Abstract
Purpose
To develop and validate an automated segmentation tool for COVID-19 lung CTs. To combine it with densitometry information in identifying Aerated, Intermediate and Consolidated Volumes in admission (CT1) and follow up CT (CT3).
Materials and Methods
An Atlas was trained on manually segmented CT1 of 250 patients and validated on 10 CT1 of the training group, 10 new CT1 and 10 CT3, by comparing DICE index between automatic (AUTO), automatic-corrected (AUTOMAN) and manual (MAN) contours. A previously developed automatic method was applied on HU lung density histograms to quantify Aerated, Intermediate and Consolidated Volumes. Volumes of subregions in validation CT1 and CT3 were quantified for each method.
Results
In validation CT1/CT3, manual correction of automatic contours was not necessary in 40% of cases. Mean DICE values for both lungs were 0.94 for AUTOVsMAN and 0.96 for AUTOMANVsMAN. Differences between Aerated and Intermediate Volumes quantified with AUTOVsMAN contours were always < 6%. Consolidated Volumes showed larger differences (mean: −95 ± 72 cc). If considering AUTOMANVsMAN volumes, differences got further smaller for Aerated and Intermediate, and were drastically reduced for consolidated Volumes (mean: −36 ± 25 cc). The average time for manual correction of automatic lungs contours on CT1 was 5 ± 2 min.
Conclusions
An Atlas for automatic segmentation of lungs in COVID-19 patients was developed and validated. Combined with a previously developed method for lung densitometry characterization, it provides a fast, operator-independent way to extract relevant quantitative parameters with minimal manual intervention.
Keywords: Automatic segmentation Atlas-based, Quantitative imaging computed tomography, Lung segmentation, Covid-19
Introduction
COVID-19 is an infectious disease characterized by several and non-specific clinical manifestations, as fever, cough, dyspnea and fatigue [1] which can cause from very mild to severe illness, including Acute Respiratory Distress Syndrome (ARDS) [2]. Computed tomography (CT) plays a key role in the clinical classification and management of COVID-19 patients especially for its high sensitivity in identifying COVID-19 pneumonia (up to 97% when having RT-PCR as reference standard) [3], [4], [5], [6]. Moreover, the quantitative analysis of the CT images for the extraction, analysis and interpretation of quantitative data has become widespread especially because of the experience acquired on ARDS [7], [8], [9]. Lung quantitative CT analysis embraces several techniques, for example the extraction of parameters from the intensity histogram [10], [11], [12], [13], [14], texture-based parameters detailing spatial relationship between voxels [15], [16] and also includes the development of predictive models based on AI tools [17], [18], [19]. Several studies used threshold measures to quantify the healthy and the more compromised, high-density consolidated volumes as predictors of the disease severity and its complications, such as the need for oxygenation support or ICU admission or the risk of death [9], [10], [11], [13], [14], [16]. Segmentation plays a key role in quantitative CT analyses, consisting in the identification and delineation of the entire lung volume [20]. This is a critical step because time-consuming if pursued manually, and on the other hand difficult to perform automatically without incorrected results. In fact, as long as on healthy lungs the automatic segmentation algorithms work well being low-density and high-contrast regions [21], conversely, the presence of abnormalities, like pleural effusion or parenchymal consolidations, which have attenuation characteristics similar to the pleural margin and the thoracic soft tissues, often leads to inaccurate output [22], [23]. This is also well reflected in lung density histograms of COVID patients with ARDS, which often appear largely modified with respect to healthy individuals because of the presence of a typical peak in the region around 0 HU. Therefore, manual contouring remains the actual reference standard for lung segmentation [24]. As reported in the Internet Analysis Tools Registry [25] there several softwares developed for lung segmentation. Despite manual contouring is actually the standard, the automatic approach has the potential to minimize the operator variability, reduce the time for the contouring and extending studies to a larger quantity of images to be segmented [26]. Deep learning-based or atlas-based algorithms performances constantly improve, so it is expected that they will replace manual methods to become the standard [27], [28], [29], [30]. Some of these methods have been proved to be very effective to produce accurate contours requiring minimal editing by physicians, but their implementation and training is very demanding. Even when neural networks are implemented in commercial software, hospitals usually do not have the possibility to collect an adequate training set of studies. In this context Atlas-based segmentation remains a reasonable option and it is implemented by several vendors. Beyond automatic segmentation, another critical aspect for quantitative CT consists in quantifying the impact of segmentations uncertainties on the quantitative CT results [31], [32], [33], even when automatic [34]. Accordingly, the purposes of this work are:
-
1)
To develop and validate an Atlas-based automated segmentation tool for COVID-19 lungs CT scans.
-
2)
To extract quantitative parameters [13], [14] from histograms of manual and automatic contours in order to test if significant differences were present.
-
3)
To apply the suggested approach to quantify variations of densitometry parameters along time from the admission to follow-up at few months after the discharge in a prospectively followed cohort of patients.
Materials and methods
Atlas-based automated segmentation tool development
The Atlas workflow was developed using the MIM Assistant package of the MIM Maestro software (MIM 6 v 6.9.6) and trained with lungs on CTs of 250 first wave COVID-19 patients manually revised by experts [14] during the first phase of hospitalization. Importantly, all CTs were acquired at maximum inspiration. The Atlas development was based on the registration of each CT to a reference CT chosen as Atlas representative subject and also named “template”. The registration was performed using a rigid algorithm, to determine a similarity index, which aims to quantify the anatomical affinity of each Atlas subject to the template. Once all the 250 subjects were registered with the template, the Atlas was ready to automatically extract contours for a new set of patients, but a validation was required. The workflow used to invoke the Atlas can be customize with the following settings: the deformable registration method, the finalization algorithm, and the number of subjects used by the multi-subject Atlas [35]. The chosen finalization method is the Majority Vote (MV): a voxel is assigned to a certain structure if that voxel belongs to the same structure for most k subjects. Five consecutive image registrations are performed for every patient (k = 5), to find the best matches within the Atlas images database. In order to regularize any odd shape of the contours, it is convenient to add some post-processing functions to the workflow. In this case smooth and fill holes tools were used. In Fig. 1S is reported the workspace of MIM for the Atlas workflow editor. During the segmentation procedure, before proceeding with each deformable image registration, the software asks users to confirm the rigid registration found by the algorithm. In this phase, when not satisfied with the performance of the software, the operator can interact and manually modify the overlap between the template and the new patient image. This step can also be skipped and the workflow would choose all the rigid registrations independently, without any confirmation request. About this, Casati et al. [35] showed that the approach used to register the Atlas’ subjects to the template did not influence the accuracy of auto-contours.
Atlas-based automated segmentation tool validation
To evaluate the performance of the Atlas automatic segmentation, a validation cohort of patients was defined. Three different samples of patients’ CT have been chosen: 10 CTs already used to build the Atlas (CT1 internal validation set), 10 new CTs of patients at the hospital admission (CT1 external validation set) and 10 CTs of new patients scanned during a follow-up exam, few months after the discharge (CT3 external validation set). This last set was considered with the aim of extending its use, based on the assumption that an atlas trained on CTs of hospitalized patients should work in identifying lungs whose appearance is more near to normal lungs, as the ones referred to CT3.
