Abstract
COVID-19 is marked by extensive damage to the respiratory system, often accompanied by systemic manifestations, due to both viral cytopathic effects and hyperinflammatory syndrome. Therefore, the development of new therapeutic strategies or drug repurposing aiming to control virus replication and inflammation are required to mitigate the impact of the disease. Hydroxypropyl-beta-cyclodextrin (HP-BCD) is a cholesterol-sequestering agent with antiviral activity that has been demonstrated against enveloped viruses in in vitro and in vivo experimental models. We also demonstrated that HP-BCD has an immunomodulatory effect, inhibiting the production of selected proinflammatory cytokines induced by microbial products. Importantly, this drug has been used in humans for decades as an excipient in drug delivery systems and as a therapeutic agent in the treatment of Niemann pick C disease. The safety profile for this compound is well established. Here, we investigated whether HP-BCD would affect SARS-CoV-2 replication and virus-induced inflammatory response, using established cell lines and primary human cells. Treating virus or cells with HP-BCD significantly inhibited SARS-CoV-2 replication with a high selective index. A broad activity against distinct SARS-CoV-2 variants was evidenced by a remarkable reduction in the release of infectious particles. The drug did not alter ACE2 surface expression, but affected cholesterol accumulation into intracellular replication complexes, lowering virus RNA and protein levels, and reducing virus-induced cytopathic effects. Virus replication was also impaired by HP-BCD in Calu-3 pulmonary cell line and human primary monocytes, in which not only the virus, but also the production of proinflammatory cytokines were significantly inhibited. Given the pathophysiology of COVID-19 disease, these data indicate that the use HP-BCD, which inhibits both SARS-CoV2 replication and production of proinflammatory cytokines, as a potential COVID-19 therapeutic warrants further investigation.
Keywords: SARS-COV-2, COVID-19, Beta-cyclodextrin, Virus replication, Inflammation, Cholesterol
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiologic agent of COVID-19, first emerged in 2019 and has rapidly spread to infect more than 500 million people and causing more than 6 million deaths around the globe (WHO COVID-19 Dashboard). Representing an unprecedent human threat in this century, therapeutic interventions are urgently needed along with currently available vaccines to alleviate patients’ symptoms, lower case fatality, and relieve overburdened health care systems. A few pharmacology interventions have been approved (De Clercq, 2021; Assadiasl et al., 2021; Bernal et al., 2021), but most of them do not show a robust impact on disease progression. The use of monoclonal antibodies has been one of the important strategies available so far, but their use is restricted to early acute disease (Kreuzberger et al., 2021). In addition, given the rapid virus evolution with the emergence of new variants of concern, the development and testing of specific antibodies may need constant development at high cost.
COVID-19 is a complex and multifactorial disease marked by respiratory damage with diffuse alveolar edema in the lungs, increased inflammatory infiltrates and thrombi, and systemic inflammatory response (Huang et al., 2020; Bradley et al., 2020; Yao et al., 2021). The clinical outcome results from both direct cellular effects due to virus replication, and from an exacerbated host response, triggering an hyperinflammatory and hypercoagulative syndrome with systemic consequences (Morris et al., 2020; Lucas et al., 2020; Song et al., 2020). Epithelial cells from the respiratory tract are predicted to be major targets of SARS-CoV-2 infection, resulting in epithelial damage and dysfunction (Sungnak et al., 2020; Bridges et al., 2021; Melms et al., 2021). In addition, several other cell types such as endothelial cells, hepatocytes and monocytes have been reported to be susceptible to SARS-CoV-2 and infection of these cells may contribute to tissue lesions and/or inflammation (Hoffmann et al., 2020; Hikmet et al., 2020; Pontelli et al., 2020). Increased cytokine levels, including IL-6, IL10, and TNF-α, are positively correlated with disease severity (Tang et al., 2021; Liu et al., 2021). Indeed, several lines of evidence suggest that hyperinflammation may be triggered by virus intracellular sensing pathways in cellular targets, and by activation of infiltrated monocytic and polymorphonuclear cells (Pantazi et al., 2021; Melms et al., 2021; Ackermann et al., 2021).
Cellular infection initiates by the interaction of the S (Spike) protein present at the virus envelope with host ACE2 receptor (Hoffmann et al., 2020). Additional receptors have also been reported to facilitate virus adsorption or entry, such as Neuropilin-1 and CD147 (Daly et al., 2020; Wang et al., 2020). After adsorption, cellular proteases, present at the surface or at endocytic compartments, cleave and prime S1/S2 protein, which allows virus fusion and release of the virus RNA into the cell cytoplasm (Hoffmann et al., 2020; Peacock et al., 2021; Shang et al., 2020). Although the exact uncoating compartment had not been clearly identified, once the genomic RNA (gRNA) is released, it is translated into the replicases polyproteins. Those nonstructural (NS) proteins and the gRNA are assembled into double-membrane replication–transcription complexes (RTCs), where the transcription of shorter subgenomic RNAs (sgRNAs) and replication of gRNA occur. The sgRNA codifies for the structural and other accessory proteins, and virus components then form new budding virions (Malone et al., 2022). Cell-cell fusion might be another important mechanism for SARS-CoV-2 dissemination, and extensive syncytia formation has been detected in patients’ lungs (Bussani et al., 2020; Sanders et al., 2021). Cholesterol depletion strongly affected Spike-mediated fusion in an in vitro model (Sanders et al., 2021), suggesting that cholesterol-rich regions are essential to syncytia formation. The importance of lipid metabolism to SARS-CoV-2 replication was also evidenced by the accumulation of lipid droplets and cholesterol-rich membranes in cell structures containing virus RNA, indicating that those might function as platforms for virus maturation (Sanders et al., 2021; Dias et al., 2020).
2-Hydroxypropyl-beta-cyclodextrin (HP-BCD) is a cholesterol-sequestering drug, which is being used in humans for several pharmaceutical applications, mostly to optimize drug delivery (Davis and Brewster, 2004). In addition, HP-BCD has been used for a decade for the treatment of Niemann Pick C disease (NPCD), administered intravenously, intrathecally, or by a combination of both routes, with a safe profile (Ottinger et al., 2014; Tanaka et al., 2015; Ory et al., 2017; Hastings et al., 2019). Notably, the antiviral effect of different isoforms of beta-cyclodextrins (BCD) was first reported by Liao et al., in 2001 who showed that HP-BCD blocked HIV-1 infection. Subsequently different derivatives of BCD have been shown to inhibit a number of enveloped viruses associated to systemic and respiratory diseases, including SIV, Influenza, Dengue, and SARS-CoV (Liao et al., 2001; Graham et al., 2003; Sun and Wang et al., 2020; Li et al., 2007; Lu et al., 2008; Glende et al., 2008; Verma et al., 2018). Treatment of cell-free virions or host cells with BCD demonstrated that this compound could mediate both virus inactivation (Graham et al., 2003; Liao et al., 2003; Sun and Whittaker, 2003), and impaired virus entry and budding in vitro (Liao et al., 2001; Verma et al., 2018). Preclinical studies also showed that vaginal application of HP-BCD reduced HIV transmission in mice models by as much as 91% (Khanna et al., 2002) and could protect monkeys from SIV infection upon the first contact with the virus (Ambrose et al., 2008). More recently, we reported that HP-BCD also showed immunomodulatory activity and suppressed the secretion of inflammatory cytokines by monocytes obtained from chronically infected HIV patients, when challenged by microbial products (Matassoli et al., 2018).
Here, we addressed whether HP-BCD would affect SARS-CoV-2 replication and impact virus-induced cellular activation using in vitro models of cell lines and human primary cells. We observed that HP-BCD significantly reduced virus replication and the release of infectious SARS-CoV-2 particles derived from different variants. Strikingly, treatment of lung epithelial cells and human primary monocytes with HP-BCD not only reduced the viral load, but also significantly impaired the production of inflammatory cytokines and chemokines. We propose HP-BCD as a potential therapeutic for COVID-19 given its dual ability to block SARS-CoV2 replication and virus-induced proinflammatory cytokines, which are both strongly associated with pathophysiology of the disease.
2. Materials and methods
2.1. Cell lines and viruses
All cell lines were cultured in Dulbecco's Modified Eagle Medium containing 4.5 g/L or 1 g/L (Calu-3 cells) D-Glucose (DMEM; Thermo Fisher Scientific Inc., Pittsburgh, PA, USA). African green monkey epithelial kidney cells (VERO C1008, Vero 76, clone E6, ATCC CRL-1586) (Vero E6) and VERO E6 expressing Transmembrane Protease Serine 2 and Human Angiotensin-Converting Enzyme 2 (Vero E6-TMPRSS2-T2A-ACE2 cells; NR-54970) were maintained in DMEM supplemented with 10% fetal bovine serum (FBS; Gibco); human lung adenocarcinoma epithelial cells (Calu-3, ATCC HTB-55) were maintained in DMEM supplemented with 20% FBS; ACE2 transfected 293T (kindly given by PhD. Paul D. Bieniasz, The Rockfeller University, NY) were maintained in DMEM supplemented with 7.5% fetal bovine serum. Cells were cultured at 37 °C with 5% CO2.
SARS-CoV-2 A2 (GISAID: 528539), Gamma, and Delta variants were isolated from symptomatic individuals cared at Universidade Federal do Rio de Janeiro (Rio de Janeiro, Brazil) COVID-19 trial center. Isolation and sequencing of each variant were performed, as previously described (Voloch et al., 2021). The study was approved by the National Committee of Research Ethics (CAAE-30161620.0.1001.5257). Virus isolation, stock production and titration were performed on Vero E6 cells.
2.2. Ethical statement and isolation of human primary monocytes
Blood samples (fresh buffy coats) from healthy donors were obtained from the Hemotherapy Service at the Hospital Universitario Clementino Fraga Filho (HUCFF) of Universidade Federal do Rio de Janeiro (UFRJ). The study protocol was approved by the Experimental Ethics Committee of UFRJ (permit 105/07). Peripheral blood mononuclear cells (PBMC) were isolated by Histopaque-1077 density gradient centrifugation and were cultured at a concentration of 107 cells/ml for 2 h for monocyte adhesion. Non adherent cells were discarded, and enriched monocyte population was then removed with cell scraper, washed, and cultured in RPMI-1640 (Thermo Fisher Scientific Inc.) supplemented with 10% FBS.
2.3. Cellular infection with SARS-COV-2
Cell lines or primary monocytes were incubated with the different isolates of SARS-CoV-2, at an MOI of 0.1, for 1 h at 37 °C for virus adsorption. The cells were then washed and cultured for different time periods according to the experimental protocol. For the analysis of initial steps of virus replication, virus adsorption was performed for 1 h at 4 °C. A portion of the wells were harvested, and this time point was considered 0 hpi. Other wells were transferred to 37 °C and incubated for 1 h, being designated as 1 hpi. Culture supernatants were harvested to evaluate virus replication by RT-qPCR or plaque assay. Cell lysates were obtained to measure intracellular virus RNA or protein, and cytokine expression.
2.4. Treatment of cells and viruses with HP-BCD
Cells were cultured with different concentrations of 2-hydroxypropyl-beta-cyclodextrin (HP-BCD) (Cyclodextrin Technologies Development, Inc. [CTD], High Springs, FL), in serum-free medium, for 1 h at 37 °C and 5% CO2. The cells were then washed with PBS and maintained in the appropriate culture medium (without HP-BCD) for the subsequent assays (pre-treatment assays). Alternatively, the cells were infected, as described and, after 1 hpi, treated with 20 mM HP-BCD for 1 h. Cells were washed and maintained in HP-BCD-free culture medium for additional 48 h (post treatment assays). Virus treatment was performed by incubating virus stocks with HP-BCD for 1 h. The suspensions were added to the cell cultures for adsorption for 1 h at 37 °C. Then the cells were washed and cultured for different time points according to the experimental assay. In a set of experiments, HP-BCD-treated virus or virus stock samples were serially diluted and directly titrated in Vero E6 cells, to avoid residual inoculation of the cells with the drug.
