Abstract
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2), has spread over the world causing a pandemic which is still ongoing since its emergence in late 2019. A great amount of effort has been devoted to understanding the pathogenesis of COVID-19 with the hope of developing better therapeutic strategies. Transcriptome analysis using technologies such as RNA sequencing became a commonly used approach in study of host immune responses to SARS-CoV-2. Although substantial amount of information can be gathered from transcriptome analysis, different analysis tools used in these studies may lead to conclusions that differ dramatically from each other. Here, we re-analyzed four RNA-sequencing datasets of COVID-19 samples including human bronchoalveolar lavage fluid, nasopharyngeal swabs, lung biopsy and hACE2 transgenic mice using the same standardized method. The results showed that common features of COVID-19 include upregulation of chemokines including CCL2, CXCL1, and CXCL10, inflammatory cytokine IL-1β and alarmin S100A8/S100A9, which are associated with dysregulated innate immunity marked by abundant neutrophil and mast cell accumulation. Downregulation of chemokine receptor genes that are associated with impaired adaptive immunity such as lymphopenia is another common feather of COVID-19 observed. In addition, a few interferon-stimulated genes but no type I IFN genes were identified to be enriched in COVID-19 samples compared to their respective control in these datasets. These features are in line with results from single-cell RNA sequencing studies in the field. Therefore, our re-analysis of the RNA-seq datasets revealed common features of dysregulated immune responses to SARS-CoV-2 and shed light to the pathogenesis of COVID-19.
Keywords: COVID-19, Immune response, Host-virus interactions, RNA-seq datasets, Innate immunity, Cytokines
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a new virus that causes coronavirus disease 2019 (COVID-19). Since late 2019 the virus started to spread over the world and caused a pandemic with unprecedented health and economic consequences. The World Health Organization has reported a total of 429,189,439 confirmed cases and 6,159,474 confirmed deaths as of 7th of April 2022 (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). 11,054, 362,790 vaccine doses were administered worldwide by then. Individuals infected by SARS-CoV-2 showed a large variety of disease manifestations. Some infected individuals were asymptomatic or had only flu-like symptoms, whereas others needed supplemental oxygen or even succumbed to the disease (1,2). Quarantine, social isolation, and infection-control measures remain the main ways to prevent disease spread (3), highlighting the urgent need for effective therapies against the disease. Understanding virus-host interaction including how SARS-CoV-2 viruses damage host cells and tissues, how various components of the host immune system work cooperatively to control the spreading of the virus, and how dysregulated immune activation contributes the development of the disease will provide opportunities for development of therapeutic strategies against COVID-19.
Numerous studies on immune response after SARS-CoV-2 infection have demonstrated that severe COVID-19 disease is associated with elevated plasma levels of inflammatory cytokines/chemokines and unrecoverable lymphopenia. For instance, analysis of forty-one COVID-19 patients who had a history of direct exposure to the Huanan seafood market in Wuhan, China, Huang et al. (1) found that COVID-19 patients had higher plasma levels of various cytokines and chemokines including IL1β, IL1RA, IL-7, IL8, IL9, G-CSF, GM-CSF, IFNγ, IP10 (CXCL10) and MCP1 (CCL2) compared to healthy adults. In addition, patients with severe symptoms requiring intensive care unit admission had higher concentrations of IL2, IL-7, IL-10, G-CSF, IP10, MCP1, MIP1A and TNFα than those with milder symptoms, indicating that severe COVID-19 disease is likely associated with the development of cytokine storm. Interestingly, patients with severe symptoms had increased neutrophil counts (10.6×109 per L vs. 4.4×109 per L) but more severe lymphopenia (0.4×109 per L vs. 1.0×109 per L) compared to patients with mild disease upon admission to hospital. In another study, by comparing patients who survived COVID-19 (137 patients) against those who died of COVID-19 disease (54 patients) from two hospitals in Wuhan, Zhou et al. (2) discovered that severe lymphopenia was associated with the non-survivors, whereas survivors had lymphopenia with the lowest lymphocyte count observed on day 7 after disease onset followed by recovery during hospitalization. In addition, the non-survivors had significantly elevated levels of IL-6 compared to the survivors. These two studies demonstrated that unrecoverable lymphopenia and exaggerated inflammation are two factors associated with severe COVID-19 and even death from the disease. These observations were supported by analysis of COVID-19 patients in other parts of the world. For instance, by immune cell profiling of 113 COVID-19 patients admitted to Yale New Heaven Hospital, USA, ranged between day 3 to day 51 after disease onset, Lucas et al. (4) observed that both CD4+ and CD8+ T cells were markedly reduced, whereas granulocytes including macrophages, neutrophils and eosinophils were increased in PBMCs in COVID-19 patients compared to healthy control, and the levels of granulocytes correlated with disease severity. In addition, although increased levels of inflammatory cytokines, including IL-1α, IL-1β, IL-17A, IL-12 and IFNα were observed in all COVID-19 patients compared to healthy control, patients with severe symptoms had increased levels of additional inflammatory mediators including IL16, IL-21, IL-23, IL33 and IFNl when compared to those with mild diseases. Interestingly, patients with severe symptoms had sustained increased IFNα and IFNl levels as well as elevated levels of IL-1β and IL-18 in plasma compared with patients with mild disease, suggesting that in addition to elevated levels of proinflammatory cytokines and lymphopenia, dysregulated innate immune cell recruitment and impaired type I immunity are markers for severe COVID-19. Another study on two cohorts of COVID-19 patients from two different locations, Hong Kong, China and Atlanta, USA, performed by Arunachalam et al. (5) showed that although plasma cytokine including IL-6, MCP-3 and CXCL10 were significantly up-regulated in COVID-19 patients compared to healthy control, only the levels of three inflammatory mediators, namely TNFSF14, EN-RAGE and OSM, were strongly correlated with disease severity. Furthermore, pDCs from COVID-19 patients were found to be impaired in production of IFNα, while monocytes and mDCs had reduced expression of CD86 and HLA-DR. Lymphopenia characterized by reduced CD4+ and CD8+ T cells in COVID-19 patients was also observed by Giamarellos-Bourboulis et al. (6) in their fifty-four COVID-19 patient cohort in Athens, Greece, and it was found to be more pronounced in patients with sudden respiratory failure (SRF). Majority of their patients with SRF had significant decrease of HLA-DR expression on CD14 monocytes accompanied by profound depletion of CD14+ lymphocytes, CD19+ lymphocytes and NK cells. The low HLA-DR expression on monocytes was likely due to increased plasma IL-6 and C-reactive protein levels in patients, as opposed to TNFa and IFNγ, and blocking IL-6 partially restored the expression of HLA-DR on monocytes. Therefore, impaired antigen presentation in severe COVID-19 patients might contribute to the development of lymphopenia. However, the monocytes remain potent for production of inflammatory cytokines. Together, these studies showed that host immune responses to SARS-CoV-2 infection may vary in different individuals, but dysregulated host immune response characterized by impaired antigen presentation and exaggerated inflammatory cytokine production by the innate immune system and lymphopenia are key factors contributing to severe COVID-19 disease. However, the underlying mechanisms requires further investigation.
Transcriptome analysis using next-generation RNA-sequencing (RNA-seq) enables researchers to determine the presence and quantity of RNA, i.e. gene expression, in biological samples on a genome-wide scale. In addition, alternatively spliced transcripts, mutations and post-transcriptional modifications in different experimental conditions can be identified. Therefore, comparison of the transcriptome using RNA-seq between experimental conditions can reveal a wealth of information about the underlying biological mechanisms. Several studies utilized RNA-seq to analyze COVID-19 patient samples to profile global gene expression changes triggered by SARS-CoV-2 infection, unravelling molecular insights underlying host response to the virus and pathogenesis of the disease. For example, by RNA sequencing profiling of nasopharyngeal swabs from 430 individuals infected with SARS-CoV-2 and 53 negative controls, Lieberman et al. (7) showed a strong antiviral response upon SARS-CoV-2 infection with upregulation of antiviral factors such as OAS1-3 and IFIT1-3. Blanco-Melo et al. (8) used cells and animal models of SARS-CoV-2 infection in joint with transcriptional and serum profiling of COVID-19 patients to reveal a unique immune response against COVID-19 and disease progression, characterized by low levels of type I and III interferons and elevated levels of chemokines and IL-6. By metatranscriptome sequencing of bronchoalveolar lavage fluid (BALF) of 8 COVID-19 patients and 20 healthy controls, Zhou et al. (9) showed the presence of hypercytokinemia and a robust upregulation of interferon stimulated genes in COVID-19 patients.
Various tools and approaches are available for RNA-seq data analysis, which could potentially lead to differential interpretation of the results. Some of the data analytic tools that are currently used in some of the RNA-seq studies were originally developed for microarray analyses, therefore may be outdated and unsuitable for analysis of RNA-seq datasets. For instance, quantile normalization used in some RNA-seq studies was designed to work on data where only a small proportion of genes is differentially expressed. When this assumption is unmet, results from analysis of the data using this method will not be accurate. Hence, more modern normalization methods using scaling factors would be preferred. Here, we re-analyzed four SARS-CoV-2 RNA-seq datasets using the same standardized approach and compared our results with results of the original studies, aiming to identify common immune features in response to SARS-CoV-2 infection. For one of the datasets, we found that the majority of reads in some samples are mapped to the virus, rather than to the human host, causing lower read counts for thousands of genes in the COVID-19 samples compared to the control samples. Furthermore, we compared the differentially expressed genes over datasets and report similarities in transcriptomic changes after SARS-CoV-2 infection. In addition, we compared the results from our re-analysis of RNA-seq datasets with results of scRNA-seq analysis of COVID-19 studies in the literature and found high concordance in differentially expressed genes between COVID-19 and control samples revealed by both technologies.