As already reported above, during the run of the Atlas on validation cohorts, the patient’s CT study was registered on the Atlas template and the similarity index was evaluated. This value was compared to the similarity indices of all the Atlas subjects in order to find the 5 subjects, which best matches the patient anatomy. For each validation cohort, the following material was available: two manual contours performed by two in-training radiologists (MAN), an automatic contour traced by the Atlas (AUTO), a manual revision of the automatic contour (AUTOMAN). In this last case, the same operator who generated the automatic contours corrected them, if necessary, using the 3D brush tool provided by MIM, which allows to operate a correction on more slices simultaneously, and the smoothing tool. Right and left lungs were delineated separately for each patient. Importantly, the two radiologists providing MAN were not previously involved in the training of the Atlas, aiming to make the validation fully independent. For the same reason, the operator making corrections after running the automatic segmentation was not one of the two radiologists providing MAN.
DICE computation for volumes segmented comparison
Volumes defined by the different segmentation methods were compared computing Dice coefficients using the dedicated function “dice” in Matlab. For each validation subset, Dice indexes were computed between manual and automatic segmentation (MAN-AUTO), manual and corrected automatic segmentation (MAN-AUTOMAN), automatic and corrected automatic segmentation (AUTO-AUTOMAN). All the indexes were computed separately for right and left lungs. Having two references for the manual segmentation, a mean value of the two Dice coefficients was computed when MAN contour was involved. Moreover, to represent the volume agreement between the three methods, the DICE mean among all patients for each couple of methods was computed for the right and left lung separately.
Lung sub-regions definition: Parameters extraction comparison
To accomplish the second aim of the work, quantitative parameters were extracted from the density histograms, once lungs were segmented using the three approaches in order to quantify the differences derived in the case of MAN, AUTO and AUTOMAN segmentation. HU histograms of the segmented contours were exported from MIM and quantitative parameters were extracted by applying a previously developed and tested method [12], [13]. Having two references for the manual segmentation, the mean of the two histograms corresponding to each observer was computed when MAN contours were involved. The method refers to the typical HU-distribution of the lungs of COVID-19 patients, generally characterized by the presence of two peaks, one next to the air HU (−1000 HU), which defines the aerated and therefore properly “functioning” lung (Aerated Volume); and one next to the water HU (0 HU) corresponding to the lung component with consolidated disease (Consolidated Volume). Between these two peaks there is a quite evident and pronounced region corresponding to lung affected by the disease with highly variable patterns from patient to patient (Intermediate Volume). These volumes were extracted by the search of the HU thresholds which better separated the peaks from the intermediate plateau; other extracted parameters which are absent in healthy histograms were the height and width of the intermediate volume area under the HU histogram (Height Intermediate, Width Intermediate), the shift of the Aerated Peak (Apeak) with respect to −1000 HU (ShiftAirPeak) and the shift of Consolidated Peak (Cpeak) with respect to 0 HU (ShiftWaterPeak). Moreover, Aerated, Intermediate and Consolidated Volumes were also computed for the combined lungs (CL) together with their ratios (ConsolidatedVolume/AreatedVolume and IntermediateVolume/AreatedVolume). Differences between all these extracted parameters for left, right and CL lungs were tested with the Wilcoxon and T-test (significance level p = 0.05). In order to quantify the differences of Volumes between AUTO vs MAN and AUTOMAN vs MAN for CT1 and CT3 the absolute and percentage differences of the volume parameters were computed.
Longitudinal densitometric study
After the validation phase, the Atlas was exploited to obtain lungs segmentation of a sample of 50 patients with SARS-CoV-2 infection, confirmed by RT-PCR on nasopharyngeal swab. Of note, half part of this group of patients was used to train the Atlas. These patients referred to a secondary analysis of the COVID-BioB study (Clinical trials govNCT04318366) concerning the analysis of longitudinal changes of prospectively collected radiological, immunologic and medical features with time of survived patients. All patients underwent a first chest CT (CT1) in the first few days of hospitalization: in all cases, a follow up CT (CT3) acquired few months after the discharge was available. For a sub-group of 30 patients also a second CT (CT2) made during the hospitalization period was available. Time interval between CT1-CT2, CT2-CT3, CT1-CT3 were quantified and represented in histograms with mean, median times and standard deviations. Time intervals between consecutive CTs (CT1-CT2, CT2-CT3 and CT1-CT3) were also quantified. Qualitative and quantitative analysis of HU histograms variation and lung sub-regions through time were performed, by applying the suggested AUTOMAN method. Total volumes of combined lungs were evaluated for CT1, CT2, and CT3 in order to follow variations and test their significance with T-test (significance level p = 0.05). Mean density variation was computed along time. Other parameters quantified were the aerated and the consolidated peak position variation and significance was tested with the two-tailed T-test (significance level p = 0.05).
Results
Atlas validation and critical points
The time required for the automatic segmentation of COVID-19 patients’ lungs, comprehensive of an optional manual “start registration”, was of about 2 min with a workstation with a 2.10 GHz Intel® processor, 32 GB RAM, Windows 10 Enterprise. Although Atlas automatic segmentation recognized quite well the consolidated region of the lungs that is generally badly identified by most common region grow or thresholds-based segmentation algorithms, this tool presented some recurrent critical points, in particular:
-
•
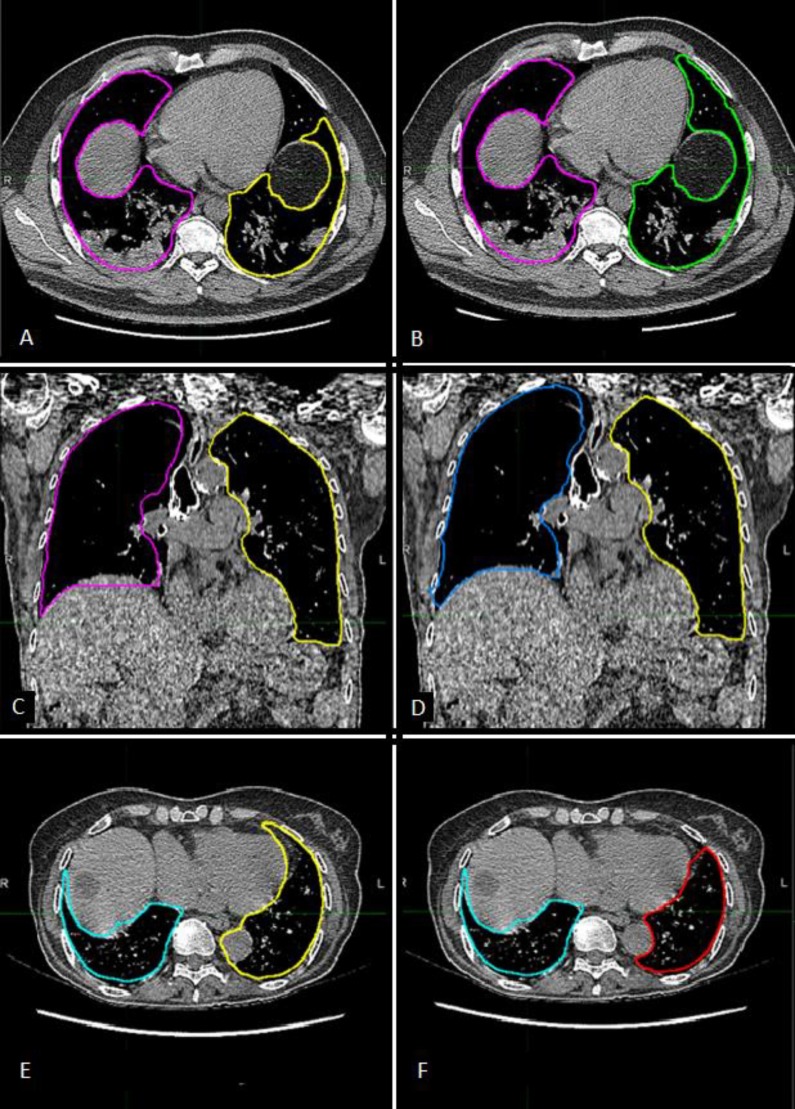
Type 1 failure: Anterior segments of lower lobes were often not properly included in the automatic contour (Fig. 1 A).
