LNPs to endogenously target hepatocytes in the liver, as exploited in Onpattro® [34] (Fig. 3C). That said, other plasma components, in addition to apoE, could potentially also facilitate uptake of LNPs in hepatocytes or any other cell type.
As discussed above, PEG desorption rate can be controlled by formulating LNPs using PEG-lipids with different acyl chain lengths [24], [49], [51]. Relatedly, Mui et al. [51] showed that increasing the molar ratio of the PEG-lipid from 1.5 to 2.5 mol% compromised transfection efficiency in hepatocytes when the PEG chain was attached to a C18 lipid tail but not a C14 lipid tail. This indicates that only a C14 lipid tail facilitates PEG-lipid desorption followed by association with apoE, thereby promoting receptor-mediated uptake in the liver, and that the molar ratio of the PEG-lipid therefore has no impact on transfection efficiency as most PEG-lipids desorb rapidly following administration. Mui et al. [51] also showed that for 1.5 mol% PEG-lipids, the in vivo transfection level was almost independent of the carbon length of the PEG-lipids. Hence, the expected longer circulation time of LNPs coated with C18 compared to C14 PEG-lipids do not change final transfection efficacy and/or apoE adsorption levels. These data lead one to question whether PEG-lipid desorption (when using 1.5 mol% PEG-lipid) is essential for apoE adsorption to LNPs.
In addition to the indirect active targeting properties obtained via the association between apoE and LNPs in blood, more passive targeting could be attained by preserving the steric barrier of the LNP in vivo. A PEG-lipid like the PEG-DSPE (C18) (Fig. 3B) used in Doxil® inhibits rapid desorption and thus limits protein binding and clearance by the mononuclear phagocytic system, thereby prolonging circulation time of the PEGylated nanocarrier. Approaches [72], [73], [74] using a persistent PEG-lipid to improve LNP accumulation elsewhere employ a formulation with compromised activity and therefore require higher doses to achieve silencing at the target tissue. A slow PEG desorption rate will also minimize specific uptake in the liver and further increase circulation time, enhancing LNP accumulation in tumor tissues [51].
Taken together, the research detailed above shows that the choice of PEG-lipid greatly influences key LNP properties such as size, stability, in vivo distribution, and transfection efficiency. PEG-lipids stabilize LNPs during preparation and storage by providing a steric barrier that both drives self-assembly and prevents aggregation. Further, the type of PEG-lipid partly controls LNP circulation time and cellular interactions. Which PEG-lipid to choose highly depends on therapeutic purpose, target organ and/or cell type, and administration route, and should include considerations regarding both molar ratios and length of the akyl/acyl chain(s) constituting the lipid tail, as these parameters have all shown to influence key properties of LNPs.
3.2.1.2. Do PEG-lipids stimulate an immune response?
Grafting PEG to long chained lipids onto liposomes grants them stealth-like properties that prolong their half-life in circulation. The PEG-coating was thought to hinder antibody and complement protein binding, known as opsonins, that elicit phagocytosis of foreign entities they identify. However, many studies have challenged the notion that PEGylated liposomes are inert to opsonins and thus non-immunogenic. One type of immunogenic response leads to rapid clearance of PEGylated drug delivery systems upon repeated injections. This effect, called the accelerated blood clearance (ABC) phenomenon [75], is mediated by antibodies raised against the foreign drug delivery system after the first injection. The ABC phenomenon has been observed across animal species [76]. Factors that affect ABC clearance of PEGylated liposomes are dose (in a reciprocal way), time interval between injections, and liposomal physicochemical properties including lipid composition, size, surface charge, and drug cargo [76]. However, the ABC phenomenon has not seriously impacted the clinical use of PEGylated liposomal anticancer drug formulations such as Doxil® [76]. Such anticancer drugs may impair anti-PEG antibody production in B-cells [76], [77]. Yet rapid clearance as in the ABC phenomenon could pose a challenge for the safety and efficacy of non-anticancer drugs loaded into PEGylated drug delivery systems. Besin et al. [78] showed in mice studies that LNPs can trigger ABC and that this immune response is based on production of both anti-phosphatidylcholine and anti-PEG antibodies.
Another observed immune response to PEGylated and non-PEGylated liposomes is a hypersensitivity or infusion reaction, termed complement activation-related pseudoallergy (CARPA), that can cause mild to severe hypersensitivity. This effect, which opposite to the ABC phenomenon, is revealed at the first treatment and the symptoms usually decrease or disappear in subsequent treatments. In the clinic, a protocol using a slow infusion rate and corticosteroid pre-treatment is mitigates this effect in Doxil®-based treatments [79]. Whether this response is directly linked to PEG-lipids is unclear.
PEG-lipid-derived antibodies have also been observed, though in a very small number of cases, with i.v. administered LNP-siRNA formulations [80], [81]. A recent study showed that serious adverse events are uncommon and occur similarly in active vaccine and control groups following a third (booster) dose, e.g. of Comirnaty® [82]. Further, this study confirmed improved immunogenicity against the SARS-CoV-2 virus following this third dose. These results stem from the combination of several aspects including, but not limited, to the very low dose administered, the long time between vaccine injections, and the ability of the PEG-lipids to desorb rapidly from LNPs when injected into a biological environment. Regarding the latter effect, Suzuki et al. [49] showed that IgM antibody production against PEG correlated with the PEG-lipids’ acyl chains lengths, so that the LNPs with the fastest-desorbing PEG-lipids induced less anti-PEG IgM. Another study [83] showed that substituting slow-desorbing PEG-lipids with fast-desorbing PEG-lipids abrogate a strong immune response to pegylated liposomes containing nucleic acid cargo.
Screening for immune-triggered adverse effects while developing LNPs for therapeutic applications could be key to LNP clinical success beyond vaccines. A recent study [84] shows that LNP size influences the immunogenicity of LNP-mRNA vaccines in mice, though these size effects were not observed in non-human primates. This study highlights the need to identify immunogenic animal models that translate well to humans. An older study [85] demonstrated that inserting gangliosides (a family of natural lipids containing an oligosaccharide-based headgroup) into PEGylated liposomes attenuates the ABC effect. Münter et al. [86] observe that the degree of antibody and complement 3 protein binding to liposomes depend on liposome surface chemistry. The authors also reveal that opsonin binding is highly heterogenous across individual liposomes in a given formulation.
In addition to adjusting dose level, infusion rate, time interval between treatments, and immunosuppressant co-treatment, synthesizing new stabilizing lipids to replace PEG-lipids or introducing additional components to the four lipids traditionally used to prepare LNPs (see Section 5.2) could circumvent potential adverse immunogenic events from single and multiple LNP-based treatments or improve vaccine potency via adjuvant effects. A recent review [87] describes the impact of PEG on immunological properties of nanomedicine in more details.
3.3. Role of helper lipids
The pervading hypothesis regarding the role of helper lipids is that they support stability during storage and circulation. As LNP formulations used for nucleic acid delivery (except for ionizable lipids) are often derived from the lipids used to form liposomes for small molecule therapeutics, the expectation of the roles of these lipids in LNPs is similar to their liposomal counterpart.
The term “helper lipids” broadly defines a range of lipids such as sterols, phospholipids, and glycerolipids that are typically non-cationic, although the term has also been used to describe surfactant and PEG-lipids [88]. In this Review, we discuss sterol and phospholipid components as helper lipids.
3.3.1. Role of cholesterol
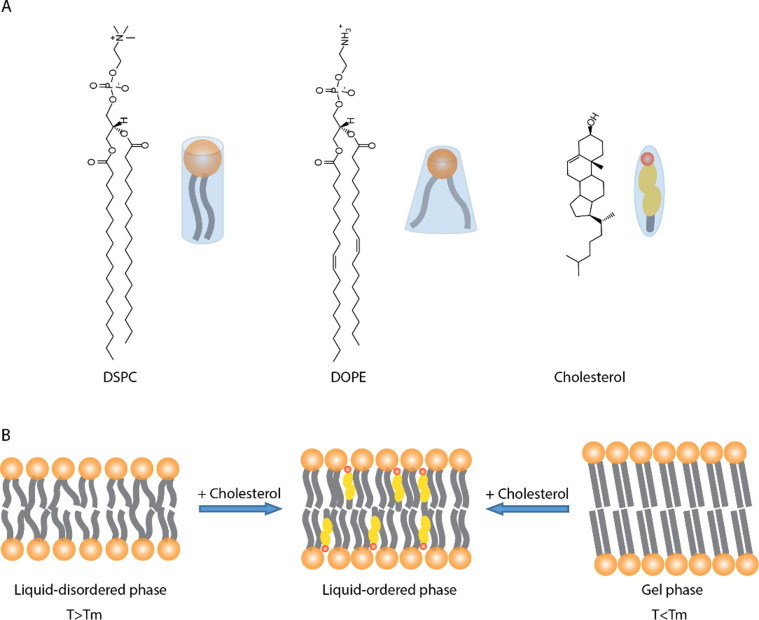
The role of cholesterol in membranes largely depends on context. When combined with phospholipids with low gel-liquid crystalline phase transitions (Tm), cholesterol helps formation of the liquid-ordered phase which is characterized by decreased membrane fluidity and increased bilayer thickness (Fig. 4 B). Cholesterol and low Tm lipids undergo a “condensation” whereby the cross-sectional area of the lipid and cholesterol is lower than the sum of the individual cross-sectional areas. However, when combined with high Tm lipids, cholesterol boosts membrane fluidity and narrows the bilayer. In both cases, cholesterol pulls the lipids towards a liquid-ordered phase [89] (Fig. 4B).
Fig. 4.
A) The molecular shape of standard helper lipids, including DSPC, DOPE, and cholesterol, used in LNPs. Current commercial LNPs use both DSPC and cholesterol. B) The three different lipid-membrane phases: liquid disordered, liquid-ordered, and gel phase.
In LNP formulations containing nucleic acid, incorporating cholesterol is largely based on two main findings obtained with liposomal formulations of small molecule therapeutics: 1) cholesterol is an exchangeable molecule and can accumulate within a liposome during circulation [90], and 2) cholesterol dramatically reduces the amount of surface-bound protein and improves circulation half-lives [91]. Therefore, an equimolar amount of cholesterol is included in LNP formulations relative to endogenous membranes; this prevents net efflux or influx and maintains membrane integrity. Following these pioneering studies on the role of cholesterol in membranes, it was found to be essential to particle stability and subsequently debuted in stable antisense lipid particles (SALP) [92].
