Abstract
As an enveloped virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) delivers its viral genome into host cells via fusion of the viral and cell membranes. Here, we show that ANO6/TMEM16F-mediated cell surface exposure of phosphatidylserine is critical for SARS-CoV-2 entry and that ANO6-selective inhibitors are effective against SARS-CoV-2 infections. Application of the SARS-CoV-2 Spike pseudotyped virus (SARS2-PsV) evokes a cytosolic Ca2+ elevation and ANO6-dependent phosphatidylserine externalization in ACE2/TMPRSS2-positive mammalian cells. A high-throughput screening of drug-like chemical libraries identifies three different structural classes of chemicals showing ANO6 inhibitory effects. Among them, A6-001 displays the highest potency and ANO6 selectivity and it inhibits the single-round infection of SARS2-PsV in ACE2/TMPRSS2-positive HEK 293T cells. More importantly, A6-001 strongly inhibits authentic SARS-CoV-2-induced phosphatidylserine scrambling and SARS-CoV-2 viral replications in Vero, Calu-3, and primarily cultured human nasal epithelial cells. These results provide mechanistic insights into the viral entry process and offer a potential target for pharmacological intervention to protect against coronavirus disease 2019 (COVID-19).
Keywords: ANO6/TMEM16F, phosphatidylserine, SARS-CoV-2, virus-cell fusion
Graphical abstract
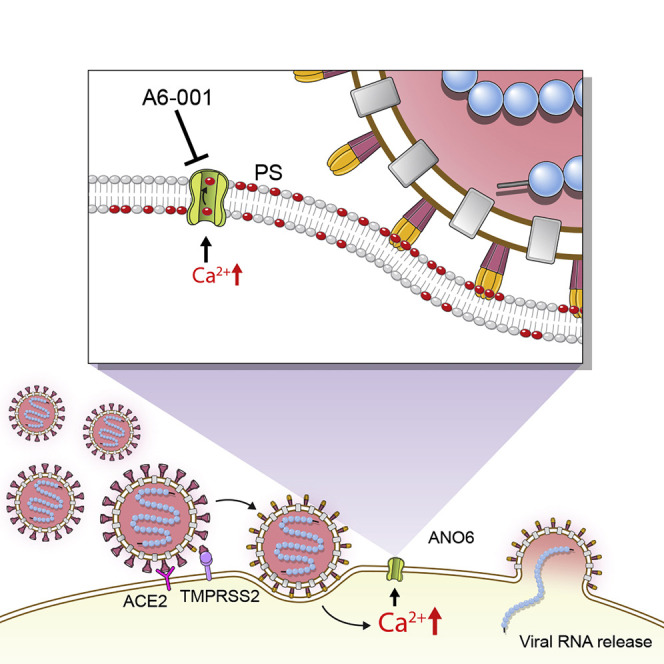
Sim et al. show that ANO6/TMEM16F-mediated phosphatidylserine scrambling participates in the SARS-CoV-2 entry into host cells and ANO6 inhibitors are effective against SARS-CoV-2 infection. These findings provide mechanistic insights into the viral entry process as well as a potential target for the development of drugs to treat COVID-19.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), belongs to the β-coronavirus genus and has the Spike (S) glycoprotein, a class I viral fusion protein, on the virion envelope (Bosch et al., 2003; V'Kovski et al., 2021). The SARS-CoV-2 S protein is the primary determinant of cell tropism and mediates binding to angiotensin-converting enzyme 2 (ACE2), the viral entry receptor on the host cells, which constitutes the initial step of the membrane fusion process (Hoffmann et al., 2020). Here, we show that SARS-CoV-2-S-induced activation of ANO6 phospholipid scramblase plays an important role in the fusion of the viral and host cell membranes.
Anoctamin 6 (ANO6), also known as TMEM16F, is a cell membrane protein functioning as a Ca2+-activated ion channel and Ca2+-dependent phospholipid scramblase (Kim et al., 2015; Kunzelmann et al., 2014; Suzuki et al., 2010; Yang et al., 2012). Phosphatidylserine (PS), an anionic phospholipid, is mostly found in the inner leaflet of the cell membrane under normal conditions. The exposure of PS at the external surface of the plasma membrane is associated with diverse physiological and pathological events, such as platelet aggregation, innate immunity, and apoptotic cell death (Amara and Mercer, 2015; Doktorova et al., 2020). In addition, a number of studies have shown that the cell surface exposure of PS is associated with various membrane-fusion events, not only between mammalian cell membranes, such as myotube formation, but also between the viral envelope and host-cell membrane, which facilitates the virus entry into host cells (Coil and Miller, 2005; Whitlock and Chernomordik, 2021; Younan et al., 2018; Zaitseva et al., 2017). This cell surface externalization of PS is mediated by the inactivation of lipid flippases that build membrane PS asymmetry or by the activation of phospholipid scramblases, such as ANO6, that enhance the translocation of anionic phospholipids (Amara and Mercer, 2015).
We initially performed a drug screen for ANO6 inhibitors, which could be used to develop antiplatelet agents. Interestingly, recent reports suggest that ivermectin, which was identified to have ANO6 inhibitory effects in our screen, has an antiviral effect against SARS-CoV-2 infections (Caly et al., 2020; Chaccour et al., 2021). Therefore, we aimed to characterize the antiviral effects of ANO6 inhibitors identified in our chemical screen. In the present study, we report that ANO6-induced PS externalization plays a critical role in the SARS-CoV-2 viral entry into the host cells. Furthermore, ANO6 phospholipid scramblase inhibition using the chemical inhibitors exhibits antiviral activities against the SARS-CoV-2 infection.
Results
High-throughput screening and characterization of ANO6/TMEM16F inhibitors
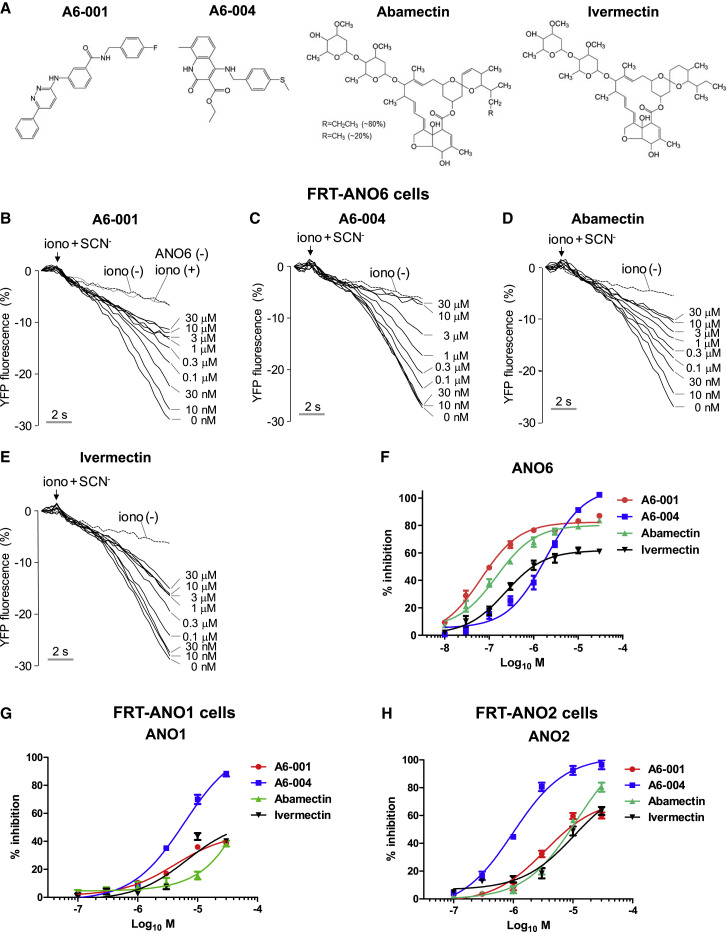
Initially, we performed a high-throughput screening of the chemical libraries composed of 56,000 drug-like small molecules to discover potential ANO6 inhibitors using a YFP fluorescence quenching-based anion channel assay with ionomycin-induced Ca2+ activation (Namkung et al., 2011). The Ca2+-activated ion channel assay in FRT cells expressing ANO6 (FRT-ANO6) (Figure S1A) identified three different structural classes of chemicals with inhibitory effects on ANO6 (Figure 1A). Among them, A6-001 and A6-004 displayed the highest potency (half-maximal inhibitory concentration [IC50] = 0.09 μM; A6-001) and maximal efficacy (100% at 30 μM; A6-004), respectively (Figures 1B, 1C, and 1F). Interestingly, ivermectin and its close chemical analog abamectin, which had been initially included in the chemical library, also exhibited inhibitory effects on the ANO6 ion channel activity (Figures 1D–1F).
Figure 1.
Characterization of ANO6/TMEM16F inhibitors
(A) Chemical structure of ANO6 inhibitors.
(B–F) ANO6 anion channel activities were measured in FRT cells expressing human ANO6 and a halide sensor YFP. Compounds were applied 10 min prior to ANO6 activation by 10 μM ionomycin (iono). Permeation of SCN− through ANO6 anion channels quenches the YFP fluorescence. Representative YFP fluorescence traces show inhibition of ANO6 activity by A6-001 (B), A6-004 (C), abamectin (D), and ivermectin (E) at the indicated concentrations. The control fluorescence quenching curve from non-transfected FRT cells with ionomycin stimulation is shown in (B) (ANO6 (−) iono (+)). A summary of the dose response of ANO6 inhibitors is shown in (F) (mean ± SEM; n = 6).
(G) A summary of the dose response of ANO6 inhibitors on the ANO1 anion channel activity is shown (mean ± SEM; n = 6). ANO1 anion channel activities were measured in FRT cells expressing human ANO1 and a halide sensor YFP. Representative YFP fluorescence traces with the indicated concentrations of A6-001, A6-004, abamectin, and ivermectin are shown in Figures S1B–S1E.
(H) A summary of the dose response of ANO6 inhibitors on the ANO2 anion channel activity is shown (mean ± SEM; n = 6). ANO2 anion channel activities were measured in FRT cells expressing human ANO2 and the halide sensor YFP. Representative YFP fluorescence traces with the indicated concentrations of A6-001, A6-004, abamectin, and ivermectin are shown in Figures S1F–S1I.
The anoctamin/TMEM16 proteins constitute a 10-membered family functioning as ion channels or lipid scramblases (Pedemonte and Galietta, 2014). Among them, ANO1, ANO2, and ANO6 are known to have calcium-activated ion channel activities. While ANO6 is permeable to both anions and cations depending on the experimental conditions, ANO1 and ANO2 are preferentially permeable to anions known as Ca2+-activated Cl− channels (CACCs) (Hartzell et al., 2009; Jung et al., 2013; Yang et al., 2008, 2012). To characterize the selectivity of the identified ANO6 inhibitors, we analyzed their effects on ANO1 and ANO2 CACCs (Figures 1G, 1H, and S1), as well as on the cytosolic Ca2+ signals and CFTR, a major Cl− channel in the respiratory epithelial cells (Figure S2). Among the four chemicals, A6-001 showed the highest ANO6 selectivity, exhibiting 460-fold and 90-fold selectivity against ANO1 and ANO2, respectively (Table S1). In contrast, A6-004 inhibited all examined ANOs with an IC50 of micromolar ranges, indicating that A6-004 is a pan-ANO inhibitor (Figures 1G, 1H, and S1B–S1I; Table S1). A6-001 did not affect ionomycin-induced cytosolic Ca2+ signals up to 30 μM; however, A6-004, abamectin, and ivermectin at 30 μM partially reduced the Ca2+ signals (Figures S2A–S2D). In addition, A6-001 up to a 30-μM concentration did not affect CFTR currents (Figures S2E and S2F). None of the four chemicals altered either cell morphology or cytoskeletal structure (Figure S2G).
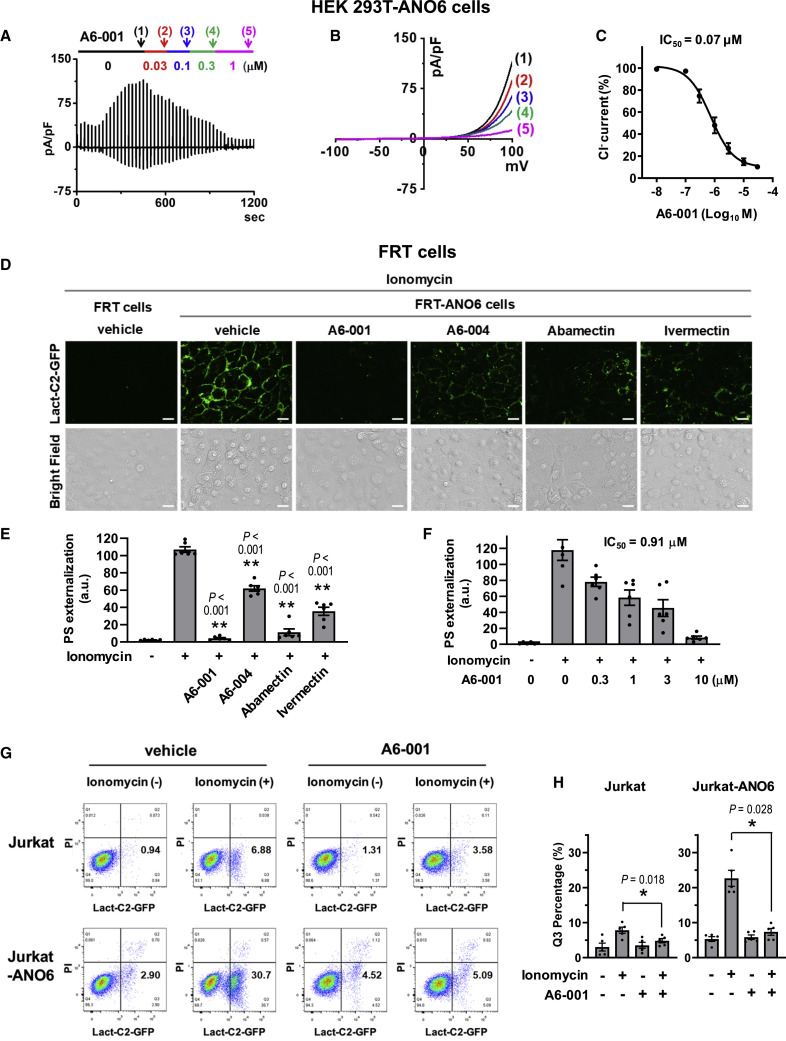
The inhibitory effect of A6-001 was further verified by whole-cell current measurements, showing that A6-001 potently inhibits ANO6 ion channel activity with an IC50 of 0.07 μM in HEK 293T-ANO6 cells (Figures 2A–2C), which is comparable to that of YFP-based assay in FRT-ANO6 cells. We then examined whether these ANO6 ion channel inhibitors also inhibit ANO6 phospholipid scramblase activity using the fluorescent-protein-labeled C2 domain of lactadherin (Lact-C2), a sensitive probe for PS (Yeung et al., 2008). At 10 μM, each of the four ANO6 ion channel inhibitors significantly reduced the ionomycin-induced PS externalization in FRT-ANO6 cells, with A6-001 being the most effective (Figures 2D and 2E). The IC50 of A6-001 for ANO6 scramblase was 0.91 μM (Figures 2F and S2H), which is approximately 10-fold larger than that for ANO6 ion channel activity. The inhibitory effect of A6-001 on PS scrambling was further verified using the four-quadrant dot blot fluorescence-activated cell sorting (FACS) analysis in Jurkat cells with exogenous ANO6 expression. The ionomycin-induced Ca2+ elevation evoked an increase in the Lact-C2-positive, propidium iodide (PI)-negative cell population, potentially due to an activation of endogenous ANO6 (see Figure S1A), which was reversed via the addition of 10 μM A6-001 (Jurkat; Figures 2G and 2H). Notably, the exogenous ANO6 expression strongly increased the Lact-C2-positive, PI-negative population in cells treated with ionomycin, and this was again reversed by 10 μM A6-001 (Jurkat-ANO6; Figures 2G and 2H).