MATERIALS AND METHODS
RNA-sequencing data
RNA-seq data of 8 COVID-19 patients and 20 healthy control samples of bronchoalveolar lavage fluid (BALF) as reported by Zhou et al. (9) were downloaded from the Genome Sequencing Archive, National Genomics Data Center, China National Center for Bioinformation (https://bigd.big.ac.cn/gsa/, project code PRJCA002273). For re-analysis of the Lieberman et al. (7) data of nasopharyngeal swabs of 430 COVID-19 cases and 54 healthy controls, we used the raw counts table as deposited in NCBI Gene Expression Omnibus under accession number GSE152075. For the Blanco-Melo dataset (8), we used the raw counts table of RNA-seq data of one lung biopsy of a COVID-19 patient and 2 lung biopsies of healthy controls available in the Gene Expression Omnibus under accession number GSE147507. RNA-seq data of a mouse model of SARS-CoV-2 based on the adeno-associated virus–mediated expression of hACE2 of 2 infected mice and 2 mock infected mice as reported in Israelow et al. (10) were downloaded from NCBI GenBank, accession number PRJNA646535.
Two computational approaches for RNA-seq data analysis
For the Zhou and Israelow data, we used the Nextflow nf-core RNA-sequencing pipeline (11) to perform adapter trimming using Cutadapt (Martin et al., 2011), alignment of reads to the genome using STAR (12) and quantification of read counts per gene using featureCounts (13). Human reads were aligned to the GRCh38 genome and mouse reads were aligned to GRCm38. For the Lieberman and Blanco-Melo data we used the raw counts tables from NCBI Gene Expression Omnibus.
Once count tables per sample and per gene were obtained, we processed each of the four datasets in two ways. As a first method, also referred to as “our” method, we removed all genes having a total read count <100 over all samples. Next, we used DESeq2 (14) for normalization and identification of differentially expressed genes (DEGs). DEGs were defined as those genes having an adjusted p value <0.05 and an absolute log2(FoldChange)>=2. As second method, similar to Zhou et al.(9), we removed the genes that were present in less than 50% of samples in both groups (COVID-19 and healthy) and genes with an average count per million below five in both groups. Then gene counts were normalized using quantile normalization and the voom method. The limma package was used to identify DEGs, that were defined as having an adjusted p value <0.05 and an absolute log2(FoldChange)>=2 (ref).
Statistical analyses
Enrichment analysis of KEGG pathways was performed using the enrichKEGG function of the clusterProfiler package (15) of the R statistical software (R Core Team, 2020). Heatmaps were drawn using the R package pheatmap. The combinations of TMM and DESeq2 normalization with edgeR or DESeq2 for DEG identification were calculated using the TCC R package, using 1 and 3 iterations (16).
The composition of immune cells was inferred using CIBERSORT v1.06 (17) using 100 permutations and the original gene signature file LM22. The calculated proportions of 22 cell types were summarized into 9 major cell types. Cell composition analysis was performed on raw read counts per gene converted into the amount of transcripts per million.
RESULTS
Human COVID-19 RNA-seq data may contain large proportions of viral reads
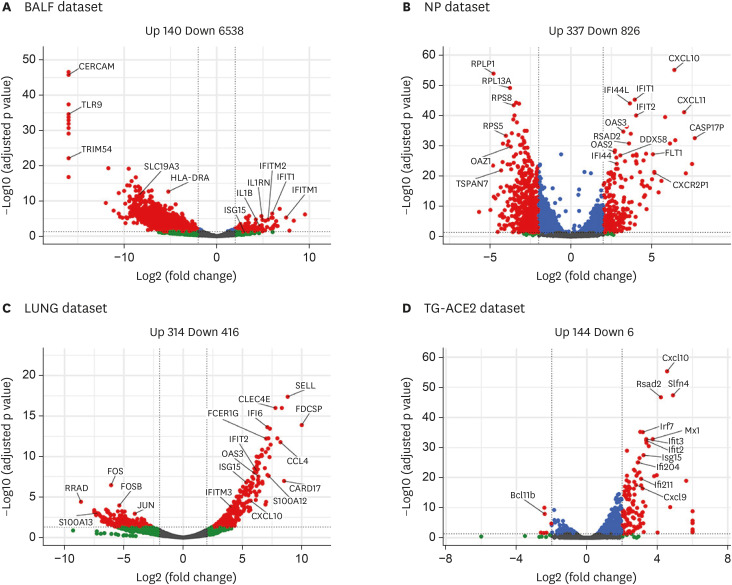
To increase our understanding of the host response after SARS-CoV-2 infection, we re-analyzed four RNA-seq datasets of COVID-19 samples versus healthy controls. We started our re-analysis with the dataset on bronchoalveolar lavage fluid (BALF) samples (BALF dataset) of 8 COVID-19 patients and 20 healthy controls (9). The original analysis identified 1014 up-regulated genes and 739 down-regulated genes in COVID-19 patient samples compared to healthy controls. However, our analysis on differential gene expression using DESeq2 identified 140 up-regulated genes and 6538 down-regulated genes (Fig. 1A, Supplementary Table 1). Compared to original analysis results, we found a much larger number of differentially expressed gene in COVID-19 samples (6,678 vs. 1,753) compared to healthy control samples, with about 7-fold reduction in the number of up-regulated genes and a 7.8-fold increase in the number of down-regulated genes (more than 6,000 genes).
Figure 1. Differentially expressed genes in re-analyzed four RNA-seq datasets of COVID-19 studies. (A) Regulated genes for Zhou et al. dataset on BALF of 8 COVID-19 patients and 20 healthy controls (BALF dataset). (B) Regulated genes for Lieberman et al. dataset on nasopharyngeal swabs of 430 COVID-19 cases and 54 healthy controls (NP dataset). (C) Regulated genes for Blanco-Melo et al. dataset on one lung biopsy of a COVID-19 patient and two lung biopsies of healthy controls (LUNG dataset). (D) Regulated genes for Israelow et al. dataset on a mouse model for SARS-CoV-2 of two virus-infected and two mock-infected mice (TG-ACE2 dataset).
To find out if the difference was caused by different analysis method used, we applied four combinations of TMM and DESeq2 normalization with edgeR and DESeq2 for finding DEGs using the TCC software package (TCC). Similar results were obtained, with a few hundred up-regulated and thousands of down-regulated genes (Table 1). During the analysis, we found that some of the COVID-19 samples in this dataset had very low amounts of human reads. Six out of the eight COVID-19 samples were found to contain only 2.0, 6.5, 13.4, 1.1 and 6.9 million human reads each, although they were sequenced at a total read depth of 20 million reads. In contrast, the 20 control samples have an average of 16.8 million human reads with a minimum of 11.6 human reads. It appeared that in these six COVID-19 samples, viral RNA takes up the remainder of the sequencing reads, and that only the human reads were deposited as supplementary data to the original paper (9). The large amount of down-regulated genes which we detected in COVID-19 samples using newer normalization techniques was not detected in the original paper where quantile normalization was used (9), as explained in Supplementary Fig. 1.
Table 1. Numbers of up- and down-regulated genes found using the BALF dataset and four combinations of TMM/DESeq2 normalization and edgeR/DESeq2 DEG identification and either 1 or 3 iterations.
| Normalization | Testing | Iteration | Total No. of gene | No. of DEG | No. of Upregulated gene | No. of downregulated gene |
|---|---|---|---|---|---|---|
| tmm | edger | 1 | 16,416 | 8,014 | 1,058 | 6,873 |
| tmm | deseq2 | 1 | 16,416 | 3,749 | 334 | 3,399 |
| deseq2 | edger | 1 | 16,416 | 10,166 | 810 | 9,295 |
| deseq2 | deseq2 | 1 | 16,416 | 5,875 | 136 | 5,685 |
| tmm | edger | 3 | 16,416 | 7,391 | 1,132 | 6,177 |
| tmm | deseq2 | 3 | 16,416 | 3,800 | 312 | 3,471 |
| deseq2 | edger | 3 | 16,416 | 4,057 | 1,790 | 2,231 |
| deseq2 | deseq2 | 3 | 16,416 | 4,441 | 243 | 4,146 |
Next, we re-analyzed another dataset from nasopharynx swab samples (NP dataset) of SARS-CoV-2 infected individuals and healthy controls (7). The original analysis of this dataset identified a total of 83 DEGs with 41 up-regulated and 42 down-regulated genes (P adj<0.1 and absolute log2 fold change >1) in the nasopharynx as a result of SARS-CoV-2 infection (7). Our analysis identified a total of 1,163 DEGs with 337 up-regulated and 826 down-regulated genes (p<0.05 and absolute log2 fold change ≥2) (Fig. 1B, Supplementary Table 2). Similarly, we identified a total of 730 DEGs with 314 up-regulated and 416 down-regulated genes in the dataset of post-mortem lung samples (LUNG dataset) of COVID-19 patients and biopsied healthy lung tissues from uninfected individuals (Fig. 1C, Supplementary Table 3), compared to ~2000 up- or down-regulated genes in the original analysis (8). In addition, re-analyzing a dataset of hACE2 transgenic mouse infected with SARS-CoV-2 viruses (TG-ACE2 dataset) identified a total of 150 DEGs with 144 up-regulated and 6 down-regulated genes (Fig. 1D, Supplementary Table 4). Table 2 gives an overview of the amount of DEGs identified using DESeq2 we used and the combination of voom and quantile normalization as used by Zhou et al. (9). Therefore, the analysis tools used by different studies may cause potential misinterpretation of RNA-Seq data in understanding host response to SARS-CoV-2 infection.