Lipids such as cholesterol are also essential for encapsulating nucleic acid. As cholesterol increase membrane rigidity, it serves to reduce drug leakage from the liposomal core. This effect may however not be considered important for large cargo such as nucleic acids. As ionizable lipid designs improved, providing more potency and less toxicity, the total fraction of helper lipid decreased. However, a recent study [52] noted that some threshold amount of helper lipid is required to facilitate stable encapsulation. Specifically, a concentration of at least 40 mol% cholesterol (in the absence of any phospholipid) was needed to achieve near-complete siRNA encapsulation. Interestingly, two independent studies [93], [94] determined that LNP formulations do not retain high cholesterol content in a soluble form. Both studies suggested that the molar amount of cholesterol, being substantially higher than can be stably retained in a membrane, likely results in insoluble cholesterol crystallite formation in the LNP core alongside deprotonated ionizable lipid.
3.3.2. Role of phospholipids
The most prevalent phospholipids in LNP formulations include DSPC and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) (Fig. 4A). Saturated phosphatidylcholine (PC) lipids like DSPC in LNP formulations originate from small-molecule liposomal delivery systems which required high Tm lipids for longer circulation times and overall stability. Comparatively, unsaturated lipids like DOPE (which has a truncated cone-shaped structure, due to the two kinks in the acyl chains introduced by the double bonds, and a smaller headgroup than the tertiary (trimethylated) amine group in PC (Fig. 4A)) improve intracellular delivery of nucleic acid by promoting Hii phase (hexagonal structure, shown Fig. 2C) formation. Current commercial LNP systems only include DSPC, likely due to its known stability factor in commercial liposomes and the fact that the ionizable lipids efficiently disrupt the endosomal membrane. Phospholipids have several different roles in LNP including improving encapsulation (as noted with cholesterol [52]) and cellular delivery [6].
Interestingly, phospholipids such as DSPC, as compared to PE-containing lipids, improved endocytosis of LNP formulations in vitro [6]. This study on plasmid DNA delivery also reported that PC-containing formulations displayed higher uptake levels than the PE-containing systems but not better intracellular delivery. Replacing DSPC with unsaturated equivalents, namely DOPC and SOPC, resulted in increased particle uptake and more intracellular delivery as measured by luciferase expression in several unrelated cell lines. The results, however, also suggested that replacing fetal bovine serum with murine serum changed uptake and expression profiles. Comparatively, lipidoids formulated for mRNA delivery displayed higher expression levels when combined with DOPE rather than DSPC [24]. Such results have not been observed when using MC3 or KC2-like lipids.
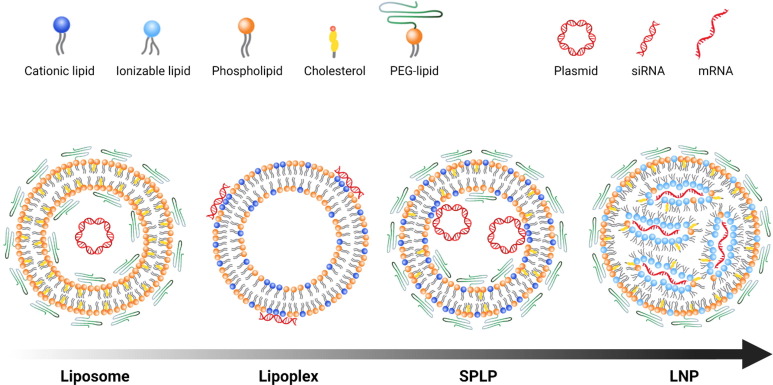
The proportion of DSPC in nucleic acid delivery systems has steadily declined. In Doxil®, the mol% of fully hydrogenated soy PC (mainly DSPC) is 56 with roughly 38 mol% cholesterol. By contrast, some of the older plasmid delivery formulations contained up to 85 mol% phospholipid, such as phosphatidylcholine or dioleoylphosphatidylethanolamine [95], where the DNA molecules were internalized into the aqueous core of the nanoparticles during the detergent-depletion process used to form these nanoentities called stabilized plasmid-lipid particles (SPLP) [95], [96]. The PC content decreased substantially when formulations incorporated cholesterol to extend circulation times and limit protein binding. SALP formulations contained only 25 mol% DSPC which further dropped to 20 mol% in stable nucleic acid lipid particle (SNALP) siRNA formulations. Current commercial LNPs only contain 10 mol% PC, in the form of DSPC, because a high proportion of ionizable lipids (50 mol%) is needed to bind and encapsulate the nucleic acid cargo. An illustration of the most commonly used lipid-based nucleic acid delivery systems, listed according to their time of invention, is presented in Fig. 5 . The illustration also highlights the declining content of phospholipids for each new generation of lipid-based nucleic acid delivery system.
Fig. 5.
Simplistic illustration of some of the most tested lipid-based nucleic acid delivery systems in the order of their invention (indicated by the time arrow), starting with liposomes, lipoplexes, SPLPs and finally the LNPs. All structures are proposed structures and can vary depending on lipid composition, nucleic acid cargo and preparation method. The illustration also highlights the trend of decreasing mol% of phospholipids and increasing mol% of cationic or ionizable lipids, introduced to facilitate active nucleic acid encapsulation, which culminates with LNPs containing typically 10 mol% phospholipids and 50 mol% ionizable lipids. Representative types of nucleic acid cargos are presented in each type of delivery system. Plasmids were typically loaded into liposomes [97] and SPLPs [95] (the similar particles SALP and SNALP contain ASO and siRNA, respectively), while lipoplexes [98] as well as LNPs have been loaded with several nucleic acid cargo types including siRNA, mRNA, microRNA, DNA, ASOs in case of LNPs. The figure was created with BioRender.com.
4. LNP structure: One does not fit all
LNP structure is often presented as in Fig. 1, with ionizable lipids arranged in an inverted micellular structure around the nucleic acid cargo [28]. However, the true structures of lipid arrangement in the core, at the surface, and around the nucleic acid cargo are not known in detail. Further, the actual spatial distribution of the different lipid components in the LNP also remains unclear. We do know the LNP structure in a specific formulation highly depends on the molecular structure of the lipid components, lipid composition, type of cargo, and intraparticle heterogeneity [13]. Hence, one LNP structural model likely does not fit all. Recent studies have investigated LNP structure/lipid arrangement by using cryo-TEM [94], [99], [100], molecular dynamics (MD) [101], small-angle neutron scattering (SANS) [54], small-angle x-ray scattering (SAXS) [54], [94], fluorescence microscopy [102], and NMR methods [103]. Here, we will discuss some key findings from these studies.
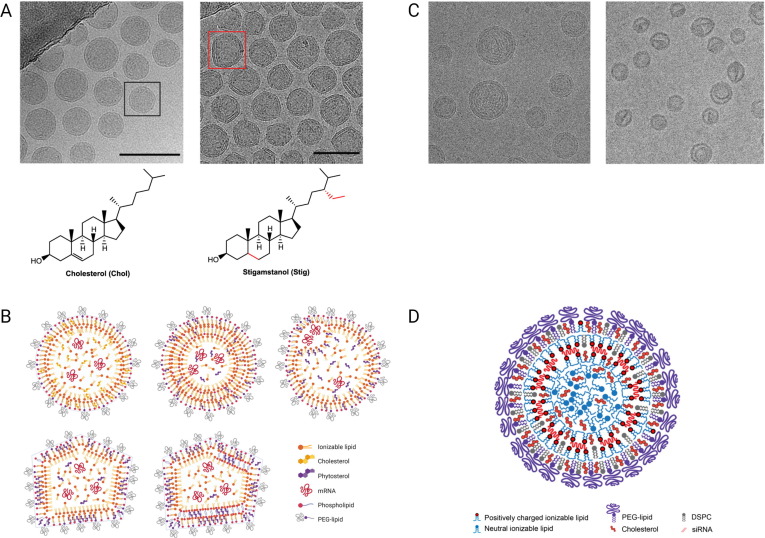
Cryo-TEM provides information about LNP size, shape, and internal morphology and can reveal whether molecular arrangement is ordered, disordered, or phase separated. A study by Eygeris et al. [99] shows that the proportion of multi- versus single-lipid bilayered LNPs largely depends on the type of cholesterol derivatives being used. In case of cholesterol LNPs contain a single lipid bilayer (Fig. 6 A), while most LNPs containing a saturated version of cholesterol (stigamstanol), with an ethyl attached to the carbon chain (Fig. 6A), have a multilamellar structure. This work highlights that even small changes in the molecular structure of a single lipid component (albeit a component comprising nearly 40 mol% of the formulation) can greatly impact the overall LNP structure. In this study, the authors also quantified the proportion of internal defects in terms of areas with varying electron densities (contrast) inside the LNPs and presented structural models of the different LNP types carrying mRNA cargo (Fig. 6B). Interestingly, the LNPs with multilamellar and faceted structures, as well as a lamellar lipid phase, showed higher gene transfection [99]. Whether these structural features induced by the cholesterol derivative or the cholesterol derivative component itself drive the improved transfection may, however, be difficult to unfold. SAXS is becoming a popular technique for characterizing LNPs. SAXS data can determine LNP size and structure and indicate whether LNPS contain ordered structures like multilamellar lipid packing and cholesterol crystallites.
Fig. 6.
A) Cryo-TEM images of LNPs prepared with cholesterol, (adopt a single lipid bilayer structure (left)) and stigamstanol instead of cholesterol (adopt a multilamellar structure (right)). Reprinted (and adapted) with permission from [99]. Copyright 2020 American Chemical Society. B) Structural models of different LNP types carrying mRNA cargo. Reprinted (and adapted) with permission from [99]. Copyright 2020 American Chemical Society. C) Cryo-TEM images of LNPs encapsulating siRNA sandwiched between closely apposed monolayers (left) and LNPs prepared in the absence of siRNA (right). The uncharged ionizable lipids that are not interacting with siRNA can adopt an amorphous oil phase when pH is raised to 7.4. Reprinted (and adapted) with permission from [94]. Copyright 2018 American Chemical Society. D) Structural model of LNP-siRNA showing two distinct phases, including a proposed oil-like phase containing the uncharged ionizable lipids and an aqueous phase. Reprinted (and adapted) with permission from [94]. Copyright 2018 American Chemical Society.
