Abstract
Background
Research has shown that some microbiomes are linked to cancer. Hence, we hypothesize that alterations in the respiratory microbiome might be associated with lung cancer.
Methods
Through droplet digital polymerase chain reaction analysis, we investigated the abundance of Acidovorax in surgically resected primary tumors and corresponding nontumor lung tissues obtained from 50 Japanese patients with non‐small cell lung cancer.
Results
The rate of positivity for Acidovorax in tumor and nontumor tissues was 44 and 26%, respectively. The abundance of Acidovorax in tumor tissues was significantly higher in patients with nonsquamous cell carcinoma complicated by chronic obstructive pulmonary disease (COPD) and those who relapsed after surgical resection (p < 0.05). In tumor tissues, the results of the univariate and multivariate analyses revealed that only COPD exerted a direct effect on the abundance of Acidovorax (p < 0.05). Furthermore, the presence of Acidovorax was high in lung cancer patients with COPD comorbidity (65%) and TP53 gene mutation; only one of the nontumor tissues was positive for Acidovorax. In patients with lung cancer complicated by COPD, Acidovorax tended to be present in both the tumor and nontumor areas.
Conclusions
This study identified novel microbiota involved in lung cancer with COPD comorbidity. The results suggested that Acidovorax may be a useful biomarker in the screening for lung cancer. Further studies are warranted to validate the clinical significance of the microbiome in a larger independent patient cohort.
Keywords: COPD, ddPCR, lung cancer, microbiome
This study identified Acidovorax, a novel microbiota involved in lung cancer with COPD comorbidity and TP53 gene mutation using droplet digital polymerase chain reaction. The abundance of Acidovorax in tumor tissues was significantly higher in patients with nonsquamous cell carcinoma complicated by COPD and those who relapsed after surgical resection (p < 0.05). Alterations in the lung microbiome may be a useful biomarker in the screening for lung cancer.

INTRODUCTION
In individuals with a normal immune status, the lower respiratory tract is generally a sterile environment. However, recent data from 16S rRNA and metagenomic analyses have completely changed the previous perceptions, showing that bacteria, such as Proteobacteria, Firmicutes, and Bacteroidetes, are localized even in the lungs of healthy individuals. 1 , 2 , 3 Moreover, an increasing number of studies reveal an association between pulmonary microbiota (or dysbiosis) and lung diseases. These diseases include chronic obstructive pulmonary disease (COPD), asthma, cystic fibrosis, and idiopathic pulmonary fibrosis (IPF). 4 , 5 , 6 , 7
Furthermore, some microbiomes have been linked to cancer. For example, infection with Helicobacter pylori (H. pylori), Fusobacterium nucleatum, human papilloma virus, and Escherichia coli can lead to various types of cancer. 8 Particularly, H. pylori and Fusobacterium nucleatum are related to a high risk of gastric cancer and poor prognosis in patients with colorectal cancer, respectively. 9 , 10 Nevertheless, further investigation of the relationship between the microbiome and lung cancer is warranted. Although limited, available evidence supports a potential link between lung cancer and alterations in the microbiome found in bronchoalveolar lavage fluid (BALF), lung tissue, sputum, saliva, and fecal samples. 11 , 12 , 13 , 14 There are notable differences in the microbiomes found in the upper and lower respiratory tracts. 15 , 16 , 17 Also, the analysis of saliva and sputum samples may be influenced by the presence of oral microflora. Therefore, microbiomes should be examined in lung tissues or BALF samples. Previous studies analyzing lung tissue samples have reported that several microbiomes, including Acidovorax, are associated with lung cancer. 18 , 19 Thus, additional research is required to examine the mechanisms involved in the association of the microbiome with lung cancer. Based on its stability in the long term, the microbiome may be valuable as a diagnostic biomarker and therapeutic target. 20
The present study focused on the specific microbiome associated with lung cancer. Using droplet digital polymerase chain reaction (ddPCR), we analyzed the abundance of Acidovorax in surgically resected primary tumors and nontumor lung tissues obtained from 50 patients with non‐small cell lung cancer (NSCLC). Since low biomass in the bacterial lung tissue microbiome is at the limit of detection, we utilized quantitative PCR (qPCR) 16S bacterial assays in this investigation.
METHODS
Patients and tissue samples
We performed a retrospective analysis of 100 patients with NSCLC who underwent surgery at Nippon Medical School Hospital between 2013 and 2018 prior to specimen collection. We also analyzed 50 patients with NSCLC to investigate the presence of an epidermal growth factor receptor (EGFR) gene mutation or ALK fusion gene. Tissue samples were stored at −80°C in liquid nitrogen. In terms of EGFR mutation status, 10 and six patients harbored a deletion in exon 19 and an L858R missense mutation in exon 21, respectively. Histological analysis indicated 35 adenocarcinomas, 11 squamous cell carcinoma (SCC), two large cell carcinoma, and two NSCLC not otherwise specified. A total of 17 patients had COPD comorbidity; 12 and five patients with stages I and II Global Initiative for Chronic Obstructive Lung Disease (GOLD), respectively. The TNM staging system and the American Joint Committee on Cancer Staging Manual (seventh edition) were utilized for TNM staging.