Table 2. Numbers of DEGs identified in the four SARS-CoV-2 RNA-seq datasets using two computational approaches.
| Dataset | Method | No. of upregulated genes | No. of downregulated genes | Total No. of regulated genes |
|---|---|---|---|---|
| Zhou | Voom and quantile normalization | 740 | 747 | 1,487 |
| DESeq2 | 140 | 6,538 | 6,678 | |
| Lieberman | Voom and quantile normalization | 97 | 66 | 163 |
| DESeq2 | 337 | 826 | 1,163 | |
| Blanco-Melo | Voom and quantile normalization | 28 | 138 | 166 |
| DESeq2 | 314 | 416 | 730 | |
| Israelow | Voom and quantile normalization | 2 | 55 | 57 |
| DESeq2 | 144 | 6 | 150 |
Functional analyses
To gain insight into biological pathways that are involved in immune response to SARS-CoV-2 infection, we performed gene enrichment analysis for KEGG pathways for the up- and down-regulated genes in each of the four datasets we re-analyzed. In the BALF dataset, the top enriched pathways include the “Ribosome,” “IL-17 signaling pathway,” “TNF signaling pathway” and “Non-alcoholic fatty liver disease” pathways (q values <0.05, Fig. 2A and C, Supplementary Table 5), whereas the original analysis identified “Interferon signaling,” “EIF2 signaling,” “Role of IL-17F in allergic airway inflammatory airway diseases,” and “Cholecystokinin/Gastrin-mediated signaling” as the top enriched pathways (7). Of note, “IL-17 signaling pathway” was also found to be up-regulated in our analysis of the LUNG dataset (Fig. 2A). Our analysis of the NP and LUNG datasets revealed commonly enriched pathways including “Viral protein interaction with cytokine and cytokine receptor,” “Cytokine-cytokine receptor interaction,” “Chemokine signaling pathway,” “NOD-like receptor signaling pathway,” and “Influenza A” in these two datasets. The “Influenza A” pathway is enriched in interferon-stimulated genes (ISGs) including OAS1, OAS2, OAS3, MX2, IFIH1, DDX58, CXCL10 and CCL2 (Fig. 2A, Supplementary Table 5). In the TG-ACE2 dataset, the top up-regulated pathways include “Viral protein interaction with cytokine and cytokine receptor,” “NOD-like receptor signaling pathway,” and “Chemokine signaling pathway” (q values <0.05) (Fig. 2C, Supplementary Table 5). Therefore, our analysis of both human and murine datasets showed that SARS-CoV-2 virus infection triggered strong cytokine and chemokine responses in hosts.
Figure 2. Enrichment analysis of KEGG pathways on regulated genes after SARS-CoV-2 infection. (A) Enrichment analysis on up-regulated genes for three human datasets including BAFL (A), NP (B) and LUNG (C). (B) Enrichment analysis on down-regulated genes for the same three human datasets. (C) Enrichment analysis for up-regulated genes on murine KEGG pathways of the TG-ACE2 dataset. (D) Enrichment analysis on down-regulated genes in the TG-ACE2 dataset.
Enrichment analysis of down-regulated genes showed that in both the BALF dataset and the TG-ACE2 dataset, “ECM-receptor interaction” is the top down-regulated pathway (q value <0.05) (Fig. 2B, Supplementary Table 6), whereas in the NP dataset, “Ribosome” is the pathway that was most down-regulated (q value <0.05) (Fig. 2D, Supplementary Table 6). In addition, top down-regulated pathways in the BALF dataset included “Dilated cardiomyopathy,” “Arrhythmogenic right ventricular cardiomyopathy,” “Hypertrophic cardiomyopathy,” “Focal adhesion,” and “Axon guidance” (q values <0.05). Interestingly, “Axon guidance” was the pathway that was commonly identified as one of the top down-regulated pathways in the BALF, LUNG and the TG-ACE2 datasets (q values <0.05) (Fig. 2B, D, Supplementary Table 6).
Differentially expressed chemokines, cytokines and ISGs
Dysregulated cytokine production is believed to play an important role in pathogenesis of severe COVID-19 disease (8). For the BALF dataset, the original analysis, which classified the 218 cytokine-related genes into seven categories including ILs, chemokines, IFNs, TNF, CSF, other cytokines and receptors, showed that chemokines were predominant among the up-regulated genes in SARS-CoV-2 samples among the 35 significant DEGs (9). Among the 14 significantly DE chemokines, CXCL17 was ranked the first in the up-regulated chemokines followed by CXCL8 and CCL2 (9). In our re-analysis, the number of total cytokine-related genes is 167 with 44 significant DEGs (Fig. 3A, Supplementary Table 1). Only 6 chemokine genes were found to be significantly differentially expressed between SARS-CoV-2 and healthy controls, with 4 genes, namely CXCL1, CXCL17, CXCL10 and CCL2, being up-regulated and two genes, CXCL16 and CCL22, being down-regulated (Fig. 3A, Supplementary Tables 5 and 6). For ILs, in contrast to the original analysis showing 3 cytokines including IL1RN, IL1B and IL4 being significantly up-regulated in SARS-CoV-2 samples, we identified 6 IL genes that are significantly differentially expressed. Among them, IL-1B was the only one that was significantly up-regulated. The other 5 ILs including IL-16, IL-13, IL-19, IL-36B and IL-24 were down-regulated. IL-1B is produced by activated macrophages and is an important mediator of the inflammatory response. IL-16 is a chemoattractant and modulator of T cell activation, whereas IL-19 polarizes macrophages into an anti-inflammatory phenotype, and IL-24 can, for example, dampen the first rounds of CD8+ T cell expansion. The most striking difference between the original analysis and our analysis was at the receptor category. Instead of 14 significantly differentially expressed receptors that were identified with 10 of them up-regulated and 4 of them down-regulated in the original analysis, we identified 24 significantly differentially expressed receptors with only 1 (CCR1) being up-regulated (Fig. 3A, Supplementary Table 5). The down-regulated receptor genes include IL2RA, IL2RB, IL7R, IL12RB1, IL12RB2, and IL21R which play important roles in T cell and NK cell growth, differentiation and function. In total, we observed up-regulation of only six genes, including IL1B, CXCL1, CXCL17, CXCL10, CCL2 and CCR1, and a larger number of other cytokines, chemokines and related genes being significantly down-regulated (Fig. 3A, Supplementary Table 6).
Figure 3. Significantly regulated cytokines and related genes after SARS-CoV-2 infection. (A) Heatmap showing log2 fold-changes of genes in seven groups. Column SARS2 shows the fold-changes calculated using DESeq2 using all samples and columns C1 until C8 show fold-changes for individual patients. (B) Regulated cytokines for the NP dataset on nasopharyngeal swabs of 430 COVID-19 cases and 54 healthy controls. (C) Regulated cytokines for the LUNG dataset on one lung biopsy of a COVID-19 patient and two lung biopsies of healthy controls. (D) Regulated cytokines for the TG-ACE2 dataset on a mouse model for SARS-CoV-2 on two virus-infected and two mock-infected mice. (E) Increased expression of S100A8 and S100A9 in the BALF and LUNG datasets in COVID-19 samples compared to their respective controls.
We also analyzed the differentially expressed cytokine-related genes in the other three datasets. We identified 37 significantly differentially expressed genes in the NP dataset, of which 17 being up-regulated (Fig. 3B, Supplementary Table 5). These 17 genes belong to three categories, with 1 IL gene (IL10), 7 chemokine genes with CXCL11 as the top up-regulated gene followed by CXCL10, CXCL9, CCL8, CCL2, CXCL13 and CXCL12, and 9 cytokine and chemokine receptor genes. Similarly, analysis of differentially expressed cytokine-related genes in the LUNG and TG-ACE2 datasets demonstrated that a small number of IL genes including IL1B which was significantly up-regulated in COVID-19 samples. In addition, the main group of significantly differentially expressed cytokine-related genes consists of chemokines (9 out of 37 in the LUNG dataset and 8 out of 30 in the TG-ACE2 dataset) followed by receptor genes (Fig. 3C and D, Supplementary Table 5). Importantly, IL1B is the only IL gene that was commonly up-regulated in the BALF, Lung and TG-ACE2 datasets, while CCL2 and CXCL10 were observed to be up-regulated in all the four datasets in our analysis (Fig. 3). Therefore, SARS-CoV-2 infection up-regulates a small number of cytokine and chemokine genes including IL1B, CCL2, CXCL10, and CXCL11 in both humans and mice. In addition, we observed increased expression of S100A8 and S100A9 in both BALF and LUNG datasets compared to their respective controls (Fig. 3E, Supplementary Tables 1 and 3).
Type I IFNs and ISGs are regarded as a key component of the innate immune response to virus infection (18). The original analysis of the BALF dataset identified 83 ISGs including IFIT1, IFITM1,2,3, IFIH1, TANK, IRF7 and STAT1 that were significantly elevated, suggesting a robust IFN response to SARS-CoV-2 infection (7). In our re-analysis, only 16 ISGs were identified to be significantly up-regulated, whereas 176 ISGs were down-regulated (Fig. 4A, Supplementary Table 1). Among the top up-regulated ISGs are IFITM1, IFIT1, IFITM2, and IFITM3 which are known to have direct antiviral activity, and CCL2, CXCL10 and CCR1 which regulate inflammation. These findings are inconsistent with the original analysis results. In our re-analysis of the NP, LUNG, and TG-ACE2 datasets, more ISGs were found to be significantly up-regulated compared to the results of the original analysis (Fig. 4B-D, Supplementary Tables 2 and 3 and 4). Tens of ISGs were up-regulated in these datasets, and there is a striking overlap of the strongly up-regulated ISGs between these three datasets (Fig. 4B-D). It was identified that CXCL10, IFIT1, IFIT2 and IFIT3 are in the top twenty up-regulated genes in all three datasets, whereas OAS2, OAS3, ISG15, IFI44 and CXCL9 are in two of the three top 20 lists. However, neither IFNβ nor IFNα genes are among the significantly upregulated genes. These results demonstrate that although elevated levels of IFNα and IFNβ may not be ready detectable, SARS-CoV-2 does trigger a robust IFN response.