Another recent cryo-TEM based study [100] on the morphology of LNP-mRNA formulations determined that a few different populations of particles exist. Specifically, using a thionine stain for RNA in cryo-TEM, the authors demonstrate that the RNA appears to localize at the outer edges of the formulation (and likely sandwiched between closely apposed monolayers of lipid) or in aqueous compartments that are surrounded by bilayer protrusions. This study remains to be the most direct analysis of the structure and organization of LNP with a very limited physical intervention to the structure of the LNP. The use of thionine is a relatively benign approach to probing the structure and localization of the LNP-RNA particle compared to the use of cryoprotectants combined with ultra-low temperatures and uncontrolled freezing.
In an LNP model based on encapsulated siRNA sandwiched between closely apposed lipid monolayers, proposed by Kulkarni et al. [94], the ionizable lipid not interacting with siRNA can adopt an amorphous oil phase located in the center of the LNP (Fig. 6C). Deprotonated ionizable lipids are almost entirely hydrophobic compared to amphiphilic molecules such as phospholipid and cholesterol. Once the ionizable lipid deprotonates, it behaves similarly to triglycerides and thus is likely to phase separate into fat-droplets. Note that the free-base derivatives of ionizable lipids, such as the ones described in Fig. 2A, exist as an oil at room temperature. Two distinct phases (as visualized by cryo-TEM), including a proposed oil-like phase containing the uncharged ionizable lipids and an aqueous phase, are pronounced for LNPs containing 20 mol% of the ionizable lipid KC2, 31.5 mol% DSPC, 47 mol% cholesterol, and 1.5 mol% PEG-lipid (Fig. 6D). That neutral ionizable lipids tend to phase separate into a purely hydrophobic environment is supported by MD simulations of a bilayer containing KC2 and POPC in various ratios at pH ∼ 4 and ∼ 7.5 pH [101]. In this study, the neutral ionizable lipids intercalate between the ends of the hydrophobic chains of POPC. An interesting single particle fluorescent-microscopy study [102] indicates siRNA clusters aggregate within the LNP in a non-uniform distribution, in close agreement with the above model (Fig. 6D).
The different spatial distributions of the ionizable lipid, DSPC, and cholesterol in LNPs have recently been investigated using SANS [54]. Exploiting the different scattering properties of the deuterated and non-deuterated lipid components shows that DSPC lipids are mainly located at the LNP surface, cholesterol is broadly distributed both at the surface and in the core, and the ionizable lipid (MC3) is primarily in the core but also to some extent present at the surface. This unique study evaluates lipid distribution in LNPs; that said, it is challenging to derive multi-parametric information about multi-component and polydisperse LNPs from SANS data. A study [103] employing dynamic nuclear polarization-enhanced NMR Spectroscopy also claims to be able to determine the spatial location of various LNP components. The SANS study [54] and NMR-based study [103] both support the same general understanding of LNP structure: the different lipid types do not solely associate with either the surface or core of the LNP (in opposition to the simplistic model shown in Fig. 1); DSPC and the PEG-lipid are enriched at the surface; and the ionizable lipid is principally present in the core. Their shared model further suggests that cholesterol is also mainly located in the core.
Taken together, these studies illustrate that different LNPs possess very different structures. Small physiochemical changes to cholesterol [99] and whether the ionizable lipids are charged or neutral [94] greatly impact LNP structure. Further, individual lipid components significantly affect the structure, and thus the biological properties, of LNPs. Complex and large MD simulations, advanced NMR, or other techniques may in the future provide even more detailed information about the distribution and arrangement of lipid components and nucleic acid cargo in LNPs. Ultimately, the correlations between this information and biological effect will make it possible to improve the therapeutic applications of LNPs through rational design.
5. Liver targeting and beyond
Designing nanocarriers that target organs and cell types beyond the liver remains a key challenge. Over the past 20–30 years, huge efforts have been put into tailoring liposomes to feature targeting capabilities beyond passive accumulation into tumor tissues followed by extensive liver clearance. From a clinical perspective, these efforts have all failed including a novel HER2-antibody–(PEGylated)-liposomal conjugate loaded with doxorubicin from Merrimack Pharmaceuticals. Hopefully, some of these clinical failures can be considered lessons, so that future work in designing targeting LNPs will avoid unnecessary missteps.
5.1. Liver targeting
The most successful liver-targeting LNPs is likely Onpattro®, which was designed to exploit apoE binding to LNP in circulation to deliver siRNA to hepatocytes via the LDLR receptor. Hepatocytes comprise most of the liver, which also contains Kupffer cells and liver sinusoidal endothelial cells (LSECs). The liver is the main organ to take up synthetic nanoparticles administered systemically. Studies including [63] have shown that preference for hepatocytes over the reticuloendothelial system (RES), which includes Kupffer cells and LSECs, can be tuned by making LNP sizes either smaller or larger than the diameter of the fenestrations in the liver (50–300 nm [104] and 107 ± 1.5 nm [57]). Kim et al. [105] show that grafting mannose onto LNPs allowed for selective delivery of RNA to LSECs while minimizing unwanted cellular uptake by hepatocytes. Interestingly, Saunders et al. [106] exploited the dimensions of the fenestrations in the liver, in an alternate way, to increase LNP accumulation in cells other than the Kupffer cells and LSECs that usually take up a significant portion of nanoparticles administered systemically [107]. Specifically, Saunders et al. [106] administered non-toxic liposomes, termed nanoprimers, that were larger than the fenestrae of the liver capillaries and designed to transiently occupy RES cells in the liver prior to LNP administration. This strategy improved LNP nucleic acid-mediated knockdown and translation efficiency of targets in hepatocytes.
5.2. LNP targeting beyond the liver
Along the lines above, Ouyang et al. [108] reported that doses above 1 trillion nanoparticles in mice overwhelmed Kupffer cell uptake rates, decreased liver clearance, prolonged circulation, and increased nanoparticle tumor delivery. This enabled up to 12% tumor delivery efficiency to 93% of cells in tumors, thereby boosting the therapeutic efficacy of Doxil®. A study that administered cationic and ionizable lipid LNPs carrying siRNA to apoE−/− mice demonstrated that hepatic uptake of ionizable, but not cationic, LNPs was apoE dependent. This observation suggests that LNP charge plays a role in LNP biodistribution [34]. Surface charge indeed controls organ-specific delivery [109]. A strategy called Selective ORgan Targeting (SORT) that adds a supplemental component (termed a SORT molecule) to the four conventional LNP lipids allowed LNPs to be systematically engineered to deliver nucleic acid therapies to the lungs, spleens, and livers of mice following i.v. administration. The SORT molecule varied from permanently positively charged ionizable lipids to negatively charged lipids. LNPs modified to include a significant mol% of permanently charged lipids sorted toward the lungs. Previous studies have shown that positively charged nanoparticles tend to accumulate in the lungs [110]. Additionally, highly positively charged particles generally aggregate in biological fluids and these agglomerates could accumulate in the narrow lung capillaries. Another explanation could be that the first highly vascularized organ that nanoparticles encounter after an i.v. tail vein injection is the lungs, in which blood flow is slower due to capillary circulation [110]. Thus, the nonspecific interactions between the negatively charged sulfated proteoglycans and glycosaminoglycans displayed by the plasma membranes of capillary endothelial cells and the positively charged nanoparticles could trap the nanoparticles in the lungs [110]. Adding permanently negatively charged SORT molecules to LNPs directed them to the spleen, while the conventional (with close to neutral surface charge) LNPs homed to the liver. It may be that changes in the apparent pKa values endow distinct protein coronas [34], [111] that could alter LNP fate. Along these lines, studies of polystyrene nanoparticles have determined that positively charged particles preferentially adsorb proteins with isoelectric points less than 5.5 (such as albumin), while negatively charged particles (and particles with surfaces bearing acidic functional groups) predominantly bind proteins with isoelectric points greater than 5.5 (such as IgG) [112]. To achieve true organ-specific delivery, however, the SORT approach still needs improvements to prevent leakage to other organs. Further, using whole-organ IVIS imaging for quantitative analysis, as in the SORT study, requires caution, because scattering and absorption effects from incoming and emitted light as well as quenching effects may impact the readout from the isolated organs, and these effects will hamper the quantitative value of these measurements [113]. Further, the liposome field has learned that successfully clinical translating liposomes requires low surface charge [56].
Another lesson learned from the liposome field is that enhanced circulation time can improve accumulation in cancer tissue. Defects in cancer tissue’s rapidly growing vasculature may lead to passive extraversion/accumulation of liposomes in tumors, an effect sometimes referred to as the enhanced permeability and retention (EPR) effect [18]. This proposed phenomenon is, however, heavily debated in the nanomedicine field [116]. Both the nanoprimer approach described above [106] and including long-chained PEG-DSPE lipids (known from the Doxil® formulation) that are less prone to dissociation than PEG-DMG [51] are strategies that could enhance LNP circulation time and thus improve LNP accumulation in non-liver organs.
As explored above, size and surface charge are parameters that partly control LNP biodistribution. Whether the biodistribution of LNPs without active-targeting ligands is associated with passive targeting is hard to determine, as Onpattro® shows that active targeting via apoE can be achieved by controlling the interaction between LNPs and proteins in blood.
In the fast-growing field of cancer immunotherapy, the focus is on modifying and activating immune cells. A recent study [117] showed that conjugating CD4 antibody to LNPs enables specific targeting to and mRNA interventions in CD4 + cells, including T cells. After systemic injection in mice, CD4-targeted radiolabeled mRNA-LNPs accumulated in spleen, providing ∼ 30-fold higher signal from reporter mRNA in splenic T cells, as compared with non-targeted mRNA-LNPs. Another recent study specifically targeted gut-homing leukocytes in experimental colitis. To do this, they fused two integrin-binding domains of MAdCAM-1, the natural ligand of α4β7 integrin, to an IgG–Fc and devised a strategy to efficiently conjugate this fusion protein to the LNP surface [118].
