Abstract
Purpose
Chromosomal instability (CIN) contributes to intercellular genetic heterogeneity and has been implicated in paclitaxel (PTX) resistance in breast cancer. In this study, we explored polo-like kinase 1 (PLK1) as an important regulator of mitotic integrity and as a useful predictive biomarker for PTX resistance in breast cancer.
Methods
We performed PTX resistance screening using the human kinome CRISPR/Cas9 library in breast cancer cells. In vitro cell proliferation and apoptosis assays and in vivo xenograft experiments were performed to determine the effects of PLK1 on breast cancer cells. Immunofluorescence microscopy was used to measure the degree of multipolar cell division.
Results
Kinome-wide CRISPR/Cas9 screening identified various kinases involved in PTX resistance in breast cancer cells; among these, PLK1 was chosen for further experiments. PLK1 knockdown inhibited the proliferation of MDA-MB-231 and MDA-MB-468 cells in vitro and in vivo. Moreover, PLK1 silencing sensitized breast cancer cells and mouse xenograft tumor models to PTX cytotoxicity. Silencing of PLK1 induced the formation of multipolar spindles and increased the percentage of multipolar cells. In addition, PLK1 silencing resulted in the downregulation of BubR1 and Mad2 in breast cancer cells. Furthermore, PLK1 upregulation in primary breast cancer was associated with decreased overall patient survival based on the analysis of The Cancer Genome Atlas and Molecular Taxonomy of Breast Cancer International Consortium databases.
Conclusion
PLK1 plays an important role in PTX resistance by regulating CIN in breast cancer cells. Targeting PLK1 may be an effective treatment strategy for PTX-resistant breast cancers.
Keywords: Breast Neoplasms, CRISPR-Cas Systems, Paclitaxel, Polo-Like Kinase 1, Spindle Poles
INTRODUCTION
Breast cancer was the most commonly diagnosed cancer globally in 2020 [1] and is the leading cause of cancer-related deaths in women [2]. Breast cancer is a heterogeneous malignancy with diverse molecular features that can be classified into several subtypes [3]. Molecular classification not only reflects the biology of breast cancer, but also reflects treatment options and oncologic outcomes [4]. Among the clinical breast cancer subtypes, the triple-negative breast cancer (TNBC) subtype represents more aggressive tumors with a high risk of recurrence and deaths [4,5]. Patients with TNBC are often treated with cytotoxic chemotherapy in addition to adequate local control measures [4,6,7].
Paclitaxel (PTX) is a commonly used cytotoxic chemotherapeutic agent for TNBC [6,7]. PTX induces cancer cell death by stabilizing microtubules and multinucleation [8,9]. However, most patients with TNBC who experience distant metastasis eventually develop resistance to PTX and show disease progression [10]. Although several mechanisms of PTX resistance have been identified in breast cancer, such as alterations to tubulin structures, defects in the spindle assembly checkpoint (SAC), and dysregulation of several proteins, including P-glycoprotein and TP53, strategies to overcome these remain challenging [8,10,11,12]. Recently, emerging evidence has suggested that chromosomal instability (CIN) in breast cancer might serve as a predictor of PTX response [8,9,12,13]. Furthermore, recent studies have shown that CIN is associated with various malignant features of multiple types of human cancers [14,15].
Kinases often perform critical cellular functions that cancer cells require to proliferate and metastasize [16]. Recent technical advances, such as CRISPR/Cas9, have enabled comprehensive screening of the kinome, a collection of kinases, to identify therapeutic targets [17]. Kinome-wide screening has identified key kinases that mediate PTX resistance in ovarian and breast cancers [18,19].
In the present study, we identified polo-like kinase 1 (PLK1) as a potential regulator of PTX resistance in breast cancer using CRISPR/Cas9-based kinome screening. Our data also demonstrated that PLK1 can modulate the response to PTX by regulating CIN in breast cancer cells.
METHODS
Breast cancer cell lines and small interfering RNA (siRNA) treatment
Breast cancer cell lines were purchased from the Korean Cell Line Bank (Seoul, Korea). MCF10A and MDA-MB-453 cells were obtained from the American Type Culture Collection (ATCC; Manassas, USA). Non-tumorigenic mammary epithelial MCF10A cells were cultured in a 1:1 mixture of Dulbecco’s modified Eagle’s medium (DMEM; Biowest, Riverside, USA) and Ham’s F12 medium (Biowest), containing 5% horse serum (Gibco, Waltham, USA), 20 ng/mL epidermal growth factor (EGF; Sigma-Aldrich, St. Louis, USA), 10 μg/mL insulin (Sigma-Aldrich), and 500 ng/mL hydrocortisone (Sigma-Aldrich). MCF7, MDA-MB-231, MDA-MB-468, HS578T, and T47D cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Gibco). SK-BR3, ZR-75-1, BT474, MDA-MB-453, BT20, HCC38, and HCC70 cells were maintained in RPMI 1640 (Biowest) supplemented with 10% FBS and 1% penicillin/streptomycin. For the siRNA experiments, commercially available PLK1 (Gene ID: 5347) siRNA was purchased from Dharmacon Inc. (Lafayette, USA). Cells were transfected with siRNA (10 nM) using the ON-TARGETplus human PLK1 siRNA-SMARTpool siRNA transfection reagent (Dhamacon Inc.), according to the manufacturer’s instructions.
Human kinome CRISPR/Cas9 knockout library screening
Lentiviral production was induced in HEK293-FT cells as described previously [20]. The human kinome CRISPR/Cas9 pooled library (Addgene #1000000083), psPAX2 (Addgene #12260), and pCMV-VSV-G (Addgene #8454) plasmids were kindly provided by John Doench, David Root, Didier Trono, and Bob Weinberg, respectively [21]. Transduction of the CRISPR lentiviral library into the MDA-MB-231 and MDA-MB-468 cells was performed as previously described [20]. Cells were divided into two groups, with vehicle or PTX (IC20 concentration) and maintained for 14 days. Genomic DNA from residual cells was extracted using the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany), and single-guide RNA (sgRNA) was amplified by polymerase chain reaction (PCR) with Illumina primers [20]. PCR amplicons were sequenced using the HiSeq 2500 (Illumina, San Diego, USA), and sgRNA frequencies were analyzed using the MAGeCK algorithm [22].
Cell transfection
The pLKO.1-Puro lentiviral vector was constructed to contain sequences of specific short hairpin RNA (shRNA) targeting human PLK1 (point 1, 5′-CACAGTCCTCAATAAAGGCTT-3′; and point 2, 5′-GTTCTTTACTTCTGGCTATAT-3′). Constructs containing pLKO.1-PLK1-shRNA were transfected into HEK-293FT cells using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, USA). The medium containing the lentivirus was incubated at 37°C and 5% CO2 for 48 hours after transfection. Then, it was harvested and used for infecting MDA-MB-231 and MDA-MB-468 cells. Cells with pLKO.1-Puro scramble shRNA (shRNA against negative control; sh-NC) were used as negative controls.