Figure 4. Significantly regulated interferon-stimulated genes after SARS-CoV-2 infection. (A) Heatmap showing log2 fold-changes of ISGs in our global DESeq2 analysis (column SARS2) and for the individual patients C1 until C8. (B) Regulated ISGs for the NP dataset on nasopharyngeal swabs of 430 COVID-19 cases and 54 healthy controls. (C) Regulated ISGs for the LUNG dataset on one lung biopsy of a COVID-19 patient and two lung biopsies of healthy controls. (D) Regulated ISGs for the TG-ACE2 dataset on a mouse model for SARS-CoV-2 on two virus infected and two mock infected mice.
Cell composition analyses
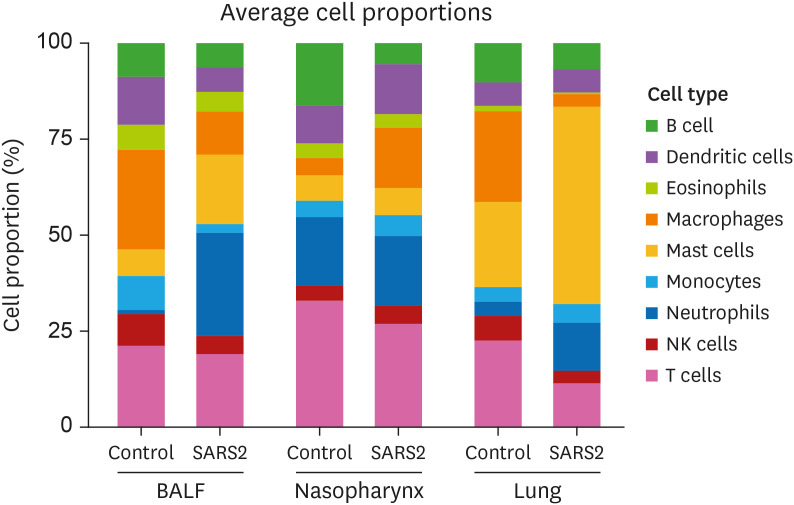
Whole transcriptome data can be used to access the abundances of immune cell types in a mixed cell population. To further understand the immune response to SARS-CoV-2, we used CIBERSORT, a program that combines support vector regression with prior knowledge of gene expression profiles from purified leukocyte subsets (19), to analyze the abundance of various immune cell types in the BALF, Nasopharynx and Lung datasets. Reduced frequency of B cells, T cells and NK cells are observed in all the three datasets, confirming that SARS-CoV-2 infection results in lymphopenia in humans (Fig. 5). In addition, increased frequency of neutrophils, especially in the BALF and the Lung datasets are identified. Interestingly, great increase of mast cell frequency is also observed in both the BALF and the Lung datasets. These findings demonstrate that in addition to lymphopenia, dysregulated innate immune cell infiltration, featured by abnormally abundant mast cells and neutrophils, contribute to the pathogenesis of COVID-19.
Figure 5. Immune cell composition analysis in healthy and COVID-19 patients. Proportion of nine major immune cell types calculated by CIBERSORT using the BALF, NP and LUNG datasets.
ns, not significant; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
DISCUSSION
After nearly two years since its emergence at the end of 2019, SARS-CoV-2 infection and COVID-19 disease remain a major health problem globally and effective treatment is yet to develop. Understanding the interaction between the virus and the host would provide opportunities for the development of therapeutic strategies against the disease. To better understand the host immune response after SARS-CoV-2 infection, we performed a re-analysis of four COVID-19 RNA-seq datasets on samples from nasopharynx, the lung and the BALF as well as a human ACE2 transgenic mice infected with SARS-CoV-2 viruses using standardized methods. We found that in the infected human BALF samples which was sequenced at a library depth of 20 million reads per sample, up to 95% of the reads can map to the virus, rather than to the human host. This means that the samples contain 18 or 19 million viral reads and only 1 or 2 million human reads. Subsequently, we found that this impacted the DEGs analysis. Due to the zero and low read counts in the COVID-19 samples, thousands of “down-regulated” genes were detected. However, due to the “undersequenced” of the host RNA since the majority of sequencing reads map to the virus in the COVID-19 samples, those genes probably only obtained fewer or no reads, rather than being down-regulated. However, this effect is masked by quantile normalization, since it equalizes the statistical distribution of the read counts of all samples, infected and not infected. Use of normalization methods that use one multiplication factor for all read counts per sample reveals that between 5,000 to 10,000 genes are detected as “down-regulated” in DEG analysis. It is impossible to discriminate between down-regulated genes and “undersequenced” genes in the COVID-19 group based on the RNA-seq data alone. One possible solution is to do a deeper sequencing for a fair comparison. Simultaneous sequencing of host and viral RNA can lead to large proportions of viral reads in sample, hence, substantially affects downstream analyses, such as detection of DEGs. For this BALF dataset, we believe that the majority of the ~6000 down-regulated genes are actually “undersequenced” genes in the COVID-19 group and that quantile normalization used in the original analysis masked this effect.
Our pathway analysis reflected a distinct difference between BALF dataset and NP/LUNG datasets. The discrepancy could firstly be due to significant levels of viral RNA sequenced from collected BALF samples, hence contributing to reduced number of human reads after clean-up. It could also be attributed to sample collection method, anatomical locations, and corresponding tissue residential immune cells.
NP and LUNG dataset samples were collected through nasopharyngeal swabs and lung biopsy respectively. The nasopharynx is an anatomical area of the upper respiratory tract, comprises of primarily epithelial cells and mucus-secreting goblet cells. It represents an immunologically distinct compartment, termed the nasopharynx-associated lymphoid tissue (NALT), enriched in specialised T cells and B cells together with tissue-specific resident macrophages and dendritic cells, act as an inductive site against inhaled pathogens (20). Within the lung, the tissue is populated with similar but specialised tissue-resident alveolar and interstitial macrophages, dendritic cells, natural killer cells, tissue memory T cells, and B cells (21). On the other hand, bronchoalveolar lavage describes a procedure by which sterile saline was introduced into the bronchoalveolar space and subsequently collected. Therefore, the main constituent of BALF recovered are of alveoli origin, the primary site of COVID-19 infection whereby not only tissue-residential immune cells but also infiltrating immune cells upon infection would be collected. Hence, nasopharynx swab and lung biopsy samples collected from COVID-19 patients and NP/LUNG datasets derived were inclined representation of the responses from tissue-resident immune cells, whereas BALF samples and corresponding datasets were reflective of the active pathways from both tissue-resident and infiltrating immune cells. Therefore, the means and site of sampling could have influenced the sequencing results and contributed to the differences observed between NP/LUNGS and BALF datasets.
Compiling to variations between datasets is potential disease stage-specific immune responses. Host response to SARS-CoV-2 can be categorised clinically into anti-viral stage, pulmonary stage and hyperinflammation stage, each with differing population of immune cells and outcome (22). In the early stage or onset of COVID-19, innate immune cell such as the tissue-resident alveolar macrophages and adaptive memory T cells will induce anti-viral responses. The patient will advance to pulmonary and hyperinflammation stage should the infection remains unresolved, experiencing cytokine storm and hyperinflammation aggravated by neutrophils activities (23). Consequently, samples collected at either stage of disease will have different immune cell population and therefore influence downstream pathway analysis to be representative of those adopted by the predominant immune cells.
KEGG pathway analysis of human and murine datasets revealed similar upregulation of anti-viral inflammatory pathways and downregulation of genes that correlate to SARS-CoV-2 infection progression. Our analysis identified upregulation of distinct genes that can be categorised into major pathways pertinent to COVID-19, namely ribosomal, IL-17 signalling, TNF signalling, cytokine/chemokine pathways, and Nod-like receptor (NLR) signalling. A clinical determinant of disease severity is the suppression of anti-viral responses, which was later identified as one of the immune evasion strategies of SARS-CoV-2. The viral non-structural proteins (NSPs) demonstrated binding affinity towards various ribosome RNAs, causing inhibition of global translation of host proteins, thereby antagonizing essential cellular processes to suppress host defence (24,25). This disrupts the function of ribosomes and the non-functional ribosomes could be degraded through ubiquitination (26). As SARS-CoV-2 persists in the host, cells at site of infection may divert resources to compensate for degraded ribosome by upregulation of associated genes, in attempt to restore translation capacity for the synthesis of anti-viral proteins.
Pathway analysis conducted has identified upregulation of several pathways, notably IL-17, TNF, and NLR associated genes and signalling. The roles of these upregulated pathways in COVID-19 infection have been investigated in multiple studies. Released from activated TH17 cells, IL-17 has been identified as a potent effector cytokine towards pulmonary hyperinflammation during COVID-infection. IL-17 exerts its effect through activation of multiple signalling pathways and transcription factors, particularly NFkB, promoting additional pro-inflammatory cytokines and chemokines of alveolar and epithelial origin, hence leading to potentially lethal cytokine storm (27,28). Akin to IL-17, significant expression of TNFα by infected tissues, innate immune cells, and TH17 detected in serum showed a strong positive correlation to COVID-19 severity and mortality (29,30). In the context of SARS-CoV-2 infection, TNF has been attributed with pro-inflammatory effector function, partially mediating cytokine shock syndrome through the Ikk, MAPK, and herein identified upregulated NLR associated inflammatory pathways (31,32,33). Recent investigation has also linked PANoptosis, an innate immune inflammatory programmed cell death pathway dependent on PANoptosome which triggers pyroptosis, apoptosis and necroptosis, to TNFα-IFNγ synergism, contributing significantly to the perpetuation of cytokine storm, tissue damage, and mortality (34). Therefore, pathways of interest highlighted in this study are in alignment with current understanding of COVID-19 pathology.
