Abstract
A recJ homolog was cloned from the extremely thermophilic bacterium Thermus themophilus HB8. It encodes a 527 amino acid protein that has 33% identity to Escherichia coli RecJ protein and includes the characteristic motifs conserved among RecJ homologs. Although T.thermophilus RecJ protein (ttRecJ) was expressed as an inclusion body, it was purified in soluble form through denaturation with urea and subsequent refolding steps. Limited proteolysis showed that ttRecJ has a protease-resistant core domain, which includes all the conserved motifs. We constructed a truncated ttRecJ gene that corresponds to the core domain (cd-ttRecJ). cd-ttRecJ was overexpressed in soluble form and purified. ttRecJ and cd-ttRecJ were stable up to 60°C. Size exclusion chromatography indicated that ttRecJ exists in several oligomeric states, whereas cd-ttRecJ is monomeric in solution. Both proteins have 5′→3′ exonuclease activity, which was enhanced by increasing the temperature to 50°C. Mg2+, Mn2+ or Co2+ ions were required to activate both proteins, whereas Ca2+ and Zn2+ had no effects.
INTRODUCTION
All living cells have DNA repair systems to recover from damage to their DNA (1). The DNA repair systems of bacteria are photoreactivation, recombinational repair, base excision repair (BER), methyl-directed mismatch repair (MMR) and nucleotide excision repair. Of the various proteins involved in such repair systems, RecJ protein is related to three DNA repair systems; recombinational repair, BER and MMR (2–5). RecJ is a 5′→3′ exonuclease specific for single-stranded DNA (ssDNA) and requires Mg2+ ions for its exonuclease activity (6). In recombinational repair, RecJ degrades the DNA unwound by RecQ helicase at the double-strand break (2). RecJ eventually forms a 3′-projection of ssDNA, which enables homologous recombination by RecA (2). In addition, recent research has revealed that RecJ and RecQ play key roles in the recombination–replication relationship (7). When a replication fork encounters DNA damage, RecJ and RecQ interrupt replication by degradation of the lagging strand. After repair of DNA damage, replication is resumed with RecF and other proteins. RecJ cannot be substituted by other exonucleases for this interruption of replication. In BER, RecJ is considered to remove a 5′-terminal 2′-deoxyribose-5-phosphate residue, whereas in MMR, it removes the ssDNA region with the mismatch lesion (3–5).
RecJ protein and its homologs have been found to contain five conserved motifs (8). Proteins having these motifs exist from prokaryotes to higher eukaryotes (8). These members include not only putative RecJ homologs, but also functionally different proteins or functionally unknown proteins; a putative poly(A) polymerase of Synechocystis sp., the PRUNE gene product of Drosophila melanogaster and an exopolyphosphatase (PPX1) of Saccharomyces cerevisiae. All the conserved motifs of RecJ protein are essential for its exonuclease activity (9). Because these motifs include aspartate and histidine residues, some of them are believed to contribute to the binding of metal ions essential for exonuclease activity (9). The molecular mechanism of RecJ exonuclease activity and the structural and/or functional roles of these motifs, however, are still unclear. Enzymatic, physicochemical and structural analyses of RecJ protein should provide clues to the structure–function relationship of other homologs with different or unknown functions. Such analyses have been hampered by the limited amounts of RecJ protein available; the only RecJ protein purified so far is that from Escherichia coli and the amount of overexpressed E.coli RecJ protein (ecRecJ) was small (6,9–11). Partially purified preparations or crude ce1l extracts, therefore, have been used in previous studies of ecRecJ (3,9).
Thermus thermophilus HB8 is a Gram-negative eubacterium that grows at temperatures >75°C (12). It is the most thermophilic bacterium for which a gene manipulation system has been established (12–14). Proteins from this bacterium are stable against heat, extreme pH and denaturants. These properties are suitable for physicochemical studies, including X-ray crystallography (15). For use in extensive structural and functional studies, we prepared a large amount of RecJ protein from T.thermophilus HB8.
We describe here the overexpression and purification of T.thermophilus RecJ protein (ttRecJ). ttRecJ contains a stable core domain. Truncated ttRecJ, which corresponds to the core domain (cd-ttRecJ), was also expressed and purified. The biochemical properties of ttRecJ and truncated ttRecJ are also described.
MATERIALS AND METHODS
Materials
DNA modification enzymes, including restriction enzymes, were purchased from Takara Shuzo and New England Biolabs. Isopropyl-β-d-thiogalactopylanoside (IPTG) was from Wako Pure Chemicals, [γ-32P]ATP from ICN, MonoQ HR5/5 and HiPrep 16/60 Sephacryl S-200 HR columns were from Amersham Pharmacia Biotech and the DNA oligomers used were from BEX Co. All the other reagents used were of the highest grade commercially available.
Plasmid constructions
Preliminary sequence data for the T.thermophilus HB8 recJ gene was provided by the Thermus thermophilus HB8 genome project (16,17). The recJ gene was amplified by PCR using LA-Taq polymerase. The forward and reverse primers were 5′-ATCATATGAGAGACCGGGTCCGCTGGCGGGT-3′ and 5′-ATAGATCTTTACAGGTCCACCGCCTGGACCTC-3′ (the underlining indicates the NdeI and BglII sites, respectively). Buffer conditions were as recommended by the manufucturer except for the addition of 5% DMSO. The reaction mixture was incubated at 95°C for 5 min. The following PCR cycle was repeated 25 times: denaturation at 95°C for 30 s, annealing of primers at 55°C for 30 s and synthesis at 72°C for 2 min. This was followed by a 7 min incubation at 72°C. The PCR-amplified gene was ligated into the pCR2.1 TOPO vector using TOPO TA cloning kits (Invitrogen), creating the plasmid pCR-ttRecJ. Sequence analysis verified that the construct was error free. The recJ gene was digested with the restriction enzymes NdeI and BglII and ligated into the compatible sites of the expression vector pET-19b (Novagen), creating the plasmid pET-19b-ttRecJ.