Figure 2.
Measurements of ANO6 Ca2+-activated ion channel and phospholipid scramblase activities
(A–C) The inhibitory effect of A6-001 on the ANO6 ion channel activity was analyzed using a whole-cell patch clamp in HEK 293T cells expressing ANO6. The pipette solution contained 10 μM free Ca2+. Voltage ramps spanning a range of −100 to +100 mV were delivered from a holding potential of −60 mV every 20 s (A). The current-voltage (I-V) relationships at indicated times (1–5) with increasing concentrations of A6-001 are shown in (B). The numbers in parentheses represent the time points to measure I-V relationships. The inward currents in (A) represent ANO6 tail currents. The half-maximal inhibitory concentration (IC50) of A6-001 on the ANO6 ion channel activity is depicted in (C) (n = 4 at each concentration).
(D–F) The effects of ANO6 inhibitors on the surface exposure of phosphatidylserine (PS) were assayed in FRT-ANO6 cells. Cells were pretreated with compounds for 10 min, before ANO6 was activated with 10 μM ionomycin. Exposed PS was stained with Lact-C2-GFP. Representative fluorescence images of A6-001 (10 μM), A6-004 (10 μM), abamectin (10 μM), and ivermectin (10 μM) are shown in (D) (scale bars: 10 μm), and fluorescence intensity values calculated from Lact-C2-GFP images are summarized in (E) (mean ± SEM; n = 6). The dose responses of A6-001 on ANO6 PS scramblase inhibition are analyzed in (F) (mean ± SEM; n = 6). Representative fluorescence images of A6-001 dose response are shown in Figure S2H.
(G and H) The inhibitory effect of A6-001 on PS scrambling was assayed using the four-quadrant dot blot FACS analysis in Jurkat cells, some of which were transfected with hANO6-expressing plasmids using a NEPA21 Super Electroporator (Jurkat-ANO6). Ionomycin evoked an increase in the Lact-C2-positive and propidium iodide (PI)-negative cell population (Q3), which represented the PS-exposed and non-apoptotic cells, respectively. The ionomycin-induced Q3 increase was augmented considerably in cells expressing exogenous ANO6. A6-001 (10 μM) reversed the Q3 increases in Jurkat and Jurkat-ANO6 cells. Representative FACS analyses are shown in (G), and a summary of the multiple experiments is presented in (H) (mean ± SEM; n = 5). ∗p < 0.05 and ∗∗p < 0.01: differences from lane 2.
Data were analyzed using one-way analysis of variance, followed by Tukey’s multiple comparison test. See also Figure S2.
SARS-CoV-2 Spike evokes ANO6-dependent PS exposure and membrane fusion
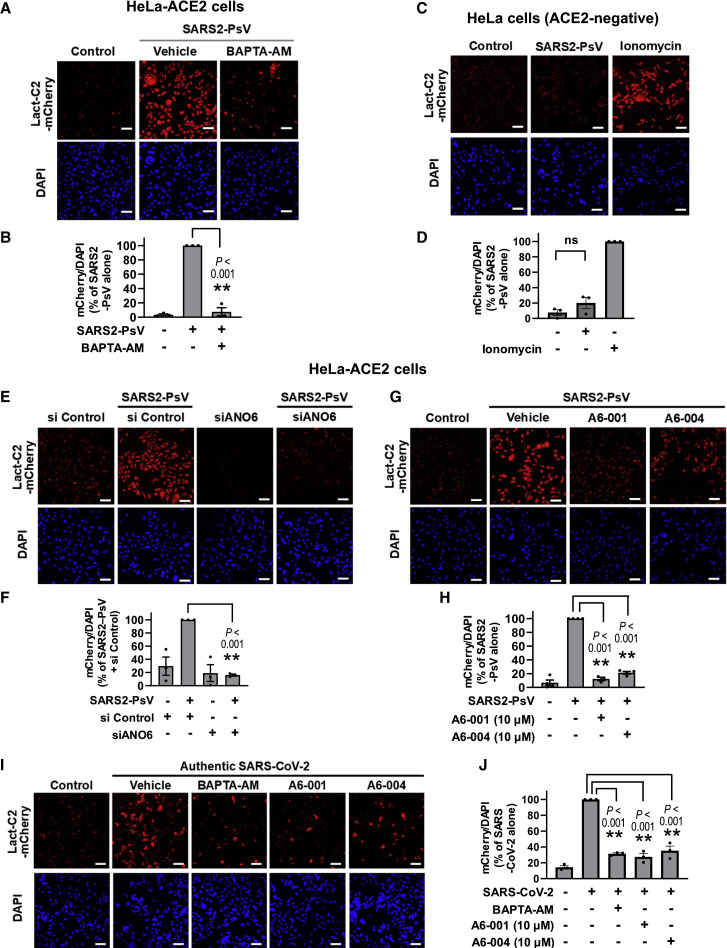
It has been shown that the binding of some enveloped viruses, such as HIV-1, to target cell induces PS externalization in the plasma membrane (Zaitseva et al., 2017). We thus next explored whether SARS-CoV-2 induces PS externalization, using the SARS-CoV-2 S pseudotyped virus (SARS2-PsV). In the ACE2-expressing HeLa (HeLa-ACE2) cells, application of a lentivirus-based SARS2-PsV evoked PS scrambling with the PS externalization being nullified by the intracellular Ca2+ chelator BAPTA-AM (Figures 3A and 3B). The same SARS2-PsV did not evoke PS scrambling in ACE2-negative cells (Figures 3C and 3D), indicating that S-ACE2 interaction is required for this to occur. Notably, the SARS2-PsV-induced PS scrambling was significantly reduced by ANO6 silencing (Figures 3E and 3F). Small interfering RNA (siRNA)-mediated ANO6 knockdown was verified by qPCR analysis (Figures S3A and S3B). This SARS2-PsV-induced, Ca2+- and ANO6-dependent PS externalization was strongly inhibited by ANO6 inhibitors, A6-001 and A6-004 (10 μM; Figures 3G and 3H). Furthermore, treatment with the authentic SARS-CoV-2 virus again evoked PS scrambling, which was in turn suppressed by BAPTA-AM and ANO6 inhibitors (Figures 3I and 3J).
Figure 3.
ANO6 is responsible for phosphatidylserine externalization evoked by pseudotyped SARS-CoV-2 S virus (SARS2-PsV)
HeLa cells expressing ACE2 (HeLa-ACE2) were incubated with a lentivirus-based SARS2-PsV (100 ng p24/mL; 20 MOI) or an authentic SARS-CoV-2 (10 MOI) for 15 min and then with Lact-C2-mCherry for 45 min.
(A and B) Chelation of cytosolic Ca2+ (BAPTA-AM; 10 μM; 1 h) inhibits SARS2-PsV-induced PS externalization. Representative images are shown in (A), and the quantification results of multiple experiments are summarized in (B) (n = 3, each from 3 to 5 fields per experimental condition).
(C and D) Control experiments in ACE2-negative HeLa cells. PS externalization was induced by ionomycin (10 μM; 10 min). Representative images are shown in (C), and the quantification results of multiple experiments are summarized in (D) (n = 3). ns, not significant.
(E and F) Silencing of ANO6 (siRNAs against ANO6; 100 nM; 24 h) inhibits the phosphatidylserine (PS) externalization evoked by SARS2-PsV. Cell nuclei were stained with DAPI. Representative images are shown in (E), and the quantification results of multiple experiments are summarized in (F) (n = 3, each from 3 to 5 fields per experimental condition).
(G and H) ANO6 inhibitors (A6-001 and A6-004, each 10 μM, 1 h) inhibit SARS2-PsV-induced PS externalization. Representative images are shown in (G), and the quantification results of multiple experiments are summarized in (H) (n = 3–4).
(I and J) Authentic SARS-CoV-2 virus evokes Ca2+- and ANO6-dependent PS externalization. The HeLa-ACE2 cells were pretreated with compounds (10 μM) for 1 h and infected with SARS-CoV-2 for 15 min. Representative images are shown in (I), and the quantification results of multiple experiments are summarized in (J) (n = 3).
Bar graph data are shown as the mean ± SEM. ∗p < 0.05 and ∗∗p < 0.01. Data were analyzed using one-way analysis of variance, followed by Tukey’s multiple comparison test. Scale bars: 50 μm. See also Figure S3.
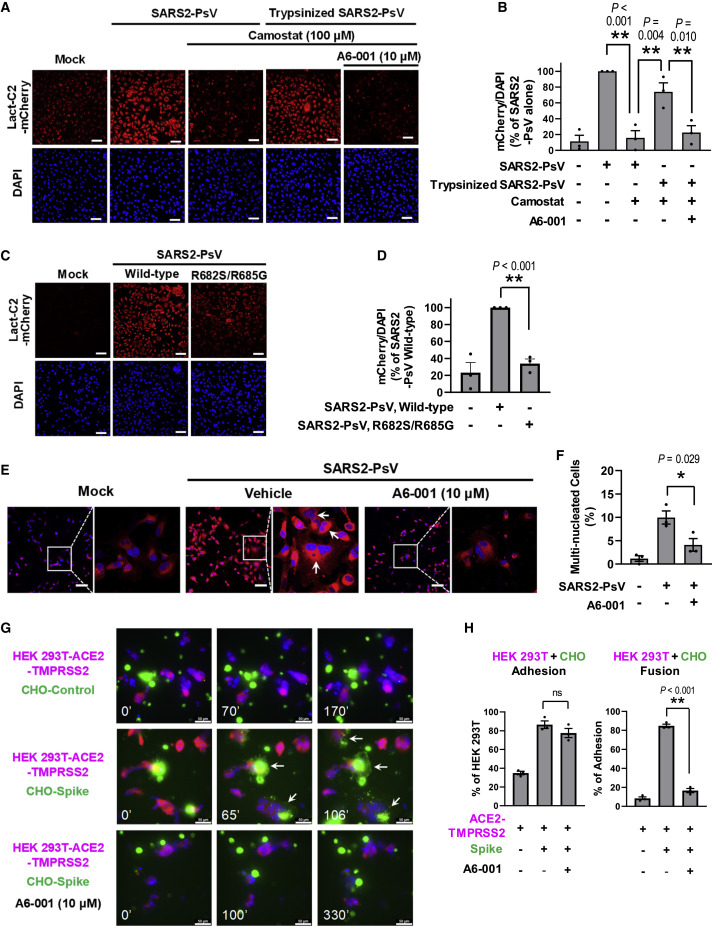
A serine protease inhibitor, camostat, prevents activation of the SARS-CoV-2 S fusion peptide (FP) by inhibiting the proteolytic cleavage of the S protein, which is mediated by TMPRSS2 and other trypsin-like enzymes (Hoffmann et al., 2020, 2021; Lee et al., 1996). Interestingly, treatment with camostat (100 μM) reduced the SARS2-PsV-induced PS externalization, yet this was reversed by the application of trypsin-pretreated SARS2-PsV (Figures 4A and 4B), suggesting that proteolytic activation of SARS-CoV-2 S contributes to the ANO6-mediated PS externalization. Camostat did not affect the PS scrambling induced by ionomycin (Figures S3C and S3D), implying that camostat reduces SARS2-PsV-induced PS scrambling by inhibiting cleaved S-mediated Ca2+ signaling. Also of note, A6-001 inhibited the PS externalization evoked by the trypsin-pretreated SARS2-PsV (Figures 4A and 4B). In addition, the R682S/R685G SARS2-PsV mutant bearing a defective protease cleavage site (S1/S2 site, Figure S3E; Belouzard et al., 2009) exhibited a significant loss in its PS-scrambling activity (Figures 4C and 4D), further implying that the proteolytic cleavage of SARS-CoV-2 S participates in the ANO6-mediated PS scrambling.
Figure 4.
Mechanisms involved in the SARS-CoV-2-Spike-mediated PS scrambling and membrane fusion
(A and B) The trypsin inhibitor camostat (100 μM; 1 h) inhibits the PS externalization evoked by SARS2-PsV, but not by trypsinized SARS2-PsV (trypsin 10 μg/mL; 10 min; 37°C). A6-001 inhibits the PS externalization evoked by trypsinized SARS2-PsV. Representative images are shown in (A), and the quantification results of multiple experiments are summarized in (B) (n = 3).
(C and D) The S1/S2 protease cleavage site defective mutation (R682S/R685G) reduces the PS-scrambling effect of SARS2-PsV. Representative images are shown in (C), and the quantification results of multiple experiments are summarized in (D) (n = 3).
(E and F) Incubation with SARS2-PsV for 30 min evokes the appearance of multinucleated cells. A6-001 inhibits the multinucleated cell formation. Representative images are shown in (E), and the quantification results of multiple experiments are summarized in (F) (n = 3).
(G and H) Time-lapse imaging of membrane fusion events (8 h; 20 frame/min) between the SARS-CoV-2-Spike-expressing CHO cells labeled with the membrane lipid probe Vybrant-DiO (CHO-Spike, green) and the cytosolic mCherry-labeled ACE2-expressing HEK 293T cells (HEK 293T-ACE2-TMPRSS2, red). The expression of Spike induces the cell-cell adhesion and membrane fusion with ACE2-expressing cells. A6-001 abolishes the Spike-ACE2-interaction-mediated membrane fusion events, while it does not affect the cell-cell adhesion. Representative images are shown in (G), and the quantification results of multiple experiments are summarized in (H) (n = 3).