-
•
Type 2 failure: Right lung contour sometimes included a not negligible slice of the liver (Fig. 1C).
-
•
Type 3 failure: In few cases, the left lung contour wrongly included the aorta (Fig. 1D).
Fig. 1.
A) Automatic Contour. Anterior segments of lower lobes are often not included in the automatic contours, as happens for the left lung in this image. Type 1 error. B) Manually corrected contour. C) Automatic contour. The right lung contour wrongly includes an important portion of the liver. Type 2 error. D) Manually corrected contour. E) Automatic contour. The left lung contour wrongly includes the aorta. Type 3 error. F) Manually corrected contour.
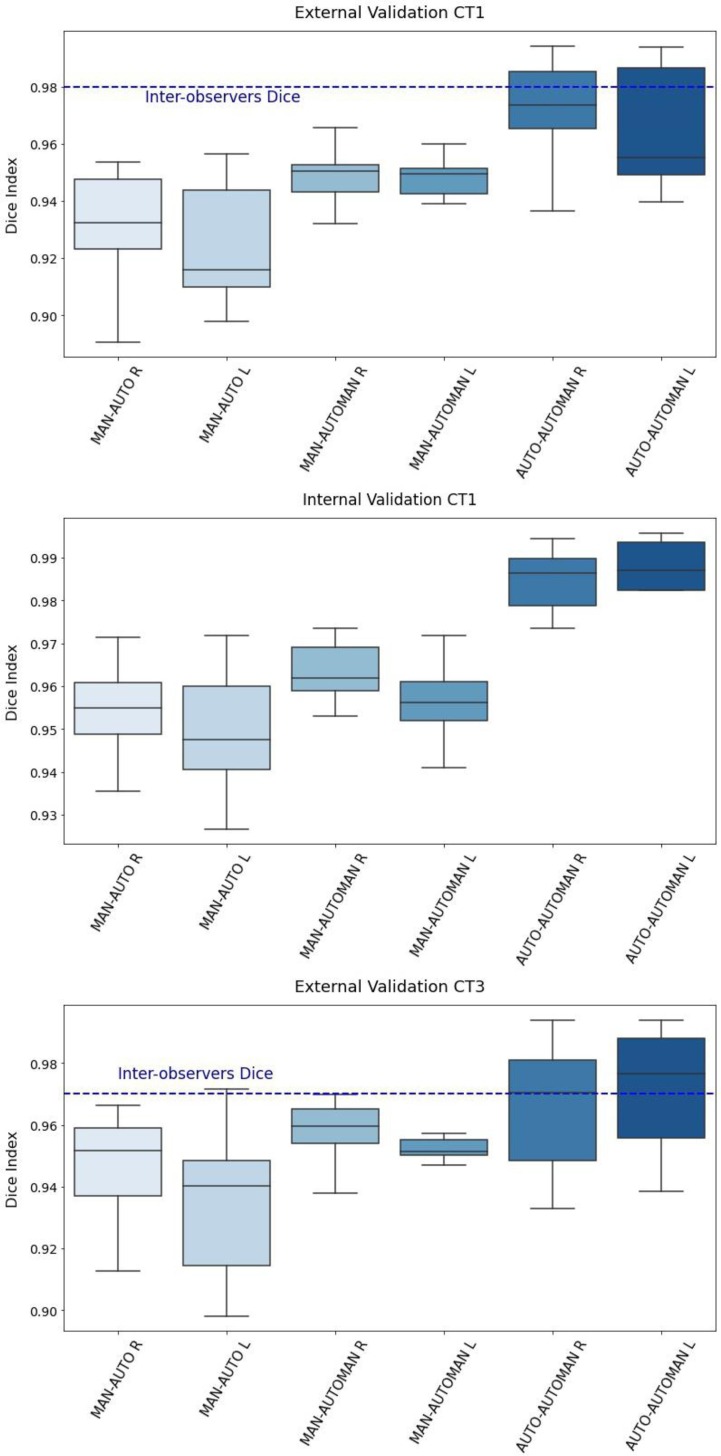
For these reasons, the manual correction was applied to the automatic contours extracted by the Atlas workflow (Fig. 1B, D, F). The time required for the corrections, when needed, was about 5 ± 2 min for both lungs included. Statistically, the most frequent error regarded the type 2 failure, namely in the segmentation of the lung near the liver, even if for few slices (55% of cases). At the second place for frequency was registered the type1 failure, in the segmentation of the anterior lobes (25% of cases), and last the type 3 failure surely more rare (15% of cases). In 40% of cases, manual correction was not considered to be necessary. Dice coefficients were computed to compare volumes defined by the different segmentation methods. First, Dice indices were computed between the two observers: mean values were 0.98, SD = 0.01 and 0.97, SD = 0.02 for the CT1 and CT3 external validation groups respectively. Results are reported in Table 1 for CT1 external and CT3 external validation group and showed that the Dice coefficients were always > 0.90, excepting one case for CT1 and one case for CT3 external validation. Values were found to be a little higher for the CT1 internal validation with respect to the CT1 and CT3 external subsets, as expected. Instead, the lowest values were those related to the comparison between manual and automatic contours in the CT1 external validation (0.93 for the right lung, 0.92 for the left lung). Moreover, the mean Dice index for the left lung in most cases resulted lower than the value for the right lung. In Fig. 2 distributions of DICE coefficients were represented for each validation cohort. Of note, the differences between MAN-AUTO vs MAN-AUTOMAN were statistically significant for the two-tailed T-test (significance level p = 0.05) for the external validation sets.
Table 1.
Dice coefficients for comparisons between various contouring methods in CT1 internal, CT1 and CT3 external validation (R = right, L = left).
| CT1 internal validation | CT1 external validation | CT3 external validation | ||||
|---|---|---|---|---|---|---|
| MAN-AUTO R | 0.95 | 0.01 | 0.93 | 0.02 | 0.95 | 0.02 |
| MAN-AUTO L | 0.95 | 0.02 | 0.92 | 0.02 | 0.94 | 0.02 |
| MAN-AUTOMAN R | 0.96 | 0.01 | 0.95 | 0.01 | 0.96 | 0.01 |
| MAN-AUTOMAN L | 0.96 | 0.01 | 0.95 | 0.01 | 0.96 | 0.01 |
| AUTO-AUTOMAN R | 0.98 | 0.01 | 0.96 | 0.02 | 0.97 | 0.02 |
| AUTO-AUTOMAN L | 0.98 | 0.02 | 0.96 | 0.02 | 0.97 | 0.02 |
| Inter-observer variability | 0.98 | 0.02 | 0.97 | 0.02 | ||
Fig. 2.