On the other spectrum, datasets analysis identified downregulation of pathways linked to ECM, cardiomyopathy, and focal adhesion. The downregulation of these pathways summates to the respiratory failure and lung injury in COVID-19 patients. ECM and focal adhesion pathways are responsible for the maintenance of extracellular microenvironment required for respiratory functionality (35). KEGG pathway analysis on samples collected from COVID-19 patients also revealed downregulation of ECM and focal adhesion pathways and lung tissue staining showed corresponding structural dysregulation, resulting in respiratory complications (36). Similarly, cardiomyopathy observed here is well documented amongst COVID-19 patients, accompanied by a recent longitudinal study on COVID-19 patients as having an increased risk of cardiovascular disease (37). With reference to the cardiomyopathy KEGG pathways highlighted, the molecular components downregulated are often structural proteins, concurring with clinical observation of cardiomyopathy experiences in COVID-19 patients (38,39).
Enrichment of SARS-CoV-2 RNA in immune cells including monocytes and macrophages in COVID-19 patients have been confirmed by single-cell transcriptome analysis studies. For instance, a single-cell atlas of multiple organs and tissues from donors who died of COVID-19 demonstrated that myeloid cells, particularly inflammatory monocytes and macrophages, were the cell categories most enriched for SARS-CoV-2 RNA (40). In addition, SARS-CoV-2 RNA reads were also detected in mast cells, B and plasma cells, and multiple other cell types who did not co-express ACE2 and TMPRSS2, or other hypothesized entry cofactors. Single-cell sequencing analysis of BALF and sputum samples from severe COVID-19 patients in the disease progression stage also detected SARS-CoV-2 RNA in a diverse set of immune cells, including neutrophils, macrophages, plasma B cells, T cells and NK cells (41). In addition, SARS-CoV-2 transcripts were detected in alveolar macrophage transcriptomes for 67% of BALF samples collected from 88 patients with severe COVID-19 disease (42). Subsequent single-cell RNA-seq analysis of BAL samples detected both positive- and negative-strand SARS-CoV-2 transcripts in migratory CCR7+ dendritic cells, monocyte-derived alveolar macrophages and tissue resident macrophages, which do not express ACE2. Therefore, the possibility of viral RNA reads in the data should be taken into consideration when choosing methods to analyze microarray data from virus infected samples such as those from COVID-19 patients.
Here, we applied a standardized method to re-analyze the four RNA-seq datasets of SARS-CoV-2 infection to probe immune action in comparison to their respective controls. Gene enrichment analysis showed that genes involved in “Cytokine-cytokine receptor interaction” and “Chemokine signaling pathway” were enriched in both Nasopharynx, Lung and TG-ACE2 datasets (Fig. 2). Analysis of DEGs further showed that up-regulation of cytokine/chemokine genes are a common feature of all four datasets. Excitingly, many of the most differentially regulated genes between COVID-19 samples and their controls in these RNA-seq datasets were also identified by single-cell RNA-seq studies in literature. For example, we identified that inflammatory chemokine CXCL9, CXCL10 and CXCL11 are strongly upregulated in both human and mouse SARS-COV-2 infected samples. Specifically, CXCL9 is upregulated in both the Nasopharynx and TG-ACE2 datasets, CXCL10 is upregulated in all the four datasets and CXCL11 is upregulated in the Nasopharynx, the Lung and the TG-ACE2 datasets (Fig. 3). This is in consistent with results from single-cell transcriptome studies, showing that CXCL10 and CXCL11 were specifically expressed in subtype of hyper-inflammatory macrophages (Macro_c2-CCL3L1) in the BALF (41), in SARS-CoV-2 RNA+ CD14high CD16high inflammatory monocytes in the lung (40), and in infected alveolar macrophages in the BALF (42), from either severe SARS-CoV-2 pneumonia or individuals who were died of COVID-19. These results demonstrate that increased expression of CXCL10 and CXCL11 likely by inflammatory monocytes/macrophages is a common feather of COVID-19 and is positively associated with severity of COVID-19 disease. The inflammatory chemokines CXCL9, CXCL10 and CXCL11 are predominantly induced by IFNγ (43,44), mainly secreted by monocytes/macrophages, endothelial cells, keratinocytes and fibroblasts. They act on the common receptor CXCR3 which is preferentially expressed on TH1 cells, CTLs, NK cells, NKT cells and monocytes/macrophages to regulate their migration. In addition, The CXCL9, 10, 11/CXCR3 signaling promotes the differentiation of CD4+ T cells into TH1 cells. Therefore, the higher expression of these chemokines suggests the potential to mount strong type I antiviral immune response to SARS-CoV-2 infection. However, in severe COVID-19, it goes derailed, leading to severe inflammation. Previous study has shown that CXCL10 expression was increased in the lung in viral acute respiratory distress syndrome (45). The increased levels of CXCL10 was originated to a large extent from infiltrated neutrophils who were also express CXCR3. The CXCL10-CXCR3 acted in an autocrine fashion on the oxidative burst and chemotaxis in the inflamed neutrophils, leading to fulminant pulmonary inflammation and lung injury (45). Therefore, it is possible that the increased expression of CXCL9, CXCL10 and CXCL11 in COVID-19 could lead to increased accumulation and activation of neutrophils in the lung which would contribute to the excessive inflammation and lung injury observed in severe COVID-19.
In our analysis, another commonly enriched gene group identified was ISGs, namely IFIT1/3, IFITM1 and IFITM2, which were strongly induced in SARS-CoV-2 infected samples in the four RNA-seq datasets (Fig. 4). The IFN family of cytokines have great importance in innate and adaptive antiviral immunity. Numerous studies have shown that impaired type I IFN response was associated with severe COVID-19. However, there are contradicting findings on IFN response in COVID-19 currently. For instance, a study including 50 COVID-19 patients with various disease severity showed that severe and critical patients were highly impaired in type I IFN response characterized by no IFNβ and low IFNα expression and activity (17). Consistently, a striking downregulation of ISGs including MX1, IFITM1 and IFIT2 was observed in whole blood cells from critical COVID-19 patients compared to moderate patients. Another study focusing on antigen-presenting cells (APCs) in PBMCs from COVID-19 patients showed that IFNα and IFNγ pathways were enriched in APCs from moderate but not these from severe COVID-19 patients (46). In addition, a decreased expression of ISGs including MX2, ISG15 and IFITMs was observed in APCs from severe COVID-19. Another scRNA-seq analysis of SARS-CoV-2 viral RNA positive immune cells including macrophages and neutrophils which were only found in BALF from severe COVID-19 had increased expression of ISGs compared to the same type virus-negative immune cells (41). Furthermore, scRNA-seq study of BAL samples from severe COVID-19 patients by Grant et al. (42) found enrichment of interferon-responsive genes in alveolar macrophages. However, no expression of type I IFNs was detected. Hence, the authors proposed that the interferon-response gene signature observed in alveolar macrophages may be due to IFNγ derived from activated T cells. Similarly, SARS-CoV-2 virus infection in rhesus macaques did not induce type I IFN expression in the lung (47). These observations suggesting that SARS-CoV-2 viruses have developed mechanisms to suppress the expression of IFNs and hence ISGs, which has also been associated with the disease severity. For example, Lei et al. (48) and Yuen et al. (49) have identified that SARS-CoV-2 nsp1, 12, 13, 15, and the M protein are potent inhibitors of the mitochondrial antiviral-signaling protein (MAVS) pathway which promotes IFNβ production. Moreover, Shin et al. (50) have reported that SARS-CoV-2 also uses its papain-like protease (PLpro) to cleave ISG15, which is a potent antiviral gene induced by IFNβ, representing another level of immune-suppressing strategy employed by the virus. On the host side, functional mutations in the type I IFN pathway was associated with severe COVID-19 (51). In addition, 10% of 987 life-threatening COVID-19 had neutralizing antibody against type I IFNs. Therefore, impaired type I IFN response, possibly due to viral evasion strategies and/or host genetic mutations contributes to the pathogenesis of severe COVID-19. However, certainly ISGs may be enriched in COVID-19 samples possibly due to high IFNγ.
Across the 4 datasets we analyzed, the number of differentially expressed ILs was much smaller than that of chemokines (Fig. 3). For instance, IL-1B is the only significantly upregulated IL in the BALF dataset. It is also found to be significantly upregulated in the Lung and TG-ACE2 datasets. Interestingly, scRNA-seq studies demonstrated that the hyper-inflammatory Macro_c2-CCL3L1 cells in both PBMCs and the BALF or the infected alveolar macrophages in the lung uniquely expressed high levels of IL1β (41,42). In addition, scRNA-seq analysis of blood APCs from COVID-19 patients and healthy controls showed that IL-1B was the top upregulated cytokine gene in APCs from severe COVID-19 patients compared to that from mild COVID-19 or healthy control (46). Furthermore, severe COVID-19 patients had higher levels of IL-1β, in addition to TNF and IL6, in their plasma (41). These studies demonstrated that high production of IL-1β is associated with severe COVID-19. IL-1β is considered as a master regulator of inflammation, having diverse physiological functions and pathological significances (52). IL-1β may contribute to acute inflammation in patients by stimulating the monocytes to differentiate into M1-like macrophages and conventional dendritic cells to produce other inflammatory cytokines including IL-6, as well as sustaining the proliferation and promoting the differentiation of activated B lymphocytes into plasma cells (53,54). IL-1β could also regulate both antigen-specific CD4+ and antigen-specific CD8+ T cells to enhance their survivability and proliferation (55). The IL-1β stimulated CD4+ cells are frequently IL-17-producing or IFNγ-producing cells, which corresponds well with the upregulated “IL-17 signaling pathway” in the functional analyses in both the BALF and Lung datasets, and the elevated IFNγ commonly reported in COVID-19 patients. Interestingly, although IL-1β is found to promote the expansion of T cells, cell composition analysis showed that T cells in COVID-19 patients were found to be significantly decreased compared to healthy control. Although the mechanism underlying the decrease of T cells in some patients is unclear, it is possible that excessive stimulation of T cell might lead to T cell exhaustion, resulting in low T cell count (56). Moreover, IL-1β has also been found to contribute to the recruitment of myeloid cells, either directly or indirectly (57,58,59). Directly, IL-1β recruits macrophages to the site of infection, which can cause tissue damage through classical activation by IFNγ and TNFα, both of which has been identified to be upregulated in COVID-19 patients (57,60). Moreover, IL-1β also recruits neutrophils by stimulating the endothelial cells to express surface proteins such as members of the integrin family, P-selectin, and E-selectin, which promote the adhesion of neutrophils via the rolling mechanism (61,62). Alternatively, IL-1β can also induce the expression of CXCL1, which subsequently recruits neutrophils (58). Neutrophils are the first immune cells to be recruited to the site of inflammation and dysregulated neutrophil recruitment and activation could lead to damage to the lungs during a pulmonary infection (63). Importantly, IL-1 receptor blockade in severe COVID-19 patients with anakinra, an antagonist of IL-1 receptor, resulted in a rapid fall in fever and CRP, reduced oxygen requirements and decreased deaths (64); (65), supporting the role of IL-1 in the pathogenesis of severe COVID-19.