Full time-lapse images are provided in Videos S1, S2, and S3. Bar graph data are shown as the mean ± SEM. ∗p < 0.05 and ∗∗p < 0.01. Data were analyzed using one-way analysis of variance, followed by Tukey’s multiple comparison test. Scale bars: 50 μm. See also Figure S3 and Videos S1, S2, and S3.
Interestingly, incubation with SARS2-PsV for 30 min resulted in the appearance of multinucleated cells, a sign of membrane fusion. Treatment with A6-001 significantly reduced the multinucleated cell formation (Figures 4E and 4F), implying that ANO6 inhibition downregulates the SARS2-PsV-induced membrane-fusion events. The membrane-fusion process comprises a series of events involving virus-host adhesion, hemifusion, and fusion pore formation. In order to more precisely analyze the S-induced membrane fusion mechanisms, we performed an 8-h time-lapse imaging of the membrane-fusion events occurring between the S-expressing cells labeled with the membrane lipid probe Vybrant-DiO (Chinese hamster ovary [CHO]-S, green) and the cytosolic mCherry-labeled ACE2-expressing cells (HEK 293T-ACE2-TMPRSS2, red; Videos S1, S2, and S3). As summarized in Figures 4G and 4H, expression of S increased cell adhesion and membrane fusion between these two types of cells. Notably, A6-001 profoundly reduced the occurrence of membrane-fusion events, despite not affecting cell-cell adhesion (Figures 4G and 4H). In time-lapse imaging of HEK 293T-ACE2-TMPRSS2/CHO-S co-cultures, all cells showing initial lipid mixing, a sign of membrane hemifusion, proceeded to develop into fully fused cells (Video S2). Interestingly, treatment with A6-001 eliminated the initial lipid-mixing event as well as the cell-cell fusion (Video S3). Collectively, these results imply that the PS-dependent stage takes place after S-ACE2 binding (virus-host adhesion) but before the membrane hemifusion process.
Viral entry of SARS-CoV-2 S pseudotyped virus is Ca2+- and ANO6-dependent
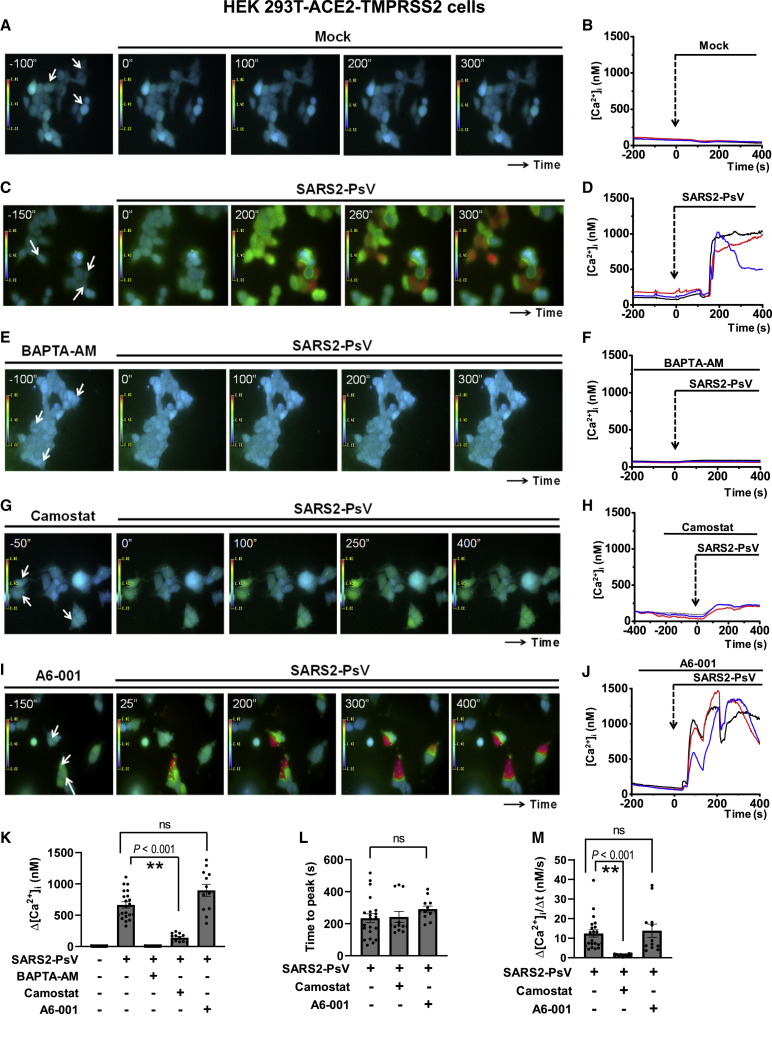
Because ANO6 is a Ca2+-dependent PS scramblase (Suzuki et al., 2010), we then investigated whether SARS-CoV-2 induces cytosolic Ca2+ signals using the ratiometric Ca2+ probe Fura-2 (Figure 5 ; Videos S4, S5, S6, S7, and S8). In the ACE2- and TMPRSS2-expressing HEK 293T (HEK 293T-ACE2-TMPRSS2) cells, application of SARS2-PsV evoked a sustained elevation of cytosolic Ca2+ levels (Figures 5C, 5D, and 5K; Video S5), which was nullified by pretreatment with the intracellular Ca2+ chelator BAPTA-AM (Figures 5E, 5F, and 5K; Video S6). Camostat significantly reduced the intensity of SARS2-PsV-induced Ca2+ signals (Figures 5G, 5H, and 5K; Video S7), implying that the proteolytic activation of S is responsible for cytosolic Ca2+ signaling in the host cells. Notably, A6-001 did not affect SARS2-PsV-induced cytosolic Ca2+ signaling in regards to Δ[Ca2+]i change, lag time (time to peak), or [Ca2+]i increase slope (Δ[Ca2+]i/Δt) (Figures 5I–5M; Video S8). Furthermore, A6-001 did not alter the purinergic agonist (ATP)-induced Ca2+ response in HEK 293T-ACE2-TMPRSS2 and Calu-3 cells (Figure S4). These results indicate that alterations in cytosolic Ca2+ signals are not responsible for the inhibitory effects of A6-001 on SARS2-PsV-induced PS scrambling.
Figure 5.
The pseudotyped SARS-CoV-2 S virus (SARS2-PsV) evokes a sustained intracellular Ca2+ elevation
[Ca2+]i was measured in HEK 293T cells expressing ACE2 and TMPRSS2 with a ratiometric Ca2+ probe (Fura-2-AM; 5 μM; 30 min). Data are presented as fluorescence emission ratios from the 340/380 nm excitation. Representative [Ca2+]i images with a rainbow scale and the [Ca2+]i values of the indicated regions (arrows) are shown.
(A and B) Mock vehicle. The 0-s time represents the point of SARS2-PsV or vehicle application.
(C and D) SARS2-PsV alone.
(E and F) BAPTA pretreatment (BAPTA-AM; 3 μM) plus SARS2-PsV.
(G and H) Camostat pretreatment (10 μM) plus SARS2-PsV.
(I and J) A6-001 pretreatment (10 μM) plus SARS2-PsV.
(K) A summary of maximum Δ[Ca2+]i is presented (three cells from each experimental replicate). Data are shown as mean ± SEM (n = 12–27).
(L) A summary of the time taken to reach the [Ca2+]i peak after SARS2-PsV application.
(M) A summary of the slope of the rising phase of [Ca2+]i transients (Δ[Ca2+]i/Δt, nM/s).
Full time-series images are provided in Videos S4, S5, S6, S7, and S8. ∗∗p < 0.01: difference from lane 2. ns, not significant. Data were analyzed using one-way analysis of variance, followed by Tukey’s multiple comparison test. See also Figure S4 and Videos S4, S5, S6, S7, and S8.
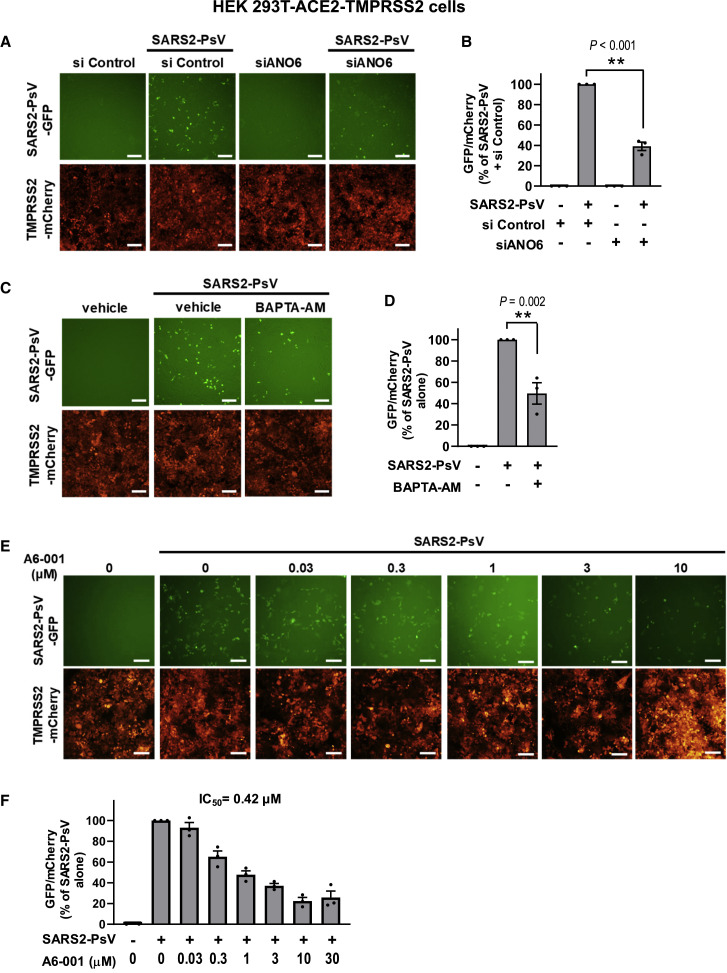
Next, the importance of PS externalization for viral infectivity was investigated via the analysis of single-round SARS2-PsV infections. Transduction with a commercially available SARS2-PsV for 48 h in HEK 293T-ACE2-TMPRSS2 cells produced a stable level of expression of the viral reporter protein GFP, which was significantly reduced by ANO6 silencing (Figures 6A, 6B, S3A, and S3B) and pretreatment with BAPTA-AM (Figures 6C and 6D), indicating that ANO6 and cytosolic Ca2+ signals are involved in the viral infection. Furthermore, treatment with A6-001 dose dependently inhibited the single-round infection with an IC50 of 0.42 μM (Figures 6E and 6F).
Figure 6.
The single-round infection of SARS2-PsV is Ca2+- and ANO6-dependent
(A and B) The single-round infection of SARS2-PsV was performed using a lentivirus-based SARS2-PsV encoding GFP. The cell viability was assessed using the mCherry fluorescence of HEK 293T-ACE2-TMPRSS2 cells. Silencing of ANO6 (siRNAs against ANO6; 100 nM; 24 h) diminished the single-round infection of SARS2-PsV (n = 3). ∗∗p < 0.01: difference from lane 2.
(C and D) Chelation of intracellular Ca2+ (BAPTA-AM; 10 μM) reduced the single-round infection of SARS2-PsV (n = 3). ∗∗p < 0.01: difference from lane 2.
(E and F) A6-001 dose dependently inhibited the single-round infection of SARS2-PsV (n = 3). A6-001 was administered during the transduction period (1-h pretreatment, 2-h incubation at 4°C, and 4-h incubation at 37°C). Then, cells were incubated without A6-001 throughout the 42-h post-transduction period. Data are shown as mean ± SEM. Data were analyzed using one-way analysis of variance followed by Tukey’s multiple comparison test. IC50, the half-maximal inhibitory concentration. Scale bars: 100 μm.
ANO6 inhibition is effective against authentic SARS-CoV-2 infections
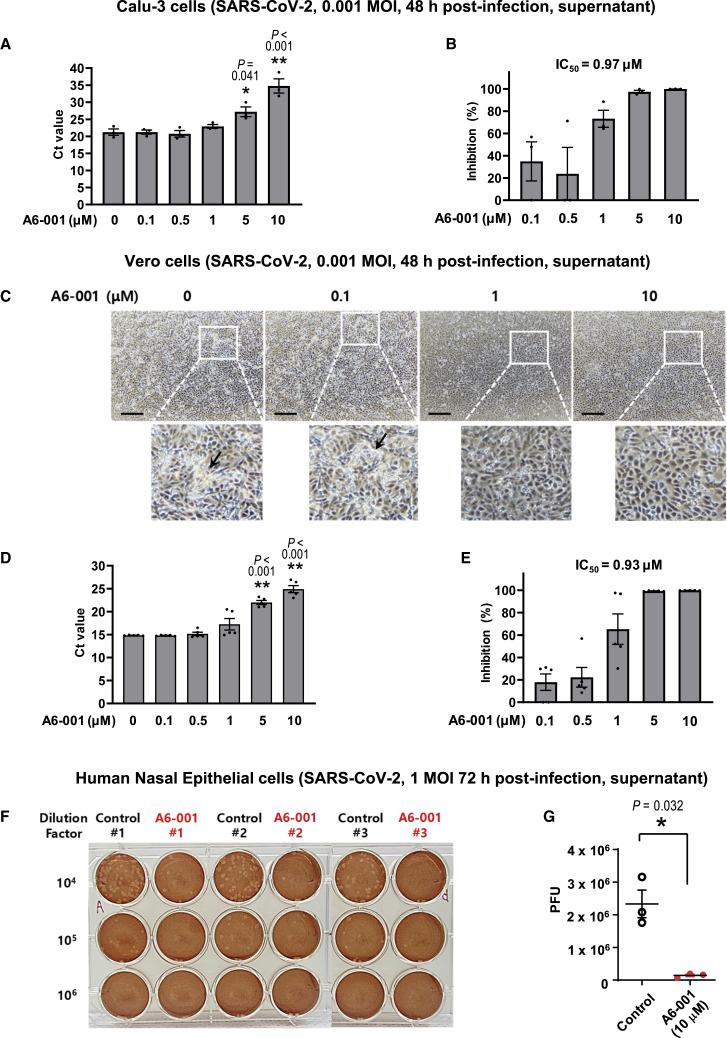
To further investigate the antiviral effects of A6-001, we performed viral replication assays using authentic SARS-CoV-2 virus. We first analyzed the antiviral effects in Calu-3 human airway epithelial cells that are TMPRSS2 positive. The qPCR quantification of virion mRNAs revealed that treatment with A6-001 (10 μM; 48 h) induced a 1/12,000 reduction in viral titers (ΔCt = −13.5; 0.001 multiplicity of infection [MOI]; Figure 7A). The IC50 value of A6-001 in Calu-3 cells was estimated as 0.97 μM (Figure 7B). SARS-CoV-2 FP activation can be achieved by several trypsin- and cathepsin-like enzymes as well as TMPRSS2 (Coutard et al., 2020). Interestingly, A6-001 exhibits antiviral activity against SARS-CoV-2 in the TMPRSS2-negative Vero cells, suggesting that ANO6 inhibition is also effective in FP activation by other enzymes (see discussion). In the light microscopic examinations, treatment with A6-001 for 48 h dose dependently reduced the virus-induced cytolysis in Vero cells (Figures 7C and S5A). A6-001 (10 μM; 48 h) induced a 1/3,500 reduction in viral titers (ΔCt −11.8; 0.001 MOI) with an IC50 value of 0.93 μM (Figures 7D and 7E). The IC50 value fell within a comparable range when cells were infected with a higher infection dose (0.01 MOI; 0.74 μM; Figure S5B).
