Abstract
The relationships between glucose metabolism and exopolysaccharide (EPS) production in a Lactococcus lactis strain containing the EPS gene cluster (Eps+) and in nonproducer strain MG5267 (Eps−) were characterized. The concentrations of relevant phosphorylated intermediates in EPS and cell wall biosynthetic pathways or glycolysis were determined by 31P nuclear magnetic resonance. The concentrations of two EPS precursors, UDP-glucose and UDP-galactose, were significantly lower in the Eps+ strain than in the Eps− strain. The precursors of the peptidoglycan pathway, UDP-N-acetylglucosamine and UDP-N-acetylmuramoyl-pentapeptide, were the major UDP-sugar derivatives detected in the two strains examined, but the concentration of the latter was greater in the Eps+ strain, indicating that there is competition between EPS synthesis and cell growth. An intermediate in biosynthesis of histidine and nucleotides, 5-phosphorylribose 1-pyrophosphate, accumulated at concentrations in the millimolar range, showing that the pentose phosphate pathway was operating. Fructose 1,6-bisphosphate and glucose 6-phosphate were the prominent glycolytic intermediates during exponential growth of both strains, whereas in the stationary phase the main metabolites were 3-phosphoglyceric acid, 2-phosphoglyceric acid, and phosphoenolpyruvate. The activities of relevant enzymes, such as phosphoglucose isomerase, α-phosphoglucomutase, and UDP-glucose pyrophosphorylase, were identical in the two strains. 13C enrichment on the sugar moieties of pure EPS showed that glucose 6-phosphate is the key metabolite at the branch point between glycolysis and EPS biosynthesis and ruled out involvement of the triose phosphate pool. This study provided clues for ways to enhance EPS production by genetic manipulation.
The exopolysaccharides (EPS) include a diverse range of molecules that play vital roles in a variety of biological processes. Insight into how these molecules are synthesized and exported is crucial for exploitation of microorganisms in order to produce EPS of industrial or medical importance (24). Over the last few decades, studies on EPS biosynthesis have focused mainly on gram-negative bacteria, such as Escherichia coli, Xanthomonas campestris, Klebsiella spp., and Pseudomonas spp. (35). However, EPS-producing lactic acid bacteria have received growing attention in recent years because the EPS which they produce are food grade and have applications as food stabilizers, gelling agents, or immunostimulants (4, 7, 11). Biosynthesis of EPS includes assembly of the repeating monosaccharide unit on a lipid carrier by sequential transfer of monosaccharides from sugar nucleotides by glycosyltransferases and subsequent polymerization and export (27, 35). Lactococcus lactis subsp. cremoris B40 produces an EPS composed of glucose, galactose, rhamnose, and phosphate at a ratio of 2:2:1:1 (18, 28). The genetic clusters for production of EPS by L. lactis B40 and Streptococcus thermophilus Sfi6 were characterized and found to encode proteins that have significant similarity to proteins implicated in EPS biosynthesis in other bacteria (26, 33). Recently, homologous and heterologous expression of the glycosyltransferase genes of the eps gene cluster of L. lactis B40 allowed determination of the order of assembly of the trisaccharide backbone of the EPS (32).
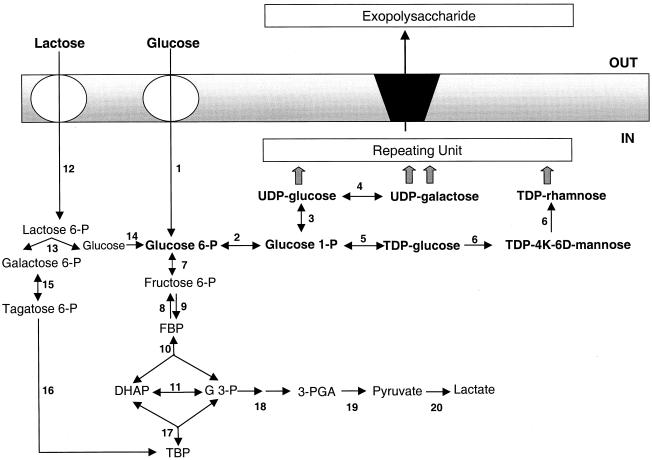
Despite the recent advances in the genetics of EPS production in lactic acid bacteria, reliable physiological data on the regulation of carbon flux from primary metabolism to EPS production are still scarce. The proposed pathway for EPS biosynthesis in L. lactis B40 is shown in Fig. 1 (6). The early steps, leading from the hexose phosphate substrate to sugar nucleotides, represent the interface between primary metabolism and secondary metabolism and, therefore, constitute an important target of research if metabolic engineering strategies to increase EPS production and/or modify EPS composition are to be implemented. No open reading frames with homology to genes involved in the synthesis of sugar nucleotide precursors were found in the eps clusters of either L. lactis or S. thermophilus (26, 33). Since these precursors of EPS are also used for maintaining primary cellular functions, such as cell wall biosynthesis, they have to be diverted from central metabolism to EPS production. Therefore, a comprehensive view of the metabolic events resulting in channeling of sugar carbon from central metabolism to the EPS biosynthetic pathway is required.
FIG. 1.
Proposed pathway for EPS biosynthesis in L. lactis. The reactions are catalyzed by the following enzymes: 1, glucose phosphoenolpyruvate:phosphotransferase system; 2, α-phosphoglucomutase; 3, UDP-glucose pyrophosphorylase; 4, UDP-galactose-4-epimerase; 5, TDP-glucose pyrophosphorylase; 6, TDP-rhamnose biosynthetic system; 7, phosphoglucose isomerase; 8, 6-phosphofructokinase; 9, fructose bisphosphatase; 10, fructose-1,6-bisphosphate aldolase; 11, triose phosphate isomerase; 12, lactose phosphoenolpyruvate:phosphotransferase system; 13, phospho-β-galactosidase; 14, glucokinase; 15, galactose-6-phosphate isomerase; 16, tagatose-6-phosphate kinase; 17, tagatose-1,6-bisphosphate aldolase; 18, glyceraldehyde-3-phosphate dehydrogenase and phosphoglycerate kinase; 19, phosphoglyceromutase, enolase, and pyruvate kinase; and 20, lactate dehydrogenase. Abbreviations: TBP, tagatose 1,6-bisphosphate; G 3-P, glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; TDP-4K-6D-mannose, TDP-4-keto-6-deoxymannose.
