Abstract
The 26S proteasome irreversibly hydrolyzes polyubiquitylated substrates to maintain protein homeostasis; it also regulates immune responses by generating antigenic peptides. An alternative form of the 26S proteasome is the immunoproteasome, which contains substituted catalytic subunits (β1i/PSMB9, β2i/PSMB10, and β5i/PSMB8) instead of constitutively expressed counterparts (β1/PSMB6, β2/PSMB7, and β5/PSMB5). The immunoproteasome expands the peptide repertoire presented on MHC class I molecules. However, how its activity changes in this context is largely elusive, possibly due to the lack of a standardized methodology to evaluate its specific activity. Here, we describe an assay protocol that measures the immunoproteasome activity of whole-cell lysates using commercially available fluorogenic peptide substrates. Our results showed that the most accurate assessment of immunoproteasome activity could be achieved by combining β5i-targeting substrate Ac-ANW-AMC and immunoproteasome inhibitor ONX-0914. This simple and reliable protocol may contribute to future studies of immunoproteasomes and their pathophysiological roles during viral infection, inflammation, and tumorigenesis.
Keywords: Proteasome, Immunoproteasome, Fluorogenic substrates, Proteasome inhibitors
INTRODUCTION
The 26S proteasome is an ATP-dependent multi-subunit protease found in eukaryotes, responsible for intracellular proteolysis of polyubiquitinated proteins (1). In addition to regulating individual protein homeostasis and maintaining cellular proteome quality, the proteasome is also involved in a diverse array of immune responses by generating antigenic peptides, which are then presented on the MHC-I (2). The constitutive, or conventional, 26S proteasome (hereafter referred to as the constitutive proteasome [c-proteasome]) is compartmentalized into distinct 20S catalytic and 19S regulatory subunits. Significant recent advances in cryo-electron microscopy improved our understanding of the structural and mechanistic details of the 26S proteasome (3,4,5). The barrel-shaped 20S proteasome is a C2-symmetric complex that contains 7 different α-type (α1–α7) and 7 β-type subunits (β1–β7) assembled in a 4-stacked ring (αββα) structure. While all 14 β subunits have active proteolytic activities in archaeal and prokaryotic 20S proteasomes, only three subunits (β1/PSMB6, β2/PSMB7, and β5/PSMB5) are active in eukaryotes (please note that proteasome subunits have multiple nomenclatures; hereafter, the common names with β annotation will be used instead of the official names from the HUGO Consortium) (6).
In higher eukaryotes, proinflammatory cytokines, including immunomodulatory cytokine IFN-γ, induce the expression of analogous catalytic subunits such as β1i/PSMB9/LMP2, β2i/PSMB10/MECL1, and β5i/PSMB8/LMP7, replacing the c-proteasome with the immunoproteasome (i-proteasome) (7,8). β2i and β5i retain similar proteolytic activities as their cognate β2 and β5 subunits, while β1i has chymotrypsin-like activity, which is different from β1 having caspase-like activity. As a result, i-proteasomes preferentially produce peptide fragments with hydrophobic C-terminal amino acids, readily bound to MHC-I (9,10). In humans, β1i and β5i genes are located within the MHC-II genomic region on chromosome 6, along with the genes encoding TAPs. The inducible βi subunits are not indispensable for class I Ag presentation, but the diverse species of the 26S proteasome can contribute to generating a broader repertoire of antigenic peptides for presentation to cytotoxic T cells (11). In addition to their role in the adaptive immune response, i-proteasomes also contribute to the clearance of proteins damaged by ROS (12).
In most cell types, c-proteasomes are highly abundant (>200 nM) and stable (half-life 12–15 days) (13,14,15,16). Nuclear factor erythroid-derived 2-related factor 1 (NRF1) functions as the major transcription factor for c-proteasome genes and is generally maintained at a low basal level through endoplasmic reticulum-associated protein degradation. If proteasome activity is inhibited, NRF1 escapes from proteasomal degradation, becomes cleaved to the mature form, and binds to the antioxidant response element (ARE). All 33 proteasome subunit genes and their assembly factors contain this response element in their promoter regions and, therefore, their expression is upregulated upon NRF1 binding, forming a “bounce-back” negative feedback system (17). Unlike conventional proteasomes, the expression of i-proteasomes (in somatic cells) is acutely upregulated by immune stimuli. Cytokines such as IFN-γ activate the JAK/STAT pathway, often initiating the gene expression of immunoproteasome subunits (18). Other cellular signaling pathways, such as TNF-α/NF-κB and TGF-β/SMAD pathways, also influence i-proteasome induction (19).