The Institutional Review Board of Nippon Medical School Hospital approved the protocol of this study. All patients provided informed consent, and the study was conducted in accordance with the tenets of the Declaration of Helsinki, 2008.
Cell culture
Human normal lung fibroblasts, namely human fetal lung fibroblast (HFL‐1), HLF were obtained from the RIKEN RBC Cell Bank and the American Type Culture Collection (ATCC), respectively. In addition, human normal bronchial epithelium, namely BET‐2A and BEAS‐2B were obtained from the ATCC. Cells were amplified and frozen, and one aliquot was thawed for this study. HFL‐1 and HLF cells were routinely screened for the absence of mycoplasma and maintained in RMPI 1640 medium (Sigma–Aldrich) with 10% heat‐inactivated fetal bovine serum and 1% penicillin/streptomycin at 37°C in an incubator containing 5% CO2. BET‐2A and BEAS‐2B cells were maintained in BEGM Bullet Kit (Takara B3170) at 37°C in an incubator containing 5% CO2.
DNA preparation
The QIAamp DNA Mini Kit (Qiagen) was used to extract genomic DNA from cell pellets or tissue samples, as previously described. 21 , 22 A NanoDrop 2000 spectrophotometer (Thermo Scientific) was employed to quantify the genomic DNA.
Bacteria and bacterial culture
Acidovorax temperans was obtained from BCCM/LMG (Laboratory of Microbiology, Ghent University) for use in qPCR positive control experiments. The bacteria were cultured and maintained in American Type Culture Collection medium 3 nutrient agar/broth for 3 days.
Detection and quantification of bacterial abundance using ddPCR
The QX200 Droplet Digital PCR system (Bio‐Rad) was employed to quantify the copies and total DNA (RPP30 reaction) of Acidovorax. Prime PCR reagents were used in the design of primers and probes (Bio‐Rad). The Unique Assay ID used for Acidovorax was dCNS492472816. The MQE Context sequence, which is a peripheral reference sequence containing the amplicon sequence of the assay system, was GGATTAGATACCCTGGTAGTCCACGCCCTAAACGATGTCAACTGGTTGTTGGGTCTTCACTGACTCAGTAACGAAGCTAACGCGTGAAGTTGACCGCCTGGGGAGTACGGCCGCAAGGTTGAA. Similarly, the Unique Assay ID used for RPP30 was dHsaCP2500350. The MQE context sequence was TCGGCCATCAGAAGGAGATGAAGATTGTCTTCCAGCTTCCAAGAAAGCCAAGTGTGAGGGCTGAAAAGAATGCCCCAGTCTCTGTCAGCACTCCCTTCTTCCCTTTTATAGTTCATCAGCCAC. Each reaction (total volume: 20 μl) contained forward and reverse primers/probe sets (1 μl), 1× ddPCR SuperMix for Probes (Bio‐Rad) (10 μl), and bisulfite‐converted DNA. The amount of DNA per well was adjusted to 500 ng. Samples were analyzed using automated droplet generator cartridges (Bio‐Rad) with automated droplet generation oil for probes (Bio‐Rad).
Droplets were added to a 96‐well PCR plate. The reaction was performed using a C1000 touch thermal cycle with 96‐deep well reaction module (Bio‐Rad) as follows: DNA polymerase activation (10 min at 95°C), denaturation (40 cycles of 30 s at 94°C), annealing and extension (1 min at 63°C), DNA polymerase deactivation (10 min at 98°C), and cooling (4°C). The PCR plates were subsequently loaded into a QX200 Droplet Reader (Bio‐Rad), and the presence of Acidovorax or RPP30 was determined. The QuantaSoft software (QuantaSoft v1.7.4; Bio‐Rad) was employed for data analysis. All reactions included a blank DNA template control well and bisulfite‐converted control DNA (500 ng). The amount of DNA used for each sample was 10 or 20 ng. For quality control, samples that yielded <100 positive droplets in the RPP30 reaction were excluded. In this study, the number of copies of Acidovorax or RPP30 was used for statistical analysis.
TP53 gene sequencing and mutation analysis
Genomic DNA extracted from 47 lung cancer tissues was submitted for TP53‐targeted sequencing using Sanger sequencing. This was performed following standard procedures, as previously described. 23 Table S1 lists sequences for forward and reverse primers used in the sequencing assay. Because only the tumor was sequenced, during the scoring of somatic mutations, those deemed to be germline were excluded. These included single‐nucleotide variants present in the single nucleotide polymorphism database with high reported allele frequency (i.e., common polymorphisms). In addition, single‐nucleotide variants in untranslated regions and introns were not considered as their somatic status and functional implications are unclear. The presence of putative somatic exonic and splicing variants was verified using database from The Cancer Genome Atlas and Catalogue of Somatic Mutations in Cancer.