In this work we examined the relationship between glycolysis and EPS biosynthesis in L. lactis. Phosphorous-31 nuclear magnetic resonance (NMR) spectroscopy was used as the main analytical technique, which allowed simultaneous identification and quantification of various phosphorylated metabolites. Furthermore, 13C enrichment of EPS was determined after growth of an Eps+ strain in chemically defined medium containing [1-13C]glucose, and the activities of relevant enzymes in both EPS and glycolytic pathways were measured.
MATERIALS AND METHODS
Microbial strains and growth conditions.
L. lactis MG5267 (a lactose-proficient strain derived from L. lactis MG1363), referred to below as the Eps− strain (34), and a derivative strain carrying plasmid pNZ4030 containing the EPS gene cluster, referred to below as the Eps+ strain (33), were grown at 30°C in a 2-liter fermentor. The defined medium described by Poolman and Konings (21) was modified as follows: K2HPO4 was omitted, the concentration of KH2PO4 was decreased to 0.2 g · liter−1, and erythromycin (10 μg · ml−1) was added for selection of the Eps+ strain. Glucose or lactose was added at a final concentration of 1% (wt/vol). The medium was gassed with argon during the 10 min preceding inoculation (4% inoculum from a culture grown overnight); the pH was kept at 6.5 by automatic addition of 10 N NaOH, and a low agitation rate (70 rpm) was used. Growth was monitored by measuring the optical density at 600 nm (OD600) and calibrating with the protein concentration. The specific growth rate of each strain was calculated from three independent experiments.
The protein concentration was determined by the method of Bradford (3) by using bovine serum albumin as the standard. To determine proteins in cell suspensions, cells were disrupted by treatment with 1 M NaOH and incubation at 80°C for 2 min. The concentrations of glucose in the supernatants obtained before ethanol extraction were determined enzymatically.
Preparation of ethanol extracts for analysis by 31P-NMR.
Extracts were obtained as follows. At different growth phases, a portion of culture was withdrawn from the fermentor, the medium was removed by centrifugation (6,000 × g, 5 min, 4°C), and the cells were suspended in 20 ml of 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer, pH 7.0 (Sigma Chemical Co., St. Louis, Mo.). The volume of culture withdrawn from the fermentor at each growth phase was adjusted in order to obtain approximately 30 mg of cell protein for each extract. Each cell suspension was transferred to 100 ml of cold 70% (vol/vol) ethanol in an ice bath, and extraction was performed for 30 min with vigorous agitation. Cell debris was removed by centrifugation (30,000 × g, 30 min, 4°C), the ethanol was evaporated, and the residue was lyophilized. The extracts were each dissolved in 3 ml of ultrapure H2O containing 5 mM EDTA and stored at −20°C. The pH values of the extracts were approximately 6.9.
Quantification of intracellular metabolites. (i) 31P-NMR.
The concentrations of intracellular metabolites were calculated from the areas of their resonances in 31P spectra by comparison with the area of the resonance due to methylphosphonic acid (Aldrich), added as an internal standard, and after application of an appropriate factor for correcting saturation of resonances. The limit of detection for intracellular metabolites under the conditions used to acquire 31P spectra (2,000 transients) was 0.2 mM. Resonances were assigned after addition of pure compounds to the extracts or on the basis of comparison with previous studies (23).
(ii) Enzymatic methods.
Glucose 6-phosphate (G6P), fructose 6-phosphate, and fructose 1,6-bisphosphate (FBP) were also quantified in extracts by using enzymatic methods based on spectrophotometric determination of NAD(P)H (16, 17).
(iii) Purification and identification of UDP-N-acetylmuramoyl-pentapeptide (UDP-NAcMur-pentapeptide).
A culture of the Eps− strain was grown until the stationary phase, and an ethanol extract was prepared as described above. The extract was applied to a Sephadex G-10 column (Pharmacia-LKB, Uppsala, Sweden) that was equilibrated with water and eluted at a flow rate of 0.5 ml · min−1. Aliquots of each fraction were tested to determine the total phosphorus content as described by Ames (1), and the aliquots giving a positive response were concentrated by lyophilization and analyzed by 1H- and 31P-NMR. The fractions containing UDP-sugars were further purified with a Resource column (Pharmacia-LKB) that was equilibrated with 10 mM ammonium bicarbonate buffer, pH 8.0. Elution was performed with a linear 10 mM to 1 M NH4HCO3 gradient at a flow rate of 2 ml · min−1. Fractions containing phosphorus were concentrated by lyophilization, dissolved in 2H2O or 1H2O, and analyzed by one-dimensional and two-dimensional NMR. Sequential assignment of the peptide moiety was done by using total correlation and heteronuclear multiple bond correlation spectroscopy in 1H2O. Heteronuclear multiple quantum coherence and 1H-1H correlation spectra were obtained to complete elucidation of the structure. Intracellular metabolite concentrations were calculated by using 2.9 μl · mg of protein−1 for the intracellular volume of L. lactis (22).
Extraction and partial purification of 13C-enriched EPS.
The Eps+ strain was grown in a bioreactor (working volume, 50 ml) containing defined medium supplemented with 1% (wt/vol) [1-13C]glucose (99% enrichment; Campro Scientific, Veenendaa, The Netherlands). The medium was gassed with argon for 10 min before inoculation (4% inoculum), and the pH was kept at 6.5 by addition of 1 N NaOH. The culture was removed when the stationary growth phase was reached (OD600, approximately 5), and the 13C-enriched EPS was extracted as follows. Proteins were precipitated by addition of tricholoroacetic acid (final concentration, 20% [wt/vol]) and incubation in an ice bath for 2 h. After centrifugation (25,000 × g, 20 min, 4°C), 2 volumes of cold ethanol was added to the supernatant. The precipitated EPS was recovered by centrifugation (30,000 × g, 20 min, 4°C), the pH of the solution was adjusted to 7 with KOH before dialysis against water for 24 h, and the preparation was freeze-dried. The EPS was dissolved in 1 ml of ultrapure H2O and stored at −20°C until it was analyzed by 13C- and 31P-NMR.
NMR spectroscopy.
Carbon-13 or phosphorus-31 spectra were acquired with a Bruker DRX500 spectrometer. 31P-NMR spectra of ethanol extracts were acquired with a quadruple nucleus probe head at 28°C with a pulse width of 13 μs (flip angle, 60°) and a recycle delay of 3 s; the number of transients was 2,000. Saturation of resonances due to fast pulsing conditions was calculated by comparison with spectra acquired under fully relaxed conditions (recycle delay, 30 s). To monitor product formation during measurement of enzyme activity by 31P-NMR, the recycle delay was decreased to 0.5 s. Two-dimensional homonuclear and heteronuclear NMR data were acquired as previously described (13).