2.5. Cell viability
Cell lines were incubated with different concentrations of HP-BCD for 1 h, washed and cultured for additional 72 h. Cell viability was then assessed using Cell-Titer blue kit (Promega, Madison, USA), according to the manufacturer's protocol. Dose–response curves (non-linear regression) was performed to calculate CC50 (concentration decreasing cell viability by 50% in comparison to control cells) using GraphPad Prism software (v8.0.1).
2.6. Cholesterol measurement
Cells were treated with the indicated concentrations of HP-BCD for 1h in serum-free medium at 37 °C and 5% CO2, as described. The cells were washed once with PBS and the cholesterol content was measured by Amplex Red reagent (Thermo Fisher Scientific Inc.), following manufacturer's protocol.
2.7. Analysis of virus replication by RT-qPCR
Culture supernatants and cell lysates were obtained from different cell types, treated or not with HP-BCD, mock-cultured or infected with SARS-CoV-2, as described. Total RNA was purified using TRIzol reagent (Thermo Fisher Scientific Inc.), following manufacturer's instructions. Complementary DNA (cDNA) was then synthesized from 1 μg RNA using a High-Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific Inc.), according to manufacturer's protocol. The obtained cDNA was subjected to qPCR reaction using SARS-CoV-2 N2 primers (synthesized as primetime chemistry by Integrated DNA Technologies (IDT) according to sequences obtained from the US CDC) and conducted in StepOne™ Real-Time PCR System using the TaqMan Mix (ThermoFisher Scientific). The cycling parameters were: 95 °C for 2 min; 45 cycles of 95 °C for 3 s; and 55 °C for 30 s. CT values were used to calculate SARS-CoV-2 copy number, according to a standard curve derived from 10-fold dilutions of 2019-nCoV_N positive control gene transcript previously generated (IDT – ref #10006625). Analysis of intracellular subgenomic virus RNA were performed using specific primers for N gene subgenomic product and a probe for N gene, with the same cycling parameters. Housekeeping gapdh expression was also measured, using Power SYBR Green PCR master mix reagent (Thermo Fisher Scientific Inc.) and the cycling parameters described in the analysis of cytokine expression subsection. All the primers and probe used are described in Table 1 . Intracellular subgenomic RNA levels were normalized by GAPDH CT values. Dose–response curves (non-linear regression) was performed to calculate IC50 using GraphPad Prism software (v8.0.1).
Table 1.
Evaluated genes and corresponding primer and probe sequences used for qPCR.
| Gene | Primer sequence (5′-3′) | |
|---|---|---|
| Subgenomic N | Forward | CGA TCT CTT GTA GAT CTG TTC TCT AAA CGA ACT TAT GTA CTC |
| Reverse | ATA TTG CAG CAG TAC GCA CAC A | |
| Genomic N | Forward | TTA CAA ACA TTG GCC GCA AA |
| Reverse | GCG CGA CAT TCC GAA GAA | |
| GAPDH | Forward | GTG GAC CTG ACC TGC CGT CT |
| Reverse | GGA GGA GTG GGT GTC GCT GT | |
| IL-6 | Forward | TGT GAA AGC AGC AAA GAG GCA CTG |
| Reverse | ACA GCT CTG GCT TGT TCC TCA CTA | |
| TNF-α | Forward | CAG AGG GAA GAG TTC CCC AGG GAC C |
| Reverse | CCT TGG TCT GGT AGG AGA CGG C | |
| IL-10 | Forward | AAT AAG GTT TCT CAA GGG GCT |
| Reverse | AGA ACC AAG ACC CAG ACA TCA A | |
| CCL2 | Forward | CAG CCA GAT GCA ATC AAT GCC |
| Reverse | TGG AAT CCT GAA CCC ACT TCT | |
| Gene | Probe sequence (5′-3′) | |
| N | ACA CTA GCC ATC CTT ACT GCG CTT CG |
2.8. Plaque assay
Virus stocks and infectious virus particles released in the cultures were titrated by plaque assay using Vero E6 cells. Briefly, the cells were incubated with 10-fold dilutions of the samples for 1 h at 37 °C and 5% CO2. After adsorption, medium was replaced by fresh DMEM supplemented with 1% FBS and 1.4% carboxymethylcellulose (Sigma-Aldrich). The cells were cultured for 4 days at 37 °C/5% CO2, then fixed with 4% formaldehyde, and stained with 1% crystal violet diluted in 20% methanol for plaque visualization and quantification. Viral titers were expressed as plaque forming units (PFU) per milliliter (mL) (PFU/mL).
2.9. Analysis of ACE2, CD147 and spike proteins expression by flow cytometry and western blotting
The surface expression of ACE-2 in Calu-3 cells, and HEK-293 or Vero cells overexpressing ACE2 (293-ACE2+ and Vero-ACE2+) were evaluated before and after 1h treatment with 20 mM HP-BCD. For this, flow cytometry analysis of intact nonpermeabilized cells was performed using two different primary antibodies (MA5-31395, Thermo Fisher Scientific Inc.- Fig. 3; or neutralizing Ab272500, Abcam-Fig. S2), followed by FITC-conjugated secondary antibody. Cells were acquired in a FACSCanto ™ flow cytometer and analyzed using FlowJo software (v10.8.1; Becton Dickson Immunocytometry System).
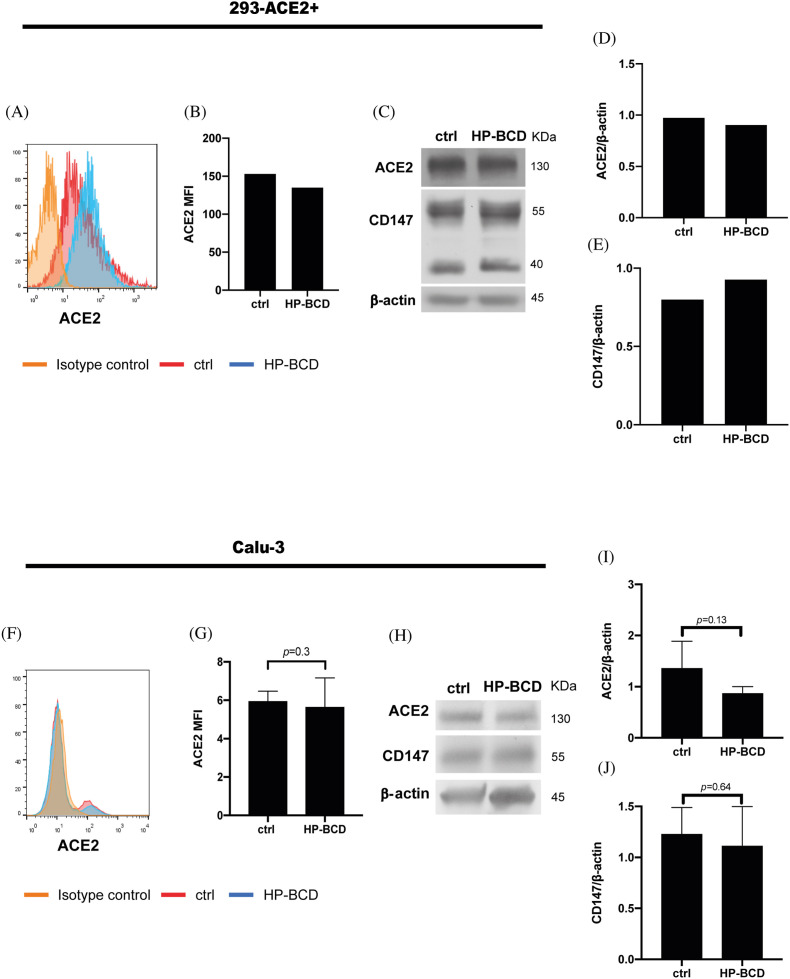
Fig. 3.
HP-BCD does not affect the expression of SARS-CoV-2 receptors. ACE-2-expressing HEK-293 cells or Calu-3 cells were treated with 20 mM HP-BCD for 1 h. The expression of ACE2 in intact nonpermeabilized cells was analyzed by flow cytometry, whereas total expression of ACE2 and CD147 were evaluated in the cell lysates by western blotting. Representative histograms of ACE2 staining and flow cytometry analysis are depicted in (A) and (F); and the MFI average obtained from independent experiments represented in (B) and (G). For the western blotting analysis, the membranes were stained with anti-ACE2, anti-CD147, and anti-β-actin, as a loading control. Representative blots are demonstrated in (C) and (H); the expression level of ACE-2 (D, I) and CD147 (E, J) were estimated using ImageJ software and normalized according to β-actin expression. Three experiments were run with Calu-3 cells and statistical analyses were performed by paired t-test.
Total expression of ACE-2 and CD147 were analyzed in 293-ACE2+, Vero-ACE-2, or Calu-3 by western blotting. The cells were maintained in culture medium or treated with 20 mM HP-BCD for 1 h and then, harvested in RIPA buffer (10 mM Tris-Cl [pH 8.0]; 1 mM EDTA; 0.5 mM EGTA; 1% Triton X-100; 0.1% sodium deoxycholate; 0.1% SDS; 140 mM NaCl) with 0.1% protease inhibitors cocktail (Sigma-Aldrich). Protein extracts were subjected to polyacrylamide gel electrophoresis (SDS-PAGE), followed by transfer to nitrocellulose membrane (Thermo Fisher Scientific Inc.). Membranes were blocked using 5% nonfat dried milk diluted in 1X Tris Buffered Saline with 1% Tween 20 (Isofar, Rio de Janeiro, Brazil) to reduce background. Membranes were incubated with anti-ACE2 (Ref.: MA5-31395, Thermo Fisher Scientific Inc.- Fig. 3; or Ref.: Ab108252, Abcam-Fig. S2), anti-CD147 (Ref.: 34–5600, Thermo Fisher Scientific Inc.), and anti-β-actin (Ref.: A2228, Sigma-Aldrich), followed by incubation with HRP-conjugated secondary antibodies (Santa Cruz Biotechnology). To measure SARS-COV-2 spike (S) expression, Calu-3 cells were treated with 20 mM HP-BCD for 1 h, infected with SARS-CoV-2 (MOI = 0.1) in HP-BCD free medium, and collected at 48 hpi. Cell lysates were submitted to electrophoresis, followed by membrane transfer and blocking, as described. Then, the membranes were stained with anti-SARS-CoV-2 S protein (Ref.: 569965, Cell Signaling Technology, Inc), followed by HRP-secondary antibody. Super Signal West Pico chemiluminescent substrate (Thermo Fisher Scientific Inc.) was used for protein detection following manufacturer's instructions. The ratio of the protein of interest and β-actin was determined using ImageJ software.
2.10. Evaluation of cholesterol and dsRNA content by fluorescence microscopy
Vero E6 cells were treated or not with 20 mM HP-BCD and infected with SARS-CoV-2 (gamma strain, MOI 0.1), as described. After 48 h, the cells were fixed with 4% formaldehyde, and stained with filipin (0.05 mg/mL in 3% BSA; Ref.: F9765, Sigma Aldrich) for 2h. Then, cells were incubated with J2 antibody (0,05 mg/mL in 3% BSA, Ref.: RNT-SCI-10010200, Jena Bioscience GmbH), for 2 h, followed by AlexaFluor594-conjugated anti-mouse IgG (1 μg/mL, Ref.: A32744, Invitrogen). The cells were analyzed by fluorescence microscopy, using OLYMPUS IX81 equipment. Five fields from each experimental situation, including a total of about 500 cells were randomly selected in each experiment; the mean fluorescence intensity (MFI) and frequency of J2 positive staining were calculated using ImageJ software.