Statistical analysis
A logistic regression analysis was performed to investigate the associations of bacterial abundances with the clinicopathological characteristics of patients. The correlation between the abundances of bacterial genera was evaluated using Spearman rank correlation analysis. Logistic regression was utilized for the construction of prediction models. To evaluate the diagnostic significance of potential biomarkers, we performed receiver operating characteristic (ROC) curve analysis and calculated the area under the ROC curve (AUC). Statistically significant differences (i.e., p‐values < 0.05) versus control were determined using the standard Student's t‐test. The SPSS (version 25.0; IBM Corporation) software was used for the analyses.
RESULTS
Analysis of Acidovorax using the ddPCR assay
The initial experiments aimed to accurately identify the abundance of Acidovorax using ddPCR. The sensitivity and analytical range of this method were determined by identifying the lower limit of detection. For this assay, 10‐fold serial dilutions of control DNA (RPP30) in water were prepared. An input DNA amount ≥0.02 ng was sufficient to perform the assay, which showed linearity over the titration series. Through this assay, Acidovorax was detectable at a minimum level of 0.025% (Figure S1). Figure 1 shows the DNA abundances of bacteria genera. All the bacterial genera tested using ddPCR generated ≥10 000 droplets per well; therefore, their presence could be quantified in the tissue specimens.
FIGURE 1.
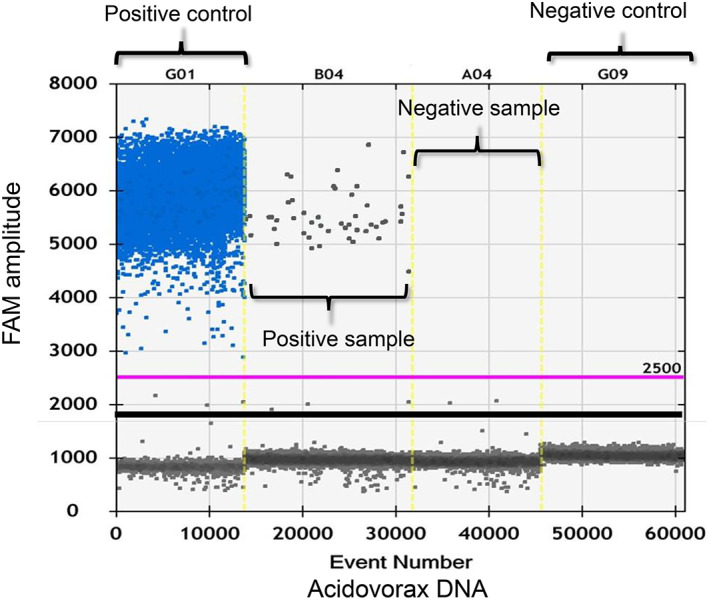
Plot of the ddPCR analysis showing the presence of Acidovorax (blue and gray dots indicate positive and negative amplification droplets, respectively). The reaction was specific to the abundance of the Acidovorax genera. ddPCR, droplet digital polymerase chain reaction.
Abundance of Acidovorax in NSCLC
We first investigated the abundance of both Acidovorax and total DNA (RPP30) in 50 primary lung tumors and nontumor tissues using ddPCR analysis. Representative results are shown in Figure 1, and the positivity rates for all clinicopathological subtypes are provided in Table 1 (median age: 68 years; males: 33 [66%]; females: 17 [34%]; smokers: 42 [84%]). The histological subtypes were adenocarcinoma (n = 35, 70%), SCC (n = 11, 22%), and others (n = 4, 8%). All patients, except for four patients with pathological stage IV disease, underwent complete tumor resection. Seventeen patients (34%) had lung cancer with COPD comorbidity. Treatment with antibiotics prior to surgery was not administered to any of the patients.
TABLE 1.