Box-plots of Dice coefficient distributions among patients from the CT1 internal (A), CT1 external (B) and CT3 external validation subsets (C) for right and left lungs. The horizontal blue line represents the value of the mean inter-observers Dice coefficient found for CT1 (0.98 ± 0,01) and CT3 (0.97 ± 0.02) external validation subsets. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
HU histograms analysis
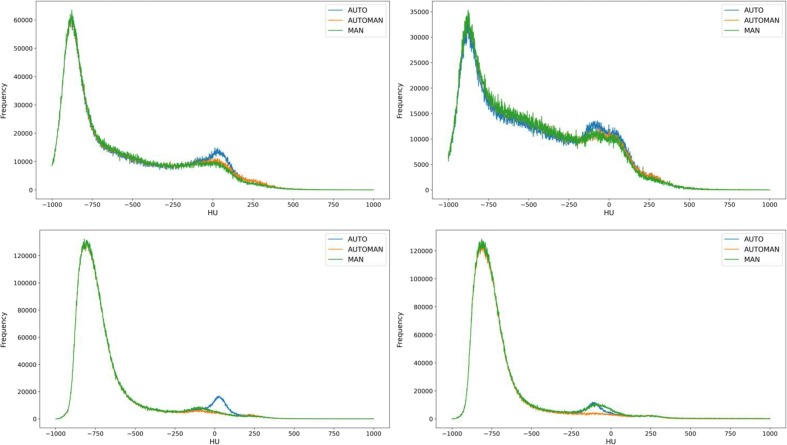
With regard to the comparison between MAN, AUTOMAN and AUTO contours, having two references for the manual segmentation, the mean of the two histograms corresponding to each observer was computed when MAN contours were involved. The extracted histograms of a CT1 were reported in Fig. 3 A (right) and B (left). Similarly, the extracted histograms of a CT3 were reported in Fig. 3C (right) and B (left). Aside from differences between segmentations, in CT1 histograms the Consolidated Volume is clearly visible at densities around 0 HU. Differently it disappears in CT3 histograms after the recover. In most cases especially for the right lung (12 and 6 cases out of 20 for right and left lung respectively), the Cpeak for the AUTO contour tended to be higher with respect to MAN and AUTOMAN. This may be due to the already exposed Atlas critical point regarding the wrong inclusion of the liver for the right lung, which can normally be associated with Hounsfield Units within 40–60 as reported in Fig. 3A. Regarding the left lung the peak in the consolidated region was present for all contours, without differences as reported in Fig. 3B. Instead for CT3 no evidence of a peak for the consolidated region were reported in both MAN and AUTOMAN. In fact, several months after the critical phase of the infection, the lung prevalently restores its normal or almost normal functionality. Here, again, the automatic contour showed an enhanced peak compatible with the improper segmentation of the liver (Fig. 3C). In Fig. 3D a particular case of CT3 was reported, showing a residual Consolidated Volume. Mean values and standard deviation for absolute and percentage variation of the Aerated Volume, Intermediate Volume and Consolidated Volume for CT1 and CT3 validation samples were computed. As reported in Table 2 , values of Δ and Δ % obtained for Consolidated Volume CL in the CT1 validation sample are shown. There is an improvement moving from AUTO to AUTOMAN because the absolute value of Δ Consolidated Volume CL and Δ % Consolidated Volume CL were smaller for MAN vs AUTOMAN respect to MAN vs AUTO: all the values found for variation of consolidated volume were on average negative. This means that, as already reported in the qualitative analysis of the HU histograms, automatic segmentation tended to overestimate the ConsolidatedVolume, including in the contours some tissues which do not belong to the lung. The values obtained for the percentage variation between MAN and AUTOMAN were −19% (−36 cc) for CT1 and –32% (–32 cc) for CT3. For Aerated and Intermediate Volumes, differences between AUTO and AUTOMAN in approaching the manual contour were less evident and an improvement was constantly observed as reported in Table 2 (from 5% to 2% and from 6% to 4% for Aerated and Intermediate Volume respectively). In Table 1S-4S of the Supplementary Material are reported the p-values obtained for Wilcoxon and two tails T-test between AUTOMAN and MAN, and between AUTO and MAN, for the validation samples.
Fig. 3.
A) CT1 example of right lung, with Consolidated region overestimated by AUTO probably because of error type 2 B) CT1 example of left lung C) CT3 example of right lung: the Consolidated region decreased for MAN and AUTOMAN because of recovery, instead was overstimated by AUTO probably because of error type 2 D) CT3 particular case of left lung still showing a residual Consolidated Volume. In this case the manual revision with AUTOMAN has removed the Cpeak, instead the histogram extracted by AUTO segmentation reproduced as well the MAN reference histogram.
Table 2.
Percentage differences in consolidated, aerated, and intermediate volumes for CT1 and CT3 between different segmentation methods (Man = manual; AUTO = atlas-based automatic; AUTOMAN = atlas-based automatic + manual correction); for consolidated volume, also absolute changes are shown. *p < 0.05. CL= (Combined Lungs).
| MAN vs AUTO CT1 (Δ (Δ%)) | MAN vs AUTOMAN CT1 (Δ (Δ%)) | MAN vs AUTO CT3 (Δ (Δ%)) | MAN vs AUTOMAN CT3 (Δ (Δ%)) | |
|---|---|---|---|---|
| Consolidated Volume CL | ||||
| mean | −53%* (−74 cc) | −19%* (−36 cc) | −89%* (−95 cc) | –32%* (–32 cc) |
| SD | 59% (49 cc) | 12% (25 cc) | 63% (72 cc) | 22% (24 cc) |
| Aerated Volume CL | ||||
| mean | 5%* | 2%* | 3%* | 2%* |
| SD | 3% | 1% | 2% | 1% |
| Intermediate Volume CL | ||||
| mean | 6% | 4%* | 2% | 6%* |
| SD | 6% | 3% | 7% | 3% |
Longitudinal densitometric study
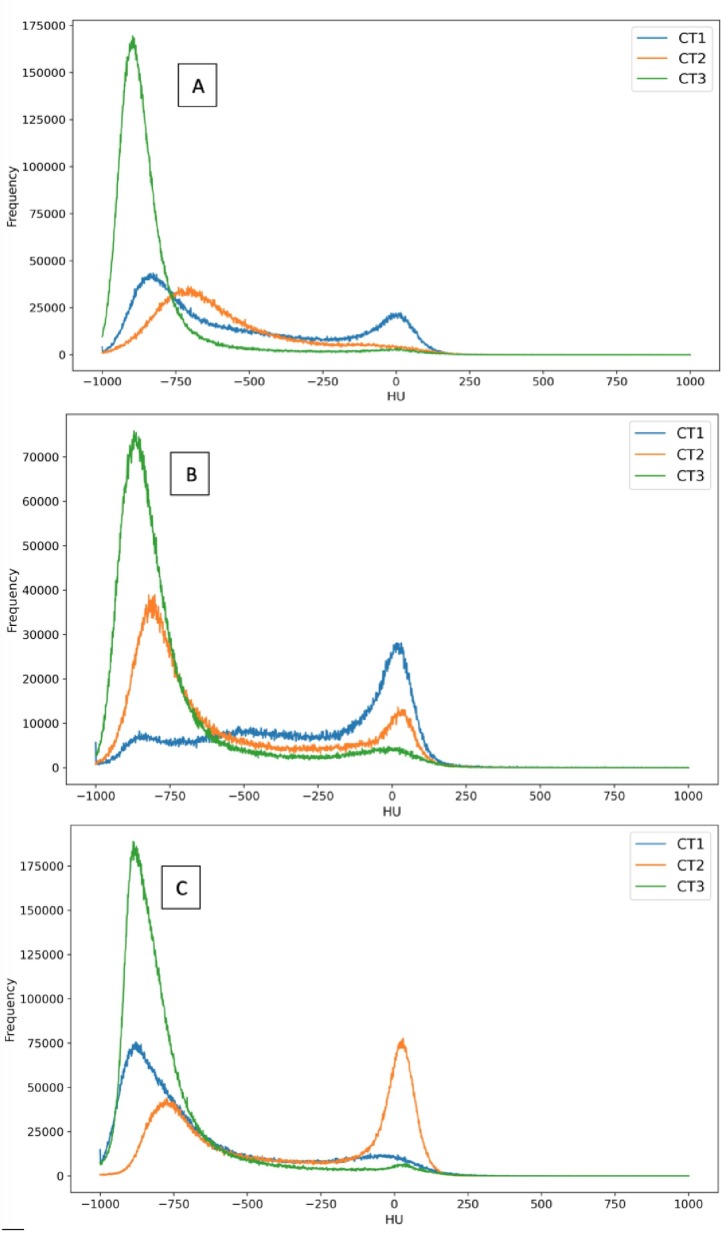
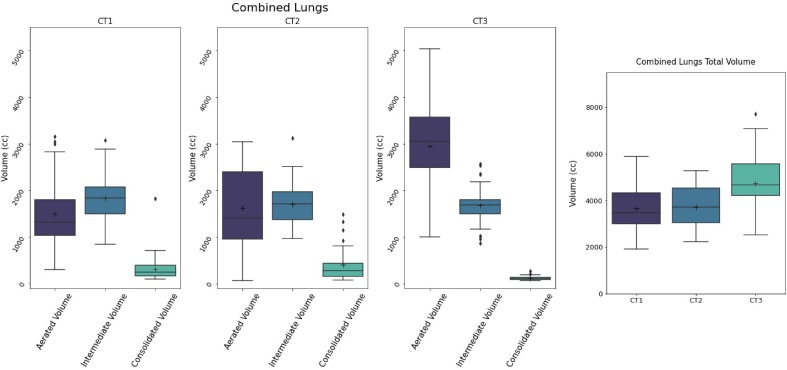
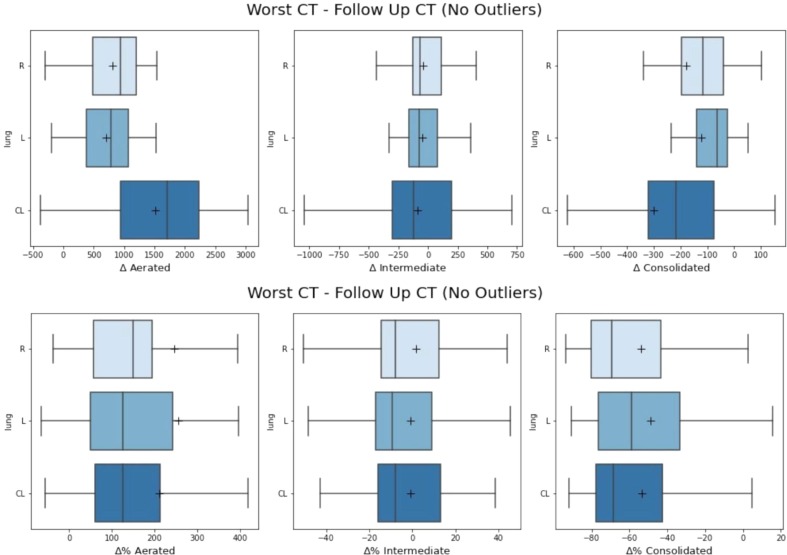