2.11. Analysis of cytokine expression by RT-qPCR
Calu-3 cells or human primary monocytes were treated with HP-BCD and infected with SARS-CoV-2, as described. In some experiments, cells were stimulated with LPS (100 ng/ml) for 4 h, as a positive control. RNA and cDNA from cell lysates were obtained as previously described and the expression of il6, tnfa, il10, and ccl2 were evaluated by real-time PCR using Power SYBR Green PCR master mix reagent (Thermo Fisher Scientific Inc.) and the primers described in Table 1. The reaction was performed in a StepOnePlus real-time PCR system (Thermo Fisher Scientific Inc.), using the following cycling parameters: 95 °C for 2 min, and 40 cycles composed by 1 cycle consisting of denaturation (95 °C, 15 s) and primer annealing/extension (60 °C, 1 min); finally, samples were subjected to a melting curve analysis to eliminate primer dimers: 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s. The comparative CT method (ΔΔCt) was used to quantify gene expression levels, using GAPDH for normalization.
2.12. Statistical analysis
Data were analyzed using the GraphPad Prism software (GraphPad Software, San Diego, CA, USA). Comparisons were performed by one way ANOVA, followed by Dunnett's multiple comparison test or by t-test when two groups were analyzed. The tests are indicated in every figure legend; p < 0.05 were considered statistically significant.
3. Results
3.1. HP-BCD presents a broad antiviral activity against SARS-CoV-2 variants in vitro
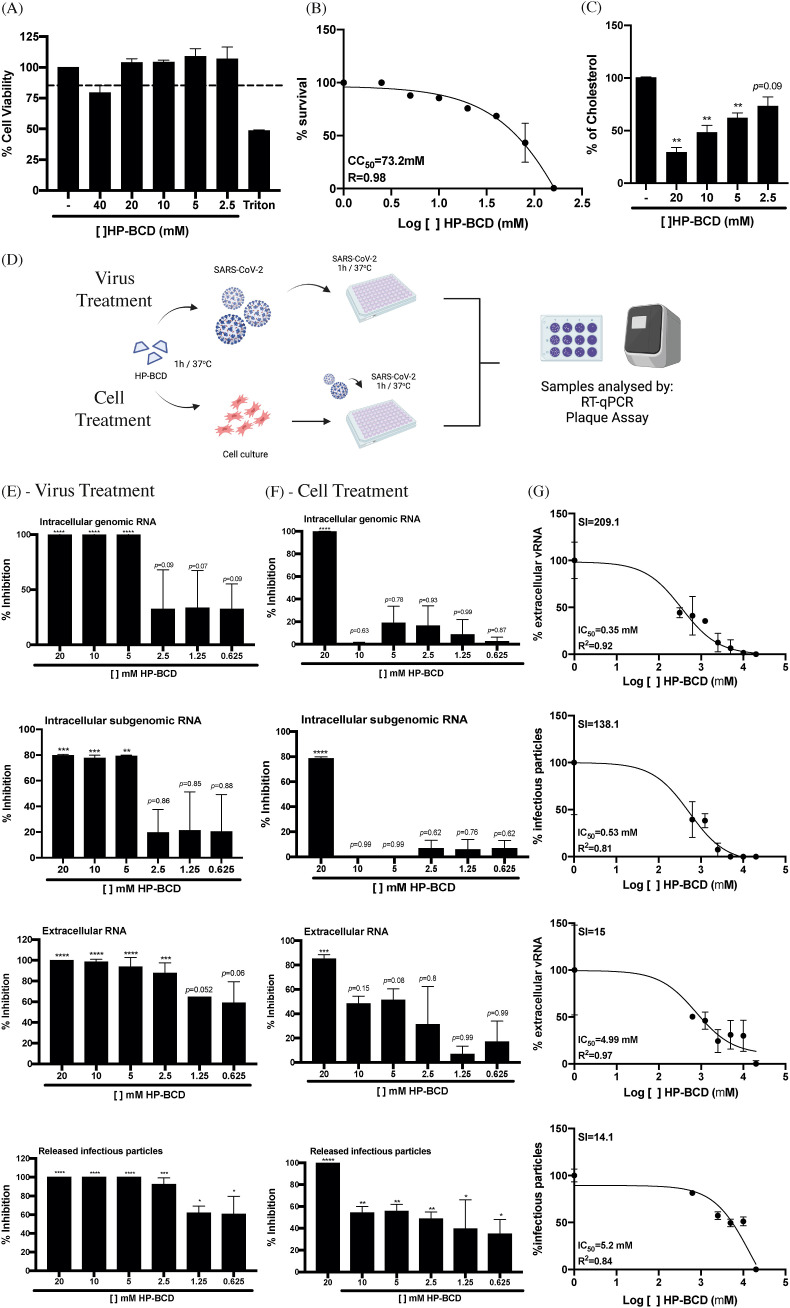
The impact of HP-BCD treatment on SARS-CoV-2 replication was initially evaluated using the well-established Vero E6 cell infection model. First, the cells were treated with different concentrations of HP-BCD, for 1 h, washed and cultured in HP-BCD-free culture medium; cholesterol levels were promptly measured, and cell viability was analyzed at 72 h post treatment. We did not detect any cytotoxic effect after treating the cells with HP-BCD concentration up to 20 mM (Fig. 1 A), and an estimated CC50 of 73.2 mM was determined (Fig. 1B). Significant cholesterol depletion was observed in the cells treated with 5–20 mM HP-BCD, with a maximum of 70% reduction (Fig. 1C).
Fig. 1.
HP-BCD inhibits SARS-CoV-2 replication in Vero cells. A-C) Vero cells were treated with the indicated concentrations of HP-BCD for 1h, the cells were washed and then cultured in complete culture medium. A-B) After 72 h, cell viability was measured using cell-titer blue kit (A); the data was analyzed and the CC50 was calculated using GraphPad Prisma software (B). C) Total cholesterol concentration was measured by amplex red reagent after 1 h treatment. D) Schematic representation of the following assays. Top (Virus treatment): SARS-CoV-2 stock samples (A2 variant) were treated or not with HP-BCD for 1 h, and then inoculated in Vero cells at a MOI of 0.1. After 1 h of virus adsorption, the cells were washed and cultured in HP-BCD-free culture medium for additional 72 h. Bottom (Cell treatment): Vero cells were treated with HP-BCD for 1 h, the cells were washed, and cultured in HP-BCD-free culture medium; then, virus was inoculated at a MOI of 0.1, as described. This figure was created with BioRender.com. E, F) SARS-CoV-2 stock samples or Vero cells were treated with HP-BCD and the infection proceeded, as indicated in (D). After 72 hpi, the cell lysates and culture supernatants were harvested. The concentration of intracellular genomic and subgenomic virus RNA, and released genomic RNA were measured by RT-qPCR; titration of released infectious virus particles was performed by plaque assay. The bars indicate the percentage inhibition of RNA copy numbers or PFU/ml, in relation to untreated virus or cells, obtained from five independent experiments. Statistical analyses were performed by one-way anova, followed by Dunnet's multiple comparison tests; * represents p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. G, H) The released virus RNA and infectious particles data obtained after virus treatment (G) or cell treatment (H) were analyzed, and the IC50 was calculated using GraphPad Prisma software. The selectivity indexes (SI) were calculated using the CC50 data obtained in (B).
Since HP-BCD may function as an antiviral drug by altering host cell cholesterol content and membrane domains, and by inactivating enveloped virus particles through disruption of the lipid bilayer (Liao et al., 2001; Verma et al., 2018; Liao et al., 2003; Graham et al., 2003; Carro and Damonte, 2013), we either treated the cells or the SARS-CoV-2 stock samples with the drug to verify its impact on virus replication (Fig. 1D). SARS-CoV-2 from A2 variant was incubated with different concentrations of HP-BCD for 1 h and then inoculated onto Vero E6 cells at an MOI of 0.1. After 1 h for virus adsorption, the cells were washed and cultured for further 72 h (Virus treatment). In another set of experiments, the cells were incubated with the indicated concentrations of HP-BCD, for 1 h, washed, and then the virus samples were inoculated, using the same MOI (Cell treatment). At 72 hpi, the cell lysates and culture supernatants obtained from each experimental setting were harvested and intracellular genomic and subgenomic virus RNA, as well as released gRNA and infectious particles were measured. Treatment of SARS-CoV-2 with concentrations as low as 5 mM significantly inhibited both gRNA and sgRNA levels in cells (Fig. 1E). Furthermore, the drug was more potent to reduce released virus RNA and infectious particles by these cells. Notably, no PFU was detected in the medium when the viruses were treated with 5–20 mM HP-BCD (Fig. 1E). Raw virus RNA copy numbers and PFU data, as well as a representative plaque assay are demonstrated in Fig. S1. IC50 of 0.35 mM and 0.53 mM were calculated based on the amount of virus RNA and infectious particles released, given selectivity indexes of 209.1 and 138.1, respectively (Fig. 1G). Similarly, HP-BCD pretreatment of the cells resulted in lower intracellular and released virus RNA and no PFU was detected when the cells were treated with 20 mM HP-BCD (Fig. 1F). Based on the IC50 values of cell treatment (4.99 mM, based on released virus RNA data, and 5.2 mM, when considered infectious particles data), we obtained estimated SIs of 15 and 14.1 (Fig. 1H). These data suggest that HP-BCD may act on the host cells and on free virus particles, affecting SARS-CoV-2 replication.
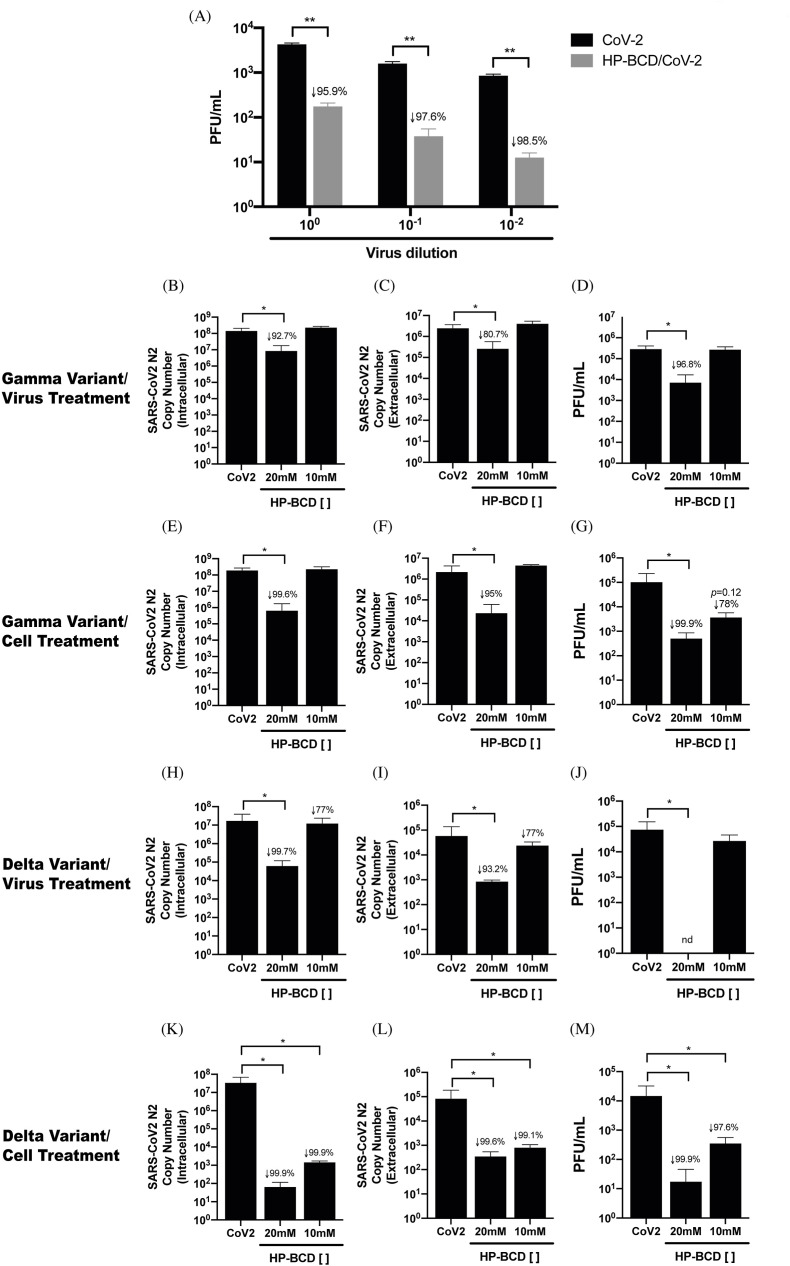
To confirm that HP-BCD was indeed acting on the virus particles, and not only in the host cells, we performed serial dilutions of the HP-BCD-treated virus suspension and compared their titers with equivalent dilutions of HP-BCD-free SARS-CoV-2 samples. The percentage inhibition of virus replication was maintained at every dilution analyzed, indicating that virus treatment effect did not result from residual drug in the cell culture (Fig. 2 A).