The fragment corresponding to cd-ttRecJ was amplified by PCR. The forward and reverse primers were 5′-GGAATTCCATATGTTTAGGCGCAAGGAGGACCTG-3′ and 5′-AAGATCTTTAGAGGAAGAGGGGCTCGGGGT-3′ (the underlining indicates the NdeI and BglII sites, respectively). The design of these primers was based on the results of limited proteolysis and multiple sequence alignment. PCR was performed under the conditions described above. The PCR product was cloned into the pCR2.1 vector, creating pCR2.1-cd-ttRecJ, and the fragment for cd-ttRecJ was further subcloned into the expression vector pET-19b, resulting in pET-19b-cd-ttRecJ. Cloning of the DNA fragment of cd-ttRecJ was as described above.
Expression and purification of ttRecJ and cd-ttRecJ
Escherichia coli BL21(DE3) (Novagen) was transformed with pET-19b-ttRecJ or pET-19b-cd-ttRecJ and the transformant was grown at 30°C in 5 l of 2× YT medium containing 100 µg/ml ampicillin (18). When the cultures reached an optical density at 600 nm of 0.4, IPTG was added to 0.4 mM. Cells were grown at 30°C for 3 h after induction, harvested by centrifugation and stored at –20°C. ttRecJ was expressed in insoluble form, cd-ttRecJ in soluble form.
ttRecJ was purified as follows. All steps were performed at 4°C or on ice. Frozen cells (10 g) were thawed and suspended in 80 ml buffer A (20 mM Tris–HCl, 500 mM NaCl, 10% glycerol, 5 mM imidazole, 0.5 mM phenylmethanesulfonyl fluoride, pH 7.9), then disrupted by sonication at maximum power with the standard probe. The cell lysate was centrifuged at 20 000 g for 15 min and the pellet collected and washed with 80 ml buffer A. ttRecJ was extracted from the pellet in 80 ml buffer B (6 M urea, 20 mM Tris–HCl, 500 mM NaCl, 10% glycerol, 5 mM imidazole, pH 7.9) and the extract incubated at room temperature for 60 min. After centrifugation at 39 000 g for 60 min, the supernatant was loaded onto a 5 ml Ni2+–nitriloacetate–agarose column (Qiagen) equilibrated with buffer B. The column was then washed with 60 ml of buffer B and RecJ was eluted with 20 ml of buffer B containing 100 mM imidazole. The denatured ttRecJ was refolded by gradual removal of urea by multi-step dialysis. It was successively dialyzed against the following buffers: buffer B containing 4, 2 and 1 M urea and buffer C (20 mM Tris–HCl, 0.1 mM EDTA, 100 mM NaCl, 10% glycerol, pH 7.5). After centrifugation at 39 000 g for 60 min, the supernatant, which contained the refolded ttRecJ, was collected. This fraction was loaded onto a MonoQ HR5/5 column equilibrated with buffer C. ttRecJ was eluted with a linear gradient of NaCl ranging from 100 to 500 mM in a total volume of 20 ml of buffer C. The fraction was dialyzed against buffer C and concentrated to ∼10 mg/ml by CentriPrep ultrafiltration (Millipore).
cd-ttRecJ was purified as follows. Frozen cells (10 g) were thawed, suspended in 80 ml buffer A and disrupted by sonication at maximum power with the standard probe. The cell lysate was centrifuged (48 000 g) for 60 min. The supernatant was collected and loaded onto a 10 ml Ni2+–nitriloacetate–agarose column (Qiagen) equilibrated with buffer A. The column was washed with 50 ml buffer A and 50 ml buffer A containing 20 mM imidazole. cd-ttRecJ was eluted with a linear gradient of imidazole from 20 to 200 mM in a total volume of 200 ml of buffer A. The fraction containing cd-ttRecJ was dialyzed against buffer D (10 mM potassium phosphate, 0.1 mM EDTA, 10% glycerol, pH 6.8) and loaded onto a Bio-Gel HTP gel column (Bio-Rad) equilibrated with buffer D. The column was washed with 15 ml of buffer D and cd-ttRecJ was eluted with a linear gradient of potassium phosphate from 10 to 300 mM in a total volume of 200 ml of buffer D. The peak fractions, eluted with ∼50 mM potassium phosphate, were directly loaded onto a MonoQ HR5/5 column equilibrated with buffer C. cd-ttRecJ was eluted with a linear gradient of NaCl from 100 to 500 mM in a total volume of 20 ml of buffer C. The peak fraction was loaded onto a HiPrep 16/60 Sephacryl S-200 HR column equilibrated with buffer C. cd-ttRecJ was eluted as a single peak and concentrated to ∼10 mg/ml by CentriPrep ultrafiltration.
The molar extinction coefficient of ttRecJ at 278 nm was calculated as 65 200 M–1 cm–1 and that of cd-ttRecJ as 33 400 M–1 cm–1 by a procedure reported previously (19).