The hydrolysis of fluorogenic peptides, either from purified forms or whole-cell lysates (WCLs), has been used as a standard method to measure the activity of c-proteasomes. In principle, different types of active sites can be analyzed with relatively specific cognate substrates, such as carbonylbenzyl-Leu-Leu-Glu-7-amino-4-methylcoumarin (Z-LLE-AMC for caspase-like β1 activity; The italic denotes the hydrolyzing residue by cognate catalytic subunit.), ter-butyloxycarbonyl-Leu-Arg-Arg-AMC (Boc-LRR-AMC for trypsin-like β2 activity), and succinyl-Leu-Leu-Leu-Val-Tyr-AMC (suc-LLVY-AMC for chymotrypsin-like β5 activity) (20). In addition, several commercially available substrates are heavily used to measure the specific activities of i-proteasomes, such as acetyl-Pro-Ala-Leu-AMC (Ac-PAL-AMC for β1i activity) and acetyl-Ala-Asn-Trp-AMC (Ac-ANW-AMC for β5i). Although using these i-proteasome substrates is generally accepted, our mechanistic understanding of their identification, substrate specificity, or inhibition kinetics is incomplete. To measure i-proteasome activity, investigators use the WCLs of cells treated with cytokines and other stress inducers because the selective purification of i-proteasomes lacking c-proteasomes is technically demanding. However, there has been no standardized protocol for independent reproduction and cross-validation. Here, we evaluated these substrates to provide an optimizing and reliable assay for measuring specific i-proteasome activity.
MATERIALS AND METHODS
Materials
Fluorogenic peptides suc-LLVY-AMC, Boc-LRR-AMC, and Z-LLE-AMC were purchased from Bachem (Bubendorf, Switzerland; I-1395), Enzo Life Sciences (Farmingdale, NY, USA; BW8515), and Cayman Chemical (Ann Arbor, MI, USA; 10008117), respectively. Ac-ANW-AMC was purchased from AdipoGen Life Sciences (San Diego, CA, USA; AG-CP3-0037), while Ac-WLA-WLA and Ac-PAL-AMC were purchased from R&D Systems (Minneapolis, MN, USA; S-330 and S310, respectively). In addition, we have tested other brands of peptide substrates and obtained similar results. Sources of other major chemical and biochemical reagents were as follows: ONX-0914 (MedKoo Biosciences, Cary, NC, USA; 205730), recombinant human IFNγ (PeproTech, Rocky Hill, NJ, USA; 300-02), EDTA (Daejung Chemicals, Siheung, Korea; 4000-4405), BSA (Bovogen Biologicals, Keilor, VIC, Australia; BSA025), ATP (Bio Basic, Markham, ON, Canada; AB0020), and DTT (Geogiachem, Suwanee, GA, USA; CDT10).
Cell culture and immunoblotting
HeLa cells were cultured in DMEM supplemented with 10% FBS, 2 mM glutamine, and 100 U/ml penicillin/streptomycin in 6-well plates. Upon reaching up to 30% confluency, cells were treated with 500 U/ml of IFN-γ for 48 h, and then lysed using the proteasome activity lysis buffer (25 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 10% glycerol, 1 mM ATP, 1 mM DTT, 1× protease inhibitor cocktail). For immunoblotting, 2× sample buffer was added to the lysates (final concentration of 6.3% glycerol, 80 mM Tris-HCl [pH 6.8], 62.5 µg/ml bromophenol blue, and 10 mg/ml SDS) and samples were heated at 85°C for 10 min. Samples were separated using SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. Next, the membranes were blocked with 5% nonfat milk and probed with primary Abs. Ab sources and dilution factors were as follows: anti-GAPDH (Santa Cruz Biotechnology, Dallas, TX, USA; sc-32233, 1:10,000), anti-PSMB5 (β5) (Invitrogen, Waltham, MA, USA; PA1-977, 1:3,000), anti-PSMB6 (β1) (Invitrogen; PA1-978, 1:3,000), anti-PSMB7 (β2) (Invitrogen; PA5-30988, 1:1,000), anti-PSMB8 (β5i) (Invitrogen; PA1-972, 1:3,000), anti-PSMB9 (β1i), (Invitrogen; PA1-1960, 1:3,000), anti-PSMB10 (β2i) (Abcam, Cambridge, UK; ab183506, 1:3,000), anti-PSMB2 (β4) (Santa Cruz Biotechnology; SC-54676, 1:3,000), anti-PSMA4 (α3) (ENZO Life Sciences; PW8115, 1:10,000), anti-PSMD1 (Santa Cruz Biotechnology; sc-514809, 1:3,000), anti-PSMDC2 (Santa Cruz Biotechnology; sc-166972, 1:3,000), and anti-Ub (P4D1 monoclonal, Santa Cruz Biotechnology; sc-8017, 1:10,000).