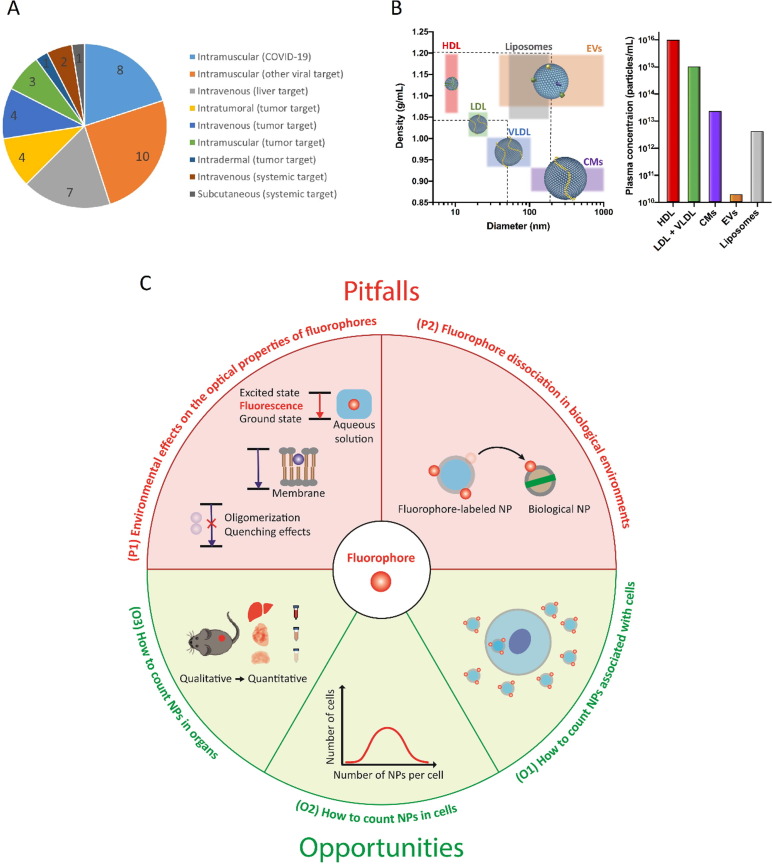
The current LNP-based clinical trials, including the approved LNP products, illustrate both the general challenges related to targeting LNPs beyond the liver and the lack of effective targeting strategies (Fig. 7 A). Searching https://www.clinicaltrials.gov for “lipid nanoparticle” shows, after filtering, 14 LNP hits. Since not all LNP-based clinical trials include information on the LNPs, some trials do not appear in this search. Additional LNP-based clinical trials were therefore identified from Moderna, BioNTech, and CureVac pipelines as well as current literature [14], [114]. These searches identified a total of 40 LNP clinical trials. Of these 40 clinical trials, 22 used local injection, including i.m. for vaccines and intratumoral for cancer. Three trials used i.m. injection to treat cancer. Eleven clinical studies used i.v. administration, seven targeting the liver and four to treat cancer. The numeric breakdown of these trials supports the notion that non-liver targeting using non-local administration routes such as i.v. is challenging.
Fig. 7.
A) Administration routes and disease targets used in clinical trials based on LNP delivery of nucleic acid therapies/vaccines. Trials were identified on clinicaltrials.gov using the search string “lipid nanoparticles” and excluding non-LNP-based hits (search date 18.01.2022). Additional trials were identified from Moderna, BioNTech, and CureVac pipelines (search date 18.01.2022), as well as current literature [14], [114]. B) Mapping the physical properties (size and density) of biological (high-density lipoproteins (HDL), low-density lipoproteins (LDL), very low-density lipoproteins, chylomicrons (CM), and extracellular vesicles (EVs)) and liposome-based nanoparticles including their particle concentrations in human plasma (the liposome concentration is based on 1 mM lipid concentration and a size of 100 nm with an average lipid footprint of 0.425 nm2). The LNP density may likely be slightly higher than liposomes due to its nucleic acid cargo [115]. Reprinted with permission from WILEY. J. B. Simonsen, R. Münter, Angew. Chem. Int. Ed. 2020, 59, 12584. [115]. C) Illustration of some of the pitfalls and opportunities in quantitative fluorescence-based nanomedicine studies discussed in [113]. Reprinted from [113] under the terms of Creative Commons license. Copyright 2021 Journal of Controlled Release.
5.3. Technical and biological challenges in LNP uptake/biodistribution studies
We have learned from the liposome field that PEGylated liposomes can undergo ABC and CARPA triggered by the immune system in reaction to PEG upon repeated administrations [76]. Hence, carefully investigating whether LNPs containing targeting ligands or any new surface component triggers this immune response is advisable early in the LNP development process.
Investigating LNP and other lipid-based drug delivery systems’ protein corona, which in the case of Onpattro® is essential for efficient hepatic uptake, has proven challenging because body fluids contain biological nanoparticles, including extracellular vesicles and lipoproteins, with sizes, densities, and lipid-based surfaces similar to synthetic lipid-based nanocarriers (Fig. 7B) [115]. These similarities make it difficult to isolate pure lipid-based nanomedicines [115], including LNPs, from biological fluids. Eventually, this leads to unwanted protein contaminations in protein corona studies of lipid-based nanomedicines like LNPs and liposomes.
Another challenge linked to the endogenous nanoparticles that the Onpattro® formulation exploits is that PEG-lipids dissociate in biological environments due to their short hydrophobic tails. This type of dynamic can, however, also complicate the use of lipid-like labels, containing fluorophores or radiolabels [113], [119], to track LNPs during in vitro and in vivo uptake/accumulation studies. Indeed, exchange of lipids/lipid-like labels between exogenous and endogenous lipid nanoparticles in biological environments could lead to wrong conclusions about the targeting features of lipid-based drug delivery systems including LNPs [113], [120]. Properly validating the probe type used to track LNPs is therefore highly recommended for both in vitro and in vivo studies [119] (Fig. 7C). As mentioned above, the whole-body fluorescence imaging is only considered a semi-quantitative method compared to the analysis of organ homogenates. These challenges along with the optical changes of some fluorophores due to change in the local surroundings (including self-aggregation/quenching) are discussed in detail in [113] and illustrated in Fig. 7C. This article, including Fig. 7C, also presents opportunities on how to improve the quantitative information from nanomedicine studies based on fluorophores, including fluorescence microscopy and flow cytometry approaches to count nanoparticles in cells and bulk-fluorescence measurements of cell solutions to estimate the proportion of nanoparticles taken up by cells. That said, the biological response to nucleic acid therapies in terms of knockdown, protein expression, and gene editing can often be measured and thus qualitatively assess whether LNPs end up in the cell of interest.
5.4. Future targeting strategies
The nanomedicine field constantly seeks novel organ-targeting compounds. Huge peptide/protein libraries including phage display [121] and bar-coding assays [122] are used to identify strong affinity binders. These and other efforts will hopefully reveal effective active targeting ligands that will advance the LNP targeting capabilities beyond the liver.
In addition to LNP composition and overall design, administration route also affects LNP biodistribution and which cell types are exposed to LNPs [30]. Relatedly, current clinical trials highlight that local administration, when possible, is a simple and efficient way to deliver LNPs to the right tissues and cells (Fig. 7A).
In sum, several new strategies for enhancing LNP uptake beyond liver cells are being developed and tested, with more expected to come. The success of these targeting strategies will be linked not only to how strongly the targeting ligand (either synthesized or adsorbed in the biological fluid, as in Onpattro®) binds to its receptor but also how well the ligand integrates into the matrix of conventional LNP lipids and how well these new multi-component LNPs evade the immune system.
6. Summary and outlook
Although four different lipid components in a conventional LNP ‘cocktail’ seem manageable, variations in nucleic acid cargo types and lipid modifications lead to countless distinct LNP formulations. To navigate this huge matrix of possible LNP designs requires a fundamental understanding of the physicochemical properties of the various lipid-based building blocks that make up LNPs.
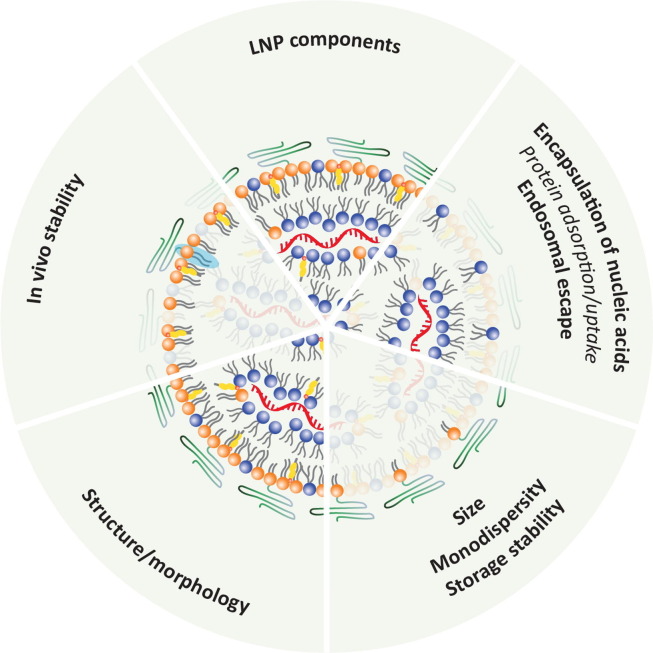
The aim of this Review was to exploit how or what the physicochemical features of different lipids add to the physical and biological properties of LNPs. We have highlighted challenges related to in vitro/in vivo studies of nanomedicines including LNPs and summarized the key functions of different lipid components (Fig. 8). Further, we discussed immunological concerns associated with certain lipid components and provided examples from the literature of possible strategies for directing LNPs beyond the liver. The latter two aspects ultimately define the in vivo fate of LNPs, and thus, solutions to these will put us in a stronger position to realize some of the therapeutic potential that nucleic acid-based modalities hold.