13C-NMR spectra of isolated EPS were acquired at 60°C with a dual 13C-1H 5-mm-diameter probe head with a 6-μs pulse width (flip angle, 60°) and a recycle delay of 2 s. 31P-NMR spectra were acquired with a broadband 5-mm-diameter probe head for indirect detection by using a pulse width of 16 μs (flip angle, 75°) and a recycle delay of 1 s. Carbon and phosphorus chemical shifts were referenced to the resonances of external methanol and 85% H3PO4 (49.3 and 0.0 ppm, respectively).
Preparation of crude cell extracts.
Cells were cultivated as described above until the mid-exponential phase of growth (in defined medium containing glucose), harvested, washed, and suspended in 50 mM potassium phosphate buffer (pH 6.5). For UDP-glucose pyrophosphorylase determination, cells were suspended in 100 mM Tris-HCl buffer (pH 7.5) containing 1 mM 2-mercaptoethanol, 1 mM EDTA, a cocktail of protease inhibitors (2 μg of leupeptin per ml, 2 μg of antipain per ml, and 2.5 μM phenylmethylsulfonyl fluoride), and 20% (vol/vol) glycerol. Cell extracts were prepared by mechanical disruption with a French press (three passages at 120 MPa), debris was removed by centrifugation (30,000 × g, 20 min, 4°C), and the extracts were used immediately for determination of enzyme activity.
Enzyme assays.
The reverse reactions of phosphoglucomutase (EC 5.4.2.2), phosphoglucose isomerase (EC 5.3.1.9), and UDP-galactose-4-epimerase (EC 5.1.3.2) were monitored as described previously (10). 6-Phosphofructokinase (EC 2.7.1.11) activity was assayed by the method of Fordyce et al. (8), and fructose bisphosphatase (EC 3.1.3.11) activity was measured as described by Babul and Guixé (2). l-Lactate dehydrogenase (EC 1.1.1.27) activity was determined as described previously (9).
The forward reactions catalyzed by UDP-glucose pyrophosphorylase (EC 2.7.7.9) and phosphoglucomutase were assayed at 30°C by 31P-NMR spectroscopy. For the pyrophosphorylase assay, the 3-ml reaction mixtures contained 50 mM Tris-HCl buffer (pH 8.0), 10 mM MgCl2, 6 mM α-glucose 1-phosphate, 6 mM UTP, and cell extract. For the phosphoglucomutase assay, the reaction mixtures contained 50 mM Tris-HCl buffer (pH 7.5), 5 mM MgCl2, 12 mM α-G6P or 12 mM β-G6P, 50 μM glucose 1,6-bisphosphate, and cell extract. A sequence of spectra was acquired following the addition of one of the substrates (glucose 1-phosphate and G6P for the pyrophosphorylase assay and the phosphoglucomutase assay, respectively). The reaction rates were determined by monitoring the formation of the product (UDP-glucose or glucose 1-phosphate). Resonance intensities were compared to that of a known amount of methylphosphonate. Control experiments were performed by omitting the individual substrates or the crude cell extracts.
RESULTS
Identification and quantification of phosphorylated metabolites by 31P-NMR.
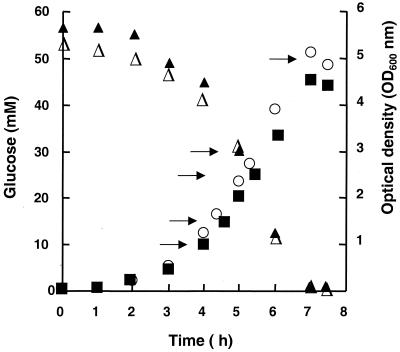
The growth-associated nature of EPS production in L. lactis hampered the use of in vivo NMR, since the high cell densities required are generally achieved by using nongrowing cells (25). In fact, the glycolytic kinetics of nongrowing cells of the Eps+ strain were identical to those found previously for plasmid-free parental strain MG5267 (the Eps− strain) (19). Glucose was metabolized mainly to lactate (and small amounts of acetate and aspartate), and no incorporation of 13C label in EPS was observed (data not shown). Therefore, we resorted to ethanol extracts to characterize the pools of phosphorylated intermediates in growing cells. Figure 2 shows the growth of both strains in defined medium, the consumption of glucose, and the OD600 values at which samples for ethanol extraction were withdrawn. No differences in glucose consumption were found; however, the growth rate of the Eps+ strain (1.19 ± 0.03 h−1) was consistently slightly lower than that of the Eps− strain (1.29 ± 0.02 h−1).
FIG. 2.
Growth of the Eps− (○) and Eps+ (■) strains in chemically defined medium at pH 6.5. The consumption of glucose in the Eps− (▵) and Eps+ (▴) strains is also shown. The arrows indicate the optical densities at which samples were withdrawn for ethanol extraction and quantification of intracellular metabolites. Three independent extractions for each growth phase were performed.