Limited proteolysis
ttRecJ (10 µM) was treated at 25°C with 0.5 µg/ml thermolysin in 20 mM Tris–HCl and 2 mM CaCl2, pH 8.0. Proteolysis was stopped by the addition of SDS gel loading buffer (18). The sample was subjected to 12% SDS–PAGE and the products stained with Coomassie Brilliant Blue R-250.
The proteolytic fragments were electroblotted onto a polyvinylidene difluoride membrane (Immobilon-PSQ transfer membrane; Millipore) and the bands stained with Coomassie Brilliant Blue R-250 were excised (20). The N-terminal amino acid sequence was determined with a gas phase protein sequencer (model 473A; Applied Biosystems).
Circular dichroism (CD) measurements
CD spectra were measured with a Jasco spectropolarimeter, model J720W. Spectra of 1.6 µM ttRecJ and of 1.5 µM cd-ttRecJ were measured in a 1 mm cell in the far-UV region between 200 and 250 nm. The reaction mixtures contained 50 mM potassium phosphate, 100 mM KCl, 0.1 mM dithioerythritol (DTE) and 0.1 mM EDTA, pH 7.2.
Size exclusion chromatography
Size exclusion chromatography was carried out using Superdex 200 HR 10/60 column. The sample was dissolved in 20 mM Tris–HCl, 100 mM NaCl, 0.1 mM EDTA, 10 mM 2-mercaptoethanol and 10% glycerol, pH 7.5, then loaded onto the column equilibrated with the same buffer. The molecular sizes of ttRecJ and cd-ttRecJ were estimated using a Molecular Weight Marker Kit (Sigma)
Assay for exonuclease activity
A 49mer synthetic oligonuleotide (5′-ACTACTTGGTACACTGACGCGAGCACGCAGGAGCTCATTCCAGTGCGCA-3′) was used as the substrate for ttRecJ and cd-ttRecJ. This ssDNA was radiolabeled at the 5′-end with [γ-32P]ATP using polynucleotide kinase. The 10 nM and 1 nM 5′-32P-labeled ssDNAs were denatured at 95°C for 2 min, then mixed with various concentrations of ttRecJ and cd-ttRecJ, respectively. The reaction mixture (20 µl), which contained 20 mM Tris–HCl, 10 mM MgCl2, 100 mM KCl and 1 mM dithiothreitol (DTT), pH 7.5, was incubated at various temperatures. For each time point, 4.5 µl aliquots were taken from the reaction mixture and quenched by immediate addition to 1 µl of 10× loading buffer (1% SDS, 50% glycerol, 0.05% bromophenol blue) (TaKaRa). The samples were loaded onto a 15% (w/v) acrylamide gel containing 7 M urea and 1× TBE buffer, then electrophoresed in 1× TBE buffer (18). The gel was dried and placed in contact with an imaging plate. The bands were visualized and analyzed using a BAS2000 image analyzer (Fuji Photo Film). To investigate the effect of metal ions on exonuclease activity, 20 nM 5′-32P-labeled ssDNA was incubated at 25°C for 30 min with ttRecJ or cd-ttRecJ. The reaction mixture contained 20 mM Tris–HCl, 100 mM KCl, pH 7.5, and 10 mM MgCl2, MnCl2, CoCl2, CaCl2 or ZnCl2. Activity was analyzed as described above.
The percentage of undegraded DNA was plotted for each incubation period. As the plot showed a linear region, the initial rate of the reaction could be obtained. In our assay, the substrate was mixed with a large excess of the enzyme. Assuming that enzyme and substrate react according to the scheme described below, the second step (kapp) is considered to be the rate limiting step when the enzyme-bound intermediate is in the steady state. The initial rate was plotted against the concentration of protein. The kinetic parameters were calculated by fitting the data to equation 4 using the software Igor Pro 3.14 (Wave Metrics).
k1 kcat
E + S ↔ ES → E + P
k–1
[E]0 = [E] + [ES] 1
For the initial reaction time, the following equation should be assumed.
[S]0 = [S] + [ES] 2
where [S] is the free concentration of the substrate and [E] the free concentration of the enzyme.
KS = (kcat + k–1)/k1 3
As the initial rate (v) is defined as kcat[ES], v is expressed as:
v = kcat[ES] = kcat[S]0[E]/(KS + [E]) 4
Equations 1–3 can be combined with equation 4.
[ES] = [S]0[E]/(KS + [E])
In our assays, the enzyme concentration was in excess of that of the substrate, [E] being nearly equal to [E]0.