Fig. 2.
HP-BCD presents a broad antiviral activity against different SARS-CoV-2 variants of concern. A) SARS-COV-2 stock samples were incubated or not with HP-BCD for 1 h, serially diluted and inoculated onto Vero E6 cells for virus titration. After 48 h, virus titers were calculated and indicated as PFU/ml. Statistical analysis of two independent experiments were performed using t-test; ** represents p < 0.01, and insert numbers indicate the percentage inhibition in relation to non treated viruses. B-M) Vero E6 cells were infected with SARS-CoV-2 sequenced and characterized as Gamma (B–G) or Delta (H–M) variants, at a MOI of 0.1. Cell-free virus or host cell treatments were performed as described in Fig. 1. After 72 h, cell lysates and culture supernatants were harvested. The concentration of intracellular (B, E, H, K) and released virus RNA (N2 copy numbers) (C, F, I, L) were measured by RT-qPCR; titration of released infectious virus particles was performed by plaque assay and represented as PFU/ml (D, G, J, M). The bars indicate the average and SD of two independent experiments, in triplicate. Statistical analyses were performed by one-way anova, followed by Dunnet's multiple comparison; * represents p < 0.05, and insert numbers indicate the percentage inhibition in relation to infected non treated cells; nd-not detected.
We then evaluated the HP-BCD effect upon infection with different SARS-CoV-2 variants. Vero E6 cells and SARS-CoV-2 from Gamma or Delta variants were treated with 10 or 20 mM HP-BCD, as before. We observed that both virus and cell treatment with 20 mM HP-BCD significantly inhibited the accumulation of intracellular virus RNA and substantially reduced the release of virus RNA and infectious particles (Fig. 2B–M), with inhibition indexes above 90%. Notably, the reduction of Delta variant replication was even more pronounced and no PFU was detected upon virus treatment with 20 mM HP-BCD (Fig. 2 J). These data clearly demonstrate that HP-BCD presents a broad antiviral activity against SARS-CoV-2 variants, acting on both the host cell and virus particles.
3.2. HP-BCD antiviral activity does not involve ACE2 depletion
SARS-CoV-2-host cell entry primarily depends on virus Spike protein interaction with ACE2 receptor (Hoffmann et al., 2020), and a few studies suggested that other receptors, like CD147, and mechanisms such as cell-cell fusion may be alternative entry pathways (Hoffmann et al., 2020; Wang et al., 2020; Sanders et al., 2021). Since ACE2 may be expressed in cholesterol-rich domains (Lu et al., 2008), we evaluated whether HP-BCD downregulates virus receptor’ expression. Initially, we analyzed ACE2 expression in 293 cells modified to overexpress the human receptor (293-ACE2+). Flow cytometry and western blotting analyses demonstrated that ACE2 expression were unaffected by cellular treatment with HP-BCD (Fig. 3A–D). Analysis of CD147 expression by western blotting showed that treatment did not alter expression of this receptor either (Fig. 3C, E). Given the overexpression status of transfected cells, we performed a similar assay using Calu-3 cells, which also express both receptors, but at lower physiological levels. Like 293 cells, no significant alteration of ACE2 nor CD147 was detected after HP-BCD treatment (Fig. 3F–J). To confirm the data, Calu-3 cells were also stained with other antibodies, which were targeted against different epitopes and suitable for either flow cytometry or western blotting assays. Vero cells overexpressing ACE2 (Vero-ACE2+) were also evaluated, as controls. Consistently with the previous data, no difference was detected in ACE2 expression after HP-BCD treatment, irrespective of the assay and cell type (Fig. S2).
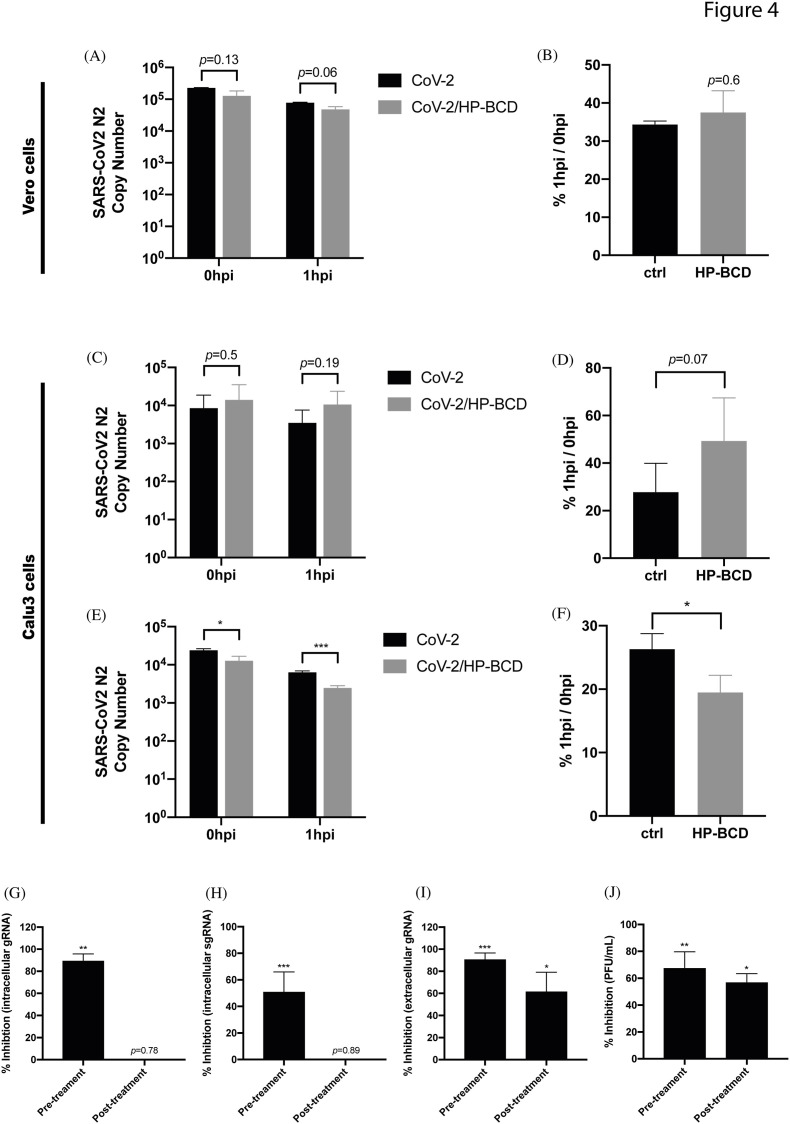
Even though receptor expression was not impacted by HP-BCD treatment, lipid raft disruption by the compound could affect receptor distribution and, therefore, virus adsorption and entry (Lu et al., 2008). Therefore, we set up another experimental approach to investigate these initial steps of SARS-CoV-2 infection. Vero E6 cells were pretreated with HP-BCD for 1 h before virus inoculation. Cells were then cultured with SARS-CoV-2, for 1 h, at 4 °C for virus adsorption. Part of the cells were harvested (0 hpi) and part were further incubated for 1 h, at 37 °C, to allow virus entry (1 hpi). Evaluation of virus RNA obtained at those time points revealed no difference between treated and untreated cells (Fig. 4 A), although a slight, but not significant decrease had been detected at 1 hpi in HP-BCD-treated cultures. These data indicate that virus adsorption or early entrance events into Vero E6 cells might not be the major targets of HP-BCD. In addition, HP-BCD treatment did not affect the efficiency of virus entrance, as calculated by the ratio between virus RNA copies detected at 1 and 0 hpi (Fig. 4B). To confirm this finding, the experiments were also performed using Calu-3 cells. Like the previous results, we did not detect any significant alteration in the number of virus RNA copies in cells treated or not with HP-BCD, after adsorption and after 1 h incubation (Fig. 4C and D). On the other hand, when the viruses were incubated with the drug, we detected a small, but significant difference in the number of RNA copies after virus adsorption and 1 h later (Fig. 4E). Also, a lower entry efficiency was evidenced when the cells were inoculated with HP-BCD-pretreated viruses (Fig. 4F).
Fig. 4.
SARS-CoV-2 incubation, but not cell treatment, with HP-BCD reduces SARS-CoV-2 adsorption and entry. A–B) Vero cells were treated with 20 mM HP-BCD for 1h, washed, and cultured in HP-BCD-free culture medium. SARS-CoV-2 (A2 strain) was inoculated at an MOI of 0.1 for 1 h/4 °C for virus adsorption and harvested (0 hpi) or further incubated for 1 h/37 °C for virus entry (1 hpi). Virus RNA was measured by qRT-PCR (A) and the frequency of virus entry in relation to virus adsorption (%1 hpi/0 hpi) was calculated (B). C-D) Calu-3 cells were treated with HP-BCD, infected with SARS-CoV2, and virus adsorption and entry were analyzed as in (A–B). E-F) SARS-CoV-2 stock samples were incubated with 20 mM HP-BCD and then inoculated onto Calu-3 cells cultures. Analysis of virus RNA copy numbers after virus adsorption (0 hpi) and virus entry (1 hpi) was measured as in (A, C) and the entrance ratio (1 hpi/0 hpi) was calculated as in (B, D). The bars indicate the average and SD from three independent experiments; statistical analyses were performed by unpaired t-test. G-J) Vero cells were treated or not with HP-BCD and infected with SARS-CoV-2 as previously described (pre-treatment). In some wells, the cells were incubated with SARS-CoV-2 for 1 h, for virus adsorption, washed, and maintained in culture for 1 h more. Then, the cells were treated with 20 mM HP-BCD (post treatment). After 48 hpi, the analysis of intracellular virus gRNA (G), sgRNA (H), and extracelular gRNA (I) were performed by RT-qPCR, and titration of released infectious particles (J) was performed by plaque assay. The bars indicate the percentage inhibition of RNA copy numbers or PFU/ml, in relation to untreated virus or cells, obtained from two independent experiments; statistical analyses were performed by one-way anova, followed by Dunnet's multiple comparison tests; * represents p < 0.05; **p < 0.01; ***p < 0.001.
To investigate drug activity after those initial steps, cells were infected and then treated with HP-BCD after 1 h post adsorption (1 hpi). We observed that treating the cells with HP-BCD after virus adsorption and initial entry steps, although had been slightly less efficient than the pre-treatment protocol, still resulted in significant reduction of released virus RNA (p = 0.021) and infectious particles (p = 0.023), but not of intracellular gRNA no sgRNA (Fig. 4G–J). Taken together, these data suggest that HP-BCD does not prevent SARS-CoV-2 binding and initial entry into host cells but affects virus infectivity and production of infectious virus.