Measurement of proteasome activity from WCLs
For the proteasome activity assay, HeLa cells were lysed in the proteasome activity lysis buffer. Next, cells were homogenized by passing the lysates 15 times through a 26G×1/2″ needle attached to a 1-ml syringe. The lysates were then centrifuged at 12,000 rpm for 15 min using a benchtop centrifuge. Protein concentrations of the samples were determined using the Bradford assay. Proteasome activity was determined using model peptide substrates by measuring free AMC fluorescence on a TECAN infinite m200 fluorometer) as previously described (21). Briefly, using a black 96-well plate, 10 µl of WCLs and fluorogenic peptides (concentrations as indicated) were mixed with 100 µl proteasome activity assay buffer (50 mM Tris-HCl [pH 7.5], 1 mg/ml BSA, 1 mM EDTA, 1 mM fresh ATP, and 1 mM fresh DTT). Our pilot studies demonstrated that the final 12.5 µM X-AMC concentration in a microplate-format was cost-effective and generated strong signals with good linearity. The AMC fluorescence unquenched after hydrolysis by proteasomes was monitored every three minutes at 380 nm excitation and 460 nm emission wavelengths at 30°C. Each sample was assayed in triplicate. The fluorescence intensity from i-proteasome substrates was normalized to the basal activity (e.g., X-AMC cleavage by potential endopeptidases in WCLs), which was determined separately by treating samples with i-proteasome inhibitor ONX-0914.
Native gel electrophoresis
Non-denaturing (native) gel analysis using HeLa WCL was performed as previously described (22,23). Samples were resolved using NuPAGE™ 3%–8% Tris-Acetate Protein Gels (Thermo Fisher Scientific, Waltham, MA, USA) at 150 V for 3–4 h. Native gels were transferred from the gel to PVDF membranes for immunoblotting analysis with indicated proteasome subunit Abs.
Statistical analysis
All analyses were performed using GraphPad Prism (ver. 5.03, GraphPad Software, San Diego, CA, USA). Results are presented as the mean±SD. Statistical significance of differences between groups was determined by Student’s t-test or one-way ANOVA with Bonferroni’s multiple comparison test.
RESULTS
Induction of i-proteasomes after IFN-γ treatment
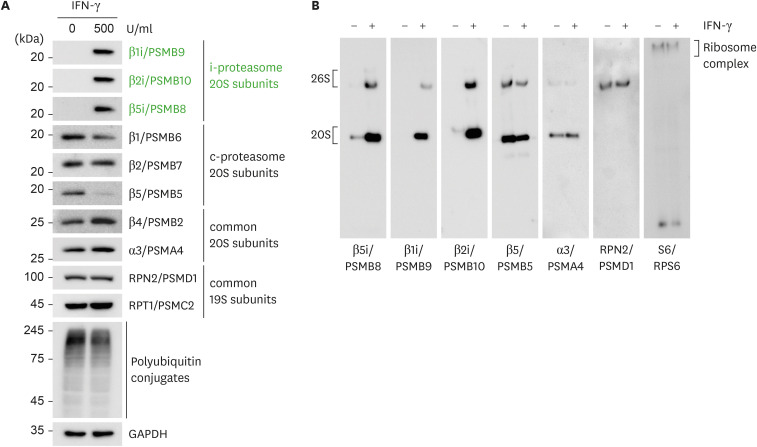
First, we evaluated the induction of β1i, β2i, and β5i subunit expression in HeLa cells in response to IFN-γ treatment. While the basal levels of these inducible βi subunits in the cells were virtually undetectable by immunoblotting, IFN-γ treatment (500 U/ml for 48 h) significantly increased the protein levels of all βi subunits (Fig. 1A). The unprocessed βi subunits were virtually undetectable. The effect of IFN-γ was highly variable depending on treatment conditions and cell types. For example, we found that the treatment of HEK293 cells with IFN-γ had little effect on the induction of βi subunits, while their expression was significantly induced in A549 and HK2 cells (data not shown). Most non-inducible proteasome subunits from catalytic 20S complex and regulatory complex did not increase by IFN-γ treatment (Fig. 1A). In addition, the levels of polyubiquitin conjugates were also comparable between the samples, suggesting that total proteasome activity (as well as overall proteasome content) in the cells remained largely unchanged (Fig. 1A). Native gel analysis with a similar protein loading amount revealed that the induced βi subunits were well incorporated into the assembly of the 20S proteasome to form the i-proteasome holoenzyme (Fig. 1B). Based on the results, we used WCLs from HeLa cells treated with IFN-γ to investigate the specificity of commonly used fluorogenic peptide substrates.
Figure 1. Induction of i-proteasome subunits in HeLa cells after IFN-γ treatment. HeLa cells (up to 30% confluency) were treated with 500 U/ml IFN-γ for 48 h. WCLs were collected in a detergent-free buffer and subjected to conventional SDS-PAGE (A) or non-denaturing (native) PAGE (B) for subsequent immunoblotting with indicated Abs.