RESULTS
Cloning of the T.thermophilus recJ gene
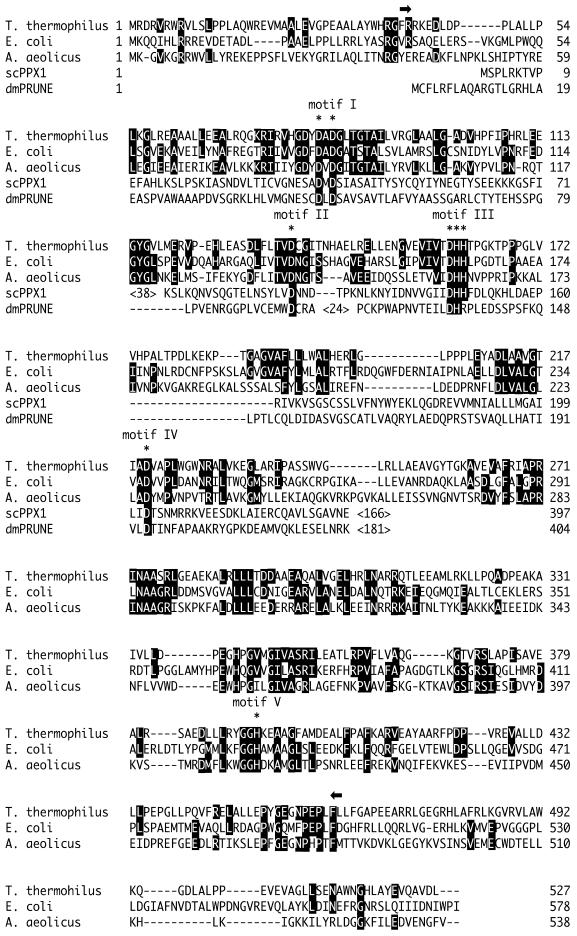
Using information obtained from the genome project of T.thermophilus HB8, we identified an open reading frame homologous to the recJ gene and cloned this region as the T.thermophilus recJ gene (DDBJ/EMBL/GenBank accession no. AB064549). ttRecJ encoded 527 amino acids with a molecular mass of 57.4 kDa. This amino acid sequence is similar to the sequences of other RecJ homologs (Fig. 1). It has 33% identity and 48% similarity to ecRecJ (10). As shown in Figure 1, ttRecJ protein contains five characteristic motifs conserved among RecJ homologs (8).
Figure 1.
Amino acid sequence of the T.thermophilus, E.coli and Aquifex aeolicus RecJ proteins. PPX1 of S.cerevisiae and the PRUNE gene product of D. melanogaster are also aligned according to Aravind and Koonin (8). Identical amino acid residues are indicated by white letters on black. The conserved aspartate and hisitidine residues of five motifs are indicated at the top by asterisks. The cd-ttRecJ region (40–463) is indicated at the top by two arrows. ScPPX1, PPX1 of S.cerevisiae; dmPRUNE, PRUNE gene product of D.melanogaster. Motifs I–V indicate the five conserved motifs described in Aravind and Koonin (8).
Overproduction and purification of ttRecJ
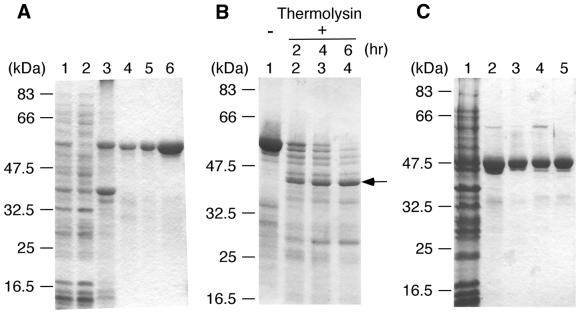
ttRecJ was expressed in E.coli as an N-terminal (His)10-tagged protein under the control of an IPTG-inducible T7 promoter. The induced band was observed at ∼57 kDa (Fig. 2A), which corresponds to the molecular mass calculated from the amino acid sequence of ttRecJ. As ttRecJ was expressed in an insoluble form, it was made soluble by denaturation with urea and effectively isolated by affinity chromatography. After gradual removal of the urea by dialysis, the refolded ttRecJ protein was purified to homogeneity, as judged by SDS–PAGE. Approximately 3 mg ttRecJ was purified from 10 g cells.
Figure 2.
Purification of ttRecJ and cd-ttRecJ and limited proteolysis of ttRecJ. (A) Purification of ttRecJ. Lane 1, total cell extract; lane 2, supernatant of the total cell extract; lane 3, extract from the pellet; lane 4, Ni2+–nitriloacetate–agarose; lane 5, refolding fraction; lane 6, MonoQ HR5/5. (B) Limited proteolysis of ttRecJ with thermolysin. An aliquot of 10 µM ttRecJ was reacted with 0.5 µg/ml thermolysin in 20 mM Tris–HCl buffer, pH 8.0, containing 2 mM CaCl2. This reaction mixture was incubated at 25°C for the indicated incubation periods. Lane 1, ttRecJ; lanes 2–4, digests incubated for the periods indicated above each lane. The arrow indicates the core domain of ttRecJ. (C) Purification of cd-ttRecJ. Lane 1, supernatant of the total cell extract; lane 2, Ni2+–nitriloacetate–agarose; lane 3, Bio-Gel HTP gel; lane 4, MonoQ HR5/5; lane 5, HiPrep 16/60 Sephacryl S-200 HR. Each step was analyzed by staining the SDS–PAGE gel (12% w/v acrylamide) with Coomassie brilliant blue. The molecular mass markers were: MBP–paramyosin, 83 kDa; glutamic acid dehydrogenase, 62 kDa; aldolase, 47.5 kDa; triosephosphate isomerase, 32.5 kDa; β-lactoglobulin A, 25 kDa; lysozyme, 16.5 kDa.
Domain structure of ttRecJ
The five conserved motifs are distributed throughout the entire sequence of RecJ proteins (Fig. 1). Motifs I–IV are in the N-terminal to middle regions, at relatively short intervals of 30–50 amino acid residues. Motif V is separated from the other motifs, the distance between motifs IV and V being ∼170 amino acid residues. Of the RecJ homologs, the PRUNE gene product of D.melanogaster and S.cerevisiae PPX1 do not contain motif V (8). This suggests that RecJ has at least two domains; one includes motifs I–IV while another includes motif V.