3.3. Host cell treatment with HP-BCD reduces the accumulation of SARS-CoV-2 RNA and S protein and restrains virus-induced CPE
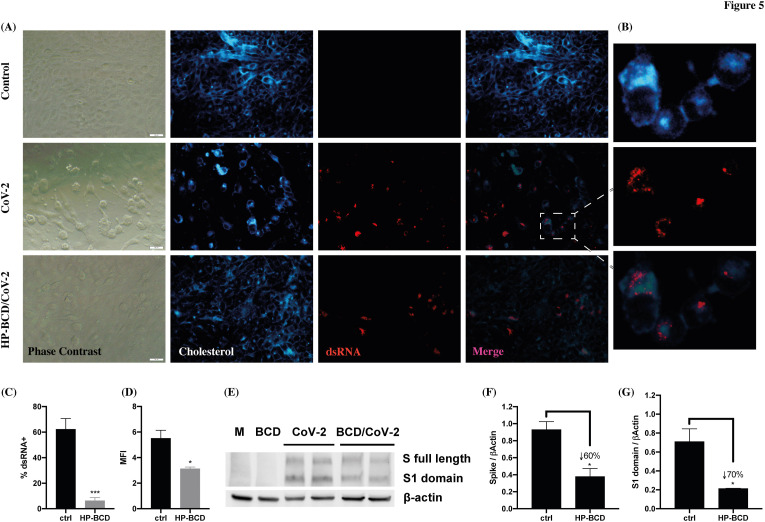
To further investigate HP-BCD effect on host cells, infected Vero E6 cells, treated or not with HP-BCD, were double stained with J2 antibody and filipin to evaluate the expression and distribution of dsRNA and cholesterol. Cell death was not detected at this time point (data not shown), but clear cytopathic effect was observed in the SARS-COV-2-infected cultures, evidenced by alterations in the cell morphology and in the monolayer confluency (Fig. 5 A – phase contrast). Infected cells showed altered distribution in the cellular cholesterol, which accumulated at the proximity of dsRNA enriched compartments (Fig. 5A and B – cholesterol and dsRNA). A remarkable reduction in the frequency of dsRNA positive cells was observed when the cells were pretreated with HP-BCD (Fig. 5C–D). Also, despite similar overall content, the cholesterol distribution and its accumulation close to dsRNA was barely detected, suggesting that the drug may affect assembly of the virus replication complex. Western blotting analysis of Calu-3 cells showed an estimated 50% reduction in Spike protein expression, and it was even more pronounced when only S1 domain was evaluated (Fig. 5E–G). This finding suggests that protein synthesis and/or maturation could also be affected by HP-BCD. Alternatively, reduced RNA and protein detection may be a consequence of reduced virus infectivity revealed after multiple cycles in the culture.
Fig. 5.
HP-BCD treatment inhibits virus RNA and protein expression and reduces SARS-CoV-2-mediated CPE. Vero cells were treated with 20 mM HP-BCD for 1 h. The cells were washed, and mock-treated (control) or infected with SARS-CoV-2 (gamma isolate; MOI = 0.1) for 48 h. A-C) Expression and distribution of dsRNA and cholesterol were evaluated by staining the cells with J2 antibody and filipin, followed by fluorescence microscopy analysis. Representative images are demonstrated in (A; scale bar = 20 μm), with a zoom of infected cell depicted in (B); mean fluorescence intensity (MFI) and frequency of J2 positive staining (% dsRNA+) of 5 randomly selected fields, with a total of about 500 cells, were calculated using ImageJ software and are represented in (C) and (D), respectively. E-G) Calu-3 cells were lysed and the expression of Spike protein was analyzed by western blotting. A representative blot is demonstrated in (E) and the expression level of whole S (F) or S1 (G) was calculated based on β-actin loading control staining, using ImageJ software. Statistical analyses were performed by unpaired t-test; * represents p < 0.05; ***p < 0.001; insert numbers indicate the percentage inhibition in relation to untreated cells.
3.4. HP-BCD restricts SARS-CoV-2 replication and inhibit cytokine production in human alveolar epithelial cell line and primary monocytes
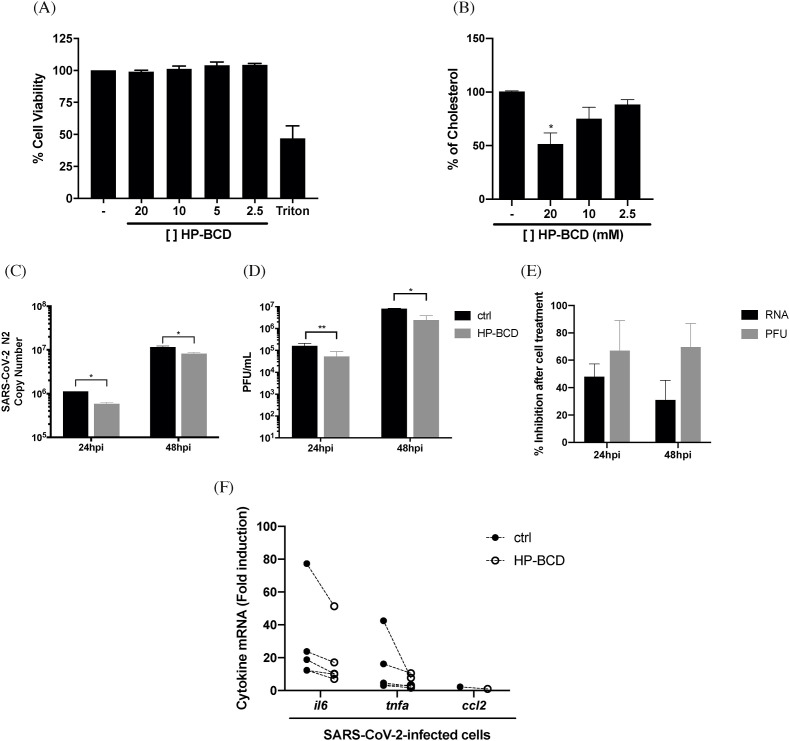
Airway epithelial cells are major targets of SARS-CoV-2 infection and, together with monocytes and other immune cells, play a central role in triggering innate immune response and inflammation during COVID-19. Therefore, we next tested if HP-BCD treatment would affect viral replication in these relevant cell types. Calu-3 cells were used as a model of lung epithelial cells, whereas human primary monocytes were isolated from healthy donor's blood. Regarding Calu-3 cells, we initially performed a toxicity assay with HP-BCD concentrations ranging from 2.5 to 20 mM and none were toxic (Fig. 6 A). Cholesterol depletion was observed at higher concentrations tested, although less efficiently than observed in Vero E6 cells (Fig. 6B). Virus replication was accessed at 24 and 48 hpi in cells pretreated or not with 20 mM HP-BCD. We observed that the release of both virus RNA and infectious particles were significantly reduced (Fig. 6C–D), with inhibition levels above 50% and 60%, respectively (Fig. 6E), corresponding to the estimated frequency of cholesterol depletion. Infected cell lysates were also harvested, and the expression of relevant cytokines were analyzed. Reduction in TNF-α, IL-6 and CCL2 expression was detected in all experiments, as indicated in Fig. 6F.
Fig. 6.
Treatment of Calu3 cells with HP-BCD inhibit SARS-CoV-2 replication and cytokine production. Calu-3 cells were treated with the indicated concentrations of HP-BCD for 1 h, the cells were washed and then cultured in HP-BCD-free culture medium. A) After 72 h, cell viability was measured using cell-titer blue kit. B) Total cholesterol concentration was measured after 1 h treatment by amplex red reagent. C-E) The cells were treated or not with 20 mM HP-BCD for 1 h, washed, and the medium substituted by culture medium. Then, the cells were infected with SARS-CoV-2 (A2 isolate; MOI of 0.1). After 24 or 48 hpi, and the concentration of virus RNA (N2 log copy numbers; C) or infectious particles (PFU) (D) released in the supernatants were measured by qRT-PCR and plaque assay, respectively. The percentage inhibition of both RNA and PFU released by HP-BCD treated, in relation to untreated cells, was calculated and demonstrated in (E). F) mRNA expression corresponding to IL-6, TNF-α and CCL2 were evaluated in the cell lysates, at 48 hpi, by qRT-PCR; individual mRNA fold induction obtained from infected untreated cells (closed symbols) and infected HP-BCD-treated cells (open symbols), in each individual experiment, are indicated. The data are representative of four independent experiments. * represents p < 0.05; **p < 0.01, according to paired t-test statistical analysis.
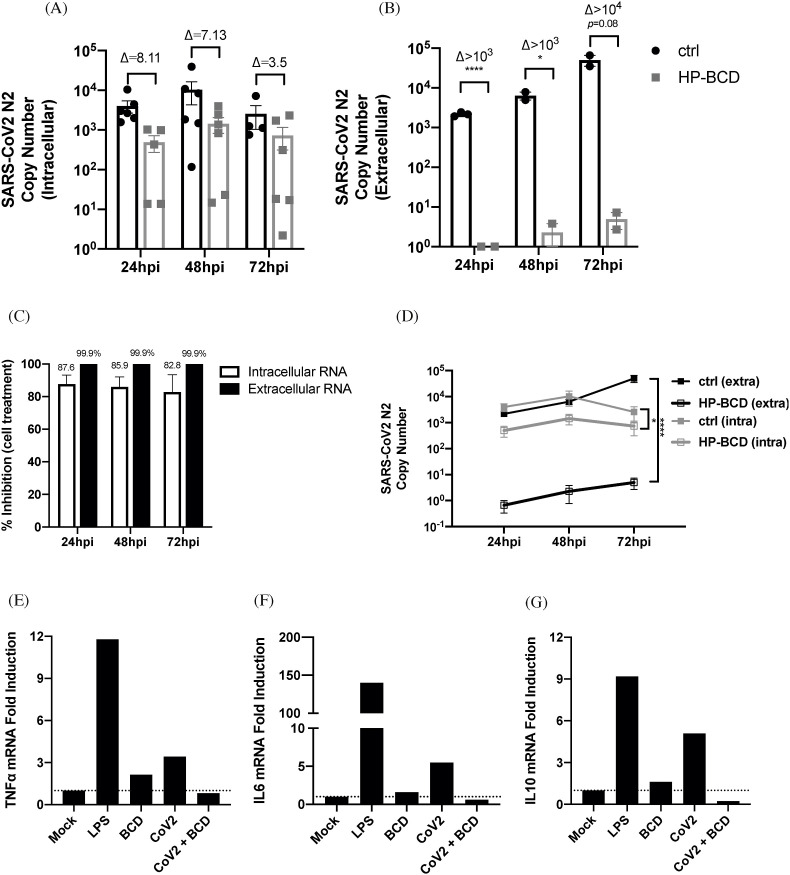
Similar results were observed when primary human monocytes were analyzed, using the previously established nontoxic HP-BCD concentrations (Matassoli et al., 2018). SARS-CoV-2 replication was not as efficient in monocytes as observed in the cell lines and PFU was not detected in all donors evaluated (data not shown). We detected a remarkable reduction in the concentration of intracellular and released virus RNA, with a 4log reduction at 72 hpi when the cells were pretreated with HP-BCD (Fig. 7 A and B). Inhibition above 80% and 99% was detected when N2 copy number was analyzed into the cells and in the culture medium respectively (Fig. 7C). Importantly, a kinetic comparison between untreated and HP-BCD-treated infected cells demonstrated a significant inhibition over the culture period (Fig. 7D). As a proof of concept, we analyzed the cytokine expression in cells obtained from one of these donors. LPS was added to the cultures as a positive control. As demonstrated with Calu-3 cells, treatment of primary human monocytes with HP-BCD diminished virus-induced cytokine expression to the same levels of mock-cultured cells, as evidenced by TNF-α, IL-6 and IL-10 RT-qPCR (Fig. 7E–G).
Fig. 7.