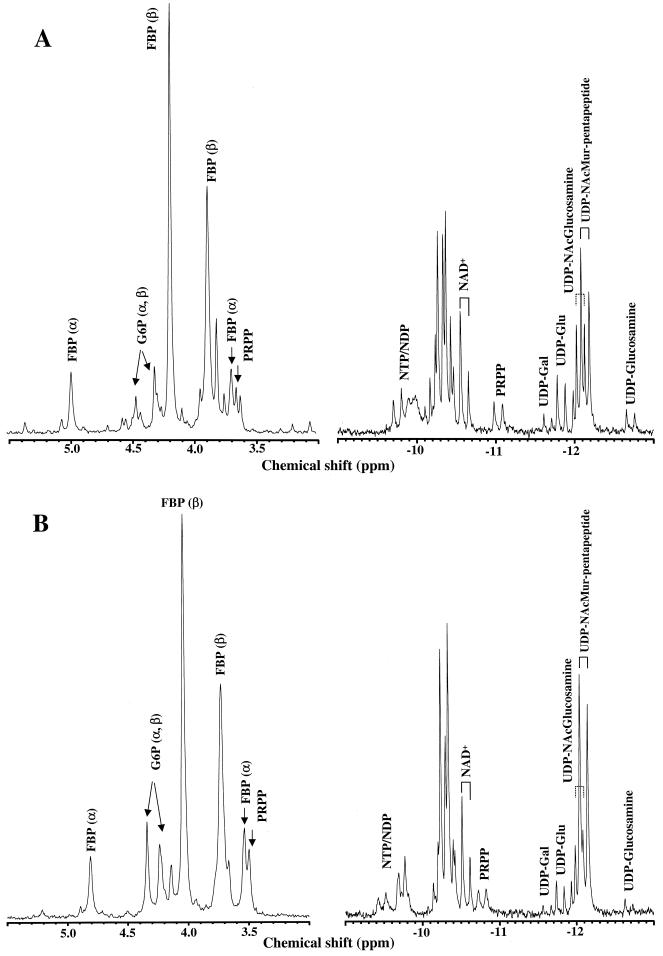
Figure 3 shows the 31P-NMR spectra of ethanol extracts obtained from Eps− and Eps+ cells at the mid-logarithmic growth phase (OD600, 2.5). In the phosphomonoester region (5.5 to 3.0 ppm), the major resonances due to glycolytic intermediates in both strains were assigned to G6P and FBP (α and β anomers). Glucose 1-phosphate did not accumulate at detectable concentrations in these extracts. In the diphosphodiester region (−9.0 to −13.0 ppm), the following resonances were identified by addition of pure compounds: UDP-glucose, UDP-galactose, UDP-N-acetylglucosamine, NAD+ (Sigma Chemical Co.), and UDP-glucosamine. Since UDP-glucosamine was not commercially available, it was synthesized enzymatically from glucosamine 1-phosphate and UTP (Sigma Chemical Co.) by using UDP-glucose pyrophosphorylase as described previously (20). The doublet centered at −11 ppm was identified as the α-phosphate of the pyrophosphate moiety in 5-phosphorylribose 1-pyrophosphate (PRPP), an intermediate in the biosynthesis of histidine and nucleotides. The corresponding doublet due to the β-phosphate of this metabolite was also clearly evident at −5.5 ppm (data not shown). The assignment of PRPP was confirmed by the detection at 3.1 ppm of the resonance due to the phosphoryl group at position 5 of ribose. The doublets at −10.8 and −12.9 ppm, which were present in extracts from both strains, could not be assigned to any of the following metabolites, which were added as pure compounds: GDP-glucose, ADP-ribose, UDP-N-acetylgalactosamine, UDP-glucuronic acid, and TDP-glucose (Sigma Chemical Co.). TDP-rhamnose and UDP-N-acetylmuramic acid were not tested, since these compounds were not available from commercial sources. Those doublets were assigned to UDP-NAcMur-pentapeptide, a precursor of peptidoglycan, by two-dimensional NMR. The pentapeptide sequence (attached to the lactyl residue of UDP-N-acetylmuramic acid) was found to be AlaGluLysAlaAla. While this paper was in preparation, the proton and carbon chemical shifts of UDP-NAcMur-pentapeptide isolated from Anabaena cylindrica appeared, and they are in complete agreement with our data (12). Since all the resonances due to nucleotide sugars were identified, we concluded that TDP-rhamnose could not be detected, due to its intrinsic low level and/or to chemical instability. It is worth mentioning that to minimize the degradation of TDP-rhamnose under alkaline conditions, the extracts were obtained at pH values below 7.0.
FIG. 3.
31P-NMR spectra of ethanol extracts obtained from growing cells of the Eps− (A) and Eps+ (B) strains in the mid-log phase. The chemical shifts of the relevant resonances in the phosphomonoester region (5.5 to 3.0 ppm) were as follows: G6P, 4.5 and 4.3 ppm; FBP (β anomer), 4.1 and 3.8 ppm; FBP, (α anomer), 4.8 and 3.6 ppm; and the phosphorus at position 5 of PRPP, 3.6 ppm. In the diphosphodiester region (−9.0 to −13.0 ppm) the following resonances were identified: UDP-glucose (UDP-Glu), UDP-galactose (UDP-Gal), UDP-N-acetylglucosamine (UDP-NAcGlucosamine), UDP-glucosamine, the α-phosphate of PRPP, UDP-NAcMur-pentapeptide, NAD+, nucleoside triphosphate (NTP), and nucleoside diphosphate (NDP). Resonances were assigned by spiking the extracts with pure compounds except for UDP-NAcMur-pentapeptide, which was identified by two-dimensional NMR spectroscopy.
Analysis of an extract obtained during the exponential growth phase of the Eps+ strain in lactose-containing medium showed that galactose 6-phosphate accumulated at a high intracellular concentration (14 mM) along with G6P (5.5 mM) and FBP (25 mM). Accumulation of galactose 6-phosphate is consistent with operation of the tagatose pathway during lactose fermentation. The pattern of resonances in the diphosphodiester region remained unchanged compared to that obtained when glucose was the energy source, and the concentrations of UDP-glucose (1.8 mM), UDP-galactose (trace amounts), and UDP-N-acetylglucosamine (4.2 mM) were also similar (data not shown).
Figure 4 shows the evolution in the concentrations of glycolytic intermediates in glucose-grown cells of the Eps+ and Eps− strains. Again, FBP and G6P were the prominent metabolites throughout the exponential growth phase, whereas 3-phosphoglyceric acid (3-PGA), phosphoenolpyruvate, and 2-phosphoglyceric acid started to dominate towards the end of the exponential growth phase and were the only metabolites detected in the stationary phase. It is interesting that this change in the pattern of intracellular metabolites occurred at an earlier growth state in the Eps− strain than in the Eps+ strain. For instance, at an OD600 of 2.5, 3-PGA, a metabolite typical of starvation, was detected in the Eps− strain but not in the Eps+ strain. Moreover, at an OD600 of 3, when both strains were still growing exponentially (Fig. 2), the ratio 3-PGA to FBP was considerably higher in the Eps− strain, indicating that there was a significant shift in the transition from a typical exponential pattern to typical starvation status. One could hypothesize that this distinct behavior was due to different growth rates and/or different rates of glucose consumption. However, determinations of the glucose concentrations in the supernatants obtained prior to ethanol extraction showed no differences between the strains (Fig. 2).