Limited proteolysis of the refolded ttRecJ was carried out to investigate the domain architecture of RecJ protein (Fig. 2B). Treatment of ttRecJ (57 kDa) with thermolysin produced a 43 kDa band even at a low protease concentration. The 43 kDa band was relatively resistant to further digestion, indicative of the presence of a stable core domain in ttRecJ protein. The N-terminal amino acid sequence of the 43 kDa band was determined to be Phe-Arg-Arg-Lys-Glu-Asp-Leu-Asp-Pro, identical to the sequence starting from Phe40. As the molecular mass of the fragment from Phe40 to the C-terminus is calculated to be 53 kDa, the 43 kDa band is thought to be produced by another proteolysis on the C-terminal side in addition to cleavage at Phe40. Based on the mass of the fragment and the protease specificity, the C-terminus of the 43 kDa protein is presumed to be at or near Leu439. This region (40–439) contains all five motifs and signifies the presence of the core domain in RecJ protein.
Multiple sequence alignment of RecJ proteins showed high homology for the amino acid sequence from the N-terminus to Leu463 of ttRecJ, but poor homology in the extended C-terminal region from Leu463. The calculated molecular mass of the region from 40 to 463 is 45.7 kDa. Although slightly larger than 43 kDa, this region should include the stable core domain.
As the core domain contains all the characteristic motifs, it should retain the exonuclease activity of the full-length protein. To confirm this, we produced an expression plasmid for the 40–463 fragment of ttRecJ (cd-ttRecJ). cd-ttRecJ was expressed in soluble form with an N-terminal (His)10 tag, then highly purified by several runs of column chromatography (Fig. 2C). cd-ttRecJ (4 mg) was purified from 10 g cells.
Secondary structure and thermostability
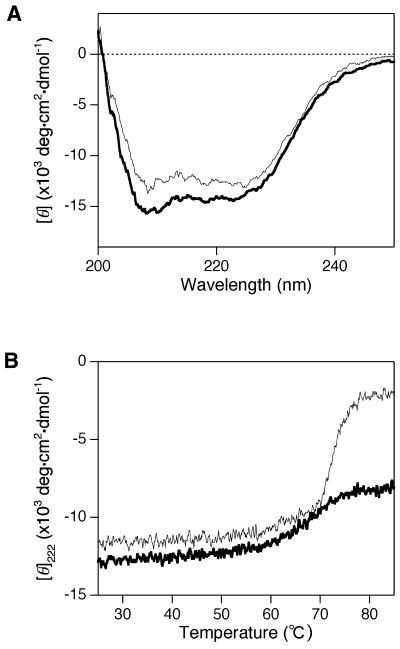
The far-UV CD spectra of purified ttRecJ and cd-ttRecJ were examined to obtain information on the conformations of their polypeptide backbones. The spectra of each showed negative double maxima at ∼210 and 220 nm (Fig. 3A), characteristic of an α-helical structure. Compared to ttRecJ, cd-ttRecJ showed less intensity, probably due to the truncation at each terminus. These results indicate that both recombinant proteins were folded.
Figure 3.
CD spectra and thermostabilities of ttRecJ and cd-ttRecJ. (A) Far-UV CD spectra. Measurements were performed at 25°C in buffer solution at pH 7.2 containing 1.6 µM ttRecJ or 1.5 µM cd-ttRecJ, 50 mM potassium phosphate, 100 mM KCl, 0.1 mM DTE and 0.1 mM EDTA. (B) Temperature dependence of the molar ellipticity at 222 nm ([θ]222). Measurements were perfomed in the same solutions used for the far-UV CD spectra measurements. The heating rate was 1°C/min. Thick line, ttRecJ; thin line, cd-ttRecJ.
The thermostabilities of ttRecJ and cd-ttRecJ were examined based on the residue molar ellipticities at 222 nm ([θ]222). As shown in Figure 3B, ttRecJ was stable up to ∼60°C, evidence of proper folding of the protein. cd-ttRecJ also showed thermostability up to 60°C, although it unfolded drastically above 70°C.
Oligomeric states of ttRecJ and cd-ttRecJ
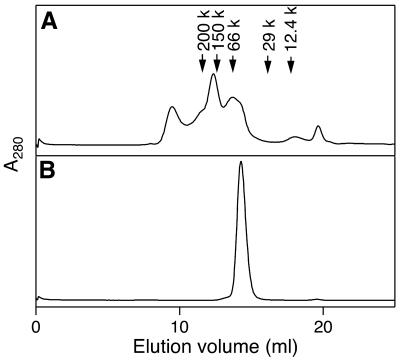
The oligomeric states of ttRecJ and cd-ttRecJ in solution were investigated by size exclusion chromatography. The elution profile of ttRecJ protein had three main peaks (Fig. 4A). The first peak corresponded to the void volume. The apparent molecular masses of the second and third peaks were estimated to be ∼155 and ∼58 kDa, respectively. This indicates that the solution is a mixture of the monomeric, trimeric and highly aggregated states of ttRecJ. In contrast, cd-ttRecJ eluted as a single peak at ∼50 kDa (Fig. 4B), indicating that cd-ttRecJ exists as a monomer. These results also suggest that regions external to the core domain are involved in aggregation.
Figure 4.