Cell viability assay and 3D cell culture
Briefly, the cells were seeded in 96-well plates (3 × 103 cells/well) and treated with various concentrations of PTX for 72 hours. The cells were incubated with 0.5 mg/mL thiazolyl blue tetrazolium bromide (MTT; Sigma-Aldrich) for 3 hours at 37°C. The medium was discarded and 200 μL of dimethyl sulfoxide (Duchefa Biochemie, Harriem, Netherlands) was added to each well to dissolve the formazan crystals in the cells. The absorbance was measured at 570 nm using a microplate reader (BioTek Instruments, Winooski, USA). For 3D cell culture, cells (5 × 103 cells/well) were suspended and seeded in 24-well plates in growth factor-reduced Matrigel (BD Biosciences, San Jose, USA). Spheroid growth and dimensions were measured as previously described [23].
Western blotting, real-time PCR, and immunohistochemistry
Cell lysates were harvested using RIPA buffer (Thermo Scientific), protease, and phosphatase inhibitor (Thermo Scientific), incubated for 10 minutes on ice, and centrifuged at 14,000 rpm for 15 minutes at 4°C. Protein concentration was measured using a BCA assay kit (Thermo Scientific), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene fluoride (PVDF; Sigma-Aldrich) membranes. After blocking with 5% bovine serum albumin (BSA; Biosesang, Seongnam, Korea) solution, membranes were incubated with primary antibody overnight at 4°C. The secondary antibody was diluted (1:3,000) in a 5% BSA solution. Western blotting bands were detected using an Amersham Imager 680 (GE Healthcare Life Sciences, Piscataway, USA). The following antibodies were used: β-actin (#sc-47778; Santa Cruz Biotechnology, Dallas, USA), PLK1 (#ab17056; Abcam, Cambridge, UK), Mad2L1 (#ab97777; Abcam), and BubR1 (#ab172584; Abcam).
Total RNA was extracted from the cells using TRIzol reagent (Favorgen, Pingtung, Taiwan). The Prime Script 1st strand cDNA Synthesis Kit (Takara, Osaka, Japan) was used for reverse transcription of RNA, and qPCR assays were performed using Power SYBR Green PCR Master mix (Thermo Scientific). The reactions were performed using an ABI7500 real-time PCR system (Thermo Scientific). To compare the relative mRNA expression levels, the expression levels of PLK1 were expressed as ratios to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The primer sequences were as follows: PLK1 forward:5′-CAGCAAGTGGGTGGACTATT-3′, reverse:5′-GTAGAGGATGAGGCGTGTTG-3′; GAPDH forward:5′-TTTCTAGACGGCAGGTCAGG-3′, reverse:5′-ACCCAGAAGACTGTGGATGG-3′.
Immunohistochemistry was performed using an IHC staining kit (Agilent, Santa Clara, USA). Tissue sections were deparaffinized in xylene and rehydrated in a series of graded alcohol solutions, and the antigen was retrieved in an antigen unmasking solution (Vector Laboratories, Inc., Burlingame, USA). The sections were then incubated with 3% hydrogen peroxide to inhibit endogenous peroxidase and blocked with normal goat serum (#AAR-6591–02; ImmunoBioScience Corp., Mukilteo, USA). Subsequently, sections were incubated with primary antibodies at a dilution of 1:1,000 or 1:2,000 at 4°C overnight. Next, the sections were incubated with a secondary anti-rabbit/mouse antibody, followed by incubation with a peroxidase solution. Finally, sections were developed with diaminobenzidine and hydrogen peroxide solution and counterstained with hematoxylin.
Immunofluorescence microscopy
Cells on coverslips were fixed with methanol at −20°C for 30 minutes. Alternatively, cells were extracted using BRB80-T buffer (80 mM PIPES, pH 6.8, 1 mM MgCl2, 5 mM EGTA, and 0.5% Triton X-100) and fixed with 4% paraformaldehyde for 15 minutes at room temperature. Fixed cells were permeabilized and blocked with phosphate-buffered saline (PBS)-BT (1× PBS, 3% BSA, and 0.1% Triton X-100) for 30 minutes at room temperature. The coverslips were then incubated with primary and secondary antibodies diluted in PBS-BT. Images were acquired using stimulated emission depletion (STED) at 3× super-resolution (Leica Microsystems GmbH, Mannheim, Germany) under a Leica TCS SP8 confocal microscope and a 63× oil immersion lens. Data from all studies were analyzed using the Leica Application Suite X (LAS X) software (Leica Microsystems GmbH, Mannheim, Germany). The primary antibodies used were mouse anti-r-tubulin (#T6557; Sigma) and rabbit anti-pericentrin (#ab4448; Abcam). The secondary antibodies used were Alexa Fluor 488 and 594 (Invitrogen, Waltham, USA).
Cell apoptosis and cell cycle arrest assay
The cells were inoculated into 6-well plates (2 × 105 cells/well) and transfected with siPLK1 in FBS-free DMEM for 6 hours. The cells were then cultured in PTX-containing 10% FBS DMEM at 37°C for 48 hours and stained with annexin V-FITC and propidium iodide (PI; BD Biosciences) according to the apoptosis detection kit’s instructions. For the analysis of cell cycle arrest, the transfection of siPLK1 was conducted as described above, and the cells were treated with PTX, fixed with 75% ethanol, and stained with PI at 4°C for 30 minutes. Finally, apoptosis and cell cycle arrest were detected using BD FACSCanto and BD FACSCalibur (BD Biosciences).
Xenograft murine model and drug treatment
MDA-MB-231 cells stably transfected with sh-NC and shPLK1 were injected into the fourth mammary fat pad of 6-week-old athymic nude female mice. The mice were cared for according to the institutional guidelines for animal care. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Seoul National University (No. 18-0127-C1A1). Drug treatment was initiated after the tumors reached approximately 100 mm3. Mice were randomly divided according to tumor size into four treatment groups (five mice per group): 1) sh-NC group treated with vehicle, 2) sh-NC group treated with PTX, 3) sh-PLK1 group treated with vehicle, and 4) sh-PLK1 group treated with PTX. For each murine xenograft model, either PBS (200 μL/mouse; 5 sh-NC mice, 5 shPLK1 mice) or PTX (15 mg/kg; 5 sh-NC mice, 5 shPLK1 mice) was intraperitoneally (IP) injected twice weekly until tumors reached 1,000 mm3. The length and width of each tumor were measured using calipers, and the volumes were calculated using the following equation: V = (length × width2)/2.
Statistical analysis
GraphPad Prism version 8.02 (GraphPad Software, San Diego, USA) was used to generate graphs and perform statistical tests. Most data values are presented as the mean ± standard deviation (SD) and represent three independent experiments, except for CRISPR/Cas9 screening, 3D spheroid growth, and in vivo tumor growth experiments. Student’s t-tests or Mann-Whitney U tests were used to compare the means between the groups. For in vivo drug responses, we used multiple t-tests to compare tumor volumes. Kaplan-Meier survival analyses were performed using log-rank tests to assess the time to progression and survival.
