Abstract
Atopic dermatitis (AD) is a relapsing inflammatory skin disease; filaggrin (FLG) variation has been consistently associated with its pathogenesis. Filaggrin-2 (FLG2) and trichohyalin-like-1 (TCHHL1) are members of the same protein family (S100 fused type proteins), are similar in structure to FLG, and may be involved in AD pathogenesis. We sought to evaluate the association between variation in FLG2, TCHHL1 and AD remission. We sequenced FLG2 and TCHHL1 in a longitudinal AD cohort using targeted capture based massively parallel sequencing. Association between individual alleles and AD remission was evaluated with generalized estimating equations for binary outcomes. Association between groups of alleles and AD remission was evaluated using a genetic algorithm to group alleles. We identified 2 loss-of-function (LoF) mutations in FLG2 (Ser2377Ter,Arg2207Ter) and 2 LoF mutations in TCHHL1 (Gln656Ter,Gln294Ter), none of which were associated with AD remission. Common (MAF>5%) alleles in FLG2 were similarly unassociated with AD. No common alleles in TCHHL1 were associated with AD remission after multiple testing correction. Among self-described whites, a group of 34 uncommon alleles in FLG2 were associated with increased AD remission (OR 7.64e17; 95%CI 4.41e17–1.32e18; adjusted p<1.0e-16). 12 uncommon alleles in TCHHL1 were associated with increased AD remission (OR 23.46; 95%CI 7.07–77.89; adjusted p=0.064). Among self-described African Americans, 13 uncommon FLG2 alleles were associated with increased AD remission (OR 21.01; 95%CI 11.90–37.09; adjusted p<1.0e-16). No TCHHL1 uncommon allele groups were associated with AD remission among African Americans. Our study supports the role of uncommon alleles in FLG2 and TCHHL1 in AD pathogenesis.
Keywords: atopic dermatitis, epidemiology, epidermal biology, epidermal barrier function, filaggrin 2, trichohyalin-like-1, population genetics, S100 fused type proteins
INTRODUCTION
Atopic dermatitis (AD) is a common chronic inflammatory skin disease which typically presents with red itchy patches on the flexural parts of the extremities [1,2,3,4]. AD is common, affecting up to 20% of children and 3% of adults in the industrialized world [5]. As with many common diseases, the pathogenesis of AD likely varies among individuals, and is influenced by both genetic and environmental factors [4,6,7].
Genetic studies of AD have suggested that both barrier dysfunction and immunodysregulation are key in disease pathogenesis [4,8]. Genetic variation in cytokines involved in the TH2 response, such as thymic stromal lymphopoietin (TSLP), and epidermal surface barrier proteins, primarily filaggrin (FLG), have been associated with variation in AD onset and persistence [5,8,15]. Of these, the most consistently associated and widely studied is FLG. Loss-of-function mutations in FLG lead decreased protein levels and are thought to produce a barrier defect which predisposes an individual to AD [17].
FLG protein belongs to a family of S100 fused-type proteins (SFTPs), many of which are part of the development of the skin’s cornified envelope [5,9,23]. The genes cluster in a region of chromosome 1 called the epidermal differentiation complex (EDC) [5,9,23]. Other proteins in this family include filaggrin-2 and trichohyalin-like 1 protein [5,9,10,11]. Due to these proteins’ structural and functional similarity to FLG, it is possible that variation in any of the SFTP genes could be associated with variation in skin barrier function and thus increase AD susceptibility and persistence [5,9,10,11,17,23].
Filaggrin-2 is an SFTP encoded for by the FLG2 gene. It is located very close to FLG. Previous studies suggest that the FLG2 protein is involved in maintenance of the epidermal barrier and cell-cell adhesion in the cornified layers of skin [11,13]. Decreased expression of FLG2 has been associated with a thinner epidermis; decreased levels of filaggrin-2 have been observed in the lesional skin of patients with AD and psoriasis [5,10,11,12]. Study of FLG2 variation in AD has been limited and largely involved studies in African Americans. In 2014, Margolis et al. identified three notable FLG2 variants in African American individuals, two of which were associated with increased AD persistence [10]. In contrast, in 2015, Pellerin et al. found numerous single nucleotide polymorphisms (SNPs) in FLG2, but found no variants associated with the presence of AD [5].