Size exclusion chromatography of ttRecJ (A) and cd-ttRecJ (B). ttRecJ and cd-ttRecJ were loaded onto a Superdex 200 HR 10/30 column equilibrated with 20 mM Tris–HCl, 100 mM NaCl, 0.1 mM EDTA, 10 mM 2-mercaptoethanol and 10% glycerol, pH 7.5. ttRecJ and cd-ttRecJ were analyzed by absorbance at 280 nm with a flow rate of 0.5 ml/min. The molecular size markers (Sigma) are: potato β-amylase, 200 kDa; yeast alcohol dehydrogenase, 150 kDa; bovine serum albumin, 66 kDa; bovine carbonic anhydrase, 29 kDa; horse heart cytochrome c, 12.4 kDa.
Exonuclease activity
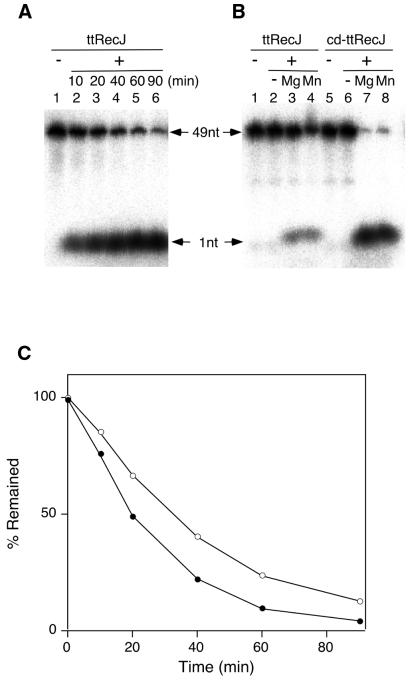
Exonuclase activity was analyzed with 5′-end-labeled ssDNA as the substrate. If degradation of ssDNA by ttRecJ proceeds in the 5′→3′ direction, the reaction mixture should contain the products of the 5′-labeled mononucleotide excised from the 5′-end and the unlabeled excised nucleotide, as well as the undegraded 5′-labeled substrate ssDNA. When these products are separated by gel electrophoresis, two radioactive bands should appear; one corresponding to the mononucleotides excised from the 5′-end and the other to the undegraded substrate ssDNA. As expected, the reaction mixture of the labeled substrate DNA and ttRecJ gave two radioactive bands on the gel (Fig. 5A). As a control, the pellet from cells transformed with pET-19b without a recJ insert was treated in the same manner with ttRecJ and had no exonuclease activity (data not shown). cd-ttRecJ also showed exonuclease activity in the 5′→3′ direction (Fig. 5B), indicating that the core domain preserves the exonuclease activity. In the absence of magnesium ions, neither protein exhibited activity (Fig. 5B). As shown in Figure 5B, Mn2+ could substitute for Mg2+, whereas the Mn2+ ions inhibited the activity of ecRecJ (6). The effects of the metal ions Ca2+, Co2+ and Zn2+ on exonuclease activity were also investigated. Co2+ enhanced the exonuclease activity of both ttRecJ and cd-ttRecJ, but Ca2+ and Zn2+ had no effect (data not shown). The metal ion requirements of ttRecJ and cd-ttRecJ are the same as those of scPPX1, one of the RecJ homologs that requires Mg2+, Mn2+ or Co2+ but not Ca2+, Zn2+ or Cu2+ (21). Figure 5C shows the time course of ssDNA degradation. As the plot is linear for the first 20 min, the initial rate of exonulease activity was calculated from the slope in this region.
Figure 5.
Exonuclease activity of ttRecJ and cd-ttRecJ. (A) An aliquot of 50 nM 5′-32P-labeled ssDNA was reacted with 0.1 µM ttRecJ at 37°C. The reaction mixture contained 20 mM Tris–HCl, 10 mM MgCl2, 100 mM KCl and 1 mM DTT, pH 7.5. The degradation of ssDNA was analyzed by denaturing PAGE (15% polyacrylamide). Lane 1, ssDNA; lanes 2–6, ssDNA reacted with ttRecJ for the incubation period indicated above each lane. The 49nt arrow indicates the substrate 49mer ssDNA and the 1 nt arrow the released 5′-end nucleotide. (B) An aliquot of 20 nM 5′-32P-labeled ssDNA was reacted with 0.1 µM ttRecJ and cd-ttRecJ at 25°C for 30 min. The reaction mixture contained 20 mM Tris–HCl and 100 mM KCl, pH 7.5, with the addition of 10 mM MgC12 or MnCl2. Lanes 1 and 5, ssDNA; lane 2, with ttRecJ; lane 3, with ttRecJ in the presence of MgC12; lane 4, with ttRecJ in the presence of MnCl2; lane 6, with cd-ttRecJ; lane 7, with cd-ttRecJ in the presence of MgC12; lane 8, with cd-ttRecJ in the presence of MnCl2. (C) Time course of ttRecJ exonuclease activity. The reaction mixture was as in (A). The percentage of undegraded DNA was plotted against incubation time. Open circle, 37°C; closed circle, 50°C.