RESULTS
CRISPR/Cas9-based kinome-wide screening identified PLK1 as a candidate gene regulating PTX resistance
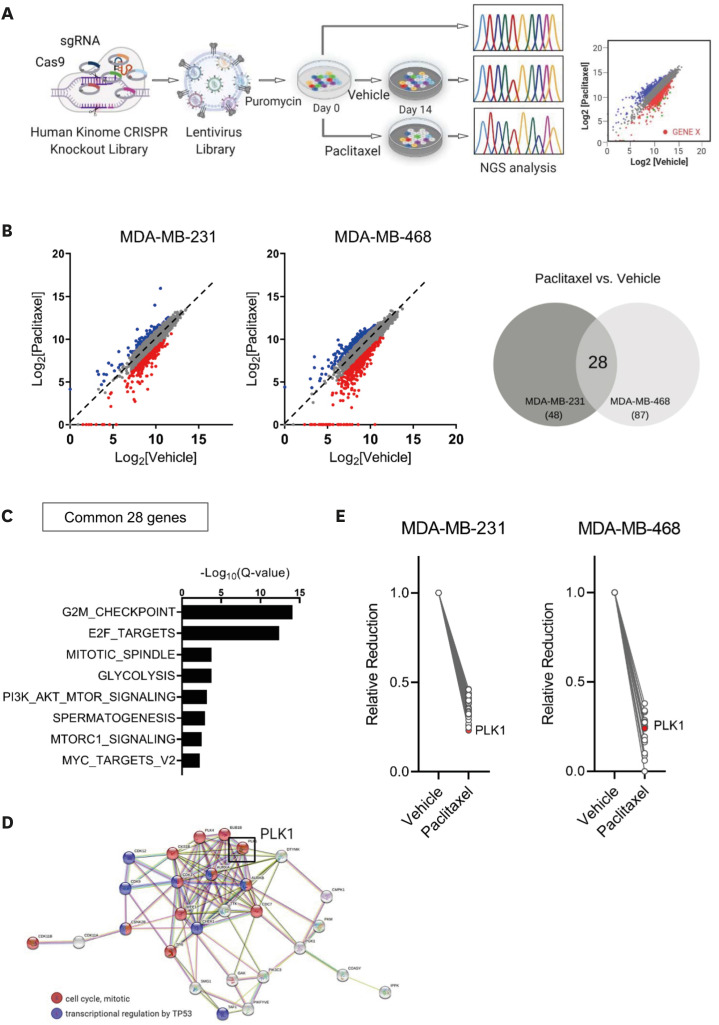
First, we conducted kinome-wide screening of genes associated with PTX resistance using the human kinome CRISPR/Cas9 in breast cancer cells. To this end, we generated an sgRNA library using a lentivirus to knockout 763 human kinases. After transfecting MDA-MB-231 and MDA-MB-468 breast cancer cells, they were treated with the vehicle or a 20% inhibitory concentration of PTX for 14 days. We sequenced the cells on day 0 (no treatment) and day 14 (vehicle- or PTX-treated) and estimated the differences in sgRNA frequencies using the MAGeCK algorithm [22]. We assumed that genes that confer resistance to PTX would show a lower sgRNA frequency in cells treated with PTX than in those treated with vehicle. The comparison data from day 0 and day 14 for vehicle-treated cells were used to exclude kinases associated with survival fitness for in vitro cell survival in the absence of PTX pressure (Figure 1A).
Figure 1. Human kinome CRISPR/Cas9 screening for the identification of candidate therapeutic target genes related to paclitaxel resistance in breast cancer.
(A) Schematic illustration showing the human kinome CRISPR/Cas9 knockout screens used to identify genes associated with PTX resistance in MDA-MB-231 and MDA-MB-468 cells. (B) Scatterplots of normalized sgRNA counts for PTX- versus vehicle-treatment at day 14 (left panel). Red dots show that the sgRNA frequencies were depleted in PTX-treated cells (log2 [fold change] ≤ −1). Venn diagram of 28 genes that overlapped in MDA-MB-231 and MDA-MB-468 cells (right panel). (C) Significantly enriched gene sets (FDR Q < 0.01) for the 28 genes from the PTX-treated group. (D) STRING network analysis of the 28 genes depleted in the PTX-treated group. Genes involved in cell cycle progression and transcriptional regulation by TP53 are indicated in red and blue, respectively. PLK1 indicated by a black box is a one of the hub nodes in the network. (E) Changes in the normalized sgRNA counts in key hub genes between the vehicle- and PTX-treated groups.
FDR = false discovery rate; NGS = next-generation sequencing; PLK1 = polo-like kinase 1; PTX = paclitaxel; sgRNA = single-guide RNA.
We identified 48 and 87 kinase genes from MDA-MB-231 cells and MDA-MB-468 cells, respectively, for which the sgRNA frequencies were decreased by more than half after treatment with PTX compared with vehicle treatment (Figure 1B, Supplementary Table 1). A total of 28 (22.9%) out of the 135 genes overlapped in both the cell types. For the comparison between day 0 and day 14 for the vehicle-treated cells, 95 genes showed substantial reduction, with only the ATR gene overlapping between the two cell types (Supplementary Figure 1A, Supplementary Table 2). The 28 genes were significantly enriched with genes involved in cell division, such as the G2M checkpoint or E2F targets (Figure 1C). In addition, reactome pathway analysis (http://reactome.org) revealed several key hub genes among these 28 genes (Figure 1D). Among the key hub genes, we selected PLK1 for further investigation because it showed the largest reduction in MDA-MB-231 cells (Figure 1E).