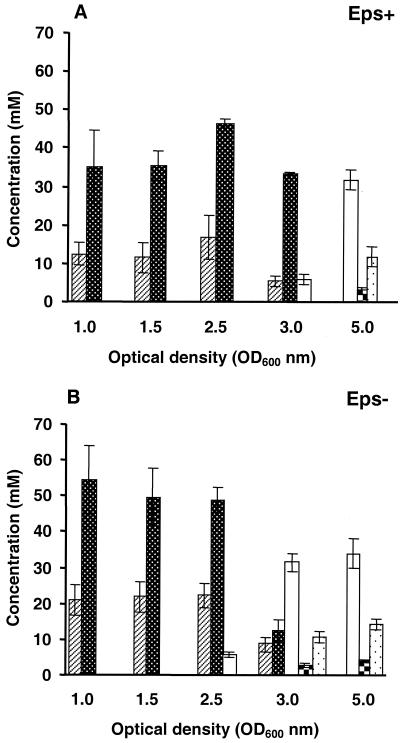
FIG. 4.
Intracellular concentrations of glycolytic intermediates in ethanol extracts of the Eps+ (A) and Eps− (B) strains obtained at different growth phases by using glucose as the energy source. The intracellular concentrations shown are averages based on three independent extracts obtained under identical conditions. ▨, G6P; ▩, FBP; □, 3-phosphoglyceric acid; , 2-phosphoglyceric acid; , phosphoenolpyruvate.
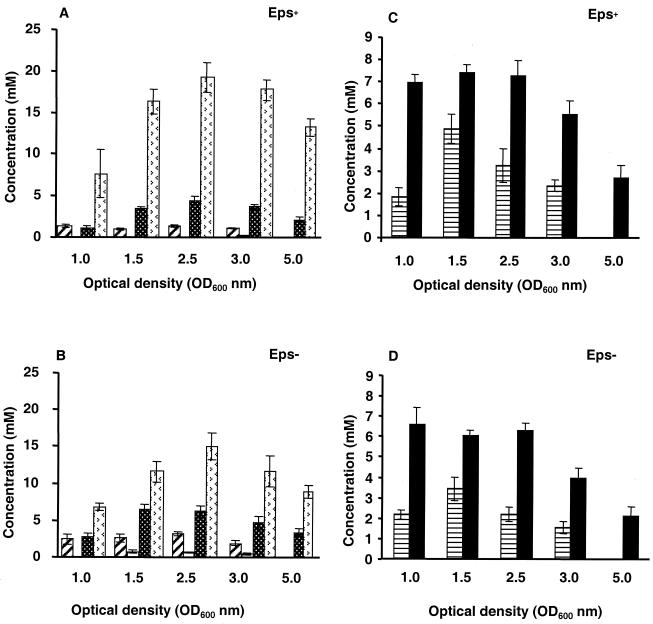
Fructose 6-phosphate accumulated to a maximal concentration of 3 mM during exponential growth in both strains (data not shown). The concentrations of nucleoside diphosphosugars and related metabolites throughout growth are shown in Fig. 5 for the Eps+ and Eps− strains. The concentrations of UDP-glucose and UDP-galactose reached maximum values during exponential growth and decreased towards the late-logarithmic phase, and these compounds were absent in the stationary phase. The intracellular concentrations of UDP-glucose and UDP-galactose, which are precursors of B40 EPS, were significantly lower in the Eps+ strain than in the parental strain. For instance, the concentration of UDP-glucose in the mid-exponential growth phase was threefold higher in the Eps− strain. The concentrations of UDP-N-acetylglucosamine and UDP-NAcMur-pentapeptide (precursors of peptidoglycan) also reached maximum values during exponential growth, but these compounds were still present at high levels in the extracts obtained in the stationary phase. The UDP-N-acetylglucosamine concentration was the same in cells of both strains, whereas throughout growth the concentration of UDP-NAcMur-pentapeptide was on average 1.5-fold higher in the Eps+ strain. The concentrations of NAD+ and PRPP during growth are also shown in Fig. 5 for the Eps+ and Eps− strains. The NAD+ concentrations were comparable in extracts obtained from the two strains and remained fairly constant during growth, decreasing considerably when the stationary phase was reached. The PRPP concentrations were also the same in Eps+ and Eps− cells, reaching the maximum level during exponential growth and decreasing to undetectable levels in the stationary phase. It is worth mentioning that the resonances due to PRPP were observed only in extracts derived from growing cells, and accumulation of this compound did not occur following addition of glucose to resting cells of L. lactis (data not shown).
FIG. 5.
Intracellular concentrations of UDP-glucose,
UDP-galactose, UDP-N-acetylglucosamine, and
UDP-NAcMur-pentapeptide in ethanol extracts of Eps+ (A) and
Eps− (B) cells obtained at different growth phases. The
concentrations of NAD+ and PRPP throughout growth are also
shown for the Eps+ (C) and Eps− (D) strains.
The intracellular concentrations shown are averages based on three
independent extracts obtained under identical conditions. ▨,
UDP-glucose; □, UDP-galactose; ▩, UDP-N-acetylglucosamine;
 , UDP-NAcMur-pentapeptide; ▤,
phosphorybosyl pyrophosphate; ■, NAD+.
, UDP-NAcMur-pentapeptide; ▤,
phosphorybosyl pyrophosphate; ■, NAD+.
Carbon-13 enrichment of the EPS and NMR analysis.
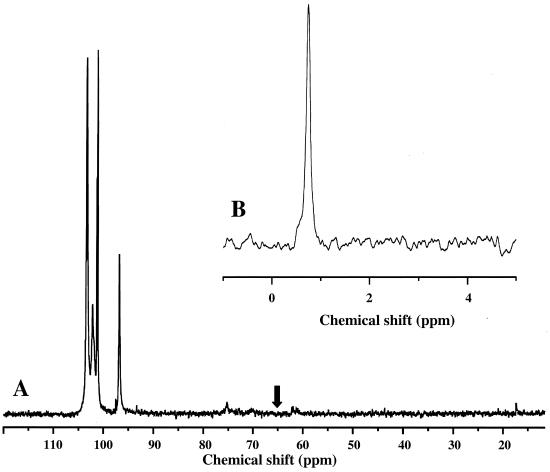
It was known from our previous studies with nongrowing cells of the Eps− strain that the carbon-13 label supplied at carbon 1 in glucose is scrambled in the triose phosphate pool and that back-flux through aldolase results in accumulation of FBP labeled at both carbon 1 and carbon 6 (19). This could be relevant for production of EPS; therefore, we decided to verify whether back-flux through aldolase and fructose bisphosphatase was an important source of carbon for the EPS biosynthetic pathway. To do this, the producer strain was grown in defined medium containing [1-13C]glucose, and the EPS was extracted, partially purified, and analyzed by 13C- and 31P-NMR (Fig. 6). The 31P spectrum had a single resonance in the phosphodiester region (δ = −0.7 ppm), which is consistent with the presence of a single phosphate group in the repeating unit (18, 28). In the carbon spectrum, four high-intensity resonances were detected in the anomeric carbon region, with the following chemical shifts: 103.3, 102.1, 101.2, and 96.8 ppm. These resonances corresponded to carbon 1 of glucose, galactose, rhamnose, and galactose phosphate, respectively, in the repeating unit of EPS (18). No resonance was detected in the region where the resonances due to carbon 6 of the sugars were expected to appear (around 70 ppm).