Association of the Acidovorax abundances with clinicopathological characteristics of NSCLC patients in tumor tissues
| Characteristic | Total | Acidovorax of tumor tissue | p‐value* | |
|---|---|---|---|---|
| Positive (%) | Negative (%) | |||
| Gender | ||||
| Male | 33 | 14 (42%) | 19 (58%) | 0.760 |
| Female | 17 | 8 (47%) | 9 (53%) | |
| Age | ||||
| <65 years | 19 | 6 (32%) | 13 (68%) | 0.173 |
| ≥65 years | 31 | 16 (55%) | 15 (45%) | |
| Smoking status | ||||
| Never smoked | 16 | 6 (37%) | 10 (63%) | 0.535 |
| Current or former smoker | 34 | 16 (47%) | 18 (53%) | |
| Histological subtype | ||||
| Adenocarcinoma | 35 | 16 (46%) | 19 (54%) | 0.041 |
| Squamous cell carcinoma | 11 | 2 (18%) | 9 (82%) | |
| Others | 4 | 4 (100%) | 0 (0%) | |
| Pathological stage † | ||||
| I, II | 30 | 13 (43%) | 17 (57%) | 0.910 |
| III, IV | 20 | 9 (45%) | 11 (55%) | |
| COPD | ||||
| Absent | 33 | 11 (33%) | 22 (67%) | 0.040 |
| Present | 17 | 11 (65%) | 6 (35%) | |
| EGFR gene mutation/ALK fusion gene | ||||
| Wild‐type | 34 | 16 (47%) | 18 (53%) | 0.535 |
| Mutant | 16 | 6 (38%) | 10 (62%) | |
| Recurrence | ||||
| Absent | 20 | 5 (25%) | 15 (75%) | 0.046 |
| Present | 26 | 14 (54%) | 12 (46%) | |
| TP53 gene mutation | ||||
| Absent | 41 | 14 (34%) | 27 (66%) | 0.022 |
| Present | 6 | 5 (83%) | 1 (17%) | |
Abbreviations: AC, adenocarcinoma; NSCLC, non‐small cell lung cancer; SCC, squamous cell carcinoma.
Fisher's exact test. p‐values of <0.05 are shown in bold.
According to the International Union Against Cancer (UICC) TNM Classification of Malignant Tumors, seventh edition (2010).
Next, the ROC analysis of tumor tissues showed that the diagnostic threshold for the concentration of Acidovorax used to screen NSCLC in patients with and without COPD comorbidity was 12.3 copies/well. The sensitivity and specificity of this value were 64.7% and 66.7%, respectively (Figure 2). The AUC was 0.670 (standard error = 0.061; 95% confidence interval = 0.053–0.833; p = 0.050). Therefore, we used this threshold as a positive criterion in this analysis. Using the threshold of Acidovorax abundance as the cutoff point, its levels were converted into binary variables (i.e., positive and negative).
FIGURE 2.
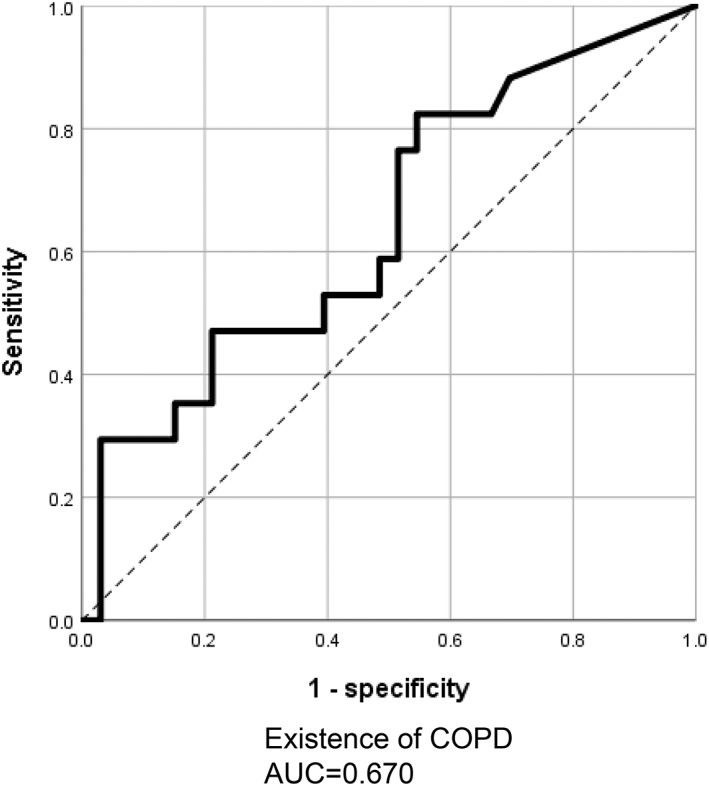
Receiver‐operating characteristic (ROC) curve analysis of the abundance of Acidovorax genera in lung tissues obtained from patients with NSCLC. The use of Acidovorax as a lung tissue biomarker assisted in detecting lung cancer with COPD comorbidity (AUC: 0.670). AUC, area under the curve; COPD, chronic obstructive pulmonary disease; NSCLC, non‐small cell lung cancer.
Acidovorax was detected in 46% (n = 16/35) and 18% (n = 2/11) of patients with adenocarcinoma and SCC, respectively (Table 1). It was also detected in 47% (n = 16/34) of current or former smokers, 38% (n = 6/16) of patients with an EGFR mutation, and 54% (n = 14/26) of those with disease recurrence. The difference in the frequency of Acidovorax in SCC and non‐SCC patients was statistically significant (p = 0.041, Fisher's exact test). Moreover, the difference in the frequency of relapse after surgical resection was also statistically significant (p = 0.046, Fisher's exact test). Furthermore, there was also a statistically significant difference between COPD and non‐COPD patients. (p = 0.040, Fisher's exact test). Nevertheless, in nontumor part tissues, there was no significant correlation detected between clinicopathological features and the presence of Acidovorax (Table S2). In this study, we investigated TP53 gene mutations in 47 patients for whom re‐extraction of DNA was possible. The abundance of Acidovorax correlated with the TP53 mutation (Table 1), as 26% (5/19) and 3.6% (1/28) of Acidovorax‐positive and Acidovorax‐negative cases, respectively, had only the TP53 mutation.