PLK1 silencing inhibits breast cancer cell growth and sensitizes cells to PTX cytotoxicity
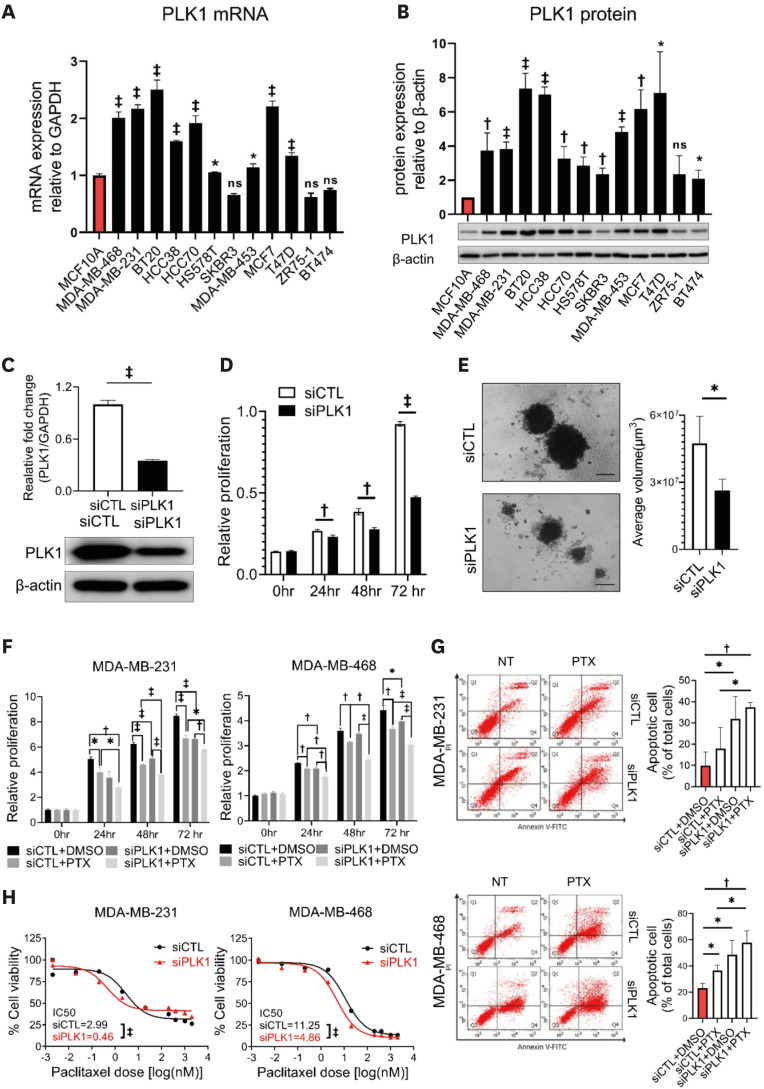
To determine the role of PLK1 in breast cancer, we first evaluated PLK1 expression levels in normal mammary epithelial cells (MCF10A) and 10 breast cancer cell lines. Breast cancer cells often showed significant upregulation of PLK1 mRNA and protein expression compared with MCF10A cells (Figure 2A and B). Next, we investigated the role of PLK1 in the breast cancer cell phenotype in vitro. Treatment of cells with siRNA against PLK1 significantly reduced the expression of PLK1 in MDA-MB-231 and MDA-MB-468 cells (Figure 2C, Supplementary Figure 1B). Silencing of PLK1 resulted in a significant reduction in cell viability (Figure 2D, Supplementary Figure 1C) and growth in 3D culture (Figure 2E).
Figure 2. PLK1 expression in breast cancer cells and the effect of PLK1 knockdown on cell proliferation and PTX resistance.
(A) qPCR analysis and (B) western blotting gel images showing the expression levels of PLK1 mRNA and protein in breast cancer cells and mammary normal epithelial cells. (C) mRNA and protein levels of PLK1 after siPLK1 treatment. At 72 hours after transfection, cell proliferation was examined using MTT (D) and 3D Matrigel (E) assays (n = 10 spheroids) Scale bar = 200 μm; Student’s t-tests. (F) An MTT assay and (G) analysis of representative flow cytometry plots of annexin V staining in MDA-MB-231 and MDA-MB-468 cells after si-PLK1 and PTX treatment. Cells were treated for 72 hours with 10 nM PLK1 siRNA, 5 nM PTX, and both in combination before annexin V and MTT staining (n = 3); Mann–Whitney U test (F) and Student’s t-tests (G). (H) IC50 values and response curves against PTX in a panel of MDA-MB-231 and MDA-MB-468 cells according to si-PLK1 treatment (n = 3); Mann–Whitney U test. This applies to all figures shown.
IC50 = half maximal inhibitory concentration; MTT = thiazolyl blue tetrazolium bromide; ns = non-significant; PLK1 = polo-like kinase 1; PTX = paclitaxel; qPCR = qualitative polymerase chain reaction; siRNA = small interfering RNA.
*p < 0.05, †p < 0.01, and ‡p < 0.001.
Since PLK1 is a potential regulator of PTX resistance, we evaluated whether silencing of PLK1 resulted in increased PTX sensitivity. In both MDA-MB-231 and MDA-MB-468 cells, combining si-PLK1 and PTX resulted in increased cell death compared with PTX treatment alone (Figure 2F). The annexin V assay also demonstrated that the combination of si-PLK1 and PTX was associated with increased apoptosis in both cell types, which occurred by inducing G2/M cell cycle arrest (Figure 2G, Supplementary Figure 1D). Additionally, silencing of PLK1 resulted in a significant reduction in the IC50 value of PTX in vitro (Figure 2H). We also confirmed a decreased IC50 value for PTX after PLK1 blockade by treatment with the PLK1 inhibitor volasertib (Supplementary Figure 1E).
Increased multipolar spindles and impaired SACs in PLK1-silenced breast cancer cells
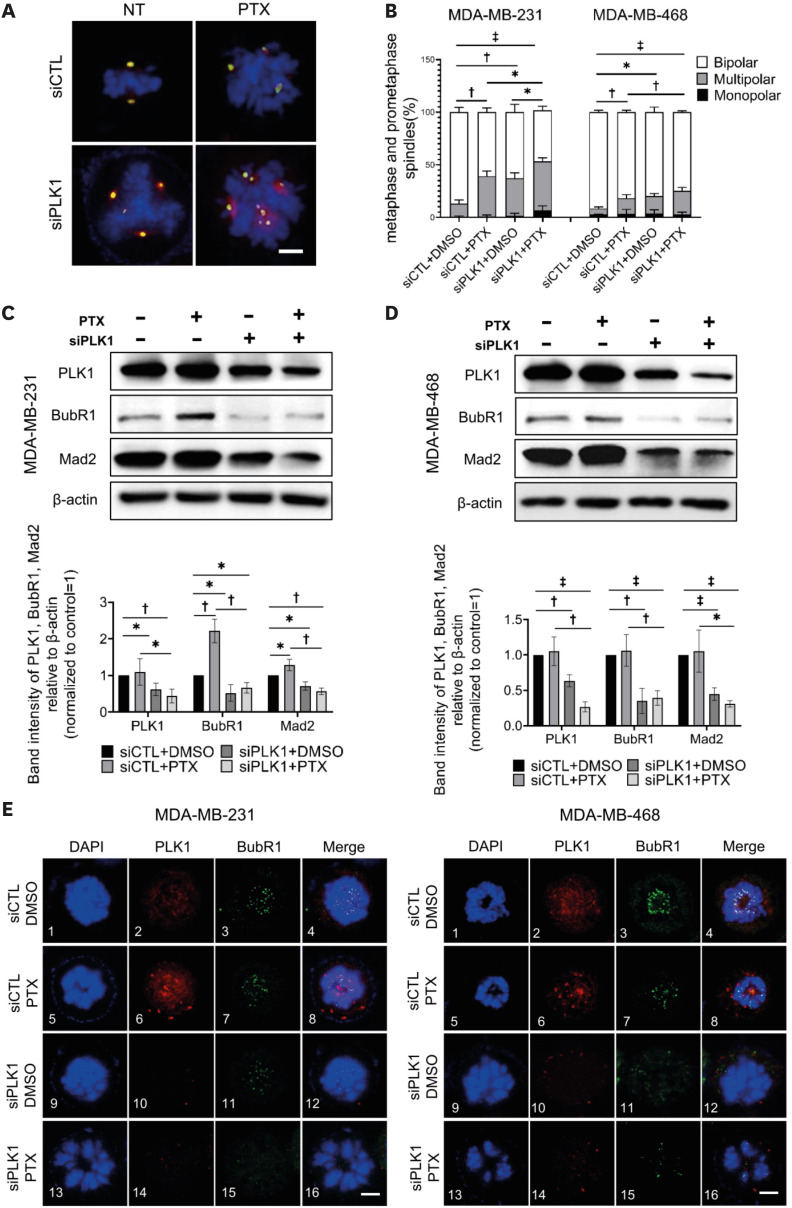
Recent studies have suggested that PTX sensitivity is regulated by the degree of CIN in cancer [8,9,13]. To explore the association between PLK1 expression and CIN in breast cancer, we examined multipolar cell division using DNA, centrosome, and microtubule staining (Supplementary Figure 2A and B). As shown in Figure 3A, PTX treatment increased the incidence of multipolar spindles in breast cancer cells. PLK1 silencing resulted in an increased incidence of multipolar spindles. Furthermore, the combined use of si-PLK1 and PTX resulted in a significantly higher incidence of multipolar spindles in MDA-MB-231 cells than PTX treatment alone (Figure 3B). These data indicate that the downregulation of PLK1 promotes multipolar spindle formation, which can lead to increased PTX sensitivity.