We observed that standard lysis buffers containing up to 1% detergents, such as SDS, Triton X-100, and NP-40, affected both the AMC fluorescence signal and the cleavage between the peptides and the AMC group (24). Therefore, we collected HeLa cells in a detergent-free buffer (see MATERIALS AND METHODS) and homogenized these cells using a 26-gauge needle to prepare WCLs. Human proteasomes affinity-purified from mammalian cells were stable in the detergent-free buffer (15,25). Protein samples were carefully collected from the soluble fraction after centrifugation and were then aliquoted to avoid freeze-thaw cycles. The hydrolysis of the fluorogenic peptides by the 20S proteasome is independent of ATP; therefore, this assay cannot discriminate substrate hydrolysis from free 20S proteasomes and 26S holoenzymes. Therefore, assays using peptide substrates measure the overall output of cellular proteasomal activity from both 20S and 26S proteasomes.
Measurement of c- and i-proteasome activity using WCLs and fluorogenic peptide substrates
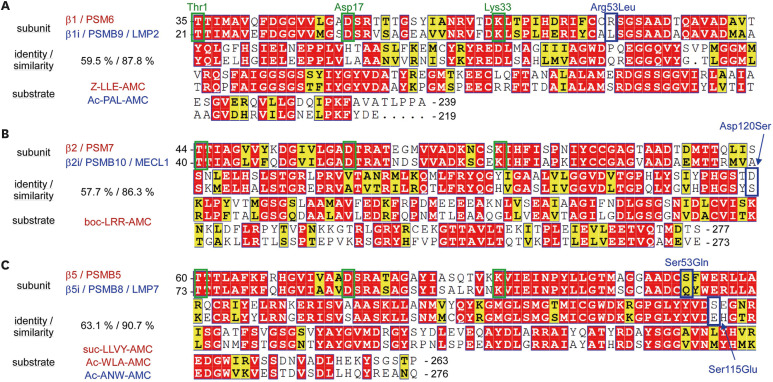
To compare c- and i-proteasome activity, caspase-like β1 activity, and chymotrypsin-like β1i activity were measured using Z-LLE-AMC and Ac-PAL-AMC, respectively. All active β subunits from c-proteasomes utilize the same biochemical reaction that involves the catalytic triad Thr1, Asp17, and Lys33 in their mature forms (after the autocatalytic cleavage of the conserved Gly residue in the C-termini adjacent to the Thr1 residue). Furthermore, the amino acid sequences between cognate β subunits (e.g., β1 vs. β1i) are highly homologous (Fig. 2). Therefore, the substrate specificity towards i-proteasomes is expected to be determined by the smallest amino acid substitution in S1 substrate binding pockets: Arg53Leu substitution alters the S1 binding pocket charge from positive to neutral in β1i (Fig. 2). The C-terminal Glu residue in Z-LLE-AMC is preferentially recognized by acidic Arg53 in β1, while Leu in Ac-PAL-AMC is preferentially recognized by Leu53 in β1i. These differences in the peptidase activity of i-proteasomes also appear to be beneficial, if not obligatory, for generating peptides with hydrophobic C-termini, which are the better anchoring motifs for binding to MHC-I (26,27).
Figure 2. Sequence alignment, characteristics, and the model substrates of catalytic β-subunits from the c- and i-proteasome. Mature form sequences of human β1/β1i (A), β2/β2i (B), and β5/β5i (C) subunits are aligned using UniProt (https://www.uniprot.org/align/) and sequence similarities are rendered by ESPript 3.0 (https://espript.ibcp.fr/ESPript/ESPript/). Residue numbers are assigned based on the forms of the unprocessed (before autocatalytic cleavage). The percentage of amino acid identity and similarity between the cognate subunits is shown on the left. Similar residues are presented in yellow, identical residues in red, and key residues forming proteasomal catalytic triads (Thr1, Asp17, Lys33 after cleavage) are in green boxes. Potentially critical amino acid changes in S1 binding pockets, which could contribute to the substrate specificity of inducible βi subunits, are shown in blue boxes. Conventional fluorogenic peptide substrates are presented in red for c-proteasomes and in blue for i-proteasomes.