Comparison of Acidovorax abundance in NSCLC tumors and nontumor tissues
Next, we examined the presence of Acidovorax in tumors and nontumor tissues obtained from patients with NSCLC. The positivity rate for Acidovorax was 44% (n = 22/50) and 26% (n = 13/50), respectively; the rate of positivity in both types of samples was 18% (n = 9/50) (Figure 3a). We confirmed a statistically significant difference in the number of positive and negative cases of Acidovorax/RPP30 between the tumor tissues (p < 0.05) and nontumor tissues (p < 0.0001) (Figure 3b).
FIGURE 3.
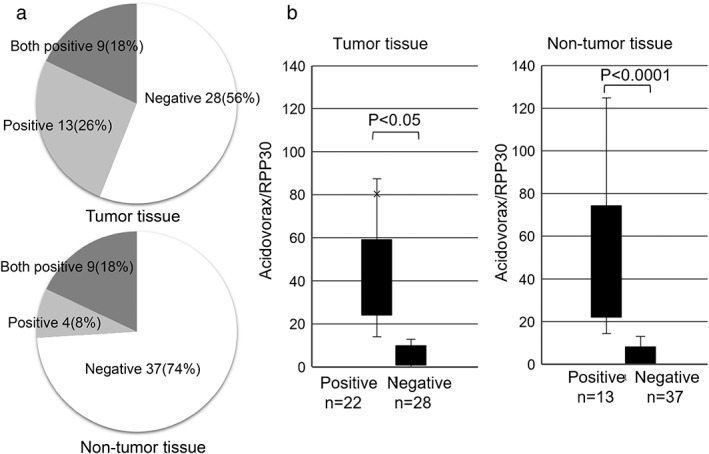
Comparison of the abundance of Acidovorax in tumor and nontumor tissues. (a) The rate of positivity for Acidovorax in tumor and nontumor tissues. (b) The ratio of Acidovorax/RPP30 abundance in tumor and nontumor tissues. The paired t‐test was used to evaluate the mean values.
Abundance of Acidovorax is related to NSCLC with COPD comorbidity
Finally, logistic regression analysis was employed to investigate clinical parameters that may affect the abundance of Acidovorax. A total of six variables (i.e., sex, age, histology, pathological stage, respiratory comorbidities, and gene mutation) were included as dependent variables (Table 2). The smoking status was excluded as a confounding factor in this analysis. This is because COPD has been closely related to the smoking status, and all registered patients with COPD comorbidity were smokers. In tumor tissues, the results of the univariate analysis showed that COPD (p = 0.038) tended to affect the abundance of Acidovorax. The multivariate analysis also showed that COPD (p = 0.048) and histological subtype (non‐SCC) (p = 0.029) had a direct effect on the abundance of Acidovorax. In the multivariate analysis of nontumor tissues, COPD (p = 0.045) was also associated with the abundance of Acidovorax (Table S3). For COPD, positivity for Acidovorax was detected in 65% (n = 11/17) of tumor tissues, 29% (n = 5/17) of both tumor and nontumor tissues, and only one of the nontumor tissues (n = 1/17) (Figure S2). Among the lung cancer patients with COPD comorbidity who were positive for Acidovorax, seven and four had GOLD stages I and II disease, respectively. Therefore, the abundance of Acidovorax in patients with lung cancer may be involved in early‐stage COPD.
TABLE 2.
Logistic regression in patients with NSCLC in tumor tissues
| Variable | n | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | p‐value | HR | 95% CI | p‐value | ||
| Gender | |||||||
| Male/female | 33/17 | 0.829 | 0.256–2688 | 0.755 | 0.404 | 0.091–1.787 | 0.232 |
| Age | |||||||
| <65/≥65 years | 19/31 | 2.311 | 0.698–7.647 | 0.170 | 4.502 | 0.969–20.914 | 0.055 |
| Histological subtype | |||||||
| SCC/non‐SCC | 11/39 | 0.211 | 0.040–1.106 | 0.066 | 0.118 | 0.018–0.799 | 0.029 |
| Pathological stage | |||||||
| I, II/III, IV | 30/20 | 1.070 | 0.342–3.342 | 0.907 | 1.838 | 0.463–7.297 | 0.387 |
| COPD | |||||||
| Absent/present | 33/17 | 3.667 | 1.072–12.55 | 0.038 | 4.501 | 1.014–19.989 | 0.048 |
| Gene mutation | |||||||
| Absent/present | 34/16 | 0.675 | 0.200–2.277 | 0.526 | 0.592 | 0.129–2.709 | 0.499 |
Abbreviations: CI, confidence interval; HR, hazard ratio; SCC, squamous cell carcinoma. P‐values of <0.05 are shown in bold.