Trichohyalin-like 1 protein is encoded for by the TCHHL1 gene. Similarly to filaggrin-2, trichohyalin-like 1 protein is an SFTP located near FLG in the EDC. This gene, and its protein’s function, is not well-understood [18,19]. A previous investigation, from a small subset of the present cohort, identified loss of function (LoF) mutations in TCHHL1, but did not find an association with AD [20]. Another report identified mutations in TCHHL1 associated with AD, but the sample size was small with no correction for multiple comparisons [21]. Due to TCHHL1’s proximity to FLG and limited prior studies, further studies are needed to determine if variation in TCHHL1 is associated with AD.
Our goal was to examine genetic variation in two filaggrin-like proteins, previously known to have loss-of-function variants, in a longitudinal cohort of individuals with AD, and to better appreciate the relationship between variation in filaggrin-2, trichohyalin-like 1, and AD remission.
METHODS
Genetic data were obtained from a subset of the Pediatric Eczema Elective Registry (PEER) study for which DNA samples were available. Both the overall PEER cohort and the subset with genetic data have been previously described [7,8]. Analyses were conducted on both the overall cohort, and after stratification by ancestry. Determination of ancestry was based on self-report. Self-described ancestry previously has been determined to strongly correlate with genetic markers of ancestry within this cohort [15]. Written informed consent was obtained from all participants. This study was approved by the institutional review board of the University of Pennsylvania.
DNA was collected with Oragene DNA collection kits (DNA Genotek, Ottawa, Canada). Sequencing of FLG2 and TCHHL1 was conducted using targeted capture based Massively Parallel Sequencing (MPS) on PEER individuals with sufficient DNA. Sequencing read alignment and variant calling were accomplished as previously described [8,22].
Disease clearance was defined using a self-reported outcome of whether or not a child’s skin was symptom-free during the previous six months. As children in PEER could be followed for up to 10 years, PEER participants can have multiple reports of this outcome over time. The association between these outcomes and individual SNPs were evaluated with generalized estimating equations (GEE) for binary outcomes, assuming an exchangeable working correlation structure with empirical standard errors [8]. This GEE model incorporates all individual reports to provide a single point estimate of the likelihood of AD healing over time, which we interpret as a measure of AD remission. An OR>1 indicates an association with greater AD remission, whereas an OR<1 indicates an association with decreased AD remission. GEE models were implemented via the geeglm function from the R package geepack 1.2–1.
Our analysis plan is demonstrated in Figure 1. FLG2 and TCHHL1 were evaluated independently, and analyses were conducted in both the pooled cohort and race-stratified cohorts. Association of variants differed depending on whether the SNPs had MAF ≥ 5%. MAF <5% is a commonly used cutoff for distinguishing common from low-frequency and rare alleles, and has been used by our group in previous work [8,22,26]. We a priori decided that SNPs with MAF ≥ 5% would be independently assessed for association with AD remission. SNPs with MAF < 5% were considered to have insufficient data for reliable GEE estimates when evaluated independently, so these variables were grouped according to an algorithm previously developed and validated [22]. Thus, groups of variants with MAF < 5%, rather than individual variants, were evaluated for association with AD remission. All analyses were implemented in R, version 3.6.1.