Figure 3. Increased incidence of multipolar divisions and impaired SAC after PLK1 depletion.
(A) Images of mitotic spindles with the indicated number of poles in breast cancer cells after treatment with si-PLK1 and PTX. Images are maximum projections from z stacks of representative cells stained for DNA (DAPI, blue), centrosomes (pericentrin, green), and MTs (γ-tubulin, red). Scale bar = 5 μm. (B) Quantification of multipolar spindles in MDA-MB-231 and MDA-MB-468 cells in prometaphase/metaphase (n > 50 cells in each of three replicates). (C) and (D) Top: Western blotting of si-PLK1- and PTX-treated MDA-MB-231 and MDA-MB-468 cells. Loading control, β-actin. Bottom: Quantification of BubR1 and Mad2 expression levels normalized to β-actin. (E) Localizations of BubR1 and PLK1 in prometaphase shown by immunofluorescence staining. DAPI (blue), BubR1 (green), and PLK1 (red). Scale bar = 5 μm. Error bars are mean ± SD (n = 3); Student’s t-tests (B) and Mann–Whitney U test (C and D).
DAPI = 4′,6-diamidino-2-phenylindole; PLK1 = polo-like kinase 1; PTX = paclitaxel; SD = standard deviation.
*p < 0.05, †p < 0.01, and ‡p < 0.001.
Next, we investigated the expression levels of BubR1 and Mad2, which are key regulatory proteins for SAC activity in prometaphase, as SAC controls the dynamic interaction between spindle microtubules and kinetochores [24]. BubR1 expression was upregulated when breast cancer cells were treated with PTX (Figure 3C). In contrast, cells treated with si-PLK1 showed significant downregulation of BubR1 expression, which was not attenuated by the PTX treatment (Figure 3D). Similar findings were observed in Mad2. Furthermore, kinetochore localization of BubR1 was significantly reduced in PLK1-silenced breast cancer cells (Figure 3E). These observations suggest that sPLK1 plays a crucial regulatory role in SAC activity, which can lead to CIN by regulating the expression levels of BubR1 and Mad2 in breast cancer cells.
Effect of PLK1 on breast cancer growth and PTX resistance in vivo
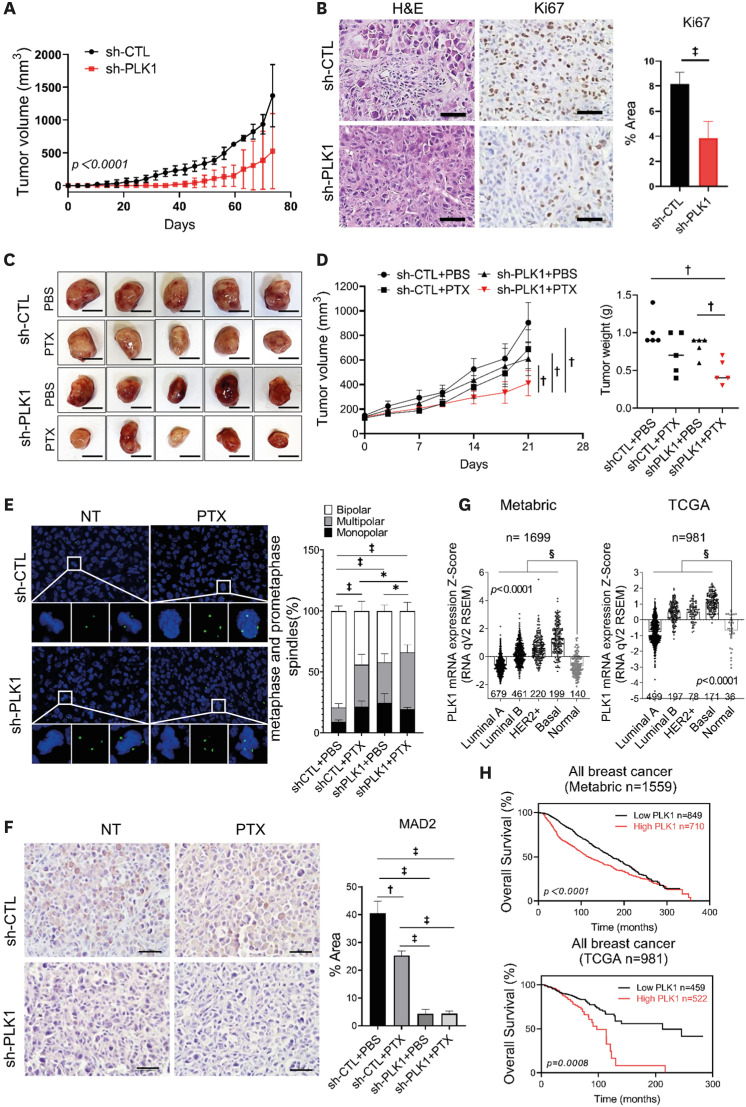
To investigate the role of PLK1 in tumorigenesis, we established a stable PLK1-knockdown MDA-MB-231 cell line (sh-PLK1) using a lentiviral shRNA (Supplementary Figure 2C). We evaluated xenograft tumor growth in mice by injecting cancer cells into the fat pad. PLK1-knockdown cells showed significantly decreased tumor growth rates compared to control cells (non-targeting shRNA; sh-CTL) (Figure 4A). PLK1-knockdown tumor cells showed low PLK1 expression (Supplementary Figure 2D) and significantly fewer Ki-67 positive cells (Figure 4B). Next, we treated mice harboring sh-CTL or sh-PLK1 MDA-MB-231 cells with DMSO or PTX, respectively. Treatment with PTX resulted in modest and statistically non-significant tumor growth inhibition in control xenograft tumors (Figure 4C). However, treatment of PLK1-silenced xenograft tumors with PTX resulted in significant inhibition of tumor growth (Figure 4C and D). Similar to the results of the in vitro experiments, the PLK1-silenced xenograft tumors showed a significantly increased incidence of multipolar spindles to a similar degree to that of the PTX-treated xenograft tumors (Figure 4E). In addition, Mad2 expression was significantly reduced in PLK1-silenced tumors (Figure 4F). Next, we investigated the clinical implications of PLK1 expression using data from The Cancer Genome Atlas (TCGA) and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) databases. In both datasets, PLK1 expression was significantly upregulated in tumor tissues, with the basal type showing the highest expression level (Figure 4G). Furthermore, high PLK1-expressing tumors showed significantly worse survival than low PLK1-expressing tumors in both datasets (Figure 4H).