Time intervals between CT1-CT2, CT2-CT3 and CT1-CT3 were found to have a mean/SD of 30/30 days for CT1-CT2, 193/92 for CT2-CT3 and 202/94 for CT1-CT3. Distributions are reported in the Fig. 2S. Most of the changes during time were similar to the three examples reported in Fig. 4 ; in a fraction of patients, with a mean interval of 17 days between CT1-CT2, Cpeak (next to HU = 0) was more evident in CT2 (Fig. 4C) scans than in CT1, proving that CT1 not always represented the most critical phase of the infection. Second, the position of Apeak shifts from higher HU values to lower ones, as attended when the disease regresses (Fig. 4A, B, C). In almost all cases, Cpeak completely or almost disappeared in CT3. As reported in Table 3 , the mean CL volume was 3549 ± 979 cc, 3685 ± 947 cc and 4621 ± 1136 cc for CT1, CT2 and CT3 respectively: the difference between CT1 and CT2 was not significant (p = 0.42) while the differences between CT1 and CT3 and between CT2 and CT3 were significant (p < 0.0001). The changes of Aerated, Intermediated and Consolidated Volumes between CT1 and CT2 had not a clear trend, as shown in Fig. 5 . Then, in order to better point out the differences against CT3, the “Worst CT” between CT1 and CT2 (corresponding to the CT with the smaller Aerated Volume) was considered for each patient and named “Worst CT”. Instead CT3 was named “Follow UP CT”. Aerated, Intermediate and Consolidated mean CL Volumes mean variations between Worst CT and Follow UP CT were + 1517 ± 967 cc, −88 ± 436, and −302 ± 369. Absolute and percentage variations are reported in Fig. 6 . Median/mean variation of total CL volume between Worst CT and Follow UP CT was 1265 cc (39%)/1127 cc (36%) (p < 0.0001): only 6/50 patients showed a decrease of −636 cc (−15%). Median/mean variations of mean CL density were −177HU/−186HU (p < 0.0001). Histograms showing the distributions of mean density absolute and percentage variations for both left and right lung are shown in Fig. 3S. From the T-test performed for the total volume of CL between Worst CT and Follow Up CT, p-values were found significative different (p < 0.001). In Table 3, values of Apeak obtained for CL in the Worst and Follow Up CTs are shown and distributions are depicted in Fig. 4S. The differences were found statistically significant (T-Test p = 0.0001 and 0.017 for right and left lung respectively). Regarding the Cpeak position, the T-Test performed between distributions in Worst CT and Follow Up CT did not give significant differences. In Table 3 values of Cpeak position are reported and distributions are depicted in Fig. 5S.
Fig. 4.
Example of HU histograms time evolution for three patients in the longitudinal study population, A), B), C).
Table 3.
. Combined Lung (CL) mean total volume for CT1, CT2 and CT3. Position of Areated Peak (Apeak) and Consolidated Peak (Cpeak) in the Worst CT and in the Follow UP CT.
| CL Parameters | CT1 | CT2 | CT3 |
|---|---|---|---|
| Mean volume (±SD) | 3459 (±979) cc | 3685 (±947) cc |
4261 (±1136) cc |
| Worst CT | Follow UP CT | ||
| Apeak (±SD) | −836 (±55) HU | 862 (±40) HU | |
| Cpeak (±SD) | 8 (±32) HU | 4 (±39) HU | |
Fig. 5.
Aerated, Intermediate, Consolidated and total lung Volume temporal variations in combined lungs from CT1, CT2 and CT3. The changes between CT1 and CT2 had not a clear trend.
Fig. 6.
A) Absolute and B) percentage variations in Aerated, Intermediate and Consolidated Volume between Worst CT and Follow UP CT. Boxplots for right, left and combined lungs are shown. Outliers were removed to simplify the graphic representation, but still contribute to the mean (black cross) and median (black line) values.
Discussion
In the current study, an Atlas for the automatic segmentation of lungs in chest CT images of COVID-19 patients was developed with the aim of assessing its accuracy and suitability in a fully integrated workflow of image analysis. The impact of auto-segmentation uncertainty on quantitative densitometry analyses was also computed, aiming to make available an almost completely automatic framework to extract parameters characterizing the severity of pulmonary symptoms of COVID-19 patients as well as functioning as robust imaging-based predictors. Concomitantly, the temporal changes of parameters from the admission to the follow-up at few months after the discharge in a prospectively followed cohort of patients were quantified as an example of relevant application of the suggested approach. Particularly the total CL volume between Worst CT and Follow UP CT was found to increment on average of 1265 cc (39%)/1127 cc (36%) (p < 0.0001): only 6/50 patients showed a decrease of −636 cc (−15%). The CL volume distributions along time were reported in the last plot of Fig. 5. This difference was probably due to a different ability in respiration, improved at the follow up respect to previous times as clear evidence of the functionality recovery. The decrease of CL volume is likely associated to a compromised lung functionality.