Figure 7.
A6-001, an ANO6 inhibitor, inhibits the viral replication of SARS-CoV-2 in Calu-3, Vero, and human nasal epithelial (HNE) cells
(A and B) Viral replication of authentic SARS-CoV-2 (0.001 MOI) was assayed in Calu-3 cells. The qPCR results of virion mRNAs with the indicated concentrations of A6-001 are shown in (A) (∗p < 0.05 and ∗∗p < 0.01: difference from 0 μM; one-way analysis of variance followed by Tukey’s multiple comparison test), and the IC50 values of A6-001 at 48 h post-infection are presented in (B) (n = 3 at each concentration).
(C–E) Viral replication of SARS-CoV-2 (0.001 MOI) was assayed in Vero cells. In the light microscopic analyses, A6-001 dose dependently reduced the virus-induced cytolysis (C) (48 h post-infection; five independent experiments showed similar results; arrows indicate cytolytic cells). The qPCR results of virion mRNAs with the indicated concentrations of A6-001 are shown in (D) (∗∗p < 0.01: difference from 0 μM; one-way analysis of variance followed by Tukey’s multiple comparison test), and the IC50 values of A6-001 at 48-h post-infection are presented in (E) (n = 5 at each concentration). Scale bars: 200 μm.
(F and G) Viral replication of SARS-CoV-2 (1 MOI) was assayed in HNE cells. The passage no. 2 HNE cells were cultured under air-liquid interface conditions and apically infected with SARS-CoV-2. A6-001 (10 μM) was administered to the basolateral compartments. Representative plaque assay results at 72-h post-infection are shown in (F), and the summary of multiple experiments is presented in (G) (n = 3; ∗p < 0.05; two-tailed Student’s t test). Bar graph data are shown as the mean ± SEM. See also Figures S5–S7.
We then analyzed the antiviral effects of A6-001 using the side-by-side parallel data for RNA accumulations and output virus titers by plaque assay. In Vero cells infected with a higher infection dose of SARS-CoV-2 (0.01 MOI), the plaque assay results indicated that A6-001 (10 μM; 48 h) induced a 1/500 viral reduction (9.8 × 106 versus 2.0 × 104 plaque-forming units [PFUs]/mL), while the qPCR results represented a 1/150 reduction (9.6 × 106 versus 6.4 × 104 PFUs/mL; Figures S5C and S5D). A lower viral titer reduction rate in the qPCR analysis was also observed in the case of experiments using Calu-3 cells infected with an increased viral dose (0.01 MOI). While A6-001 (10 μM; 48 h) induced a 1/14,000 viral reduction in the plaque assay, the same A6-001 evoked a 1/50 reduction in the qPCR analysis (Figure S5E). The contamination by non-viable virus genomes in the cell culture supernatant appears to be a reason for the lower viral titer reduction rate in the qPCR analysis, since qPCR also detects non-viable viruses (Figure S5F). Notably, the A6-001-induced viral reduction rate in the qPCR analysis in Calu-3 cells with a low infection dose (0.001 MOI, a 1/12,000 reduction; Figure 7A) was closely comparable to that in the plaque assay with a higher infection dose (0.01 MOI, a 1/14,000 reduction; Figure S5E). A6-001 did not affect replications of the non-enveloped adenovirus (Figure S6A). The half-maximal cytotoxic concentration (CC50) of A6-001 was calculated as >100 μM in both Calu-3 and Vero cells (Figure S7), indicating that the safety margin of A6-001 is >100.
Lastly, the antiviral effects of A6-001 on SARS-CoV-2 were analyzed in primary cultures of human nasal epithelial (HNE) cells (Yoon et al., 1999). The passage no. 2 HNE cells were cultured under air-liquid interface conditions to retain airway epithelial properties, including endogenous expressions of ANO1 and ANO6 (Figure S3A). Notably, treatments with A6-001 (10 μM) to the basolateral compartment of HNE cells for 72 h, in which the apical side was infected with authentic SARS-CoV-2 virus, induced an average reduction of 94% in viral replications when determined by the plaque assay (Figures 7F and 7G).
Discussion
In the present study, we show that SARS-CoV-2 S evokes cytosolic Ca2+ signals and a subsequent activation of ANO6-mediated PS externalization in the target cells, which facilitates fusion between the viral envelope and target cell membrane. Furthermore, treatment with an ANO6-selective inhibitor resulted in antiviral effects against SARS-CoV-2 infection. These findings provide mechanistic insight into the viral entry process as well as a potential target for the development of drugs to treat SARS-CoV-2 infection. The potent ANO6 inhibitor, A6-001, and its derivatives optimized for higher potency and lower toxicity would be candidates for the treatment of COVID-19.
Interestingly, the ANO6 inhibitors identified in this study bore differential effects on ANO protein functions. For example, while A6-001 primarily inhibited ANO6, A6-004 potently inhibited ANO1 and ANO2 as well as ANO6 (Table S1). In addition, A6-004 was less efficacious in inhibiting PS scrambling than it was at inhibiting ANO6 ion channel current, whereas abamectin appeared to be relatively efficacious in inhibiting PS scrambling (Figures 1 and 2). Recent structural analyses have suggested that the ion channel and lipid-scrambling functions of ANO6/TMEM16F are mediated either by alternate protein conformations or separate pathways, despite both functions being activated by a common mechanism (Alvadia et al., 2019; Feng et al., 2019). Therefore, a plausible explanation would be that binding of inhibitors to the different regions of ANO6 may induce differential effects. For example, A6-004 may bind to the common ion channel pore region, which is conserved also in ANO1 and ANO2, while abamectin may bind to a region that is more critical to the scramblase activity. Exploration of these differences in future studies should contribute to the development of individual function- and paralog-specific inhibitors.
The exponential decay of multiple-round viral entries appears to be primarily responsible for the potent effects of A6-001 on authentic virus infection in the 48-h treatment (3,500- to 14,000-fold reduction; Figures 7 and S5). Application of A6-001 (10 μM) during the infection period alone yielded similar cellular viral reductions in authentic virus (86%; ΔΔCt = 2.84; Figure S6B) and pseudotyped virus (78%; Figure 6F), suggesting little difference between the two virus types in the effects of A6-001 on viral entry. Interestingly, A6-001 treatment for 4 h post-infection induced a 98% reduction in cellular viral RNAs for authentic virus infection (ΔΔCt = 5.58; Figure S6B), which corresponds to the overall reduction rate when the in vitro SARS-CoV-2 infecting time (time required for a single virus-producing cell to infect one more) is 2–4 h. In fact, the infecting time of Ebola virus, an enveloped RNA virus, has been estimated to be as short as 2 h (Liao et al., 2020). However, the in vitro replication time of SARS-CoV-2 appeared to be longer than this and was estimated as 6–8 h (Eymieux et al., 2021). Therefore, A6-001 may also exhibit antiviral effects against SARS-CoV-2 at the post-entry stages, although it did not affect replications of the non-enveloped adenovirus (Figure S6A).
Since the initial discovery in the 1970s that PS is involved in the membrane fusion of phospholipid vesicles (Papahadjopoulos et al., 1974), a number of studies have reported a role of exofacial PS in diverse biomembrane fusion events. The cell surface exposure of PS is associated with various types of mammalian cell fusion, such as the multinucleated cell formation of myoblasts and osteoclasts, fertilization of sperm and oocytes, and cancer cell fusion and progression (Whitlock and Chernomordik, 2021). Furthermore, exofacial PS on either the viral envelope or the target cell membrane is required for the viral entry of many enveloped viruses, such as the Ebola, HIV-1, and vesicular stomatitis viruses (Coil and Miller, 2005; Younan et al., 2018; Zaitseva et al., 2017). The PS-induced alterations in the biophysical properties of a membrane, including changes in lipid packing, lipid bilayer curvature, and membrane fluidity, may contribute to the fusion events (Coil and Miller, 2005). However, the negative charge of PS itself is thought to be an inhibitory factor in biomembrane fusion (Tarafdar et al., 2012). Instead, protein factors, such as PS-binding proteins and PS receptors, appear to play a major role in most biomembrane fusion events. For example, the exofacial PS may trigger fusogenic restructuring of protein fusogens or direct the assembly of protein machinery at the site of fusion (Whitlock and Chernomordik, 2021). PS on target cell membranes has been suggested to draw out positively charged regions of the fusogens of dengue virus, vesicular stomatitis virus (VSV), and HIV-1 to facilitate their fusogenic restructuring and oligomerization (Whitlock and Chernomordik, 2021). Therefore, it is possible that PS may interact with positively charged regions around SARS-CoV-2 FP or Ca2+ ions that bind to FP (Lai et al., 2017), in order to promote fusogenic reconstruction of the S protein.
In the plasma membrane of dead and dying cells, externally exposed PS can serve as “eat-me” signals for phagocytes to clear dead cells via phagocytosis. Clearance of apoptotic cells is initiated by engagement of exposed PS with PS receptors, such as TIM1, on the phagocyte membrane and has the physiological function of preventing autoimmune reactions and inflammation (Amara and Mercer, 2015). Interestingly, many enveloped viruses, such as HIV-1, vaccinia virus, dengue virus, and Ebola virus, expose PS on their viral envelope and associate with PS receptors on the target cells to facilitate viral entry without evoking immune reactions, which is known as viral apoptotic mimicry (Amara and Mercer, 2015). It has been suggested that Ebola virus envelopes acquire PS via ANO6 scramblase activation during viral budding from the host cells (Younan et al., 2018). However, it is unknown whether SARS-CoV-2 viral envelopes are rich in PS. In addition, it has been shown that SARS-CoV-1 does not use viral apoptotic mimicry for its infection process (Jemielity et al., 2013). Therefore, it is unlikely that the apoptotic mimicry-related PS receptors expressed on the host cell membrane play a dominant role in the fusion between the SARS-CoV-2 envelope and the host cell membrane. Instead, PS receptors may play a major role in the SARS2-PsV-induced fusion between cell membranes of HeLa-ACE2 cells (Figures 4E and 4F). While the present study was in the submission process, a manuscript was published online suggesting a potential role of ANO/TMEM16 proteins in the SARS-CoV-2-S-induced syncytia formation (Braga et al., 2021). Although this paper also expands on the role of ANO6/TMEM16F protein in SARS-CoV-2 infection and related pathologies, it differs from our study in terms of the therapeutic potentials and mechanistic details. Among the chemicals that they investigated, only niclosamide had a partial inhibitory effect on the endogenous Cl− currents in HEK 293 cells, which were in part reduced by the silencing of ANO6. The mechanisms for the remaining chemicals, such as salinomycin and clofazimine, remain unknown since they did not affect the Cl− channel activities (Braga et al., 2021). In addition, niclosamide reduced the cytosolic Ca2+ signals, indicating that niclosamide is not an ANO6-specific inhibitor. Nonspecific inhibition of Ca2+ signaling and other ion channels could be associated with undesired effects during the clinical application.
Although the TMPRSS2-dependent direct fusion of the viral envelope with the cellular membrane appears to be a major viral entry route in the human respiratory tract, SARS-CoV-2 can also enter into cells via the cathepsin-dependent endocytic pathway (Ou et al., 2021). Camostat inhibits TMPRSS2 and many other trypsin-like serine proteases (Hoffmann et al., 2021; Lee et al., 1996). The finding that camostat reduced the SARS2-PsV-induced cytosolic Ca2+ signaling in HEK 293T-ACE2-TMPRSS2 cells indicates the importance of the S protein cleavage for Ca2+-induced activation of ANO6 (Figures 5G and 5H). Interestingly, camostat also inhibited SARS2-PsV-induced PS externalization in HeLa-ACE2 cells (Figures 4A and 4B). An earlier study reported that camostat inhibits the SARS-CoV-2 cell entry in HeLa-ACE2 cells, and at that time, the authors interpreted the results as suggesting that HeLa cells may endogenously express TMPRSS2 (Shang et al., 2020). However, HeLa cells are TMPRSS2 negative in RNA expression datasets of the Human Protein Atlas (https://www.proteinatlas.org/ENSG00000184012-TMPRSS2/cell). In addition, the ANO6 inhibitor A6-001 inhibited the SARS-CoV-2 replication in Vero cells that were also TMPRSS2 negative (Figures 7C–7E). A plausible explanation for these findings is that cleavage of the S protein by other peptidases at the cell surface may contribute to the ANO6 activation. In fact, a number of enzymes at the cell surface and extracellular space, such as transmembrane serine proteases other than TMPRSS2 (i.e., TMPRSS11D, and TMPRSS13), trypsin, HAT, elastase, and ADAM17, are shown to induce the cleavage of SARS-CoV-1 and SARS-CoV-2 S proteins (Hoffmann et al., 2021; Lu et al., 2015). Alternatively, the cathepsin-mediated cleavage may also induce ANO6 activation. Therefore, it will be intriguing to examine whether the endocytosed SARS-CoV-2 can evoke a cathepsin-dependent ANO6 activation in future investigations.