FIG. 6.
13C-NMR (A) and 31P-NMR (B) spectra of isolated EPS after growth of the Eps+ strain in [1-13C]glucose until the stationary growth phase. In spectrum A, the resonances due to anomeric carbons of glucose, galactose, rhamnose, and galactose phosphate in the EPS repeating unit were detected. The arrow indicates the region where the resonances due to carbon 6 of these sugars would appear.
Enzymatic activities in the Eps+ and Eps− strains.
The activities of enzymes that were expected to exert control on the glycolytic flux or be involved in modulation of the carbon flux to the EPS biosynthetic route were measured. A comparison of the results obtained with cell extracts obtained from mid-exponential Eps+ and Eps− cultures is shown in Table 1. No significant differences were found when phosphoglucomutase, phosphoglucose isomerase, UDP-galactose-4-epimerase, 6-phosphofructokinase, fructose bisphosphatase, and l-lactate dehydrogenase activities were compared. The activity of fructose bisphosphatase was very low, 5 nmol · mg of protein−1 · min−1, a result which is consistent with the aforementioned absence of incorporation of the 13C label at carbon 6 in sugar moieties in the EPS. The ratios of phosphoglucose isomerase activity to phosphoglucomutase activity were 6.0 and 7.5 in the Eps+ and Eps− strains, respectively, as evaluated from the rates of the reverse reactions. The forward reaction catalyzed by phosphoglucomutase was monitored by 31P-NMR, and a rate of 7 nmol · mg of protein−1 · min−1 was found. Furthermore, only the α-anomer of glucose 1-phosphate was produced. The in vitro activity of UDP-glucose pyrophosphorylase (forward reaction) was also low in both strains, approximately 10 nmol · mg of protein−1 · min−1.
TABLE 1.
Comparison of enzyme activities in extracts of the Eps− and Eps+ strains
| Enzyme | Activity
(nmol · mg of
protein−1 · min−1)a
in:
|
|
|---|---|---|
| Eps+ strain | Eps− strain | |
| Phosphoglucose isomerase (r)b | 1,326 ± 26 | 1,449 ± 19 |
| α-Phosphoglucomutase (r) | 220 ± 8 | 190 ± 5 |
| α-Phosphoglucomutase (f) | 7 ± 0.2 | 7 ± 0.3 |
| UDP-glucose pyrophosphorylase (f) | 9 ± 0.3 | 10 ± 0.5 |
| UDP-galactose-4-epimerase (r) | 108 ± 5 | 112 ± 4 |
| 6-Phosphofructokinase | 820 ± 33 | 1,070 ± 42 |
| Fructose bisphosphatase | 4 ± 0.1 | 5 ± 0.2 |
| l-Lactate dehydrogenase | 15,271 ± 861 | 12,150 ± 570 |
Mean ± standard deviation (n ≥ 4).
r, reverse reaction; f, forward reaction.
DISCUSSION
The growing interest in food grade polysaccharides for industrial applications has triggered an increase in research to elucidate regulation of the carbon flux from glycolysis to EPS biosynthesis in lactic acid bacteria. However, the variable and low levels of production achieved thus far with these bacteria impose severe limitations on utilization of EPS (11, 31). For example, the EPS-producing strain studied in this work produces only 50 mg of EPS per liter when it is grown in defined medium.
Detailed knowledge concerning the relevant metabolic parameters is essential if improved EPS-producing strains are to be successfully designed. The present work was planned to contribute to our understanding of regulation of EPS production in L. lactis, primarily by characterization of intracellular pools of phosphorylated intermediates involved in biosynthesis of EPS and cell walls and in glycolysis.
The concentrations of UDP-glucose and UDP-galactose, precursors of EPS, were significantly lower in the producer strain than in the Eps− strain. Several authors have found that limiting concentrations of sugar nucleotides (namely, UDP-glucose and UDP-galactose) restrict polysaccharide biosynthesis. In fact, a L. lactis mutant deficient in UDP-galactose-4-epimerase was unable to produce EPS when it was grown on medium containing glucose as the sole carbon source (I. C. Boels, M. Kleerebezem, J. Hugenholtz, and W. M. de Vos, Abstr. 5th Am. Soc. Microbiol. Meet. Genet. Mol. Biol. Streptococci, Enterococci, Lactococci, p. 66, 1998); increasing the expression of UDP-glucose pyrophosphorylase greatly enhanced heterologous expression of Streptococcus pneumoniae type 3 polysaccharide in L. lactis (C. Gilbert, K. Robinson, R. W. F. LePage, and J. M. Wells, Abstr. 5th Am. Soc. Microbiol. Meet. Genet. Mol. Biol. Streptococci, Enterococci, Lactococci, p. 67, 1998), and overexpression of fructose bisphosphatase from E. coli in L. lactis led to increased production of EPS from fructose, a result that also indicated the important role of sugar nucleotides in this process (14). However, it should be pointed out that carbon fluxes are not simply dependent on the pools of precursors and that a more global approach is required to achieve reliable predictions concerning metabolic shifts.
The slightly lower growth rate of the Eps+ strain compared to the Eps− strain can be explained by diversion of sugar carbon towards EPS biosynthesis. Several authors have reported higher production of EPS at suboptimal temperatures, a result that supports the hypothesis that the availability of the lipid carrier undecaprenyl-phosphate is increased at lower growth rates (4, 5, 29). Our data showed that the concentration of UDP-NAcMur-pentapeptide, the last cytoplasmic precursor of peptidoglycan, was higher in cells of the Eps+ strain, which is consistent with the existence of competition between growth and EPS production caused by limitation of the lipid carrier. In E. coli cells, the pool level of undecaprenyl-phosphate is considered the main limiting factor in the membrane-associated steps of peptidoglycan biosynthesis (30). Interestingly, only the first and last cytoplasmic nucleotide precursors of the peptidoglycan pathway (UDP-N-acetylglucosamine and UDP-NAcMur-pentapeptide) accumulated in L. lactis. These biosynthetic precursors are also predominant in acid extracts derived from growing cells of E. coli (15).