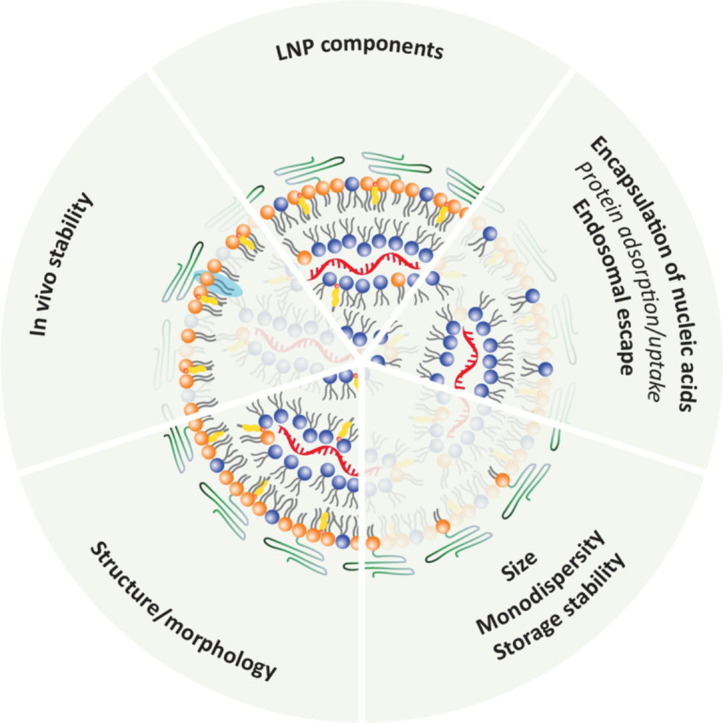
Fig. 8.
Summary of key functions of the different lipid components in commercial LNPs. Whether the ionizable surface lipids drive the “Protein adsorption/uptake” has not yet been thoroughly investigated but they are likely involved in protein/apoE corona formation on the LNP surface. Note that many structural and biological properties of LNPs should be ascribed not to a single lipid component but rather a combination of lipids, as illustrated above. This is illustrated by all the LNP components that contribute to the structure/morphology of LNPs. The PEG-lipids dictate the LNP size, and thus, influence the ratio of the surface to core distribution of the remaining components, and thereby ultimately their packing motif. The light blue highlight illustrates that PEG-lipids containing a long lipid tail improve the in vivo stability of LNPs. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
That all said, it is important that the LNP field focused on translational research is not sidetracked by very complex LNP designs, which are likely to fail in clinical trials. The field also needs to embrace basic research to increase our fundamental understanding on e.g., LNP uptake processes, endosomal escape mechanisms, the interplay between LNP structures/components and the immune system, and safe ligands that can enrich LNPs in certain organ/tissues. These ‘small’ but solid steps, including learning from the more mature liposome field, will hopefully allow us to take a giant leap towards making more clinically successful nucleic acid-based vaccines and therapies that employ LNP delivery.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We acknowledge Kaley Joyes for her detailed proofreading and Pia Pernille Søgaard and Kasper Bendix Johnsen for their valuable inputs.
References
- 1.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Jr, Hammitt L.L., Tureci O., Nell H., Schaefer A., Unal S., Tresnan D.B., Mather S., Dormitzer P.R., Sahin U., Jansen K.U., Gruber W.C. C.C.T. Group, Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B.S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T. C.S. Group, Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams D., Gonzalez-Duarte A., O'Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L., Lin K.P., Vita G., Attarian S., Plante-Bordeneuve V., Mezei M.M., Campistol J.M., Buades J., Brannagan T.H., 3rd, Kim B.J., Oh J., Parman Y., Sekijima Y., Hawkins P.N., Solomon S.D., Polydefkis M., Dyck P.J., Gandhi P.J., Goyal S., Chen J., Strahs A.L., Nochur S.V., Sweetser M.T., Garg P.P., Vaishnaw A.K., Gollob J.A., Suhr O.B. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka H., Takata N., Sakurai Y., Yoshida T., Inoue T., Tamagawa S., Nakai Y., Tange K., Yoshioka H., Maeki M., Tokeshi M., Akita H. Delivery of oligonucleotides using a self-degradable lipid-like material. Pharmaceutics. 2021;13 doi: 10.3390/pharmaceutics13040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gokita K., Inoue J., Ishihara H., Kojima K., Inazawa J. Therapeutic potential of LNP-mediated delivery of miR-634 for cancer therapy. Mol Ther Nucleic Acids. 2020;19:330–338. doi: 10.1016/j.omtn.2019.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kulkarni J.A., Myhre J.L., Chen S., Tam Y.Y.C., Danescu A., Richman J.M., Cullis P.R. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomedicine. 2017;13:1377–1387. doi: 10.1016/j.nano.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 7.Gillmore J.D., Gane E., Taubel J., Kao J., Fontana M., Maitland M.L., Seitzer J., O'Connell D., Walsh K.R., Wood K., Phillips J., Xu Y., Amaral A., Boyd A.P., Cehelsky J.E., McKee M.D., Schiermeier A., Harari O., Murphy A., Kyratsous C.A., Zambrowicz B., Soltys R., Gutstein D.E., Leonard J., Sepp-Lorenzino L., Lebwohl D. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N Engl J Med. 2021;385:493–502. doi: 10.1056/NEJMoa2107454. [DOI] [PubMed] [Google Scholar]
- 8.Yin H., Kanasty R.L., Eltoukhy A.A., Vegas A.J., Dorkin J.R., Anderson D.G. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 9.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., Rajeev K.G., Hafez I.M., Akinc A., Maier M.A., Tracy M.A., Cullis P.R., Madden T.D., Manoharan M., Hope M.J. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem Int Ed Engl. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitehead K.A., Dorkin J.R., Vegas A.J., Chang P.H., Veiseh O., Matthews J., Fenton O.S., Zhang Y., Olejnik K.T., Yesilyurt V., Chen D., Barros S., Klebanov B., Novobrantseva T., Langer R., Anderson D.G. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel P., Ibrahim N.M., Cheng K. The Importance of Apparent pKa in the Development of Nanoparticles Encapsulating siRNA and mRNA. Trends Pharmacol Sci. 2021;42:448–460. doi: 10.1016/j.tips.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., Crommelin D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601 doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulkarni J.A., Cullis P.R., van der Meel R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther. 2018;28:146–157. doi: 10.1089/nat.2018.0721. [DOI] [PubMed] [Google Scholar]
- 15.Kulkarni J.A., Witzigmann D., Chen S., Cullis P.R., van der Meel R. Lipid nanoparticle technology for clinical translation of siRNA therapeutics. Acc Chem Res. 2019;52:2435–2444. doi: 10.1021/acs.accounts.9b00368. [DOI] [PubMed] [Google Scholar]
- 16.Barenholz Y. Doxil(R)–the first FDA-approved nano-drug: lessons learned. J Control Release. 2012;160:117–134. doi: 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 17.Gabizon A., Catane R., Uziely B., Kaufman B., Safra T., Cohen R., Martin F., Huang A., Barenholz Y. Prolonged circulation time and enhanced accumulation in malignant exudates of doxorubicin encapsulated in polyethylene-glycol coated liposomes. Cancer Res. 1994;54:987–992. [PubMed] [Google Scholar]
- 18.Matsumura Y., Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 19.Bailey A.L., Cullis P.R. Membrane fusion with cationic liposomes: effects of target membrane lipid composition. Biochemistry. 1997;36:1628–1634. doi: 10.1021/bi961173x. [DOI] [PubMed] [Google Scholar]
- 20.Heyes J., Palmer L., Bremner K., MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Lin P.J., Tam Y.Y., Hafez I., Sandhu A., Chen S., Ciufolini M.A., Nabi I.R., Cullis P.R. Influence of cationic lipid composition on uptake and intracellular processing of lipid nanoparticle formulations of siRNA. Nanomedicine. 2013;9:233–246. doi: 10.1016/j.nano.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 22.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W., Stebbing D., Crosley E.J., Yaworski E., Hafez I.M., Dorkin J.R., Qin J., Lam K., Rajeev K.G., Wong K.F., Jeffs L.B., Nechev L., Eisenhardt M.L., Jayaraman M., Kazem M., Maier M.A., Srinivasulu M., Weinstein M.J., Chen Q., Alvarez R., Barros S.A., De S., Klimuk S.K., Borland T., Kosovrasti V., Cantley W.L., Tam Y.K., Manoharan M., Ciufolini M.A., Tracy M.A., de Fougerolles A., MacLachlan I., Cullis P.R., Madden T.D., Hope M.J. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 23.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., Mihai C., Lynn A., McFadyen I., Moore M.J., Senn J.J., Stanton M.G., Almarsson O., Ciaramella G., Brito L.A. Optimization of lipid nanoparticles for intramuscular administration of mRNA Vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kauffman K.J., Dorkin J.R., Yang J.H., Heartlein M.W., DeRosa F., Mir F.F., Fenton O.S., Anderson D.G. Optimization of lipid nanoparticle formulations for mRNA Delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 25.Israelachvili J.N. second ed. Academic Press; London: 1992. Intermolecular and surface forces. [Google Scholar]
- 26.Gruner S.M., Cullis P.R., Hope M.J., Tilcock C.P. Lipid polymorphism: the molecular basis of nonbilayer phases. Annu. Rev. Biophys. Biophys. Chem. 1985;14:211–238. doi: 10.1146/annurev.bb.14.060185.001235. [DOI] [PubMed] [Google Scholar]
- 27.Leung A.K., Tam Y.Y., Chen S., Hafez I.M., Cullis P.R. Microfluidic mixing: a general method for encapsulating macromolecules in lipid nanoparticle systems. J. Phys. Chem B. 2015;119:8698–8706. doi: 10.1021/acs.jpcb.5b02891. [DOI] [PubMed] [Google Scholar]