DISCUSSION
We determined the abundance of Acidovorax in samples of lung tissues derived from patients using qPCR 16S bacterial assays. We confirmed the abundance of Acidovorax through ddPCR analysis using its DNA. Subsequently, we investigated the abundance of Acidovorax in a variety of NSCLC samples using a newly established ddPCR primer/probe (Figure 1). In tumor and nontumor tissues, the rate of positivity for Acidovorax was 44% and 26%, respectively. In the present study, we used DNA extracted from tumor and nontumor areas instead of bronchoalveolar lavage samples from patients with lung cancer. Therefore, it is important to verify the rate of positivity in the future. There was no association between the abundance of Acidovorax and the smoking history (Brinkman Index). However, the abundance of Acidovorax was linked to an increase in the recurrence of lung cancer after surgery (Table 1). Therefore, we concluded that the abundance of Acidovorax is correlated with the risk of lung cancer. In patients with COPD comorbidity, the levels of Acidovorax were significantly higher in patients with COPD comorbidity (65%, p < 0.05). Furthermore, in this subset of patients, Acidovorax was detected even in nontumor tissues (n = 5/17, 29%). Our results showed that non‐SCC had a more pronounced effect on the abundance of Acidovorax than SCC (p = 0.029) (Table 2); however, Acidovorax was not significantly increased in SCC. These results indicated that the abundance of Acidovorax may not be a biomarker for a specific histological subtype; instead, it appears to be associated with the malignant progression of NSCLC with COPD comorbidity.
It has been reported that the levels of Acidovorax are elevated in NSCLC, including cases involving the TP53 mutation pathway. 18 Furthermore, a significant increase was observed in the volume of lung tumors in mice inoculated with Acidovorax temperans. Based on this finding, Acidovorax may contribute to lung carcinogenesis in the presence of activated Kras and mutant p53. Therefore, this microbiome may promote the occurrence and progression of disease. 18 Studies analyzing BALF samples detected Acidovorax in both patients with lung cancer and nonmalignant pulmonary diseases. 24 Notably, alterations in the microbiome may be partly responsible for tumorigenesis or the occurrence of secondary tumors. The present study further suggests that Acidovorax may be a biomarker for lung cancer with COPD comorbidity.
Smokers with COPD are at an increased risk (3–10‐fold higher) of developing lung cancer versus those without emphysema. 25 Numerous epidemiological studies have associated COPD with lung cancer, regardless of the extent of smoking. 26 SCC is the most frequent histological type among patients with COPD comorbidity. However, when stratified according to the Global Initiative for GOLD stage of COPD, adenocarcinoma was a more frequent histological subtype than SCC in patients with GOLD stage I disease. 27 In this study, the number of SCC patients with COPD comorbidity who were positive for Acidovorax was low due to the greater proportion of patients with GOLD stage I versus stage II disease. Genome‐wide association studies revealed overlapping genetic susceptibility loci for lung cancer, smoking behavior, COPD, and pulmonary function. 28 However, there is considerable unexplained variation among individuals in terms of susceptibility for progression from COPD to lung cancer. Thus far, there are no studies investigating the coexistence of lung cancer and COPD using lung cancer tissues; nevertheless, sequencing of the 16S rRNA gene has been conducted using nontumor tissues. Previous research reported that the abundance of Proteobacteria (i.e., Acinetobacter and Acidovorax) was significantly lower, whereas that of Firmicutes (Streptococcus) and Bacteroidetes (Prevotella) was higher in patients with lung cancer with or without emphysema versus those with emphysema. 19 Several studies revealed that the gut microbiota affects the response of patients with cancer to immunotherapy. 29 , 30 According to Tsay et al., the lower airway microbiota is commonly enriched by oral commensals, and some of these bacteria affect tumor progression and prognosis by triggering host transcriptomic signatures associated with carcinogenesis. 31
Most bacterial genera were not detectable by conventional PCR, suggesting that the levels of the microbiome were low in the clinical specimens. Therefore, we used ddPCR, which is more sensitive than conventional PCR, to measure the bacteria. Based on the ddPCR data, the abundance of Acidovorax has almost zero copy in normal and lung cancer cells (Figure S3). In contrast, it was >12.3 copies/well in lung cancer tissues obtained from patients with lung cancer, which we judged to be Acidovorax‐positive. Similarly, Leng et al. reported that the sputum microbiome may provide noninvasive biomarkers for the early detection and classification of NSCLC using ddPCR. 32 Therefore, the bacteria may not be present in the normal specimens. The presence of this microbiome could be used as a biomarker of lung cancer.