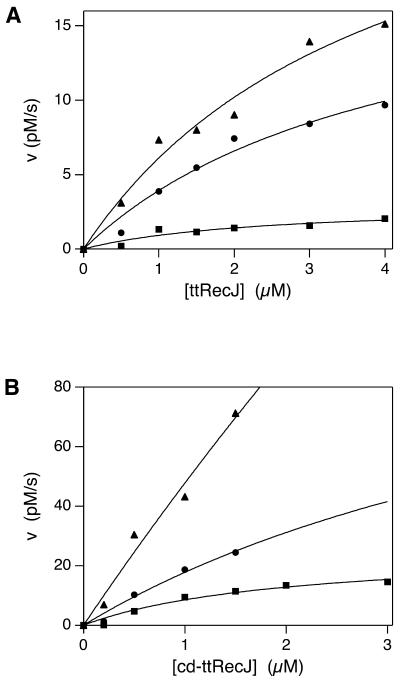
Figure 6A and B shows the apparent rates of exonuclease activity of ttRecJ and cd-ttRecJ at the indicated concentrations of protein. They also show the dependence of the exonuclease activities of both proteins on temperature. Kinetics parameters are given in Table 1. For high concentrations of protein, the time course of reaction with cd-ttRecJ was non-linear at 37 and 50°C. Thus, the kinetic parameters under these conditions were underestimated. The exonuclease activities of both proteins were enhanced at high temperature. The activity of ttRecJ at 50°C was 10 times higher than that at 37°C. cd-ttRecJ showed a 40-fold enhancement between 25 and 50°C. These findings are consistent with the thermophilic properties of T.thermophilus. cd-ttRecJ exhibited higher activity than ttRecJ at the temperatures examined; at 25°C its activity was 100 times higher than that of ttRecJ.
Figure 6.
Kinetic analysis of the exonuclease activities of ttRecJ (A) and cd-ttRecJ (B). An aliquot of 10 nM 5′-32P-labeled ssDNA was reacted with ttRecJ and 1 nM 5′-32P-labeled ssDNA with cd-ttRecJ. The reaction mixture containing 20 mM Tris–HCl, 10 mM MgCl2, 100 mM NaCl and 1 mM DTT, pH 7.5, was incubated at 25, 37 and 50°C. The rates of ssDNA degradation by both proteins were plotted against each concentration of protein. Closed square, 25°C; closed circle, 37°C; closed triangle, 50°C. Solid lines show the theoretical curves (see Materials and Methods).
Table 1. Kinetic parameters of the exonuclease activities of ttRecJ and cd-ttRecJ.
| Temperature (°C) | Kcat (×10–3 s–1) | KS (µM) | |
|---|---|---|---|
| ttRecJ | 25 | 0.3 ± 0.1 | 2.4 ± 1.5 |
| 37 | 2.0 ± 0.5 | 4.1 ± 1.6 | |
| 50 | 3.0 ± 0.6 | 4.0 ± 1.4 | |
| cd-ttRecJ | 25 | 25 ± 5.5 | 2.0 ± 0.8 |
| 37 | >130 | (6.1) | |
| 50 | >900 | (17.9) |
DISCUSSION
We cloned the recJ gene from T.thermophilus and purified its gene product (ttRecJ). Although the RecJ protein had previously been purified from E.coli, its yield was very low (6,10). Partially purified RecJ protein or crude cell extract has therefore been used in most previous studies (3,9). The present expression/purification system provides purified ttRecJ in sufficient quantity to allow more extensive characterization and further structural studies.
Expression of a recombinant protein under the control of a strong promoter sometimes leads to inclusion body formation, a very troublesome problem (22). ttRecJ overexpressed in an insoluble fraction was successfully recovered to active ttRecJ by denaturation with urea and subsequent refolding steps. Its efficient refolding may be due to the high structural stability of ttRecJ. Moreover, we could overproduce cd-ttRecJ as a soluble protein. Size exclusion chromatography results indicate that cd-ttRecJ exists as a monomer, while ttRecJ exists as an oligomer. It is unclear whether the oligomeric state of ttRecJ is an intrinsic property or an artifact of the denaturation and refolding steps. It is, however, certain that regions external to the core domain are involved in the aggregation of ttRecJ. These results indicate that this method of refolding and limited proteolysis is useful for obtaining the recombinant protein in soluble form.
ttRecJ and cd-ttRecJ exhibited 5′→3′ exonuclease activity, which was enhanced by increasing the temperature, indicating that both proteins are properly folded. It should be noted that ttRecJ is present as a mixture of the monomeric and oligomeric states, whereas cd-ttRecJ is in the monomeric state. Because ecRecJ is a monomer in solution, RecJ exonulease may be activite in the monomeric form (6). A possible reason why the exonuclease activity of cd-ttRecJ was higher than that of ttRecJ is that the external region of ttRecJ, improper aggregation or both mask the active site. We determined the kinetic parameters of ttRecJ protein. The maximum rate of ecRecJ is reported to be ∼16 nt/s/RecJ molecule (6). The kcat value of cd-ttRecJ at 50°C is relatively close to that of ecRecJ. The metal ion requirement of ttRecJ is the same as scPPX1, which suggests that the mechanisms of metal binding of RecJ and scPPX1 are similar (21). Because scPPX1 contains motifs I–IV, RecJ and scPPX1 may bind metal ions using these motifs (8).
Limited proteolysis showed that RecJ protein has a stable core domain. This is the first report of the presence of a core domain in RecJ protein. This domain contains the motifs conserved among RecJ and functionally different proteins (8,11). These have the conserved aspartate and histidine residues. Other nuclease families also contain conserved aspartate residues in several motifs. Multiple sequence alignment using PSI-BLAST shows that motifs I and II are similar to the Exo I and II motifs of the PolI and Fen1 group of 5′-exonucleases, which are involved in metal binding (9). Furthermore, the DnaQ superfamily has three conserved stretches which have conserved aspartate and glutamate residues (23). Recently, the crystal structure of exonuclease I from E.coli, a member of the DnaQ superfamily, was determined (23). Its conserved motifs, which are separated by 50–100 amino acids in the sequence, are clustered and form an active site in the tertiary structure. Although the DnaQ motifs differ from those of RecJ, the conserved aspartate and histidine residues of the RecJ family are also thought to be involved in catalysis and metal binding.