- 28.Leung A.K., Hafez I.M., Baoukina S., Belliveau N.M., Zhigaltsev I.V., Afshinmanesh E., Tieleman D.P., Hansen C.L., Hope M.J., Cullis P.R. Lipid Nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured core. J. Phys. Chem. C Nanomater. Interfaces. 2012;116:18440–18450. doi: 10.1021/jp303267y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., Fan H., Levorse D.A., Crocker L.S. Ionization behavior of amino lipids for siRNA delivery: determination of ionization constants, SAR, and the impact of lipid pKa on cationic lipid-biomembrane interactions. Langmuir. 2011;27:1907–1914. doi: 10.1021/la104590k. [DOI] [PubMed] [Google Scholar]
- 30.Carrasco M.J., Alishetty S., Alameh M.G., Said H., Wright L., Paige M., Soliman O., Weissman D., Cleveland T.E.t., Grishaev A., Buschmann M.D. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun. Biol. 2021;4:956. doi: 10.1038/s42003-021-02441-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryantsev V.S., Diallo M.S., Goddard W.A., 3rd pKa calculations of aliphatic amines, diamines, and aminoamides via density functional theory with a Poisson-Boltzmann continuum solvent model. J. Phys. Chem. A. 2007;111:4422–4430. doi: 10.1021/jp071040t. [DOI] [PubMed] [Google Scholar]
- 32.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J., Zhang X., Wang Q., Panesar S., Hutabarat R., Carioto M., Hettinger J., Kandasamy P., Butler D., Rajeev K.G., Pang B., Charisse K., Fitzgerald K., Mui B.L., Du X., Cullis P., Madden T.D., Hope M.J., Manoharan M., Akinc A. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koitabashi K., Nagumo H., Nakao M., Machida T., Yoshida K., Sakai-Kato K. Acidic pH-induced changes in lipid nanoparticle membrane packing. Biochim. Biophys. Acta Biomembr. 2021;1863 doi: 10.1016/j.bbamem.2021.183627. [DOI] [PubMed] [Google Scholar]
- 34.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R., Butler J.S., Qin L., Racie T., Sprague A., Fava E., Zeigerer A., Hope M.J., Zerial M., Sah D.W., Fitzgerald K., Tracy M.A., Manoharan M., Koteliansky V., Fougerolles A., Maier M.A. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper A.D. Hepatic uptake of chylomicron remnants. J. Lipid Res. 1997;38:2173–2192. [PubMed] [Google Scholar]
- 36.Yan X., Kuipers F., Havekes L.M., Havinga R., Dontje B., Poelstra K., Scherphof G.L., Kamps J.A. The role of apolipoprotein E in the elimination of liposomes from blood by hepatocytes in the mouse. Biochem. Biophys. Res. Commun. 2005;328:57–62. doi: 10.1016/j.bbrc.2004.12.137. [DOI] [PubMed] [Google Scholar]
- 37.Francia V., Schiffelers R.M., Cullis P.R., Witzigmann D. The biomolecular corona of lipid nanoparticles for gene therapy. Bioconjug Chem. 2020;31:2046–2059. doi: 10.1021/acs.bioconjchem.0c00366. [DOI] [PubMed] [Google Scholar]
- 38.Alameh M.G., Tombacz I., Bettini E., Lederer K., Sittplangkoon C., Wilmore J.R., Gaudette B.T., Soliman O.Y., Pine M., Hicks P., Manzoni T.B., Knox J.J., Johnson J.L., Laczko D., Muramatsu H., Davis B., Meng W., Rosenfeld A.M., Strohmeier S., Lin P.J.C., Mui B.L., Tam Y.K., Kariko K., Jacquet A., Krammer F., Bates P., Cancro M.P., Weissman D., Luning Prak E.T., Allman D., Locci M., Pardi N. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity. 2021;54:2877–2892 e2877. doi: 10.1016/j.immuni.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paramasivam P., Franke C., Stoter M., Hoijer A., Bartesaghi S., Sabirsh A., Lindfors L., Arteta M.Y., Dahlen A., Bak A., Andersson S., Kalaidzidis Y., Bickle M., Zerial M. Endosomal escape of delivered mRNA from endosomal recycling tubules visualized at the nanoscale. J. Cell Biol. 2022;221 doi: 10.1083/jcb.202110137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hafez I.M., Maurer N., Cullis P.R. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 41.Sahay G., Querbes W., Alabi C., Eltoukhy A., Sarkar S., Zurenko C., Karagiannis E., Love K., Chen D., Zoncu R., Buganim Y., Schroeder A., Langer R., Anderson D.G. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stoter M., Epstein-Barash H., Zhang L., Koteliansky V., Fitzgerald K., Fava E., Bickle M., Kalaidzidis Y., Akinc A., Maier M., Zerial M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 43.Heuts J., Jiskoot W., Ossendorp F., van der Maaden K. Cationic nanoparticle-based cancer vaccines. Pharmaceutics. 2021;13 doi: 10.3390/pharmaceutics13050596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pizzuto M., Gangloff M., Scherman D., Gay N.J., Escriou V., Ruysschaert J.M., Lonez C. Toll-like receptor 2 promiscuity is responsible for the immunostimulatory activity of nucleic acid nanocarriers. J. Control Release. 2017;247:182–193. doi: 10.1016/j.jconrel.2016.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terada T., Kulkarni J.A., Huynh A., Tam Y.Y.C., Cullis P. Protective effect of edaravone against cationic lipid-mediated oxidative stress and apoptosis. Biol. Pharm. Bull. 2021;44:144–149. doi: 10.1248/bpb.b20-00679. [DOI] [PubMed] [Google Scholar]
- 46.Cui S., Wang Y., Gong Y., Lin X., Zhao Y., Zhi D., Zhou Q., Zhang S. Correlation of the cytotoxic effects of cationic lipids with their headgroups. Toxicol. Res. (Camb) 2018;7:473–479. doi: 10.1039/c8tx00005k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lv H., Zhang S., Wang B., Cui S., Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 48.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., Almarsson O., Stanton M.G., Benenato K.E. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki T., Suzuki Y., Hihara T., Kubara K., Kondo K., Hyodo K., Yamazaki K., Ishida T., Ishihara H. PEG shedding-rate-dependent blood clearance of PEGylated lipid nanoparticles in mice: faster PEG shedding attenuates anti-PEG IgM production. Int J Pharm. 2020;588 doi: 10.1016/j.ijpharm.2020.119792. [DOI] [PubMed] [Google Scholar]
- 50.Holland J.W., Hui C., Cullis P.R., Madden T.D. Poly(ethylene glycol)–lipid conjugates regulate the calcium-induced fusion of liposomes composed of phosphatidylethanolamine and phosphatidylserine. Biochemistry. 1996;35:2618–2624. doi: 10.1021/bi952000v. [DOI] [PubMed] [Google Scholar]
- 51.Mui B.L., Tam Y.K., Jayaraman M., Ansell S.M., Du X., Tam Y.Y., Lin P.J., Chen S., Narayanannair J.K., Rajeev K.G., Manoharan M., Akinc A., Maier M.A., Cullis P., Madden T.D., Hope M.J. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol Ther Nucleic Acids. 2013;2 doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kulkarni J.A., Witzigmann D., Leung J., Tam Y.Y.C., Cullis P.R. On the role of helper lipids in lipid nanoparticle formulations of siRNA. Nanoscale. 2019;11:21733–21739. doi: 10.1039/c9nr09347h. [DOI] [PubMed] [Google Scholar]
- 53.Kurimoto S., Yoshinaga N., Igarashi K., Matsumoto Y., Cabral H., Uchida S. PEG-OligoRNA hybridization of mRNA for developing sterically stable lipid nanoparticles toward in vivo administration. Molecules. 2019;24 doi: 10.3390/molecules24071303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sebastiani F., Yanez Arteta M., Lerche M., Porcar L., Lang C., Bragg R.A., Elmore C.S., Krishnamurthy V.R., Russell R.A., Darwish T., Pichler H., Waldie S., Moulin M., Haertlein M., Forsyth V.T., Lindfors L., Cardenas M. Apolipoprotein E binding drives structural and compositional rearrangement of mRNA-containing lipid nanoparticles. ACS Nano. 2021;15:6709–6722. doi: 10.1021/acsnano.0c10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Judge A., McClintock K., Phelps J.R., Maclachlan I. Hypersensitivity and loss of disease site targeting caused by antibody responses to PEGylated liposomes. Mol. Ther. 2006;13:328–337. doi: 10.1016/j.ymthe.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 56.Akinc A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., Du X., Hope M.J., Madden T.D., Mui B.L., Semple S.C., Tam Y.K., Ciufolini M., Witzigmann D., Kulkarni J.A., van der Meel R., Cullis P.R. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 57.Lokugamage M.P., Vanover D., Beyersdorf J., Hatit M.Z.C., Rotolo L., Echeverri E.S., Peck H.E., Ni H., Yoon J.K., Kim Y., Santangelo P.J., Dahlman J.E. Optimization of lipid nanoparticles for the delivery of nebulized therapeutic mRNA to the lungs. Nat. Biomed. Eng. 2021;5:1059–1068. doi: 10.1038/s41551-021-00786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang H., Leal J., Soto M.R., Smyth H.D.C., Ghosh D. Aerosolizable lipid nanoparticles for pulmonary delivery of mRNA through design of experiments. Pharmaceutics. 2020;12 doi: 10.3390/pharmaceutics12111042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.E.M. Agency, Summary of product characteristics - Comirnaty, 2021, updated 2022.
- 60.Dolgin E. COVID-19 vaccines poised for launch, but impact on pandemic unclear. Nat Biotechnol. 2020 doi: 10.1038/d41587-020-00022-y. [DOI] [PubMed] [Google Scholar]
- 61.E.M. AGENCY, EMA'S SUMMARY OF PRODUCT CHARACTERISTICS - ONPATTRO 2018 (Updated 2022).