Treatment of human primary monocytes with HP-BCD inhibits SARS-CoV-2 replication and cytokine expression. A-D) Human primary monocytes were treated or not with 10 mM HP-BCD for 1 h, the cells were washed and then mock treated or infected with SARS-CoV-2 (A2 strain; MOI = 0.1). After 24–72 hpi, the cell lysates and supernatants were harvested, and virus RNA concentration (N2 log copy numbers) was evaluated by qRT-PCR. A-B) Bars indicate the average and SD of SARS-CoV-2-N2 copy numbers detected in the cell lysates (intracellular; A) or in the supernatants (extracellular; B), and dots indicate the data obtained from individual donors; the ratio between treated and untreated cells were indicate as (D). C) The percentage inhibition of viral RNA levels was calculated from the data obtained in (A) and (B). D) Lines indicate the kinetic accumulation of intracellular and extracellular virus RNA in HP-BCD-pretreated (HP-BCD) or untreated (ctrl) cells; statistical difference between the obtained AUCs were calculated and *p < 0.05. E-G) Monocytes were pretreated or not with 10 mM HP-BCD, then, the cells were mock-treated or infected with SARS-CoV-2 at a MOI of 0.5. LPS was added as a positive control. After 24 h, the cell lysates were harvested and TNF-α (E), IL-6 (F), and IL-10 (G) mRNA levels were measured by qRT-PCR and calculated by ΔΔCt method, based on GAPDH mRNA expression; bars indicate fold induction in relation to mock-treated cells.
4. Discussion
In the present study we demonstrated that HP-BCD is a broad and potent antiviral drug, inhibiting the replication of distinct SARS-CoV-2 variants, in different cell types, and significantly suppressing virus-induced production of inflammatory cytokines.
The COVID-19 pandemic represents an unprecedented public health challenge, and a number of approaches are required to minimize its consequences. The development of safe, effective vaccines has notably improved the epidemiological scenario, with significantly lower rates of severe and fatal cases among vaccinated people. Still, although vaccination reduces the likelihood of disease, it does not completely prevent transmission. Furthermore, the vaccination ratio is unequal around the globe, allowing continuous virus circulation. This results in a fast virus evolution and emergence of new variants, which may eventually escape host and vaccine-mediated immune responses causing sequential infection waves. Also, the elderly and other patients with comorbidities may still develop severe symptoms, which will require alternative therapeutic strategies, ideally working against any circulating or emergent virus variant.
Beta-cyclodextrins (BCD) have been demonstrated to have antiviral properties against several enveloped viruses. Its predicted mechanism of action involves cholesterol depletion and disruption of lipid rafts, cholesterol-rich membrane domains. Therefore, BCD may potentially inhibit the infection by viruses that requires cholesterol or lipid rafts to maintain its infectivity and/or complete its biosynthesis. Indeed, we and others have shown that treating viruses, like HIV, SIV, Influenza, DENV, SARS-CoV, or their respective host cells with different derivatives of BCD blocks virus replication in in vitro and in vivo experimental models (Liao et al., 2001; Khanna et al., 2002; Graham et al., 2003; Ambrose et al., 2008; García Cordero et al., 2014; Verma et al., 2018; Goncharova et al., 2019; Jones et al., 2020). Most important, HP-BCD has been used for more than a decade for the treatment of NPCD, in adults and children, with efficacy and safety (Ory et al., 2017; Hastings et al., 2019).
Recent studies demonstrated that the intracellular SARS-CoV-2 replication complex appears to be localized in cholesterol-rich membranes, in which accumulation of virus RNA was detected (Sanders et al., 2021). Furthermore, cholesterol depletion reduced Spike-mediated cell-cell fusion and syncytia formation in vitro (Sanders et al., 2021), a process likely to hinder virus transmission. A loss-of-function screening assay identified the cholesterol transporter Nieman-Pick disease type C1 (NPC1) receptor, as one of the host factors required for virus replication (Zhu et al., 2021). Although the exact function of NPC1 had not been addressed, its interaction with SARS-CoV-2 N protein has been demonstrated (García-Dorival et al., 2021). Also, NPC1 gene editing or incubation of infected cells with NPC1-interacting compounds inhibited virus replication, possibly through impairing late endosome-lysosome fusion and virus uncoating (García-Dorival et al., 2021; Zhu et al., 2021).
Our data demonstrated that treatment of SARS-CoV-2 or Vero E6 cells with HP-BCD significantly inhibited virus replication, by acting on the viruses and host cells. Also, the determined IC50 was lower than the predicted plasma concentrations achieved in the intravenous treatment of NPCD patients (Hastings et al., 2019). Despite cholesterol depletion, treatment of host cells with HP-BCD did not reduce ACE2 expression, which could be harmful for the patient, given the important physiological roles of this receptor controlling renin-angiotensin and kallikrein-kinin systems’ functions (Garvin et al., 2020). Although there are indications that ACE2 may be expressed in raft domains, previous studies regarding SARS-CoV infection corroborate our data. Vero cells treatment with methyl-BCD (MBCD) inhibited the replication of SARS-CoV, but no alteration in ACE2 expression or distribution was detected (Li et al., 2007). Another work reported that ACE2 was present in caveolin-enriched fractions, and it was relocated to nonraft fractions after treating Vero cells with MBCD. Still, ACE2 surface expression or SARS-CoV spike binding were also not affected by MBCD treatment, despite reduced virus replication (Lu et al., 2008).
We did not detect a significant effect in virus binding after treating Vero E6 or Calu-3 cells with HP-BCD, unless the virus particles were treated. In addition, virus gRNA copies were not significantly affected after 1 hpi, suggesting that initial entry events are not affected by the drug either, although we cannot discard that gRNA detected at 1 hpi might include residual attached viruses. Our result partially contrasts with previous data obtained with a pseudovirus model of SARS-CoV-2, in which pretreatment of 293-ACE2+ cells with BCD inhibited infection (Li et al., 2021). It is important to note, however, that the authors maintained the drug for a total 5 h during pseudovirus infection, before washing the cells. Therefore, other early steps could also be affected in that assay, such as virus uncoating in late endosomes/lysosomes, or assembly of membrane structured virus replication complexes. Also, one cannot rule out that depending on the construction and preparation, pseudovirus may not fully serve as surrogate for native virus, in terms of binding and fusion mechanisms (Chen et al., 2021).
SARS-CoV-2 entrance relies on different pathways, including membrane fusion or endocytosis, depending on the cell type and on the expression of TMPRSS2 protease (Hoffmann et al., 2020; Dittmar et al., 2021). Our data demonstrated that HP-BCD did not reduce virus binding/entry in cell lines that expresses (Calu-3) or not (Vero-E6) the protease (Dittmar et al., 2021), but virus production was significantly inhibited in both cell types. These findings support the hypothesis that other mechanisms downstream of initial virus-host cell interaction might be important for the activity of the drug. Accordingly, cell treatment after 1 hpi still reduced the levels of gRNA and infectious particles released, although did not significantly affect the intracellular vRNA concentration. These data support the hypothesis that HP-BCD might impact virus biosynthesis and/or affect the infectivity of released viruses, what worth further investigation. For instance, blocking of virus release would consequently imply accumulation of viral gRNA. Also, in every investigated cell type, the drug was more potent in reducing the released virus RNA and infectious particles, in comparison with intracellular RNA, further suggesting that later steps of virus biosynthesis could have been affected. Similar data was reported in the SARS-CoV studies, in which the treatment of host cells with MBCD at 3 hpi also resulted in diminished expression of virus mRNA, although with lower efficiency, when compared to pre-treatment (Li et al., 2007).
Our group has been mostly working with 2-hydroxi-propyl-beta-cyclodextrin (HP-BCD), instead of MBCD, which is less toxic than MBCD, but may be less efficient in depleting cholesterol and thus could act differently than MBCD. We demonstrated that HP-BCD inhibited HIV replication in treated primary CD4T cells (Liao et al., 2001). In addition, virus treatment with the drug did not affect virus morphology nor its binding to host cells, but inhibited virus fusion and induced viral membrane permeabilization with loss of selected virus proteins and consequent inactivation (Graham et al., 2003; Liao et al., 2003). Similar data were obtained after treatment of the gammaretrovirus XMRV (Tang et al., 2012). In this model, pretreatment of host cell with HP-BCD remarkably inhibited virus release, but not intracellular capsid content, suggesting that the drug functioned at assembly and/or budding steps.
Vero E6 cells infected with SARS-CoV-2 showed an accumulation of cholesterol at the proximity with dsRNA-enriched intracellular regions. When the cells were pretreated with HP-BCD, we noticed a reorganization of intracellular cholesterol distribution and a remarkable reduction of dsRNA staining, suggesting that the drug may interfere with replication complex assembly. However, since those assays were performed at 48 hpi, we cannot exclude that decreased dsRNA and protein resulted from lower infectivity of released virus particles, reducing the infection rates at subsequential replication cycles. Indeed, by comparing the inhibition of intracellular virus genomic and subgenomic RNA, released virus RNA, and released infectious particles, one can assume that HP-BCD mostly impaired the secretion of infectious particles. Remarkably, no PFU was detected after cell treatment with 20 mM HP-BCD, even when virus RNA was detected in the culture medium. Clarifying this aspect of HP-BCD effect on SARS-CoV2 replication will require further investigation.
Similar findings were reported regarding dengue and influenza infections. Dengue NS3 protease interacts with caveolin, in a predicted association with cholesterol-rich membranes. Host cell treatment with MBCD inhibited the release of infectious particles, possibly due to reduced DENV polyprotein processing (García Cordero et al., 2014). Also, other studies had demonstrated that DENV treatment with MBCD affected virus infectivity but had no effect on virus binding or entry (Carro and Damonte, 2013; Carro et al., 2018). In vitro models of Influenza infection showed that MBCD inhibited the transport of viral proteins HA from trans-Golgi network to the apical membrane and also disrupted virus membrane integrity, lowering the infectivity of the budding Influenza virus (Keller and Simons, 1998; Barman and Nayak, 2007). Accordingly, a sulfonated β-cyclodextrin derivative did not affect earlier steps of virus replication, but inhibited influenza infection in vitro and in vivo (Goncharova et al., 2019).
A major finding revealed from our study is that HP-BCD treatment also suppressed SARS-CoV-2-induced cellular activation and cytokine production. Lung inflammation, sometimes accompanied with systemic syndromes, are central issues associated to severe COVID-19 outcomes (Yao et al., 2021). Patients with severe disease present with extensive infiltration of leukocytes, and virus transcripts are detected in monocytes in the lung. Increased BAL and plasma cytokine levels are markers of severe disease and might contribute to exacerbated inflammation (Coperchini et al., 2020; Lucas et al., 2020; Yao et al., 2021). We had previously demonstrated that monocyte treatment with HP-BCD downregulated the expression and secretion of certain inflammatory cytokines, especially TNF-α, IL-10, and IL-6 (Matassoli et al., 2018). Those cytokines are also upregulated by monocyte infection with SARS-CoV-2, although at much lower level in comparison to LPS. HP-BCD treatment reduced cytokine production to the levels of mock-treated cultures. Treatment of lung epithelial cell line Calu-3 cells also inhibited TNF-α and IL-6 expression, as well as CCL2 chemokine. The latter may contribute to lower monocytes and neutrophils recruitment and activation, diminishing airway respiratory inflammation and COVID-19 symptoms. This effect may be a consequence of lower virus replication and RNA sensing but could also be related to altered lipid metabolism. Inhibition of lipid droplet formation decreased IL-6, TNF-α and IL-10 produced by monocytes infected with SARS-CoV-2 (Dias et al., 2020). In addition, our previous study showed that HP-BCD impaired signal transduction pathways triggered by toll like receptors activation (Matassoli et al., 2018). Even though we can still not define whether decreased cytokine production was a direct effect of HP-BCD on cytokine gene expression or a consequence of lower virus replication and sensing, reduced inflammation might benefit the patients, if clinical trials proceed.