Figure 4. Increased PTX cytotoxicity after PLK1 depletion in vivo.
(A) Inhibition of tumor growth generated by xenografted MDA-MB-231 cell lines in nude mice after silencing of PLK1. Tumor growth curves are shown (n = 4 mice/group); two-way ANOVA with Turkey’s post hoc test. (B) Representative H&E and Ki-67-stained images of tumor and the expression levels of Ki-67. The area percentage was measured from 10 different images; Student’s t-tests. (C) PTX treatment of nude mice bearing MDA-MB-231 sh-CTL and sh-PLK1 xenograft tumors. Tumors removed from five mice in each group are shown. (D) Left: Measured tumor volume from days 0 to 21 after treatment plotted versus time. Right: Statistical analysis of the weights of dissected tumors (n = 5 mice/group); multiple t-testing. (E) Images of mitotic spindles with the indicated number of poles in MDA-MB-231 sh-CTL and sh-PLK1 xenograft tumors after PTX treatment. Quantification of multipolar spindles in MDA-MB-231 xenograft tumors (n > 30 cells/mice); Mann–Whitney U test. (F) Representative images of MAD2 staining of tumors derived from MDA-MB-231 with PLK1 depletion and treatment PTX. (G) Expression of PLK1 in the METABRIC BRCA and TCGA breast cancer databases. (H) Overall survival of patients with breast cancer based on PLK1 transcription levels using the Kaplan-Meier plotter online tool. Scale bars = 1 cm (C), 50 μm (B and F). Error bars are mean ± SD.
METABRIC = Molecular Taxonomy of Breast Cancer International Consortium; PLK1 = polo-like kinase 1; PTX = paclitaxel; SD = standard deviation; sh-CTL = non-targeting shRNA; TCGA = The Cancer Genome Atlas.
*p < 0.05, †p < 0.01, ‡p < 0.001, and §p < 0.0001.
DISCUSSION
In the present study, CRISPR/Cas9 screening was performed, which revealed several kinases involved in PTX resistance in breast cancer, including PLK1. Our study demonstrated that the use of an anti-PLK1 treatment strategy could potentially reverse PTX resistance in breast cancer. In addition, PLK1 downregulation increased the incidence of multipolar spindles, thereby exacerbating mitotic abnormalities and ultimately leading to cell death in response to treatment. Our data suggest that PLK1 downregulation affects PTX sensitivity in breast cancer cells by increasing the rate of CIN.
PLK1 is a serine/threonine-protein kinase that plays multiple roles in the cell cycle, such as in mitotic entry and at the G2/M checkpoint, coordinates the centrosome and cell cycle, regulates spindle assembly and chromosome segregation, and facilitates DNA replication [25]. PLK1 has been reported to be highly expressed in many types of cancer [26], including TNBC [27]. PTX is a cytotoxic microtubule-targeting agent that stabilizes microtubules, suppresses tubulin dynamics, and induces mitotic arrest, resulting in apoptotic cell death [10]. In the present study, sgRNA-mediated screening of kinases was performed to identify PLK1, whose depletion sensitized breast cancer cells to PTX and potentiated the effects of the drug on mitotic arrest and apoptosis. These observations suggested that PLK1 can be used as a biomarker to predict PTX response, highlighting the importance of further mechanistic studies.
Extensive published data have shown that CIN is associated with PTX sensitivity in breast cancer [12,13,28]. CIN has recently been shown to increase PTX sensitivity in breast cancer cells [13]. CIN refers to errors in mitosis, including multipolar spindles, defects in mitotic spindle assembly, and improper kinetochore-microtubule attachment [29]. In cell cultures, mitotic divisions on multipolar spindles result in chromosome missegregation and increase cell death [30]. Genetic ablation of PLK1 or its chemical inhibition induces G2/M arrest, creates multipolar cell division, and eventually induces apoptotic cell death [31,32]. Accordingly, the current study found that PLK1 depletion induced the formation of multipolar spindles and increased the percentage of multipolar cells. However, PLK1 inhibition did not increase PTX-induced multipolar division. Thus, PLK1 silencing before PTX exposure resulted in transient CIN and improved breast cancer sensitivity to treatment.
Our data further demonstrated that PLK1 regulates CIN, which is associated with PTX resistance. PLK1 activity stabilizes kinetochore-microtubule attachments by reducing microtubule dynamics at the kinetochores [33]; however, overactive PLK1 enhances stabilization of microtubules and promotes misattachments, leading to CIN [34]. Furthermore, dysregulation of PLK1 prematurely generates kinetochore–microtubule attachments, leading to CIN on chromosome missegregation [35,36]. Therefore, dysregulation of PLK1 in either direction results in erroneous kinetochore–microtubule attachments and chromosome missegregation [37]. Additionally, PLK1 can dysregulate mitotic entry and impair mitotic checkpoints, resulting in CIN [38]. Notably, we found that PLK1 depletion impaired SAC, which monitors kinetochore-microtubule attachments. Our data suggest that the inhibition of PLK1 weakens the mitotic checkpoint and causes CIN during multipolar division.
Our study had several limitations. The distinction between aneuploidy and CIN was first recognized when the former was denoted as a state of abnormal chromosome number and morphology, whereas the latter as chromosome mis-segregation [15]. The abnormal chromosome number and morphology of PLK1 depleted cells could not be determined. Although aneuploidy is frequently deleterious to cell fitness, it has a selective advantage in certain tumor environments. Second, mitotic spindles and centrosomes were counted using staining to confirm the presence of CIN. However, the correlation between PLK1 and CIN-associated genes could not be analyzed. There are several methods for measuring CIN, including in situ hybridization [39], flow and DNA image cytometry [40], CIN70 signatures [41], and comparative genomic hybridization [42]. Finally, a time-lapse analysis of cell division was not performed. Analysis of the percentage of cell death in multipolar spindle cells further demonstrated that PLK1-induced multipolar cell division directly resulted in cell death.
In conclusion, our data indicated that PLK1 may induce CIN to improve PTX sensitivity in breast cancer. Targeting PLK1 in PTX-resistant TNBC with CIN is an effective therapeutic strategy.