- 62.Li S.D., Huang L. Pharmacokinetics and biodistribution of nanoparticles. Mol Pharm. 2008;5:496–504. doi: 10.1021/mp800049w. [DOI] [PubMed] [Google Scholar]
- 63.Chen S., Tam Y.Y.C., Lin P.J.C., Sung M.M.H., Tam Y.K., Cullis P.R. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Control Release. 2016;235:236–244. doi: 10.1016/j.jconrel.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 64.Belliveau N.M., Huft J., Lin P.J., Chen S., Leung A.K., Leaver T.J., Wild A.W., Lee J.B., Taylor R.J., Tam Y.K., Hansen C.L., Cullis P.R. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol Ther Nucleic Acids. 2012;1 doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harvie P., Wong F.M., Bally M.B. Use of poly(ethylene glycol)-lipid conjugates to regulate the surface attributes and transfection activity of lipid-DNA particles. J. Pharm. Sci. 2000;89:652–663. doi: 10.1002/(SICI)1520-6017(200005)89:5<652::AID-JPS11>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 66.Johnsen K.B., Gudbergsson J.M., Andresen T.L., Simonsen J.B. What is the blood concentration of extracellular vesicles? Implications for the use of extracellular vesicles as blood-borne biomarkers of cancer. Biochim. Biophys. Acta Rev. Can. 1871;2019:109–116. doi: 10.1016/j.bbcan.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 67.Song L.Y., Ahkong Q.F., Rong Q., Wang Z., Ansell S., Hope M.J., Mui B. Characterization of the inhibitory effect of PEG-lipid conjugates on the intracellular delivery of plasmid and antisense DNA mediated by cationic lipid liposomes. Biochim Biophys Acta. 2002;1558:1–13. doi: 10.1016/s0005-2736(01)00399-6. [DOI] [PubMed] [Google Scholar]
- 68.Gabizon A., Shmeeda H., Barenholz Y. Pharmacokinetics of pegylated liposomal Doxorubicin: review of animal and human studies. Clin Pharmacokinet. 2003;42:419–436. doi: 10.2165/00003088-200342050-00002. [DOI] [PubMed] [Google Scholar]
- 69.Safra T., Muggia F., Jeffers S., Tsao-Wei D.D., Groshen S., Lyass O., Henderson R., Berry G., Gabizon A. Pegylated liposomal doxorubicin (doxil): reduced clinical cardiotoxicity in patients reaching or exceeding cumulative doses of 500 mg/m2. Ann. Oncol. 2000;11:1029–1033. doi: 10.1023/a:1008365716693. [DOI] [PubMed] [Google Scholar]
- 70.Seynhaeve A.L.B., Dicheva B.M., Hoving S., Koning G.A., Ten Hagen T.L.M. Intact Doxil is taken up intracellularly and released doxorubicin sequesters in the lysosome: evaluated by in vitro/in vivo live cell imaging. J Control Release. 2013;172:330–340. doi: 10.1016/j.jconrel.2013.08.034. [DOI] [PubMed] [Google Scholar]
- 71.Hatakeyama H., Akita H., Harashima H. The polyethyleneglycol dilemma: advantage and disadvantage of PEGylation of liposomes for systemic genes and nucleic acids delivery to tumors. Biol. Pharm. Bull. 2013;36:892–899. doi: 10.1248/bpb.b13-00059. [DOI] [PubMed] [Google Scholar]
- 72.Lee J.B., Zhang K., Tam Y.Y., Tam Y.K., Belliveau N.M., Sung V.Y., Lin P.J., LeBlanc E., Ciufolini M.A., Rennie P.S., Cullis P.R. Lipid nanoparticle siRNA systems for silencing the androgen receptor in human prostate cancer in vivo. Int. J. Can. 2012;131:E781–E790. doi: 10.1002/ijc.27361. [DOI] [PubMed] [Google Scholar]
- 73.Yamamoto Y., Lin P.J., Beraldi E., Zhang F., Kawai Y., Leong J., Katsumi H., Fazli L., Fraser R., Cullis P.R., Gleave M. siRNA lipid nanoparticle potently silences clusterin and delays progression when combined with androgen receptor cotargeting in enzalutamide-resistant prostate cancer. Clin. Can. Res. 2015;21:4845–4855. doi: 10.1158/1078-0432.CCR-15-0866. [DOI] [PubMed] [Google Scholar]
- 74.Lee J.B., Zhang K., Tam Y.Y., Quick J., Tam Y.K., Lin P.J., Chen S., Liu Y., Nair J.K., Zlatev I., Rajeev K.G., Manoharan M., Rennie P.S., Cullis P.R. A Glu-urea-Lys ligand-conjugated lipid nanoparticle/siRNA system inhibits androgen receptor expression in vivo. Mol. Ther. Nucleic Acids. 2016;5 doi: 10.1038/mtna.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dams E.T., Laverman P., Oyen W.J., Storm G., Scherphof G.L., van Der Meer J.W., Corstens F.H., Boerman O.C. Accelerated blood clearance and altered biodistribution of repeated injections of sterically stabilized liposomes. J Pharmacol Exp Ther. 2000;292:1071–1079. [PubMed] [Google Scholar]
- 76.Mohamed M., Abu Lila A.S., Shimizu T., Alaaeldin E., Hussein A., Sarhan H.A., Szebeni J., Ishida T. PEGylated liposomes: immunological responses. Sci. Technol. Adv. Mater. 2019;20:710–724. doi: 10.1080/14686996.2019.1627174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ishida T., Atobe K., Wang X., Kiwada H. Accelerated blood clearance of PEGylated liposomes upon repeated injections: effect of doxorubicin-encapsulation and high-dose first injection. J. Control Release. 2006;115:251–258. doi: 10.1016/j.jconrel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 78.Besin G., Milton J., Sabnis S., Howell R., Mihai C., Burke K., Benenato K.E., Stanton M., Smith P., Senn J., Hoge S. Accelerated blood clearance of lipid nanoparticles entails a biphasic humoral response of B-1 followed by B-2 lymphocytes to distinct antigenic moieties. Immunohorizons. 2019;3:282–293. doi: 10.4049/immunohorizons.1900029. [DOI] [PubMed] [Google Scholar]
- 79.E.M. Agency, Summary of product charateristics - Caelyx pegylated liposomal, 2009, updated 2021.
- 80.Zhang X., Goel V., Attarwala H., Sweetser M.T., Clausen V.A., Robbie G.J. Patisiran pharmacokinetics, pharmacodynamics, and exposure-response analyses in the phase 3 APOLLO trial in patients with hereditary transthyretin-mediated (hATTR) amyloidosis. J. Clin. Pharmacol. 2020;60:37–49. doi: 10.1002/jcph.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang X., Goel V., Robbie G.J. Pharmacokinetics of patisiran, the first approved RNA interference therapy in patients with hereditary transthyretin-mediated amyloidosis. J. Clin. Pharmacol. 2019 doi: 10.1002/jcph.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Munro A.P.S., Janani L., Cornelius V., Aley P.K., Babbage G., Baxter D., Bula M., Cathie K., Chatterjee K., Dodd K., Enever Y., Gokani K., Goodman A.L., Green C.A., Harndahl L., Haughney J., Hicks A., van der Klaauw A.A., Kwok J., Lambe T., Libri V., Llewelyn M.J., McGregor A.C., Minassian A.M., Moore P., Mughal M., Mujadidi Y.F., Murira J., Osanlou O., Osanlou R., Owens D.R., Pacurar M., Palfreeman A., Pan D., Rampling T., Regan K., Saich S., Salkeld J., Saralaya D., Sharma S., Sheridan R., Sturdy A., Thomson E.C., Todd S., Twelves C., Read R.C., Charlton S., Hallis B., Ramsay M., Andrews N., Nguyen-Van-Tam J.S., Snape M.D., Liu X., Faust S.N. C.-B.s. group, Safety and immunogenicity of seven COVID-19 vaccines as a third dose (booster) following two doses of ChAdOx1 nCov-19 or BNT162b2 in the UK (COV-BOOST): a blinded, multicentre, randomised, controlled, phase 2 trial. Lancet. 2021;398:2258–2276. doi: 10.1016/S0140-6736(21)02717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Semple S.C., Harasym T.O., Clow K.A., Ansell S.M., Klimuk S.K., Hope M.J. Immunogenicity and rapid blood clearance of liposomes containing polyethylene glycol-lipid conjugates and nucleic Acid. J. Pharmacol. Exp. Ther. 2005;312:1020–1026. doi: 10.1124/jpet.104.078113. [DOI] [PubMed] [Google Scholar]
- 84.Hassett K.J., Higgins J., Woods A., Levy B., Xia Y., Hsiao C.J., Acosta E., Almarsson O., Moore M.J., Brito L.A. Impact of lipid nanoparticle size on mRNA vaccine immunogenicity. J. Control Release. 2021;335:237–246. doi: 10.1016/j.jconrel.2021.05.021. [DOI] [PubMed] [Google Scholar]
- 85.Mima Y., Abu Lila A.S., Shimizu T., Ukawa M., Ando H., Kurata Y., Ishida T. Ganglioside inserted into PEGylated liposome attenuates anti-PEG immunity. J. Control Release. 2017;250:20–26. doi: 10.1016/j.jconrel.2017.01.040. [DOI] [PubMed] [Google Scholar]
- 86.Munter R., Stavnsbjerg C., Christensen E., Thomsen M.E., Stensballe A., Hansen A.E., Parhamifar L., Kristensen K., Simonsen J.B., Larsen J.B., Andresen T.L. Unravelling heterogeneities in complement and antibody opsonization of individual liposomes as a function of surface architecture. Small. 2022;18 doi: 10.1002/smll.202106529. [DOI] [PubMed] [Google Scholar]
- 87.Shi D., Beasock D., Fessler A., Szebeni J., Ljubimova J.Y., Afonin K.A., Dobrovolskaia M.A. To PEGylate or not to PEGylate: immunological properties of nanomedicine's most popular component, polyethylene glycol and its alternatives. Adv. Drug. Deliv. Rev. 2022;180 doi: 10.1016/j.addr.2021.114079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cheng X., Lee R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug. Deliv. Rev. 2016;99:129–137. doi: 10.1016/j.addr.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 89.Krause M.R., Regen S.L. The structural role of cholesterol in cell membranes: from condensed bilayers to lipid rafts. Acc. Chem. Res. 2014;47:3512–3521. doi: 10.1021/ar500260t. [DOI] [PubMed] [Google Scholar]