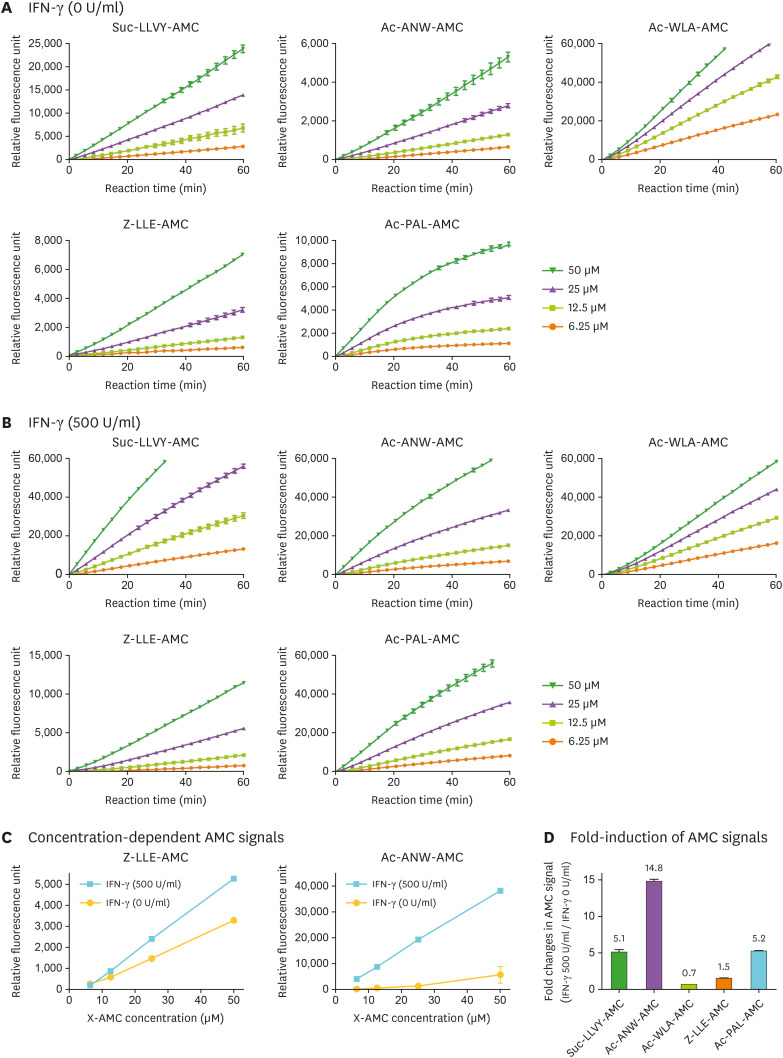
To measure c- and i-proteasome activity, we performed kinetic assays over time using a wide range of X-AMC concentrations (6.25, 12.5, 25, and 50 µM) in a 100 µl hydrolysis reaction which contains up to 15 µg of total protein. In the absence of IFN-γ induction, c-proteasome substrates (Z-LLE-AMC for β1, and both suc-LLVY-AMC and Ac-WLA-AMC for β5) showed strong AMC signals, with Ac-WLA-AMC exhibiting the highest fluorescence intensity (Fig. 3A). Signal intensities from i-proteasome substrates (Ac-PAL-AMC for β1i and Ac-ANW-AMC for β5i) were generally weaker, but were not negligible, than those of c-proteasome substrates. In response to IFN-γ treatment, both c- and i-proteasome substrates, except Ac-WLA-AMC, showed increased AMC signal intensities (Fig. 3B) compared to the untreated samples. In addition, our protocol produced a high degree of linearity between X-AMC concentration and AMC fluorescence intensity, showing that our assay was performed in a reliable analytical range (Fig. 3C).
Figure 3. Measurement of i-proteasome activity in WCLs using fluorogenic reporter substrates. (A) The hydrolysis of fluorogenic peptide substrates (final 6.25, 12.5, 25, and 50 µM in 100 µl hydrolysis reaction) was continuously measured every 3 min for 60 min. WCLs (10 µl, corresponding up to 15 µg total protein) from untreated HeLa cells were added at time 0. Representative raw fluorescence graphs from three independent experiments are shown. (B) The hydrolysis of peptide substrates described in (A) was measured in WCLs from IFN-γ-treated HeLa cells (500 U/ml for 48 h). (C) The fluorescence intensities of the c-proteasome substrate (Z-LLE-AMC) and i-proteasome substrate (Ac-ANW-AMC) at 25 min time point; data are presented as the mean±SD (n=3). (D) The i-proteasome specificity of the X-AMC substrates was evaluated at 12.5 µM concentration in IFN-γ-treated samples. The fold induction of AMC fluorescence intensity over IFN-γ-untreated controls is shown.
It should be noted that the AMC fluorescence signal from suc-LLVY-AMC increased more than 5-fold (3,121 relative fluorescence unit [RFU] at 0 U/ml IFN-γ vs. 15,857 RFU at 500 U/ml IFN-γ) in response to IFN-γ treatment (Fig. 3D), indicating that this substrate was hydrolyzed by both c- and i-proteasomes. The AMC signal from other c-proteasome substrates showed more c-proteasome-specific activity: for example, the AMC signal from Z-LLE-AMC increased only up to 1.5-fold (from 581 RFU to 855 RFU). Signals from β5-specific Ac-WLA-AMC increased in IFN-γ-treated WCLs (from 13,880 RFU to 21,164 RFU), potentially reflecting our observations that β5i upregulation by IFN-γ was accompanied by β5 downregulation (Fig. 2). Collectively, our data suggest that Ac-WLA-AMC, but not suc-LLVY-AMC, is a specific c-proteasome substrate. In addition, our results indicate that, for Ac-ANW-AMC and Ac-PAL-AMC, fluorescence intensity less than 5,000 RFU should be considered as a background signal rather than a weak i-proteasome activity. Both i-proteasome substrates showed a dramatic signal increase in IFN-γ-treated samples: 14.8-fold increase for Ac-ANW-AMC (from 590 RFU to 8,716 RFU) and 5.2-fold increase for Ac-PAL-AMC (from 1,694 RFU to 8,854 RFU) (Fig. 3D). In the absence of IFN-γ induction, Ac-ANW-AMC demonstrated a lower basal signal; however, AMC intensity was higher upon the induction of i-proteasomes. Thus, these results collectively indicated that, in response to IFN-γ, Ac-ANW-AMC generates more dynamic ranges when used to measure the activity of the i-proteasome.