Footnotes
Funding: This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2019R1A2C2005277, 2019R1C1C1006898). This study was also partly supported by a research donation to Seoul National University Hospital by Mr. Dae Woo Chang and Mrs. Bok Rye Kim. This study was also supported in part by the grant from Bertis Inc. (0620193060).
Conflict of Interest: The authors declare that they have no competing interests.
- Conceptualization: Moon HG.
- Data curation: Quan M, Oh Y, Cho SY, Kim JH, Moon HG.
- Formal analysis: Quan M, Oh Y, Cho SY, Kim JH.
- Funding acquisition: Moon HG, Kim JH.
- Investigation: Quan M, Moon HG.
- Validation: Cho SY.
- Visualization: Quan M.
- Writing - original draft: Quan M, Moon HG.
- Writing - review & editing: Kim JH, Moon HG.
SUPPLEMENTARY MATERIALS
List of genes derived from the CRISPR/Cas9 library screenings
List of genes represented by the Venn diagrams
The effect of PLK1 knockdown on cell proliferation and PTX resistance. (A) Scatterplots of normalized sgRNA counts on day 14 versus normalized sgRNA counts on day 0 for cells grown in control media. Red dots represent the sgRNAs for which normalized counts were depleted at day 14 (log2 [fold-change] ≤ −1). The Venn diagram of one gene overlapped in MDA-MB-231 and MDA-MB-468 cells (right panel). (B) mRNA and protein level of PLK1 after siPLK1 treatment in MDA-MB-468 cells. (C) Cell proliferation determined by an MTT assay 72 hours after transfection, and cell proliferation was examined with MTT assay. (D) Cell cycle progression of PLK1 depletion cells was analyzed after PTX treatment. Cells were treated for 72 hours with 10 nM PLK1 siRNA, 5 nM PTX, and both in combination before PI staining, followed by flow cytometric analysis. (E) IC50 values and response curves against PTX are shown in a panel of MDA-MB-231 and MDA-MB-468 cells according to PLK1 inhibitor (volasertib) treatment. Error bars are mean ± SD.
Images of mitotic stages, mitotic spindles, and PLK1 depletion in vivo. (A) Mitotic stages visualized with DNA, centrosome, and MTs staining. Mitotic MDA-MB-231 cells in prophase (panel 1), prometaphase (panels 2-4), metaphase (panel 5), anaphase (panel 6), and telophase (panel 7). Images are maximum projections from z stacks of representative cells stained for DNA (DAPI, blue), centrosomes (pericentrin, green), and MTs (γ-tubulin, red). Scale bar = 5 μm. (B) Images of mitotic spindles with the indicated number of poles in MDA-MB-231 and MDA-MB-468 cells after treatment with siPLK1 and PTX. Monopolar spindle (panel 1), bipolar spindle (panel 2), and multipolar spindle (panels 3–6). Scale bar = 5 μm. (C) Protein level of PLK1 after sh-PLK1 transfection in MDA-MB-231 cells. (D) Representative images of PLK1 staining of tumors derived from MDA-MB-231 with PLK1 depletion. Error bars are mean ± SD.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, et al. Cancer statistics for the year 2020: an overview. Int J Cancer. 2021;149:778–789. doi: 10.1002/ijc.33588. [DOI] [PubMed] [Google Scholar]
- 3.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Waks AG, Winer EP. Breast cancer treatment: a review. JAMA. 2019;321:288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 5.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26:2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 6.Senkus E, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rutgers E, et al. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(Suppl 5):v8–30. doi: 10.1093/annonc/mdv298. [DOI] [PubMed] [Google Scholar]
- 7.Cardoso F, Costa A, Norton L, Senkus E, Aapro M, André F, et al. ESO-ESMO 2nd international consensus guidelines for advanced breast cancer (ABC2) Ann Oncol. 2014;25:1871–1888. doi: 10.1093/annonc/mdu385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.A’Hern RP, Jamal-Hanjani M, Szász AM, Johnston SR, Reis-Filho JS, Roylance R, et al. Taxane benefit in breast cancer--a role for grade and chromosomal stability. Nat Rev Clin Oncol. 2013;10:357–364. doi: 10.1038/nrclinonc.2013.67. [DOI] [PubMed] [Google Scholar]
- 9.Weaver BA, Cleveland DW. Decoding the links between mitosis, cancer, and chemotherapy: The mitotic checkpoint, adaptation, and cell death. Cancer Cell. 2005;8:7–12. doi: 10.1016/j.ccr.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Murray S, Briasoulis E, Linardou H, Bafaloukos D, Papadimitriou C. Taxane resistance in breast cancer: mechanisms, predictive biomarkers and circumvention strategies. Cancer Treat Rev. 2012;38:890–903. doi: 10.1016/j.ctrv.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Sudo T, Nitta M, Saya H, Ueno NT. Dependence of paclitaxel sensitivity on a functional spindle assembly checkpoint. Cancer Res. 2004;64:2502–2508. doi: 10.1158/0008-5472.can-03-2013. [DOI] [PubMed] [Google Scholar]
- 12.Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li Q, et al. Chromosomal instability determines taxane response. Proc Natl Acad Sci U S A. 2009;106:8671–8676. doi: 10.1073/pnas.0811835106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scribano CM, Wan J, Esbona K, Tucker JB, Lasek A, Zhou AS, et al. Chromosomal instability sensitizes patient breast tumors to multipolar divisions induced by paclitaxel. Sci Transl Med. 2021;13:eabd4811. doi: 10.1126/scitranslmed.abd4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasudevan A, Schukken KM, Sausville EL, Girish V, Adebambo OA, Sheltzer JM. Aneuploidy as a promoter and suppressor of malignant growth. Nat Rev Cancer. 2021;21:89–103. doi: 10.1038/s41568-020-00321-1. [DOI] [PubMed] [Google Scholar]
- 15.Bakhoum SF, Cantley LC. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell. 2018;174:1347–1360. doi: 10.1016/j.cell.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhullar KS, Lagarón NO, McGowan EM, Parmar I, Jha A, Hubbard BP, et al. Kinase-targeted cancer therapies: progress, challenges and future directions. Mol Cancer. 2018;17:48. doi: 10.1186/s12943-018-0804-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurata M, Yamamoto K, Moriarity BS, Kitagawa M, Largaespada DA. CRISPR/Cas9 library screening for drug target discovery. J Hum Genet. 2018;63:179–186. doi: 10.1038/s10038-017-0376-9. [DOI] [PubMed] [Google Scholar]