A limitation of our study is that we did not address virus replication in lung primary cells, which could further strength the findings. Still, in conclusion, we showed that HP-BCD potently inhibited the replication of SARS-CoV-2 variants, and downregulated cytokine production by major host cell models, suggesting that it may function as a broad-spectrum compound, diminishing viral load and inflammatory condition in COVID-19. HP-BCD warrants further investigation as a potential COVID-19 therapeutic agent in pre-clinical and clinical investigations.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by Rede Corona-ômica BR MCTI/FINEP affiliated to RedeVírus/MCTI (FINEP = 01.20.0029.000462/20, CNPq 404096/2020-4), Brazil; Coordination for the Improvement of Higher Education Personnel (CAPES), Brazil; Brazilian National Council for Scientific and Technological Development (CNPq), Brazil; Carlos Chagas Filho Research Support Foundation (FAPERJ; LBA E-26/201.206/2021, E-26/210.371/2019), Brazil; Fundação Serrapilheira, Brazil; and National Institute of Health (#2013/21719-3), USA. BBB, SVAC and BMM were recipients of a FAPERJ fellowship. GPDS is recipient of a FUJB fellowship. AT, LJC, LBA, IAC and MRMS were recipients of a CNPq fellowship. The authors wish to thank Franklin Nouvet, (Meharry Medical College) and Ronaldo Rocha (UFRJ) for technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105373.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ackermann M., Anders H.J., Bilyy R., Bowlin G.L., Daniel C., De Lorenzo R., Egeblad M., Henneck T., Hidalgo A., Hoffmann M., Hohberger B., Kanthi Y., Kaplan M.J., Knight J.S., Knopf J., Kolaczkowska E., Kubes P., Leppkes M., Mahajan A., Manfredi A.A., Maueröder C., Maugeri N., Mitroulis I., Muñoz L.E., Narasaraju T., Naschberger E., Neeli I., Ng L.G., Radic M.Z., Ritis K., Rovere-Querini P., Schapher M., Schauer C., Simon H.U., Singh J., Skendros P., Stark K., Stürzl M., van der Vlag J., Vandenabeele P., Vitkov L., von Köckritz-Blickwede M., Yanginlar C., Yousefi S., Zarbock A., Schett G., Herrmann M. Patients with COVID-19: in the dark-NETs of neutrophils. Cell Death Differ. 2021:1–15. doi: 10.1038/s41418-021-00805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose Z., Compton L., Piatak M., Jr., Lu D., Alvord W.G., Lubomirski M.S., Hildreth J.E., Lifson J.D., Miller C.J., KewalRamani V.N. Incomplete protection against simian immunodeficiency virus vaginal transmission in rhesus macaques by a topical antiviral agent revealed by repeat challenges. J. Virol. 2008;82(13):6591–6599. doi: 10.1128/JVI.02730-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assadiasl S., Fatahi Y., Mosharmovahed B., Mohebbi B., Nicknam M.H. Baricitinib: from rheumatoid arthritis to COVID-19. J. Clin. Pharmacol. 2021;61(10):1274–1285. doi: 10.1002/jcph.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman S., Nayak D.P. Lipid raft disruption by cholesterol depletion enhances influenza A virus budding from MDCK cells. J. Virol. 2007;81(22):12169–12178. doi: 10.1128/JVI.00835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal A.J., da Silva M.M.G., Musungaie D.B., Kovalchuk E., Gonzalez A., Reyes V.D., Martín-Quirós A., Caraco Y., Williams-Diaz A., Brown M.L., Du J., Pedley A., Assaid C., Strizki J., Grobler J., Shamsuddin H.H., Tipping R., Wan H., Paschke A., Butterton J.R., Johnson M.G., De Anda C. MOVe-OUT study group. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N. Engl. J. Med. 2021;16 doi: 10.1056/NEJMoa2116044. NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley B.T., Maioli H., Johnston R., Chaudhry I., Fink S.L., Xu H., Najafian B., Deutsch G., Lacy J.M., Williams T., Yarid N., Marshall D.A. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet. 2020;396(10247):320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges J.P., Vladar E.K., Huang H., Mason R.J. Respiratory epithelial cell responses to SARS-CoV-2 in COVID-19. Thorax. 2021;17 doi: 10.1136/thoraxjnl-2021-217561. thoraxjnl-2021-217561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussani R., Schneider E., Zentilin L., Collesi C., Ali H., Braga L., Volpe M.C., Colliva A., Zanconati F., Berlot G., Silvestri F., Zacchigna S., Giacca M. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine. 2020;61 doi: 10.1016/j.ebiom.2020.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro A.C., Damonte E.B. Requirement of cholesterol in the viral envelope for dengue virus infection. Virus Res. 2013 Jun;174(1–2):78–87. doi: 10.1016/j.virusres.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Carro A.C., Piccini L.E., Damonte E.B. Blockade of dengue virus entry into myeloid cells by endocytic inhibitors in the presence or absence of antibodies. PLoS Neglected Trop. Dis. 2018 Aug 9;12(8) doi: 10.1371/journal.pntd.0006685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.Y., Huang C., Tian L., Huang X., Zhang C., Llewellyn G.N., Rogers G.L., Andr’sen K., O'Gorman M.R.G., Chen Y.W., Cannon P.M. Cytoplasmic tail truncation of SARS-CoV-2 spike protein enhances titer of pseudotyped vectors but masks the effect of the D614G mutation. J. Virol. 2021 Oct 27;95(22) doi: 10.1128/JVI.00966-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020 Jun;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Antón-Plágaro C., Shoemark D.K., Simón-Gracia L., Bauer M., Hollandi R., Greber U.F., Horvath P., Sessions R.B., Helenius A., Hiscox J.A., Teesalu T., Matthews D.A., Davidson A.D., Collins B.M., Cullen P.J., Yamauchi Y. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020 Nov 13;370(6518):861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.E., Brewster M.E. Cyclodextrin-based pharmaceutics: past, present and future. Nat. Rev. Drug Discov. 2004 Dec;3(12):1023–1035. doi: 10.1038/nrd1576.PMID:15573101. [DOI] [PubMed] [Google Scholar]
- De Clercq E. Remdesivir. Quo vadis? Biochem. Pharmacol. 2021 Oct 19;193 doi: 10.1016/j.bcp.2021.114800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S.S.G., Soares V.C., Ferreira A.C., Sacramento C.Q., Fintelman-Rodrigues N., Temerozo J.R., Teixeira L., Nunes da Silva M.A., Barreto E., Mattos M., de Freitas C.S., Azevedo-Quintanilha I.G., Manso P.P.A., Miranda M.D., Siqueira M.M., Hottz E.D., Pão C.R.R., Bou-Habib D.C., Barreto-Vieira D.F., Bozza F.A., Souza T.M.L., Bozza P.T. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog. 2020 Dec 16;16(12) doi: 10.1371/journal.ppat.1009127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar M., Lee J.S., Whig K., Segrist E., Li M., Kamalia B., Castellana L., Ayyanathan K., Cardenas-Diaz F.L., Morrisey E.E., Truitt R., Yang W., Jurado K., Samby K., Ramage H., Schultz D.C., Cherry S. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 2021 Apr 6;35(1) doi: 10.1016/j.celrep.2021.108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Cordero J., León Juárez M., González-Y-Merchand J.A., Cedillo Barrón L., Gutiérrez Castañeda B. Caveolin-1 in lipid rafts interacts with dengue virus NS3 during polyprotein processing and replication in HMEC-1 cells. PLoS One. 2014 Mar 18;9(3) doi: 10.1371/journal.pone.0090704. ornal1/journal.pone.0090704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Dorival I., Cuesta-Geijo M.Á., Barrado-Gil L., Galindo I., Garaigorta U., Urquiza J., Puerto A.D., Campillo N.E., Martínez A., Gastaminza P., Gil C., Alonso C. Identification of Niemann-Pick C1 protein as a potential novel SARS-CoV-2 intracellular target. Antivir. Res. 2021 Oct;194 doi: 10.1016/j.antiviral.2021.105167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin M.R., Alvarez C., Miller J.I., Prates E.T., Walker A.M., Amos B.K., Mast A.E., Justice A., Aronow B., Jacobson D. A mechanistic model and therapeutic interventions for COVID-19 involving a RAS-mediated bradykinin storm. Elife. 2020 Jul 7;9 doi: 10.7554/eLife.59177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glende J., Schwegmann-Wessels C., Al-Falah M., Pfefferle S., Qu X., Deng H., Drosten C., Naim H.Y., Herrler G. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology. 2008 Nov 25;381(2):215–221. doi: 10.1016/j.virol.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham D.R., Chertova E., Hilburn J.M., Arthur L.O., Hildreth J.E. Cholesterol depletion of human immunodeficiency virus type 1 and simian immunodeficiency virus with beta-cyclodextrin inactivates and permeabilizes the virions: evidence for virion-associated lipid rafts. J. Virol. 2003 Aug;77(15):8237–8248. doi: 10.1128/jvi.77.15.8237-8248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncharova E.P., Kostyro Y.A., Ivanov A.V., Zenkova M.A. A novel sulfonated derivative of β-cyclodextrin effectively inhibits influenza A virus infection in vitro and in vivo. Acta Naturae. 2019 Jul-Sep;11(3):20–30. doi: 10.32607/20758251-2019-11-3-20-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings C., Vieira C., Liu B., Bascon C., Gao C., Wang R.Y., Casey A., Hrynkow S. Expanded access with intravenous hydroxypropyl-β-cyclodextrin to treat children and young adults with Niemann-Pick disease type C1: a case report analysis. Orphanet J. Rare Dis. 2019 Oct 21;14(1):228. doi: 10.1186/s13023-019-1207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikmet F., Méar L., Å Edvinsson, Micke P., Uhlén M., Lindskog C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020 Jul;16(7) doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 Apr 16;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb 15;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.T., Cagno V., Janeček M., Ortiz D., Gasilova N., Piret J., Gasbarri M., Constant D.A., Han Y., Vuković L., Král P., Kaiser L., Huang S., Constant S., Kirkegaard K., Boivin G., Stellacci F., Tapparel C. Modified cyclodextrins as broad-spectrum antivirals. Sci. Adv. 2020 Jan 29;6(5) doi: 10.1126/sciadv.aax9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P., Simons K. Cholesterol is required for surface transport of influenza virus hemagglutinin. J. Cell Biol. 1998 Mar 23;140(6):1357–1367. doi: 10.1083/jcb.140.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna K.V., Whaley K.J., Zeitlin L., Moench T.R., Mehrazar K., Cone R.A., Liao Z., Hildreth J.E., Hoen T.E., Shultz L., Markham R.B. Vaginal transmission of cell-associated HIV-1 in the mouse is blocked by a topical, membrane-modifying agent. J. Clin. Invest. 2002 Jan;109(2):205–211. doi: 10.1172/JCI13236. PMID: 11805132; PMCID: PMC150835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzberger N., Hirsch C., Chai K.L., Tomlinson E., Khosravi Z., Popp M., Neidhardt M., Piechotta V., Salomon S., Valk S.J., Monsef I., Schmaderer C., Wood E.M., So-Osman C., Roberts D.J., McQuilten Z., Estcourt L.J., Skoetz N. SARS-CoV-2-neutralising monoclonal antibodies for treatment of COVID-19. Cochrane Database Syst. Rev. 2021 Sep 2;9(9) doi: 10.1002/14651858.CD013825.pub2. PMID: 34473343; PMCID: PMC8411904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.M., Li Y.G., Yamate M., Li S.M., Ikuta K. Lipid rafts play an important role in the early stage of severe acute respiratory syndrome-coronavirus life cycle. Microb. Infect. 2007 Jan;9(1):96–102. doi: 10.1016/j.micinf.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhu W., Fan M., Zhang J., Peng Y., Huang F., Wang N., He L., Zhang L., Holmdahl R., Meng L., Lu S. Dependence of SARS-CoV-2 infection on cholesterol-rich lipid raft and endosomal acidification. Comput. Struct. Biotechnol. J. 2021;19:1933–1943. doi: 10.1016/j.csbj.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Z., Cimakasky L.M., Hampton R., Nguyen D.H., Hildreth J.E. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res. Hum. Retrovir. 2001 Jul 20;17(11):1009–1019. doi: 10.1089/088922201300343690. [DOI] [PubMed] [Google Scholar]