An additional point to discuss is that the cell-surface exposure of PS is a well-identified signal for initiating an inflammatory response and platelet aggregation (Bevers and Williamson, 2016). Critically ill patients with COVID-19 often suffer from severe inflammation known as cytokine storm and blood coagulopathy defined as disseminated intravascular coagulation, which are the leading causes of morbidity and mortality (Connors and Levy, 2020). Therefore, it is expected that reduced PS externalization via ANO6 inhibition in platelets and immune cells may alleviate the coagulopathy and cytokine storm associated with SARS-CoV-2 infections. A thorough in vivo examination studying the effectiveness of ANO6 inhibitors at the various stages of SARS-CoV-2 infection would provide further information on whether ANO6 inhibitors are effective not only in inhibiting viral replications but also in ameliorating severe inflammation and coagulopathy.
Limitations of the study
Although we have adopted a multidisciplinary approach to verify the role of ANO6 in the SARS-CoV-2 viral infection, some limitations remain. First, we have focused on the TMPRSS2-dependent viral entry mechanism in this study. However, SARS-CoV-2 can also enter into cells via other pathways, particularly via the cathepsin-dependent endocytic pathway. The results in TMPRSS2-negative Vero cells (Figures 7C and 7D) indicate that ANO6 is also involved in the TMPRSS2-independent process, whose mechanisms need to be further explored. Second, this study did not determine the A6-001 binding regions of ANO6. The identification of inhibitor-binding regions of A6-001 and other agents will contribute not only to the development of a highly PS scramblase-specific inhibitor but also to greater understanding of the ion- and PS-transporting mechanisms of ANO6. Lastly, in vivo pharmacokinetic analyses of A6-001 and its chemical analogs are required for the development of antiviral agents that are effective in human SARS-CoV-2 infections.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-ANO6 antibody | This study | Epitope from Yang et al., 2012 |
| Anti-SARS-CoV/SARS-CoV-2 (COVID-19) spike antibody [1A9] | GeneTex | Cat#GTX632604; RRID: AB_2864418 |
| Anti-Aldolase A (C-10) | Santa Cruz | Cat#SC-390733 |
| Anti-HIV-p24 antibody | GeneTex | Cat#GTX64128 |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s modified Eagle’s medium-high glucose | Gibco | Cat#11995-065 |
| DMEM/Ham's F-12 (1:1) medium | WELGENE | Cat#LM 001-05, Cat#LM 010-02 |
| L-glutamine | WELGENE | Cat#LS 002-01 |
| Fetal Bovine Serum (FBS) | Gibco | Cat#26140-079 |
| Penicillin-streptomycin | Gibco | Cat#15140122 |
| RPMI 1640 with L-Glutamine | Capricorn Scientific | RPMI-A |
| Opti-MEM™ Reduced Serum Medium | Gibco | Cat#31985-070 |
| Minimum Essential Medium (MEM) with Earle’s Balanced Salts containing 2.2 g/L sodium bicarbonate | HyClone | Cat#SH30024.01 |
| Fetal Bovine Serum (FBS) | HyClone | Cat#SH30084.03 |
| Penicillin-streptomycin | HyClone | Cat#SV300010 |
| Poly-D-lysine Hydrobromide | Sigma-Aldrich | Cat#P6407 |
| 8-well Cell Culture Chamber slide | SPL Life Science | Cat#30108 |
| BEBM™ Bronchial Epithelial Cell Growth Basal Medium | Lonza | Cat#CC-3171 |
| Dulbecco’s Modified Eagle Medium (DMEM) with 1.0 g/L glucose and without L-Glutamine | Lonza | Cat#12-707F |
| BEGM™ Bronchial Epithelial Cell Growth Medium SingleQuots™ Supplements and Growth Factors | Lonza | Cat#CC-4175 |
| Bovine Serum Albumin | Sigma-Aldrich | Cat#A7030-50G |
| 12mm Transwell® with 0.45μm Pore Polyester Membrane Insert, Sterile | Corning | Cat#3460 |
| Costar® 12-well Clear TC-treated Multiple Well Plates, Individually Wrapped, Sterile | Corning | Cat#3513 |
| SmartTM library | Chemdiv | Cat#903OTP09 |
| 1,700 approved drugs | TargetMol | Cat#L1000 |
| A6-001 | Chemdiv | Cat#F575-0482 |
| A6-004 | Chemdiv | Cat#F474-2423 |
| Fluo-4 NW | Invitrogen | Cat#F36206 |
| Ionomycin | Alomone Labs | Cat#I-700 |
| Cisplatin | Tokyo chemical industry | Cat#D3371 |
| Amphotericin B solution | Sigma-Aldrich | Cat#A2942 |
| Fura-2 AM | Invitrogen | Cat#F1201 |
| cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail | Sigma-Aldrich, Roche | Cat#04693159001 |
| IPTG | Duchefa | Cat#I1401.0025 |
| Ni-NTA protein purification system | QIAGEN | Cat#30230 |
| Formaldehyde solution for molecular biology, 36.5–38% in H2O | Sigma-Aldrich | Cat#F8775 |
| Paraformaldehyde | Biosesang | Cat#PC2031-050-00 |
| Triton X-100 | Biosesang | Cat#T1020 |
| Bovine Serum Albumin | Millipore | Cat#82-100-6 |
| Alexa Fluor 488 Phalloidin | Invitrogen | Cat#A12379 |
| DAPI | Sigma-Aldrich | Cat#D9542 |
| Fluorescence Mounting Medium | Dako | Cat#S3023 |
| VECTASHIELD mounting medium with DAPI | Vector Laboratories | Cat#H-1500 |
| TransIT-X2 Dynamic Delivery System | Mirus Bio LLC | Cat#MIR6006 |
| Polybrene | Merck-Millipore | Cat#TR-1003-G |
| Blasticidin S HCl | Gibco | Cat#A1113903 |
| Puromycin dihydrochloride | Gibco | Cat#A1113803 |
| Hygromycin B | Invitrogen | Cat#10687010 |
| Abamectin | Cayman Chemical | Cat#19201 |
| Ivermectin | Sigma-Aldrich | Cat#I8898 |
| Camostat mesylate | Sigma-Aldrich | Cat#SML0057 |
| BAPTA-AM | Sigma-Aldrich | Cat#A1076 |
| Trypsin from porcine pancreas | Sigma-Aldrich | Cat#T6567 |
| SeaPlaque™ Agarose | Lonza | Cat#50101 |
| Neutral Red Solution | Sigma-Aldrich | Cat#2889 |
| (2-Hydroxypropyl)-beta-cyclodextrin | Sigma-Aldrich | Cat#H107-5G |
| Lipofectamine® 2000 | Invitrogen | Cat#11668-019 |
| 16:0-06:0 NBD PS | Avanti Polar Lipids | Cat# 810192 |
| Lenti-X™ Concentrator | Clontech Laboratories | Cat#631232 |
| Vybrant™ DiO Cell-Labeling Solution | Invitrogen | Cat#V22886 |
| Propidium Iodide | Abcam | Cat#ab14083 |
| Critical commercial assays | ||
| BioFACT™ Pfu DNA Polymerase | BioFACT | Cat#PD116-250 |
| BioFACT™ Taq DNA Polymerase | BioFACT | Cat#ST116-500 |
| NEBuilder® HiFi DNA Assembly Master Mix | NEB Biolabs | Cat#E2621 |
| Tri-RNA reagent | Favorgen Biotech | Cat#FATRR 001 |
| RNA to cDNA EcoDry Premix | Takara Bio | Cat#639549 |
| TB Green™ Premix Ex Taq™ (Tli RNaseH Plus) | Takara Bio | Cat#RR420L |
| Pierce™ BCA Protein Assay Kit | Thermo Scientific | Cat#23227 |
| CellTiter 96® AQueous One Solution Cell Proliferation Assay kit | Promega | Cat#G3581 |
| Cell counting kit-8 | Dojindo Molecular Technologies Inc. | Cat#CK04 |
| QIAamp Viral RNA Mini kit | QIAGEN | Cat#52906 |
| QIAamp DNA mini kit | QIAGEN | Cat#51304 |
| Luna® Universal One-Step RT-qPCR Kit | NEW ENGLAND BioLabs®Inc | Cat#E3006E |
| Luciferase assay system | Promega | Cat#E1501 |
| KAPA SYBR FAST qPCR Kits | Sigma-Aldrich | Cat#KK4605 |
| Experimental models: Cell lines | ||
| CHO-K1 | ATCC | CCL-61 |
| HeLa | ATCC | CCL-2 |
| HEK293T | ATCC | CRL-3216 |
| HeLa-ACE2 | This study | N/A |
| FRT-YFP(H148Q/I152L/F46L)-ANO1 | This study | N/A |
| FRT-YFP(H148Q/I152L/F46L)-ANO2 | This study | N/A |
| FRT-YFP(H148Q/I152L/F46L)-ANO6 | This study | N/A |
| Vero | Ministry of Food and Drug Safety, Republic of Korea | VERO01WCB-1201 |
| Calu-3 | Korean Cell Line Bank | KCLB #30055 |
| HEK293T-ACE2-TMPRSS2 (mCherry-expressing) | Genecopoeia | Cat#SL222 |
| Human nasal epithelial (HNE) cells | This study | N/A |
| Jurkat | Korean Cell Line Bank | KCLB #40152 |
| Experimental models: Bacterial and virus strains | ||
| HIT Competent Cells-BL21(DE3) strains (Value 107) | RBC | Cat#RH217 |
| HIT Competent Cells-DH5 alpha strains (Value 108) | RBC | Cat#RH617 |
| Lentifect™ SARS-CoV-2 ER retention signal removed Spike-pseudotyped Lentivirus | Genecopoeia | Cat#SP001 |
| SARS-CoV-2 (BetaCoV/korea/KUMC-2) | Clinical isolate | GISAID accession#: EPI_ISL_413018 |
| Recombinant eGFP adenovirus (Ad-eGFP) | Vector Biolabs | Cat#1060 |
| Oligonucleotides | ||
|
AccuTargetTM pre-designed human TMEM16F-specific siRNA : 5′-GAG AAC AGC CUC ACC AUG A-3′ and 5′-UCA UGG UGA GGC UGU UCU C-3′ |
Bioneer | 196527-1 |
|
AccuTargetTM pre-designed human TMEM16F-specific siRNA : 5′-CAG CUU UCC CAC UCC AUG A-3′ and 5′-UCA UGG AGU GGG AAA GCU G-3′ |
Bioneer | 196527-2 |
| AccuTargetTM pre-designed human TMEM16F-specific siRNA: 5′-CAC GAU GUA CUC ACG UAG U-3′ and 5′-ACU ACG UGA GUA CAU CGU G-3′ | Bioneer | 196527-3 |
| hGAPDH qPCR forward primer: 5′-AAT CCC ATC ACC ATC TTC CA-3′ | This study | N/A |
| hGAPDH qPCR reverse primer: 5′-TGG ACT CCA CGA CGT ACT CA-3′ | This study | N/A |
| hTMEM16A qPCR forward primer: 5′-AGA GGA GGC TGT CAA GGA TCA-3′ | This study | N/A |
| hTMEM16A qPCR reverse primer: 5′- ATA TGC CGG CGC TTC TCT TT -3′ | This study | N/A |
| hTMEM16F qPCR forward primer: 5′-AGA AAA GAT AAC CCC ACG AT-3′ | This paper | N/A |
| hTMEM16F qPCR reverse primer: 5′-GAC GAA CCC AAA CTG AAT AA-3′ | This paper | N/A |
| SARS-CoV-2 nsp targeting forward primer: 5′- TGG GGY TTT ACR GGT AAC CT-3′ |
Chu et al., 2020 | N/A |
| SARS-CoV-2 nsp targeting reverse primer: 5′- AAC RCG CTT AAC AAA GCA CTC -3′ | Chu et al., 2020 | N/A |
| probe 5′-56-FAM- TAGTTGTGA/ ZEN/TGCWATCATGACTAG-3IABkFQ-3′ |
Chu et al., 2020 | N/A |
| (Ad-)eGFP qPCR forward primer: 5′-AGT CCG CCC TGA GCA AAG A | SignaGen® Laboratories |
N/A |
| (Ad-)eGFP qPCR reverse primer: 5′- TCC AGC AGG ACC ATG TGA TC | SignaGen® Laboratories |
N/A |
| Recombinant DNA | ||
| Plasmid: pcDNA3.1-ANO6(V5) | Lin et al., 2019 | N/A |
| Plasmid: pcDNA3.1-hACE2 | Addgene | #145033 |
| Plasmid: pcDNA3.1-SARS2-Spike | Addgene | #145032 |
| Plasmid: pcDNA3.1-SARS2-Spike-R682SR685G | This study | Modified from Addgene #145032 |
| Plasmid: lentiCas9-Blast | Addgene | #52962 |
| Plasmid: pLV-mcherry | Addgene | #36084 |
| Plasmid: psPAX2 | Addgene | #12260 |
| Plasmid: pMD2.G | Addgene | #12259 |
| Plasmid: Lact-C2-GFP | Addgene | #22852 |
| Plasmid: pET28c(+) | Novagen | #69866 |
| Software and algorithms | ||
| SnapGene (Version 2.3.2) | Snapgene | N/A |
| Primer Blast (A tool for finding specific primers) | NCBI | https://www.ncbi.nlm.nih.gov/tools/primer-blast/ |
| MetaFluor | Molecular Devices | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy |
| Meta Morph microscopy analysis software (version 7.1) | Molecular Devices | https://www.moleculardevices.com/ |
| pCLAMP software v.10.7 | Molecular Devices | https://www.moleculardevices.com/products/axon-patch-clamp-system/acquisition-and-analysis-software/pclamp-software-suite#gref |
| Digidata-1440A | Molecular Devices | https://www.moleculardevices.com/products/axon-patch-clamp-system |
| Clampfit v. 10.7 | Molecular Devices | https://www.moleculardevices.com/products/axon-patch-clamp-system |
| Origin v. 8.0 | Origin Lab Inc. | https://www.originlab.com/index.aspx?go=PRODUCTS/Origin |
| Python3 | Python Software Foundation | https://www.python.org/downloads/ |
| OpenCV | Open Source Computer Vision Library | https://opencv.org/ |
| NIS-elements AR | Nikon | |
| ZEN 2012 black (2012 version) | Carl Zeiss | https://www.zeiss.com/microscopy/ |
| Graph Pad Prism (version 8) | Graph Pad Software | https://www.graphpad.com/scientific-software/prism/ |
| Fiji: an open-source platform for biological-image analysis | http://fiji.sc/ | N/A |
| SoftMax Pro v 5.4 | Molecular Devices | https://www.moleculardevices.com/products/microplate-readers/acquisition-and-analysis-software/softmax-pro-software |
| Leica Application Suite X software | Leica Microsystems | https://www.leica-microsystems.com/products/light-microscopes/p/leica-las-x-widefield-systems/ |
| Flowjo (version 10) | Tree Star Inc. | https://www.flowjo.com/solutions/flowjo/downloads |
| Other | ||
| FLUOstar Omega microplate reader | BMG Labtech | https://www.bmglabtech.com/fluostar-omega/ |
| Lionheart FX Automated Microscope | BioTek | https://www.biotek.com/products/imaging-microscopy-automated-cell-imagers/lionheart-fx/ |
| Infinite M200 microplate reader | Tecan | https://lifesciences.tecan.com/plate_readers/infinite_200_pro |
| Zeiss LSM780, confocal laser scanning microscope | Carl Zeiss | https://www.zeiss.com/microscopy/ |
| Zeiss Axio microscope | Carl Zeiss | https://www.zeiss.com/microscopy/ |
| Ultra-fast switching monochromator- Polychrome V | TILL Photonics | https://www.labwrench.com/manufacturer/1363/till-photonics |
| ECLIPSE Ti-2 microscope | Nikon | https://www.nikon.com/about/technology/design/works/eclipse_ti2.htm |
| Patch-clamp amplifier | Molecular Devices | https://www.moleculardevices.com/products/axon-patch-clamp-system |
| Applied Biosystem StepOne System | Applied Biosystems | https://www.fishersci.ca/shop/products/stepone-real-time-pcr-system-4/4376357 |
| Applied Biosystem Quantstudio 3 detection system | Applied Biosystems | https://www.thermofisher.com/kr/ko/home/life-science/pcr/real-time-pcr/real-time-pcr-instruments/quantstudio-3-5-real-time-pcr-system.html |
| Tissue Lyzer II | Qiagen | Cat#85300 |
| Centro XS3 Luminescence Microplate Reader LB 960 | EG & G Berthold | https://www.berthold.com/en/bioanalytic/products/microplate-readers/centro-lb963/ |
| THUNDER Imager Live Cell | Leica Microsystems | https://www.leica-microsystems.com/products/thunder-imaging-systems/ |
| NEPA21 Super Electroporator | Nepa Gene Co. | http://www.nepagene.jp/e_products_nepagene_0001.html |
| BD FACSCelesta™ cell Analyzer | BD Biosciences | https://www.bdbiosciences.com/ko-kr/products/instruments/flow-cytometers/research-cell-analyzers/facscelesta |
| Deposited data | ||
| Raw datasets and additional supplemental items | This study | https://doi.org/10.17632/8vgrh6dyfp.1 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Min Goo Lee (mlee@yuhs.ac).