This study was characterized by several limitations. First, the study involved a small sample size. In the future, we plan to prospectively validate the present findings in a large cohort. Second, we only assessed Acidovorax genera that had been previously associated with lung cancer. Although the results are meaningful, the exact association of this bacterial flora with lung cancer remains unclear. Third, we found that the non‐SCC histological subtype was associated with the abundance of Acidovorax. However, there was no correlation between the abundance of Acidovorax and the SCC subtype (i.e., the most common histological type in lung cancer patients with COPD comorbidity). In this study, the positive rate for Acidovorax in patients with SCC was low (18%), and most patients had early GOLD stage disease. Acidovorax may also be a preferable risk factor for lung cancer with COPD versus others, such as the extent of smoking and the SCC histological subtype. Finally, this study included only Japanese patients; hence, these findings may not be generalizable to other ethnic groups.
Based on the present results, alterations in the lung microbiome may be a useful biomarker in the screening for lung cancer. Further investigation is warranted to validate the present evidence and identify additional bacterial biomarkers.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
Supporting information
Figure S1. Plot of the ddPCR analysis showing Acidovorax in serial dilutions. Blue and gray dots denote positive and negative amplification droplets, respectively. ddPCR, droplet digital polymerase chain reaction
Figure S2. Rate of positivity for Acidovorax in tumor and nontumor tissues. (A) 50 cases of lung cancer. (B) 17 cases of lung cancer with COPD comorbidity. COPD, chronic obstructive pulmonary disease
Figure S3. Plot of the ddPCR analysis showing the presence of Acidovorax (blue and gray dots indicate positive and negative amplification droplets, respectively) in human normal lung fibroblasts and bronchial epithelium. ddPCR, droplet digital polymerase chain reaction
Table S1. List of primers and targets for TP53 sequences
Table S2. Association of the Acidovorax abundances with clinicopathological characteristics of NSCLC patients in nontumor tissues
Table S3. Logistic regression in patients with NSCLC in nontumor tissues
ACKNOWLEDGMENTS
This study was supported by a KAKENHI Grant‐in‐Aid for Scientific Research (C) (20K08552 to A. Miyanaga), and the Clinical Rebiopsy Bank Project for Comprehensive Cancer Therapy Development from the Ministry of Education, Culture, Sports, Science and Technology Supported Program for the Strategic Research Foundation at Private Universities (grant S1311022 to A. Gemma and M. Seike).
Shimizu M, Miyanaga A, Seike M, Matsuda K, Matsumoto M, Noro R, et al. The respiratory microbiome associated with chronic obstructive pulmonary disease comorbidity in non‐small cell lung cancer. Thorac Cancer. 2022;13(13):1940–1947. 10.1111/1759-7714.14463
Funding information Strategic Research Foundation, Grant/Award Number: S1311022; Ministry of Education, Culture, Sports, Science and Technology; KAKENHI Grant‐in‐Aid for Scientific Research (C), Grant/Award Number: 20K08552
REFERENCES
- 1. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One 2010; 5: e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maddi A, Sabharwal A, Violante T, Manuballa S, Genco R, Patnaik S, et al. The microbiome and lung cancer. J Thorac Dis. 2019;11:280–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ubags NDJ, Marsland BJ. Mechanistic insight into the function of the microbiome in lung diseases. Eur Respir J. 2017;50:1602467. [DOI] [PubMed] [Google Scholar]
- 4. Garcia‐Nuñez M, Garcia‐Gonzalez M, Pomares X, Montón C, Millares L, Quero S, et al. The respiratory microbiome in cystic fibrosis: compartment patterns and clinical relationships in early stage disease. Front Microbiol. 2020;11:1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coburn B, Wang PW, Diaz Caballero J, Clark ST, Brahma V, Donaldson S, et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep. 2015;5:10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takahashi Y, Saito A, Chiba H, Kuronuma K, Ikeda K, Kobayashi T, et al. Impaired diversity of the lung microbiome predicts progression of idiopathic pulmonary fibrosis. Respir Res. 2018;19:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun‐Fahrländer C, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–9. [DOI] [PubMed] [Google Scholar]
- 8. Garrett WS. Cancer and the microbiota. Science. 2015;348:80–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Polk DB, Peek RM Jr. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10:403–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee SH, Sung JY, Yong D, Chun J, Kim SY, Song JH, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer. 2016;102:89–95. [DOI] [PubMed] [Google Scholar]