- 18.Lai TC, Fang CY, Jan YH, Hsieh HL, Yang YF, Liu CY, et al. Kinase shRNA screening reveals that TAOK3 enhances microtubule-targeted drug resistance of breast cancer cells via the NF-κB signaling pathway. Cell Commun Signal. 2020;18:164. doi: 10.1186/s12964-020-00600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y, Gaillard S, Phillip JM, Huang TC, Pinto SM, Tessarollo NG, et al. Inhibition of spleen tyrosine kinase potentiates paclitaxel-induced cytotoxicity in ovarian cancer cells by stabilizing microtubules. Cancer Cell. 2015;28:82–96. doi: 10.1016/j.ccell.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S, Kim TM, Cho SY, Kim S, Oh Y, Kim M, et al. Combined blockade of polo-like kinase and pan-RAF is effective against NRAS-mutant non-small cell lung cancer cells. Cancer Lett. 2020;495:135–144. doi: 10.1016/j.canlet.2020.09.018. [DOI] [PubMed] [Google Scholar]
- 21.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMillan KS, McCluskey AG, Sorensen A, Boyd M, Zagnoni M. Emulsion technologies for multicellular tumour spheroid radiation assays. Analyst (Lond) 2016;141:100–110. doi: 10.1039/c5an01382h. [DOI] [PubMed] [Google Scholar]
- 24.Khodjakov A, Pines J. Centromere tension: a divisive issue. Nat Cell Biol. 2010;12:919–923. doi: 10.1038/ncb1010-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zitouni S, Nabais C, Jana SC, Guerrero A, Bettencourt-Dias M. Polo-like kinases: structural variations lead to multiple functions. Nat Rev Mol Cell Biol. 2014;15:433–452. doi: 10.1038/nrm3819. [DOI] [PubMed] [Google Scholar]
- 26.King SI, Purdie CA, Bray SE, Quinlan PR, Jordan LB, Thompson AM, et al. Immunohistochemical detection of polo-like kinase-1 (PLK1) in primary breast cancer is associated with TP53 mutation and poor clinical outcom. Breast Cancer Res. 2012;14:R40. doi: 10.1186/bcr3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maire V, Némati F, Richardson M, Vincent-Salomon A, Tesson B, Rigaill G, et al. Polo-like kinase 1: a potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res. 2013;73:813–823. doi: 10.1158/0008-5472.CAN-12-2633. [DOI] [PubMed] [Google Scholar]
- 28.Lukow DA, Sausville EL, Suri P, Chunduri NK, Wieland A, Leu J, et al. Chromosomal instability accelerates the evolution of resistance to anti-cancer therapies. Dev Cell. 2021;56:2427–2439.e4. doi: 10.1016/j.devcel.2021.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giam M, Rancati G. Aneuploidy and chromosomal instability in cancer: a jackpot to chaos. Cell Div. 2015;10:3. doi: 10.1186/s13008-015-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- 32.Wachowicz P, Fernández-Miranda G, Marugán C, Escobar B, de Cárcer G. Genetic depletion of polo-like kinase 1 leads to embryonic lethality due to mitotic aberrancies. BioEssays. 2016;38(Suppl 1):S96–106. doi: 10.1002/bies.201670908. [DOI] [PubMed] [Google Scholar]
- 33.Liu D, Davydenko O, Lampson MA. Polo-like kinase-1 regulates kinetochore-microtubule dynamics and spindle checkpoint silencing. J Cell Biol. 2012;198:491–499. doi: 10.1083/jcb.201205090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumitru AM, Rusin SF, Clark AE, Kettenbach AN, Compton DA. Cyclin A/Cdk1 modulates Plk1 activity in prometaphase to regulate kinetochore-microtubule attachment stability. eLife. 2017;6:e29303. doi: 10.7554/eLife.29303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raab M, Sanhaji M, Matthess Y, Hörlin A, Lorenz I, Dötsch C, et al. PLK1 has tumor-suppressive potential in APC-truncated colon cancer cells. Nat Commun. 2018;9:1106. doi: 10.1038/s41467-018-03494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cunningham CE, MacAuley MJ, Vizeacoumar FS, Abuhussein O, Freywald A, Vizeacoumar FJ. The CINs of polo-like kinase 1 in cancer. Cancers (Basel) 2020;12:2953. doi: 10.3390/cancers12102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simonetti G, Bruno S, Padella A, Tenti E, Martinelli G. Aneuploidy: cancer strength or vulnerability? Int J Cancer. 2019;144:8–25. doi: 10.1002/ijc.31718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Speicher MR, Carter NP. The new cytogenetics: blurring the boundaries with molecular biology. Nat Rev Genet. 2005;6:782–792. doi: 10.1038/nrg1692. [DOI] [PubMed] [Google Scholar]
- 40.Darzynkiewicz Z, Halicka HD, Zhao H. Analysis of cellular DNA content by flow and laser scanning cytometry. Adv Exp Med Biol. 2010;676:137–147. doi: 10.1007/978-1-4419-6199-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Habermann JK, Doering J, Hautaniemi S, Roblick UJ, Bündgen NK, Nicorici D, et al. The gene expression signature of genomic instability in breast cancer is an independent predictor of clinical outcome. Int J Cancer. 2009;124:1552–1564. doi: 10.1002/ijc.24017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat Genet. 2005;37(Suppl):S11–S17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes derived from the CRISPR/Cas9 library screenings
List of genes represented by the Venn diagrams
The effect of PLK1 knockdown on cell proliferation and PTX resistance. (A) Scatterplots of normalized sgRNA counts on day 14 versus normalized sgRNA counts on day 0 for cells grown in control media. Red dots represent the sgRNAs for which normalized counts were depleted at day 14 (log2 [fold-change] ≤ −1). The Venn diagram of one gene overlapped in MDA-MB-231 and MDA-MB-468 cells (right panel). (B) mRNA and protein level of PLK1 after siPLK1 treatment in MDA-MB-468 cells. (C) Cell proliferation determined by an MTT assay 72 hours after transfection, and cell proliferation was examined with MTT assay. (D) Cell cycle progression of PLK1 depletion cells was analyzed after PTX treatment. Cells were treated for 72 hours with 10 nM PLK1 siRNA, 5 nM PTX, and both in combination before PI staining, followed by flow cytometric analysis. (E) IC50 values and response curves against PTX are shown in a panel of MDA-MB-231 and MDA-MB-468 cells according to PLK1 inhibitor (volasertib) treatment. Error bars are mean ± SD.
Images of mitotic stages, mitotic spindles, and PLK1 depletion in vivo. (A) Mitotic stages visualized with DNA, centrosome, and MTs staining. Mitotic MDA-MB-231 cells in prophase (panel 1), prometaphase (panels 2-4), metaphase (panel 5), anaphase (panel 6), and telophase (panel 7). Images are maximum projections from z stacks of representative cells stained for DNA (DAPI, blue), centrosomes (pericentrin, green), and MTs (γ-tubulin, red). Scale bar = 5 μm. (B) Images of mitotic spindles with the indicated number of poles in MDA-MB-231 and MDA-MB-468 cells after treatment with siPLK1 and PTX. Monopolar spindle (panel 1), bipolar spindle (panel 2), and multipolar spindle (panels 3–6). Scale bar = 5 μm. (C) Protein level of PLK1 after sh-PLK1 transfection in MDA-MB-231 cells. (D) Representative images of PLK1 staining of tumors derived from MDA-MB-231 with PLK1 depletion. Error bars are mean ± SD.