Many unsolved problems remain concerning the molecular mechanisms of substrate specificity for ssDNA, direction, functional residues and the roles of metal ions. As a large amount of stable cd-ttRecJ is now available, further biochemical analysis and the study of the structural biology of RecJ protein should be facilitated.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by a Grant-in-Aid for Scientific Research (no. 12680655) to K.F. from the Ministry of Education, Culture, Sports, Science and Technology of Japan. A.Y. is the recipient of a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists (no. 809).
DDBJ/EMBL/GenBank accession no. AB064549
REFERENCES
- 1.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. American Society of Microbiology, Washington, DC.
- 2.Kowalczykowski S.C. (2000) Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci., 25, 156–165. [DOI] [PubMed] [Google Scholar]
- 3.Dianov G., Sedgwick,B., Daly,G., Olsson,M., Lovett,S.T. and Lindahl,T. (1994) Release of 5′-terminal deoxyribose-phosphate residues from incised abasic sites in DNA by the Escherichia coli RecJ protein. Nucleic Acids Res., 22, 993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper D.L., Lahue,R.S. and Modrich,P. (1993) Methyl-directed mismatch repair is bidirectional. J. Biol. Chem., 268, 11823–11829. [PubMed] [Google Scholar]
- 5.Burdett V., Baitinger,C., Viswanathan,M., Lovett,S.T. and Modrich,P. (2001) In vivo requirement for RecJ, ExoVII, ExoI and ExoX in methyl-directed mismatch repair Proc. Natl Acad. Sci. USA, 98, 6765–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lovett S.T. and Kolodner,R.D. (1989) Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc. Natl Acad. Sci. USA, 86, 2627–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courcelle J. and Hanawalt,P.C. (1999) RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet., 262, 543–551. [DOI] [PubMed] [Google Scholar]
- 8.Aravind L. and Koonin,E.V. (1998) A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem. Sci., 23, 17–19. [DOI] [PubMed] [Google Scholar]
- 9.Sutera V.A. Jr, Han,E.S., Rajman,L.A. and Lovett,S.T. (1999) Mutational analysis of the RecJ exonuclease of Escherichia coli: identification of phosphoesterase motifs. J. Bacteriol., 181, 6098–6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lovett S.T. and Kolodner,R.D. (1991) Nucleotide sequence of the Escherichia coli recJ chromosomal region and construction of recJ-overexpression plasmids. J. Bacteriol., 173, 353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajman L.A. and Lovett,S.T. (2000) A thermostable single-stranded DNase from Methanococcus jannaschii related to the RecJ recombination and repair exonuclease from Escherichia coli. J. Bacteriol., 182, 607–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oshima T. and Imahori,K. (1974) Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int. J. Syst. Bacteriol., 24, 102–112 [Google Scholar]
- 13.Yamagishi A., Tanimoto,N., Suzuki,T. and Oshima,T. (1996) Pyrimidine biosynthesis genes (pyrE and pyrF) of an extreme thermophile, Thermus thermophilus.Appl. Environ. Microbiol., 62, 2191–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoseki J., Yano,T., Koyama,Y., Kuramitsu,S. and Kagamiyama,H. (1999) Directed evolution of thermostable kanamycin-resitance gene: a convenient selection marker for Thermus thermophilus. J. Biochem. (Tokyo), 126, 951–956. [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa N., Sugahara,M., Masui,R., Kato,R., Fukuyama,K. and Kuramitsu,S. (1999) Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair. J. Biochem. (Tokyo), 126, 986–990. [DOI] [PubMed] [Google Scholar]
- 16.Yokoyama S., Hirota,H., Kigawa,T., Yabuki,T., Shirouzu,M., Terada,T., Ito,Y., Matsuo,Y., Kuroda,Y., Nishimura. Y. et al. (2000) Structural genomics projects in Japan. Nature Struct. Biol., 7, 943–945. [DOI] [PubMed] [Google Scholar]
- 17.Yokoyama S., Matsuo,Y., Hirota,H., Kigawa,T., Shoruzu,M., Kuroda,Y., Kurumizaka,H., Kawaguchi,S., Ito,Y., Shibata,T. et al. (2000) Structural genomics project in Japan. Prog. Biophys. Mol. Biol., 73, 363–376. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Kuramitsu S., Hiromi,K., Hayashi,H., Morino,Y. and Kagamiyama,H. (1990) Pre-steady-state kinetics of Escherichia coli aspartate aminotransferase catalyzed reactions and thermodynamic aspects of its substrate specificity. Biochemistry, 29, 5469–5476. [DOI] [PubMed] [Google Scholar]
- 20.Matsudaira P. (1987) Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem., 262, 10035–10038. [PubMed] [Google Scholar]
- 21.Wurst H. and Kornberg,A. (1994) A soluble exopolyphosphatase of Saccharomyces cerevisiae. J. Biol. Chem., 269, 10996–11001. [PubMed] [Google Scholar]
- 22.Clark E.D. (2001) Protein refolding for industrial processes. Curr. Opin. Biotechnol., 12, 202–207. [DOI] [PubMed] [Google Scholar]
- 23.Breyer W.A. and Matthews,B.W. (2000) Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nature Struct. Biol., 7, 1125–1128. [DOI] [PubMed] [Google Scholar]