- 12. Peters BA, Hayes RB, Goparaju C, Reid C, Pass HI, Ahn J. The microbiome in lung cancer tissue and recurrence‐free survival. Cancer Epidemiol Biomarkers Prev. 2019;28:731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cameron SJS, Lewis KE, Huws SA, Hegarty MJ, Lewis PD, Pachebat JA, et al. A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer. PLoS One. 2017;12:e0177062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang J, Mu X, Wang Y, Zhu D, Zhang J, Liang C, et al. Dysbiosis of the salivary microbiome is associated with non‐smoking female lung cancer and correlated with immunocytochemistry markers. Front Oncol. 2018;8:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu G, Gail MH, Consonni D, Carugno M, Humphrys M, Pesatori AC, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bassis CM, Erb‐Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015;6:e00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang D, Su X, Yuan M, Zhang S, He J, Deng Q, et al. The characterization of lung microbiome in lung cancer patients with different clinicopathology. Am J Cancer Res. 2019;9:2047–63. [PMC free article] [PubMed] [Google Scholar]
- 18. Greathouse KL, White JR, Vargas AJ, Bliskovsky VV, Beck JA, von Muhlinen N, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Y, O'Brien JL, Ajami NJ, Scheurer ME, Amirian ES, Armstrong G, et al. Lung tissue microbial profile in lung cancer is distinct from emphysema. Am J Cancer Res. 2018;8:1775–87. [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng Y, Wang T, Tu X, Huang Y, Zhang H, Tan D, et al. Gut microbiome affects the response to anti‐PD‐1 immunotherapy in patients with hepatocellular carcinoma. J Immunother Cancer. 2019;7:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miyanaga A, Masuda M, Tsuta K, Kawasaki K, Nakamura Y, Sakuma T, et al. Hippo pathway gene mutations in malignant mesothelioma: revealed by RNA and targeted exon sequencing. J Thorac Oncol. 2015;10:844–51. [DOI] [PubMed] [Google Scholar]
- 22. Miyanaga A, Masuda M, Motoi N, Tsuta K, Nakamura Y, Nishijima N, et al. Whole‐exome and RNA sequencing of pulmonary carcinoid reveals chromosomal rearrangements associated with recurrence. Lung Cancer. 2020;145:85–94. [DOI] [PubMed] [Google Scholar]
- 23. Miyanaga A, Matsumoto M, Beck JA, Horikawa I, Oike T, Okayama H, et al. EML4‐ALK induces cellular senescence in mortal normal human cells and promotes anchorage‐independent growth in hTERT‐transduced normal human cells. BMC Cancer. 2021;21:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jin J, Gan Y, Liu H, Wang Z, Yuan J, Deng T, et al. Diminishing microbiome richness and distinction in the lower respiratory tract of lung cancer patients: a multiple comparative study design with independent validation. Lung Cancer. 2019;136:129–35. [DOI] [PubMed] [Google Scholar]
- 25. Etzel CJ, Kachroo S, Liu M, D'Amelio A, Dong Q, Cote ML, et al. Development and validation of a lung cancer risk prediction model for African‐Americans. Cancer Prev Res. 2008;1:255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Houghton AM. Mechanistic links between COPD and lung cancer. Nat Rev Cancer. 2013;13:233–45. [DOI] [PubMed] [Google Scholar]
- 27. de Torres JP, Marín JM, Casanova C, Cote C, Carrizo S, Cordoba‐Lanus E, et al. Lung cancer in patients with chronic obstructive pulmonary disease‐‐ incidence and predicting factors. Am J Respir Crit Care Med. 2011;184:913–9. [DOI] [PubMed] [Google Scholar]
- 28. Young RP, Hopkins RJ, Hay BA, Epton MJ, Black PN, Gamble GD. Lung cancer gene associated with COPD: triple whammy or possible confounding effect? Eur Respir J. 2008;32:1158–64. [DOI] [PubMed] [Google Scholar]
- 29. Sivan A, Corrales L, Hubert N, Williams JB, Aquino‐Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti‐PD‐L1 efficacy. Science. 2015;350:1084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA‐4 blockade relies on the gut microbiota. Science 2015; 350: 1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsay JJ, Wu BG, Sulaiman I, et al. Lower airway Dysbiosis affects lung cancer progression. Cancer Discov. 2021;11:293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leng Q, Holden VK, Deepak J, Todd NW, Jiang F. Microbiota biomarkers for lung cancer. Diagnostics. 2021;11(3):407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Plot of the ddPCR analysis showing Acidovorax in serial dilutions. Blue and gray dots denote positive and negative amplification droplets, respectively. ddPCR, droplet digital polymerase chain reaction
Figure S2. Rate of positivity for Acidovorax in tumor and nontumor tissues. (A) 50 cases of lung cancer. (B) 17 cases of lung cancer with COPD comorbidity. COPD, chronic obstructive pulmonary disease
Figure S3. Plot of the ddPCR analysis showing the presence of Acidovorax (blue and gray dots indicate positive and negative amplification droplets, respectively) in human normal lung fibroblasts and bronchial epithelium. ddPCR, droplet digital polymerase chain reaction
Table S1. List of primers and targets for TP53 sequences
Table S2. Association of the Acidovorax abundances with clinicopathological characteristics of NSCLC patients in nontumor tissues
Table S3. Logistic regression in patients with NSCLC in nontumor tissues