The Atlas developed and validated at our Centre could be easy adopted on CTs coming from other Institutes, because the only input required consists in the CT image scanned at maximum inspiration. There are many papers in the literature on automatic segmentations for lung abnormalities [36], [37], [38], [39], [40], [41], [42], several also dedicated to the topic in COVID-19 diagnostic imaging field, mostly based on AI-based approaches including neural networks [43], [44], [45], [46], [47]. On the other hand, atlas-based approaches were rarely reported for COVID-19 lung segmentation [48]. Differently from AI-based methods, Atlas-based segmentation may be easily implemented using commercially available solutions, as the one we used.
The recent study by Berta et al. [48] reported the accuracy of several segmentation tools applied on COVID-19 lungs with qualitative and quantitative assessment, including an Atlas-based approach, using a commercial system different from ours. Authors reported a non-inferiority result in terms of contouring accuracy between Atlas-based against several AI-based methods in an internal validation loop. Unfortunately, the limited number of patients and the lack of external validation limits the possibility of comparing these results with ours, despite the quite positive findings with using an Atlas-based approach. Interestingly, the cited article reported similar issues for Atlas-based tools, unveiling the weak point of including in segmentation more dense areas (e.g. part of the liver).
Focusing on our major results, the high values of the Dice coefficients (even in the “different” situation of follow-up CTs) are very encouraging, but they may be also attributed to the fact that lungs are in general very large anatomical volumes. Therefore, relatively large differences between automatic and manual segmentation may go “unnoticed” computing the Dice coefficient. These differences are indeed still small compared to the total volume of the lung but they can translate into relatively large errors when estimating the consolidated part of the lungs, mostly due to the failures occurring at the interface between lung apex and liver and, secondarily, at the edge between lung and aorta. In other words, the Atlas developed for COVID-19 automatic segmentation is still not completely stand-alone for all patients and frequently (roughly in 60% of cases) needs manual correction to be consistent with the manual lungs’ delineations. However, the manual correction is fast (around 5 min, both lungs included) making the approach suitable for clinical use. Improving the performances of the methods by automatizing at least in part the corrections of the automatically generated contours could be a further refining task for the future. A solution could consist in running the Atlas on the entire set of CT available after selecting only better segmentations without or with minimal errors of type 1, 2, 3. With this smaller set of images more correctly segmented, the Atlas could be re-trained to better learn the correct anatomy of COVID-19 lungs.
A limit of this work regards the relatively small number of patients involved in the validation procedure especially because the differences in Consolidated Volume between the AUTOMAN and the reference segmentation method (MAN) were found significative. On the other hand, these differences effectively represented by tens cubic centimeters and included in the uncertainties of vascular structures volume are systematic and had little relevance to characterize the severity of pulmonary symptoms of COVID-19 patients (considering that the total lungs volume for the CT1 sample is on average about 3500 cc). As reported by Fig. 6S of Supplementary Material the higher Consolidated Volumes resulted associated to the smaller percentual differences between AUTOMAN and MAN segmented and vice-versa, proving that the manual correction introduced a systematic error which is percentage significatively large only in the less serious cases of small Consolidated Volumes. To corroborate this, in our previous work [14] when the same procedure to extract histogram parameters was used and a model for mortality prediction was developed and validated with 80% of accuracy (90% when combining histogram parameters with clinical parameters), patients with high risk of mortality were found to have too much high consolidated fractions in CT1. Surely the systematic error reported of about 36 cc in CT1 could not impact the predictive power of the model.
Moreover, it is worth to underline that the combination of the Atlas-based segmentation with our previously developed fully automatic method to quantify the fractions of Aerated, Intermediate and Consolidated Volumes, makes the whole process (from CT acquisition to assessment of densitometry quantitative features) fast and robust.
Thanks to this, the approach was applied to a longitudinal study based on data of 50 prospectively followed patients. Although it was referred to a relatively small population, different and interesting aspects were investigated. A first, exploratory comparison between Worst CT and Follow Up CTs was performed. It was pointed out that the Aerated Volume largely increased with time, as expected [49], [50], while Apeak position shifted towards air reference HU value.
In most cases especially for the right lung, the Cpeak for the AUTO contour tended to be higher with respect to MAN and AUTOMAN in both Worst and Follow UP CTs (infact differences in in Cpeak were found not significative at the T-Test). This may be due to the already exposed Atlas critical point regarding the wrong inclusion of the liver for the right lung, which can normally be associated with Hounsfield Units within 40–60 as reported in Fig. 3A. Regarding the left lung the peak in the consolidated region was present for all contours, without differences as reported in Fig. 3B. Instead for Follow UP CT for the right lung (Fig. 3C) no evidence of a peak for the consolidated region were reported in both MAN and AUTOMAN. In fact, several months after the critical phase of the infection, the lung prevalently restores its normal or almost normal functionality. In Fig. 3D a particular case of Follow UP CT was reported, showing a residual Consolidated Volume. In this case the manual revision with AUTOMAN has removed the Cpeak, instead the histogram extracted by AUTO segmentation reproduced as well the MAN reference histogram. The recovery trend was obtained also by mean lung density which evolved toward air density values. Total volume variation, mean density variation, Apeak and Cpeak position shift, together with other possible parameters extracted from HU histograms, could be used in the future to assess a quantitative relationship between CT findings and clinical outcomes in patients suffering from the so called “Long Covid”. The quantitative approach used to follow changes in COVID-19 lungs in terms of Aerated, Intermediate, Consolidated Volumes, mean density, Apeak position shift etc. differs from the methods generally used in other studies reported by literature. In fact, they are often strictly related to the temporal changes in CT of specific findings described by using internationally standard nomenclature defined by the Fleischner Society glossary and peer-reviewed literature on viral pneumonia, using terms including ground-glass opacity, crazy-paving pattern, and consolidation [51], [52]. Moreover, the assessment of temporal changes of these findings in general are reported by a qualitative definition of visual scores [53], [54], [55]; instead, in this work the temporal changes of parameters extracted are objective and operator-independent.
In summary, AI-based medical imaging has played an important role in fighting against COVID-19 and image segmentation is the first step for each study, so it needs to be accurate and fast to permit the analysis of large amounts of data.