Figure 1:
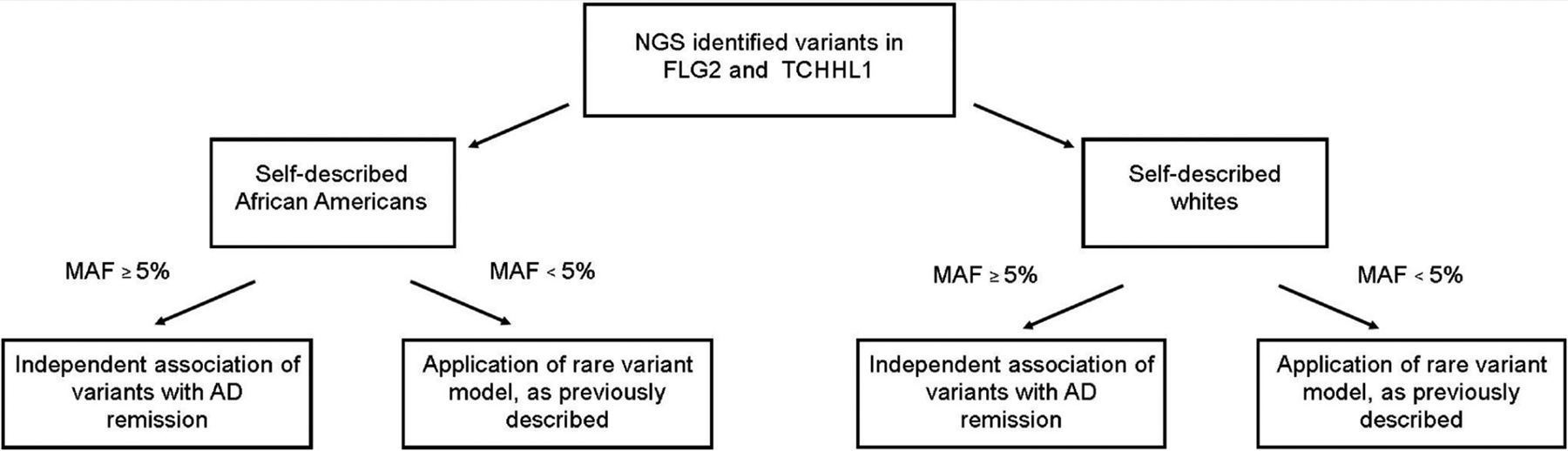
Schematic of analysis flow.
RESULTS
We obtained genotyping of FLG2 and TCHHL1 on 705 PEER individuals; 326 were self-described African Americans and 379 were self-described whites. As of June 2020, over 12,000 follow-up surveys were completed within the PEER study, following participants for a mean of 102 months. Demographic information for the PEER genetic cohort is provided in Table 1.
Table 1:
Participant Demographics
| Whole cohort, n(%) | African American, n(%) | White, n (%) | |
|---|---|---|---|
| Number | 705 | 326 | 379 |
| Age of AD onset, mean (SD) | 1.89 (2.65) | 2.14 (2.85) | 1.84 (2.65) |
| Sex: male, n (%) | 332 (47.1) | 136 (42.0) | 196 (51.7) |
| Asthma, n (%) | 382 (54.1) | 182 (56.2) | 200 (52.8) |
| Seasonal Allergies, n (%) | 494 (70.1) | 228 (70.4) | 266 (70.2) |
163 unique variants were identified in FLG2, of which three were LoF. 120 unique variants were identified in the African American cohort, and 99 unique variants were identified in the white cohort. Of these, 23 variants in the pooled cohort had MAF ≥ 5%. Supplementary Table S1 presents the unadjusted association of these FLG2 SNPs with AD persistence. No variants reached significance at a p-threshold of 0.05. Two LoF variants were identified in our cohort; they are presented in Table 2. The previously studied SNP rs12568784 was not associated with AD persistence (OR, 0.98, 95% CI, 0.82–1.16, unadjusted p-value=0.790). Independent GEE estimates for rs377218292 (NC_000001.10:g.152323643G>A) could not be obtained because of insufficient sample size. One stop-loss variant was identified in our sample, NP_001014364.1:p.Ter2392Ser (rs150529054). This variant was only present in three individuals, so it was not studied further.
Table 2:
Loss of function variants in FLG2 and TCHHL1, and their association with AD remission.
| Pooled cohort (n = 705) | African American (n = 379) | White (n = 379) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | RSID | Location | Odds of Clearing (95% CI) | raw p value | n | Odds of Clearing (95% CI) | raw p value | n | Odds of Clearing (95% CI) | raw p value | n |
| FLG2 | rs12568784* | NC_000001.10: g.152323132G>T | 0.977 (0.822,1.161) | 0.790 | 281 | 0.944 (0.768,1.16) | 0.583 | 161 | 1.092 (0.792,1.507) | 0.592 | 120 |
| FLG2 | rs377218292 | NC_000001.10: g.152323643G>A | *** | *** | 1 | *** | *** | 1 | *** | *** | 0 |
| TCHHL1 | rs150014958 | NP_001008536.1: p.Gln656Ter | *** | *** | 12 | *** | *** | 4 | *** | *** | 8 |
| TCHHL1 | rs61749316** | NP_001008536.1: p.Gln294Ter | *** | *** | 16 | *** | *** | 6 | *** | *** | 10 |
Previously identified in Margolis et al. 2014b.