Materials availability
All unique/stable materials and reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Cell lines
FRT cells were stably transfected with both a yellow fluorescent protein (YFP) variant (H148Q/I152L/F46L) and human ANO1 (abc isoform), ANO2 or ANO6 (variant 5), and the cells were cultured in DMEM/Ham’s F-12 (1:1) medium with 10% FBS, 2 mM L-glutamine, 100 units/mL penicillin and 100 μg/mL streptomycin at 37°C and 5% CO2. HEK293T cells and HeLa cells were obtained from the American Type Culture Collection, Manassas, VA, USA. For the generation of stable ACE2 overexpressing HeLa cells, hACE2 and P2A-BSD encoding sequences were amplified from pcDNA3.1-hACE2 (Addgene #145033) and lentiCas9-Blast (Addgene #52962), respectively. The amplicons were cloned into the lentiviral pLV-mCherry vector using a NEBuilder HiFi DNA Assembly Kit (NEB Biolabs). HeLa cells were transduced with lentiviral particles encoding hACE-P2A-BSD in the presence of 8 μg/mL of polybrene (Merck-Millipore, Burlington, MA, USA) for one day. Cells were maintained at a low density in the presence of 10 μg/mL of blasticidin for the selection. CHO cells, HEK293T cells, HeLa cells and their derivatives, were cultured in Dulbecco’s modified Eagle’s medium-high glucose (Gibco #11995-065, Carlsbad, CA) and supplemented with 10% heat-inactivated fetal bovine serum (Gibco #26140-079), and 100 U/mL penicillin and 100 μg/mL streptomycin at 37°C in a 5% CO2-humidified incubator. The HEK293T-ACE2-TMPRSS2 cells bearing mCherry fluorescence were commercially purchased from Genecopoeia, Rockville, MD, USA, and maintained in culture media in the presence of 1 μg/mL of puromycin and 100 μg/mL of Hygromycin B (USA #10687010; Invitrogen, Carlsbad, CA).
Calu-3 and Vero cells were maintained in Minimum Essential Medium (MEM) with Earle’s Balanced Salts containing 2.2 g/L sodium bicarbonate (HyClone #SH30024.01, Logan, UT) supplemented with 10% FBS (HyClone #SH30084.03) and 1% penicillin-streptomycin (HyClone #SV300010). Cells were grown at 37°C in a 5% CO2 incubator. BHK-21/WI-2 (Kerafast, Boston, MA) cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco) supplemented with 10% FBS (Gibco), Penicillin (100 units/mL) (Gibco), and Streptomycin (100 ug/mL) (Gibco) in an 8% CO2 incubator at 37°C and passaged every 2–3 days.
Primary culture of human nasal epithelial (HNE) cells
All experiments using HNE cells were approved by the Institutional Review Board of Yonsei University College of Medicine (4-2016-0902). The primary cells were obtained from both male and female subjects aged 29–54 years. The cells were isolated from nasal polyps from patients with chronic rhinosinusitis and who had no clinical history of asthma, aspirin sensitivity, or cystic fibrosis; and cultured under air–liquid interface (ALI) culture conditions as previously described (Yoon et al., 1999). Briefly, passage #2 HNE cells were seeded at a density of 1 × 105 cells/well on a 12-mm, 0.45-μm pore transwell-clear culture insert (3460; Corning, Kennebunk, ME) and cultured in a 1:1 mixture of bronchial epithelial cell growth media (BEGM) and DMEM, supplemented with growth factors according to the manufacturer’s instructions (Lonza, Walkersville, MD).
Method details
Plasmid cloning and siRNA transfection
The mammalian expressible pcDNA3.1-Spike plasmid was generated by inserting a stop codon before the C9 tag of a pcDNA3.1-SARS2-Spike plasmid (Addgene #145032). To generate the plasmids encoding the S1/S2 cleavage mutants of the Spike protein, the cleavage site was abolished by amino acid substitution at R682S and R685G using an NEBuilder HiFi DNA Assembly Kit. The lactadherin-C2 (Lact-C2-eGFP) encoding plasmid (Addgene #22852) was subcloned into a pET28a bacterial expression vector for His6-fusion protein purification. For the generation of mCherry-Lact-C2, fragments of mCherry encoding sequences (Addgene #36084) and Lact-C2 encoding sequences were PCR amplified and cloned into the pET28a plasmid using an NEBuilder HiFi DNA Assembly Kit. AccuTarget™, pre-designed human TMEM16F-specific (gene ID:196527, a cocktail of three different siRNAs) and control scrambled siRNAs (Bioneer #SN-1002) were commercially purchased from Bioneer (Daejeon, Korea). Transfection of siRNAs into HEK293T or HeLa cells was performed using the TransIT-X2 Dynamic Delivery System (#MIR6006, Mirus Bio LLC, Madison, WI, US) following the manufacturer’s protocol.
Chemical reagents, lentiviral particles, and antibodies
A6-001 (ChemDiv; Cat# F575-0482, N-(4-fluorobenzyl)-3-((6-phenylpyridazin-3-yl)amino)benzamide), A6-004 (ChemDiv; Cat# F474-2423, ethyl 8-methyl-4-((4-(methylthio)benzyl)amino)-2-oxo-1,2-dihydroquinoline-3-carboxylate), abamectin (Cayman Chemical, Ann Arbor, MI, USA), ivermectin (Sigma-Aldrich, St. Louis, MO, USA), camostat mesylate (Sigma-Aldrich, #SML0057), BAPTA-AM (Sigma-Aldrich, #A1076), amphotericin B solution (Sigma-Aldrich, #A2942), and trypsin (Sigma-Aldrich, #T6567) were purchased commercially. Lentifect™ SARS-CoV-2 Spike-pseudotyped Lentivirus used in the single-round infection (GFP reporter) was commercially acquired from Genecopoeia (cat# SP001). The following antibodies were acquired commercially: anti-SARS-CoV-2 S2 (GeneTex #GTX632604), anti-aldolase A (Santa Cruz Biotechnology #SC-390733), and anti-HIV-p24 (GeneTex #GTX64128).
Generation of pseudotyped SARS-CoV-2 S virus particles
For generation of lentivirus-based SARS2-PsV used in the PS externalization and [Ca2+]i measurements, HEK 293T cells were plated in a 6-well plate and transfected the next day when they were approximately 80% confluent with a combination of the following plasmids: 1 μg of pEGIP (Addgene #26777), 0.75 μg of psPAX2, and 2.25 μg of pcDNA3.1-SARS2-Spike (Addgene #145032). The TransIT-X2 Dynamic Delivery System (#MIR6006; Mirus Bio LLC, Madison, WI, US) was used for transfection, following the manufacturer’s protocol. The next day, the transfection medium was replaced with fresh culture medium and cells were cultured for 48 h. The pseudotyped virus-containing culture medium was collected 48 h post transfection, centrifuged at 500 ×g for 5 min, and stored at −80°C.
Western blot analysis
For immunoblotting of ANO6, FRT and Jurkat cells were lysed in lysis buffer (50 mM Tris-HCl (pH 7.4), 1 mM EDTA, 1 mM Na3VO4, 150 mM NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, protease inhibitor mixture). The cell lysates were centrifuged at 13,000 rpm for 20 min at 4°C and protein concentrations were measured with a Bradford assay kit (Sigma) in accordance with the manufacturer’s instructions. The supernatant proteins of FRT (40 μg) and FRT-ANO6 (40 μg) were separated using 4–12% Tris-glycine precast gel (KOMA BIOTECH) and transferred to a nitrocellulose filter membrane and then blocked with 5% non-fat skim milk in Tris-buffered saline containing 0.1% Tween 20 (TBST) for 1 h. Membranes were incubated in a solution containing primary antibodies against ANO6. Antibodies against mANO6 were generated in this study using the same epitope (67–81 a.a, DFRTPEFEEFNGKPD-C) appearing in a previous study (Yang et al., 2012), which retains a high homology among human, rat, and mouse ANO6s. Membranes were then incubated with HRP-conjugated anti-secondary IgG antibody (Enzo Life Sciences) and visualized using the ECL Plus Western Blotting detection system (Pierce).
For immunoblotting of the SARS-CoV-2 Spike glycoprotein, the cell lysates of wild-type and R682S/R685G SARS2-PsV infected HEK 293T cells were homogenized with sonication for 20 s (1-second pulses) followed by centrifugation at 16,000 x g for 20 min at 4°C. The samples were separated by a 4–12% SDS-PAGE gel electrophoresis and transferred to nitrocellulose filter membranes. After being blocked by 5% skim milk, the membranes were blotted with primary antibodies against the S2 domain of the Spike protein (GeneTex #GTX632604) and aldolase A (Santa Cruz #SC-390733), then incubated with horseradish peroxidase (HRP) conjugated secondary antibodies (1:2000) and detected with Chemiluminescent Reagent (Amersham #RPN2105).
YFP quenching assay
FRT cells expressing a YFP variant (H148Q/I152L/F46L) with ANO1, ANO2 or ANO6 were plated in 96-well black-walled microplates (Corning Inc., Corning, NY, USA) at a density of 2 × 104 cells/well. After a 48-h incubation, each well of 96-well plates was washed twice in phosphate-buffered solution (PBS, 200 μL/wash) and test compounds containing 50 μL PBS were added to each well. For the high-throughput screening of a library of 54,000 drug-like small-molecule compounds (Chemdiv, San Diego, CA, USA) and a library of 1,700 approved drugs (TargetMol, Boston, MA, USA), FRT-ANO6 cells were treated with 25 μM compounds. After a 10-min incubation at 37°C, the 96-well plates were transferred to a FLUOstar Omega microplate reader (BMG Labtech, Ortenberg, Germany) for fluorescence assay. Each well was assayed individually for ANO6-mediated thiocyanate (SCN−) influx by monitoring YFP fluorescence continuously (0.4 s/point) for 1 s (baseline), then 140 mM SCN− solution containing 10 μM ionomycin was added at 1 s before YFP fluorescence was recorded for 7 s. The initial rate of SCN− influx was computed from fluorescence data by nonlinear regression.
Cytoplasmic calcium measurements
FRT cells were plated in 96-well black-walled microplates at a density of 2 × 104 cells/well. After a 48-h incubation, the cells were loaded with Fluo-4 NW (Invitrogen) as per the manufacturer’s protocol. Cells were incubated with 100 μL assay buffer containing Fluo-4 NW. After a 1-h incubation, cells were treated with test compounds for 10 min and the 96-well plates were transferred to a FLUOstar Omega microplate reader (BMG Labtech) equipped with syringe pumps and custom Fluo-4 excitation/emission filters (485/538 nm). Intracellular calcium signaling was induced via application of 10 μM ionomycin.
The [Ca2+]i imaging in HEK 293T-ACE2-TMPRSS2 cells was performed with a ratiometric Ca2+ probe, Fura-2 (Fura-2 AM; Molecular Probes, Eugene, Oregon, USA). The ACE2 and TMRPSS2 stable cells grown on glass cover slips were loaded with cell permeable Fura-2 AM (5 μM, 30 min, 37°C) in HEPES-buffered physiological salt solution (PSS) containing (mM) 145 NaCl, 5 KCl, 10 HEPES, 5 D-Glucose, 1 MgCl2 and 1 CaCl2 (adjusted to pH 7.4 NaOH). Some cells were additionally treated with BAPTA-AM (3 μM, 20 min) for the chelation of intracellular Ca2+. Glass coverslips were placed into an experimental chamber (Live Cell Instrument, Seoul, Korea) with a perfusion system at 37°C and then the cells were transferred to the perfusion chamber. Ca2+ measurements were performed using a Zeiss Axio microscope attached to a charge coupled device camera in a temperature-controlled chamber set to 37°C. For the illumination of Fura-2, an ultra-fast switching monochromator (Polychrome V, Till Photonics) was used. Fura-2 was excited alternatively at 340 nm and 380 nm and the emission light was simultaneously collected at 510 nm. Images were acquired at 2 frames/s using MetaFluor (Molecular devices). In each recording, three cells showing the maximum cytosolic Ca2+ response were chosen for the analysis of [Ca2+]i dynamics. A region of interest (ROI) of [Ca2+]i increasing microdomain in each cell was monitored to determine the cytoplasmic Fura-2 fluorescence changes during SARS2-PsV or ATP application. The [Ca2+]i values were estimated from the Fura-2 fluorescence ratiometric values using the following equation (Grynkiewicz et al., 1985)
where Kd is the indicator’s dissociation constant for Ca2+ and R is the intensity ratio for fluorescence at the two chosen wavelengths (340, 380 nm). The Rmax and Rmin are ratios at zero and saturating [Ca2+]i, respectively, and Sf2/Sb2 is the ratio of excitation efficiencies for free and bound Fura-2. For calibration, 20 mM EGTA was added to obtain the minimum fluorescence ratio and 5 μM ionomycin with 10 mM CaCl2 was added to obtain the maximum fluorescence ratio. In each recording, three cells showing the maximum Ca2+ response were chosen for statistical analyses.