The S100 protein family members S100A8 and S100A9 were found to be specifically upregulated in the lungs by coronaviruses, but not other viruses including influenza A virus, encephalomyocarditis virus and herpes simplex virus 1 in mice (47). In addition, massive release of S100A8/S100A9 was associated with severe disease among COVID-19 patients (66,67). Furthermore, single-cell transcriptome analysis of samples from SARS-CoV-2 infected individuals with various disease severity showed significant upregulation of S100A8/S100A9 in almost all cell clusters of both BALF and PBMCs from severe COVID-19 patients in the disease progression stage (41). In line with these findings, we also observed significant upregulation of both S100A8 and S100A9 in the BALF and LUNG datasets (Fig. 3E, Supplementary Tables 1 and 3). S100A8 and S100A9 are two Ca2+ binding proteins which are constitutively expressed in neutrophils and monocytes (68). These two proteins comprise approximately 45% of the cytoplasmic proteins in neutrophils. The expression of S100A8/S100A9 is intensely upregulated by many inflammatory processes including infection and stress, and these two molecules in turn magnify the inflammatory response by stimulating leukocyte recruitment and accelerating release of more cytokines from neutrophils and monocytes/macrophages likely through TLR4-MyD88 signaling. Therefore, S100A8/A9 are considered as hallmarks of numerous inflammation-associated pathological conditions such as rheumatoid arthritis, systemic lupus erythematosus, cystic fibrosis, chronic inflammatory bowel diseases, and Psoriasis (69).
Interestingly, S100A8/A9 could stimulates both the transcription and release of IL-1β in macrophages and neutrophils through TLR4-MyD88 and NLRP3 inflammasome pathways respectively (68). Therefore, the increase of both S100A8/S100A9 and IL-1β in severe COVID-19 suggests dysregulation of neutrophil recruitment and function, which likely contribute to the pathogenesis of the disease. Indeed, accumulating evidences demonstrate that aberrant neutrophil accumulation and function in the lung is a feature of severe COVID-19. For instance, Banerjee et al. (24) showed that both S100A8 and neutrophil marker genes were upregulated in post-mortem lung samples from COVID-19 patients, which was in line with robust upregulation of S100A8 expression and neutrophil chemotaxis in both rhesus macaques and human ACE2 (hACE2) transgenic mice infected with SARS-CoV-2 (47). The high expression of S100A8 and neutrophil chemotaxis were associated with the accumulation of a population of aberrant neutrophils with surface characteristics of CD45+CD11b+Ly6Gvariable. Treatment of SARS-CoV-2 infected hACE2 transgenic mice with Paquinimod, a S100A8/A9 inhibitor, resulted in suppressed accumulation of aberrant neutrophils and a remarkably 100% survival of the mice. Together, these studies indicated that S100A9/S100A9 are markers of aberrant accumulation and dysregulated function of neutrophils in the lung, which contribute to the development of severe COVID-19. This is in line with our cell component analysis of the four datasets, showing that an increase in neutrophil proportion in both the BALF and LUNG datasets (8,9). Interestingly, stronger upregulation of S100A12 than S100A8/S100A9 was observed in the LUNG dataset (Fig. 1C, Supplementary Table 3). S100A12 is primarily expressed by neutrophils and serum levels of this protein were found to be elevated in numerous inflammatory diseases including glomerulonephritis, inflammatory bowel disease, rheumatoid and psoriatic arthritis (70). When secreted extracellularly, it mediates chemotactic activity, induces the of production of inflammatory cytokines including TNFa and IL-1β and provokes mast cell degranulation (71). Therefore, the increased expression of S100A8/S100A9 and S100A12 suggests the aberrant neutrophil accumulation and function in triggered by SARS-CoV-2 infection.
Interestingly, our cell component analysis also identified increased proportion of mast cells in severe COVID-19 patients compared to healthy control, especially in the BALF and LUNG datasets (Fig. 5). Similarly, a scRNA-seq study demonstrated that mast cells were enriched in BALF samples from severe COVID-19 patients at disease progression stage (41). Mast cells are derived from hematopoietic progenitor cells in the bone marrow, and migrate and complete their maturation in most vascularized tissues (72). They also reside in certain body cavities including the peritoneum and the pulmonary cavities. IgE-mediated mast cell activation is a key reaction in allergy and anaphylaxis. In addition, they can be activated by numerous stimuli including components of complement activation, agonists of TLRs, and a variety of endogenous peptides including vasoactive intestinal peptide and substance P. Upon activation, mast cells release various biologically active compounds including biogenic amines (e.g. histamine), proteases (e.g. trypase and chymase), arachidonic acid products (e.g. prostaglandins and leukotrienes), growth factors, and cytokines and chemokines, thereby participating in many types of innate and adaptive immune responses (73). Mast cells are an important source of proinflammatory cytokines including TNFα and IL-1β. For instance, mast cell-derived IL-1 including IL1α and IL1β in synovial joint contributes to the initiation of autoimmune inflammation in autoantibody-mediated arthritis (74). The production of IL1 by mast cells could increases the number of infiltrating neutrophils with their release of proteases and rising inflammation (73). In addition, it has also been shown that stabilizing mast cells prevents apoptosis and sepsis (75), both of which exacerbate COVID-19 symptoms.23 In COVID-19, multiple retrospective and observational clinic studies suggest that the use of famotidine, a histamine H2 receptor antagonist, was associated with lower incidence of adverse clinical outcomes in hospitalized patients with COVID-19 and could significantly reduce the fatality (76,77). Together, these studies suggest that dysregulated mast cell activation is part of dysregulated innate immune response to SARS-CoV-2 infection in the pathogenesis of severe COVID-19.
One striking feature we observed is that more genes were downregulated compared to genes that were upregulated in human COVID-19 datasets in our reanalysis (Fig. 1A-C). Cytokine receptor genes are a major group of genes that were downregulated. For example, in the BALF dataset, both IL2RA (also known as CD25) and IL2RB are among the top downregulated genes, consistent with reduced T cells in severe COVID-19. In addition, IL2RA and IL2RB together with IL2RG, constitute a high-affinity receptor for IL-2, which plays an essential role in maintaining the proper protective suppressor function of CD4+ regulatory T (Treg) cells (78). IL21R was also among the top downregulated genes in the BALF dataset. IL-21 not only promotes the proliferation of T cells and the proliferation and maturation of B cells, but also plays a role in the proliferation and maturation of NK cells from bone marrow (27). Receptor for IL-27 (Il27ra) was found to be downregulated in the BALF, the LUNG and the TG-ACE2 datasets (Fig. 4). IL-27 not only promotes TH1 differentiation and IFNγ production in cooperation with IL-12 and increases CTL generation, but also inhibits TH2 and TH17 differentiation (79,80). In addition, it promotes the growth and survival of Tregs. Therefore, the downregulation of these cytokine receptors is in line with impaired adaptive immunity against SARS-CoV-2 viruses, leading to uncontrolled inflammation and severe outcome.
In summary, re-analysis of four COVID-19 RNA-seq datasets using the same standardized method revealed common immune features of COVID-19, which include increased expression of CCL2, CXCL10, and CXCL11 chemokines, heightened expression of IL-1β and S100A8/A9, reduced expression of receptors for chemokines/cytokines that are important for T cell growth, differentiation and function which is in line with lymphopenia of COVID-19 patients especially those with severe disease. In addition, overabundance of neutrophils and mast cells in the lung is part of dysregulated innate immune responses in COVID-19. These features are verified by latter scRNA-sequencing studies in the field. This study demonstrates that RNA-seq technology, despite its limitations such as being unable to analyze immune features at single cell level, is powerful in revealing specific biological features caused by microbial infection or in other pathological conditions, providing with the correct analysis tools which can be commonly adopted and close collaborations within the scientific communicate.
ACKNOWLEDGEMENTS
This work was supported by grants from the Singapore National Medical Research Council (NMRC/OFIRG/0059/2017 to Y. Z.) and the National University Health System (NUHSRO/2020/113/T1/Seed-Mar/09 to Y. Z.).
Abbreviations
- APC
antigen-presenting cell
- BALF
bronchoalveolar lavage fluid
- COVID-19
Coronavirus disease 2019
- DEG
differentially expressed gene
- ISG
interferon-stimulated gene
- NSP
non-structural protein
- RNA-seq
RNA-sequencing
- SARS-CoV-2
severe acute respiratory syndrome-coronavirus-2
Footnotes
- Conceptualization: Zhang Y, Alberts R.