- Liao Z., Graham D.R., Hildreth J.E. Lipid rafts and HIV pathogenesis: virion-associated cholesterol is required for fusion and infection of susceptible cells. AIDS Res. Hum. Retrovir. 2003 Aug;19(8):675–687. doi: 10.1089/088922203322280900. [DOI] [PubMed] [Google Scholar]
- Liu K., Yang T., Peng X.F., Lv S.M., Ye X.L., Zhao T.S., Li J.C., Shao Z.J., Lu Q.B., Li J.Y., Liu W. A systematic meta-analysis of immune signatures in patients with CverD-19. Rev. Med. Virol. 2021 Jul;31(4) doi: 10.1002/rmv.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Liu D.X., Tam J.P. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem. Biophys. Res. Commun. 2008 May 2;369(2):344–349. doi: 10.1016/j.bbrc.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., Takahashi T., Tokuyama M., Lu P., Venkataraman A., Park A., Mohanty S., Wang H., Wyllie A.L., Vogels C.B.F., Earnest R., Lapidus S., Ott I.M., Moore A.J., Muenker M.C., Fournier J.B., Campbell M., Odio C.D., Casanovas-Massana A., Impact Team Yale, Herbst R., Shaw A.C., Medzhitov R., Schulz W.L., Grubaugh N.D., Dela Cruz C., Farhadian S., Ko A.I., Omer S.B., Iwasaki A. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020 Aug;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone B., Urakova N., Snijder E.J., Campbell E.A. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022 Jan;23(1):21–39. doi: 10.1038/s41580-021-00432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassoli F.L., Leão I.C., Bezerra B.B., Pollard R.B., Lütjohann D., Hildreth J.E.K., Arruda L.B. Hydroxypropyl-beta-cyclodextrin reduces inflammatory signaling from monocytes: possible implications for suppression of HIV chronic immune activation. mSphere. 2018 Nov 7;3(6) doi: 10.1128/mSphere.00497-18. e00497-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melms J.C., Biermann J., Huang H., Wang Y., Nair A., Tagore S., Katsyv I., Rendeiro A.F., Amin A.D., Schapiro D., Frangieh C.J., Luoma A.M., Filliol A., Fang Y., Ravichandran H., Clausi M.G., Alba G.A., Rogava M., Chen S.W., Ho P., Montoro D.T., Kornberg A.E., Han A.S., Bakhoum M.F., Anandasabapathy N., Suárez-Fariñas M., Bakhoum S.F., Bram Y., Borczuk A., Guo X.V., Lefkowitch J.H., Marboe C., Lagana S.M., Del Portillo A., Tsai E.J., Zorn E., Markowitz G.S., Schwabe R.F., Schwartz R.E., Elemento O., Saqi A., Hibshoosh H., Que J., Izar B. A molecular single-cell lung atlas of lethal COVID-19. Nature. 2021 Jul;595(7865):114–119. doi: 10.1038/s41586-021-03569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G., Bortolasci C.C., Puri B.K., Olive L., Marx W., O'Neil A., Athan E., Carvalho A.F., Maes M., Walder K., Berk M. The pathophysiology of SARS-CoV-2: a suggested model and therapeutic approach. Life Sci. 2020 Oct 1;258 doi: 10.1016/j.lfs.2020.118166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory D.S., Ottinger E.A., Farhat N.Y., King K.A., Jiang X., Weissfeld L., Berry-Kravis E., Davidson C.D., Bianconi S., Keener L.A., Rao R., Soldatos A., Sidhu R., Walters K.A., Xu X., Thurm A., Solomon B., Pavan W.J., Machielse B.N., Kao M., Silber S.A., McKew J.C., Brewer C.C., Vite C.H., Walkley S.U., Austin C.P., Porter F.D. Intrathecal 2-hydroxypropyl-β-cyclodextrin decreases neurological disease progression in Niemann-Pick disease, type C1: a non-randomised, open-label, phase 1-2 trial. Lancet. 2017 Oct 14;390(10104):1758–1768. doi: 10.1016/S0140-6736(17)31465-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottinger E.A., Kao M.L., Carrillo-Carrasco N., Yanjanin N., Shankar R.K., Janssen M., Brewster M., Scott I., Xu X., Cradock J., Terse P., Dehdashti S.J., Marugan J., Zheng W., Portilla L., Hubbs A., Pavan W.J., Heiss J., Vite C.H., Walkley S.U., Ory D.S., Silber S.A., Porter F.D., Austin C.P., McKew J.C. Collaborative development of 2-hydroxypropyl-β-cyclodextrin for the treatment of Niemann-Pick type C1 disease. Curr. Top. Med. Chem. 2014;14(3):330–339. doi: 10.2174/1568026613666131127160118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazi I., Al-Qahtani A.A., Alhamlan F.S., Alothaid H., Matou-Nasri S., Sourvinos G., Vergadi E., Tsatsanis C. SARS-CoV-2/ACE2 interaction suppresses IRAK-M expression and promotes pro-inflammatory cytokine production in macrophages. Front. Immunol. 2021 Jun 23;12 doi: 10.3389/fimmu.2021.683800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T.P., Goldhill D.H., Zhou J., Baillon L., Frise R., Swann O.C., Kugathasan R., Penn R., Brown J.C., Sanchez-David R.Y., Braga L., Williamson M.K., Hassard J.A., Staller E., Hanley B., Osborn M., Giacca M., Davidson A.D., Matthews D.A., Barclay W.S. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021 Jul;6(7):899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- Pontelli M.C., Castro I.A., Martins R.B., Veras F.P., Serra L., Nascimento D.C., Cardoso R.S., Rosales R., Lima T.M., Souza J.P., Caetité D.B., de Lima M.H.F., Kawahisa J.T., Giannini M.C., Bonjorno L.P., Lopes M.I.F., Batah S.S., Siyuan L., Assad R.L., Almeida S.C.L., Oliveira F.R., Benatti M.N., Pontes L.L.F., Santana R.C., Vilar F.C., Martins M.A., Cunha T.M., Calado R.T., Alves-Filho J.C., Zamboni D.S., Fabro A., Louzada-Junior P., Oliveira R.D.R., Cunha F.Q., Arruda E. Vol. 7. 2020. (Infection of Human Lymphomononuclear Cells by SARS-CoV-2). 07.28.225912. [DOI] [Google Scholar]
- Sanders D.W., Jumper C.C., Ackerman P.J., Bracha D., Donlic A., Kim H., Kenney D., Castello-Serrano I., Suzuki S., Tamura T., Tavares A.H., Saeed M., Holehouse A.S., Ploss A., Levental I., Douam F., Padera R.F., Levy B.D., Brangwynne C.P. SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation. Elife. 2021 Apr 23;10 doi: 10.7554/eLife.65962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020 May 26;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.W., Zhang C., Fan X., Meng F.P., Xu Z., Xia P., Cao W.J., Yang T., Dai X.P., Wang S.Y., Xu R.N., Jiang T.J., Li W.G., Zhang D.W., Zhao P., Shi M., Agrati C., Ippolito G., Maeurer M., Zumla A., Wang F.S., Zhang J.Y. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 2020 Jul 8;11(1):3410. doi: 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Whittaker G.R. Role for influenza virus envelope cholesterol in virus entry and infection. J. Virol. 2003 Dec;77(23):12543–12551. doi: 10.1128/jvi.77.23.12543-12551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L., HCA Lung Biological Network SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020 May;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Yamada Y., Ishitsuka Y., Matsuo M., Shiraishi K., Wada K., Uchio Y., Kondo Y., Takeo T., Nakagata N., Higashi T., Motoyama K., Arima H., Mochinaga S., Higaki K., Ohno K., Irie T. Efficacy of 2-Hydroxypropyl-β-cyclodextrin in niemann-pick disease type C model mice and its pharmacokinetic analysis in a patient with the disease. Biol. Pharm. Bull. 2015;38(6):844–851. doi: 10.1248/bpb.b14-00726. Erratum in: Biol Pharm Bull. 2016;39(3):455. [DOI] [PubMed] [Google Scholar]
- Tang Y., George A., Taylor T., Hildreth J.E. Cholesterol depletion inactivates XMRV and leads to viral envelope protein release from virions: evidence for role of cholesterol in XMRV infection. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0048013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Sun J., Pan H., Yao F., Yuan Y., Zeng M., Ye G., Yang G., Zheng B., Fan J., Pan Y., Zhao Z., Guo S., Liu Y., Liao F., Duan Y., Jiao X., Li Y. Aberrant cytokine expression in COVID-19 patients: associations between cytokines and disease severity. Cytokine. 2021 Jul;143 doi: 10.1016/j.cyto.2021.155523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D.K., Gupta D., Lal S.K. Host lipid rafts play a major role in binding and endocytosis of influenza A virus. Viruses. 2018 Nov 18;10(11) doi: 10.3390/v10110650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloch C.M., da Silva Francisco R., Jr., de Almeida L.G.P., Cardoso C.C., Brustolini O.J., Gerber A.L., Guimarães A.P.C., Mariani D., da Costa R.M., Ferreira O.C., Jr., Workgroup Covid19-UFRJ., Workgroup L.N.C.C., Cony Cavalcanti Adriana, Frauches T.S., de Mello C.M.B., Leitão I.C., Galliez R.M., Faffe D.S., Castiñeiras T.M.P.P., Tanuri A., de Vasconcelos A.T.R. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021 Mar 1;95(10):e00119–e00121. doi: 10.1128/JVI.00119-21. Epub ahead of print. PMID: 33649194; PMCID: PMC8139668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Chen W., Zhang Z., Deng Y., Lian J.Q., Du P., Wei D., Zhang Y., Sun X.X., Gong L., Yang X., He L., Zhang L., Yang Z., Geng J.J., Chen R., Zhang H., Wang B., Zhu Y.M., Nan G., Jiang J.L., Li L., Wu J., Lin P., Huang W., Xie L., Zheng Z.H., Zhang K., Miao J.L., Cui H.Y., Huang M., Zhang J., Fu L., Yang X.M., Zhao Z., Sun S., Gu H., Wang Z., Wang C.F., Lu Y., Liu Y.Y., Wang Q.Y., Bian H., Zhu P., Chen Z.N. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Targeted Ther. 2020 Dec 4;5(1):283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.H., Luo T., Shi Y., He Z.C., Tang R., Zhang P.P., Cai J., Zhou X.D., Jiang D.P., Fei X.C., Huang X.Q., Zhao L., Zhang H., Wu H.B., Ren Y., Liu Z.H., Zhang H.R., Chen C., Fu W.J., Li H., Xia X.Y., Chen R., Wang Y., Liu X.D., Yin C.L., Yan Z.X., Wang J., Jing R., Li T.S., Li W.Q., Wang C.F., Ding Y.Q., Mao Q., Zhang D.Y., Zhang S.Y., Ping Y.F., Bian X.W. A cohort autopsy study defines COVID-19 systemic pathogenesis. Cell Res. 2021 Aug;31(8):836–846. doi: 10.1038/s41422-021-00523-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Feng F., Hu G., Wang Y., Yu Y., Zhu Y., Xu W., Cai X., Sun Z., Han W., Ye R., Qu D., Ding Q., Huang X., Chen H., Xu W., Xie Y., Cai Q., Yuan Z., Zhang R. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 2021 Feb 11;12(1):961. doi: 10.1038/s41467-021-21213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.