Previously identified in Margolis et al. 2014c.
Could not be estimated, or insufficient sample size for estimation.
60 unique variants were identified in TCHHL1; there were 41 unique variants in the African American cohort and 26 unique variants in the white cohort. Of these, only three variants had MAF ≥ 5% in the pooled population. Supplementary Table S2 presents the unadjusted association of these TCHHL1 SNPs with AD persistence. None were statistically significantly associated with AD remission after correction for multiple comparisons. Two loss-of-function mutations were identified; they are presented in Table 2. Neither of the LoF variants in TCHHL1 (rs150014958 and rs61749316) were present at a high enough frequency to assess for association with AD persistence independently.
The rare variant algorithm was run independently on rare variants in FLG2 and TCHHL1, and independently run on white and African American individuals. All p-values were corrected for 250,000 independent tests. Within the white population, thirty-four rare variants were identified in FLG2; having any of these was associated with increased AD remission (OR, 7.64e17, 95% CI 4.41e17–1.32e18, adjusted p-value < 1.0e-16). 26 individuals, representing 6.9% of the study population, carried at least one of these variants. These variants are presented in Table 3. Within the self-described African American population, 13 uncommon FLG2 alleles were associated with increased AD remission (OR 21.01; 95%CI 11.90–37.09; adjusted p<1.0e-16). These variants are presented in Table 3. In TCHHL1, in the white population, twelve rare variants were identified, which together trended towards association with increased AD remission (OR, 23.46, 95% CI 7.07–77.89, adjusted p-value 0.064). These variants were identified in 13 individuals, representing 3.4% of the study population. These variants are presented in Table 3. Within the African American population, no TCHHL1 uncommon allele groups were associated with AD remission.
Table 3:
Uncommon variants in uncommon variant groupings.
| FLG2, whites | FLG2, African Americans | TCHHL1, whites | ||||||
|---|---|---|---|---|---|---|---|---|
| Location | Nucleotide change | Amino acid change | Location | Nucleotide change | Amino acid change | Location | Nucleotide change | Amino acid change |
| 152328471 | C>T | p.G597G | 152322250 | G>A | intronic | 152057347 | G>A | |
| 152329677 | G>A | p.D195D | 152322365 | T>G | intronic | 152059461 | G>T | p.Q233K |
| 152331902 | G>C | 152323269 | A>G | p.H2331H | 152057769 | A>G | p.Y797H | |
| 152331264 | G>A | p.L33L | 152324043 | A>G | p.H2073H | 152058358 | A>G | p.P600P |
| 152330789 | G>A | 152325368 | G>A | p.H1632Y | 152058563 | C>T | p.G532E | |
| 152329534 | C>T | p.G243E | 152325410 | C>T | p.G1618R | 152058633 | A>T | p.S509T |
| 152329530 | C>G | p.L244F | 152325444 | A>G | p.H1606H | 152058648 | G>T | p.P504T |
| 152329370 | A>G | p.C298R | 152325742 | C>T | p.S1507N | 152059278 | G>A | p.Q294X |
| 152328924 | T>C | p.V446V | 152326154 | G>A | p.H1370Y | 152059570 | T>A | p.I196I |
| 152328475 | T>C | p.H596R | 152326696 | A>G | p.F1189S | 152059638 | C>T | p.E174K |
| 152328308 | A>G | p.S652P | 152327295 | C>G | p.Q989H | 152061454 | C>T | |
| 152328190 | G>A | p.S691F | 152330196 | C>T | intronic | 152061533 | C>T | |
| 152327955 | G>A | p.S769S | 152330801 | C>T | intronic | |||
| 152326902 | C>T | p.S1120S | ||||||
| 152326715 | C>A | p.V1183L | ||||||
| 152326620 | T>C | p.Q1214Q | ||||||
| 152325965 | G>A | p.H1433Y | ||||||
| 152325911 | C>T | p.G1451R | ||||||
| 152325766 | G>T | p.S1499Y | ||||||
| 152325522 | A>G | p.T1580T | ||||||
| 152325455 | C>T | p.G1603R | ||||||
| 152324325 | A>G | p.A1979A | ||||||
| 152324316 | G>T | p.H1982Q | ||||||
| 152324315 | A>G | p.Y1983H | ||||||
| 152324312 | G>C | p.P1984A | ||||||
| 152324311 | G>C | p.P1984R | ||||||
| 152324146 | T>G | p.H2039P | ||||||
| 152324114 | C>T | p.A2050T | ||||||
| 152324109 | A>G | p.H2051H | ||||||
| 152324106 | G?A | p.G2052G | ||||||
| 152323176 | C>T | p.G2362G | ||||||
| 152322465 | C>T | |||||||
| 152322324 | C>T | |||||||
| 152322292 | G>A | |||||||
DISCUSSION