Electrophysiology
The Ca2+-activated Cl− channel activities were measured in HEK 293T cells using the whole-cell clamp techniques. Cells were transferred into a bath mounted on a stage with an inverted microscope (Ti-2, Nikon). The whole-cell clamp was achieved by rupturing the patch membrane after forming a giga-seal. The bath solution was perfused at 5 mL/min. The voltage and current recordings were performed at room temperature (22–25°C). Patch pipettes with a free-tip resistance of approximately 2–5 MΩ were connected to the head stage of a patch-clamp amplifier (Axopatch-200B, Axon Instruments). pCLAMP software v.10.7 and Digidata-1440A (Axon Instruments) were used to acquire data and apply command pulses. Voltage and current traces were stored and analyzed using Clampfit v. 10.7 and Origin v. 8.0 (Origin Lab Inc.). To record ANO6 (TMEM16F) currents, voltage ramps spanning a range of −100 to +100 mV were delivered from a holding potential of −60 mV every 20 s. Currents were sampled at 5 kHz. All data were low-pass filtered at 1 kHz. The standard pipette solution for whole-cell clamp contained (in mM) 141.8 NMDG-Cl, 5 HEPES, 0.5 MgCl2, 10 HEDTA, and 1 Mg-ATP. An appropriate amount of CaCl2 was added to the pipette solution to obtain a pipette Ca2+ concentration of 10 μM (adjusted to pH 7.2 with NMDG-OH). The bath solution contained (in mM) 146 NMDG-Cl, 1 CaCl2, 1 MgCl2, 10 HEPES, and 5 Glucose (adjusted to pH 7.2 with NMDG-OH).
For the CFTR short-circuit current measurements, Snapwell cell culture inserts containing FRT cells expressing human WT-CFTR were mounted in Ussing chambers (Physiological Instruments, San Diego, CA). The apical hemichamber was filled with buffer solution containing (in mM): 60 NaCl, 60 Na-gluconate, 5 KCl, 1 MgCl2, 1 CaCl2, 10 D-glucose, 2.5 HEPES, and 25 NaHCO3 (pH 7.4). The basolateral hemichamber was filled with buffer solution containing (in mM): 120 NaCl, 5 KCl, 1 MgCl2, 1 CaCl2, 10 D-glucose, 2.5 HEPES, and 25 NaHCO3 (pH 7.4). The basolateral membrane was permeabilized using 250 μg/mL amphotericin B, and then the cells were incubated for 20 min and aerated with 95% O2/5% CO2 at 37°C. The apical membrane current was recorded using an EVC4000 Multi-Channel V/I Clamp (World Precision Instruments, Sarasota, FL) and PowerLab 4/35 (AD Instruments, Castle Hill, Australia).
PS externalization assay and confocal microscopy
The Lact-C2-GFP and Lact-C2-mCherry fusion proteins were produced in the BL-21 (DE3) Escherichia coli strain. Cells were grown in an LB medium at 30°C in 120 μg/mL kanamycin until the culture reached A600 = 0.6. After the addition of 0.5 mM IPTG, the culture was incubated for 6 h at 30°C. Cells were lysed in lysis buffer containing 50 mM sodium phosphate (pH 7.4), 200 mM NaCl, 0.5% NP-40, 10 mM imidazole and protease inhibitor cocktail (Roche Applied Science, Mannheim, Germany). The cell lysates were homogenized with sonication for 3 min (1-s pulse) followed by centrifugation at 12,000 × g for 20 min at 4°C. After centrifugation, His6-mcherry-Lact-C2 was purified with a nickel-nitrilotriacetic acid (Ni-NTA) protein purification system (QIAGEN) by following the manufacturer’s instructions. Stock solutions of 1–3 mg/mL were stored in the dialysis buffer (100 mM KCl, 1 M HEPES-KOH pH 8.0, 0.2 M EDTA, 10% glycerol, 1 M DTT, 200 mM PMSF) and were used at 1–3 μg/mL.
FRT cells stably expressing ANO6 (variant 5) were plated in 96-well black-walled microplates at a density of 2 × 104 cells/well. After a 48-h incubation, cells were treated with the test compound for 10 min, then 10 μM of ionomycin was applied and finally each well was washed with 200 μL PBS after 10 min. Cells were fixed with 4% paraformaldehyde (100 μL/well) for 10 min at room temperature. After washout, the phosphatidylserine and nuclei were stained with PBS containing 500 nM Lact-C2-GFP, then cells were washed with 200 μL PBS and microscopic images were acquired with a Lionheart FX Automated Microscope (BioTek, Winooski, VT, USA). For morphological analysis, some cells were stained with fluorescently labelled phalloidin and DAPI (Sigma-Aldrich) for 15 min at room temperature. Quantitative analysis of the fluorescence intensity of Lact-C2-GFP was performed with Python3 (Python Software Foundation, Wilmington, DE, USA) and OpenCV (Open Source Computer Vision Library). OpenCV was used to remove background and noise pixels, and the sum of all remaining pixel values was used to evaluate the fluorescence intensity of Lact-C2-GFP. The original microscopic images contained approximately 130 to 160 cells per image, and these images were used for the calculation of fluorescence intensity. Figures 2D and S2H show magnified images cropped to show the cells more clearly, and these images contain approximately 50 cells per image.
For SARS2-PsV-induced PS externalization assay, HeLa-ACE2 cells (2 × 105 cells/mL) were plated on 12-mm round coverslips and incubated with the lentivirus-based SARS2-PsV (p24 100 ng/mL, 20 MOI) for 15 min. For authentic SARS-CoV-2 virus-induced PS externalization, the cells were infected with SARS-CoV-2 with an MOI of 10 for 15 min. After infection, the cells were fixed with 2% formaldehyde for 5 min at room temperature, and then incubated with solutions containing Lact-C2-mCherry for 45 min at 37°C. For nucleus staining, cells were also stained with DAPI. Fluorescent images were obtained with a Zeiss LSM confocal microscope (LSM 780; Carl Zeiss, Berlin, Germany) with a 20× or 63× (1.4 numerical aperture) objective lens. For image quantification using the open-source software ImageJ Fiji, 24-bit confocal images including red, green, and blue components were converted into three 8-bit mono-channel images. To quantify PS externalization, pixels above a threshold level of 50 were defined as externalized PS. The percentage of Lact-C2 labeled cells in DAPI-stained cells (100 to 200 cells) was calculated. To minimize variations between experimental replicates, the recorded fluorescence values were normalized to a maximum value in each set of experiments. For example, in Figure 3B, the maximum Lact-C2-mCherry fluorescence value was observed in cells treated with SARS2-PsV alone (lane 2). Then, the fluorescence values of Control (lane 1) and SARS2-PsV + BAPTA-AM (lane 3) were normalized to the value of SARS2-PsV alone (lane 2) in each experimental replication set.
PS externalization assay using 4-quadrant dot blot analysis (flow cytometry)
Jurkat cells cultured in RPMI 1640 (Capricorn Scientific GmbH, Ebsdorfergrund, Germany) were washed with Opti-MEM™ (Thermo Fisher Scientific Cat#31985-070, Waltham, MA, USA) twice, then transfected with 10 μg of pcDNA3.1-ANO6(V5) plasmids per 1 × 106 cell using a NEPA21 Super Electroporator (NEPAGENE #NPG-NEPA21, Japan) following the manufacturer’s protocol. After a 24-h incubation, the cells were treated with A6-001 (10 μM) or vehicle for 30 min. Some cells were co-treated with 10 μM ionomycin for 20 min to activate ANO6. Subsequently, cells were washed with PBS and resuspended in 100 μL binding buffer containing propidium iodide (PI, Abcam, cat #ab14083) and Lact-C2-GFP. The reaction was stopped by adding 400 μL of PBS and the flow cytometry was performed immediately using a FACSCelesta cell analyzer (BD Biosciences) with the FACSDiva software. Data profiles were analyzed using the Flowjo flow cytometry software package (TreeStar Inc. Ashland, OR, USA).
Membrane fusion assay
For analysis of SARS2-PsV-induced cell fusion, HeLa-ACE2 cells were plated on 12-mm round coverslips and incubated with SARS2-PsV (p24 100 ng/mL, 20 MOI) for 30 min. To quantify the number of fusion events, Lact-C2-positive multinucleated cells containing more than two nuclei were considered to be membrane-fused multinucleated cells. We analyzed micrograph images of 3 to 5 fields (∼150 cells per field) in each experimental replication set and counted the number of Lact-C2-positive multinucleated cells per field. The percentage of multinucleated cells/total cells (DAPI-positive) was calculated.
For analysis of Spike-induced membrane fusion mechanisms, time-lapse imaging of membrane fusion events occurring between the Spike-expressing CHO cells and ACE2-expressing HEK 293T-ACE2-TMPRSS2 cells was performed. CHO cells were transfected with plasmids expressing Spike envelope proteins (2 μg/2 mL, pcDNA3.1-SARS2-Spike). Two days after transfection, Spike-expressing CHO cells at a density of 2 × 105 cells/well were labeled with the fluorescent lipophilic tracer Vybrant DiO (2 μM) in serum-free medium for 20 min at 37°C, washed, and added to the cytosolic mCherry-expressing HEK 293T-ACE2-TMPRSS2 cells (2 × 105 cells/well) labeled with Hoechst 33342 (1 μg/mL). For the examination of the multistep processes of Spike-mediated cell-cell fusion, live imaging was performed using Thunder Imager Live cell fluorescence microscopy (Leica microsystems, Wetzlar, Germany). An 8-h time-lapse image acquired at 20 frames/min was analyzed using the Leica Application Suite X software. The cell-cell adhesion was quantified by counting the number of GFP-positive cells in contact with mCherry-positive cells during the initial 1 h co-incubation period, and the cell-cell fusion was quantified by counting the membrane fusion events between the GFP-positive and mCherry-positive cells during the overall 8-h coincubation period.
Single-round infection
The cytosolic mCherry-expressing HEK293T-ACE2-TMPRSS2 cells were seeded to 96-well plates at a density of 5 × 104 cells/well. After a 24-h incubation, the cells were pretreated with different concentrations of inhibitors for 1 h. Subsequently, 5 μL of Lentifect SARS-CoV-2 Spike-pseudotyped lentivirus (1.88 × 107 IFU/mL) was added to the cells and incubated first at 4°C for 2 h and subsequently at 37°C for 4 h. After the 6-h incubation with pseudotyped virus, cells were fed with fresh culture medium and further incubated for 42 h. We quantified the amount of infection by counting the number of GFP-positive cells (threshold level of 32 in 8-bit mono channel images) using fluorescence microscopy with a software package (NIS-elements AR; Nikon, Tokyo, Japan); and the percentage of GFP-positive cells/total cells (mCherry-positive) was calculated.
Reverse-transcription (RT) and quantitative PCR (qPCR)
Twenty-four hours after transfection with siRNAs, the total cellular RNA was extracted using Tri-RNA reagent (#FATRR 001; Favorgen Biotech Corp., Taiwan) according to the manufacturer’s protocol. Complementary DNA (cDNA) was synthesized using an RNA to cDNA EcoDry premix (#639549; Takara Bio Inc., Shiga, Japan). Mixtures were incubated at 42°C for 1 h, followed by 70°C for 10 min. qPCR was performed with an Applied Biosystem StepOne System (Applied Biosystems, Forster City, CA, USA).
Cell viability assay
FRT, Calu-3 and Vero cells were seeded at ∼20% confluency in 96-well plates. After a 24-h incubation, cells were treated with different concentrations (0.3, 1, 3, 10, 30 and 100 μM) of compounds. After incubation for 48 h, the effects of the compounds on cell viability were evaluated via 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay using a CellTiter 96® AQueous One Solution Cell Proliferation Assay kit (Promega) following the manufacturer’s instructions. The soluble formazan produced by the cellular reduction of MTS was quantified by measuring the absorbance at 490 nm with a microplate reader (Infinite M200; Tecan, Grödig, Austria).
Virus propagation, quantification, and infection of SARS-CoV-2
A clinically isolated SARS-CoV-2 KUMC-2 strain (GISAID accession#: EPI_ISL_413018) was used. For viral propagation, Vero cells were infected with SARS-CoV-2 (0.01 MOI), then culture supernatant was harvested at 48 h post-infection. The harvested supernatant was filtered and stocked at −80°C. For plaque formation assay, one of the stock vials was serially diluted and used to infect a Vero cell for 1 h, which was then incubated with culture media containing 1% low melting temperature agarose (#50101; Lonza, Rockland, ME) for 48 h. Viral plaques were visualized with Neutral Red Solution (Sigma #2889) for an additional 24 h and were counted to determine the plaque-forming unit of the viral stock. To evaluate drug efficacy against virus infection, Calu-3 and Vero cells were pretreated with the indicated concentrations of drugs for 2 h, prior to infection with the virus (0.01 or 0.001 MOI) for 1 h. After washing with PBS, cells were replenished with culture media supplemented with 2% FBS together with the indicated concentrations of drugs and then incubated for 48 h. All experiments with infectious virus were approved by the Institutional Biosafety Committee of the Yonsei University Health System (IBC 2020-003) and conducted in the Biosafety Level 3 facility at Yonsei University College of Medicine.
For the quantification of the SARS-CoV-2 virus secretion, viral RNAs in cell culture supernatant were extracted using a QIAamp Viral RNA Mini kit (QIAGEN #52906, Valencia, CA), as per the manufacturer’s instructions. Quantitative PCR was performed with a Luna® Universal One-Step RT-qPCR Kit (NEW ENGLAND BioLabs®Inc, Ipswich, MA, USA) using the Applied Biosystem Quantstudio 3 detection system (Applied Biosystems, Waltham, MA, USA). For amplification, 2 μL of extracted RNA was added to the mixture of 10 μL of 2X Luna Universal One-Step Reaction Mix, 0.4 μL of 10 μM primers, 0.2 μL of 10 μM probe and 1 μL of 20X Luna WarmStart® RT Enzyme Mix adjusted to a total reaction volume of 20 μL with RNase-free water. Amplification was carried out using the optimal thermocycling conditions described in the manufacturer’s protocol for 40 cycles. The standard curve was generated by conducting qRT-PCR using serially diluted virus stock. Ct values were then converted into viral titer (PFU/ml, Figure S5D).