- Data curation: Zhang Y, Alberts R, Meng QF, Rao L.
- Formal analysis: Zhang Y, Alberts R, Meng QF, He S, Rao L.
- Funding acquisition: Zhang Y.
- Investigation: Zhang Y, Alberts R.
- Methodology: Alberts R, He S.
- Project administration: Zhang Y.
- Resources: Zhang Y.
- Software: Alberts R.
- Supervision: Zhang Y, Rao L.
- Validation: Alberts R, Liu X.
- Writing - original draft: Zhang Y, Alberts R, Chan SC, He S.
- Writing - review & editing: Zhang Y, Alberts R, Chan SC, Rao L, Liu X.
SUPPLEMENTARY MATERIALS
DEGs_dataset_1
DEGs_dataset_2
DEGs_dataset_3
DEGs_dataset_4
DEGs_dataset_5
DEGs_dataset_6
Quantile normalization with and without meeting the assumption. Quantile normalization should be used when only a small proportion of genes, i.e., a few tens or hundreds out of around 20,000 human genes in total, are differentially expressed. (A) Log2 gene expression values for three samples in condition A and three samples in condition B are shown before and after quantile normalization. Quantile normalization equalizes the statistical distribution of all samples in the experiment (right panel). The yellow line represents the expression of a differentially expressed gene. Global differences between samples are removed by quantile normalization, while the yellow example gene in the figure is still identified as differentially expressed after normalization. (B) Here a large proportion of around 20,000 genes have a higher expression value in condition B compared to condition A. After quantile normalization, the statistical distribution of all samples is equalized. Hence, the majority of differentially expressed genes before normalization will not be seen after normalization. The yellow example genes are not differentially expressed in the right panel, whereas they are in the left panel.
References
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silva TF, Tomiotto-Pellissier F, Sanfelice RA, Gonçalves MD, da Silva Bortoleti BT, Detoni MB, Rodrigues AC, Carloto AC, Concato VM, Siqueira ED, et al. A 21st century evil: immunopathology and new therapies of COVID-19. Front Immunol. 2020;11:562264. doi: 10.3389/fimmu.2020.562264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucas C, Wong P, Klein J, Castro TB, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arunachalam PS, Wimmers F, Mok CK, Perera RA, Scott M, Hagan T, Sigal N, Feng Y, Bristow L, Tak-Yin Tsang O, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giamarellos-Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, Antonakos N, Damoraki G, Gkavogianni T, Adami ME, Katsaounou P, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lieberman NA, Peddu V, Xie H, Shrestha L, Huang ML, Mears MC, Cajimat MN, Bente DA, Shi PY, Bovier F, et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020;18:e3000849. doi: 10.1371/journal.pbio.3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R, Jordan TX, Oishi K, Panis M, Sachs D, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, Guo L, Yang J, Wang C, Jiang S, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, Alfajaro MM, Wei J, Dong H, Homer RJ, et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med. 2020;217:e20201241. doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, Garcia MU, Di Tommaso P, Nahnsen S. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020;38:276–278. doi: 10.1038/s41587-020-0439-x. [DOI] [PubMed] [Google Scholar]
- 12.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 14.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun J, Nishiyama T, Shimizu K, Kadota K. TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics. 2013;14:219. doi: 10.1186/1471-2105-14-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Péré H, Charbit B, Bondet V, Chenevier-Gobeaux C, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tacchi L, Musharrafieh R, Larragoite ET, Crossey K, Erhardt EB, Martin SA, LaPatra SE, Salinas I. Nasal immunity is an ancient arm of the mucosal immune system of vertebrates. Nat Commun. 2014;5:5205. doi: 10.1038/ncomms6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ardain A, Marakalala MJ, Leslie A. Tissue-resident innate immunity in the lung. Immunology. 2020;159:245–256. doi: 10.1111/imm.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X, Bucci E, Piacentini M, Ippolito G, Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinha S, Rosin NL, Arora R, Labit E, Jaffer A, Cao L, Farias R, Nguyen AP, de Almeida LG, Dufour A, et al. Dexamethasone modulates immature neutrophils and interferon programming in severe COVID-19. Nat Med. 2022;28:201–211. doi: 10.1038/s41591-021-01576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee AK, Blanco MR, Bruce EA, Honson DD, Chen LM, Chow A, Bhat P, Ollikainen N, Quinodoz SA, Loney C, et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–1339.e21. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut M, Thiel V, Mühlemann O, Ban N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 26.Wang JY, Zhang W, Roehrl MW, Roehrl VB, Roehrl MH. An autoantigen profile of human A549 lung cells reveals viral and host etiologic molecular attributes of autoimmunity in COVID-19. J Autoimmun. 2021;120:102644. doi: 10.1016/j.jaut.2021.102644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibabaw T. Inflammatory cytokine: IL-17A signaling pathway in patients present with COVID-19 and current treatment strategy. J Inflamm Res. 2020;13:673–680. doi: 10.2147/JIR.S278335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maione F, Casillo GM, Raucci F, Salvatore C, Ambrosini G, Costa L, Scarpa R, Caso F, Bucci M. Interleukin-17A (IL-17A): a silent amplifier of COVID-19. Biomed Pharmacother. 2021;142:111980. doi: 10.1016/j.biopha.2021.111980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Del Valle DM, Kim-Schulze S, Huang HH, Beckmann ND, Nirenberg S, Wang B, Lavin Y, Swartz TH, Madduri D, Stock A, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mortaz E, Tabarsi P, Jamaati H, Dalil Roofchayee N, Dezfuli NK, Hashemian SM, Moniri A, Marjani M, Malekmohammad M, Mansouri D, et al. Increased serum levels of soluble TNF-α receptor is associated with ICU mortality in COVID-19 patients. Front Immunol. 2021;12:592727. doi: 10.3389/fimmu.2021.592727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merad M, Martin JC. Author correction: pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:448. doi: 10.1038/s41577-020-0353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kox M, Waalders NJ, Kooistra EJ, Gerretsen J, Pickkers P. Cytokine levels in critically ill patients with COVID-19 and other conditions. JAMA. 2020;324:1565. doi: 10.1001/jama.2020.17052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah A. Novel coronavirus-induced NLRP3 inflammasome activation: a potential drug target in the treatment of COVID-19. Front Immunol. 2020;11:1021. doi: 10.3389/fimmu.2020.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karki R, Sharma BR, Tuladhar S, Williams EP, Zalduondo L, Samir P, Zheng M, Sundaram B, Banoth B, Malireddi RK, et al. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184:149–168.e17. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgstaller G, Oehrle B, Gerckens M, White ES, Schiller HB, Eickelberg O. The instructive extracellular matrix of the lung: basic composition and alterations in chronic lung disease. Eur Respir J. 2017;50:1601805. doi: 10.1183/13993003.01805-2016. [DOI] [PubMed] [Google Scholar]
- 36.Leng L, Cao R, Ma J, Mou D, Zhu Y, Li W, Lv L, Gao D, Zhang S, Gong F, et al. Pathological features of COVID-19-associated lung injury: a preliminary proteomics report based on clinical samples. Signal Transduct Target Ther. 2020;5:240. doi: 10.1038/s41392-020-00355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie Y, Xu E, Bowe B, Al-Aly Z. Long-term cardiovascular outcomes of COVID-19. Nat Med. 2022;28:583–590. doi: 10.1038/s41591-022-01689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahranavard M, Akhavan Rezayat A, Zamiri Bidary M, Omranzadeh A, Rohani F, Hamidi Farahani R, Hazrati E, Mousavi SH, Afshar Ardalan M, Soleiman-Meigooni S, et al. Cardiac complications in COVID-19: a systematic review and meta-analysis. Arch Iran Med. 2021;24:152–163. doi: 10.34172/aim.2021.24. [DOI] [PubMed] [Google Scholar]
- 39.Ramadan MS, Bertolino L, Marrazzo T, Florio MT, Durante-Mangoni E, Durante-Mangoni E, Iossa D, Bertolino L, Ursi MP, D’Amico F, et al. Cardiac complications during the active phase of COVID-19: review of the current evidence. Intern Emerg Med. 2021;16:2051–2061. doi: 10.1007/s11739-021-02763-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delorey TM, Ziegler CG, Heimberg G, Normand R, Yang Y, Segerstolpe Å, Abbondanza D, Fleming SJ, Subramanian A, Montoro DT, et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595:107–113. doi: 10.1038/s41586-021-03570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren X, Wen W, Fan X, Hou W, Su B, Cai P, Li J, Liu Y, Tang F, Zhang F, et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184:1895–1913.e19. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grant RA, Morales-Nebreda L, Markov NS, Swaminathan S, Querrey M, Guzman ER, Abbott DA, Donnelly HK, Donayre A, Goldberg IA, et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S, McSkane M, Baba H, Lenz HJ. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - a target for novel cancer therapy. Cancer Treat Rev. 2018;63:40–47. doi: 10.1016/j.ctrv.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metzemaekers M, Vanheule V, Janssens R, Struyf S, Proost P. Overview of the mechanisms that may contribute to the non-redundant activities of interferon-inducible CXC chemokine receptor 3 ligands. Front Immunol. 2018;8:1970. doi: 10.3389/fimmu.2017.01970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ichikawa A, Kuba K, Morita M, Chida S, Tezuka H, Hara H, Sasaki T, Ohteki T, Ranieri VM, dos Santos CC, et al. CXCL10-CXCR3 enhances the development of neutrophil-mediated fulminant lung injury of viral and nonviral origin. Am J Respir Crit Care Med. 2013;187:65–77. doi: 10.1164/rccm.201203-0508OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saichi M, Ladjemi MZ, Korniotis S, Rousseau C, Ait Hamou Z, Massenet-Regad L, Amblard E, Noel F, Marie Y, Bouteiller D, et al. Single-cell RNA sequencing of blood antigen-presenting cells in severe COVID-19 reveals multi-process defects in antiviral immunity. Nat Cell Biol. 2021;23:538–551. doi: 10.1038/s41556-021-00681-2. [DOI] [PubMed] [Google Scholar]