FLG LoF variation is an important risk factor for AD and AD persistence. Several genes that code for filaggrin-like proteins (S100FTPs) cluster on chromosome 1 in the EDC region. However, there have been very few studies examining FLG2 and TCHHL1, and prior studies focused on African American individuals. This report is the largest sequencing study of FLG2 and TCHHL1 in AD to date, and includes both African American and white individuals. This study is further distinguished by the use of remission, rather than presence of AD, as the primary outcome measure.
Taken together, our analyses of both FLG2 and TCHHL1 imply that LoF mutations in S100FTPs are not necessarily associated with AD persistence in either whites or African Americans. Since LoF mutations are generally thought to significantly decrease protein production, and since these LoF mutations in FLG2 and TCHHL1 are not associated with more persistent AD, it is tempting to speculate that the barrier function defect found in AD is relatively specific—that is, only specific types of skin-barrier alteration, namely those induced by filaggrin loss-of-function mutations, influence AD pathogenesis. Conversely, recent research into filaggrin suggests that filaggrin’s contribution to keratohyalin granules, and the skin’s barrier function, is dependent upon liquid-liquid phase separation, and that only relatively small amounts of protein are necessary for adequate granule formation and function [24,25]. Extending this hypothesis to FLG2 and TCHHL1, it is possible that the identified loss-of-function mutations within these genes do not sufficiently decrease protein quantity to alter their function.
That said, missense variants have been found to be important in other genes associated with AD remission, so it is reasonable to hypothesize that other variation in FLG2 and TCHHL1 may be associated with AD remission [8]. We identified no relatively common variants (MAF ≥ 5%) in FLG2 and TCHHL1 associated with AD remission after correction for multiple comparisons. This finding, coupled with the analysis of LoF variants, suggests that individual, common variants in FLG2 and TCHHL1 are not significantly associated with AD remission.
Unlike common variants, uncommon alleles, particularly in FLG2, were associated with AD remission. We found 34 rare variants in the white population that, when analyzed as a group, are associated with increased AD remission. This represents the first association of FLG2 variation with AD in white individuals, and the first association of FLG2 non-LoF variants with AD remission. These variants represent 6.9% of the study population, a considerable number of individuals for a disease with a prevalence of up to 20% of the general population [5]. Similarly, in African Americans, 13 uncommon FLG2 alleles were associated with increased AD remission. In TCHHL1, when analyzed together, 12 rare variants trended towards association with AD remission. These variants were only present in 3.4% of the study population, and the association did not reach statistical significance after adjustment for multiple testing. Future investigations, not subject to such a stringent correction factor, may identify a significant association here.
The primary limitation of our study was that we only studied African American and white individuals, so our results may not apply to AD in patients of differing ancestries. Further studies in diverse populations will be needed to confirm these findings. The incidence of several LoF variants in both genes was very small (<5%), so we were unable to evaluate independent associations between these genes and AD remission.
In this investigation, we report results from the largest sequencing study of FLG2 and TCHHL1 in whites with AD. In contrast with previous studies, we observed no common FLG2 variants associated with AD severity. We also found no common variants in TCHHL1 associated with AD severity. However, we did find that uncommon allele composites within both FLG2 and TCHHL1 were associated with AD remission, implying that these genes may be relevant in certain subsets of AD individuals. This study provides further insight into the genetics of AD, demonstrating that uncommon alleles within non-FLG skin barrier proteins may be involved in AD pathogenesis.