Authentic virus infection of HNE cells
The cells were cultured using the media in both the apical and basolateral compartments until the cells became confluent (approximately 3 days). Once confluent, the cells were placed under ALI culture conditions; the apical compartments were exposed to air and only the basolateral compartments were supported by the media. The media were changed on alternate days for 21 days. ANO6 inhibiting drugs were administered to the basolateral compartments of the fully differentiated HNE cells 2 h prior to infection with the virus. SARS-CoV-2 virus (1.0 MOI) was added to the apical compartments and removed after incubation for 3 h. At 3 days after infection, 150 μL of 1x PBS was added to the apical compartments, incubated for 10 min at 37°C, and collected to recover progeny viruses. The plaque assay for viral quantification was performed using Vero cell culture at 85–95% confluence in 12-well plates.
Adenovirus infection and quantification
HEK 293T cells were seeded to 12-well plates. After a 24-h incubation, the cells were pretreated with 10 μM of A6-001 or vehicle (0.1% DMSO) for 2 h. Subsequently, 1 MOI of recombinant eGFP adenovirus (Ad-eGFP; Vector Biolabs #1060, Malvern, PA, USA) was added to the cells and incubated at 37°C. At 48 h post-infection, all attached and suspended cells were harvested, and DNAs including genomes of propagated Ad-eGFP were extracted using a QIAamp DNA Mini Kit (QIAGEN #51304, Valencia, CA) as per the manufacturer’s protocol. Real-time PCR was performed to measure the amount of Ad-eGFP from the extracted DNA by using SYBR green dye. For amplification, 2 μL of extracted DNA was added to the mixture of 10 μL of 2X KAPA SYBR FAST qPCR Kit Master Mix (Sigma-Aldrich #KK4605, St. Louis, MO, USA), and 1 μL of 10 μM each primer was adjusted to a total reaction volume of 20 μL with RNase-free water. Amplification was achieved using the Applied Biosystem StepOne System (Applied Biosystems, Waltham, MA, USA) under the recommended thermocycling conditions: 95°C for 10 min followed by 40 cycles at 95°C for 15 min and then 60°C for 40 s.
Quantification and statistical analysis
The results of multiple experiments are presented as the mean ± standard error of the mean (SEM). For biochemical and in vitro assays, statistical analysis was performed using a two-tailed Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test, as appropriate, using GraphPad Prism8 (GraphPad Software, Inc., La Jolla, CA, USA). A p value <0.05 was considered statistically significant.
Acknowledgments
The authors thank the Medical Illustration & Design team at Yonsei University College of Medicine for providing support with medical illustrations, Benjamin J. Latimer for his editorial assistance, and the Yonsei-Carl Zeiss Advanced Imaging Center for their technical assistance. This work was supported by grants 2013R1A3A2042197, 2022R1A2C3002917 (M.G.L.), 2018R1A6A1A03023718 (W.N.), 2022M3A9I2082293, and 2019R1A6A1A03032869 (J.M.L.) from the National Research Foundation, the Ministry of Science and ICT, and the Ministry of Education, and grant HI22C0984 (M.G.L.) from the Korea Health Industry Development Institute, the Ministry of Health and Welfare, Republic of Korea. M.G.L. was supported by the Heung Ki Kim Memorial Fund from Yonsei University, Seoul, Korea.
Author contributions
J.-R.S. designed and carried out the molecular experiments of SARS-CoV-2 and analyzed the data. D.H.S. performed patch-clamp and intracellular calcium measurements. P.-G.P. performed experiments using an infectious virus. S.-H.P. performed the high-throughput screening of chemicals. J.-Y.B. performed authentic virus isolation. Y.L., D.-Y.K., Y.J.K., and S.A. generated pseudotyped virus and immunochemistry. S.H.N. and H.-R.C. performed the four-quadrant dot blot FACS analysis. S.J.H., C.B.K., S.H.K., and S.P. performed experiments using an infectious virus. D.J. and S.C. performed YFP quenching assays. G.E.L. and J.K. performed experiments with authentic virus. Y.M. and J.-O.K. generated pseudotyped virus. J.-S.N., C.-H.K., S.M., and Y.W.C. performed experiments in HNE cells, and M.-S.P. designed and supervised experiments with authentic virus. J.-H.R. designed and supervised experiments in HNE cells. W.N. and J.M.L. designed and conceived experiments involving high-throughput screening and in vitro antiviral assays and edited the manuscript. M.G.L. designed and conceived all experiments and wrote the manuscript.
Declaration of interests
M.G.L., J.M.L., and W.N. are inventors on a pending patent related to this work filed by Yonsei University Office of Research Affairs/University Industry Foundation. The authors declare no other competing interests.
Published: July 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111117.
Supplemental information
Data and code availability
-
•
The complete sequences of SARS-CoV-2 KUMC-2 are available through GISAID: EPI_ISL_413018. Additional Supplemental Items are available from Mendeley Data: https://doi.org/10.17632/8vgrh6dyfp.1
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alvadia C., Lim N.K., Clerico Mosina V., Oostergetel G.T., Dutzler R., Paulino C. Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F. Elife. 2019;8:e44365. doi: 10.7554/eLife.44365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara A., Mercer J. Viral apoptotic mimicry. Nat. Rev. Microbiol. 2015;13:461–469. doi: 10.1038/nrmicro3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevers E.M., Williamson P.L. Getting to the outer leaflet: physiology of phosphatidylserine exposure at the plasma membrane. Physiol. Rev. 2016;96:605–645. doi: 10.1152/physrev.00020.2015. [DOI] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga L., Ali H., Secco I., Chiavacci E., Neves G., Goldhill D., Penn R., Jimenez-Guardeño J.M., Ortega-Prieto A.M., Bussani R., et al. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 Spike-induced syncytia. Nature. 2021;594:88–93. doi: 10.1038/s41586-021-03491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaccour C., Casellas A., Blanco-Di Matteo A., Pineda I., Fernandez-Montero A., Ruiz-Castillo P., Richardson M.A., Rodríguez-Mateos M., Jordán-Iborra C., Brew J., et al. The effect of early treatment with ivermectin on viral load, symptoms and humoral response in patients with non-severe COVID-19: a pilot, double-blind, placebo-controlled, randomized clinical trial. EClinicalMedicine. 2021;32:100720. doi: 10.1016/j.eclinm.2020.100720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coil D.A., Miller A.D. Enhancement of enveloped virus entry by phosphatidylserine. J. Virol. 2005;79:11496–11500. doi: 10.1128/JVI.79.17.11496-11500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors J.M., Levy J.H. COVID-19 and its implications for thrombosis and anticoagulation. Blood. 2020;135:2033–2040. doi: 10.1182/blood.2020006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doktorova M., Symons J.L., Levental I. Structural and functional consequences of reversible lipid asymmetry in living membranes. Nat. Chem. Biol. 2020;16:1321–1330. doi: 10.1038/s41589-020-00688-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymieux S., Rouillé Y., Terrier O., Seron K., Blanchard E., Rosa-Calatrava M., Dubuisson J., Belouzard S., Roingeard P. Ultrastructural modifications induced by SARS-CoV-2 in Vero cells: a kinetic analysis of viral factory formation, viral particle morphogenesis and virion release. Cell. Mol. Life Sci. 2021;78:3565–3576. doi: 10.1007/s00018-020-03745-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Dang S., Han T.W., Ye W., Jin P., Cheng T., Li J., Jan Y.N., Jan L.Y., Cheng Y. Cryo-EM studies of TMEM16F calcium-activated ion channel suggest features important for lipid scrambling. Cell Rep. 2019;28:567–579.e4. doi: 10.1016/j.celrep.2019.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grynkiewicz G., Poenie M., Tsien R.Y. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- Hartzell H.C., Yu K., Xiao Q., Chien L.T., Qu Z. Anoctamin/TMEM16 family members are Ca2+-activated Cl- channels. J. Physiol. 2009;587:2127–2139. doi: 10.1113/jphysiol.2008.163709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Hofmann-Winkler H., Smith J.C., Kruger N., Arora P., Sorensen L.K., Sogaard O.S., Hasselstrom J.B., Winkler M., Hempel T., et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine. 2021:103255. doi: 10.1016/j.ebiom.2021.103255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielity S., Wang J.J., Chan Y.K., Ahmed A.A., Li W., Monahan S., Bu X., Farzan M., Freeman G.J., Umetsu D.T., et al. TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog. 2013;9:e1003232. doi: 10.1371/journal.ppat.1003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J., Nam J.H., Park H.W., Oh U., Yoon J.H., Lee M.G. Dynamic modulation of ANO1/TMEM16A HCO3- permeability by Ca2+/calmodulin. Proc. Natl. Acad. Sci. USA. 2013;110:360–365. doi: 10.1073/pnas.1211594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Jun I., Yoon J.S., Jung J., Kim Y.K., Kim W.K., Kim B.J., Song J., Kim S.J., Nam J.H., et al. Selective serotonin reuptake inhibitors facilitate ANO6 (TMEM16F) current activation and phosphatidylserine exposure. Pflugers Arch. 2015;467:2243–2256. doi: 10.1007/s00424-015-1692-6. [DOI] [PubMed] [Google Scholar]
- Kunzelmann K., Nilius B., Owsianik G., Schreiber R., Ousingsawat J., Sirianant L., Wanitchakool P., Bevers E.M., Heemskerk J.W.M. Molecular functions of anoctamin 6 (TMEM16F): a chloride channel, cation channel, or phospholipid scramblase? Pflugers Arch. 2014;466:407–414. doi: 10.1007/s00424-013-1305-1. [DOI] [PubMed] [Google Scholar]
- Lai A.L., Millet J.K., Daniel S., Freed J.H., Whittaker G.R. The SARS-CoV fusion peptide forms an extended bipartite fusion platform that perturbs membrane order in a calcium-dependent manner. J. Mol. Biol. 2017;429:3875–3892. doi: 10.1016/j.jmb.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.G., Kim K.H., Park K.Y., Kim J.S. Evaluation of anti-influenza effects of camostat in mice infected with non-adapted human influenza viruses. Arch. Virol. 1996;141:1979–1989. doi: 10.1007/BF01718208. [DOI] [PubMed] [Google Scholar]
- Liao L.E., Carruthers J., Smither S.J., CL4 Virology Team. Weller S.A., Williamson D., Laws T.R., Hiscox J., Hiscox J., Holder B.P., et al. Quantification of Ebola virus replication kinetics in vitro. PLoS Comput. Biol. 2020;16:e1008375. doi: 10.1371/journal.pcbi.1008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Jun I., Woo J.H., Lee M.G., Kim S.J., Nam J.H. Temperature-dependent increase in the calcium sensitivity and acceleration of activation of ANO6 chloride channel variants. Sci. Rep. 2019;9:6706. doi: 10.1038/s41598-019-43162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namkung W., Yao Z., Finkbeiner W.E., Verkman A.S. Small-molecule activators of TMEM16A, a calcium-activated chloride channel, stimulate epithelial chloride secretion and intestinal contraction. FASEB J. 2011;25:4048–4062. doi: 10.1096/fj.11-191627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou T., Mou H., Zhang L., Ojha A., Choe H., Farzan M. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog. 2021;17:e1009212. doi: 10.1371/journal.ppat.1009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papahadjopoulos D., Poste G., Schaeffer B.E., Vail W.J. Membrane fusion and molecular segregation in phospholipid vesicles. Biochim. Biophys. Acta. 1974;352:10–28. doi: 10.1016/0005-2736(74)90175-8. [DOI] [PubMed] [Google Scholar]
- Pedemonte N., Galietta L.J.V. Structure and function of TMEM16 proteins (anoctamins) Physiol. Rev. 2014;94:419–459. doi: 10.1152/physrev.00039.2011. [DOI] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J., Umeda M., Sims P.J., Nagata S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 2010;468:834–838. doi: 10.1038/nature09583. [DOI] [PubMed] [Google Scholar]
- Tarafdar P.K., Chakraborty H., Dennison S.M., Lentz B.R. Phosphatidylserine inhibits and calcium promotes model membrane fusion. Biophys. J. 2012;103:1880–1889. doi: 10.1016/j.bpj.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J.M., Chernomordik L.V. Flagging fusion: phosphatidylserine signaling in cell-cell fusion. J. Biol. Chem. 2021;296:100411. doi: 10.1016/j.jbc.2021.100411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Kim A., David T., Palmer D., Jin T., Tien J., Huang F., Cheng T., Coughlin S.R., Jan Y.N., et al. TMEM16F forms a Ca2+-activated cation channel required for lipid scrambling in platelets during blood coagulation. Cell. 2012;151:111–122. doi: 10.1016/j.cell.2012.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.D., Cho H., Koo J.Y., Tak M.H., Cho Y., Shim W.S., Park S.P., Lee J., Lee B., Kim B.M., et al. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008;455:1210–1215. doi: 10.1038/nature07313. [DOI] [PubMed] [Google Scholar]
- Yeung T., Gilbert G.E., Shi J., Silvius J., Kapus A., Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- Yoon J.H., Kim K.S., Kim H.U., Linton J.A., Lee J.G. Effects of TNF-alpha and IL-1 beta on mucin, lysozyme, IL-6 and IL-8 in passage-2 normal human nasal epithelial cells. Acta Otolaryngol. 1999;119:905–910. doi: 10.1080/00016489950180261. [DOI] [PubMed] [Google Scholar]
- Younan P., Iampietro M., Santos R.I., Ramanathan P., Popov V.L., Bukreyev A. Role of transmembrane protein 16F in the incorporation of phosphatidylserine into budding Ebola virus virions. J. Infect. Dis. 2018;218:S335–S345. doi: 10.1093/infdis/jiy485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitseva E., Zaitsev E., Melikov K., Arakelyan A., Marin M., Villasmil R., Margolis L.B., Melikyan G.B., Chernomordik L.V. Fusion stage of HIV-1 entry depends on virus-induced cell surface exposure of phosphatidylserine. Cell Host Microbe. 2017;22:99–110.e7. doi: 10.1016/j.chom.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The complete sequences of SARS-CoV-2 KUMC-2 are available through GISAID: EPI_ISL_413018. Additional Supplemental Items are available from Mendeley Data: https://doi.org/10.17632/8vgrh6dyfp.1
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.