In contrast with the observed changes in the pools of sugar nucleotides, no significant differences in the activities of key enzymes were found between the two strains examined. Phosphoglucose isomerase and phosphoglucomutase, the enzymes that catalyze the reactions at the branch point between glycolysis and the EPS biosynthetic route, are expected to play a key role in governing carbon availability for EPS production in vivo. The reverse reactions catalyzed by these enzymes were similar in the two strains studied (Table 1). We determined the rates of the forward reactions of phosphoglucomutase and UDP-glucose pyrophosphorylase by resorting to 31P-NMR spectroscopy. The forward reaction catalyzed by α-phosphoglucomutase proceeded at a low rate. Moreover, the activity of the next enzyme in the biosynthetic pathway of sugar nucleotides, UDP-glucose pyrophosphorylase, was also very low, and the levels were similar in the two strains studied. Interestingly, the net rate of production of UDP-glucose monitored in vivo by 13C-NMR after addition of [1-13C]glucose to a nongrowing cell suspension of the Eps− strain was 4.8 nmol · mg of protein−1 · min−1 (A. R. Neves, A. Ramos, and H. Santos, unpublished data), a value on the same order of magnitude as the value for activity of UDP-glucose pyrophosphorylase measured in cell extracts.
The accumulation of PRPP, a precursor of nucleotides and histidine, showed that the pentose phosphate pathway was active during growth of L. lactis in defined medium, allowing the organism to fulfill the requirement for important biosynthetic intermediates. The EPS labeling pattern after growth on defined medium containing [13C]glucose confirmed the involvement of G6P as the metabolite at the branch point between glycolysis and the EPS biosynthetic route. In addition, the data ruled out involvement of the triose phosphate pool as a source of carbon for biosynthesis of EPS since the label appeared only at the C-1 positions of the sugar residues. This pattern of incorporation suggested that either a very low level of activity of fructose bisphosphatase precluded carbon back-flux from FBP to fructose 6-phosphate or no significant scrambling of the label at the triose phosphate pool occurred during growth. Either of these situations would lead to the observed lack of incorporation of 13C label at position C-6 of the EPS repeating units (Fig. 1). Work in our laboratory with the Eps− strain grown in defined medium containing [1-13C]glucose showed that scrambling between positions 1 and 6 of FBP does occur during growth (Neves et al., unpublished data). Although this experiment was not performed with the Eps+ strain, it is unlikely that the behavior of this organism is different, and the pattern of 13C incorporation in EPS is interpreted as being due to the low level of fructose bisphosphatase activity.
This study revealed significant decreases in the sizes of the pools of two EPS precursors (UDP-glucose and UDP-galactose) in the Eps+ strain, whereas the size of the pool of UDP-MurNAc-pentapeptide increased. These results indicate that there are two putative metabolic targets for genetic manipulation: the flux to production of EPS precursors could be enhanced in conjunction with an increase in the size of the pool of the lipid carrier, so that a high level of EPS productivity can occur without a severe effect on cell growth.
ACKNOWLEDGMENTS
This work was supported by BIOTECH Program contract BIO4CT-96-0498 from the Commission of the European Communities and by contracts PRAXIS/PCNA/P/BIO/39/96 and PRAXIS/P/BIA/11072/98 from Fundação para a Ciência e Tecnologia (FCT), Portugal. A. Ramos acknowledges a postdoctoral fellowship from FCT.
We thank Pedro Lamosa for help with the assignment of UDP-NAcMur-pentapeptide and Alexandra Frias for technical assistance during determination of enzymatic activities.
REFERENCES
- 1.Ames B N. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 1966;8:115–118. [Google Scholar]
- 2.Babul J, Guixé V. Fructose bisphosphatase from Escherichia coli. Purification and characterization. Arch Biochem Biophys. 1983;225:944–949. doi: 10.1016/0003-9861(83)90109-1. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Cerning J. Exocellular polysaccharides produced by lactic acid bacteria. FEMS Microbiol Rev. 1990;87:113–130. doi: 10.1111/j.1574-6968.1990.tb04883.x. [DOI] [PubMed] [Google Scholar]
- 5.Cerning J, Bouillanne C, Landon M, Desmazeaud M. Isolation and characterization of exopolysaccharides from slime-forming mesophilic lactic acid bacteria. J Dairy Sci. 1992;75:692–699. [Google Scholar]
- 6.de Vos W M. Metabolic engineering of sugar catabolism in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:223–242. doi: 10.1007/BF00395934. [DOI] [PubMed] [Google Scholar]
- 7.de Vos W M, Hols P, van Kranenburg R, Luesink E, Kuipers O, van der Oost J, Kleerebezem M, Hugenholtz J. Making more of milk sugar by engineering lactic acid bacteria. Int Dairy J. 1998;8:227–233. [Google Scholar]
- 8.Fordyce A M, Moore C H, Pritchard G G. Phosphofructokinase from Streptococcus lactis. Methods Enzymol. 1982;90:77–82. doi: 10.1016/s0076-6879(82)90109-4. [DOI] [PubMed] [Google Scholar]
- 9.Garrigues C, Loubiere P, Lindley N D, Cocaign-Bousquet M. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ratio. J Bacteriol. 1997;179:5282–5287. doi: 10.1128/jb.179.17.5282-5287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grobben G J, Smith M R, Sikkema J, de Bont J A M. Influence of fructose and glucose on the production of exopolysaccharides and the activities of enzymes involved in the sugar metabolism and the synthesis of sugar nucleotides in Lactobacillus delbrueckii subsp. bulgaricusNCBF 2772. Appl Microbiol Biotechnol. 1996;46:279–284. [Google Scholar]
- 11.Kleerebezem M, van Kranenburg R, Tuinier R, Boels I C, Zoon P, Looijesteijn E, Hugenholtz J, de Vos W M. Exopolysaccharides produced by Lactococcus lactis: from genetic engineering to improved rheological properties? Antonie Leeuwenhoek. 1999;76:357–365. [PubMed] [Google Scholar]