Obtaining specific i-proteasome activity using Ac-ANW-AMC and ONX-0914
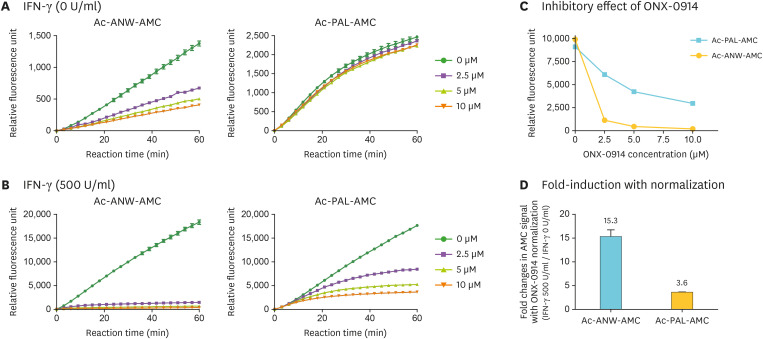
It is possible that co-existing c-proteasomes may affect the hydrolysis rate of fluorogenic i-proteasome substrates. To specifically assess i-proteasome activity and determine the contribution of the non-i-proteasomal activity to the assay, the X-AMC fluorescence intensity was measured in the presence of the i-proteasome inhibitor ONX-0914 (also known as PR-957). ONX-0914 has been reported to specifically and irreversibly target β5i (28). On the contrary, bortezomib and carfilzomib, two other well-characterized proteasome inhibitors and anticancer drugs, are known to block both c- and i-proteasomes (28). We observed that the hydrolysis of Ac-ANW-AMC in WCLs from untreated HeLa cells was delayed by ONX-0914 in a dose-dependent manner, while the Ac-PAL-AMC signal was virtually not affected by the inhibitor (Fig. 4A). The effect of ONX-0914 was very different in IFN-γ-treated WCLs: the fluorescence signal from Ac-ANW-AMC was effectively abolished with at 2.5 µM ONX-0914 (89% inhibition), while the signal from Ac-PAL-AMC was modestly decreased in response to the increasing concentrations of the inhibitor (Fig. 4B). Almost 95% inhibition of Ac-ANW-AMC signal was achieved with 5 µM ONX-0914, while up to 53% reduction was observed with Ac-Pal-AMC signal. Collectively, these results indicated that Ac-ANW-AMC hydrolysis was effectively blocked by ONX-0914 (only up to 2.0% of residual AMC signal with 10 µM ONX-0914) (Fig. 4C). In addition, these results suggested that the basal i-proteasome level in HeLa cells under normal conditions was practically negligible. When we compared the degree of i-proteasome induction, normalization using ONX-0914-treated values produced more significant differences between two i-proteasome substrates: 15.3-fold induction for Ac-ANW-AMC (from 635 RFU to 9,719 RFU) vs. 3.6-fold induction for Ac-PAL-AMC (from 1,699 RFU to 6,143 RFU) (Fig. 4D). Collectively, our findings indicate that, in order to measure i-proteasome activity in WCLs, fluorogenic substrate Ac-ANW-AMC and i-proteasome inhibitor ONX-0914 should be used since normalized values obtained in these experiments produce the most accurate and reproducible results.
Figure 4. Inhibitory effect of ONX-0914 on the hydrolysis of i-proteasome-specific substrates. (A) Hydrolysis of β5i-specific Ac-ANW-AMC and β1i-specific Ac-PAL-AMC (final concentration of 12.5 µM) in untreated WCLs was measured in the presence of i-proteasome inhibitor ONX-0914 (0, 2.5, 5, and 10 µM concentrations) for up to 60 min. (B) The hydrolysis was measured as described in (A) in IFN-γ-treated WCLs (500 U/ml for 48 h). (C) The concentration-dependent inhibitory effect of ONX-0914 on Ac-ANW-AMC and Ac-PAL-AMC was calculated based on the raw fluorescence intensity shown in (B). (D) Normalized X-AMC fluorescence intensity in samples treated with 5 µM ONX-0914 at 30 min. Relative changes in samples with IFN-γ treatment compared to samples without the treatment.