The method described has surely an evident weak point in segmenting consolidated regions but we also demonstrate that it consists in a systematic error which becomes negligible in the most serious cases of larger consolidated zones. The encouraging results of current study suggest that our Atlas-based segmentation method combined with automatic extraction of densitometry information may be a useful tool in supporting studies with large cohorts of patients. Also, differently from fully AI-based approaches, it keeps high interpretability of the results and potentially higher generalizability and clinical usability. Nevertheless, and interestingly, its performances could also be investigated in the future on patients affected by pathologies other than COVID-19 pneumonia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
M Mori is funded by an AIRC grant (IG-23015)
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmp.2022.06.018.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lechien JR, Chiesa-Estomba CM, Place S, VanLaethem Y, Cabaraux P, Mat Q, et al. Clinical and epidemiological characteristics of 1420 European patients with mild- to-moderate coronavirus disease 2019. J Intern Med 2020;288:335–44. https:// doi.org/10.1111/joim.13089. [DOI] [PMC free article] [PubMed]
- 2.Zhang J.J.Y., Lee K.S., Ang L.W., Leo Y.S., Young B.E. Risk Factors for Severe Disease and Efficacy of Treatment in Patients Infected With COVID-19: A Systematic Review, Meta-Analysis, and Meta-Regression Analysis. Clin Infect Dis. 2020;71:2199–2206. doi: 10.1093/cid/ciaa576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen Z., Chi Y., Zhang L., Liu H., Du K., Li Z., et al. Coronavirus Disease 2019: Initial Detection on Chest CT in a Retrospective Multicenter Study of 103 Chinese Subjects. Radiol Cardiothorac Imaging. 2020;2:e200092. doi: 10.1148/ryct.2020200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., et al. Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agricola E., Beneduce A., Esposito A., Ingallina G., Palumbo D., Palmisano A., et al. Heart and Lung Multimodality Imaging in COVID-19. JACC Cardiovasc Imaging. 2020;13(8):1792–1808. doi: 10.1016/j.jcmg.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Cobelli F., Palumbo D., Ciceri F., Landoni G., Ruggeri A., Rovere-Querini P., et al. Pulmonary Vascular Thrombosis in COVID-19 Pneumonia. J Cardiothorac Vasc Anesth. 2021;35(12):3631–3641. doi: 10.1053/j.jvca.2021.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang S., Wang Y.C., Ju S. Advances in medical imaging to evaluate acute respiratory distress syndrome. Chin J Acad Radiol. 2021;17:1–9. doi: 10.1007/s42058-021-00078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishiyama A, Kawata N, Yokota H, Sugiura T, Matsumura Y, Higashide T, et al. A predictive factor for patients with acute respiratory distress syndrome: CT lung volumetry of the well-aerated region as an automated method. Eur J Radiol 2020; 122:108748. 10.1016/j.ejrad.2019.108748. [DOI] [PubMed]
- 9.Esposito A., Palmisano A., Cao R., Rancoita P., Landoni G., Grippaldi D., et al. Quantitative assessment of lung involvement on chest CT at admission: Impact on hypoxia and outcome in COVID-19 patients. Clin Imaging. 2021;77:194–201. doi: 10.1016/j.clinimag.2021.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romanov A., Bach M., Yang S., Franzeck F.C., Sommer G., Anastasopoulos C., et al. Automated CT Lung Density Analysis of Viral Pneumonia and Healthy Lungs Using Deep Learning-Based Segmentation, Histograms and HU Thresholds. Diagnostics (Basel) 2021 Apr 21;11(5):738. doi: 10.3390/diagnostics11050738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomé MH, Gjini M, Zhu S, Kabarriti R, Guha C, Garg MK et al. Using Statistical Measures and Density Maps Generated From Chest Computed Tomography Scans to Identify and Monitor COVID-19 Cases in Radiation Oncology Rapidly. Cureus 2021 Aug 25;13(8):e17432. 10.7759/cureus.17432. eCollection 2021 Aug. [DOI] [PMC free article] [PubMed]
- 12.Ash SY, Harmouche R, Vallejo DLL, Villalba JA, Ostridge K, Gunville R, et al. Densitometric and local histogram based analysis of computed tomography images in patients with idiopathic pulmonary fibrosis. Respir Res 2017;18:1–11. https:// doi.org/10.1186/s12931-017-0527-8. [DOI] [PMC free article] [PubMed]
- 13.Mazzilli A., Fiorino C., Loria A., Mori M., Esposito P.G., Palumbo D., et al. An automatic approach for individual HU-based characterization of lungs in COVID-19 patients. Appl Sci. 2021;11(3):1238. [Google Scholar]
- 14.Mori M., Palumbo D., De Lorenzo R., Broggi S., Compagnone N., Guazzarotti G., et al. Robust prediction of mortality of COVID-19 patients based on quantitative, operator-independent, lung CT densitometry. Physica Med. 2021;87:115–122. doi: 10.1016/j.ejmp.2021.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei W., Hu X.W., Cheng Q., Zhao Y.M., Ge Y.Q. Identification of common and severe COVID-19: the value of CT texture analysis and correlation with clinical characteristics. Eur Radiol. 2020;30(12):6788–6796. doi: 10.1007/s00330-020-07012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen C., Yu N., Cai S., Zhou J., Sheng J., Liu K., et al. Quantitative computed tomography analysis for stratifying the severity of Coronavirus Disease 2019. J Pharm Anal. 2020;10(2):123–129. doi: 10.1016/j.jpha.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput Biol Med. 2020;121:103795. doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang L.u., Han R., Ai T., Yu P., Kang H., Tao Q., et al. Serial Quantitative Chest CT Assessment of COVID-19: A Deep Learning Approach. Radiol Cardiothorac Imaging. 2020;2(2):e200075. doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lessmann N., Sánchez C.I., Beenen L., Boulogne L.H., Brink M., Calli E., et al. Automated Assessment of COVID-19 Reporting and Data System and Chest CT Severity Scores in Patients Suspected of Having COVID-19 Using Artificial Intelligence. Radiology. 2021;298(1):E18–E28. doi: 10.1148/radiol.2020202439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mascalchi M., Camiciottoli G., Diciotti S. Lung densitometry: Why, how and when. J Thorac Dis. 2017;9(9):3319–3345. doi: 10.21037/jtd.2017.08.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pirozzi S., Horvat M., Piper J., Nelson A. SU-E-J-106: Atlas-based segmentation: eval- uation of a multi-atlas approach for lung cancer. Med Phys. 2012;39:3677. doi: 10.1118/1.4734942. [DOI] [PubMed] [Google Scholar]
- 22.Shukla-Dave A, Obuchowski NA, Chenevert TL, Jambawalikar S, Schwartz LH, Malyarenko D, et al. Quantitative imaging biomarkers alliance (qiba) recommendations for improved precision of dwi and dce-mri derived biomarkers in multicenter oncology trials. J Magn Reson Imaging, 49(7):e101–e121, 2019. 10.1002/jmri.26518. [DOI] [PMC free article] [PubMed]
- 23.Simpson S, Kay FU, Abbara S, Bhalla S, Chung JH, Chung M, et al. Radiological society of north america expert consensus document on reporting chest ct findings related to covid-19: endorsed by the society of thoracic radiology, the american college of radiology, and rsna. Radiol Cardiothor Imaging, 2(2):e200152, 2020. 10.1148/ryct.2020200152. [DOI] [PMC free article] [PubMed]
- 24.Piper J., Nelson A., Harper J. MiM Software Inc; Cleveland, OH: 2013. Deformable image registration in mim maestro evaluation and description. [Google Scholar]