Supplementary Material
Funding:
This work was supported in part by a grant from the National Institutes for Health NIAMS R01-AR070873 (PI: Margolis). The PEER study is funded as the Atopic Dermatitis Registry by Valeant Pharmaceuticals International (PI: Margolis).
Footnotes
Publisher's Disclaimer: This AM is a PDF file of the manuscript accepted for publication after peer review, when applicable, but does not reflect post-acceptance improvements, or any corrections. Use of this AM is subject to the publisher’s embargo period and AM terms of use. Under no circumstances may this AM be shared or distributed under a Creative Commons or other form of open access license, nor may it be reformatted or enhanced, whether by the Author or third parties. See here for Springer Nature’s terms of use for AM versions of subscription articles: https://www.springernature.com/gp/open-research/policies/accepted-manuscript-terms
Conflicts of interest: David Margolis is a consultant for Pfizer with respect to studies of atopic dermatitis and serves on an advisory board for the National Eczema Association. No other authors state financial conflicts of interest with respect to this investigation
Code availability: R code for this project is available upon request from the corresponding author.
IRB approval: The PEER study was approved by the Institutional Review Board of the University of Pennsylvania, and written informed consent was obtained from all participants from the participant or his or her caregiver.
Data availability:
The PEER data (source) is not currently publically available. The PEER study is an ongoing study sponsored by Valeant in response to a post marketing commitment with the FDA. A BED file of the probes used for targeted capture is available upon request from the corresponding author.
REFERENCES
- 1.Bieber T Atopic dermatitis. N Engl J Med. 2008; 358(14):1483–1494. [DOI] [PubMed] [Google Scholar]
- 2.Abramovits W Atopic dermatitis. J Am Acad Dermatol. 2005;53(1)(suppl 1):S86–S93. [DOI] [PubMed] [Google Scholar]
- 3.Akdis CA, Akdis M, Bieber T, Bindslev-Jensen C, Boguniewicz M, Eigenmann P, et al. Diagnosis and treatment of atopic dermatitis in children and adults: European Academcy of Allerology and Clinical Immunology/American Academy of Allergy, Asthma and Immunology.PRACTALL Consensus Report. Allergy. 2006; 61:969–987. [DOI] [PubMed] [Google Scholar]
- 4.Leung DY, Bieber T. Atopic dermatitis. Lancet. 2003; 361(9352):151–160. [DOI] [PubMed] [Google Scholar]
- 5.Pellerin L, Henry J, Hsu C-Y, Balica S, Jean-Decoster C, Mechin M-C, et al. Defects of filaggrin-like proteins in both lesional and nonlesional atopic skin. J Allergy Clin Immunol. 2013;131(4):1094–1102. [DOI] [PubMed] [Google Scholar]
- 6.Berna R, Mitra N, Hoffstad O, Wan J, Margolis DJ. Identifying phenotypes of atopic dermatitis in a longitudinal US cohort using unbiased statistical clustering. J Invest Dermatol. Article in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margolis JS, Abuabara K, Bilker W, Hoffstad O, Margolis DJ. Persistence of mild to moderate atopic dermatitis. JAMA Dermatol. 2014a; 150(6):593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berna R, Mitra N, Lou C, Hoffstad O, Wubbenhorst B, D’Andrea K, et al. Thymic Stromal Lymphopoietin and IL7R Variants are Associated with Persistent Atopic Dermatitis. J Invest Dermatol. Article in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z, Latendorf T, Meyer-Hoffert U, and Schroder J-M. Identification of trichohyalin-like 1, an S100 fused-type protein selectively expressed in hair follicles. J Invest Dermatol. 2011; 131(8):1761–1763. [DOI] [PubMed] [Google Scholar]