- 90.Rodrigueza W.V., Pritchard P.H., Hope M.J. The influence of size and composition on the cholesterol mobilizing properties of liposomes in vivo. Biochim Biophys Acta. 1993;1153:9–19. doi: 10.1016/0005-2736(93)90270-a. [DOI] [PubMed] [Google Scholar]
- 91.Semple S.C., Chonn A., Cullis P.R. Influence of cholesterol on the association of plasma proteins with liposomes. Biochemistry. 1996;35:2521–2525. doi: 10.1021/bi950414i. [DOI] [PubMed] [Google Scholar]
- 92.Semple S.C., Klimuk S.K., Harasym T.O., Dos Santos N., Ansell S.M., Wong K.F., Maurer N., Stark H., Cullis P.R., Hope M.J., Scherrer P. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: formation of novel small multilamellar vesicle structures. Biochim. Biophys. Acta. 2001;1510:152–166. doi: 10.1016/s0005-2736(00)00343-6. [DOI] [PubMed] [Google Scholar]
- 93.Yanez Arteta M., Kjellman T., Bartesaghi S., Wallin S., Wu X., Kvist A.J., Dabkowska A., Szekely N., Radulescu A., Bergenholtz J., Lindfors L. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. USA. 2018;115:E3351–E3360. doi: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kulkarni J.A., Darjuan M.M., Mercer J.E., Chen S., van der Meel R., Thewalt J.L., Tam Y.Y.C., Cullis P.R. On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano. 2018;12:4787–4795. doi: 10.1021/acsnano.8b01516. [DOI] [PubMed] [Google Scholar]
- 95.Wheeler J.J., Palmer L., Ossanlou M., MacLachlan I., Graham R.W., Zhang Y.P., Hope M.J., Scherrer P., Cullis P.R. Stabilized plasmid-lipid particles: construction and characterization. Gene Ther. 1999;6:271–281. doi: 10.1038/sj.gt.3300821. [DOI] [PubMed] [Google Scholar]
- 96.Tam P., Monck M., Lee D., Ludkovski O., Leng E.C., Clow K., Stark H., Scherrer P., Graham R.W., Cullis P.R. Stabilized plasmid-lipid particles for systemic gene therapy. Gene Ther. 2000;7:1867–1874. doi: 10.1038/sj.gt.3301308. [DOI] [PubMed] [Google Scholar]
- 97.Maurer N., Wong K.F., Stark H., Louie L., McIntosh D., Wong T., Scherrer P., Semple S.C., Cullis P.R. Spontaneous entrapment of polynucleotides upon electrostatic interaction with ethanol-destabilized cationic liposomes. Biophys J. 2001;80:2310–2326. doi: 10.1016/S0006-3495(01)76202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khatri N., Baradia D., Vhora I., Rathi M., Misra A. Development and characterization of siRNA lipoplexes: Effect of different lipids, in vitro evaluation in cancerous cell lines and in vivo toxicity study. AAPS PharmSciTech. 2014;15:1630–1643. doi: 10.1208/s12249-014-0193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Eygeris Y., Patel S., Jozic A., Sahay G. Deconvoluting lipid nanoparticle structure for messenger RNA delivery. Nano Lett. 2020;20:4543–4549. doi: 10.1021/acs.nanolett.0c01386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brader M.L., Williams S.J., Banks J.M., Hui W.H., Zhou Z.H., Jin L. Encapsulation state of messenger RNA inside lipid nanoparticles. Biophys J. 2021;120:2766–2770. doi: 10.1016/j.bpj.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ramezanpour M., Schmidt M.L., Bodnariuc I., Kulkarni J.A., Leung S.S.W., Cullis P.R., Thewalt J.L., Tieleman D.P. Ionizable amino lipid interactions with POPC: implications for lipid nanoparticle function. Nanoscale. 2019;11:14141–14146. doi: 10.1039/c9nr02297j. [DOI] [PubMed] [Google Scholar]
- 102.A. Kamanzi, Y. Gu, R. Tahvildari, Z. Friedenberger, X. Zhu, R. Berti, M. Kurylowicz, D. Witzigmann, J.A. Kulkarni, J. Leung, J. Andersson, A. Dahlin, F. Hook, M. Sutton, P.R. Cullis, S. Leslie, Simultaneous, Single-Particle Measurements of Size and Loading Give Insights into the Structure of Drug-Delivery Nanoparticles, ACS Nano, (2021). [DOI] [PubMed]
- 103.Viger-Gravel J., Schantz A., Pinon A.C., Rossini A.J., Schantz S., Emsley L. Structure of lipid nanoparticles containing siRNA or mRNA by dynamic nuclear polarization-enhanced NMR spectroscopy. J. Phys. Chem. B. 2018;122:2073–2081. doi: 10.1021/acs.jpcb.7b10795. [DOI] [PubMed] [Google Scholar]
- 104.Horn T., Christoffersen P., Henriksen J.H. Alcoholic liver injury: defenestration in noncirrhotic livers–a scanning electron microscopic study. Hepatology. 1987;7:77–82. doi: 10.1002/hep.1840070117. [DOI] [PubMed] [Google Scholar]
- 105.Kim M., Jeong M., Hur S., Cho Y., Park J., Jung H., Seo Y., Woo H.A., Nam K.T., Lee K., Lee H. Engineered ionizable lipid nanoparticles for targeted delivery of RNA therapeutics into different types of cells in the liver. Sci Adv. 2021;7 doi: 10.1126/sciadv.abf4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Saunders N.R.M., Paolini M.S., Fenton O.S., Poul L., Devalliere J., Mpambani F., Darmon A., Bergere M., Jibault O., Germain M., Langer R. A Nanoprimer to improve the systemic delivery of siRNA and mRNA. Nano Lett. 2020;20:4264–4269. doi: 10.1021/acs.nanolett.0c00752. [DOI] [PubMed] [Google Scholar]
- 107.Tsoi K.M., MacParland S.A., Ma X.Z., Spetzler V.N., Echeverri J., Ouyang B., Fadel S.M., Sykes E.A., Goldaracena N., Kaths J.M., Conneely J.B., Alman B.A., Selzner M., Ostrowski M.A., Adeyi O.A., Zilman A., McGilvray I.D., Chan W.C. Mechanism of hard-nanomaterial clearance by the liver. Nat. Mater. 2016;15:1212–1221. doi: 10.1038/nmat4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ouyang B., Poon W., Zhang Y.N., Lin Z.P., Kingston B.R., Tavares A.J., Zhang Y., Chen J., Valic M.S., Syed A.M., MacMillan P., Couture-Senecal J., Zheng G., Chan W.C.W. The dose threshold for nanoparticle tumour delivery. Nat Mater. 2020;19:1362–1371. doi: 10.1038/s41563-020-0755-z. [DOI] [PubMed] [Google Scholar]
- 109.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.le Masne Q., de Chermont C., Chaneac J., Seguin F., Pelle S., Maitrejean J.P., Jolivet D., Gourier M., Bessodes D.S. Nanoprobes with near-infrared persistent luminescence for in vivo imaging. Proc. Natl. Acad. Sci. USA. 2007;104:9266–9271. doi: 10.1073/pnas.0702427104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Monopoli M.P., Aberg C., Salvati A., Dawson K.A. Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 112.Gessner A., Lieske A., Paulke B.R., Muller R.H. Functional groups on polystyrene model nanoparticles: influence on protein adsorption. J. Biomed. Mater. Res. A. 2003;65:319–326. doi: 10.1002/jbm.a.10371. [DOI] [PubMed] [Google Scholar]
- 113.Simonsen J.B., Kromann E.B. Pitfalls and opportunities in quantitative fluorescence-based nanomedicine studies – a commentary. J. Control Release. 2021;335:660–667. doi: 10.1016/j.jconrel.2021.05.041. [DOI] [PubMed] [Google Scholar]
- 114.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021:1–17. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Simonsen J.B., Munter R. Pay attention to biological nanoparticles when studying the protein corona on nanomedicines. Angew. Chem. Int. Ed. Engl. 2020;59:12584–12588. doi: 10.1002/anie.202004611. [DOI] [PubMed] [Google Scholar]
- 116.Challenging paradigms in tumour drug delivery Nat Mater. 2020;19:477. doi: 10.1038/s41563-020-0676-x. [DOI] [PubMed] [Google Scholar]
- 117.Tombacz I., Laczko D., Shahnawaz H., Muramatsu H., Natesan A., Yadegari A., Papp T.E., Alameh M.G., Shuvaev V., Mui B.L., Tam Y.K., Muzykantov V., Pardi N., Weissman D., Parhiz H. Highly efficient CD4+ T cell targeting and genetic recombination using engineered CD4+ cell-homing mRNA-LNPs. Mol Ther. 2021;29:3293–3304. doi: 10.1016/j.ymthe.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dammes N., Goldsmith M., Ramishetti S., Dearling J.L.J., Veiga N., Packard A.B., Peer D. Conformation-sensitive targeting of lipid nanoparticles for RNA therapeutics. Nat Nanotechnol. 2021;16:1030–1038. doi: 10.1038/s41565-021-00928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Munter R., Kristensen K., Pedersbaek D., Larsen J.B., Simonsen J.B., Andresen T.L. Dissociation of fluorescently labeled lipids from liposomes in biological environments challenges the interpretation of uptake studies. Nanoscale. 2018;10:22720–22724. doi: 10.1039/c8nr07755j. [DOI] [PubMed] [Google Scholar]
- 120.Pedersbaek D., Kraemer M.K., Kempen P.J., Ashley J., Braesch-Andersen S., Andresen T.L., Simonsen J.B. The composition of reconstituted high-density lipoproteins (rHDL) dictates the degree of rHDL cargo- and size-remodeling via direct interactions with endogenous lipoproteins. Bioconjug Chem. 2019;30:2634–2646. doi: 10.1021/acs.bioconjchem.9b00552. [DOI] [PubMed] [Google Scholar]
- 121.Ray R.M., Hansen A.H., Taskova M., Jandl B., Hansen J., Soemardy C., Morris K.V., Astakhova K. Enhanced target cell specificity and uptake of lipid nanoparticles using RNA aptamers and peptides. Beilstein J. Org. Chem. 2021;17:891–907. doi: 10.3762/bjoc.17.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dahlman J.E., Kauffman K.J., Xing Y., Shaw T.E., Mir F.F., Dlott C.C., Langer R., Anderson D.G., Wang E.T. Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics. Proc. Natl. Acad. Sci. USA. 2017;114:2060–2065. doi: 10.1073/pnas.1620874114. [DOI] [PMC free article] [PubMed] [Google Scholar]