- 47.Guo Q, Zhao Y, Li J, Liu J, Yang X, Guo X, Kuang M, Xia H, Zhang Z, Cao L, et al. Induction of alarmin S100A8/A9 mediates activation of aberrant neutrophils in the pathogenesis of COVID-19. Cell Host Microbe. 2021;29:222–235.e4. doi: 10.1016/j.chom.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, Wang C, Wang Y, Li L, Ren L, et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuen CK, Lam JY, Wong WM, Mak LF, Wang X, Chu H, Cai JP, Jin DY, To KK, Chan JF, et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, Schulz L, Widera M, Mehdipour AR, Tascher G, et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carvalho T. Interferon linked to COVID-19 severity. Nat Med. 2020;26:1806. doi: 10.1038/s41591-020-01164-x. [DOI] [PubMed] [Google Scholar]
- 52.Dinarello CA. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev. 2018;281:8–27. doi: 10.1111/imr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lipsky PE, Thompson PA, Rosenwasser LJ, Dinarello CA. The role of interleukin 1 in human B cell activation: inhibition of B cell proliferation and the generation of immunoglobulin-secreting cells by an antibody against human leukocytic pyrogen. J Immunol. 1983;130:2708–2714. [PubMed] [Google Scholar]
- 54.Schenk M, Fabri M, Krutzik SR, Lee DJ, Vu DM, Sieling PA, Montoya D, Liu PT, Modlin RL. Interleukin-1β triggers the differentiation of macrophages with enhanced capacity to present mycobacterial antigen to T cells. Immunology. 2014;141:174–180. doi: 10.1111/imm.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ben-Sasson SZ, Hu-Li J, Quiel J, Cauchetaux S, Ratner M, Shapira I, Dinarello CA, Paul WE. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc Natl Acad Sci U S A. 2009;106:7119–7124. doi: 10.1073/pnas.0902745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, Chen L, Li M, Liu Y, Wang G, et al. Reduction and functional exhaustion of t cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rider P, Carmi Y, Guttman O, Braiman A, Cohen I, Voronov E, White MR, Dinarello CA, Apte RN. IL-1α and IL-1β recruit different myeloid cells and promote different stages of sterile inflammation. J Immunol. 2011;187:4835–4843. doi: 10.4049/jimmunol.1102048. [DOI] [PubMed] [Google Scholar]
- 58.Biondo C, Mancuso G, Midiri A, Signorino G, Domina M, Lanza Cariccio V, Mohammadi N, Venza M, Venza I, Teti G, et al. The interleukin-1β/CXCL1/2/neutrophil axis mediates host protection against group B streptococcal infection. Infect Immun. 2014;82:4508–4517. doi: 10.1128/IAI.02104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 60.Laskin DL, Sunil VR, Gardner CR, Laskin JD. Macrophages and tissue injury: agents of defense or destruction? Annu Rev Pharmacol Toxicol. 2011;51:267–288. doi: 10.1146/annurev.pharmtox.010909.105812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bruehl RE, Moore KL, Lorant DE, Borregaard N, Zimmerman GA, McEver RP, Bainton DF. Leukocyte activation induces surface redistribution of P-selectin glycoprotein ligand-1. J Leukoc Biol. 1997;61:489–499. doi: 10.1002/jlb.61.4.489. [DOI] [PubMed] [Google Scholar]
- 62.Buscher K, Riese SB, Shakibaei M, Reich C, Dernedde J, Tauber R, Ley K. The transmembrane domains of L-selectin and CD44 regulate receptor cell surface positioning and leukocyte adhesion under flow. J Biol Chem. 2010;285:13490–13497. doi: 10.1074/jbc.M110.102640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laforge M, Elbim C, Frère C, Hémadi M, Massaad C, Nuss P, Benoliel JJ, Becker C. Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat Rev Immunol. 2020;20:515–516. doi: 10.1038/s41577-020-0407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cauchois R, Koubi M, Delarbre D, Manet C, Carvelli J, Blasco VB, Jean R, Fouche L, Bornet C, Pauly V, et al. Early IL-1 receptor blockade in severe inflammatory respiratory failure complicating COVID-19. Proc Natl Acad Sci U S A. 2020;117:18951–18953. doi: 10.1073/pnas.2009017117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cavalli G, De Luca G, Campochiaro C, Della-Torre E, Ripa M, Canetti D, Oltolini C, Castiglioni B, Tassan Din C, Boffini N, et al. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2020;2:e325–e331. doi: 10.1016/S2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silvin A, Chapuis N, Dunsmore G, Goubet AG, Dubuisson A, Derosa L, Almire C, Hénon C, Kosmider O, Droin N, et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi H, Zuo Y, Yalavarthi S, Gockman K, Zuo M, Madison JA, Blair C, Woodward W, Lezak SP, Lugogo NL, et al. Neutrophil calprotectin identifies severe pulmonary disease in COVID-19. J Leukoc Biol. 2021;109:67–72. doi: 10.1002/JLB.3COVCRA0720-359R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J. S100A8/A9 in inflammation. Front Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gebhardt C, Németh J, Angel P, Hess J. S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol. 2006;72:1622–1631. doi: 10.1016/j.bcp.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 70.Meijer B, Gearry RB, Day AS. The role of S100A12 as a systemic marker of inflammation. Int J Inflamm. 2012;2012:907078. doi: 10.1155/2012/907078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang Z, Yan WX, Cai H, Tedla N, Armishaw C, Di Girolamo N, Wang HW, Hampartzoumian T, Simpson JL, Gibson PG, et al. S100A12 provokes mast cell activation: a potential amplification pathway in asthma and innate immunity. J Allergy Clin Immunol. 2007;119:106–114. doi: 10.1016/j.jaci.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 72.Galli SJ, Gaudenzio N, Tsai M. Mast cells in inflammation and disease: recent progress and ongoing concerns. Annu Rev Immunol. 2020;38:49–77. doi: 10.1146/annurev-immunol-071719-094903. [DOI] [PubMed] [Google Scholar]
- 73.Conti P, Pregliasco FE, Bellomo RG, Gallenga CE, Caraffa A, Kritas SK, Lauritano D, Ronconi G. Mast cell cytokines IL-1, IL-33, and IL-36 mediate skin inflammation in psoriasis: a novel therapeutic approach with the anti-inflammatory cytokines IL-37, IL-38, and IL-1Ra. Int J Mol Sci. 2021;22:8076. doi: 10.3390/ijms22158076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nigrovic PA, Lee DM. Synovial mast cells: role in acute and chronic arthritis. Immunol Rev. 2007;217:19–37. doi: 10.1111/j.1600-065X.2007.00506.x. [DOI] [PubMed] [Google Scholar]
- 75.Ramos L, Peña G, Cai B, Deitch EA, Ulloa L. Mast cell stabilization improves survival by preventing apoptosis in sepsis. J Immunol. 2010;185:709–716. doi: 10.4049/jimmunol.1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Freedberg DE, Conigliaro J, Wang TC, Tracey KJ, Callahan MV, Abrams JA, Sobieszczyk ME, Markowitz DD, Gupta A, O’Donnell MR, et al. Famotidine use is associated with improved clinical outcomes in hospitalized COVID-19 patients: a propensity score matched retrospective cohort study. Gastroenterology. 2020;159:1129–1131.e3. doi: 10.1053/j.gastro.2020.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mura C, Preissner S, Nahles S, Heiland M, Bourne PE, Preissner R. Real-world evidence for improved outcomes with histamine antagonists and aspirin in 22,560 COVID-19 patients. Signal Transduct Target Ther. 2021;6:267. doi: 10.1038/s41392-021-00689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chinen T, Kannan AK, Levine AG, Fan X, Klein U, Zheng Y, Gasteiger G, Feng Y, Fontenot JD, Rudensky AY. An essential role for the IL-2 receptor in Treg cell function. Nat Immunol. 2016;17:1322–1333. doi: 10.1038/ni.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fabbi M, Carbotti G, Ferrini S. Dual roles of IL-27 in cancer biology and immunotherapy. Mediators Inflamm. 2017;2017:3958069. doi: 10.1155/2017/3958069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kourko O, Seaver K, Odoardi N, Basta S, Gee K. IL-27, IL-30, and IL-35: a cytokine triumvirate in cancer. Front Oncol. 2019;9:969. doi: 10.3389/fonc.2019.00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DEGs_dataset_1
DEGs_dataset_2
DEGs_dataset_3
DEGs_dataset_4
DEGs_dataset_5
DEGs_dataset_6
Quantile normalization with and without meeting the assumption. Quantile normalization should be used when only a small proportion of genes, i.e., a few tens or hundreds out of around 20,000 human genes in total, are differentially expressed. (A) Log2 gene expression values for three samples in condition A and three samples in condition B are shown before and after quantile normalization. Quantile normalization equalizes the statistical distribution of all samples in the experiment (right panel). The yellow line represents the expression of a differentially expressed gene. Global differences between samples are removed by quantile normalization, while the yellow example gene in the figure is still identified as differentially expressed after normalization. (B) Here a large proportion of around 20,000 genes have a higher expression value in condition B compared to condition A. After quantile normalization, the statistical distribution of all samples is equalized. Hence, the majority of differentially expressed genes before normalization will not be seen after normalization. The yellow example genes are not differentially expressed in the right panel, whereas they are in the left panel.