- 10.Margolis DJ, Gupta J, Apter AJ, Ganguly T, Hoffstad O, Papadopoulos M, et al. Filaggrin-2 variation is associated with more persistent atopic dermatitis in African American subjects. J Invest Dermatol. 2014b; 133(3):784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pendaries V, Le Lamer M, Cau L, Hansmann B, Malaisse J, Kezic S, et al. In a three-dimensional reconstructed human epidermis filaggrin-2 is essential for proper cornification. Cell Death Dis. 2015; 6(2):e1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makino T, Mizawa M, Yamakoshi T, Takaishi M, and Shimizu T. Expression of filaggrin-2 protein in the epidermis of human skin diseases: a comparative analysis with filaggrin. Biochem Biophys Res Comm. 2014; 449(1):100–106. [DOI] [PubMed] [Google Scholar]
- 13.Mohamad J, Sarig O, Godsel LM, Peled A, Malchin N, Bochner R, et al. Filaggrin 2 deficiency results in abnormal cell-cell adhesion in the cornified cell layers and causes peeling skin syndrome type A. J Invest Dermatol. 2018; 138(8):1736–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolling MC, Jan SZ, Pasmooij AMG, Lemmink HH, Franke LH, Yenamandra VK, et al. Generalized ichthyotic peeling skin syndroem due to FLG2 mutations. J Invest Dermatol. 2018; 138(8):1881–1884. [DOI] [PubMed] [Google Scholar]
- 15.Lou C, Mitra N, Wubbenhorst B, D’Andrea K, Hoffstad O, Kim BS, et al. Association between fine mapping thymic stromal lymphopoietin and atopic dermatitis onset and persistence. Ann Allergy Asthma Immunol. 2019;123(6):595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler Transform. Bioinformatics. 2010; 26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margolis DJ, Gupta J, Apter AJ, Hoffstad O, Papadopoulos M, Rebbeck TR, et al. Exome sequencing of filaggrin and related genes in African-American children with atopic dermatitis. J Invest Dermatol. 2014c; 134:2272–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamakoshi T, Makino T, Rehman MU, Yoshihisa Y, Sugimori M, and Shimizu T. Trichohyalin-like 1 protein, a member of fused S100 proteins, is expressed in nromal and pathologic human skin. Biochem Biophys Res Commun. 2013; 432:66–72. [DOI] [PubMed] [Google Scholar]
- 19.Wu Z, Latendorf T, Meyer-Hoffert U, and Schroder J-M. Identification of trichohyalin-like 1, an S100 fused-type protein selectively expressed in hair follicles. J Invest Dermatol. 2011; 131:1761–1763. [DOI] [PubMed] [Google Scholar]
- 20.Margolis DJ, Gupta J, Apter AJ, Hoffstad O, Papadopoulos M, Rebbeck TR, et al. Exome sequencing of filaggrin and related genes in African-American children with atopic dermatitis. J Invest Dermatol. 2014c; 134:2272–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pigors M, Common JEA, Wong XFC, Malik S, Scott CA, Tabarra N, et al. Exome sequencing and rare variant analysis reveals multiple filaggrin mutations in Bangladeshi families with atopic eczema and additional risk genes. J Invest Dermatol. 2018; 138:2674–2677. [DOI] [PubMed] [Google Scholar]
- 22.Berna R, Mitra N, Hoffstad O, Wubbenhorst B, Nathanson K, Margolis DJ. Using a machine learning approach to identify low-frequency and rare filaggrin alleles associated with remission of atopic dermatitis. JID Innov. Article in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mischke D, Korge BP, Marenholz I, Volz A, Ziegler A. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (“Epidermal Differentiation Complex”) on human chromosome 1q21. J Invest Dermatol. 1996; 106(5):989–992. [DOI] [PubMed] [Google Scholar]
- 24.Quiroz FG, Fiore VF, Levorse J, Polak L, Wong E, Pasolli HA, et al. Liquid-liquid phase separation drives skin barrier formation. Science. 2020;367(6483):eaax9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chua Avecilla AR, Quiroz FG, Cracking the skin barrier: liquid-liquid phase separation shines under the skin, JID Innovations (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bomba L, Walter K, Soranzo N. The impact of rare and low-frequency genetic variants in common disease. Genome Biol. 2017. Apr 27;18(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PEER data (source) is not currently publically available. The PEER study is an ongoing study sponsored by Valeant in response to a post marketing commitment with the FDA. A BED file of the probes used for targeted capture is available upon request from the corresponding author.