- 25.Kennedy D.N., Haselgrove C. The Internet Analysis Tools Registry: A Public Resource for Image Analysis. Neuroinformatics. 2006;4:263–270. doi: 10.1385/NI:4:3:263. [DOI] [PubMed] [Google Scholar]
- 26.Withey D.J., Koles Z.J. A Review of Medical Image Segmentation: Methods and Available Software. IjbemOrg. 2008;10:125–148. [Google Scholar]
- 27.Cardenas C.E., Yang J., Anderson B.M., Court L.E., Brock K.B. Advances in Auto- Segmentation. Semin Radiat Oncol. 2019;29:185–197. doi: 10.1016/j.semradonc.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Xie W., Jacobs C., Charbonnier J.P., van Ginneken B. Relational Modeling for Robust and Efficient Pulmonary Lobe Segmentation in CT Scans. IEEE Trans Med Imaging. 2020;39:2664–2675. doi: 10.1109/TMI.2020.2995108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maffei N., Fiorini L., Aluisio G., D’Angelo E., Ferrazza P., Vanoni V., et al. Hierarchical clustering applied to automatic atlas based segmentation of 25 cardiac sub- structures. Phys Med. 2020;69:70–80. doi: 10.1016/j.ejmp.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Hofmanninger J., Prayer F., Pan J., Röhrich S., Prosch H., Langs G. Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur Radiol Exp. 2020;4(1) doi: 10.1186/s41747-020-00173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizzetto F., Calderoni F., De Mattia C., Defeudis A., Giannini V., Mazzetti S., et al. Impact of inter-reader contouring variability on textural radiomics of colorectal liver metastases. Eur Radiol Exp. 2020;4:62. doi: 10.1186/s41747-020-00189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haarburger C., Müller-Franzes G., Weninger L., Kuhl C., Truhn D., Merhof D. Radiomics feature reproducibility under inter-rater variability in segmentations of CT images. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-69534-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavic M., Bogowicz M., Würms X., Glatz S., Finazzi T., Riesterer O., et al. Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol. 2018;57(8):1070–1074. doi: 10.1080/0284186X.2018.1445283. [DOI] [PubMed] [Google Scholar]
- 34.Lim H, Weinheimer O, Wielpütz MO, Dinkel J, Hielscher T, Gompelmann D, et al. Fully Automated Pulmonary Lobar Segmentation: Influence of Different Prototype Software Programs onto Quantitative Evaluation of Chronic Obstructive Lung Disease. PLoS ONE 2016;11:e0151498. 10.1371/journal. pone.0151498. [DOI] [PMC free article] [PubMed]
- 35.Casati M., Piffer S., Calusi S., Marrazzo L., Simontacchi G., Di Cataldo V., et al. Methodological approach to create an atlas using a commercial auto-contouring software. J Appl Clin Med Phys. 2020;21(12):219–230. doi: 10.1002/acm2.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofmanninger J, Prayer F, Pan J, Ro ¨hrich S, Prosch H, Langs G. Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur Radiol Exp 2020;4:50. 10.1186/ s41747-020-00173-2. [DOI] [PMC free article] [PubMed]
- 37.Doel T., Gavaghan D.J., Grau V. Review of automatic pulmonary lobe segmentation methods from CT. Comput Med Imaging Graph. 2015;40:13–29. doi: 10.1016/j.compmedimag.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Cicek O, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-net: Learning dense volumetric segmentation from sparse annotation. MICCAI 2016:424–432. 10.48550/arXiv.1606.06650.
- 39.Milletari F, Navab N, and Ahmadi SA. V-net: Fully convolutional neural networks for volumetric medical image segmentation. Fourth International Conference on 3D Vision (3DV), 2016:565–571. doi: 10.1109/3DV.2016.79.
- 40.Zhou Z, Siddiquee MMR, Tajbakhsh N, and Liang J. UNet++: A nested U-net architecture for medical image segmentation. Learn Med Image Anal Multimodal Learn Clin Decis Support 2018; 3-11. 10.1007/978-3-030-00889-5_1. [DOI] [PMC free article] [PubMed]
- 41.Isensee F, Petersen J, Klein A, Zimmerer D, Jaeger PF, Kohl S et al. nnU-net: Self-adapting framework for U-Net-based medical image segmentation. 2018, arXiv:1809.10486. 10.48550/arXiv.1809.10486.
- 42.Oktay O, Schlemper L, Le Folgoc L, Lee M, Heinrich M, Misawa K, et al. Attention U-net: Learning where to look for the pancreas. 2018, arXiv:1804.03999. 10.48550/arXiv.1804.03999.
- 43.Zheng C, Xianbo Deng X, Fu Q, Zhou Q, Feng J, Ma H et al. Deep learning-based detection for COVID-19 from chest CT using weak label. 2020, medRxiv:2020.03.12.20027185. 10.1101/2020.03.12.20027185.
- 44.Cao Y., Xu Z., Feng J., Jin C., Han X., Wu H., et al. Longitudinal assessment of COVID-19 using a deep learning-based quantitative CT pipeline: Illustration of two cases. Radiol Cardiothorac Imaging. 2020;2(2):e200082. doi: 10.1148/ryct.2020200082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang L., Han R., Ai T., Yu P., Kang H., Tao Q., et al. Serial quantitative chest CT assessment of COVID-19: Deep-learning approach. Radiol Cardiothorac Imaging. 2020;2(2):e200075. doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi X, Jiang Z, Yu Q, Shao C, Zhang H, Yue H et al. Machine learning-based CT radiomics model for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection: A multicenter study. 2020, MedRxiv:2020.02.29.20029603. http://dx.doi.org/10.21037/atm-20-3026. [DOI] [PMC free article] [PubMed]
- 47.Franquet T. Imaging of pulmonary viral pneumonia. Radiology. 2011;260(1):18–39. doi: 10.1148/radiol.11092149. [DOI] [PubMed] [Google Scholar]
- 48.Berta L., Rizzetto F., De Mattia C., Lizio D., Felisi M., Colombo P.E., et al. Automatic lung segmentation in COVID-19 patients: Impact on quantitative computed tomography analysis. Phys Med. 2021;87:115–122. doi: 10.1016/j.ejmp.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Compagnone N, Palumbo D, Cremona G, Vitali G, De Lorenzo R, Calvi MR et al. Residual lung damage following ARDS in COVID-19 ICU survivors [published online ahead of print, 2021 Nov 10]. Acta Anaesthesiol Scand. 2021;10.1111. 10.1111/aas.13996. [DOI] [PMC free article] [PubMed]
- 50.Zangrillo A, Belletti A, Palumbo D, Calvi MR, Guzzo F, Fominskiy EV et al. One-Year Multidisciplinary Follow-Up of Patients With COVID-19 Requiring Invasive Mechanical Ventilation [published online ahead of print, 2021 Nov 27]. J Cardiothorac Vasc Anesth. 2021;S1053-0770(21)01036-3. 10.1053/j.jvca.2021.11.032. [DOI] [PMC free article] [PubMed]
- 51.Koo H.J., Lim S., Choe J., Choi S.H., Sung H., Do K.H. Radiographic and CT Features of Viral Pneumonia. RadioGraphics. 2018;38(3):719–739. doi: 10.1148/rg.2018170048. [DOI] [PubMed] [Google Scholar]
- 52.Hansell DM, Bankier AA, MacMahon H, McLoud TC, Müller NL, Remy J. Fleischner Society: glossary of terms for thoracic imaging. Radiology 2008;246(3):697–722. doi: 10.1148/radiol.2462070712. [DOI] [PubMed]
- 53.Wang Y., Dong C., Hu Y., Li C., Ren Q., Zhang X., et al. Temporal Changes of CT Findings in 90 Patients with COVID-19 Pneumonia: A Longitudinal Study. Radiology. 2020 Aug;296(2):E55–E64. doi: 10.1148/radiol.2020200843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan F., Ye T., Sun P., Gui S., Liang B., Liet L., et al. Novel Coronavirus (COVID-19) Pneumonia. Radiology. 2019;2020(295):715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van der Sar-Van der Brugge S, Talman S, Boonman-de Winter L, de Mol M, Hoefman E, van Etten RW et al. Pulmonary function and health-related quality of life after COVID-19 pneumonia. Respir Med 2021 Jan;176:106272. 10.1016/j.rmed.2020.106272. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.