DISCUSSION
Based on phylogenetic analysis, i-proteasomes appeared later than c-proteasomes during the evolution of the immune system. Since both types of proteasome have overlapping functionality and common target substrates, it is critical to investigate the profiles of cellular activity for each proteasome in diverse physiological and pathological conditions. We believe that there is a need for a standardized analytical method for measuring specific c- and i-proteasome activity. We discovered that the assessment of c-proteasome-specific activity (excluding i-proteasome activity) was more complex than initially envisioned, even though Z-LLE-AMC did not appear to be affected by IFN-γ treatment (Fig. 3). This appears mainly because most conventional proteasome inhibitors target both c- and i-proteasome proteolytic sites.
Both β2 and β2i cleave after basic residues (trypsin-like activity), and β2i-specific substrates are not commercially available. Currently available β2 fluorogenic substrates such as Boc-LRR-AMC were less sensitive and generated weaker signals than those for other proteolytic sites. Suc-LLVY-AMC, which has been most widely used since it was used at the first proteasome activity assays (29), targets the chymotrypsin-like activity of β5; however, it is generally considered to be able to represent the overall proteasome activity (30). To measure β5i activity, a hydrophobic residue-possessing Ac-ANW-AMC is commonly used; nevertheless, its specificity toward i-proteasomes vs. c-proteasomes in WCLs has not been convincingly reported.
Our results also suggested that it was highly unlikely that hybrid i-proteasomes (either the 20S or 26S complex) would perturb the accuracy of this assay in cultured cells since IFN-γ induced all βi subunits at the same time (Fig. 1), and even the “mixed” forms of i-proteasomes usually contained β5i, if not β1i or β2i subunits (31). Therefore, until we have highly specific β1i/β2i substrates or inhibitors, our assay protocol (Ac-ANW-AMC with ONX-0914) may provide a sound system for evaluating i-proteasome activity in WCLs, and even possibly in animal/human tissues as well. These findings will provide a practical guideline for studies at the intersection of immunology and cell biology, investigating the role of i-proteasomes in Ag presentation and stress responses. As a result, these studies will further extend the therapeutic use of proteasome inhibitors as anticancer drugs.
ACKNOWLEDGEMENTS
S.K., S.H.P, and W.H.C. were supported by the BK21 FOUR Biomedical Science Program funded by the Ministry of Education and National Research Foundation (NRF) of Korea. This work was also supported by other grants from the NRF (2020R1A5A1019023 and 2021R1A2C2008023 to M.J.L.), the Korea Health Industry Development Institute and Korea Dementia Research Center (HU21C0071 to M.J.L.), and the Creative-Pioneering Researchers Program through Seoul National University.
Abbreviations
- ARE
antioxidant response element
- c-proteasome
constitutive proteasome
- i-proteasome
immunoproteasome
- NRF1
nuclear factor erythroid-derived 2-related factor 1
- PVDF
polyvinylidene difluoride
- RFU
relative fluorescence unit
- WCL
whole-cell lysate
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
- Conceptualization: Kim S, Choi WH, Lee MJ.
- Data curation: Kim S, Park SH, Choi WH, Lee MJ.
- Formal analysis: Kim S, Park SH, Choi WH.
- Funding acquisition: Lee MJ.
- Investigation: Choi WH, Lee MJ.
- Methodology: Kim S, Choi WH.
- Project administration: Kim S, Lee MJ.
- Supervision: Choi WH, Lee MJ.
- Visualization: Park SH, Choi WH.
- Writing - original draft: Kim S, Lee MJ.
- Writing - review & editing: Lee MJ.