- 12.Kodani S, Ishida K, Murakami M. Occurrence and identification of UDP-N-acetylmuramyl-pentapeptide from the cyanobacterium Anabaena cylindrica. FEMS Microbiol Lett. 1999;176:321–325. doi: 10.1111/j.1574-6968.1999.tb13678.x. [DOI] [PubMed] [Google Scholar]
- 13.Lamosa P, Martins L O, da Costa M S, Santos H. Effects of temperature, salinity, and medium composition on compatible solute accumulation by Thermococcusspp. Appl Environ Microbiol. 1998;64:3591–3598. doi: 10.1128/aem.64.10.3591-3598.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Looijesteijn P J, Boels I C, Kleerebezem M, Hugenholtz J. Regulation of exopolysaccharide production by Lactococcus lactis subsp. cremorisby the sugar source. Appl Environ Microbiol. 1999;65:5003–5008. doi: 10.1128/aem.65.11.5003-5008.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mengin-Lecreulx D, Flouret B, van Heijenoort J. Pool levels of UDP N-acetylglucosamine and UDP N-acetylglucosamine-enolpyruvate in Escherichia coliand correlation with peptidoglycan biosynthesis. J Bacteriol. 1983;154:1284–1290. doi: 10.1128/jb.154.3.1284-1290.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michal G. d-Fructose 1,6-bisphosphate, dihydroxyacetone phosphate and d-glyceraldehyde 3-phosphate. In: Bergmeyer H U, Bergmeyer J, Graßl M, editors. Methods of enzymatic analysis. 3rd ed. Vol. 5. Weinheim, Germany: Verlag-Chemie; 1988. pp. 342–350. [Google Scholar]
- 17.Michal G. d-Glucose 6-phosphate and d-fructose 6-phosphate. In: Bergmeyer H U, Bergmeyer J, Graßl M, editors. Methods of enzymatic analysis. 3rd ed. Vol. 5. Weinheim, Germany: Verlag-Chemie; 1988. pp. 191–198. [Google Scholar]
- 18.Nakajima H, Hirota T, Toba T, Itoh T, Adachi S. Structure of the extracellular polysaccharide from slime-forming Lactococcus lactis subsp. cremorisSBT 0495. Carbohydr Res. 1992;224:245–253. doi: 10.1016/0008-6215(92)84110-e. [DOI] [PubMed] [Google Scholar]
- 19.Neves A R, Ramos A, Nunes M C, Kleerebezem M, Hugenholtz J, de Vos W M, Almeida J S, Santos H. In vivo NMR studies of glycolytic kinetics in Lactococcus lactis. Biotechnol Bioeng. 1999;64:200–212. doi: 10.1002/(sici)1097-0290(19990720)64:2<200::aid-bit9>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 20.Perlman M E, Davis D G, Gabel S A, London R E. Uridine diphosphosugars and related hexose phosphates in the liver of hexosamine-treated rats: identification using 31P-{1H} two-dimensional NMR with HOHAHA relay. Biochemistry. 1990;29:4318–4325. doi: 10.1021/bi00470a009. [DOI] [PubMed] [Google Scholar]
- 21.Poolman B, Konings W N. Relation of growth of Streptococcus lactis and Streptococcus cremoristo amino acid transport. J Bacteriol. 1988;170:700–707. doi: 10.1128/jb.170.2.700-707.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poolman B, Smid E J, Veldkamp H, Konings W N. Bioenergetic consequences of lactose starvation for continuously cultured Streptococcus cremoris. J Bacteriol. 1987;169:1460–1468. doi: 10.1128/jb.169.4.1460-1468.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramos A, Santos H. Citrate and sugar cofermentation in Leuconostoc oenos, a 13C nuclear magnetic resonance study. Appl Environ Microbiol. 1996;62:2577–2585. doi: 10.1128/aem.62.7.2577-2585.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts I S. Bacterial polysaccharides in sickness and in health. Microbiology. 1995;141:2023–2031. doi: 10.1099/13500872-141-9-2023. [DOI] [PubMed] [Google Scholar]
- 25.Santos H, Fareleira P, Ramos A, Pereira H, Miranda M. Nuclear magnetic resonance: a noninvasive technique in the study of life processes in situ. Rev Port Quim. 1995;2:3–11. [Google Scholar]
- 26.Stingele F, Neeser J-R, Mollet B. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilusSfi6. J Bacteriol. 1996;178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutherland I W. Biotechnology of microbial exopolysaccharides. Cambridge, United Kingdom: Cambridge University Press; 1990. [Google Scholar]
- 28.van Casteren W H M, Dijkema C, Schols H A, Beldman G, Voragen A G J. Characterization and modification of the exopolysaccharide produced by Lactococcus lactis subsp. cremorisB40. Carbohydr Polymers. 1998;37:123–130. doi: 10.1016/s0008-6215(99)00292-x. [DOI] [PubMed] [Google Scholar]
- 29.van den Berg D J C, Robjin G W, Jansen A C, Giuseppin M L, Vrekker R, Kamerling J P, Vliegenthart J G, Ledeboer A M, Verrips C T. Production of a novel extracellular polysaccharide by Lactobacillus sake0-1 and characterization of the polysaccharide. Appl Environ Microbiol. 1995;61:2840–2844. doi: 10.1128/aem.61.8.2840-2844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Heijenoort J. Assembly of the monomer unit of bacterial peptidoglycan. Cell Mol Life Sci. 1998;54:300–304. doi: 10.1007/s000180050155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Kranenburg R, Boels I C, Kleerebezem M, de Vos W M. Genetics and engineering of microbial exopolysaccharides for food: approaches for the production of existing and novel polysaccharides. Curr Opin Biotechnol. 1999;10:498–504. doi: 10.1016/s0958-1669(99)00017-8. [DOI] [PubMed] [Google Scholar]
- 32.van Kranenburg R, Van Swan I I, Marugg J D, Kleerebezem M, de Vos W M. Exopolysaccharide biosynthesis in Lactococcus lactisNIZO B40: functional analysis of the glycosyltransferase genes involved in synthesis of the polysaccharide backbone. J Bacteriol. 1999;181:338–340. doi: 10.1128/jb.181.1.338-340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Kranenburg R, Marugg J, van Swam I, Willem N J, de Vos W M. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol Microbiol. 1997;24:387–397. doi: 10.1046/j.1365-2958.1997.3521720.x. [DOI] [PubMed] [Google Scholar]
- 34.van Rooijen R J, de Vos W M. Molecular cloning, transcriptional analysis and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J Biol Chem. 1990;265:18499–18503. [PubMed] [Google Scholar]
- 35.Whitfield C, Valvano M A. Biosynthesis and expression of cell-surface polysaccharides in Gram-negative bacteria. Adv Microb Physiol. 1993;35:135–246. doi: 10.1016/s0065-2911(08)60099-5. [DOI] [PubMed] [Google Scholar]