References
- 1.Hershko A, Ciechanover A, Varshavsky A. Basic medical research award. The ubiquitin system. Nat Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 2.Ferrington DA, Gregerson DS. Immunoproteasomes: structure, function, and antigen presentation. Prog Mol Biol Transl Sci. 2012;109:75–112. doi: 10.1016/B978-0-12-397863-9.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci U S A. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 6.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monaco JJ, Nandi D. The genetics of proteasomes and antigen processing. Annu Rev Genet. 1995;29:729–754. doi: 10.1146/annurev.ge.29.120195.003501. [DOI] [PubMed] [Google Scholar]
- 8.Brown MG, Driscoll J, Monaco JJ. Structural and serological similarity of MHC-linked LMP and proteasome (multicatalytic proteinase) complexes. Nature. 1991;353:355–357. doi: 10.1038/353355a0. [DOI] [PubMed] [Google Scholar]
- 9.Gaczynska M, Rock KL, Goldberg AL. Role of proteasomes in antigen presentation. Enzyme Protein. 1993;47:354–369. doi: 10.1159/000468693. [DOI] [PubMed] [Google Scholar]
- 10.Gaczynska M, Rock KL, Goldberg AL. Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature. 1993;365:264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- 11.Kincaid EZ, Che JW, York I, Escobar H, Reyes-Vargas E, Delgado JC, Welsh RM, Karow ML, Murphy AJ, Valenzuela DM, et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat Immunol. 2011;13:129–135. doi: 10.1038/ni.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schröter F, Prozorovski T, Lange N, Steffen J, Rieger M, et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142:613–624. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 13.Asano S, Fukuda Y, Beck F, Aufderheide A, Förster F, Danev R, Baumeister W. Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science. 2015;347:439–442. doi: 10.1126/science.1261197. [DOI] [PubMed] [Google Scholar]
- 14.Pack CG, Yukii H, Toh-e A, Kudo T, Tsuchiya H, Kaiho A, Sakata E, Murata S, Yokosawa H, Sako Y, et al. Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat Commun. 2014;5:3396. doi: 10.1038/ncomms4396. [DOI] [PubMed] [Google Scholar]
- 15.Choi WH, Yun Y, Park S, Jeon JH, Lee J, Lee JH, Yang SA, Kim NK, Jung CH, Kwon YT, et al. Aggresomal sequestration and STUB1-mediated ubiquitylation during mammalian proteaphagy of inhibited proteasomes. Proc Natl Acad Sci U S A. 2020;117:19190–19200. doi: 10.1073/pnas.1920327117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka K, Ichihara A. Half-life of proteasomes (multiprotease complexes) in rat liver. Biochem Biophys Res Commun. 1989;159:1309–1315. doi: 10.1016/0006-291x(89)92253-5. [DOI] [PubMed] [Google Scholar]
- 17.Hamazaki J, Murata S. Er-resident transcription factor NRF1 regulates proteasome expression and beyond. Int J Mol Sci. 2020;21:3683. doi: 10.3390/ijms21103683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston-Carey HK, Pomatto LC, Davies KJ. The immunoproteasome in oxidative stress, aging, and disease. Crit Rev Biochem Mol Biol. 2015;51:268–281. doi: 10.3109/10409238.2016.1172554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamber Kaya HE, Radhakrishnan SK. Trash talk: mammalian proteasome regulation at the transcriptional level. Trends Genet. 2021;37:160–173. doi: 10.1016/j.tig.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- 21.Shin SK, Kim JH, Lee JH, Son YH, Lee MW, Kim HJ, Noh SA, Kim KP, Kim IG, Lee MJ. Docosahexaenoic acid-mediated protein aggregates may reduce proteasome activity and delay myotube degradation during muscle atrophy in vitro . Exp Mol Med. 2017;49:e287. doi: 10.1038/emm.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elsasser S, Schmidt M, Finley D. Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 2005;398:353–363. doi: 10.1016/S0076-6879(05)98029-4. [DOI] [PubMed] [Google Scholar]
- 23.Kim E, Park S, Lee JH, Mun JY, Choi WH, Yun Y, Lee J, Kim JH, Kang MJ, Lee MJ. Dual function of USP14 deubiquitinase in cellular proteasomal activity and autophagic flux. Cell Reports. 2018;24:732–743. doi: 10.1016/j.celrep.2018.06.058. [DOI] [PubMed] [Google Scholar]
- 24.Choi WH, Kim S, Park S, Lee MJ. Concept and application of circulating proteasomes. Exp Mol Med. 2021;53:1539–1546. doi: 10.1038/s12276-021-00692-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi WH, de Poot SA, Lee JH, Kim JH, Han DH, Kim YK, Finley D, Lee MJ. Open-gate mutants of the mammalian proteasome show enhanced ubiquitin-conjugate degradation. Nat Commun. 2016;7:10963. doi: 10.1038/ncomms10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heemels MT, Schumacher TN, Wonigeit K, Ploegh HL. Peptide translocation by variants of the transporter associated with antigen processing. Science. 1993;262:2059–2063. doi: 10.1126/science.8266106. [DOI] [PubMed] [Google Scholar]
- 27.Momburg F, Roelse J, Howard JC, Butcher GW, Hämmerling GJ, Neefjes JJ. Selectivity of MHC-encoded peptide transporters from human, mouse and rat. Nature. 1994;367:648–651. doi: 10.1038/367648a0. [DOI] [PubMed] [Google Scholar]
- 28.Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, Sylvain C, Ring ER, Shields J, Jiang J, et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med. 2009;15:781–787. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]
- 29.Hough R, Pratt G, Rechsteiner M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J Biol Chem. 1987;262:8303–8313. [PubMed] [Google Scholar]
- 30.Yun Y, Lee SY, Choi WH, Park JC, Lee DH, Kim YK, Lee JH, Lee JY, Lee MJ, Kim YH. Proteasome activity in the plasma as a novel biomarker in mild cognitive impairment with chronic tinnitus. J Alzheimers Dis. 2020;78:195–205. doi: 10.3233/JAD-200728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy MK, Weinberg JB. The immunoproteasome and viral infection: a complex regulator of inflammation. Front Microbiol. 2015;6:21. doi: 10.3389/fmicb.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]